Abstract
Introduction
Differences in the metabolite profiles between serum and plasma are incompletely understood.
Objectives
To evaluate metabolic profile differences between serum and plasma and among plasma sample subtypes.
Methods
We analyzed serum, platelet rich plasma (PRP), platelet poor plasma (PPP), and platelet free plasma (PFP), collected from 8 non-fasting apparently healthy women, using untargeted standard 1D and CPMG 1H NMR and reverse phase and hydrophilic (HILIC) UPLC-MS. Differences between metabolic profiles were evaluated using validated principal component and orthogonal partial least squares discriminant analysis.
Results
Explorative analysis showed the main source of variation among samples was due to inter-individual differences with no grouping by sample type. After correcting for inter-individual differences, lipoproteins, lipids in VLDL/LDL, lactate, glutamine, and glucose were found to discriminate serum from plasma in NMR analyses. In UPLC-MS analyses, lysophos-phatidylethanolamine (lysoPE)(18:0) and lysophosphatidic acid(20:0) were higher in serum, and phosphatidylcholines (PC) (16:1/18:2, 20:3/18:0, O-20:0/22:4), lysoPC(16:0), PE(O-18:2/20:4), sphingomyelin(18:0/22:0), and linoleic acid were lower. In plasma subtype analyses, isoleucine, leucine, valine, phenylalanine, glutamate, and pyruvate were higher among PRP samples compared with PPP and PFP by NMR while lipids in VLDL/LDL, citrate, and glutamine were lower. By UPLC-MS, PE(18:0/18:2) and PC(P-16:0/20:4) were higher in PRP compared with PFP samples.
Conclusions
Correction for inter-individual variation was required to detect metabolite differences between serum and plasma. Our results suggest the potential importance of inter-individual effects and sample type on the results from serum and plasma metabolic phenotyping studies.
Keywords: Metabolic phenotyping, Metabolic profiling, NMR, Plasma, Serum, UPLC-MS
1. Introduction
Human blood contains a multitude of proteins and metabolites and is commonly used in epidemiological and metabolic phenotyping studies (Tzoulaki et al. 2014). Although serum and plasma both originate from whole blood, they are drawn into distinct phlebotomy tubes, undergo different biochemical processing after collection, and contain different constituents.
Serum is obtained from whole blood collected without an anticoagulant. The blood is allowed to clot, and the fibrin clot, blood cells, and a majority of residual cellular material are removed by centrifugation (some cell fragments and microvesicles remain) (Colombo et al. 2014). Platelets are activated during this process and release proteins and metabolites into the serum (Schnabel et al. 2010; Yatomi et al. 1997). Therefore, while serum has major modifications of coagulation factors, it also contains secreted components (e.g., cytokines and growth factors) from platelets and other cells including neutrophils and monocytes. Even when inhibited with an additive, glucose utilization by platelets and leukocytes continues while the blood is clotting at a rate of ~ 5–7%/h (5–10 mg/dL) (Chan et al. 1989).
Plasma is obtained from blood collected with an anticoagulant (e.g., EDTA, heparin, citrate) and centrifugation during sample processing eliminates glucose utilization and removes the white blood cells, red blood cells, and a variable amount of platelets depending on the centrifugation protocol. Similar to serum, any microvesicles present are mostly retained by standard centrifugation protocols. Consequently, plasma contains relatively unmodified coagulation proteins with variable degrees of platelet components present in platelet rich plasma (PRP) and platelet poor plasma (PPP) but not in platelet free plasma (PFP). Freezing plasma samples results in platelet lysis and release of the components (e.g., membranes and intracellular molecules). Therefore, platelet-derived metabolites are likely to contribute to overall differences between serum and plasma metabolite profiles.
As serum and plasma possess both shared and unique components, these differences may be an important consideration for biospecimen selection and metabolite identification in metabolic phenotyping studies. A few studies in humans have compared serum and plasma samples (Teahan et al. 2006; Liu et al. 2010; Wedge et al. 2011; Denery et al. 2011; Yu et al. 2011; Ishikawa et al. 2013, 2014; Hirayama et al. 2015). These studies utilized different nuclear magnetic resonance (NMR) or mass spectrometry (MS) techniques and reported several metabolites that differentiated the sample types. To date, no single study has evaluated differences between serum and plasma using both NMR and MS analyses and to our knowledge, differences among plasma subtypes have not been investigated. We performed a comprehensive untargeted analysis of serum, PRP, PPP, and PFP using Carr-Purcell-Meiboom-Gill (CPMG), standard 1D, and lipoprotein subclass 1H NMR and hydrophilic interaction chromatography (HILIC) and reverse phase (RP) ultra-performance liquid chromatography-MS (UPLC-MS) to evaluate metabolic profile differences across blood sample types.
2. Materials and methods
2.1. Sample collection and processing
Serum, PRP, PPP, and PFP blood samples were collected from 8 apparently healthy non-fasting female volunteers (n = 32 total samples). For preparation of serum, blood was drawn into 10-mL Serum Separator Tubes (SST; containing clot activator and serum separator gel) (BD Bioscience, Franklin Lakes, NJ, USA). Samples were inverted 5 times and allowed to clot at room temperature (RT) for 45 min prior to centrifugation (2000×g 10 min at RT).
Plasma samples were prepared from blood collected into 10-mL lithium heparin tubes. PRP was prepared by one centrifugation at 200×g 10 min (no brake) and PPP was prepared by one centrifugation at 2000×g 10 min (low brake). PFP was prepared by centrifugation at 2000×g 10 min (low brake), aliquoting the plasma into 1-mL conical vials, and performing a second centrifugation at 15,000×g 7 min (all centrifugations at RT). Following centrifugation, samples were aliquoted into 1-mL conical vials and frozen at −80 °C.
In order to evaluate the day-to-day biovariability of the metabolites we identified in our analyses, we collected a separate set of serum samples from those used for the serum-plasma comparison. Samples were collected from 7 non-fasting apparently healthy female volunteers into SST on days 0, 3, and 6, and prepared using the serum sample processing methods described above. Samples were analyzed in a single batch, separate from the serum-plasma sample analysis and run in random order.
A quality control (QC) sample was generated by pooling 50 μL of each sample from the serum-plasma study. QC samples were used for the assessment of instrument stability and data quality throughout the study. For UPLC-MS, the QC spectra were viewed during the run to check that the mass correction was applied, the instrument pressure was stable, and the retention time drift between QC samples was acceptable. The coefficient of variation (CV) of the ionic features across the QC samples were used to assess the stability of the analytical run. A QC filter was applied to remove any features from the analysis with a CV > 30%. For NMR, the QC and experimental samples were assessed visually by PCA. Instrument stability was considered acceptable if QC samples had low intra-group variability compared with the inter-group variability among the experimental samples.
