Abstract
Recent studies have clearly shown that bats are the reservoir hosts of a wide diversity of novel viruses with representatives from most of the known animal virus families. In many respects bats make ideal reservoir hosts for viruses: they are the only mammals that fly, thus assisting in virus dispersal; they roost in large numbers, thus aiding transmission cycles; some bats hibernate over winter, thus providing a mechanism for viruses to persist between seasons; and genetic factors may play a role in the ability of bats to host viruses without resulting in clinical disease. Within the broad diversity of viruses found in bats are some important neurological pathogens, including rabies and other lyssaviruses, and Hendra and Nipah viruses, two recently described viruses that have been placed in a new genus, Henipaviruses in the family Paramyxoviridae. In addition, bats can also act as alternative hosts for the flaviviruses Japanese encephalitis and St Louis encephalitis viruses, two important mosquito-borne encephalitogenic viruses, and bats can assist in the dispersal and over-wintering of these viruses. Bats are also the reservoir hosts of progenitors of SARS and MERS coronaviruses, although other animals act as spillover hosts. This chapter presents the physiological and ecological factors affecting the ability of bats to act as reservoirs of neurotropic viruses, and describes the major transmission cycles leading to human infection.
Keywords: Bats, Megachiroptera, Microchiroptera, Rabies, Lyssaviruses, Hendra virus, Nipah virus, Henipaviruses, Japanese encephalitis virus, Flaviviruses, Coronaviruses
Introduction
It is now well recognized that more than 75 % of emerging diseases over the past 2–3 decades have been zoonoses. Many of these zoonotic viruses have arisen from wildlife sources and caused neurological disease, especially those emerging during this period in the Asian and Australasian regions (Mackenzie et al. 2001; Mackenzie 2005; Griffin 2010; Bale 2012; Wang and Crameri 2014). Most of the diseases emerging from wildlife have been from bats and rodents (Enria and Pinheiro 2000; Calisher et al. 2006; Goeijenbier et al. 2013; Luis et al. 2013). Bats are only second to rodents in terms of mammalian species richness (Wilson and Reeder 2005) and constitute about 20 % of all mammalian species. Thus, with their wide distribution and abundance, it is not surprising that there is growing awareness that bats are the reservoir hosts for a number of these emerging viruses (Calisher et al. 2006; van der Poel et al. 2006; Wong et al. 2007; Halpin et al. 2007; Wood et al. 2012; Smith and Wang 2013; Shi 2013) and suspected of being associated with many others on serological grounds. Not only have they been shown to be the reservoir hosts for rabies and related lyssaviruses but also for other human pathogens, or potential pathogens, such as SARS-coronavirus-like viruses (Lau et al. 2005; Li et al. 2005; Shi and Hu 2008), Ebola and Marburg viruses (Leroy et al. 2005; Gonzalez et al. 2007; Towner et al. 2007, 2009; Marí Saéz et al. 2014; Olival and Hayman 2014; Ogawa et al. 2015), Menangle virus (Philbey et al. 1998), and Hendra and Nipah viruses (Young et al. 1996; Halpin et al. 2000; Yob et al. 2001; Chua et al. 2002). This brief review looks at the biological features that make bats good reservoir hosts, and the more important neurological viruses associated with bats that are, or have the potential to be, transmitted to humans.
Bats as Reservoirs Hosts: Implications for Virus Transmission
The Evolution, Origin, Taxonomy, and Diversity of Order Chiroptera
The evolution of bats remains a controversial topic; bats have a poor fossil record and their phylogenetic relationships have been relatively understudied (Teeling et al. 2000, 2002). Historically, the mammalian Order Chiroptera has been divided into two suborders, the Megachiroptera, or Old World fruit bats, including flying-foxes, and the Microchiroptera, or echolocating bats (Simmons and Conway 2003). Taxonomic methods based on morphological cladistics of cochlea structure among Eocene fossil bats, complemented by some molecular data, led Simmons and colleagues to the conclusion that echolocation, originating secondary to flight, evolved only once and that the echolocating microchiropteran bats are monophyletic (Simmons and Conway 2003). Based on this study the 188 species of megachiropteran bats were grouped within a single family, Pteropodidae, and the other 917 species of bats were divided into 18 families.
However more recently molecular-based phylogenies suggest a far more complex paraphyletic relationship of echolocating bats suggesting echolocation evolved twice and that five families of echolocating bats (previously classified with the monophyletic microchiropteran bat families) are more closely related to the Pteropodidae (of note rhinolophid bats harbor viruses closely related to the severe acute respiratory syndrome (SARS) CoV) (Springer 2013). These data suggest that the grouping of all echolocating bats into the suborder Microchiroptera is unwarranted and new suborders of bats have been adopted; the Pteropodiformes contains the Pteropodidae, Rhinolophidae, Hipposideridae, Rhinopomatidae, Craseonycteridae, and Megadermatidae, while the suborder Vespertilioniformes contains the other 12 families formerly of the Microchiroptera (Springer 2013). Within this review we will use the suborder terms Megachiroptera and Microchiroptera as these are the most familiar to many nonspecialists.
Irrespective of evolutionary controversy, bats are believed to have originated in the late Cretaceous/early Paleocene, some 65 million years ago, with three major microchiropteran lineages traced to Lauarasia and a fourth to Gondwana (Teeling et al. 2005). The Chiroptera underwent rapid speciation with at least 24 genera of bats extant by the Eocene [52–50 million years ago (Simmons and Conway 2003; Teeling et al. 2005)]. The divergence of the Megachiroptera and Microchiroptera, irrespective of suborder status, occurred well prior to the oldest fossil record from the Eocene. Although the evolution of flight may have preceded echolocation, fossil remains from the Eocene indicate echolocation was well established (Simmons and Geisler 1998; Simmons et al. 2010). Following the early evolution of flight and echolocation, bats have changed little as a taxonomic group relative to other mammals (Jepsen 1970). Bats also have traits (e.g., flight, sheltered roosts and ability to hibernate and entire torpor) which may have allowed them to preferentially survive the Cretaceous-Tertiary (K–T) extinction, occurring ~65 million years ago following the impact of the large bolide creating the 180–300-km-wide Chicxulub crater in northern Yucatan, Mexico (for more details see Wang et al. 2011a).
Bat Population Ecology
Bats are unique with regard to the abundance and density achieved by certain cave-dwelling species. Colonies of Mexican free-tailed bats (Tadarida brasiliensis) can achieve numbers in excess of a million individuals, reaching densities of 500 individuals per square foot, and several species of Myotis achieve hibernating population densities of >300 per square foot (Constantine 1967a; Humphrey and Cope 1976; Tuttle 1976; Clawson 2002). The close proximity of numerous individuals packed into dense concentrations can obviously facilitate virus transmission by direct contact, such as biting or licking and other means, such as through respiratory transmission or contact transmission by transfer of infectious secreta and excreta. It is in caves harboring millions of closely packed free-tailed bats that airborne rabies virus transmission was documented (Constantine 1967b; Winkler 1968).
Tree roosting bats can also be highly gregarious with camps of pteropid bats containing thousands of individuals, often including more than one species, clustered within trees. In Australia, little red flying foxes (Pteropus scapulatus) hang together with up to 30 individuals clustered on a single branch (Hall and Richards 2000). Species of microchiropteran bats roost in colonies of several dozen to thousands of individuals while less gregarious species may roost in small colonies or singly, such as Lasionycterus noctivagans, an important reservoir host of a variant of rabies virus often associated with bat-associated human deaths in the United States (Noah et al. 1998; Messenger et al. 2003b; Rupprecht and Gibbons 2004).
Diet and roosting behavior have been hypothesized as influencing exposure to viruses and susceptibility to infection. A provocative study, although extremely preliminary, found that the decreasing permanence and protection offered by roosting sites of bat species increased the “soluble part of the constitutive immune function ” (as measured by the in vitro bacterial killing activity of plasma against Escherichia coli), while increases in the cellular immune potential (measured as white blood cell count) varied with and body mass and diet (carnivorous bats having higher counts than insectivorous or frugivorous species) (Schneeberger et al. 2013).
Bat Flight and Movements
Bats are the only mammals able to fly, and many species travel considerable distances from roost sites to feeding locations. The entire or some proportion of the population of 87 species within ten families of bats migrate to some degree; migratory behavior has been less studied in the genus Pteropus but has been well established for eight species (for review see Krauel and McCracken 2013). Although most frugivorous bats will travel distances <200 km during a season when shifting roosts in response to the availability of fruit production (Rosevear 1965; Fleming and Eby 2003), a few species, such as the pteropodid bat, Eidolon helvum, will travel ~1500 km in one-way migrations from forest habitats to savannahs in Africa (Fleming and Eby 2003). Pteropus species have been recorded traveling across open sea between peninsular Malaysia and Sumatra and between Australia and New Guinea (Breed et al. 2006, 2010).
Migratory behavior among temperate bat species has been categorized as sedentary, regional, and long distance (Fleming and Eby 2003). Regional migration (typically <500 km) is common among European and North American species of Myotis while the long distance, one-way migrations of the subtropical/tropical Mexican free-tailed bats, Tadarida brasiliensis, exceed 1800 km (Krauel and McCracken 2013; Cockrum 1969; Griffin 1970). Unlike birds which may migrate long distances without feeding, bats forage as they migrate. Locally abundant, but widely distributed fruit resources, may serve to aggregate species of bats and other terrestrial fruit-eating mammals, such as great apes and ungulates, at feeding sites thus potentially enhancing the risk of intra- and interspecific transmission of viruses. Temporary seasonal clustering of bats and terrestrial mammals during dry seasons in Africa has been proposed as a means of promoting interspecific transmission of Ebola virus from a putative fruit bat reservoir host (Leroy et al. 2005) to other species (Pinzon et al. 2004).
A further example of how the migratory behavior of bats may influence the geographic distribution and genetic variability of viruses is illustrated by the lyssaviral variants associated with specific bat species and their overlap with human disease (Table 1). A variant of rabies virus maintained by the silver-backed bat, Lasionycterus noctivagans, and the eastern pipistrelle bat, , and known as the Ln/Ps variant (Franka et al. 2006), has been the most commonly recognized cause of indigenously acquired human rabies in North America over the last few decades (Noah et al. 1998; Messenger et al. 2003b; Rupprecht and Gibbons 2004). The summer and winter range of L. noctivagans in North America extends from central Canada to the southern United States (Rohde et al. 2004) overlapping the distribution of human rabies cases associated with the LN/PS variant. Phylogenetic studies of European bat lyssavirus subgroup 1a (EBLV-1a) suggest that virus trafficking between migratory bat species and the sedentary Eptesicus serotinus, a principal reservoir host, has contributed to the genetic homogeneity observed among EBLV-1a isolates (Davis et al. 2005b). Within a bat species, such as T. brasiliensis, both sedentary and migratory populations may exist and intermingle seasonally (Cockrum 1969; McCraken and Gassel 1998), providing a mechanism for viruses exchange and introduction.
Table 1.
Recognized or proposed members of the genus Lyssavirus, family Rhabdoviridae
| Name | Species implicated in maintenance | Distribution | Annual human deaths | References |
|---|---|---|---|---|
| ICTV abbreviationa | ||||
| Rabies virus RABV | Dogs, wild carnivores, bats >50 spp. | Worldwide among dogs (with the exception of Australia and Antarctica, and designated rabies-free countries); Restricted to New World bats | ~55,000 (dog related) | Fooks et al. (2014), Knobel et al. (2005), Velasco-Villa et al. (2006), Mondul et al. (2003), Banyard et al. (2011), Schaefer et al (2005), and Bourhy et al. (1992) |
| European bat Lyssavirus EBLV-1 | Bats—Microchiroptera: Eptecicus serotinus, Tadarida teniotis, Myotis myotis, Myotis nattererii, Miniopterus schreibersii, Rhinolophus ferrumequinum, Pipistrellus pipistrellus, Plecotus auritus | Mainland Europe | Occasional | Serra-Cobo et al. (2002), Davis et al. (2005b), Picard-Meyer et al. (2011, 2014), Schatz et al. (2013), McElhinney et al. (2013), and Schatz et al. 2014 |
| European bat Lyssavirus EBLV-2 | Bats—Microchiroptera: Eptecicus serotinus, Myotis dasycneme; M. daubentonii | Europe, United Kingdom | Occasional | Fooks et al. (2003b), Brookes et al. (2005a), Harris et al. (2006, 2007), McElhinney et al. (2013), Schatz et al. (2013), and Miia et al. (2015) |
| Australian bat lyssavirus ABLV | Bats—Megachiroptera: Pteropus alecto, P. scapulatus, P. poliocephalus, P. conspicullatus | Australia; possibly SE Asia, including Philippines | Occasional | Allworth et al. (1996), Hooper et al. (1997), Gould et al. (1998, 2002), Hanna et al. (2000), Arguin et al. (2002), and Francis et al. (2014b) |
| Microchiroptera: Saccolaimus | ||||
| Flaviventris | ||||
| Bokeloh virus BBLV | Bats—Microchiroptera: Myotis natererii | Europe: France, Germany | None reported | Freuling et al. (2011, 2013), Picard-Meyer et al. (2013), and Nolden et al. (2014) |
| Aravan virus ARAV | Bats—Microchiroptera: Myotis blythi | Asia: Kyrghyzstan | None reported | Arai et al. (2003) |
| Irkut virus IRKV | Bats—Microchiroptera: Murina leucogaster | Asia: Eastern Siberia, China | None reported | Botvinkin et al. (2003) and Liu et al. (2013b) |
| Duvenhage virus DUVV | Bats—Microchiroptera: Miniopterus schreibersii, Nycteris thebaica | Africa: South Africa, Guinea | Occasional | Tignor et al. (1977), Paweska et al. (2006), and van Thiel et al. (2009) |
| Zimbabwe, Kenya, Swaziland | ||||
| Khujand virus KHUV | Bats—Microchiroptera: Myotis mystacinus | Asia: Tajikstan | None reported | Kuzmin et al. (2003) |
| Lagos bat virus LBV | Bats—Megachiropteran: Eidolan helvum, Micropterus pusillus, Nycteris gambiensis, Epomophourus wahlbergi, Rousettus aegyptiacus | Africa: Nigeria, Central Africa | None reported | Sureau et al. (1977), Meredith and Standing (1981), Crick et al. (1982), Markotter et al. (2006b), Kuzmin et al (2008a), Dzikwi et al. (2010), Hayman et al. (2008a), Nel and Rupprecht (2007), and Peel et al. (2013) |
| Republic, Egypt, Senegal, South Africa, Ghana, Zimbabwe, Ethiopia. Kenya | ||||
| Mokola virus MOKV | Shrew—Insectivora: Crocidura spp., Rodentia: Lopyhromys silkapusi | Africa: Cameroon, Central African Republic, Ethiopia, Nigeria, Zimbabwe, South | Occasional | Shope et al. (1970), Wiktor et al. (1984), Nel and Rupprecht (2007), Sabeta et al. (2007), and Kgaladi et al. (2013b) |
| Shimoni bat virus SHIMV | Bats—Microchiroptera: Hipposideros commersoni | Africa: Kenya | None reported | Kuzmin et al. (2010) |
| West Caucasian Bat virus WCBV | Bats—Microchiroptera: Miniopterus schreiberbersi | Europe/Asia: Caucasus | None reported | Botvinkin et al. (2003) and Kuzmin et al. (2005, 2008b) |
| Africa: Kenya? | ||||
| Ikoma lyssavirus IKOV | Civet—Carnivora: Civettictis | Africa: Tanzania | None reported | Marston et al. (2012) and Horton et al. (2014) |
| Lleida virusb LLEBV | Bats—Microchiroptera: Miniopterus schreiberbersi | Europe: Spain | None reported | Aréchiga Ceballos et al. (2013) |
a ICTV International Committee on Taxonomy of Viruses
bAs yet unclassified new lyssavirus
Long distance movements of bats may also lead to regular, but not constant, contact between individual bats from different subpopulations allowing partial connectivity between colonies of bats (e.g., Pteropus spp. in Australia). A metapopulation may exist where a spatial mosaic involves a constellation of subpopulations of which, at any given time, some are susceptible, some infected, and some immune to a particular disease. This may permit viruses to persist in a species with a total population that would otherwise be too small to maintain the pathogen (Lloyd and May 1996).
Bat Echolocation
Microchiropteran bats are the only mammals to use “sophisticated echolocation” (Simmons and Conway 2003), although Rousettus aegyptiacus, a megachiropteran bat, uses a brief, low amplitude clicking that may aid in orientation (Holland et al. 2004). The intense energy required to produce echolocation emissions (Neuweiler 2000) may promote virus transmission by aerosols or droplets when bats are aggregated and in close proximity. This transmission route has been hypothesized to occur with rabies virus as virus can be recovered from the mucous or respiratory fluids of infected bats (Constantine et al. 1972). The evidence supporting possible rabies virus transmission by an airborne route under natural conditions in caves harboring large colonies of Mexican free-tailed bats was previously mentioned.
Bat Hibernation and Torpor
Most, if not all, temperate bat species are capable of entering into regulated torpor whereby their body temperature (T b) is allowed to fall below ambient temperature (T a) (Speakman and Thomas 2003), and many species enter true hibernation during winter (Lyman 1970). Additionally, some tropical microchiropteran and megachiropteran species reduce T b, but whether this is regulated torpor or caused by extreme peripheral vasoconstriction remains to be determined (Speakman and Thomas 2003).
The impact of torpor and hibernation on the immune response and the persistence of viral infections among experimentally infected bats has been investigated for Japanese encephalitis virus (JEV) and rabies virus (Sulkin and Allen 1974). The thermogenic organ, or brown fat, of bats has been suggested as a storage depot of various viruses and rabies virus has been isolated from this tissue from experimentally infected bats kept at low temperatures (Sulkin et al. 1959), in addition to naturally infected bats (Bell and Moore 1960). JEV studies with persistence are described below in the section “Flaviviruses.”
There are data suggesting that on rare occasions bats may experience abortive infection by rabies virus or unusually long incubation or latency periods (Messenger et al. 2003a). The presence of neutralizing antibody among apparently healthy bats and the delayed development of rabies among captured bats has been interpreted as suggesting recovery from or prolonged incubation following infection (Moore and Raymond 1970; Trimarchi and Debbie 1977). Decreased rabies virus pathogenicity has been inferred by subjecting experimentally infected bats (Myotis lucifugus, T. brasiliensis, and Anthrozous pallidus) to low temperatures (4–10 °C) and then observing the onset of rabies after transferring animals to temperatures of 22–29 °C (Sulkin and Allen 1974; Sulkin et al. 1959; Sadler and Enright 1959). In a mathematical model of rabies persistence in a colony of big brown bats (Eptesicus fuscus) during hibernation, a long incubation period played a significant role in maintaining dormant infection. Subsequent transmission during spring warming avoided epizootic “fadeout” (George et al. 2011).
Apparently healthy common vampires, Desmodus rotundus, surviving experimental rabies virus challenge can excrete virus in their saliva (Aguilar-Setién et al. 2005). Similarly, apparently healthy bats have been shown to harbor low levels of EBLV RNA suggesting that there is a nonreproductive infection stage or subacute persistence of viral RNA (Serra-Cobo et al. 2002; Wellenberg et al. 2002).
Bat Longevity
Bats mature slowly and live long lives in comparison to other mammals of similar bodyweight (Barclay and Harder 2003). Several species of microchiropteran bat, M. lucifugus, Plecotus auritus, and Rhinolophus ferrumequinum, have been shown to have life spans exceeding 30 years in the wild (reviewed in Barclay and Harder 2003). This extreme longevity in a small mammal places bats well outside the traditional regression line scaling life expectancy to mammalian size (Austad 2005). The impact of extreme longevity on the potential for bats to maintain and transmit viruses could be enormous when coupled with the possibility of bats developing a tolerant, persistent infection following infection by certain viruses.
Bat Genomics and Immunology
A recurring theme in studies of bat evolution and behavior is how immune function can covary with these elements. As noted above, roost behavior and diet may influence the constitutive elements of the immune system (Schneeberger et al. 2013). To better understand the roles played by both genetic and nongenetic factors in bats’ ability to coexist and coevolve with pathogens successfully for a very long history, it is essential to do comparative genomics studies between bats and terrestrial mammals. Since 2001, whole genome sequencing has been conducted for nine species of bats, including two Pteropus spp., three Myotis spp., Rhinolphus ferrumequinum, Megaderma lyra, Pteronotus parnelli, and Eidolon helvum.