2.2. Metabolic profiling of serum and plasma samples using 1H NMR spectroscopy
Samples were prepared and analyzed in a single batch in random order using an in-house protocol adapted from standard Bruker methods (Dona et al. 2014). Samples (300 μL) were prepared in 300 μL of phosphate buffer and QC spectra were acquired after every 10 study samples. 1H NMR standard 1D (NOESYPR1D) with water pre-saturation and CPMG spectra were recorded on a 600 MHz Avance III Bruker NMR spectrometer (Bruker Biospin, Rheinstetten, Germany) operating at 600.13 MHz. All experiments were performed with 32 scans, 4 dummy scans, a 10 ms mixing time, and 96 k data points at a temperature of 310K. The 1H NMR standard 1D analyses were acquired with a sweep width of 30 ppm resulting in an acquisition time of 2.73 s. The time between the two 90° RF pulses was 3 μs and the relaxation delay between each FID acquisition was 4 s. For the CPMG NMR spectra, a sweep width of 20 ppm was employed resulting in an acquisition time of 3.07 s. A spin echo delay of 0.3 ms and 128 loops were used resulting in a total T2 relaxation delay of 76.8 ms. The relaxation delay between two FIDs was set at 4 s. Spectral processing was performed as described in the Supplementary Information. All spectra underwent a probabilistic quotient normalisation (Dieterle et al. 2006).
2.3. Lipoprotein profiles from deconvolution of NMR peaks
Quantification of lipoprotein subclasses were obtained using Bruker IVDr LIpoprotein Subclass Analysis (B.-LISA, Bruker, Biospin) (Petersen et al. 2005; Flote et al. 2016). For QC, Bland–Altman prediction errors and correlation coefficients were calculated between the NMR measurements and standard clinical chemistry values for total-cholesterol, high density lipoprotein (HDL)-cholesterol, low density lipoprotein (LDL)-cholesterol, and total triglycerides. The correlation between clinical values and Bruker measurements of total-cholesterol, LDL-cholesterol, and triglycerides were r > 0.72, therefore the analysis of 105 lipoprotein subclasses was performed. The lipoprotein densities and Bruker nomenclature for the lipoprotein components are described in the Supplementary Information (Supplementary Materials and Methods and Table S-1).
2.4. Metabolic profiling of serum and plasma samples using HILIC and RP UPLC-MS
Samples (200 μL) were treated with isopropanol (1:3), incubated at −20 °C for 24 h, centrifuged (2700×g 10 min) and aliquoted. Chromatography was performed with a Waters Acquity Ultra Performance LC system (Waters Corp., Milford, MA, USA) using both HILIC and RP chromatographic methods. HILIC separation was performed as described previously (Want et al. 2010). RP UPLC-MS procedures were according to Waters (Isaac et al. 2011). Electrospray ionization (ESI) mass spectrometry acquired in both positive (ESI+) and negative (ESI−) ionization modes was performed using a Xevo G2 Q-TOF (Waters Corp.). The capillary voltages were 1.5 kV (ESI+) and 2 kV (ESI−). The cone voltage was 20 V, desolvation temperature 600 °C, and the source temperature was 120 °C. The cone gas flow rate was 150 L/h and the desolvation gas flow rate was 1000 L/h. The Xevo G2 Q-TOF was operated in sensitivity mode with a scan time of 0.1 s. Data were collected in centroid mode with a scan range of 50–1200 m/z. Lock-mass scans were collected every 30 s to perform mass correction. Leucine Enkephalin (555.2645 amu) (20 μg/L) at a flow rate of 15 μL/min was used for lock-mass correction. The same parameters were used For MSe experiments with a high collision energy voltage of 30 eV. Samples were analyzed in a single batch in random order with QC spectral acquisition after every 5 samples. XCMS (Smith et al. 2006) with cent-Wave peak detection was used for pre-processing of the raw UPLC-MS data in R (version 3.1.2).
2.5. Statistical analysis, feature selection, metabolite identification, and biovariability assessment
NMR and MS data were analyzed using principal components analysis (PCA) with SIMCA-P software (version 13.0.3, Umetrics, Sweden) to assess the main sources of variation. Variation due to differences among individuals dominated the separation of the data, so a method to remove the contribution from inter-individual variation was employed. To achieve this, the mean spectra of all samples collected from one blood donor was subtracted from the same donor’s individual serum or plasma sample’s spectra. This approach (which we have termed “individual correction”) was used to compare serum and plasma and the different plasma subtypes.
We used pairwise discriminant analysis based on orthogonal partial least squares discriminant analysis (OPLS-DA) using SIMCA-P software to build classification models. Each variable was auto-scaled (mean-centered and scaled to unit variance) prior to OPLS-DA. Models that were significant (assessed by analysis of variance of the cross-validated residuals (CV-ANOVA) p < 0.05) based on sevenfold cross-validation were used to determine features that differentiated the sample types. From the OPLS-DA models we obtained the correlation between the feature and the predictive OPLS-DA score (correlation loadings, p(corr)) and the importance of the feature to the model (variable influence on projection (VIP)) for each feature. Features from each of the statistically significant OPLS-DA models were chosen if they had a VIP value above 1 since the influence of variables with a VIP > 1.0 on the explanation of the Y matrix is considered above average (Umetrics 2005; Galindo-Prieto et al. 2014). Next, a Wilcoxon-Mann-Whitney (WMW) test was performed on each selected NMR and MS feature and the p-values were adjusted using a false discovery rate (FDR) approach (FDR-adjusted p < 0.05) as described by Storey (Storey 2002). All statistically significant OPLS-DA models were used for metabolite identification.
Metabolite assignments of NMR features were performed using 1D and 2D NMR experiments (1H-1H homonuclear COrrelation SpectroscopY (COSY), TOtal Correlation Spectroscopy (TOSCY), and 1H-13C Single Quantum Coherence spectroscopy (HSQC)) by matching the recorded signals with NMR spectral databases (Human Metabolome Database (HMDB), Biological Magnetic Resonance Data Bank (BMRB)), and published assignments (Nicholson et al. 1995). Statistical total correlation spectroscopy (STOCSY) was used to locate potentially correlated peaks in complex spectral regions (Cloarec et al. 2005).
For UPLC-MS feature identification, molecular formulas and structural elucidation were assessed by matching accurate mass-to-charge (m/z) measurements to metabolites from available online databases (https://metlin.scripps.edu/index.php, http://www.lipidmaps.org/, and http://www.hmdb.ca). Our database searches included both adduct and parent ions. Initial searches were performed with the tolerance of the detected mass accuracy set to ±50 ppm in order to identify candidate compounds. Assignment of the candidate metabolites were subsequently made using information from fragmentation experiments and previously published studies that used the same analytical methods. To make direct comparisons of spectral data from different blood sample types, the relative levels of selected molecules were assessed using TargetLynx software (Waters Corp.).
Metabolite identification was given the following level of assignment (LoA), as adapted from Sumner et al. (Sumner et al. 2007): (1) Identified compound, confirmed by spiking sample with an authentic chemical reference, (2) MS/MS spectrum or 1H NMR chemical shifts and their multiplicity matched to compounds using publically available databases or published literature, (3) Detection of common fragment ions used to characterize compound classes and an accurate mass matched to a reference database and, (4) Accurate mass matched to a database to make a tentative assignment.
For biovariability assessment, random effects nested ANOVA was used to estimate components of variance using the aov function in R statistical software. The analytical (CVa), within-subject (CVi), and between-subject (CVg) coefficients of variation were calculated (Fraser and Harris 1989; Sakkinen et al. 1999). NMR biovariability samples were only analyzed once and CVa values could not be calculated.
3. Results
3.1. Removing inter-individual variation from metabolite profiles by mean-correction
The blood donors’ ages ranged from 27 to 62 years (mean 41.6 years; standard deviation (SD): 13.5 years). The mean (SD) body mass index (BMI) was 27.4 kg/m2 (4.4 kg/m2) (range 17.8–33.1 kg/m2). Typical spectral data from serum, PRP, PPP, and PFP samples analyzed using standard 1D 1H NMR are shown in Supplementary Fig. S-1 and reveal the presence of a range of amino acids, organic acids, lipids, and glucose.
Figure 1a, d show the PCA scores plots of standard 1D NMR and RP UPLC-MS (ESI+) data summarizing the main sources of variation among the different sample types. Serum and plasma samples were clustered by individual donor with no grouping by sample type. This indicated the main source of variation among the samples was due to inter-individual differences. A similar trend was observed for CPMG NMR and RP and HILIC UPLC-MS (ESI−) analyses (Fig. S-2). In the HILIC UPLC-MS (ESI+) analysis, the main source of variation was from the serum and plasma samples (Fig. S-2J).
Fig. 1.
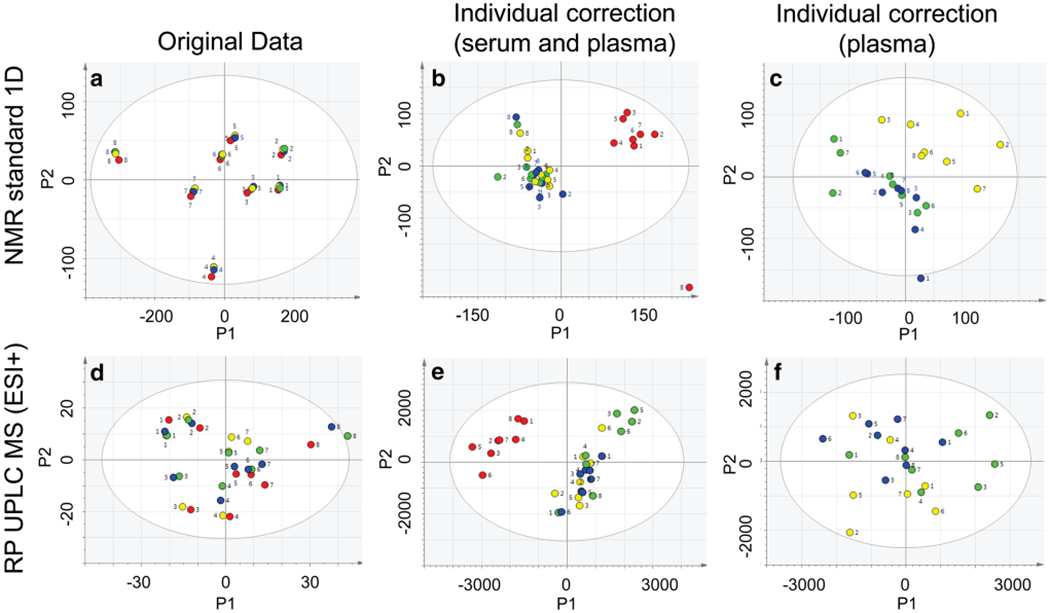
Principal components analysis (PCA) scores plots of 1H NMR standard 1D and RP UPLC-MS (ESI+) data evaluating variation among serum and plasma samples. PCA scores plots (PC1 and PC2) from serum (red), platelet rich plasma (yellow), platelet poor plasma (blue), and platelet free plasma (green) are presented before (a, d) and after applying individual correction using all sample types (b, e) or using plasma samples only (c, f). The numbers correspond to the individual donors who provided the samples
To investigate potential differences between serum and plasma samples that were not attributed to inter-individual variation, we adjusted the samples using “individual correction”, as described in Methods Section 2.5. Figure 1b, e show the PCA scores plots for the serum and plasma standard 1D NMR and RP UPLC-MS (ESI+) data after individual correction was applied. The main source of variation described by the first principal component was serum vs. plasma samples. A similar trend was observed for all NMR and UPLC-MS data sets (Fig. S-2).
The PCA scores plots in Fig. 1c, f compare the plasma subtypes alone after individual correction. Following individual correction, the PRP samples clustered together and were grouped away from PPP and PFP samples in the NMR analyses (Fig. 1c; Fig. S-2c & f). PRP samples were also grouped away from PPP and PFP samples by RP (ESI−) (Fig. S-2i), but not in the other UPLC-MS analyses (Fig. 1f; Fig. S-2l & o).
In the original (uncorrected) data, 6 pairwise OPLS-DA models significantly discriminated serum from plasma (standard 1D NMR, CPMG NMR, RP (ESI±), and HILIC (ESI±) UPLC-MS). After individual correction, all 7 OPLS-DA models were significant (Tables S-2 and S-3 in the Supplementary Information). The most significant model discriminating serum from plasma by NMR was from standard 1D data (CV-ANOVA p = 2.2 × 10−19). The RP (ESI+) data generated the most significant UPLC-MS model (CV-ANOVA p = 1.2 × 10−18). The scores plots for the cross-validated models are shown in Fig. 2 and Fig. S-3.
Fig. 2.