Although comparative genomics and transcriptomics have revealed some genetic basis of specialized traits in bats including flight, echolocation, hibernation/torpor, longevity, and antiviral immunity, additional functional studies are required to confirm that the observed genetic difference does play a role in making bats exceptionally effective reservoir hosts for a large number of viruses which can be highly lethal in other mammals.
Bats are the only flying mammals and have a smaller genome of ~2 Gb (compared to ~3 Gb for human and mouse) consistent with increased metabolic demands of flight. Interestingly, despite having smaller sizes, bat genomes contain a similar number of annotated genes to those of land mammals. Phylogenomic studies to determine evolutionary relationship of bats with other mammals remain unresolved with conflicting studies concluding either bats diverged from horses or from a sister a group within the clade of ungulates, cetaceans and carnivores (Wynne and Tachedjian 2015).
If genetic factors play a major role in bats’ ability to host viruses without clinical diseases, we can expect the following possible genetic differences: (1) Bats have unique gene/pathway(s) not present in other mammals; (2) Bats lack common gene/pathway(s) present in other mammals; or (3) Bats have similar gene/pathway(s) as in other mammals, but their expression pattern(s) is different. Although there are cases of bat-specific genetic features, such as the deletion of the AIM2 inflammasome gene family (Zhang et al. 2013) or the presence of bat-specific microRNAs (Cowled et al. 2014), to a large degree the major genetic differences seem to be at the level of gene expression and regulation. Specifically, there seems to be a trend of high basal level expression of genes involved in innate defense mechanisms (L-F Wang, unpublished results).
Flight has played an important role in shaping different aspects of the evolutionary changes of bats. The evolution of flight has been linked to immunologically important concentrations of positively selected genes in the DNA damage checkpoint and nuclear factor kB pathways (Zhang et al. 2013). Additionally, the high metabolic cost of flying requires an increased body temperature and it has been hypothesized that flight may play a role in increasing the tolerance of bats to adverse effects of viral infections in a way analogous to fever in other mammals and that flight in bats has selected for the acquisition of a diversity of viruses that may cause disease when transmitted to other mammals (O’Shea et al. 2014).
The study of regulatory genes among bats that influence their potential as reservoir hosts has greatly expanded in the last decade, although data are still limited (Schountz 2013). Bats have been shown to harbor an adaptive immune system which may provide a tolerance (see reviews by Schountz 2013; Chan et al. 2013), inhibiting demonstrable disease, permitting asymptomatic virus shedding and promoting recombination and reassortment of different RNA viruses (Chan et al. 2013). Bats behaving “normally,” such as the common vampire, Desmodus rotundus, may survive experimental challenge with rabies virus and survive to excrete virus in their saliva (Aguilar-Setién et al. 2005). Similarly, “healthy bats” have been shown to harbor low levels of EBLV RNA suggesting that there is a nonreproductive infection stage or subacute persistence of viral RNA (Serra-Cobo et al. 2002; Wellenberg et al. 2002).
Cell lines obtained from bats are now being used to investigate viral infection, tissue tropism, and the immune responses to infection. Examples include cell lines obtained from multiple tissues of Pteropus alecto and tissues of Eidolon helvum and Rousettus aegyptiacus immortalized by introducing retroviral elements. Some, but not all, of the pteropid cell lines were susceptible to infection by Hendra and Nipah viruses (Crameri et al. 2009) and the Eidolon and Rousettus cell lines were susceptible to Rift Valley Fever and O’nyong-nyong viruses, and virus titer from supernatants was decreased with IFN induction (Biesold et al. 2011). A recent study comparing the response of bat and human cell lines to a highly pathogenic zoonotic virus showed early induction of innate immune processes in bats (Wynne et al. 2014). This finding suggests there may be divergent mechanisms at a molecular level that may influence host pathogenesis.
Rabies and Other Lyssaviruses
Members of the Genus Lyssavirus , and Their Association with Specific Chiropteran Hosts
The single-stranded, negative-sense RNA viruses of the family Rhabdoviridae, Order Mononegavirales, exhibit an extraordinary host range, infecting plants, invertebrates, fish, and mammals (Kuzmin et al. 2006). However, viruses within individual genera of this family can exhibit exquisite host specification as exemplified by the 14 viruses currently classified within or proposed as members of the genus Lyssavirus (Table 1). The antigenic and genetic profiles of lyssaviruses allow segregation into phylogroups (see Fig. 1). Antibodies raised experimentally in mice against inactivated virus from one phylogroup neutralize viruses within that phylogroup but not others. With the exception of Mokola virus and Ikoma lyssavirus, each of the representative genotypes of lyssaviruses has been isolated from bats which are also believed to serve as their reservoir hosts. There is a complex relationship between lyssaviruses and bats where the viruses may cause clinical disease in bats or may circulate in the bat population without any overt disease (Banyard et al. 2011). There is also increasing evidence that rabies-related viruses are found throughout much of Africa, Europe, Asia, and Australia, but not in the Americas, whereas rabies virus is found in bats only in the Americas (Table 1). Transmission of lyssaviruses from bats to species of other mammalian orders, a process termed “spillover,” can cause fatal neurological disease among humans and other animals. The term rabies was once strictly reserved for the acute fatal encephalomyelitis caused by classical rabies virus. However, the clinical disease of rabies is now widely used to include the clinically and pathologically indistinguishable diseases caused by any Lyssavirus (Meredith et al. 1971; Lumio et al. 1986; Samaratunga et al. 1998; Nathwani et al. 2003; Fooks et al. 2003a; Banyard et al. 2011).
Fig. 1.

Phylogenetic tree of the lyssavirus phylogroups and their respective species. Nucleoprotein sequences (405 nucleotides) were aligned with ClustalW and the phylogenetic tree was visualized using TreeView version 3.2. Bootstrap values at relevant nodes are shown. According to the proposed antigenicity of each group of isolates, the viruses are divided into different phylogroups. Where available, accession numbers for sequences are rabies virus (RABV AY102999, AY062068, AY103008, AY062069, AY102993,AY352514, AY330735, AY062090, AY062070, AY062047), Lagos bat virus (LBV EF547459, EF547449, EF547447, GU170202), West Caucasian bat virus (WCBV EF614258), Shimoni bat virus (SHIBV GU170201), Mokola virus (MOKV AY062074,AY062077), Duvenhage virus (DUVV AY062079), European bat lyssavirus type 1 (EBLV-1 AY062088, EF157976), Irkut virus (IRKV EF614260), Australian bat lyssavirus (ABLV AF418014), European bat lyssavirus type 2 (EBLV-2AY062091, AY062089), Bokeloh bat lyssavirus (BBLV JF311903), Khujand virus (KHUV EF614261), Aravan virus (ARAV EF614259), and Ikoma lyssavirus (IKOV JX193798). Several sequences within the phylogeny are unpublished and as such do not have accession numbers. The scale bar represents 0.1 substitutions per nucleotide site. The number of human cases are shown next to silhouettes where reported (Reprinted with permission from Fooks AR, Banyard AC, Horton DL, et al. (2014) Current status of rabies and prospects for elimination. Lancet 384:1389–1399)
The genus has been divided into three phylogroups: phylogroup I includes rabies virus (RABV), Duvenhage virus (DUVV), European bat lyssavirus (EBLV-1), European bat lyssavirus 2 (EBLV-2), Australian bat lyssavirus (ABLV), Aravan virus (ARAV), Khujand virus (KHUV), Bokeloh bat virus (BBLV), and Irkut virus (IRKV); phylogroup II includes Lagos bat virus (LBV), Shimoni bat virus (SHIBV), and Mokola virus (MOKV); and phylogroup III includes West Caucasian bat virus (WCBV) and possibly Ikoma lyssavirus (IKOV) (see Fig. 1) (Badrane et al. 2001; Fooks et al. 2014). An additional possible member of the genus has also been described from a bat in Spain, and named Lleida virus (Aréchiga Ceballos et al. 2013). The viruses in each of the phylogroups vary in their virulence (Markotter et al. 2009; Kgaladi et al. 2013a). Specific vaccines and immunoglobulins only exist for the pre- or postexposure treatment (PET) of phylogroup I rabies virus (Anon 1999a), however, these vaccines elicit high levels of neutralizing antibodies to other phylogroup I lyssaviruses (Brookes et al. 2005b). Immunization of laboratory animals with rabies vaccine with subsequent challenge with other lyssaviruses indicates that diminishing efficacy is a function of increasing phylogenetic distance from rabies virus (Hanlon et al. 2005).
Characteristic differences in sequence variation of lyssaviral isolates have permitted identification of the primary reservoir host species. Characterization and typing of distinct virus variants has provided insights into the evolution, host species range and geographic distribution of specific genetic lineages of viruses circulating among bats, and led to the identification of bat-associated variants, which through spillover, have caused rabies in humans and animals (Badrane and Tordo 2001; McQuiston et al. 2001; Davis et al. 2005b; Loza-Rubio et al. 2005; Franka et al. 2006; Nadin-Davis and Loza-Rubio 2006; Leslie et al. 2006).
Rabies Virus and Bats
The reservoir hosts for rabies virus are mammalian species in the Orders Chiroptera and Carnivora, although virtually all of the approximately 55,000 human deaths occurring globally each year are due to virus variants maintained by domestic dogs (Knobel et al. 2005). Clinical features of rabies are described in detail in Chapter 5. The first observation linking bats to rabies was made in 1911 when the common vampire bat (Desmodus rotundus) was identified as the source of an epidemic of rabies among cattle in Brazil (Carini 1911). Human deaths attributed to bites received from vampire bats were first recorded from the island of Trinidad (Hurst and Pawan 1959; Pawan 1959). Outbreaks of human rabies due to vampire bats continue to be reported from Brazil (Batista-da-Costa et al. 1993; Sato et al. 2006), Costa Rica (Badilla et al. 2003), Peru (Lopez et al. 1992), Venezuela (Caraballo 1996), and Mexico (Martinez-Burnes et al. 1997; Velasco-Villa et al. 2006).
The recognition of insectivorous bats as reservoirs of rabies virus dates from 1953, when rabies was first described in a bat that attacked a 7-year-old boy in Florida (Scatterday 1954). Since that time, the number of rabid bats reported to CDC has increased, reaching 1680 in 2012, second in number only to raccoons (Dyer et al. 2013). The majority of indigenously acquired human rabies cases in North America over the past three decades have been caused by bat-associated variants (Messenger et al. 2003a, b). More than 30 species of North American bats have been identified as naturally infected by rabies virus (Constantine 1979; Mondul et al. 2003) and the number of bat species found to be infected in Mexico and in South America is rapidly growing (Nadin-Davis and Loza-Rubio 2006; Velasco-Villa et al. 2006; Escobar et al. 2015). Distinct virus variants of rabies virus may be associated with one or more bat species and research into species specificity and the evolution of bat-associated rabies virus is rapidly changing our knowledge-base on this subject (Hughes et al. 2005; Davis et al. 2006). Rabies virus has not been isolated from bats outside North, Central, and South America (Banyard et al. 2011).
Rabies virus variants circulating among bats are currently divided into four major groups and several additional subgroups (Nadin-Davis et al. 2001; Davis et al. 2006). Group I (four subgroups) contains virus variants primarily originating from highly colonial, migratory bats of the genera Myotis and Eptesicus, and are endemic to eastern Canada and the United States as far west as the State of Colorado (Davis et al. 2006). Group II (two subgroups) contains variants primarily originating from solitary or moderately colonial, migratory species of the genera Lasiurus and Pipistrellus and L. noctivagans, and are endemic to Canada and most of the United States; included here is a virus variant isolated from L. noctivagans and P. subflavus (Ln/Ps) which is the most common variant recovered from indigenously acquired human rabies in North America (Messenger et al. 2003b). Group III contains variants from Eptesicus fuscus, genetically distinct from group I, and is restricted to western Canada and the western United States (Nadin-Davis et al. 2001; Davis et al. 2006). Group IV (three subgroups) contains variants originating from colonial species of hematophagous bats and insectivorous bats from Central and South America. Various subgroups and paraphyletic clades within groups preclude making unequivocal statements concerning the endemic range and bat species infected by a particular group of viruses at this time (Davis et al. 2006). As additional samples become available and full genome sequencing is undertaken, finer resolution of phylogenetic relationships between virus variants and individual species can be achieved, as exemplified by recent studies of rabies virus from the genus Pipistrellus and L. noctivagans; the LN/PS variant may be two independently maintained rabies virus variants with distinct hosts (Franka et al. 2006).
African Lyssaviruses
In addition to rabies virus, five other lyssaviruses occur in Africa; DUVV, LBV, IKOV, SHIBV, and MOKV (Table 1) (Nel et al. 2000; Warrell 2010; Kuzmin et al. 2010; Banyard et al. 2011). Bats are believed to be the reservoir hosts for DUVV, LBV, and SHIBV (Markotter et al. 2006b; Kuzmin et al. 2010), but MOKV and IKOV have only been isolated from other wildlife species and no association has been found with bats.
LBV has been isolated from megachiropteran bats and domestic animals dying of rabies (Markotter et al. 2006b), with a single exception of one isolate from a water mongoose, Atilax paludinosus, Order Carnivora (Markotter et al. 2006a). The first isolate of LBV was obtained from a fruit bat in Nigeria in 1956 (Boulger and Porterfield 1958) and since that time additional isolates have been obtained from various species of fruit bats from Central African Republic, Egypt, Senegal, South Africa, Ghana, and Zimbabwe (Sureau et al. 1977; Meredith and Standing 1981; Crick et al. 1982; Markotter et al. 2006b); from single cats in South Africa and Zimbabwe (Crick et al. 1982; King and Crick 1988); and a dog in Ethiopia (Mebatsion et al. 1992). Indeed LBV may be the most common rabies-like virus in Africa and serological evidence has shown that it occurs throughout the range of the straw-colored fruit bat, Eidolon helvum, even in more isolated island colonies (Peel et al. 2013). DUVV has only been isolated a few times, with most isolates obtained from human infections and single isolates from two insectivorous bats, Miniopterus schreibersi and Nycteris thebaica (Paweska et al. 2006; Nel and Rupprecht 2007). SHIBV was isolated from a dead Commerson’s leaf-nosed bat (Hipposideros commersoni) in Kenya (Kuzmin et al. 2010).
MOKV has been isolated from shrews (genus Crocidura, Order Insectivora), rodents (Lopyhromys sikapusi, Order Rodentia), and humans (Shope et al. 1970; Kemp et al. 1972; Wiktor et al. 1984), and IKOV has been isolated from a single African civet (Civettictis civetta) (Marston et al. 2012). Although MOKV has never been recovered from a bat, but humans and domestic cats and dogs have been diagnosed with rabies caused by MOKV over an extensive geographic range including Ethiopia, Cameroon, Central African Republic, Nigeria, South Africa, and Zimbabwe (Familusi and Moore 1972; Foggin 1983; King and Crick 1988; Mebatsion et al. 1992; von Teichman et al. 1998; Nel et al. 2000; Bingham et al. 2001).
Little is known about the epidemiology of African lyssaviruses other than rabies. Domestic and feral dogs are the major reservoir for classical rabies virus in Africa with indigenous species of wild carnivores serving as reservoir hosts within several regions (Nel and Rupprecht 2007); in contrast to bats in the Americas, African bats have never been implicated in rabies virus maintenance or transmission.
DUVV and MOKV have been linked to sporadic cases of fatal encephalitis among humans (Meredith et al. 1971; Kemp et al. 1972; Familusi and Moore 1972; Familusi et al. 1972; Swanepoel et al. 1993; Paweska et al. 2006; van Thiel et al. 2009; Koraka et al. 2012). No human disease has been associated with IKOV, LBV, or SHIBV.
European Bat Lyssaviruses
EBLV-1 has been isolated from bats throughout Europe, mostly from the Serotine bat (Eptesicus serotinus) although the host range may be relatively broad, and accounts for most bat isolates in Europe. EBLV-2 is significantly less common and has been associated exclusively with Myotis bats (Myotis daubentonii and M. dasycneme). From molecular studies, EBLV- 1 and EBLV-2 have been further subdivided into two subgroups (Amengual et al. 1997; Fooks et al. 2003a; Davis et al. 2005b); EBLV-1a has been primarily isolated from the nonmigratory, colonial species E. serotinus in northern Europe; EBLV-1b has been isolated from E. serotinus obtained from northern Europe, France, and Spain; EBLV-2a has been primarily isolated from M. dasycneme from the Netherlands and M. daubentonii from the UK; EBLV-2b has been isolated from M. daubentonii from Switzerland (Serra-Cobo et al. 2002; Davis et al. 2005b).
Since 1977, four human deaths have been attributed to EBLV, two from EBLV-1 and two from EBLV-2 (Lumio et al. 1986; Roine et al. 1988; Khozinski et al. 1990; Bourhy et al. 1992; Fooks et al. 2003b; Nathwani et al. 2003). The recent case of fatal EBLV-2 infection in a Scottish bat conservationist was the first indigenously acquired case of rabies in the UK in 100 years (Nathwani et al. 2003). A photographer infected with EBLV-1 after a bite from a disoriented E. serotinus bat in Spain recovered due to previous immunization with rabies vaccine, as well as postexposure immunization (Van Gucht et al. 2013). EBLV-1 has been recovered from terrestrial mammals, five sheep in Denmark (Ronsholt 2002; Tjornehoj et al. 2006) and a stone marten (Martes foina) in Germany (Muller et al. 2004), and in captive zoo Egyptian fruit bats (R. aegyptiacus) in Denmark (Ronsholt et al. 1998), but spillover is either rare or goes undetected. It is possible that the virulence of ELBVs is lower than that of some other lyssaviruses; experimental inoculation of EBLV-1 into red foxes (Vulpes vulpes) and sheep has resulted in death in only one of 14 sheep (Soria Baltazar et al. 1988; Vos et al. 2004; Tjornehoj et al. 2006). Furthermore, ELBV-1 was detected in a range of tissues from apparently healthy bats (Schreiber’s bent-winged bats, Miniopterus schreibersii, and greater horseshoe bats, Rhinolophus ferrumequinum) in Spain (Serra-Cobo et al. 2002) and in healthy zoo fruit bats (R. aegyptiacus) (Wellenberg et al. 2002), showing that bats may survive infection with possible long-term maintenance of the virus in infected healthy animals. There have been no reported spillover cases of EBLV-2 into either wild or domestic animals.
Human cell-culture derived RABV vaccine prevented infection of mice from challenge with an EBLV-1 isolate from E. serotinus (Fekadu et al. 1988), as also demonstrated for EBLV-2 and ABLV (Brookes et al. 2005b). As in Australia, humans exposed to potentially rabid bats in Europe are treated with traditional rabies biologics (Nieuwenhuijs et al. 1992).
Two other lyssaviruses have been described in Europe; WCBV was isolated from a common bent-winged bat in south-west Russia (M. schreibersii) (Botvinkin et al. 2003; Kuzmin et al. 2005), and BBLV was isolated from a Natterer’s bat (Myotis nattererii) in Germany (Freuling et al. 2011) and subsequently from northeastern France in 2012 (Picard-Meyer et al. 2013) and a third isolation again from Germany (Freuling et al. 2013). Neither WCBV nor BBLV have been associated with human or animal disease. An addition tentative lyssavirus was recently isolated from a bent-winged bat (M. schreibersii) in Spain, but it has also not been associated with human or animal disease (Aréchiga Ceballos et al. 2013).
Australian Bat Lyssavirus
Australian bat lyssavirus ( ABLV ) was described in 1996 (Fraser et al. 1996). While Rhabdoviruses of the genus Ephemerovirus were known to occur, Australia had historically been considered free of lyssaviruses. However, St George (1989), postulating the origins of Adelaide River virus, an ephemerovirus antigenically related to rabies, had previously suggested the possibility of an undiscovered rabies-like virus in Australian bats, reflecting that the typically low prevalences of lyssaviruses in bats meant that an Australian bat lyssavirus might not become evident unless active surveillance of bats was undertaken, or unless man or a domestic animal became infected. Notwithstanding marked antigenic and genetic similarities to rabies virus, Australian bat lyssavirus is phylogenetically distinct, and represents a new lyssavirus genotype (Hooper et al. 1997; Gould et al. 1998).