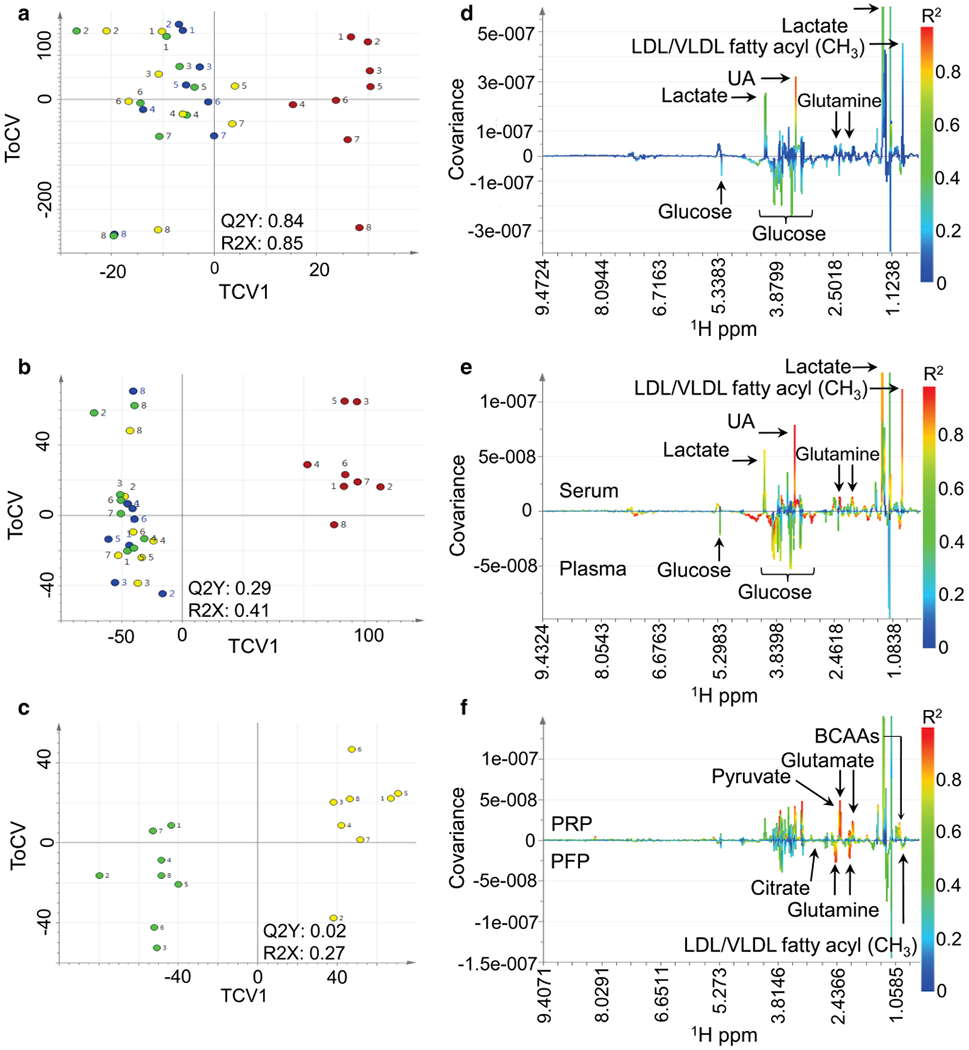
Pairwise orthogonal partial least squares discriminant analysis (OPLS-DA) of 1H NMR standard 1D serum and plasma data. Cross-validated scores (TCV1 vs. TCV2, corresponding to the orthogonal component) of the metabolic data are shown for serum (red) vs. platelet rich plasma (PRP) (yellow), platelet poor plasma (PPP) (blue), and platelet free plasma (PFP) (green) before applying individual correction (a). Comparison of serum vs. plasma is shown after applying individual correction using all samples (b) and comparison of PRP vs. PFP is shown after applying individual correction using plasma samples only (c). The numbers correspond to the individual donors who provided the samples. The corresponding OPLS-DA loadings plots (d, e, f) show the NMR data points (1H ppm) colored by their squared correlation to the sample type. The covariance of each NMR feature to the class variable is indicated by the magnitude and direction. BCAAs, branched chains amino acids; UA: unassigned
Prior to individual correction none of the OPLS-DA models comparing the plasma subtypes were predictive (all CV-ANOVA p > 0.05) (Tables S-3 & S-4). After individual correction, all 7 models significantly discriminated PRP from PPP; the most significant was CPMG NMR (CV-ANOVA p = 2.9 × 10−5). Six models significantly discriminated PRP from PFP; the most significant was RP (ESI−) UPLC-MS (CV-ANOVA p = 2.2 × 10−5). For comparisons between PPP and PFP only the CPMG (CV-ANOVA p = 0.008) and standard 1D (CV-ANOVA p = 0.01) NMR models were statistically significant.
3.2. Metabolites distinguishing serum from plasma
In standard 1D NMR analysis, we identified 4 metabolites that significantly differentiated serum from plasma samples. The methyl group of lipids in VLDL and LDL (at 0.85 ppm); the methyl group of lactate (at 1.33 ppm); and, the methylene group of glutamine (at 2.44 ppm) were higher in serum than plasma. Conversely, glucose (at 5.23 ppm) was higher in plasma than serum (Table 1). Specific features could not be assigned in the CPMG analyses, as statistically significant data points were located in the spectral baseline (Table S-5). In the NMR lipoprotein subclass analysis, 47 lipoprotein subclasses significantly differed between serum and plasma samples. Of these, 37 lipoprotein subclasses were higher in serum, including 18 subclasses from the HDL fraction (Table S-6). Some intermediate density lipoprotein (IDL), VLDL, and LDL subclasses were higher in plasma (Table S-6).
Table 1.
Metabolites discriminating serum and plasma and plasma subtypes identified by 1H NMR standard 1D analyses
| Metabolite | LoA | 1H ppm | FDR p value | p(corr) | VIP |
|---|---|---|---|---|---|
| Serum (+) vs. Plasma (−) | |||||
| Glutamine | 1 | 2.444 | 2.5 × 10−04 | 0.69 | 1.40 |
| Lipids in VLDL and LDL* | 2 | 0.852 | 3.2 × 10−04 | 0.90 | 1.75 |
| Lactate | 2 | 1.332 | 3.2 × 10−04 | 0.83 | 1.61 |
| Glucose | 2 | 5.229 | 4.8 × 10−03 | −0.52 | 1.05 |
| PRP (+) vs. PFP (−) | |||||
| Phenylalanine | 2 | 7.319 | 2.3 × 10−03 | 0.81 | 1.99 |
| Pyruvate | 2 | 2.360 | 4.6 × 10−03 | 0.95 | 2.36 |
| Glutamate | 2 | 2.338 | 4.6 × 10−03 | 0.95 | 2.35 |
| N-acetylglycoproteins | 2 | 2.036 | 4.6 × 10−03 | 0.89 | 2.20 |
| Isoleucine | 2 | 0.943 | 4.6 × 10−03 | 0.88 | 2.18 |
| Leucine | 2 | 0.953 | 4.6 × 10−03 | 0.84 | 2.08 |
| Valine | 2 | 0.954 | 7.0 × 10−03 | 0.83 | 2.05 |
| Lactate | 2 | 1.315 | 1.6 × 10−02 | 0.69 | 1.85 |
| Alanine | 2 | 1.468 | 1.6 × 10−02 | 0.79 | 1.74 |
| Glutamine | 1 | 2.452 | 4.6 × 10−03 | −0.94 | 2.34 |
| Lipids in VLDL and LDL* | 2 | 0.880 | 4.6 × 10−03 | −0.80 | 1.96 |
| Citrate | 2 | 2.512 | 1.6 × 10−02 | −0.76 | 1.88 |
| PRP (+) vs. PPP (−) | |||||
| Glutamate | 2 | 2.336 | 6.2 × 10−03 | 0.92 | 2.39 |
| Valine | 2 | 0.954 | 6.2 × 10−03 | 0.90 | 2.34 |
| N-acetylglycoproteins | 2 | 2.036 | 6.2 × 10−03 | 0.88 | 2.27 |
| Leucine | 2 | 0.965 | 6.2 × 10−03 | 0.86 | 2.24 |
| Isoleucine | 2 | 0.944 | 6.2 × 10−03 | 0.86 | 2.22 |
| Pyruvate | 2 | 2.363 | 6.2 × 10−03 | 0.83 | 2.13 |
| Phenylalanine | 2 | 7.319 | 6.2 × 10−03 | 0.79 | 2.03 |
| Glutamine | 1 | 2.444 | 6.2 × 10−03 | −0.91 | 2.34 |
| Citrate | 2 | 2.512 | 9.6 × 10−03 | −0.85 | 2.18 |
| Lipids in VLDL and LDL* | 2 | 0.852 | 9.6 × 10−03 | −0.73 | 1.86 |
| PPP (+) vs. PFP (−) | |||||
| No statistically significant features† | – | – | – | – | – |
The levels of assignment (LoA) were defined as: (1) Identified compounds, confirmed with chemical reference standards and (2) 1H NMR chemical shifts and their multiplicity matched to compounds using publically available databases or published literature. The (+) or (−) symbols listed in the sample type categories correspond to the directionality of the relationships between the OPLS-DA predictive score and sample types: a (+) symbol indicates a positive correlation between that sample type and metabolite; a (−) symbol indicates a negative correlation with that sample type. A positive correlation loading (p(corr)) value of a metabolite indicates a positive relationship with the sample type with a (+) symbol
The peak at 0.84 to 0.88 ppm is the resonance of the CH3 group of all lipids present in VLDL and LDL; the table reports information for the data point with the highest p(corr) in this range.
No features from the OPLS-DA models discriminating PPP vs. PFP were statistically significant following the false discovery rate (FDR) adjustment.
VIP, variable influence on projection of the feature to the OPLS-DA model
By UPLC-MS analyses we identified 10 metabolites that discriminated serum from plasma with an LoA ≤ 3 (n = 8 were higher in plasma) (Table 2). A majority of the metabolites were glycerophospholipids including lysophosphatidylcholine (lysoPC)16:0, phosphatidylcholines (PC)16:1/18:2, 20:3/18:0 and O-20:0/22:4, and phosphatidylethanolamine (PE)(O-18:2/20:4) which were higher in plasma than serum. LysoPE(18:0) and lysophosphatidic acid (lysoPA)(20:0) were higher in serum than plasma (Table 2). The sphingolipid sphingomyelin( 18:0/22:0), and fatty acids linoleic acid and linoleyl carnitine were higher in plasma than serum (Table 2).
Table 2.
Metabolites identified by HILIC and Reverse Phase (RP) UPLC-MS analyses as discriminant between serum and plasma and among plasma subtypes
| Metabolite | LoA | Analytical method | Adduct (as detected) | m/z | Retention time (minutes) | Chemical formula | HMDB | FDR-adjusted p value | p(corr) | VIP |
|---|---|---|---|---|---|---|---|---|---|---|
| Plasma (+) vs. Serum (−) | ||||||||||
| Linoleyl carnitine | 3 | HILIC UPLC MS (ESI+) | [M + Na]+ | 446.3244 | 3.73 | C25H45NO4 | HMDB0006469 | 4.2 × 10−05 | 0.85 | 1.30 |
| SM(18:0/22:0) | 3 | RP UPLC MS (ESI+) | [M + NH4]+ | 806.6481 | 8.00 | C45H93N2O6P | HMDB0012091 | 7.1 × 10−04 | 0.70 | 1.39 |
| PE(O-18:2/20:4) | 3 | RP UPLC MS (ESI−) | [M−H]− | 748.4611 | 6.41 | C43H76NO7P | HMDB0011452 | 1.3 × 10−03 | 0.50 | 6.21 |
| PC(16:1/18:2) | 2 | RP UPLC MS (ESI+) | [(M+3)+H]+ | 759.5613 | 5.54 | C42H78NO8P | HMDB0008006 | 1.5 × 10−03 | 0.66 | 1.55 |
| PC(20:3/18:0) | 2 | RP UPLC MS (ESI+) | [M+NH4]+ | 829.6790 | 6.96 | C46H86NO8P | HMDB0008366 | 2.3 × 10−03 | 0.58 | 1.02 |
| Linoleic acid | 3 | RP UPLC MS (ESI−) | [M−H]− | 279.2340 | 2.40 | C18H32O2 | HMDB0000673 | 3.5 × 10−03 | 0.69 | 8.89 |
| LysoPC(16:0) | 2 | RP UPLC MS (ESI+) | [M+H]+ | 496.3981 | 1.90 | C24H50NO7P | HMDB0010382 | 5.2 × 10−03 | 0.58 | 1.68 |
| PC(O-20:0/22:4) | 3 | RP UPLC MS (ESI+) | [M+H]+ | 852.6985 | 8.59 | C50H94NO7P | LMGP01020243* | 2.5 × 10−02 | 0.32 | 0.39 |
| LysoPE(18:0) | 3 | RP UPLC MS (ESI−) | [M−H]− | 480.3096 | 1.76 | C23H48NO7P | HMDB0011129 | 7.4 × 10−03 | −0.54 | 5.35 |
| LysoPA(20:0) | 2 | RP UPLC MS (ESI−) | [M−H]− | 465.3046 | 2.39 | C23H47O7P | HMDB0062315 | 8.2 × 10−03 | −0.71 | 10.82 |
| PRP (+) vs. PFP (−) | ||||||||||
| PE(18:0/18:2) | 2 | RP UPLC MS (ESI−) | [M+-FA−H]− | 788.5396 | 5.79 | C41H78NO8P | HMDB0008994 | 8.7 × 10−03 | 0.90 | 13.42 |
| PC(P-16:0/20:4) | 2 | RP UPLC MS (ESI−) | [M−H]− | 766.5372 | 6.61 | C44H80NO7P | HMDB0011220 | 9.1 × 10−03 | 0.77 | 6.95 |
Levels of assignment (LoA) were based on the following definitions: 2. MS/MS spectrum matched to compounds using publically available databases or published literature and 3. Detection of common fragment ions to characterize compound classes and accurate mass matched to a reference database (HMDB, LIPID MAPS, and/or METLIN). The (+) or (−) symbols listed in the sample type categories correspond to the directionality of the relationships between the OPLS-DA predictive score and sample types: a (+) symbol indicates a positive correlation between that sample type and metabolite; a (−) symbol indicates a negative correlation with that sample type. A positive correlation loading (p(corr)) value of a metabolite indicates a positive relationship with the sample type with a (+) symbol.
Metabolite did not contain an HMDB entry and the LIPID MAPS reference number is listed.
ACN, acetonitrile; FA, formic acid; FDR, false discovery rate (p value); HMDB, Human Metabolome Database; VIP, variable influence on projection of the feature to the OPLS-DA model
3.3. Metabolites distinguishing plasma subtypes
In comparisons of the different plasma subtypes, we identified 12 statistically significant metabolites by standard 1D NMR that discriminated PRP from PFP and 10 metabolites that differentiated PRP from PPP; 7 of these metabolites were also detected in the respective CPMG NMR analysis. Isoleucine, leucine, valine, phenylalanine, glutamate, pyruvate, and signals from N-acetylglycoproteins were consistently higher among plasma samples with increased platelet content whereas the resonances of the fatty acyl moieties in VLDL/LDL, citrate, and glutamine were consistently lower (Table 1 and Table S-5). The LDL-triglyceride subfraction, L2TG, was consistently lower among samples with higher platelet content by lipoprotein subclass analysis (Table S-6). No statistically significant differences were observed between PPP and PFP samples by standard 1D or CPMG NMR (Table 1 and Table S-5). In the RP UPLC-MS analysis, PE(18:0/18:2) and PC(P-16:0/20:4) were higher in PRP than PFP (Table 2).