ABLV has been detected in both suborders of bats in Australia (McCall et al. 2000; Gould et al. 2002; Warrilow et al. 2003). Phylogenetic analyses indicate that ABLV forms a monophyletic group which differentiates into two distinct clades, one associated with the pteropid species, and one with the insectivorous Saccolaimus flaviventris (yellow-bellied sheath-tailed bat), and that the two clades have a nucleotide divergence of up to 18.7 % (Guyatt et al. 2003). However, the ecology of ABLV is yet to be fully understood. Field (2005) found serological evidence of infection in numerous other bat species across five families, indicating that the bat–virus relationship is mature, and that additional variants may exist. Barrett (2004) reported a statistical association between species, age and health status and ABLV infection in bats, noting that most FAT-positive bats had a clinical history of generalized paresis, with a small number overtly aggressive, and others clinically indistinguishable from FAT-negative bats.
There have been three human deaths attributed to ABLV in Australia. The first case (in 1996) involved a wildlife rehabilitator who had been bitten by a yellow-bellied sheath-tailed bat in her care 5 weeks previously (Allworth et al. 1996; Speare et al. 1997; Hooper et al. 1997). The second case (in 1998) resulted from a bite from a pteropid bat following bat-initiated contact, and had an extended 2-year incubation following exposure in 1996 (Hanna et al. 2000). The third case (in 2013) also followed bat-initiated contact by a pteropid bat (Francis et al. 2014b). In all cases, the disease was similar to that caused by classical rabies (GT1) (Francis et al. 2014a). Both public health and animal health authorities in Australia strongly advise members of the general public not to handle bats and to seek medical advice should contact occur.
Standard preparations of cell-culture vaccine and human immunoglobulin against rabies virus are used to treat persons exposed or at risk of exposure to ABLV (Anon 2014). This regimen protects mice in experimental challenges (McCall et al. 2000; Brookes et al. 2005b), although the findings of Brookes et al. suggest that to ensure efficacy against ABLV, it may be prudent to maintain an antibody titer higher than that recommended for rabies virus. Bat rehabilitators and others likely to be exposed to bats are strongly encouraged to implement a preexposure vaccination strategy using cell-culture vaccine.
In 2013, two related equine cases of ABLV were reported. Virus nucleotide sequence from the horses was identical to the yellow-bellied sheath-tailed bat variant (Annand and Reid 2014). Prior to, and since this incident, there have been no reported cases of ABLV infection in terrestrial wildlife or domestic animals. In limited studies to date, experimental exposure of domestic cats and dogs to ABLV caused occasional and transient mild clinical signs, with no evidence of virus persistence. Most of the exposed animals seroconverted, and some had anti-ABLV antibodies in cerebrospinal fluid (McColl et al. 2007). Further studies are warranted to ascertain the susceptibility of terrestrial animals to bat lyssaviruses.
Rabies-Like Bat-Borne Viruses in Asia
Three lyssaviruses have been described in Asia—ARAV, which was isolated from a Lesser Mouse-eared Bat (Myotis blythi) in the Osh region of Kyrghyzstan, central Asia, in 1991 (Arai et al. 2003); KHUV, which was isolated from a female whiskered bat (M. mystacinus) in northern Tajikistan in 2001 (Kuzmin et al. 2003); and IRKV, which was isolated from the brain of a greater tube-nosed bat (Murina leucogaster) in 2002 in the town of Irkutsk in East Siberia (Botvinkin et al. 2003). IRKV was subsequently isolated from a greater tube-nosed bat in China (Liu et al. 2013b).
There have been a number of reports suggesting the presence of rabies-like viruses in bats in Asia, including serological studies in Thailand demonstrating the presence of neutralizing antibodies to ARAV, KHUV, IRKV, and ABLV largely associated with P. lylei fruit bats (Lumlertdacha et al. 2005); in China with evidence of rabies-like antibodies in various bat species but particularly in Rousettus leschenaultia fruit bats (Jiang et al. 2010); in Cambodia, with neutralizing antibodies to EBLV-1 in insectivorous bats and to ABLV in frugivorous bats (largely Cynopterus sphinx and P. lylei) (Reynes et al. 2004); and in Philippines, with neutralizing antibodies to ABLV in M. schreibersii (Arguin et al. 2002). While there have been no reports of disease due to bat-borne lyssaviruses in Asia, a number of anecdotal reports have described rabies-like illness following bat bites, particularly in China (Liu et al. 2013b).
Transmission of Lyssaviruses from and Between Bats
Transmission of bat-associated lyssaviruses occurs primarily by bite, when virus present in the saliva of an infected individual is directly inoculated into a susceptible individual. The potential for non-bite transmission of rabies virus among bats through saliva exchanged during mutual grooming has been suggested; transmission by such a mechanism may have precipitated an epidemic of rabies among kudu (Tragelaphus strepsiceros), an African ungulate (Barnard et al. 1982). Mexican free-tailed bats may transmit rabies virus in utero, as virus isolates have been obtained from cell lines established with fetal tissue (Steece and Calisher 1989). Airborne transmission of rabies virus was suggested as the possible event leading to two cases of rabies in humans visiting a cave harboring millions Mexican free-tailed bats (Irons et al. 1957; Humphrey et al. 1960). In subsequent experiments, a number of caged animals placed within caves developed rabies, and rabies virus has been isolated from air sampled from these same caves (Constantine 1967b, c; Winkler 1968). Experimental aerosol infection of mice with RABV and EBLV-2 found that mice were highly susceptible to infection by inhalation whereas ELBV-2 required direct intranasal inoculation (Johnson et al. 2006). Most recently, laboratory mice and wild-caught big brown bats (E. fuscus) and Mexican free-tailed bats were exposed to aerosolized rabies virus. All the bats and some of the mice survived exposure and produced rabies neutralizing antibody, but this antibody provided poor protection for the bats to a subsequent challenge with rabies virus 6 months later (Davis et al. 2007). Corneal transplants have been the source of human-to-human transmission of rabies virus on several occasions (Houff et al. 1979; Anon 1980, 1981; Gode and Bhide 1988), and tissues transplanted from an individual infected by a bat-associated rabies virus variant caused multiple deaths among recipients in the United States (Srinivasan et al. 2005). Most human rabies cases caused by bat-associated variants of rabies virus have involved “cryptic” exposures, as patients or family members often cannot provide a positive history of bat bite (Noah et al. 1998; Gibbons 2002; Messenger et al. 2003b; Franka et al. 2006).
Although spillover of bat-associated lyssaviruses to terrestrial mammals is rarely found in systematic surveys (McQuiston et al. 2001), clusters of bat-associated rabies have been documented in gray foxes (Urocyon cinereoargenteus) (Smith et al. 1986), red foxes (V. vulpes) (Daoust et al. 1996), and skunks (Mephitis mephitis) (Leslie et al. 2006) in North America. Such data suggest that rabies epidemics among terrestrial mammals may in rare instances be seeded by spillover from bats. Work on transmission and establishment of RABV between bat species shows diminishing frequencies of both cross-species transmission and host shifts with increasing phylogenetic distance (Streicker et al. 2010).
Understanding of the epidemiology and mechanisms of persistence of lyssavirus infection in bat populations is limited, but progress is being made through a combination of collection of field data and epidemiological modeling. Given the wide variation in life histories of different bat species, there may be significant variation in mechanisms of viral persistence in different situations. A study by George et al. (2011) of RABV in big brown bats (E. fuscus) in Colorado suggested that the slowing effects of hibernation on viral activity until susceptible individuals from the annual birth pulse become infected allows infection to maintain in the population. The persistence of EBLV-1 was investigated in a system involving multiple bat species which do not hibernate and it was found interspecies transmission was important for maintenance of infection in some species (Pons-Salort et al. 2014). Another study of LBV in a single species, Eidolon helvum, where hibernation is also absent, found a lack of increased mortality in seropositive animals suggesting infection did not result in disease after extended incubation (Hayman et al. 2012). This may suggest acute transmission of bat lyssaviruses in adapted bat hosts occurs at a much higher rate than the occurrence of disease.
Henipaviruses
The genus Henipavirus currently consists of three characterized viruses—Hendra virus, Nipah virus, and Cedar virus. Recently, evidence of related viruses has been reported in Africa, Asia, and Central and South America (Croser and Marsh 2013). Hendra and Nipah viruses emerged from fruit bats of the genus Pteropus (Order Chiroptera) to cause disease in livestock and humans (below). Both have been associated with severe neurologic disease, and both are classified as biosafety level 4 (BSL4) agents because they pose a high risk of laboratory transmission and life-threatening disease. As a consequence, laboratory work involving live virus should be done under physical containment level 4 (PC4) conditions. Hendra virus was first described in 1994 in Australia after a fatal disease outbreak in horses and humans in a horse-racing stable. Nipah virus was first described in 1999 in the investigation of a major outbreak of disease in pigs and humans in Malaysia.
Hendra Virus
In September 1994, an outbreak of acute respiratory disease of unknown etiology occurred in thoroughbred horses in a training complex in Brisbane (Queensland, Australia) (Murray et al. 1995). The syndrome was characterized by severe respiratory signs and high mortality, with 13 horses dying from acute disease. The trainer and a stable-hand suffered a concurrent severe febrile illness, with a fatal outcome for the trainer. Quarantine procedures and movement restrictions were applied, including a complete shutdown of the racing industry, and epidemiological investigations commenced (Baldock et al. 1996). The causal agent was shown to be a previously undescribed virus of the family , and initially named equine morbillivirus (EMV), but it was later renamed Hendra virus (after the Brisbane suburb where the outbreak occurred) when further characterization identified features inconsistent with morbilliviruses (Wang et al. 2000). To the end of 2014, a total of 51 incidents involving 92 confirmed or suspected equine cases have been reported in the adjoining eastern Australian states of Queensland and New South Wales (Biosecurity Queensland 2014) (Table 2; Fig. 2). The majority of incidents consist of single cases, even though in-contact horses are typically present. Case fatality rate in horses is around 75 % (Field et al. 2011); to date, all non-fatally infected horses have been euthanased because of uncertainty of the risk of recrudesence. Seven human cases have been reported, four of which were fatal; all seven cases are attributed to close and direct contact with infected horses. Two canine cases have been reported, both of which were on equine case properties (Anon 2013).
Table 2.
Identified Hendra virus spillovers in Australia since the virus was described in 1994 to 30 December 2014 (adapted from Biosecurity Queensland 2014)
| Year | Month | Location | State | Equine casesa (fatal) | Human cases (fatal) |
|---|---|---|---|---|---|
| 1994 | August | Mackay | QLDb | 2 (2) | 1 (1) |
| Sept | Hendra (Brisbane) | QLD | 20 (13) | 2 (1) | |
| 1999 | Jan | Trinity Beach (Cairns) | QLD | 1 (1) | 0 |
| 2004 | Oct | Gordonvale (Cairns) | QLD | 1 (1) | 1 (0) |
| Dec | Townsville | QLD | 1 (1) | 0 | |
| 2006 | June | Peachester | QLD | 1 (1) | 0 |
| Oct | Murwillumbah | NSWb | 1 (1) | 0 | |
| 2007 | June | Peachester | QLD | 1 (1) | 0 |
| July | Clifton Beach (Cairns) | QLD | 1 (1) | 0 | |
| 2008 | July | Redlands | QLD | 8 (7) | 2 (1) |
| Proserpine | 4 (3) | 0 | |||
| 2009 | Jul | Cawarral | QLD | 4 (3) | 1 (1) |
| Sept | Bowen | QLD | 2 | 0 | |
| 2010 | May | Tewantin | QLD | 1 | 0 |
| 2011 | June | Beaudesert, Boonah, Logan | QLD | 5 (5) | 0 |
| Wollongbar | NSW | 2 (2) | 0 | ||
| July | Park Ridge, Kuranda, Hervey Bay, Boondall, Chinchilla | QLD | 5 (5) | 0 | |
| Macksville, Lismore, Mullumbimby | NSW | 3 (3) | 0 | ||
| August | Currumbin | QLD | 1 (1) | 0 | |
| Ballinac, Mullumbimby | NSW | 5 (5) | 0 | ||
| Sept | Beachmere | QLD | 3 (1) | 0 | |
| 2012 | Jan | Townsville | QLD | 1 (1) | 0 |
| May | Rockhampton, Ingham | QLD | 2 (2) | 0 | |
| June | Mackay | QLD | 1 (1) | 0 | |
| July | Rockhampton, Cairns | QLD | 4 (4) | 0 | |
| Sept | Port Douglas | QLD | 1 (1) | 0 | |
| Oct | Ingham | QLD | 1 (1) | 0 | |
| 2013 | Jan | Mackay | QLD | 1 (1) | 0 |
| Feb | Kuranda | QLD | 1 (1) | 0 | |
| June | Macksville | NSW | 1 (1) | 0 | |
| Laidley | QLD | 1 (1) | 0 | ||
| July | Currumbin | QLD | 1 (1) | 0 | |
| Macksville, Kempsey | NSW | 2 (2) | 0 | ||
| 2014 | March | Bundaberg | QLD | 1 (1) | 0 |
| June | Beenleigh | QLD | 1 (1) | 0 | |
| July | Gladstone | QLD | 1 (1) | 0 | |
| Total | 50 incidents | 92 (82) | 7 (4) |
aIncludes confirmed cases and unconfirmed possible cases
b QLD Queensland, NSW New South Wales
cThree separate incidents
Fig. 2.
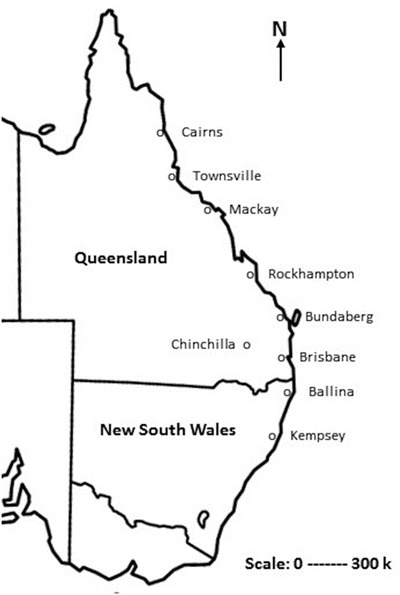
Map of the eastern Australian states of Queensland and New South Wales. The map shows the northern (Cairns), southern (Kempsey), and western (Chinchilla) extent of reported Hendra virus cases to December 2014. Additional marked locations provide an incomplete illustration of other case locations
There has been an escalating frequency of reported cases, with six incidents in the first decade, nine incidents between 2006 and 2010, then a super cluster of 18 incidents in 2011, and a further 19 between 2012 and 2014 (Biosecurity Queensland 2014). Whether this reflects an increased incidence of spillover, or increased awareness, diagnosis, and reporting is unknown. Similarly, since 2011, there has been an increased frequency of reported cases in New South Wales, and whether this reflects an expansion of a key reservoir host species is also unknown.
Nipah Virus
A major outbreak of disease in pigs and humans occurred in peninsular Malaysia between September 1998 and April 1999, resulting in the death of 106 of 265 reported human cases and the culling of over one million pigs (Chua et al. 1999, 2000; Nor et al. 2000). The outbreak spread to Singapore where a cluster of 11 cases with one death occurred at an abattoir (Paton et al. 1999). Initially attributed to Japanese encephalitis virus, the etiological agent was eventually shown to be another previously undescribed virus of the family Paramyxoviridae closely related to Hendra virus (Anon 1999b). The Malaysia outbreak primarily impacted pig and human populations, although horses, dogs, and cats were also infected. No cases of Nipah virus have been recorded in Malaysia since 1999. The disease reappeared, however, with multiple human cases diagnosed in Bangladesh in 2001 (Anon 2003). Near-annual seasonal clusters of human Nipah virus infections have been recorded in Bangladesh since (Anon 2004a, b, 2005; Hsu et al. 2004; Luby et al. 2006; Kulkarni et al. 2013; M Rahman, personal communication 2015) and occasionally in West Bengal (Chadha et al. 2006; Arankalle et al. 2011) (Table 3). In contrast to Malaysia where the majority of cases were restricted to areas where pig farming was common, the Bangladesh cases do not usually involve a domestic animal cycle (Epstein et al. 2006), and occur over a wide geographic area (Fig. 3). Food-borne transmission via date palm sap contaminated with bat saliva or bat urine is believed to be the major route of transmission (Epstein et al. 2006; Luby et al. 2006, 2009a; Rahman et al. 2012; Luby and Gurley 2012), although human-to-human transmission may also occur relatively frequently (Epstein et al. 2006; Gurley et al. 2007; Luby et al. 2009a, b; Luby and Gurley 2012).
Table 3.
Reported human cases of Nipah virus in Bangladesh and India1 since the first detection in 2001 to 30 December 2014
| Year | Month | Location | Cases | Deaths | CFR(%) |
|---|---|---|---|---|---|
| 2001 | Jan–Feb | Siliguri1 | 66 | 45 | 68 |
| April–May | Meherpur | 13 | 9 | 69 | |
| 2003 | Jan | Naogaon | 12 | 8 | 67 |
| 2004 | Jan–April | Rajbari, Faridpur | 67 | 50 | 75 |
| 2005 | Jan–March | Tangail | 12 | 11 | 92 |
| 2007 | Jan–April | Thakurgaon, Kustia, Pabna, Natore, Naogaon | 18 | 9 | 50 |
| April | Nadia1 | 5 | 5 | 100 | |
| 2008 | Feb–April | Manikgonj, Rajbari, Faridpur | 11 | 9 | 82 |
| 2009 | Jan | Gaibandha, Rangpur, Nilphamari, Rajbari | 4 | 1 | 25 |
| 2010 | Feb–Mar | Faridpur, Rajbari,Gopalganj, Madaripur | 16 | 14 | 87.5 |
| 2011 | Jan–Feb | Lalmohirhat, Dinajpur, Comilla, Nilphamari, Rangpur | 44 | 40 | 91 |
| 2012 | Feb | Joypurhat, Rajshahi, Natore, Rajbari, Gopalganj | 12 | 10 | 83 |
| 2013 | Feb–April | Gaibandha, Jhinaidaha, Kurigram, Kushtia, Magura, Manikgonj, Natore, Mymenshingh, Naogaon, Nilphamari, Pabna, Rajbari, Rajshahi | 24 | 21 | 87.5 |
| 2014 | Jan–Feb | Manikganj, Magura, Faridpur, Rangpur, Shaariatpur, Kushtia, Rajshahi, Natore, Dinajpur, Dhaka, Chapai Nawabganj, Naogaon, Madaripur | 27 | 14 | 52 |
| TOTALs | 331 | 246 | 74 % |
The table compiled from WHO (2015), the Institute of Epidemiology, Disease Control and Research for 2013–2014 (http://www.iedcr.org), and with additional information provided by Prof M Rahman, Institute of Epidemiology, Disease Control and Research, Dhaka (Personal Communication)
1Cases reported from outbreaks in India.
Fig. 3.
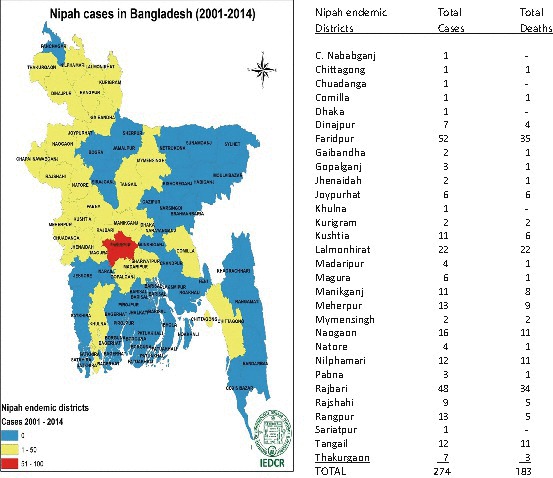
Map of Bangladesh showing districts with Nipah virus human cases, 2001–2014. Map provided by courtesy of Prof M Rahman, Institute of Epidemiology Disease Control and Research (IEDCR), Dhaka, Bangladesh
Phylogeny
Initial nucleotide sequences of the matrix (M) and fusion (F) proteins genes established that Hendra virus had a greater homology with known morbilliviruses than with other genera of the family Paramyxoviridae (Gould 1996), but the sequence comparisons also revealed substantial divergence with other morbilliviruses. Subsequent sequencing of the entire genome confirmed Hendra virus as a member of the subfamily Paramyxovirinae, but identified differences that supported the creation of a new genus. Hendra virus had a larger genome size, the replacement of a highly conserved sequence in the L protein gene, different genome end sequences, and other sequence and molecular features (Wang et al. 2000). After the characterization of the Nipah virus genome, “Henipavirus” was proposed as the new genus, with Hendra virus the type species and Nipah virus the second member. The ICTV has formally recognized the genus Henipavirus, and the virus names, Hendra virus and Nipah virus. In recent years, at least three henipa-like viruses have been identified. These are the Cedar virus from an Australian flying fox (Marsh et al. 2012), the African bat paramyxovirus M74a from Ghana (Drexler et al. 2012), and the Mojiang virus from rats in China (Wu et al. 2014). Although their formal classification into the genus Henipavirus is yet to be confirmed, their close phylogenetic relationship with Hendra and Nipah viruses strongly support this putative classification (Fig. 4).
Fig. 4.
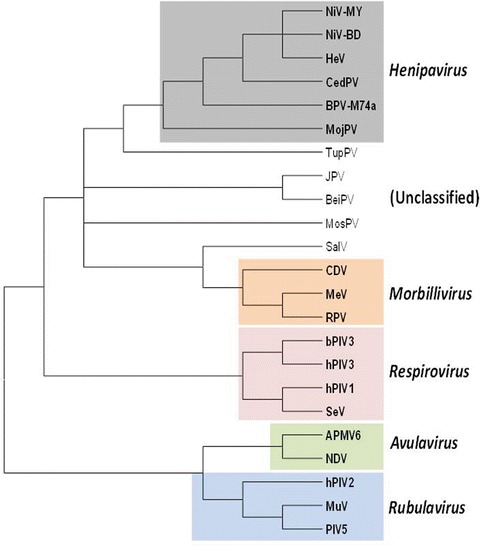
Phylogenetic tree based on the N protein sequences of selected paramyxoviruses. Viruses used in this analysis are chosen based on their relative close genetic relationship with known henipaviruses in the subfamily Paramyxovirinae. Virus name (abbreviation) and GenBank accession numbers are as follows: Bat paramyxovirus/Eid hel/GH-M74a/GHA/2009 (BatPV-M47a) HQ660129; Beilong virus (BeiPV) DQ100461; Bovine parainfluenza virus 3 (bPIV3) AF178654; Canine distemper virus (CDV) AF014953; Cedar virus (CedPV) JQ001776; Hendra virus (HeV) AF017149; Human parainfluenza virus 3 (hPIV3) Z11575; J virus (JPV) AY900001; Measles virus (MeV) AB016162; Mojiang virus (MojPV) KF278639; Mossman virus (MosPV) AY286409; Nipah virus, Bangladesh strain (NiV-BD) AY988601; Nipah virus, Malaysian strain (NiV-MY) AJ627196; Rinderpest virus (RPV) Z30697; Salem virus (SalPV) JQ697837; Sendai virus (SeV) M19661; Tupaia paramyxovirus (TupPV) AF079780
The Role of Bats
Once the virus was identified, serological screening of ubiquitous native and introduced fauna in the vicinity of the index case was undertaken to determine the origin of the virus, but no evidence of Hendra virus infection. Following the identification of a second Hendra virus outbreak in horses near the city of Mackay (1000 km north of Brisbane), the focus of the wildlife surveillance shifted to species that were common to both locations and capable of moving between locations. Mammal species were given a higher priority than avian species. Of 27 flying foxes from two species tested in an initial survey, 40 % had anti-Hendra virus neutralizing antibodies (Field 2005), and in September 1996, 2 years after the first reported outbreak, virus was isolated from a grey-headed flying fox (P. poliocephalus) (Halpin et al. 2000). A concurrent survey of over 1000 flying foxes from the four mainland species (P. poliocephalus, P. Alecto, P. conspicillatus, and P. scapulatus) identified an estimated crude seroprevalence of 47 %. In a retrospective serological survey, antibodies neutralizing Hendra virus were identified in the sera of flying foxes collected in 1982 (Field 2005).
With the demonstration that Nipah and Hendra viruses were closely related, Malaysian bat species were targeted as possible reservoirs of Nipah virus, based on the established bat–Hendra virus link in Australia. Of 324 bats from 14 species surveyed in peninsular Malaysia, neutralizing antibodies to Nipah virus were found in 21 bats from five species, but predominantly in two Pteropus species, P. vampyrus and P. hypomelanus (Johara et al. 2001). Subsequently, Nipah virus was isolated from the urine of P. hypomelanus, and from dropped partially eaten fruit (Chua et al. 2002).
The identification of pteropid bats as the primary reservoir host of both Hendra and Nipah viruses (Young et al. 1996; Halpin et al. 2000; Johara et al. 2001; Chua et al. 2002) was a major breakthrough in understanding the ecology of these “new” viruses, and not only informed management strategies (Mackenzie et al. 2003; Field et al. 2004; Breed et al. 2011; Kung et al. 2013), but precipitated further investigation of the ecology of henipaviruses and factors associated with their emergence. Subsequent studies in Australia further elaborated the ecology of Hendra virus in pteropid bats (Plowright et al. 2008; Breed et al. 2011; Field et al. 2011). There is also evidence that some species may be more significant natural reservoirs than others (Smith et al. 2014; Goldspink et al. 2015; Edson D, Field HE, Broos et al. Routes of Hendra virus excretion in naturally infected flying-foxes: implications for viral transmission and equine spillover risk, submitted for publication), and that viral excretion in bats may be seasonal (Field HE, Jordan D, Melville D et al. Spatio-temporal aspects of Hendra virus infection in pteropid bats in eastern Australia, submitted for publication), which may explain the spatiotemporal occurrence of Hendra virus spillover events and inform risk management strategies. Subsequent studies in Malaysia (Rahman et al. 2010, 2013; Sohaytati et al. 2011) and Bangladesh (Epstein et al. 2008; Khan et al. 2010; Hahn et al. 2014a, b) have elaborated the ecology of NiV in bats. More recent studies have described the occurrence of henipaviruses on a global scale (Wacharapluesadee et al. 2005; Sendow et al. 2006; Hayman et al. 2008b; Drexler et al. 2009, 2012; Chong et al. 2009; Wacharapluesadee et al. 2010; Weiss et al. 2012: Breed et al. 2013; Croser and Marsh 2013; Peel et al. 2013; Muleya et al. 2014; Ching et al. 2015).
There is little doubt that pteropid bats are major reservoir hosts of Hendra and Nipah viruses (Field et al. 2007), and it is not surprising that additional related viruses have now been detected throughout their range, which extends from the west Indian Ocean islands of Mauritius, Madagascar, and Pemba Island, along the sub-Himalayan region of Pakistan and India, through southeast Asia, The Philippines, Indonesia, to the southwest Pacific islands and Australia. There are about 60 species in total. Flying-foxes range in body weight from 300 g to over 1 kg, and in wingspan from 600 mm to 1.7 m. They are the largest bats in the world, do not echolocate, and navigate at night by eyesight and their keen sense of smell. All species eat fruits, flowers or pollen, and roost communally in trees. Flying foxes are nomadic species, capable of traveling distances of hundreds of kilometers. Where the distributions of different species overlap, roosts are shared (Hall and Richards 2000; Corbet and Hill 1992; Mickleburgh et al. 1992; Nowak 1994). Thus the potential exists for interaction between flying fox populations across much of their global distribution. Recent reports of henipa-like virus detections in non-pteropid and in other mega bat species suggest that the bat–virus relationship may be even more ancient (Li et al. 2008; Croser and Marsh 2013).
Calisher et al. (2006) review the apparent association between bats and emerging infectious diseases. They contend that information about the natural history of most viruses in bats is limited, and specifically in relation to the family Pteropodidae, that only half of the 64 genera in this family (which includes flying foxes) have been adequately studied. Thus we know relatively little about the bats from which the henipaviruses have emerged. Calisher et al. (2006) pose a number of questions in relation to the role of bats and emerging zoonoses. Do bats possess special attributes that equip them to host highly pathogenic zoonoses? Are emergences such as Hendra and Nipah viruses infrequent and incidental events, or are we detecting only the tip of the iceberg? They conclude by calling for pre-emptive potential pathogen screening in wildlife, rather than the outbreak-response surveillance that typically occurs currently.
Clinical Presentation
The clinical presentations of human and animal cases infected with Hendra and Nipah viruses are described in more detail in section “Henipaviruses.”
Hendra Virus in Animals
The putative index case in Brisbane in 1994 was a heavily pregnant thoroughbred mare at pasture. She was moved to a training stable for nursing and died within 48 h. A further 12 horses in the stable and an adjoining training stable died in the following 14 days. Clinical signs included fever, facial swelling, severe respiratory distress, ataxia, and terminally, copious frothy (sometimes blood-tinged) nasal discharge. The incubation period based on clinical observations was 8–16 days. There were four non-fatal cases, two of which exhibited mild neurological signs. A further three horses were subsequently found to have seroconverted in the absence of obvious clinical signs (Baldock et al. 1996). A small number of horses in the stable remained unaffected. A second Hendra virus outbreak in horses was retrospectively diagnosed in October 1995 after the Hendra virus-attributed death of a farmer who suffered a relapsing encephalitic disease. This second incident (1000 km north of Brisbane) chronologically preceded the Brisbane outbreak by several weeks, and resulted in the death of two horses, a 10 year old heavily pregnant thoroughbred mare and a 2 year old colt in an adjoining paddock, after a 24 h clinical course (Rogers et al. 1996; Hooper et al. 1996). Numerous other horses on the property remained unaffected.
Extensive investigations were undertaken in relation to these two outbreaks. No antibodies to Hendra virus were found in over 5000 domestic animals surveyed (including 4000 horses) (Rogers et al. 1996; Ward et al. 1996) and no epidemiological link was identified between the two outbreaks. Retrospective investigations found no evidence of previous infection in horses in Queensland.
There are no pathognomonic signs for Hendra virus infection in horses. Common features are initial depression, inappetence, and fever, rapidly progressing to fulminating neurologic and/or respiratory disease (Biosecurity Queensland 2014). The primary pathogenesis is a loss of vascular integrity associated with vasculitis, the primary location of which may determine whether the predominant clinical presentation is respiratory (Murray et al. 1995; Baldock et al. 1996) or neurological (Field et al. 2010). The majority of equine incidents involve single cases. The typical absence of transmission to in-contact horses suggests that Hendra virus is not normally highly contagious in horses, and that direct contact or mechanical transmission of infectious material is necessary for transmission to occur.
Hendra Virus in Humans
There are seven recorded human cases of Hendra virus infection, all of which are attributed to direct and close contact with infected horses. A serological survey of bat rehabilitators found no evidence of bat-to-human transmission (Selvey et al. 1996). Human-to-human transmission has not been reported. There have been no human cases since 2009 despite an increasing frequency of reported equine cases, suggesting that risk communication to at-risk groups (horse owners, veterinarians, and para-veterinarians) and the adoption of risk minimization strategies have been effective. Clinical presentation is discussed in section “Henipaviruses,” but briefly infection is characterized by an acute respiratory syndrome and/or encephalitic syndrome or relapsing encephalitis (Selvey et al. 1995; Allworth et al. 1995; Playford et al. 2010; Mahalingam et al. 2012).
Nipah Virus in Animals
Pigs on commercial pig farms were the predominant infected species in the Malaysian outbreak. Herd-level infection was typically subclinical, with estimated morbidity and mortality rates of 30 % and 5 %, respectively (Nor et al. 2000). The incubation period was estimated to be 7–14 days. Observations of clinical cases suggested a varying presentation in different classes of animals. Affected weaners and porkers (2–6 months) typically showed acute febrile illness with respiratory signs ranging from rapid and labored breathing to harsh nonproductive cough. Attributed neurological signs included trembling, twitching, muscular spasms, rear leg weakness and variable lameness or spastic paresis. Adult sows and boars typically suffered a peracute or acute febrile illness with labored breathing (panting), increased salivation and serous, mucopurulent or blood-tinged nasal discharge. Neurological signs including agitation and head pressing, tetanus-like spasms and seizures, Nystagmus, champing of mouth, and apparent pharyngeal muscle paralysis were observed. The primary means of spread between farms and between regions was the movement of pigs. The primary mode of transmission on-farm was likely oro-nasal. Secondary modes of transmission between farms within local farming communities may have included roaming infected dogs and cats. Evidence of infection (virus isolation, immunohistochemistry, serology) and neurologic disease was found in dogs and horses in the outbreak area (Nor et al. 2000). Transmission studies in pigs in Australia at the CSIRO Australian Animal Health Laboratory established that pigs could be infected orally and by parenteral inoculation, and that infection could spread quickly to in-contact pigs. Neutralizing antibodies were detectable 10–14 days postinfection (Middleton et al. 2002). In a related study, experimental infection in cats caused neurological disease (Middleton et al. 2002).
The early epidemiology of the outbreak in the northern state of Perak, and the spillover mechanism that first introduced the infection to pigs remains uncertain, however, retrospective investigations indicated that Nipah virus was responsible for sporadic disease in pigs in Perak since late 1996 (Field et al. 2001). Mathematical modeling supports the hypothesis that at least one spillover event occurred before the 1998–1999 outbreak, and that a level of residual immunity in sows provided the right herd immunological conditions for infection to become endemic in the pig index case farm in 1998, thus providing a sustained reservoir of virus from which to infect other farms (Daszak et al. 2006; Pulliam et al. 2012).
Evidence of Nipah virus infection in domestic species has recently been reported in Bangladesh (Chowdhury et al. 2014).
Nipah Virus in Humans
At least 105 people died during the course of the Malaysian outbreak. The majority of human cases had a history of direct contact with live pigs. Most were adult male Chinese pig-farmers (Chua et al. 1999; Parashar et al. 2000). Identified risk factors for human infection in Malaysia were activities requiring direct contact with pigs, with handling sick pigs and assisting with birthing posing the highest risks. Clinical presentation of Nipah virus cases is given in more detail in section “Henipaviruses,” and only briefly described below. Most patients presented with acute encephalitis characterized by fever, headache, myalgia, disorientation, dizziness, vomiting and more than 50 % had a reduced level of consciousness (Chua et al. 1999; Goh et al. 2000). The major clinical signs included areflexia, segmental myoclonus, tachycardia, hypertension, pin-point pupils, and an abnormal doll’s eye reflex. Most patients who survived acute encephalitis made a full recovery, but about 20 % had residual neurological deficits (Goh et al. 2000; Chong et al. 2002; Lim et al. 2003). Neurological sequelae included cognitive difficulties, tetraparesis, cerebellar signs, nerve palsies, and clinical depression. A number of patients developed relapse encephalitis or late onset encephalitis. About 7.5 % of patients who recovered from acute encephalitis and 3.4 % of those who experienced nonencephalitic or asymptomatic infection developed late neurological disease, presenting several months to 4 years after the initial infection (Goh et al. 2000; Tan et al. 2002). The mortality rate associated with relapse and late onset encephalitis was 18 % which was lower than the 40 % associated with acute encephalitis. However, 61 % of patients with relapse or late onset had further neurological sequelae compared with 22 % after acute encephalitis. The occurrence and frequency of clinically undetected Nipah virus infections was also notable: 6 % of persons from farms without reported encephalitis cases, and 11 % of persons from farms with reported encephalitis cases. In addition, 8 % of cases reported no contact with pigs (Parashar et al. 2000). Clinical presentation of human cases in Bangladesh and West Bengal, India, has typically been similar to that in Malaysia: fever, central nervous system signs, and a high case fatality rate (Luby et al. 2006, 2009a, b; Homaira et al. 2010a, b). Notably, a cluster of cases in the Faridpur district in 2004 exhibited an acute respiratory distress syndrome (Anon 2004b), and nosocomial and corpse to human transmission has been reported (Sazzad et al. 2013).
Evidence of Henipavirus infection in humans has recently been reported in Africa (Pernet et al. 2014), and was most frequent in those butchering fruit bats for human consumption. Indeed the most significant risk factors were butchering fruit bats and living in areas undergoing deforestation.
Flaviviruses
The family Flaviviridae contains some of the most important encephalitogenic arthropod-borne viruses, including Japanese encephalitis (JEV), West Nile (WNV), St Louis encephalitis (SLEV) (see Chapters 11 and 13), and the tick-borne encephalitis (TBEV) viruses (Gould et al. 2004). In addition, the dengue viruses (DEN1-4) have increasingly been shown to also cause encephalitis and other neurological manifestations (e.g., Solomon et al. 2000; Madi et al. 2014; Sahu et al. 2014; Tan et al. 2014) (Chapter 12). Although there are many reports describing serological evidence that some of these pathogenic flaviviruses can infect bats, and viruses have been isolated from bats, there has been relatively little direct evidence to substantiate a role for bats in virus transmission cycles (Calisher et al. 2006; Wong et al. 2007), with the possible exceptions of JEV and SLEV. Various other flavivirus species have also been isolated from bats (Gaunt et al. 2001; Calisher et al. 2006; Mackenzie and Williams 2009), but they have not been shown to cause disease in humans or animals, with the exception of Montana Myotis leukoencephalitis virus which can cause encephalitis in small rodents (Charlier et al. 2002).
Japanese Encephalitis Virus
JEV has been isolated from a number of bats of the families Pteropodidae, Rhinolophidae, Hipposideridae, and Vestpertilionidae. Early studies found that high titers of virus could be detected in the brains of microchiropteran bats infected by intracerebral inoculation, and although the titers were as high as found in fatal murine infection, the bats appeared free from disease (Ito and Saito 1952). Early studies also demonstrated that bats infected subcutaneously were capable of maintaining a latent infection with JEV in simulated hibernation for as long as 107 days, and mosquito–bat–mosquito transmission was successful at room temperature, and at 10 °C in simulated cave situations (La Motte 1958), thus making bats potential maintenance hosts of JE virus and participants in wildlife transmission cycles. These very early findings led to some extensive and elegant investigations by Sulkin, Allen, and their colleagues (reviewed in Sulkin and Allen 1974). Their studies together with those of others showed that:
Nearly 100 % of bats inoculated subcutaneously with small doses of virus developed viraemia within 24–72 h, and that some animals circulated virus for as long as 25–30 days at titers high enough to infect mosquitoes
Bats did not develop encephalitis despite significant virus titers in the brain
Following subcutaneous inoculation, replication was demonstrated in the brown adipose tissue, and this tissue was able to sequester the virus in an inactive state during hibernation, and then seed virus to provide further viraemia once hibernation ended
Transplacental transmission could readily be demonstrated, particularly in the latter stages of pregnancy, providing a mechanism for virus perpetuation in nature
Anti-JEV antibody could not be reliably detected or measured by hemagglutination-inhibition, but only by neutralization
Bats maintained at room temperature developed a viraemia in 2–3 days postinoculation in most animals which usually persisted for 10–15 days, and neutralizing antibodies developed in 3–7 weeks
Bats maintained at 37 °C developed a viraemia more rapidly than those at room temperature and reached higher titers, but the duration of viraemia was shorter and there was little evidence of replication in brown fat, brain or kidney, and neutralizing antibodies responses were faster
About 25 % of bats at both temperatures failed to develop neutralizing antibodies despite being shown to be viraemic
Studies of field-caught bats collected in different seasons yielded 24 JE virus isolates (16 from 1139 M. schreibersii and 8 from 267 Rhinolophus cornutus), with a significant number of isolates coming from collected in the fall
Neutralizing antibodies to JE were found in sera from 5 % of M. schreibersii and 9 % of R. cornutus
Isolations of JE virus from bats was also extended to China (Taiwan) and one isolate was obtained from Hipposideros armiger, and two from M. schreibersii (it is interesting to note that 9 JE virus isolates were also obtained from Cx. annulus mosquitoes at the same time and cave as the latter M. schreibersii isolates)
Neutralizing antibodies to JE virus were found in a number of other species of bats in Japan, including 21/79 R. ferrum-equinum, 9/72 Myotis macrodactylus, 4/25 Myotis mystacinus; 1/31 Pipistrellus abramus, 10/110 Vespertilio superans, and 1/22 Plecotus auritus
More recently, neutralizing antibodies were found to JE in 46 of 626 sera collected from insectivorous bats in Karnataka, India. The positive sera were from five species: H. pomona, H. speoris, H. bicolor, H. cineraceus, and Rhinolophus rouxi. The incidence of antibodies in bats was reasonably well correlated with the incidence of JE in humans in Kolar district during 1983 and 1985 (Banerjee et al. 1988), and it was suggested that bats may be involved in virus amplification.