3.4. Metabolite biovariability
We assessed biovariability of the identified metabolites by collecting serum samples from 7 donors at three time points (on days 0, 3, and 6). As observed in our experiments comparing different blood matrices, the main source of variation by PCA over the three time points was attributed to inter-individual differences (Fig. S-4). After applying individual correction the main source of variation observed across the first principal component was sampling day (Fig. S-4).
As presented in Supplementary Table S-7, we used uncorrected data to calculate the within-subject (CVi) and between-subject (CVg) coefficients of variation for these metabolites. The CVi values ranged from 0.6% (glutamate, pyruvate) to 31.5% (linoleic acid) and CVg values ranged from 1.2% (citrate) to 59.1% (linoleic acid). For the UPLC-MS measurements, run in triplicate, we calculated analytical variation (CVa); the range was 1.8% (PC(P-16:0/20:4)) to 60.5% (lysoPE(18:0)). For the metabolites, lysoPC(16:0), PC(P-16:0/20:4), linoleic acid, and linoleyl carnitine, the analytical variation values were less than half of the individual variation values (CVa < 1/2 CVi) indicating adequate precision (Fraser and Harris 1989). In general the magnitude of the CVi and CVg values were consistent with well-established biomarkers of inflammation and coagulation used in epidemiological studies (Sakkinen et al. 1999). Exceptions included some amino acids, pyruvate, and citrate which had low between-subject and within-subject variation.
4. Discussion
We used untargeted NMR, lipoprotein subclass, and UPLC-MS analyses to evaluate differences among the metabolic profiles of serum, PRP, PPP, and PFP. Our results demonstrated that differences from inter-individual variation were greater than differences by sample type. Following adjustment for inter-individual variation, we identified several metabolites, predominately glycerophospholipids, lipoproteins, and amino acids, that discriminated serum from plasma and distinguished PRP from PPP and PFP samples.
Previous studies have evaluated metabolite differences between serum and plasma using either NMR or MS profiling in both targeted (n ranging from 4 to 377 samples) or untargeted modes (n ranging from 4 to 29 samples). These studies reported heterogeneous results and are summarized in Table S-8. Some overlapping findings were reported, including higher amino acids, arachidonic acid, lactate, hypoxanthine, glycerol, myo-inositol, and lower sarcosine in serum. Our study extends findings in the literature by utilizing multiple untargeted methods and by evaluating different plasma subtypes.
Our PCA results demonstrated that inter-individual differences were greater than differences by sample type or from day-to-day biovariability. Likely sources of variability among individuals include age, sex, diet, and other environmental influences (Lenz et al. 2003, 2004; Trabado et al. 2017). In an attempt to limit such heterogeneity, all subjects in our study were women, therefore age, genetics, diet, and other external factors likely played roles in these differences. The dominance of the inter-individual variation was consistent with results from previous studies (Teahan et al. 2006; Hirayama et al. 2015; Jonsson et al. 2015; Ishikawa et al. 2013; Wedge et al. 2011) and indicated the importance of using mathematical methods to adjust for inter-individual variation in order to identify influences attributable to other sources. Results from our UPLC-MS biovariability assessment showed that many of the metabolites we measured had larger between-subject variation than within-subject plus analytical variation over the course of 3–6 days. Although only a small number of participants were used for these estimates, findings indicate a majority of the metabolites we identified are suitable for use in longitudinal epidemiological studies.
After accounting for inter-individual variation, we identified 4 metabolites and 47 lipoprotein subclasses by NMR and 10 metabolites by UPLC-MS (with an LoA ≤ 3) that significantly differentiated serum from plasma samples. In NMR analyses, VLDL/LDL fatty acyl groups, lactate, glutamine, and 37 lipoprotein subclasses (mainly from the HDL fractions) were higher in serum than plasma while glucose and some VLDL, LDL, and IDL subclasses were lower. Higher levels of several lipids in serum have been reported previously (Teahan et al. 2006). Higher lactate and lower glucose in serum was identified previously by GC-MS and attributed to continued glycolysis and metabolism in the serum samples during clot formation (Teahan et al. 2006; Dettmer et al. 2010). A separate study also using GC-MS showed that incubation of EDTA plasma samples at 37 °C resulted in decreasing glucose levels (Liu et al. 2010), consistent with ongoing cellular metabolism.
The main metabolite subclass that differentiated serum from plasma by RP UPLC-MS analyses in our study was glycerophospholipids, including (lyso)PCs, (lyso)PEs, and lysoPA, which play central roles in cell membrane structure and signalling. Previous studies among fasting blood donors identified diacyl-PC(C38:1) and lysoPCs(C16:0, C17:0, C18:0, C18:1) as higher in serum than plasma (Yu et al. 2011; Ishikawa et al. 2014). Differences were attributed to platelet phospholipase activity during serum clot formation. We observed higher PC(16:1/18:2, 20:3/18:0, and O-20:0/22:4) and lysoPC(16:0) in plasma using RP (ESI+) or HILIC UPLC-MS (ESI+). These discrepant results likely reflect differences among blood-donor fasting status (Ishikawa et al. 2013, 2014; Yu et al. 2011) or differences in analytical techniques (Yu et al. 2011). Our RP UPLC-MS (ESI−) results showed that lysoPE(18:0) and lysoPA(20:0) were higher in serum which is more consistent with studies that used fasting samples (Yu et al. 2011; Ishikawa et al. 2014; Breier et al. 2014).
To the best of our knowledge, our study is the first to evaluate metabolite differences among plasma subtypes. We hypothesized variation in platelet metabolism would contribute to metabolic phenotype differences among plasma subtypes. A previous study investigated metabolism of platelet concentrates during sample storage and reported correlations with hypoxanthine, malate, lactate, and glucose (Paglia et al. 2015). In our NMR analyses, increased plasma platelet content was associated with higher levels of the glycolysis end-product pyruvate, the amino acids leucine, isoleucine, valine, phenylalanine, and glutamate, and signals from N-acetylglycoproteins. Lower plasma platelet content was associated with higher glutamine, citrate, and VLDL/LDL lipid moieties. Ongoing platelet metabolism during sample processing may explain lower levels of citrate and pyruvate, whereas positive correlations with glutamate and other amino acids may be explained by platelet transport of these molecules (Morrell et al. 2008). By UPLC-MS, we identified 2 metabolites (with an LoA ≤ 3) that were higher in PRP compared with PFP samples: PE(18:0/18:2) and PC(P-16:0/20:4). These findings are consistent with PC and PE as the most abundant phospholipids in platelet membranes (Wang et al. 1986). Further analysis of stimulated platelet supernatants and lysates are required to confirm platelet origins of the metabolites identified in our study.