The involvement of family , or fruit bats, in the ecology of JE virus was first indicated from studies in Thailand in which neutralizing antibodies to JE were observed in 22 of 245 Cynopterus brachyotis (P.K. Russell, 1968, personal communication to Sulkin and Allen 1974), a species widely distributed in south-eastern Asia from Thailand to Lombok, and the Philippines. Experimental infection has been studied in two species of fruit bat in India, Rousettus leschenaulti (Banerjee et al. 1979) and C. sphinx (Banerjee et al. 1984). The former study demonstrated a low level of viraemia after subcutaneous inoculation of JE virus lasting up to 9 days. In the latter study, bats were infected intramuscularly with JE virus and, during the subsequent viraemic phase, Cx. bitaeniorhynchus and Cx. tritaeniorhynchus mosquitoes were allowed to feed on them. Transmission was observed between bats, from bats to chickens, and from chickens to bats. Thus frugivorous bats are potential candidates for virus maintenance and may assist in virus movement.
JEV has also been isolated from megachiropteran and microchiropteran bats in China (Wang et al. 2009; Liu et al. 2013a). All eight isolates were found to display high genetic homogeneity despite coming from different geographical areas, and were phylogenetically similar to human and mosquito isolates suggesting that bats may be involved in the natural cycle of JEV.
Experimental infection of black flying foxes, Pteropus alecto, have demonstrated that the bats could be infected with JEV after being bitten by infected Culex annulirostris mosquitoes or by subcutaneous inoculation. Anti-JEV IgG antibodies developed in the majority of the exposed bats, but only one animal exhibited low level viraemia and was able to infect recipient mosquitoes. Two further animals were also able to infect recipient mosquitoes despite their absence of viraemia (van den Hurk et al. 2009). These results demonstrate that the black flying-fox could potentially participate in natural transmission cycles of JEV.
Interestingly, a short genetic sequence (167 bp) identical to JEV strains from bats in China was observed in one pool of Cx. pipiens mosquitoes collected in Italy: further confirmation of this is urgently needed (Ravanini et al. 2012).
St Louis Encephalitis Virus
Early studies on natural and experimental infection of bats with SLEV were largely confined to the Mexican free-tailed bats (Tadarida brasiliensis) and little brown bats (Myotis lucifugus) (reviewed by Sulkin and Allen 1974). A number of isolates of SLEV were obtained from Mexican free-tailed bats during epizootic activity in Texas, and as isolations continued over winter months, these investigations indicated that SLEV could persist in this species (Allen et al. 1970). Experimental studies in Mexican free-tailed bats and big brown bats (Eptesicus fuscus) found that SLEV produced an intense and long-lasting viraemia in the Mexican free-tailed bats, and was maintained during hibernation in the big brown bats (Sulkin and Allen 1974; Herbold et al. 1983), providing further evidence of SLEV persistence in bats and suggesting a role for bats in virus spread and as potential wildlife reservoirs. Serological studies suggested that up to 9 % of big brown bats and little brown bats were seropositive for SLEV in a non-epizootic period, suggesting that these species are involved in the maintenance of SLEV in enzootic foci and could have a role in dissemination of SLEV to epizootic foci (Herbold et al. 1983). Serological evidence of SLEV in bats has been reported from Haiti (McLean et al. 1979) and Guatemala (Ubico and McLean 1995).
Other Flaviviruses
While there is evidence that bats might play a role in the persistence, over-wintering and possibly in the spread of SLEV, the evidence for bats playing a role in the natural transmission of other flaviviruses, such as WNV and DENV, is much less certain. Much of the evidence has come from serological data, but these are notoriously difficult to interpret because of cross-reacting antibodies to other members of the family from prior infections. Given these limitations, serological evidence of infection of bats with WNV has been reported from a number of countries (reviewed in Sulkin and Allen 1974), but there has only been a single report of virus isolation from a fruit bat, R. leschenaultia, in India (Paul et al. 1970). Interest in a possible role for bats in WNV transmission has increased since the emergence of WNV in North America, but although occasional bats have been found to have antibody to the virus including big brown bats, little brown bats, and Mexican free-tailed bats (Pilipski et al. 2004; Davis et al. 2005a; Bunde et al. 2006), there is no evidence to suggest they are involved in either transmission or persistence of the virus. Experimental infection has also suggested that big brown bats and Mexican free-tailed bats are unlikely to play a role as amplifying hosts of WNV (Davis et al. 2005a).
Serological investigations of several bat species at different geographic sites in the Americas have reported antibodies to DEN viruses (Platt et al. 2000; Aguilar-Setién et al. 2008; de Thoisy et al. 2009; Machain-Williams et al. 2013), and in some studies, specific viral RNA was detected by RT-PCR, especially from members of the genus Artibeus (Aguilar-Setién et al. 2008; de Thoisy et al. 2009; Sotomayor-Bonilla et al. 2014). However, as a cautionary note, experimental inoculation of Artibeus jamaicensis and A. intermedius bats showed that the bats were incapable of sustained DENV replication and were unlikely to act as reservoir hosts (Perea-Martínez et al. 2013; Cabrera-Romo et al. 2014). Thus the role of bats in dengue transmission remains uncertain, and further work is needed to resolve this issue.
Alphaviruses
A number of Alphaviuses have been associated with neurological disease, including the equine encephalitis viruses (Western, Eastern and Venezuelan), Sindbis virus, Semliki Forest virus, and less commonly Mayaro virus, chikungunya virus and possibly Ross River virus (Zacks and Paessler 2010), but the only Venezuelan equine encephalitis virus (VEEV) has an association with bats. The isolation of VEEV from wild-caught bats in Central and South America suggests that they may play a role in the ecology of VEEV. VEEV was isolated from single specimens of teapa fruit-eating bat (Artibeus turpis) and a gray short-tailed bat (Carollia subrufa) in Mexico, and experimental inoculation of Jamaican fruit bats (A. jamaicensus) resulted in viraemia as measured by infection of suckling mice (Scherer et al. 1971). VEEV has also been isolated from a vampire bat (Desmodus rotundus) in Mexico (Correa-Giron et al. 1972), a tent-making bat (Uroderma bilobatum) in Guatemala (Seymour et al. 1978), and a Seba’s short-tailed bat (Carollia perspicillata) in Brazil (Calisher et al. 1982). More recently, seroepidemiological studies in Guatemala (Ubico and McLean 1995) and Trinidad (Thompson et al. 2015), using neutralization in the former study and epitope-blocking enzyme-linked immunosorbent assay in the latter study, found antibodies to VEEV in various species of bats: Artebeus sp., including Jamaican fruit bats; Carollia sp. including Seba’s short-tailed bats, gray short-tailed bats, silky short-tailed bats (C. brevicauda); vampire bats; little yellow-shouldered bat (Sturnira lilium) and highland yellow-shouldered bat (S. ludovici); Pallas’s long-tongued bat (Glossophaga soricine); and greater bulldog bat (Noctilio leporinus). These results indicate bats may play a role as alternate hosts in the enzootic maintenance and spread of VEEV.
Although neutralizing antibodies to eastern equine encephalitis virus (EEEV) were found in bats in New England and Guatemala (Main 1979; Ubico and McLean 1995) and western equine encephalitis virus (WEEV) in Haiti (McLean et al. 1979), it is unlikely that bats play a role in either maintenance or spread of these viruses.
Coronaviruses
The neurological symptoms of coronavirus (CoV) infection in general will be covered in section “Alphaviruses.” This section discusses the role of bats in the emergence of two highly pathogenic novel zoonotic coronaviruses, severe acute respiratory syndrome (SARS) CoV, which emerged a decade ago in 2002, and Middle East respiratory syndrome (MERS) CoV, which emerged more recently in 2012.
Neurological manifestations following SARS-CoV infection were generally uncommon (Sung 2006), and consisted of isolated reports of epileptic fits, peripheral nerve disease, mental confusion, and disorientation (Hung et al. 2003; Chao et al. 2003). No focal neurological deficit or structural abnormality on computed tomography and magnetic resonance scans were found (Sung 2006). However, a number of patients developed affective psychosis during the acute phase of their illness associated with high-dose steroid use, personal vulnerability, and psychological stress (Lee et al. 2004). A chronic post-SARS condition characterized by a syndrome of chronic fatigue, pain, weakness, depression, and sleep disturbance has recently been reported (Moldofsky and Patcai 2011). Neurological manifestations have also been rare in MERS infections, with a single report of three patients who presented with severe neurologic syndrome, including altered level of consciousness ranging from confusion to coma, ataxia, and focal motor deficit (Arabi et al. 2015).
Early studies of the source of SARS-CoV indicated a possible zoonotic origin following the isolation of almost identical viruses from Himalayan palm civets (Paguma larvata) and a raccoon dog (Nyctereutes procyonoides) at a live animal market (Guan et al. 2003). Subsequent investigations, however, showed that palm civets in farms and field were largely free from SARS-CoV infection (Tu et al. 2004; Kan et al. 2005), suggesting that palm civets played a role as intermediate host rather than as a natural reservoir. Surveillance studies have revealed the presence of a diverse group of coronaviruses in bats (Drexler et al. 2014) including some closely related to the SARS-CoV in genome organization and sequence, named SARS-like coronaviruses (SL-CoVs) or SARS-CoV-like viruses, in several horseshoe bat species in the genus Rhinolophus (Lau et al. 2005; Li et al. 2005). These discoveries raised the possibility that bats could be the natural reservoirs of SARS-CoV (Wang et al. 2006).
One interesting observation was the consistent failure of PCR to detect SL-CoVs in respiratory specimens from bats whereas high levels of viral RNA could be detected in anal swabs (Li et al. 2005). This suggested that fecal-oral contact is probably the most likely route of transmission among bats and from bats to other wildlife animals. This also implied that direct contact between animals may not be a prerequisite for animal-to-animal transmission of this group of coronaviruses. Considering that bats and a diverse group of wildlife animals co-habitat in their natural environment (e.g., in caves) and that live bats are housed and traded with many different animals in live animal markets in Southern China and southeast Asian countries, there should be ample opportunities for fecal-oral transmission to occur.
Middle East respiratory syndrome (MERS) was first recognized in September 2012 when the World Health Organization (WHO) reported two fatal cases of acute respiratory syndrome with a novel CoV in the Middle East. There was no evidence of any epidemiological link between these two cases, yet the viral strains obtained from the respiratory tract specimens of these two patients shared 99.5 % nucleotide identity. The virus was initially named HCoV-EMC/2012, later changed to MERS-CoV (Bermingham et al. 2012; de Groot et al. 2013; Zaki et al. 2012). By April 2015, at least 1106 laboratory-confirmed cases of MERS-CoV infection with at least 421 deaths have been reported to WHO. All primary human infections seem to have originated from Middle East, including United Arab Emirates, Qatar, Oman, Jordan, Kuwait, Yemen, Lebanon, and Iran. Travel-mediated secondary infections have been confirmed in a number of countries, including the United Kingdom, France, Tunisia, Italy, Malaysia, Philippines, Greece, Egypt, United States, Netherlands, and Algeria. Unlike the explosive nature of the SARS outbreaks in 2002–2003, which were believed to be resulted from a single or limited spillover event(s) from animal to human, followed by human-to-human transmission, the MERS outbreaks are characterized by multiple spillover events with non-sustained human-to-human transmission, most often in health care-associated settings with a few household clusters (Mailles et al. 2013; Chan et al. 2015).
Despite rapid progress in MERS-related research since 2012, the origin of MERS-CoV and the exact route of transmission to human remains an area of on-going research. Phylogenetic analysis of different coronavirus genome sequences revealed that that MERS-CoV is most closely related to two Chinese bat coronavirus species in the genus Betacoronavirus. They are Tylonycteris bat coronavirus HKU4 (Ty-BtCoV-HKU4) and Pipistrellus bat coronavirus HKU5 (Pi-BtCoV-HKU5) first reported in 2007 (Woo et al. 2007). Furthermore, MERS-CoV related sequences were also detected in other species of bats in Europe, Africa, Central America, and Middle East (Chan et al. 2015). In one study, a short PCR fragment (182 nt) containing identical sequence to that of MERS-CoV was detected in Taphozous perforatus (Egyptian tomb bat) in Saudi Arabia (Memish et al. 2013), but due to the very short sequence obtained from a single bat specimen, this finding is generally considered inconclusive (Chan et al. 2015). To date, none of the bat CoVs detected is likely to be the direct ancestor of MERS-CoV (Lau et al. 2013). However, considering that the search for the close relative of SARS-CoV in bats took almost ten years to yield conclusive findings, it is too early to predict whether bats are the (original) source of MERS-CoV.
Epidemiology data seem to indicate that most MERS patients had a history of exposure to dromedary camels and goats (Albarrak et al. 2012; Buchholz et al. 2013). This led to several very fruitful seroepidemiological studies confirming the presence of MERS-CoV neutralizing antibodies in dromedary camels (Alagaili et al. 2014; Alexandersen et al. 2014; Hemida et al. 2013; Reusken et al. 2013a, b). Indeed a previously healthy Saudi man developed respiratory symptoms after caring for ill camels on a farm (Azhar et al. 2014; Memish et al. 2014), and virus isolated from a nasal swab from the patient was almost identical to an isolate from one of the sick camels in their genome sequences (Azhar et al. 2014). A retrospective search for MERS-CoV antibodies indicated that the virus was circulating among the camel populations in Middle East and Africa as early as 1992 and 1983 (Alagaili et al. 2014; Alexandersen et al. 2014; Meyer et al. 2014; Perera et al. 2013). Recently, the detection of a MERS-CoV conspecific virus from an African bat led the authors to hypothesize that the MERS-CoV may have originated from an African bat, followed by bat to camel transmission in Africa, then the introduction of MERS-CoV to Middle East through camel exportation/importation (Corman et al. 2014; Chan et al. 2015).
Contributor Information
Carol Shoshkes Reiss, Email: carol.reiss@nyu.edu.
John S. Mackenzie, Email: J.Mackenzie@curtin.edu.au
James E. Childs, Email: james.childs@yale.edu
Hume E. Field, Email: hume.field@ecohealthalliance.org
Lin-Fa Wang, Email: linfa.wang@duke-nus.edu.sg.
Andrew C. Breed, Email: Andrew.Breed@apha.gsi.gov.uk
References
- Aguilar-Setién A, Loza-Rubio E, Salas-Rojas M, et al. Salivary excretion of rabies virus by healthy vampire bats. Epidemiol Infect. 2005;133:517–522. doi: 10.1017/S0950268805003705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar-Setién A, Romero-Almaraz ML, Sánchez-Hernández C, et al. Dengue virus in Mexican bats. Epidemiol Infect. 2008;136:1678–1683. doi: 10.1017/S0950268808000460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alagaili AN, Briese T, Mishra N et al (2014) Middle East respiratory syndrome coronavirus infection in dromedary camels in Saudi Arabia. mBio 5: e00884–00814 [DOI] [PMC free article] [PubMed]
- Albarrak AM, Stephens GM, Hewson R, et al. Recovery from severe novel coronavirus infection. Saudi Med J. 2012;33:1265–1269. [PubMed] [Google Scholar]
- Alexandersen S, Kobinger GP, Soule G, et al. Middle East respiratory syndrome coronavirus antibody reactors among camels in Dubai, United Arab Emirates, in 2005. Transbound Emerg Dis. 2014;61:105–108. doi: 10.1111/tbed.12212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen R, Taylor SK, Sulkin SE. Studies of arthropod-borne virus infections in Chiroptera. 8. Evidence of natural St. Louis encephalitis virus infection in bats. Am J Trop Med Hyg. 1970;19:851–859. doi: 10.4269/ajtmh.1970.19.851. [DOI] [PubMed] [Google Scholar]
- Allworth A, O’Sullivan J, Selvey L, et al. Equine morbillivirus in Queensland. Commun Dis Intell. 1995;19:575. [Google Scholar]
- Allworth A, Murray K, Morgan J. A human case of encephalitis due to a Lyssavirus recently identified in fruit bats. Commun Dis Intell. 1996;20:504. [Google Scholar]
- Amengual B, Whitby JE, King A, et al. Evolution of European bat lyssaviruses. J Gen Virol. 1997;78:2319–2328. doi: 10.1099/0022-1317-78-9-2319. [DOI] [PubMed] [Google Scholar]
- Annand EJ, Reid PA. Clinical review of two fatal equine cases of infection with the insectivorous bat strain of Australian bat lyssavirus. Aust Vet J. 2014;92:324–332. doi: 10.1111/avj.12227. [DOI] [PubMed] [Google Scholar]
- Anon Human-to-human transmission of rabies via corneal transplant—France. MMWR. 1980;29:25–26. [Google Scholar]
- Anon Human-to-human transmission of rabies via corneal transplant—Thailand. MMWR. 1981;30:473–474. [PubMed] [Google Scholar]
- Anon (1999a) Human rabies prevention—United States, 1999: recommendations of the Advisory Committee on Immunization Practices (ACIP). MMWR 48(RR-1): 1–21 [PubMed]
- Anon (1999b) Outbreak of Hendra-like virus: Malaysia and Singapore, 1998-1999. CDC MMWR Wkly 48: 265–269 [PubMed]
- Anon Outbreaks of encephalitis due to Nipah/Hendra-like viruses, Western Bangladesh. Health Sci Bull. 2003;1(5):1–6. [Google Scholar]
- Anon Nipah encephalitis outbreak over wide area of western Bangladesh. Health Sci Bull. 2004;2(1):7–11. [Google Scholar]
- Anon Person-to-person transmission of Nipah virus during outbreak in Faridpur district, 2004. Health Sci Bull. 2004;2(2):5–9. [Google Scholar]
- Anon Nipah virus outbreak from date palm juice. Health Sci Bull. 2005;3(4):1–5. [Google Scholar]
- Anon (2013) Hendra virus confirmed in a dog on NSW mid north coast. [Online]. Department of Primary Industries, New South Wales Government, Sydney, Australia. http://www.dpi.nsw.gov.au/aboutus/news/all/2013/hendra-confirmed-in-dog. Accessed 28 Dec 2014
- Anon (2014) Rabies virus and other lyssavirus (including Australian bat lyssavirus) exposures and infections. CDNA National Guidelines for Public Health Units. [Online]. Department of Health, Australian Government, Canberra, Australia. http://www.health.gov.au/internet/main/publishing.nsf/Content/cdna-song-abvl-rabies.htm. Accessed 17 Apr 2015
- Arabi YM, Harthi A, Hussein J, et al. Severe neurologic syndrome associated with Middle East respiratory syndrome corona virus (MERS-CoV) Infection. 2015;43(4):495–501. doi: 10.1007/s15010-015-0720-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai YT, Kuzmin IV, Kameoka Y, et al. New lyssavirus genotype from the Lesser Mouse-eared Bat (Myotis blythi), Kyrghyzstan. Emerg Infect Dis. 2003;9:333–337. doi: 10.3201/eid0903.020252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arankalle VA, Bandyopadhyay BT, Ramdasi AY, et al. Genomic characterization of Nipah virus, West Bengal, India. Emerg Infect Dis. 2011;17:907–909. doi: 10.3201/eid1705.100968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aréchiga Ceballos N, Vázquez Morón S, Berciano JM, et al. Novel lyssavirus in bat, Spain. Emerg Infect Dis. 2013;19:793–795. doi: 10.3201/eid1905.121071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arguin PM, Murray-Lillibridge K, Miranda ME, et al. Serologic evidence of lyssavirus infections among bats, the Philippines. Emerg Infect Dis. 2002;8:258–262. doi: 10.3201/eid0803.010330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austad SN. Diverse aging rates in metazoans: targets for functional genomics. Mech Ageing Dev. 2005;126:43–49. doi: 10.1016/j.mad.2004.09.022. [DOI] [PubMed] [Google Scholar]
- Azhar EI, El-Kafrawy SA, Farraj SA, et al. Evidence for camel-to-human transmission of MERS coronavirus. N Engl J Med. 2014;370:2499–2505. doi: 10.1056/NEJMoa1401505. [DOI] [PubMed] [Google Scholar]
- Badilla X, Perez-Herra V, Quiros L, et al. Human rabies: a reemerging disease in Costa Rica? Emerg Infect Dis. 2003;9:721–723. doi: 10.3201/eid0906.020632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badrane H, Tordo N. Host switching in lyssavirus history from the Chiroptera to the Carnivora orders. J Virol. 2001;75:8096–8104. doi: 10.1128/JVI.75.17.8096-8104.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badrane H, Bahloul C, Perrin P, et al. Evidence of two lyssavirus phylogroups with distinct pathogenicity and immunogenicity. J Virol. 2001;75:3268–3276. doi: 10.1128/JVI.75.7.3268-3276.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldock FC, Douglas IC, Halpin K, et al. Epidemiological investigations into the 1994 equine morbillivirus outbreaks in Queensland, Australia. Singap Vet J. 1996;20:57–61. [Google Scholar]
- Bale JF., Jr Emerging viral infections. Semin Pediatr Neurol. 2012;19:152–157. doi: 10.1016/j.spen.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee K, Ilkal MA, Bhat HR, et al. Experimental viraemia with Japanese encephalitis virus in certain domestic and peridomestic vertebrates. Indian J Med Res. 1979;70:364–368. [PubMed] [Google Scholar]
- Banerjee K, Ilkal MA, Deshmukh PK. Susceptibility of Cynopterus sphinx (frugivorus bat) & Suncus murinus (house shrew) to Japanese encephalitis virus. Indian J Med Res. 1984;79:8–12. [PubMed] [Google Scholar]
- Banerjee K, Bhat HR, Geevarghese G, et al. Antibodies against Japanese encephalitis virus in insectivorous bats from Karnataka. Indian J Med Res. 1988;87:527–530. [PubMed] [Google Scholar]
- Banyard AC, Hayman D, Johnson N, et al. Bats and lyssaviruses. Adv Virus Res. 2011;79:239–289. doi: 10.1016/B978-0-12-387040-7.00012-3. [DOI] [PubMed] [Google Scholar]
- Barclay RMR, Harder LD. Life histories of bats: life in the slow lane. In: Kunz TH, Fenton MB, editors. Bat ecology. Chicago: The University of Chicago Press; 2003. pp. 209–253. [Google Scholar]
- Barnard BJ, Hassel RH, Geyer HJ, et al. Non-bite transmission of rabies in kudu (Tragelaphus strepsiceros) Onderstepoort J Vet Res. 1982;49:191–192. [PubMed] [Google Scholar]
- Barrett J (2004) Australian bat lyssavirus. PhD thesis, The University of Queensland, Brisbane.http://espace.library.uq.edu.au/view/UQ:9486. Accessed 1 May 2015
- Batista-da-Costa M, Bonito RF, Nishioka SA. An outbreak of vampire bat bite in a Brazilian village. Trop Med Parasitol. 1993;44:219–220. [PubMed] [Google Scholar]
- Bell JF, Moore GJ. Rabies virus isolated from brown fat of naturally infected bats. Proc Soc Exp Biol Med. 1960;103:140–142. doi: 10.3181/00379727-103-25438. [DOI] [PubMed] [Google Scholar]
- Bermingham A, Chand MA, Brown CS, et al. Severe respiratory illness caused by a novel coronavirus, in a patient transferred to the United Kingdom from the Middle East, September 2012. Euro Surveill. 2012;17(40):20290. [PubMed] [Google Scholar]
- Biesold SE, Ritz D, Gloza-Rausch F, et al. Type I interferon reaction to viral infection in interferon-competent, immortalized cell lines from the African fruit bat Eidolon helvum. PLoS One. 2011;6:e28131. doi: 10.1371/journal.pone.0028131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingham J, Javangwe S, Sabeta CT, et al. Report of isolations of unusual lyssaviruses (rabies and Mokola virus) identified retrospectively from Zimbabwe. J S Afr Vet Assoc. 2001;72:92–94. doi: 10.4102/jsava.v72i2.624. [DOI] [PubMed] [Google Scholar]
- Biosecurity Queensland (2014) What is Hendra virus [Online]. The Dept of Agriculture, Forestry & Fisheries. Queensland Government, Brisbane, Australia. http://www.daff.qld.gov.au/animal-industries/animal-health-and-diseases/a-z-list/hendra-virus/general-information/what-is-hendra-virus. Accessed 28 Dec 2014
- Botvinkin AD, Poleschuk EM, Kuzmin IV, et al. Novel lyssavirus isolated from bats Russia. Emerg Infect Dis. 2003;9:1623–1625. doi: 10.3201/eid0912.030374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulger LR, Porterfield JS. Isolation of a virus from Nigerian fruit bats. Trans R Soc Trop Med Hyg. 1958;52:421–424. doi: 10.1016/0035-9203(58)90127-5. [DOI] [PubMed] [Google Scholar]
- Bourhy H, Kissi B, Lafon M, et al. Antigenic and molecular characterization of bat rabies virus in Europe. J Clin Microbiol. 1992;30:2419–2426. doi: 10.1128/jcm.30.9.2419-2426.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breed AC, Smith CS, Epstein JH. Winged wanderers: long distance movements of flying foxes. In: Macdonald DW, editor. The encyclopedia of mammals. Oxford: Oxford University Press; 2006. pp. 474–475. [Google Scholar]
- Breed AC, Field HA, Smith CS, et al. Bats without borders: long distance movements and implications for disease risk management. Ecohealth. 2010;7:204–212. doi: 10.1007/s10393-010-0332-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breed A, Breed M, Meers J, et al. Evidence of endemic Hendra virus infection in flying-foxes (Pteropus conspicillatus)-implications for disease risk management. PLoS One. 2011;6:e28816. doi: 10.1371/journal.pone.0028816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breed AC, Meers J, Sendow I, et al. The distribution of henipaviruses in Southeast Asia and Australasia: is Wallace’s line a barrier to Nipah virus. PLoS One. 2013;8:e61316. doi: 10.1371/journal.pone.0061316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookes SM, Aegerter JN, Smith GC, et al. European bat lyssavirus in Scottish bats. Emerg Infect Dis. 2005;11:572–578. doi: 10.3201/eid1104.040920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookes SM, Parsons G, Johnson N, et al. Rabies human diploid cell vaccine elicits cross-neutralising and cross-protecting immune responses against European and Australian bat lyssaviruses. Vaccine. 2005;23:4101–4109. doi: 10.1016/j.vaccine.2005.03.037. [DOI] [PubMed] [Google Scholar]
- Buchholz U, Muller MA, Nitsche A et al (2013) Contact investigation of a case of human novel coronavirus infection treated in a German hospital, October-November 2012. Euro Surveill 18(8): pii: 20406 [PubMed]
- Bunde JM, Heske EJ, Mateus-Pinilla NE, et al. A survey for West Nile virus in bats from Illinois. J Wildl Dis. 2006;42:455–458. doi: 10.7589/0090-3558-42.2.455. [DOI] [PubMed] [Google Scholar]
- Cabrera-Romo S, Recio-Tótoro B, Alcalá AC, et al. Experimental inoculation of Artibeus jamaicensis bats with dengue virus serotypes 1 or 4 showed no evidence of sustained replication. Am J Trop Med Hyg. 2014;91:1227–1234. doi: 10.4269/ajtmh.14-0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calisher CH, Kinney RM, de Souza Lopes O, et al. Identification of a new Venezuelan equine encephalitis virus from Brazil. Am J Trop Med Hyg. 1982;31:1260–1272. doi: 10.4269/ajtmh.1982.31.1260. [DOI] [PubMed] [Google Scholar]
- Calisher CH, Childs JE, Field HE, et al. Bats: important reservoir hosts of emerging viruses. Clin Microbiol Rev. 2006;19:531–545. doi: 10.1128/CMR.00017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caraballo AJ. Outbreak of vampire bat biting in a Venezuelan village. Rev Saude Publica. 1996;30:483–484. doi: 10.1590/S0034-89101996000500012. [DOI] [PubMed] [Google Scholar]
- Carini A. Sur une grande épizootie de rage. Ann Inst Pasteur. 1911;25:843–846. [Google Scholar]
- Chadha MS, Comer JA, Lowe L, et al. Nipah virus-associated encephalitis outbreak, Siliguri, India. Emerg Infect Dis. 2006;12:235–240. doi: 10.3201/eid1202.051247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JF, To KK, Tse H, et al. Interspecies transmission and emergence of novel viruses: lessons from bats and birds. Trends Microbiol. 2013;21:544–555. doi: 10.1016/j.tim.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JFW, Lau SKP, To KKW, et al. Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin Microbiol Rev. 2015;28(2):465–522. doi: 10.1128/CMR.00102-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao CC, Tsai LK, Chiou YH, et al. Peripheral nerve disease in SARS: report of a case. Neurology. 2003;61:1820–1821. doi: 10.1212/01.WNL.0000099171.26943.D0. [DOI] [PubMed] [Google Scholar]
- Charlier N, Leysson P, Paeshuyse J, et al. Infection of SCID mice with Montana Myotis leukoencephalitis virus as a model for flavivirus encephalitis. J Gen Virol. 2002;83:1887–1896. doi: 10.1099/0022-1317-83-8-1887. [DOI] [PubMed] [Google Scholar]
- Ching PK, de los Reyes VC, Sucaldito MN, et al. Outbreak of henipavirus infection, Philippines, 2014. Emerg Infect Dis. 2015;21:328–331. doi: 10.3201/eid2102.141433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong HT, Kunjapan R, Thayaparan T, et al. Nipah encephalitis outbreak in Malaysia, clinical features in patients from Seremban. Can J Neurol Sci. 2002;29:83–87. doi: 10.1017/S0317167100001785. [DOI] [PubMed] [Google Scholar]
- Chong HT, Abdullah S, Tan CT. Nipah virus and bats. Neurol Asia. 2009;14:73–76. [Google Scholar]
- Chowdhury S, Khan SU, Crameri G, et al. Serological evidence of henipavirus exposure in cattle, goats and pigs in Bangladesh. PLoS Negl Trop Dis. 2014;8:e3302. doi: 10.1371/journal.pntd.0003302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua KB, Goh KJ, Wong KT, et al. Fatal encephalitis due to Nipah virus among pig-farmers in Malaysia. Lancet. 1999;354:1256–1259. doi: 10.1016/S0140-6736(99)04299-3. [DOI] [PubMed] [Google Scholar]
- Chua KB, Bellini WJ, Rota PA, et al. Nipah virus: a recently emergent deadly paramyxovirus. Science. 2000;288:1432–1435. doi: 10.1126/science.288.5470.1432. [DOI] [PubMed] [Google Scholar]
- Chua KB, Koh CL, Hooi PS, et al. Isolation of Nipah virus from Malaysian Island flying-foxes. Microbes Infect. 2002;4:145–151. doi: 10.1016/S1286-4579(01)01522-2. [DOI] [PubMed] [Google Scholar]
- Clawson RL. Trends in population size and current status. In: Kurta A, Kennedy J, editors. The Indiana bat: biology and management of an endangered species. Austin: Bat Conservation International; 2002. [Google Scholar]
- Cockrum EL. Migration in the guano bat, Tadarida brasiliensis. Lawerence: The University of Kansas Museum of Natural History; 1969. [Google Scholar]
- Constantine DG. Activity patterns of the Mexican free-tailed bat. Albuquerque: University of New Mexico Press; 1967. [Google Scholar]
- Constantine DG. Rabies transmission by air in bat caves. Washington: U.S. Government Printing Office; 1967. [Google Scholar]
- Constantine DG. Bat rabies in the southwestern United States. Public Health Rep. 1967;82:867–888. doi: 10.2307/4593153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantine DG. An updated list of rabies-infected bats in North America. J Wildl Dis. 1979;15:347–349. doi: 10.7589/0090-3558-15.2.347. [DOI] [PubMed] [Google Scholar]
- Constantine DG, Emmons RW, Woodie JD. Rabies virus in nasal mucosa of naturally infected bats. Science. 1972;175:1255–1256. doi: 10.1126/science.175.4027.1255. [DOI] [PubMed] [Google Scholar]
- Corbet G, Hill J. The mammals of the Indomalayan region. Oxford: Natural History Museum Publications, Oxford University Press; 1992. [Google Scholar]
- Corman VM, Ithete NL, Richards LR, et al. Rooting the phylogenetic tree of Middle East respiratory syndrome coronavirus by characterization of a conspecific virus from an African bat. J Virol. 2014;88:11297–11303. doi: 10.1128/JVI.01498-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa-Giron P, Calisher CH, Baer GM. Epidemic strain of Venezuelan equine encephalomyelitis virus from a vampire bat captured in Oaxaca, Mexico, 1970. Science. 1972;175:546–547. doi: 10.1126/science.175.4021.546. [DOI] [PubMed] [Google Scholar]
- Cowled C, Stewart CA, Likic VA, et al. Characterisation of novel microRNAs in the black flying-fox (Pteropus alecto) by deep sequencing. BMC Genomics. 2014;15:682. doi: 10.1186/1471-2164-15-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crameri G, Todd S, Grimley S, et al. Establishment, immortalisation and characterisation of pteropid bat cell lines. PLoS One. 2009;4:e8266. doi: 10.1371/journal.pone.0008266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick J, Tignor GH, Moreno K. A new isolate of Lagos bat virus from the Republic of South Africa. Trans R Soc Trop Med Hyg. 1982;76:211–213. doi: 10.1016/0035-9203(82)90277-2. [DOI] [PubMed] [Google Scholar]
- Croser EL, Marsh GA. The changing face of the henipaviruses. Vet Microbiol. 2013;167:151–158. doi: 10.1016/j.vetmic.2013.08.002. [DOI] [PubMed] [Google Scholar]
- Daoust PY, Wandeler AI, Casey GA. Cluster of rabies cases of probable bat origin among red foxes in Prince Edward Island, Canada. J Wildl Dis. 1996;32:403–406. doi: 10.7589/0090-3558-32.2.403. [DOI] [PubMed] [Google Scholar]
- Daszak P, Plowright RK, Epstein JH, et al. The emergence of Nipah and Hendra virus: pathogen dynamics across a wildlife-livestock-human continuum. In: Collinge SK, Ray C, et al., editors. Disease ecology: community structure and pathogen dynamics. Oxford: Oxford University Press; 2006. [Google Scholar]
- Davis A, Bunning M, Gordy P, et al. Experimental and natural infection of North American bats with West Nile virus. Am J Trop Med Hyg. 2005;73:467–469. [PubMed] [Google Scholar]
- Davis PL, Holmes EC, Larrous F, et al. Phylogeography, population dynamics, and molecular evolution of European bat lyssaviruses. J Virol. 2005;79:10487–10497. doi: 10.1128/JVI.79.16.10487-10497.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis PL, Bourhy H, Holmes EC. The evolutionary history and dynamics of bat rabies virus. Infect Genet Evol. 2006;6:464–473. doi: 10.1016/j.meegid.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Davis AD, Rudd RJ, Bowen RA. Effects of aerosolized rabies virus exposure on bats and mice. J Infect Dis. 2007;195:1144–1150. doi: 10.1086/512616. [DOI] [PubMed] [Google Scholar]
- de Groot RJ, Baker SC, Baric RS, et al. Middle East respiratory syndrome coronavirus (MERS-CoV): announcement of the Coronavirus Study Group. J Virol. 2013;87:7790–7792. doi: 10.1128/JVI.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Thoisy B, Lacoste V, Germain A, et al. Dengue infection in neotropical forest mammals. Vector Borne Zoonotic Dis. 2009;9:157–170. doi: 10.1089/vbz.2007.0280. [DOI] [PubMed] [Google Scholar]
- Drexler JF, Corman VM, Gloza-Rauch F, et al. Henipavirus RNA in African bats. PLoS One. 2009;4(7):e6367. doi: 10.1371/journal.pone.0006367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler JF, Corman VM, Müller MA, et al. Bats host major mammalian paramyxoviruses. Nat Commun. 2012;3:796. doi: 10.1038/ncomms1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler JF, Corman VM, Drosten C. Ecology, evolution and classification of bat coronaviruses in the aftermath of SARS. Antiviral Res. 2014;101:45–56. doi: 10.1016/j.antiviral.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer JL, Wallace R, Orciari L, et al. Rabies surveillance in the United States during 2012. J Am Vet Med Assoc. 2013;243:805–815. doi: 10.2460/javma.243.6.805. [DOI] [PubMed] [Google Scholar]
- Dzikwi AA, Kuzmin II, Umoh JU, et al. Evidence of Lagos bat virus circulation among Nigerian fruit bats. J Wildl Dis. 2010;46:267–271. doi: 10.7589/0090-3558-46.1.267. [DOI] [PubMed] [Google Scholar]
- Enria DA, Pinheiro F. Rodent-borne emerging viral zoonosis. Hemorrhagic fevers and hantavirus infections in South America. Infect Dis Clin North Am. 2000;14:167–184. doi: 10.1016/S0891-5520(05)70223-3. [DOI] [PubMed] [Google Scholar]
- Epstein JH, Field HE, Luby S, et al. Nipah virus: impact, origins, and causes of emergence. Curr Infect Dis Rep. 2006;8:59–65. doi: 10.1007/s11908-006-0036-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein JH, Prakash V, Smith CS, et al. Henipavirus infection in fruit bats (Pteropus giganteus), India. Emerg Infect Dis. 2008;14:1309–1311. doi: 10.3201/eid1408.071492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar LE, Peterson AT, Favi M, et al. Bat-borne rabies in Latin America. Rev Inst Med Trop Sao Paulo. 2015;57:63–72. doi: 10.1590/S0036-46652015000100009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Familusi JB, Moore DL. Isolation of a rabies related virus from the cerebrospinal fluid of a child with ‘aseptic meningitis’. Afr J Med Sci. 1972;3:93–96. [PubMed] [Google Scholar]
- Familusi JB, Osunkoya BO, Moore DL, et al. A fatal human infection with Mokola virus. Am J Trop Med Hyg. 1972;2:959–963. doi: 10.4269/ajtmh.1972.21.959. [DOI] [PubMed] [Google Scholar]
- Fekadu M, Shaddock JH, Sanderlin DW, et al. Efficacy of rabies vaccines against Duvenhage virus isolated from European house bats (Eptesicus serotinus), classic rabies and rabies-related viruses. Vaccine. 1988;6:533–539. doi: 10.1016/0264-410X(88)90107-7. [DOI] [PubMed] [Google Scholar]
- Field HE (2005) The ecology of Hendra virus and Australian bat lyssavirus. PhD thesis, The University of Queensland, Brisbane [Online]. http://espace.library.uq.edu.au/view.php?pid=UQ:13859
- Field H, Young P, Yob JM, et al. The natural history of Hendra and Nipah viruses. Microbes Infect. 2001;3:307–314. doi: 10.1016/S1286-4579(01)01384-3. [DOI] [PubMed] [Google Scholar]
- Field H, Mackenzie J, Daszak P. Novel viral encephalitides associated with bats (Chiroptera)—host management strategies. Arch Virol Suppl. 2004;18:113–121. doi: 10.1007/978-3-7091-0572-6_9. [DOI] [PubMed] [Google Scholar]