Our study was limited by a small sample size comprised of non-fasting women across a broad age range from whom we did not have information on current medication use or hormonal status. A prior study reported a larger number of metabolites that discriminated serum from plasma in fasting compared with non-fasting donors and suggested fasting and non-fasting blood be considered distinct matrices (Ishikawa et al. 2014). Gender-associated differences have been reported for glycerophospholipids and sphingomyelins (Ishikawa et al. 2014; Trabado et al. 2017). An additional limitation is that although several tentative metabolite assignments were made, most were not confirmed with authentic chemical references. Strengths of our study include the comprehensive instrumental analyses for untargeted metabolic phenotyping, statistical methods adjusting for inter-individual characteristics, and the evaluation of different plasma subtypes processed using standardized laboratory protocols.
5. Concluding remarks
In conclusion, we used multiple analytical methods to obtain a wide coverage of metabolites present among different blood matrices. Primary differences among blood sample types were explained by inter-individual variation and required individual correction techniques to detect differences attributed to sample type. Metabolites that discriminated serum from plasma included glycerophospholipids, lipoproteins, and energy pathway metabolites, while amino acids, glycerophospholipids, and energy pathway metabolites distinguished plasma samples with variable platelet content. As our study had only limited power, we would anticipate detection of additional metabolites in larger cohort studies. Therefore, our results underline the need for careful consideration of the potential impact of inter-individual effects (other than the main variable(s) under investigation) and sample type on the design and results of metabolic phenotyping studies. These factors become particularly important if consortia studies are being considered.
Supplementary Material
Acknowledgements
The authors are grateful to Hartmut Shaefer (Bruker Biospin) for carrying out the Bruker Lipoprotein Subclass Analysis using the 1H NMR Standard 1D spectral data acquired for this study.
Funding This work was performed as an ancillary study within the EC FP7 funded project, COMBI-BIO: Development of COMBInatorial BIOmarkers for subclinical atherosclerosis, grant number 305422 from the European Commission Seventh Framework Programme. N.C.O. was supported by K99HL129045 from the National Heart, Lung, and Blood Institute (NHLBI) of the National Institutes of Health (NIH). R.P.T. was supported by R01 HL120854 from the NHLBI. T.M.D.E. acknowledges support from the EC PhenoMeNal project (Project Reference 654241) and NIH grant R01HL133932. P.E. is Director of the MRC-PHE Centre for Environment and Health and acknowledges support from the Medical Research Council and Public Health England (MR/L01341X/1) and support from the NIHR Biomedical Research Centre at Imperial College Healthcare NHS Trust and Imperial College London, the NIHR Health Protection Research Unit in Health Impact of Environmental Hazards (HPRU-2012-10141), and UK MEDical BIOinformatics partnership (UK MED-BIO) supported by the Medical Research Council (MR/L01632X/1), and the UK Dementia Research Institute (UK DRI) at Imperial College London, funded by the Medical Research Council, Alzheimer’s Society and Alzheimer’s Research UK.
Footnotes
Electronic supplementary material The online version of this article (https://doi.org/10.1007/s11306-018-1332-1) contains supplementary material, which is available to authorized users.
Conflict of interest M.K., C.L.B., I.K., J.C.L., T.M.D.E., P.E., R.P.T., and N.C.O. declare that they have no relevant affiliations or financial involvement with any organization with a financial interest in, or conflict with, the study’s subject matter.
Ethical approval All participants provided written informed consent and the study protocol received approval by the University of Vermont’s Intuitional Review Board. All procedures involving human participants throughout the study were in accordance with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
References
- Breier M, Wahl S, Prehn C, Fugmann M, Ferrari U, Weise M, et al. (2014). Targeted metabolomics identifies reliable and stable metabolites in human serum and plasma samples. PLoS ONE, 9(2), e89728 10.1371/journal.pone.0089728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan AY, Swaminathan R, & Cockram CS (1989). Effectiveness of sodium fluoride as a preservative of glucose in blood. Clin Chem, 35(2), 315–317. [PubMed] [Google Scholar]
- Cloarec O, Dumas ME, Craig A, Barton RH, Trygg J, Hudson J, et al. (2005). Statistical total correlation spectroscopy: An exploratory approach for latent biomarker identification from metabolic 1H NMR data sets. Analytical Chemistry, 77(5), 1282–1289. 10.1021/ac048630x. [DOI] [PubMed] [Google Scholar]
- Colombo M, Raposo G, & Thery C (2014). Biogenesis, secretion, and intercellular interactions of exosomes and other extra-cellular vesicles. Annual Review of Cell and Developmental Biology, 30, 255–289. 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- Denery JR, Nunes AA, & Dickerson TJ (2011). Characterization of differences between blood sample matrices in untargeted metabolomics. Analytical Chemistry, 83(3), 1040–1047. 10.1021/ac102806p. [DOI] [PubMed] [Google Scholar]
- Dettmer K, Almstetter MF, Appel IJ, Nurnberger N, Schlamberger G, Gronwald W, et al. (2010). Comparison of serum versus plasma collection in gas chromatography-mass spectrometry-based metabolomics. Electrophoresis, 31(14), 23652373 10.1002/elps.200900778. [DOI] [PubMed] [Google Scholar]
- Dieterle F, Ross A, Schlotterbeck G, & Senn H (2006). Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Analytical Chemistry, 78(13), 4281–4290. 10.1021/ac051632c. [DOI] [PubMed] [Google Scholar]
- Dona AC, Jimenez B, Schafer H, Humpfer E, Spraul M, Lewis MR, et al. (2014). Precision high-throughput proton NMR spectroscopy of human urine, serum, and plasma for large-scale metabolic phenotyping. Analytical Chemistry, 86(19), 9887–9894. 10.1021/ac5025039. [DOI] [PubMed] [Google Scholar]
- Flote VG, Vettukattil R, Bathen TF, Egeland T, McTiernan A, Frydenberg H, et al. (2016). Lipoprotein subfractions by nuclear magnetic resonance are associated with tumor characteristics in breast cancer. Lipids in Health and Disease, 15(1), 56 10.1186/s12944-016-0225-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser CG, & Harris EK (1989). Generation and application of data on biological variation in clinical chemistry. Critical Reviews in Clinical Laboratory Sciences, 27(5), 409–437. 10.3109/10408368909106595. [DOI] [PubMed] [Google Scholar]
- Galindo-Prieto B, Eriksson L, & Trygg J (2014). Variable influence on projection (VIP) for orthogonal projections to latent structures (OPLS). Journal of Chemometrics, 28(8), 623–632. 10.1002/cem.2627. [DOI] [Google Scholar]
- Hirayama A, Sugimoto M, Suzuki A, Hatakeyama Y, Enomoto A, Harada S, et al. (2015). Effects of processing and storage conditions on charged metabolomic profiles in blood. Electrophoresis, 36, 2148–2155. 10.1002/elps.201400600. [DOI] [PubMed] [Google Scholar]
- Isaac G, McDonald S, & Astarita G (2011). Lipid Separation using UPLC with Charged Surface Hybrid Technology. http://www.waters.com/webassets/cms/library/docs/720004107en.pdf. Accessed August 10, 2017.