- Field HE, Mackenzie JS, Daszak P. Henipaviruses: emerging paramyxoviruses associated with fruit bats. Curr Top Microbiol Immunol. 2007;315:133–159. doi: 10.1007/978-3-540-70962-6_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field H, Schaaf K, Kung N, et al. Hendra virus outbreak with novel clinical features, Australia. Emerg Infect Dis. 2010;16:338–340. doi: 10.3201/eid1602.090780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field HE, De Jong C, Melville D, et al. Hendra virus infection dynamics in Australian fruit bats. PLoS One. 2011;6:e28678. doi: 10.1371/journal.pone.0028678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming TH, Eby P. Ecology of bat migration. In: Kunz TH, Fenton MB, editors. Bat ecology. Chicago: The University of Chicago Press; 2003. pp. 156–208. [Google Scholar]
- Foggin CM. Mokola virus infection in cats and a dog in Zimbabwe. Vet Rec. 1983;113:115. doi: 10.1136/vr.113.5.115. [DOI] [PubMed] [Google Scholar]
- Fooks AR, Brookes SM, Johnson N, et al. European bat lyssaviruses: an emerging zoonosis. Epidemiol Infect. 2003;131:1029–1039. doi: 10.1017/S0950268803001481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fooks AR, McElhinney LM, Pounder DJ, et al. Case report: isolation of a European bat lyssavirus type 2a from a fatal human case of rabies encephalitis. J Med Virol. 2003;71:281–289. doi: 10.1002/jmv.10481. [DOI] [PubMed] [Google Scholar]
- Fooks AR, Banyard AC, Horton DL, et al. Current status of rabies and prospects for elimination. Lancet. 2014;384:1389–1399. doi: 10.1016/S0140-6736(13)62707-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis JR, McCall BJ, Hutchinson P, et al. Australian bat lyssavirus: implications for public health. Med J Aust. 2014;201:647–649. doi: 10.5694/mja13.00261. [DOI] [PubMed] [Google Scholar]
- Francis JR, Nourse C, Vaska VL, et al. Australian bat lyssavirus in a child: the first reported case. Pediatrics. 2014;133:e1063–e1067. doi: 10.1542/peds.2013-1782. [DOI] [PubMed] [Google Scholar]
- Franka R, Constantine DG, Kuzmin I, et al. A new phylogenetic lineage of rabies virus associated with western pipistrelle bats (Pipistrellus hesperus) J Gen Virol. 2006;87:2309–2321. doi: 10.1099/vir.0.81822-0. [DOI] [PubMed] [Google Scholar]
- Fraser GC, Hooper PT, Lunt RA, et al. Encephalitis caused by a lyssavirus in fruit bats in Australia. Emerg Infect Dis. 1996;2:327–331. doi: 10.3201/eid0204.960408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freuling CM, Beer M, Conraths FJ, et al. Novel lyssavirus in Natterer’s bat, Germany. Emerg Infect Dis. 2011;17:1519–1522. doi: 10.3201/eid1708.110201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freuling CM, Abendroth B, Beer M, et al. Molecular diagnostics for the detection of Bokeloh bat lyssavirus in a bat from Bavaria, Germany. Virus Res. 2013;177:201–204. doi: 10.1016/j.virusres.2013.07.021. [DOI] [PubMed] [Google Scholar]
- Gaunt MW, Sall AA, de Lamballerie X, et al. Phylogenetic relationships of flaviviruses correlate with their epidemiology, disease association and biogeography. J Gen Virol. 2001;82:1867–1876. doi: 10.1099/0022-1317-82-8-1867. [DOI] [PubMed] [Google Scholar]
- George DB, Webb CT, Farnsworth ML, et al. Host and viral ecology determine bat rabies seasonality and maintenance. Proc Natl Acad Sci U S A. 2011;108:10208–10213. doi: 10.1073/pnas.1010875108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons RV. Cryptogenic rabies, bats, and the question of aerosol transmission. Ann Emerg Med. 2002;39:528–536. doi: 10.1067/mem.2002.121521. [DOI] [PubMed] [Google Scholar]
- Gode GR, Bhide NK. Two rabies deaths after corneal grafts from one donor. Lancet. 1988;2:791. doi: 10.1016/S0140-6736(88)92435-X. [DOI] [PubMed] [Google Scholar]
- Goeijenbier M, Wagenaar J, Goris M, et al. Rodent-borne hemorrhagic fevers: under-recognized, widely spread and preventable—epidemiology, diagnostics and treatment. Crit Rev Microbiol. 2013;39:26–42. doi: 10.3109/1040841X.2012.686481. [DOI] [PubMed] [Google Scholar]
- Goh KJ, Tan TC, Chew NK, et al. Clinical features of Nipah virus encephalitis among pig farmers in Malaysia. N Engl J Med. 2000;342:1229–1235. doi: 10.1056/NEJM200004273421701. [DOI] [PubMed] [Google Scholar]
- Goldspink LK, Edson DW, Vidgen ME, et al. Natural Hendra virus infection in flying-foxes—tissue tropism and risk factors. PLoS One. 2015;10(6):e0128835. doi: 10.1371/journal.pone.0128835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez JP, Pourrut X, Leroy E. Ebolavirus and other filoviruses. Curr Top Microbiol Immunol. 2007;315:363–387. doi: 10.1007/978-3-540-70962-6_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould AR. Comparison of the deduced matrix and fusion protein sequences of equine morbillivirus with cognate genes of the Paramyxoviridae. Virus Res. 1996;43:17–31. doi: 10.1016/0168-1702(96)01308-1. [DOI] [PubMed] [Google Scholar]
- Gould A, Hyatt AD, Lunt R, et al. Characterisation of a novel lyssavirus isolated from Pteropid bats in Australia. Virus Res. 1998;54:165–187. doi: 10.1016/S0168-1702(98)00025-2. [DOI] [PubMed] [Google Scholar]
- Gould AR, Kattenbelt JA, Gumley SG, et al. Characterisation of an Australian bat lyssavirus variant isolated from an insectivorous bat. Virus Res. 2002;89:1–28. doi: 10.1016/S0168-1702(02)00056-4. [DOI] [PubMed] [Google Scholar]
- Gould EA, Moss SR, Turner SL. Evolution and dispersal of encephalitic flaviviruses. Arch Virol Suppl. 2004;18:65–84. doi: 10.1007/978-3-7091-0572-6_6. [DOI] [PubMed] [Google Scholar]
- Griffin DR. Migration and homing in bats. In: Wimsatt WA, editor. Biology of bats. New York: Academic; 1970. pp. 233–264. [Google Scholar]
- Griffin DE. Emergence and re-emergence of viral diseases of the central nervous system. Prog Neurobiol. 2010;91:95–101. doi: 10.1016/j.pneurobio.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y, Zheng BJ, He Y, et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- Gurley ES, Montgomery JM, Hossain MJ, et al. Person-to-person transmission of Nipah virus in a Bangladeshi community. Emerg Infect Dis. 2007;13:1031–1037. doi: 10.3201/eid1307.061128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyatt KJ, Twin J, Davis P, et al. A molecular epidemiological study of Australian bat lyssavirus. J Gen Virol. 2003;84:485–496. doi: 10.1099/vir.0.18652-0. [DOI] [PubMed] [Google Scholar]
- Hahn MB, Epstein JH, Gurley ES, et al. Roosting behaviour and habitat selection of Pteropus giganteus reveals potential links to Nipah virus epidemiology. J Appl Ecol. 2014;51:376–387. doi: 10.1111/1365-2664.12212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MB, Gurley ES, Epstein JH, et al. The role of landscape composition and configuration on Pteropus giganteus roosting ecology and Nipah virus spillover risk in Bangladesh. Am J Trop Med Hyg. 2014;90:247–255. doi: 10.4269/ajtmh.13-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall L, Richards G. Flying foxes: fruit and blossom bats of Australia. Sydney: University of New South Wales Press; 2000. [Google Scholar]
- Halpin K, Young PL, Field HE, et al. Isolation of Hendra virus from pteropid bats: a natural reservoir of Hendra virus. J Gen Virol. 2000;81:1927–1932. doi: 10.1099/0022-1317-81-8-1927. [DOI] [PubMed] [Google Scholar]
- Halpin K, Hyatt AD, Plowright RK, et al. Emerging viruses: coming in on a wrinkled wing and a prayer. Clin Infect Dis. 2007;44:711–717. doi: 10.1086/511078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanlon CA, Kuzmin IV, Blanton JD, et al. Efficacy of rabies biologics against new lyssaviruses from Eurasia. Virus Res. 2005;111:44–54. doi: 10.1016/j.virusres.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Hanna JN, Carney IK, Smith GA, et al. Australian bat lyssavirus infection: a second human case, with a long incubation period. Med J Aust. 2000;172:597–599. doi: 10.5694/j.1326-5377.2000.tb124126.x. [DOI] [PubMed] [Google Scholar]
- Harris SL, Brookes SM, Jones G, et al. Passive surveillance (1987 to 2004) of United Kingdom bats for European bat lyssaviruses. Vet Rec. 2006;159:439–446. doi: 10.1136/vr.159.14.439. [DOI] [PubMed] [Google Scholar]
- Harris SL, Mansfield K, Marston DA, et al. Isolation of European bat lyssavirus type 2 from a Daubenton’s bat (Myotis daubentonii) in Shropshire. Vet Rec. 2007;161:384–386. doi: 10.1136/vr.161.11.384. [DOI] [PubMed] [Google Scholar]
- Hayman DT, Fooks AR, Horton D. Antibodies against Lagos bat virus in megachiroptera from West Africa. Emerg Infect Dis. 2008;14:926–928. doi: 10.3201/eid1406.071421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayman DT, Suu-Ire R, Breed AC, et al. Evidence of henipavirus infection in West African fruit bats. PLoS One. 2008;3(7):e2739. doi: 10.1371/journal.pone.0002739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayman DTS, Fooks AR, Rowcliffe JM, et al. Endemic Lagos bat virus infection in Eidolon helvum. Epidemiol Infect. 2012;140:2163–2171. doi: 10.1017/S0950268812000167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemida MG, Perera RA, Wang P, et al. Middle East respiratory syndrome (MERS) coronavirus seroprevalence in domestic livestock in Saudi Arabia, 2010 to 2013. Euro Surveill. 2013;18:20659. doi: 10.2807/1560-7917.ES2013.18.50.20659. [DOI] [PubMed] [Google Scholar]
- Herbold JR, Heuschele WP, Berry RL, et al. Reservoir of St. Louis encephalitis virus in Ohio bats. Am J Vet Res. 1983;44:1889–1893. [PubMed] [Google Scholar]
- Holland RA, Waters DA, Rayner JM. Echolocation signal structure in the Megachiropteran bat Rousettus aegyptiacus Geoffroy 1810. J Exp Biol. 2004;207:4361–4369. doi: 10.1242/jeb.01288. [DOI] [PubMed] [Google Scholar]
- Homaira N, Rahman M, Hossain MJ, et al. Nipah virus outbreak with person-to-person transmission in a district of Bangladesh, 2007. Epidemiol Infect. 2010;138:1630–1636. doi: 10.1017/S0950268810000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homaira N, Rahman M, Hossain MJ, et al. Cluster of Nipah virus infection, Kushtia District, Bangladesh. PLoS One. 2010;5:e13570. doi: 10.1371/journal.pone.0013570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper PT, Gould AR, Russell GM, et al. The retrospective diagnosis of a second outbreak of equine morbillivirus infection. Aust Vet J. 1996;74:244–245. doi: 10.1111/j.1751-0813.1996.tb15414.x. [DOI] [PubMed] [Google Scholar]
- Hooper P, Lunt R, Gould A, et al. A new lyssavirus-the first endemic rabies-related virus recognised in Australia. Bull Inst Pasteur. 1997;95:209–218. doi: 10.1016/S0020-2452(97)83529-5. [DOI] [Google Scholar]
- Horton DL, Banyard AC, Marston DA, et al. Antigenic and genetic characterization of a divergent African virus, Ikoma lyssavirus. J Gen Virol. 2014;95:1025–1032. doi: 10.1099/vir.0.061952-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houff SA, Burton RC, Wilson RW, et al. Human-to-human transmission of rabies virus by corneal transplant. N Engl J Med. 1979;300:603–604. doi: 10.1056/NEJM197903153001105. [DOI] [PubMed] [Google Scholar]
- Hsu VP, Hossain MJ, Parashar UD, et al. Nipath virus encephalitis reemergence, Bangladesh. Emerg Infect Dis. 2004;10:2082–2087. doi: 10.3201/eid1012.040701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes GJ, Orciari LA, Rupprecht CE. Evolutionary timescale of rabies virus adaptation to North American bats inferred from the substitution rate of the nucleoprotein gene. J Gen Virol. 2005;86:1467–1474. doi: 10.1099/vir.0.80710-0. [DOI] [PubMed] [Google Scholar]
- Humphrey SR, Cope JB. Population ecology of the little brown bat, Myotis lucifugus, in Indiana and North-Central Kentucky. Lawerence: American Society of Mammalogists; 1976. [Google Scholar]
- Humphrey GL, Kemp GE, Wood EG. A fatal case of rabies in a woman bitten by an insectivorous bat. Public Health Rep. 1960;75:317–326. doi: 10.2307/4590791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung EC, Chim SS, Chan PK, et al. Detection of SARS coronavirus RNA in the cerebrospinal fluid of a patient with severe acute respiratory syndrome. Clin Chem. 2003;49:2108–2109. doi: 10.1373/clinchem.2003.025437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst EW, Pawan JL. An outbreak of rabies in Trinidad without history of bites, and with the symptoms of acute ascending myelitis. Caribb Med J. 1959;21:11–24. [PubMed] [Google Scholar]
- Irons JV, Eads RB, Grimes JE, et al. The public health importance of bats. Tex Rep Biol Med. 1957;15:292–298. [PubMed] [Google Scholar]
- ITO Taiichi, SAITO Yutaka. Experimental studies on the susceptibility of bats to Japanese encephalitis virus. Nippon Saikingaku Zasshi. 1952;7(6):617–622. [Google Scholar]
- Jepsen GL. Bat origins and evolution. In: Wimsatt WA, editor. Biology of bats. New York: Academic; 1970. pp. 1–64. [Google Scholar]
- Jiang Y, Wang L, Lu Z, et al. Seroprevalence of rabies virus antibodies in bats from southern China. Vector Borne Zoonotic Dis. 2010;10:177–181. doi: 10.1089/vbz.2008.0212. [DOI] [PubMed] [Google Scholar]
- Johnson N, Phillpotts R, Fooks AR. Airborne transmission of lyssaviruses. J Med Microbiol. 2006;55:785–790. doi: 10.1099/jmm.0.46370-0. [DOI] [PubMed] [Google Scholar]
- Kan B, Wang M, Jing H, et al. Molecular evolution analysis and geographic investigation of severe acute respiratory syndrome coronavirus-like virus in palm civets at an animal market and on farms. J Virol. 2005;79:11892–11900. doi: 10.1128/JVI.79.18.11892-11900.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp GE, Causey OR, Moore DL, et al. Mokola virus: further studies on IbAn 27377, a new rabies-related etiological agent of zoonosis in Nigeria. Am J Trop Med Hyg. 1972;21:356–359. [PubMed] [Google Scholar]
- Kgaladi J, Nel LH, Markotter W. Comparison of pathogenic domains of rabies and African rabies-related lyssaviruses and pathogenicity observed in mice. Onderstepoort J Vet Res. 2013;80:E1–E13. doi: 10.4102/ojvr.v80i1.511. [DOI] [PubMed] [Google Scholar]
- Kgaladi J, Wright N, Coertse J, et al. Diversity and epidemiology of Mokola virus. PLoS Negl Trop Dis. 2013;7(10):e2511. doi: 10.1371/journal.pntd.0002511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MS, Hossain J, Gurley ES, et al. Use of infrared camera to understand bats’ access to date palm sap: implications for preventing Nipah virus transmission. Ecohealth. 2010;7:517–525. doi: 10.1007/s10393-010-0366-2. [DOI] [PubMed] [Google Scholar]
- Khozinski VV, Selimov MA, Botvinkin AD. Rabies virus of bat origin in the USSR. Rabies Bull Eur. 1990;14:10. [Google Scholar]
- King A, Crick J. Rabies-related viruses. In: Campbell JB, editor. Rabies. Boston: Kluwer Academic; 1988. pp. 177–199. [Google Scholar]
- Knobel DL, Cleaveland S, Coleman PG, et al. Re-evaluating the burden of rabies in Africa and Asia. Bull World Health Organ. 2005;83:360–368. [PMC free article] [PubMed] [Google Scholar]
- Koraka P, Martina BE, Roose JM, et al. In vitro and in vivo isolation and characterization of Duvenhage virus. PLoS Pathog. 2012;8(5):e1002682. doi: 10.1371/journal.ppat.1002682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauel JJ, McCracken GF. Recent advances in bat migration research. In: Adams RA, Pedersen SC, editors. Bat evolution, ecology, and conservation. New York: Springer; 2013. pp. 293–314. [Google Scholar]
- Kulkarni DD, Tosh C, Venkatesh G, et al. Nipah virus infection: current scenario. Indian J Virol. 2013;24:398–408. doi: 10.1007/s13337-013-0171-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung N, McLaughlin A, Taylor M, et al. Hendra virus and horse owners—risk perception and management. PLoS One. 2013;8(11):e80897. doi: 10.1371/journal.pone.0080897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmin IV, Orciari LA, Arai YT, et al. Bat lyssaviruses (Aravan and Khujand) from Central Asia: phylogenetic relationships according to N, P and G gene sequences. Virus Res. 2003;97:65–79. doi: 10.1016/S0168-1702(03)00217-X. [DOI] [PubMed] [Google Scholar]
- Kuzmin IV, Hughes GJ, Botvinkin AD, et al. Phylogenetic relationships of Irkut and West Caucasian bat viruses within the Lyssavirus genus and suggested quantitative criteria based on the N gene sequence for lyssavirus genotype definition. Virus Res. 2005;111:28–43. doi: 10.1016/j.virusres.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Kuzmin IV, Hughes GJ, Rupprecht CE. Phylogenetic relationships of seven previously unclassified viruses within the family Rhabdoviridae using partial nucleoprotein gene sequences. J Gen Virol. 2006;87:2323–2331. doi: 10.1099/vir.0.81879-0. [DOI] [PubMed] [Google Scholar]
- Kuzmin IV, Niezgoda M, Franka R, et al. Lagos bat virus in Kenya. J Clin Microbiol. 2008;46:1451–1461. doi: 10.1128/JCM.00016-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmin IV, Niezgoda M, Franka R, et al. Possible emergence of West Caucasian bat virus in Africa. Emerg Infect Dis. 2008;14:1887–1889. doi: 10.3201/eid1412.080750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmin IV, Mayer AE, Niezgoda M, et al. Shimoni bat virus, a new representative of the Lyssavirus genus. Virus Res. 2010;149:197–210. doi: 10.1016/j.virusres.2010.01.018. [DOI] [PubMed] [Google Scholar]
- La Motte LC. Japanese B encephalitis in bats during simulated hibernation. Am J Hyg. 1958;67:101–108. doi: 10.1093/oxfordjournals.aje.a119912. [DOI] [PubMed] [Google Scholar]
- Lau SK, Woo PC, Li KS, et al. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci U S A. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau SK, Li KS, Tsang AK, et al. Genetic characterization of Betacoronavirus lineage C viruses in bats reveals marked sequence divergence in the spike protein of pipistrellus bat coronavirus HKU5 in Japanese pipistrelle: implications for the origin of the novel Middle East respiratory syndrome coronavirus. J Virol. 2013;87:8638–8650. doi: 10.1128/JVI.01055-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DT, Wing YK, Leung HC, et al. Factors associated with psychosis among patients with severe acute respiratory syndrome: a case-control study. Clin Infect Dis. 2004;39:1247–1249. doi: 10.1086/424016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy EM, Kumulungui B, Pourrut X, et al. Fruit bats as reservoirs of Ebola virus. Nature. 2005;438:575–576. doi: 10.1038/438575a. [DOI] [PubMed] [Google Scholar]
- Leslie MJ, Messenger S, Rohde RE, et al. Bat-associated rabies virus in Skunks. Emerg Infect Dis. 2006;12:1274–1277. doi: 10.3201/eid1208.051526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Shi Z, Yu M, et al. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Li Y, Wang J, Hickey AC, et al. Antibodies to Nipah or Nipah-like viruses in bats, China. Emerg Infect Dis. 2008;14:1974–1976. doi: 10.3201/eid1412.080359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CCT, Lee WL, Leo YS, et al. Late clinical and magnetic resonance imaging follow up of Nipah virus infection. J Neurol Neurosurg Psychatry. 2003;74:131–133. doi: 10.1136/jnnp.74.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Li X, Chen Z, et al. Comparison of genomic and amino acid sequences of eight Japanese encephalitis virus isolates from bats. Arch Virol. 2013;158:2543–2552. doi: 10.1007/s00705-013-1777-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zhang S, Zhao J, et al. Isolation of Irkut virus from a Murina leucogaster bat in China. PLoS Negl Trop Dis. 2013;7:e2097. doi: 10.1371/journal.pntd.0002097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd AL, May RM. Spatial heterogeneity in epidemic models. J Theor Biol. 1996;179:1–11. doi: 10.1006/jtbi.1996.0042. [DOI] [PubMed] [Google Scholar]
- Lopez A, Miranda P, Tejada E, et al. Outbreak of human rabies in the Peruvian jungle. Lancet. 1992;339:408–411. doi: 10.1016/0140-6736(92)90088-K. [DOI] [PubMed] [Google Scholar]
- Loza-Rubio E, Rojas-Anaya E, Banda-Ruiz VM, et al. Detection of multiple strains of rabies virus RNA using primers designed to target Mexican vampire bat variants. Epidemiol Infect. 2005;133:927–934. doi: 10.1017/S095026880500405X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luby SP, Gurley ES. Epidemiology of henipavirus disease in humans. Curr Top Microbiol Immunol. 2012;359:25–40. doi: 10.1007/82_2012_207. [DOI] [PubMed] [Google Scholar]
- Luby SP, Rahman M, Hossain MJ, et al. Foodborne transmission of Nipah virus, Bangladesh. Emerg Infect Dis. 2006;12:1888–1894. doi: 10.3201/eid1212.060732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luby SP, Gurley ES, Hossain MJ. Transmission of human infection with Nipah virus. Clin Infect Dis. 2009;49:1743–1748. doi: 10.1086/647951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luby SP, Hossain MJ, Gurley ES, et al. Recurrent zoonotic transmission of Nipah virus into humans, Bangladesh, 2001-2007. Emerg Infect Dis. 2009;15:1229–1235. doi: 10.3201/eid1508.081237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luis AD, Hayman DT, O’Shea TJ, et al. A comparison of bats and rodents as reservoirs of zoonotic viruses: are bats special? Proc Biol Sci. 2013;280(1756):20122753. doi: 10.1098/rspb.2012.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumio J, Hillbom M, Roine R, et al. Human rabies of bat origin in Europe. Lancet. 1986;1:378. doi: 10.1016/S0140-6736(86)92336-6. [DOI] [PubMed] [Google Scholar]
- Lumlertdacha B, Boongird K, Wanghongsa S, et al. Survey for bat lyssaviruses, Thailand. Emerg Infect Dis. 2005;11:232–236. doi: 10.3201/eid1102.040691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyman CP. Thermoregulation and metabolism in bats. In: Wimsatt WA, editor. Biology of bats. New York: Academic; 1970. pp. 301–330. [Google Scholar]
- Machain-Williams C, López-Uribe M, Talavera-Aguilar L, et al. Serologic evidence of flavivirus infection in bats in the Yucatan Peninsula of Mexico. J Wildl Dis. 2013;49:684–689. doi: 10.7589/2012-12-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie JS. Emerging zoonotic encephalitis viruses: lessons from Southeast Asia and Oceania. J Neurovirol. 2005;11:434–440. doi: 10.1080/13550280591002487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie JS, Williams DT. The zoonotic flaviviruses of Southern, South-Eastern and Eastern Asia, and Australasia: the potential for emergent viruses. Zoonoses Public Health. 2009;56:338–356. doi: 10.1111/j.1863-2378.2008.01208.x. [DOI] [PubMed] [Google Scholar]
- Mackenzie JS, Chua KB, Daniels PW, et al. Emerging viral diseases of Southeast Asia and the Western Pacific. Emerg Infect Dis. 2001;7(3 Suppl):497–504. doi: 10.3201/eid0707.017703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie JS, Field HE, Guyatt KJ. Managing emerging diseases borne by fruit bats (flying foxes), with particular reference to henipaviruses and Australian bat lyssavirus. J Appl Microbiol. 2003;94(Suppl):59S–69S. doi: 10.1046/j.1365-2672.94.s1.7.x. [DOI] [PubMed] [Google Scholar]
- Madi D, Achappa B, Ramapuram JT, et al. Dengue encephalitis-A rare manifestation of dengue fever. Asian Pac J Trop Biomed. 2014;4(Suppl 1):S70–S72. doi: 10.12980/APJTB.4.2014C1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahalingam S, Herrero LJ, Playford EG, et al. Hendra virus: an emerging paramyxovirus in Australia. Lancet Infect Dis. 2012;12:799–807. doi: 10.1016/S1473-3099(12)70158-5. [DOI] [PubMed] [Google Scholar]
- Mailles A, Blanckaert K, Chaud P et al (2013) First cases of Middle East respiratory syndrome coronavirus (MERS-CoV) infections in France, investigations and implications for the prevention of human-to-human transmission, France, May 2013. Euro Surveill 18(24). pii: 20502 [PubMed]
- Main AJ. Virologic and serologic survey for eastern equine encephalomyelitis and certain other viruses in colonial bats of New England. J Wildl Dis. 1979;15:455–466. doi: 10.7589/0090-3558-15.3.455. [DOI] [PubMed] [Google Scholar]
- Marí Saéz A, Weiss S, Nowak K, et al. Investigating the zoonotic origin of the West African Ebola epidemic. EMBO Mol Med. 2014;7:17–23. doi: 10.15252/emmm.201404792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markotter W, Kuzmin I, Rupprecht CE, et al. Isolation of Lagos bat virus from water mongoose. Emerg Infect Dis. 2006;12:1913–1918. doi: 10.3201/eid1212.060514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markotter W, Randles J, Rupprecht CE, et al. Lagos bat virus, South Africa. Emerg Infect Dis. 2006;12:504–506. doi: 10.3201/eid1203.051306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markotter W, Kuzmin IV, Rupprecht CE, et al. Lagos bat virus virulence in mice inoculated by the peripheral route. Epidemiol Infect. 2009;137:1155–1162. doi: 10.1017/S0950268808001945. [DOI] [PubMed] [Google Scholar]
- Marsh GA, de Jong C, Barr JA, et al. Cedar virus: a novel Henipavirus isolated from Australian bats. PLoS Pathog. 2012;8(8):e1002836. doi: 10.1371/journal.ppat.1002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston D, Horton DL, Ngeleja C, et al. Ikoma lyssavirus, highly divergent novel lyssavirus in African civet. Emerg Infect Dis. 2012;18:664–667. doi: 10.3201/eid1804.111553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Burnes J, Lopez A, Medellin J, et al. An outbreak of vampire bat-transmitted rabies in cattle in northeastern Mexico. Can Vet J. 1997;38:175–177. [PMC free article] [PubMed] [Google Scholar]
- McCall BJ, Epstein JH, Neill AS, et al. Potential exposure to Australian bat lyssavirus, Queensland, 1996-1999. Emerg Infect Dis. 2000;6:259–264. doi: 10.3201/eid0603.000305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McColl KA, Chamberlain T, Lunt RA, et al. Susceptibility of domestic dogs and cats to Australian bat lyssavirus (ABLV) Vet Microbiol. 2007;123:15–25. doi: 10.1016/j.vetmic.2007.03.024. [DOI] [PubMed] [Google Scholar]
- McCraken GF, Gassel MF. Genetic structure in migratory and non-migratory populations of Brazilian free-tailed bats. J Mammal. 1998;78:348–357. doi: 10.2307/1382888. [DOI] [Google Scholar]
- McElhinney LM, Marston DA, Leech S, et al. Molecular epidemiology of bat lyssaviruses in Europe. Zoonoses Public Health. 2013;60:35–45. doi: 10.1111/zph.12003. [DOI] [PubMed] [Google Scholar]
- McLean RG, Trevino HA, Sather GE. Prevalence of selected zoonotic diseases in vertebrates from Haiti, 1972. J Wildl Dis. 1979;15:327–330. doi: 10.7589/0090-3558-15.2.327. [DOI] [PubMed] [Google Scholar]
- McQuiston JH, Yager PA, Smith JS, et al. Epidemiologic characteristics of rabies virus variants in dogs and cats in the United States, 1999. J Am Vet Med Assoc. 2001;218:1939–1942. doi: 10.2460/javma.2001.218.1939. [DOI] [PubMed] [Google Scholar]
- Mebatsion T, Cox JH, Frost JW. Isolation and characterization of 115 street rabies virus isolates from Ethiopia by using monoclonal antibodies: identification of 2 isolates as Mokola and Lagos bat viruses. J Infect Dis. 1992;166:972–977. doi: 10.1093/infdis/166.5.972. [DOI] [PubMed] [Google Scholar]
- Memish ZA, Mishra N, Olival KJ, et al. Middle East respiratory syndrome coronavirus in bats, Saudi Arabia. Emerg Infect Dis. 2013;19:1819–1823. doi: 10.3201/eid1911.131172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memish ZA, Cotten M, Meyer B, et al. Human infection with MERS coronavirus after exposure to infected camels, Saudi Arabia, 2013. Emerg Infect Dis. 2014;20:1012–1015. doi: 10.3201/eid2006.140402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meredith CD, Standing E. Lagos bat virus in South Africa. Lancet. 1981;1:832–833. doi: 10.1016/S0140-6736(81)92698-2. [DOI] [PubMed] [Google Scholar]
- Meredith CD, Prossouw AP, Koch HP. An unusual case of human rabies thought to be of chiropteran origin. S Afr Med J. 1971;45:767–769. [PubMed] [Google Scholar]
- Messenger SL, Rupprecht CE, Smith JS. Bats, emerging virus infections, and the rabies paradigm. In: Kunz TH, Fenton MB, editors. Bat ecology. Chicago: The University of Chicago Press; 2003. pp. 622–679. [Google Scholar]
- Messenger SL, Smith JS, Orciari LA, et al. Emerging pattern of rabies deaths and increased viral infectivity. Emerg Infect Dis. 2003;9:151–154. doi: 10.3201/eid0902.020083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer B, Muller MA, Corman VM, et al. Antibodies against MERS coronavirus in dromedary camels, United Arab Emirates, 2003 and 2013. Emerg Infect Dis. 2014;20:552–559. doi: 10.3201/eid2004.131746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mickleburgh S, Hutson A, Racey P. Old World fruit bats: an action plan for their conservation. Gland, Switzerland: International Union for Conservation of Nature; 1992. [Google Scholar]
- Middleton DJ, Westbury HA, Morrissy CJ, et al. Experimental Nipah virus infection in pigs and cats. J Comp Pathol. 2002;126:124–136. doi: 10.1053/jcpa.2001.0532. [DOI] [PubMed] [Google Scholar]
- Miia JV, Tiina N, Tarja S, et al. Evolutionary trends of European bat lyssavirus type 2 including genetic characterization of Finnish strains of human and bat origin 24 years apart. Arch Virol. 2015;160:1489–1498. doi: 10.1007/s00705-015-2424-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldofsky H, Patcai J. Chronic widespread musculoskeletal pain, fatigue, depression and disordered sleep in chronic post-SARS syndrome; a case-controlled study. BMC Neurol. 2011;11:37. doi: 10.1186/1471-2377-11-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondul AM, Krebs JW, Childs JE. Trends in national surveillance for rabies among bats in the United States (1993-2000) J Am Vet Med Assoc. 2003;222:633–639. doi: 10.2460/javma.2003.222.633. [DOI] [PubMed] [Google Scholar]
- Moore GJ, Raymond GH. Prolonged incubation period of rabies in a naturally infected insectivorous bat, Eptesicus fuscus (Beauvois) J Wildl Dis. 1970;6:167–168. doi: 10.7589/0090-3558-6.3.167. [DOI] [PubMed] [Google Scholar]
- Muleya W, Sasaki M, Orba Y, et al. Molecular epidemiology of paramyxoviruses in frugivorous Eidolon helvum bats in Zambia. J Vet Med Sci. 2014;76:611–614. doi: 10.1292/jvms.13-0518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller T, Cox J, Peter W, et al. Spill-over of European bat lyssavirus type 1 into a stone marten (Martes foina) in Germany. J Vet Med B Infect Dis Vet Public Health. 2004;51:49–54. doi: 10.1111/j.1439-0450.2003.00725.x. [DOI] [PubMed] [Google Scholar]
- Murray K, Selleck P, Hooper P, et al. A morbillivirus that caused fatal disease in horses and humans. Science. 1995;268:94–97. doi: 10.1126/science.7701348. [DOI] [PubMed] [Google Scholar]
- Nadin-Davis SA, Loza-Rubio E. The molecular epidemiology of rabies associated with chiropteran hosts in Mexico. Virus Res. 2006;117:215–226. doi: 10.1016/j.virusres.2005.10.006. [DOI] [PubMed] [Google Scholar]
- Nadin-Davis SA, Huang W, Armstrong J, et al. Antigenic and genetic divergence of rabies viruses from bat species indigenous to Canada. Virus Res. 2001;74:139–156. doi: 10.1016/S0168-1702(00)00259-8. [DOI] [PubMed] [Google Scholar]
- Nathwani D, McIntyre PG, White K, et al. Fatal human rabies caused by European bat Lyssavirus type 2a infection in Scotland. Clin Infect Dis. 2003;37:598–601. doi: 10.1086/376641. [DOI] [PubMed] [Google Scholar]
- Nel LH, Rupprecht CE. Emergence of lyssaviruses in the Old World: the case of Africa. In: Childs JE, Richt JA, Mackenzie JS (eds), Wildlife and emerging zoonotic diseases: the biology, circumstances and consequences of cross-species transmission. Curr Top Microbiol Immunol. 2007;315:161–193. doi: 10.1007/978-3-540-70962-6_8. [DOI] [PubMed] [Google Scholar]
- Nel L, Jacobs J, Jaftha J, et al. New cases of Mokola virus infection in South Africa: a genotypic comparison of Southern African virus isolates. Virus Genes. 2000;20:103–106. doi: 10.1023/A:1008120511752. [DOI] [PubMed] [Google Scholar]
- Neuweiler G. The biology of bats. Oxford: Oxford University Press; 2000. [Google Scholar]
- Nieuwenhuijs J, Haagsma J, Lina P. Epidemiology and control of rabies in bats in The Netherlands. Rev Sci Tech. 1992;11:1155–1161. doi: 10.20506/rst.11.4.653. [DOI] [PubMed] [Google Scholar]
- Noah DL, Drenzek CL, Smith JS, et al. Epidemiology of human rabies in the United States, 1980 to 1996. Ann Intern Med. 1998;128:922–930. doi: 10.7326/0003-4819-128-11-199806010-00012. [DOI] [PubMed] [Google Scholar]
- Nolden T, Banyard AC, Finke S, et al. Comparative studies on the genetic, antigenic and pathogenic characteristics of Bokeloh bat lyssavirus. J Gen Virol. 2014;95:1647–1653. doi: 10.1099/vir.0.065953-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nor M, Gan C, Ong B. Nipah virus infection of pigs in peninsular Malaysia. Rev Sci Off Int Epiz. 2000;19:160–165. doi: 10.20506/rst.19.1.1202. [DOI] [PubMed] [Google Scholar]
- Nowak R. Walker’s bats of the world. Baltimore: The Johns Hopkins University Press; 1994. [Google Scholar]
- O’Shea TJ, Cryan PM, Cunningham AA, et al. Bat flight and zoonotic viruses. Emerg Infect Dis. 2014;20:741–745. doi: 10.3201/eid2005.130539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa H, Miyamoto H, Nakayama E, et al. Seroepidemiological prevalence of multiple species of filoviruses in fruit bats (Eidolon helvum) migrating in Africa. J Infect Dis. 2015;212(Suppl 2):S101–S108. doi: 10.1093/infdis/jiv063. [DOI] [PubMed] [Google Scholar]
- Olival KJ, Hayman DT. Filoviruses in bats: current knowledge and future directions. Viruses. 2014;6:1759–1788. doi: 10.3390/v6041759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parashar UD, Sunn LM, Ong F, et al. Case-control study of risk factors for human infection with a new zoonotic paramyxovirus, Nipah virus, during a 1998-1999 outbreak of severe encephalitis in Malaysia. J Infect Dis. 2000;181:1755–1759. doi: 10.1086/315457. [DOI] [PubMed] [Google Scholar]
- Paton N, Leo Y, Zaki S, et al. Outbreak of Nipah virus infection among abattoir workers in Singapore. Lancet. 1999;354:1253–1256. doi: 10.1016/S0140-6736(99)04379-2. [DOI] [PubMed] [Google Scholar]
- Paul SD, Rajagopalan PK, Sreenivasan MA. Isolation of the West Nile virus from the frugivorous bat, Rousettus leschenaulti. Indian J Med Res. 1970;58:1169–1171. [PubMed] [Google Scholar]
- Pawan JL. The transmission of paralytic rabies in Trinidad by the vampire bat (Desmodus rotundus murinus Wagner) Caribb Med J. 1959;21:110–136. [PubMed] [Google Scholar]
- Paweska JT, Blumberg LH, Liebenberg C, et al. Fatal human infection with rabies-related Duvenhage virus, South Africa. Emerg Infect Dis. 2006;12:1965–1967. doi: 10.3201/eid1212.060764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peel AJ, Sargan DR, Baker KS, et al. Continent-wide panmixia of an African fruit bat facilitates transmission of potentially zoonotic viruses. Nat Commun. 2013;4:2770. doi: 10.1038/ncomms3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perea-Martínez L, Moreno-Sandoval HN, Moreno-Altamirano MM, et al. Experimental infection of Artibeus intermedius bats with serotype-2 dengue virus. Comp Immunol Microbiol Infect Dis. 2013;36:193–198. doi: 10.1016/j.cimid.2012.12.002. [DOI] [PubMed] [Google Scholar]
- Perera R, Wang P, Gomaa M, El-Shesheny R, Kandeil A, Bagato O, Siu L, Shehata M, Kayed A, Moatasim Y, Li M, Poon L, Guan Y, Webby R, Ali M, Peiris J, Kayali G. Seroepidemiology for MERS coronavirus using microneutralisation and pseudoparticle virus neutralisation assays reveal a high prevalence of antibody in dromedary camels in Egypt, June 2013. Eurosurveillance. 2013;18(36):20574. doi: 10.2807/1560-7917.es2013.18.36.20574. [DOI] [PubMed] [Google Scholar]
- Pernet O, Schneider BS, Beaty SM, et al. Evidence for henipavirus spillover into human populations in Africa. Nat Commun. 2014;5:5342. doi: 10.1038/ncomms6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philbey AW, Kirkland PD, Ross AD, et al. An apparently new virus (family Paramyxoviridae) infectious for pigs, humans, and fruit bats. Emerg Infect Dis. 1998;4:269–271. doi: 10.3201/eid0402.980214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard-Meyer E, Dubourg-Savage MJ, Arthur L, et al. Active surveillance of bat rabies in France: a 5-year study (2004-2009) Vet Microbiol. 2011;151:390–395. doi: 10.1016/j.vetmic.2011.03.034. [DOI] [PubMed] [Google Scholar]
- Picard-Meyer E, Servat A, Robardet E, et al. Isolation of Bokeloh bat lyssavirus in Myotis nattereri in France. Arch Virol. 2013;158:2333–2340. doi: 10.1007/s00705-013-1747-y. [DOI] [PubMed] [Google Scholar]
- Picard-Meyer E, Robardet E, Arthur L, et al. Bat rabies in France: a 24-year retrospective epidemiological study. PLoS One. 2014;9(6):e98622. doi: 10.1371/journal.pone.0098622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilipski JD, Pilipski LM, Risley LS. West Nile virus antibodies in bats from New Jersey and New York. J Wildl Dis. 2004;40:335–337. doi: 10.7589/0090-3558-40.2.335. [DOI] [PubMed] [Google Scholar]
- Pinzon JE, Wilson JM, Tucker CJ, et al. Trigger events: enviroclimatic coupling of Ebola hemorrhagic fever outbreaks. Am J Trop Med Hyg. 2004;71:664–674. [PubMed] [Google Scholar]
- Platt KB, Mangiafico JA, Rocha OJ, et al. Detection of dengue virus neutralizing antibodies in bats from Costa Rica and Ecuador. J Med Entomol. 2000;37:965–967. doi: 10.1603/0022-2585-37.6.965. [DOI] [PubMed] [Google Scholar]
- Playford EG, McCall B, Smith G, et al. Human Hendra virus encephalitis associated with equine outbreak, Australia, 2008. Emerg Infect Dis. 2010;16:219–223. doi: 10.3201/eid1602.090552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plowright RK, Field HE, Smith C, et al. Reproduction and nutritional stress are risk factors for Hendra virus infection in little red flying foxes (Pteropus scapulatus) Proc Biol Sci. 2008;275:861–869. doi: 10.1098/rspb.2007.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pons-Salort M, Serra-Cobo J, Jay F, et al. Insights into persistence mechanisms of a zoonotic virus in bat colonies using a multispecies metapopulation model. PLoS One. 2014;9(4):e95610. doi: 10.1371/journal.pone.0095610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulliam JR, Epstein JH, Dushoff J, et al. Agricultural intensification, priming for persistence and the emergence of Nipah virus: a lethal bat-borne zoonoses. J R Soc Interface. 2012;9:89–101. doi: 10.1098/rsif.2011.0223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman SA, Hassan SS, Olival KJ, et al. Characterization of Nipah virus from naturally infected