- Ishikawa M, Maekawa K, Saito K, Senoo Y, Urata M, Murayama M, et al. (2014). Plasma and serum lipidomics of healthy white adults shows characteristic profiles by subjects’ gender and age. PLoS ONE, 9(3), e91806 10.1371/journal.pone.0091806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa M, Tajima Y, Murayama M, Senoo Y, Maekawa K, & Saito Y (2013). Plasma and serum from nonfasting men and women differ in their lipidomic profiles. Biological and Pharmaceutical Bulletin, 36(4), 682–685. [DOI] [PubMed] [Google Scholar]
- Jonsson P, Wuolikainen A, Thysell E, Chorell E, Stattin P, Wikstrom P, et al. (2015). Constrained randomization and multivariate effect projections improve information extraction and biomarker pattern discovery in metabolomics studies involving dependent samples. Metabolomics, 11(6), 1667–1678. 10.1007/s11306-015-0818-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz EM, Bright J, Wilson ID, Hughes A, Morrisson J, Lindberg H, et al. (2004). Metabonomics, dietary influences and cultural differences: A 1H NMR-based study of urine samples obtained from healthy British and Swedish subjects. Journal of Pharmaceutical and Biomedical Analysis, 36(4), 841–849. 10.1016/j.jpba.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Lenz EM, Bright J, Wilson ID, Morgan SR, & Nash AF (2003). A 1H NMR-based metabonomic study of urine and plasma samples obtained from healthy human subjects. Journal of Pharmaceutical and Biomedical Analysis, 33(5), 1103–1115. [DOI] [PubMed] [Google Scholar]
- Liu L, Aa J, Wang G, Yan B, Zhang Y, Wang X, et al. (2010). Differences in metabolite profile between blood plasma and serum. Analytical Chemistry, 406(2), 105–112. 10.1016/j.ab.2010.07.015. [DOI] [PubMed] [Google Scholar]
- Morrell CN, Sun H, Ikeda M, Beique JC, Swaim AM, Mason E, et al. (2008). Glutamate mediates platelet activation through the AMPA receptor. Journal of Experimental Medicine, 205(3), 575–584. 10.1084/jem.20071474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson JK, Foxall PJ, Spraul M, Farrant RD, & Lindon JC (1995). 750 MHz 1H and 1H-13C NMR spectroscopy of human blood plasma. Analytical Chemistry, 67(5), 793–811. [DOI] [PubMed] [Google Scholar]
- Paglia G, Sigurjonsson OE, Rolfsson O, Hansen MB, Brynjolfsson S, Gudmundsson S, et al. (2015). Metabolomic analysis of platelets during storage: a comparison between apheresis- and buffy coat-derived platelet concentrates. Transfusion, 55(2), 301–313. 10.1111/trf.12834. [DOI] [PubMed] [Google Scholar]
- Petersen M, Dyrby M, Toubro S, Engelsen SB, Norgaard L, Pedersen HT, et al. (2005). Quantification of lipoprotein subclasses by proton nuclear magnetic resonance-based partial least-squares regression models. Clinical Chemistry, 51(8), 1457–1461. 10.1373/clinchem.2004.046748. [DOI] [PubMed] [Google Scholar]
- Sakkinen PA, Macy EM, Callas PW, Cornell ES, Hayes TE, Kuller LH, et al. (1999). Analytical and biologic variability in measures of hemostasis, fibrinolysis, and inflammation: assessment and implications for epidemiology. American Journal of Epidemiology, 149(3), 261–267. [DOI] [PubMed] [Google Scholar]
- Schnabel RB, Baumert J, Barbalic M, Dupuis J, Ellinor PT, Durda P, et al. (2010). Duffy antigen receptor for chemokines (Darc) polymorphism regulates circulating concentrations of monocyte chemoattractant protein-1 and other inflammatory mediators. Blood, 115(26), 5289–5299. 10.1182/blood-2009-05-221382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CA, Want EJ, O’Maille G, Abagyan R, & Siuzdak G (2006). XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Analytical Chemistry, 78(3), 779–787. 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- Storey JD (2002). A direct approach to false discovery rates. Journal of the Royal Statistical Society: Series B (Statistical Methodology), 64(3), 479–498. [Google Scholar]
- Sumner LW, Amberg A, Barrett D, Beale MH, Beger R, Daykin CA, et al. (2007). Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics, 3(3), 211–221. 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teahan O, Gamble S, Holmes E, Waxman J, Nicholson JK, Bevan C, et al. (2006). Impact of analytical bias in metabonomic studies of human blood serum and plasma. Analytical Chemistry, 78(13), 4307–4318. 10.1021/ac051972y. [DOI] [PubMed] [Google Scholar]
- Trabado S, Al-Salameh A, Croixmarie V, Masson P, Corruble E, Feve B, et al. (2017). The human plasma-metabolome: Reference values in 800 French healthy volunteers; impact of cholesterol, gender and age. PLoS ONE, 12(3), e0173615 10.1371/journal.pone.0173615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzoulaki I, Ebbels TM, Valdes A, Elliott P, & Ioannidis JP (2014). Design and analysis of metabolomics studies in epidemiologic research: A primer on -omic technologies. American Journal of Epidemiology, 180(2), 129–139. 10.1093/aje/kwu143. [DOI] [PubMed] [Google Scholar]
- Umetrics AB (2005). User’s guide to SIMCA-P, SIMCA-P+ (version 11.0). Umea: Umetrics. [Google Scholar]
- Wang CT, Shiao YJ, Chen JC, Tsai WJ, & Yang CC (1986). Estimation of the phospholipid distribution in the human platelet plasma membrane based on the effect of phospholipase A2 from Naja nigricollis. Biochimica et Biophysica Acta, 856(2), 244–258. [DOI] [PubMed] [Google Scholar]
- Want EJ, Wilson ID, Gika H, Theodoridis G, Plumb RS, Shockcor J, et al. (2010). Global metabolic profiling procedures for urine using UPLC-MS. Nature Protocols, 5(6), 1005–1018. 10.1038/nprot.2010.50. [DOI] [PubMed] [Google Scholar]
- Wedge DC, Allwood JW, Dunn W, Vaughan AA, Simpson K, Brown M, et al. (2011). Is serum or plasma more appropriate for intersubject comparisons in metabolomic studies? An assessment in patients with small-cell lung cancer. Analytical Chemistry, 83(17), 6689–6697. 10.1021/ac2012224. [DOI] [PubMed] [Google Scholar]
- Yatomi Y, Igarashi Y, Yang L, Hisano N, Qi R, Asazuma N, et al. (1997). Sphingosine 1-phosphate, a bioactive sphingolipid abundantly stored in platelets, is a normal constituent of human plasma and serum. The Journal of Biochemistry, 121(5), 969–973. [DOI] [PubMed] [Google Scholar]
- Yu Z, Kastenmuller G, He Y, Belcredi P, Moller G, Prehn C, et al. (2011). Differences between human plasma and serum metabolite profiles. PLoS ONE, 6(7), e21230 10.1371/journal.pone.0021230. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


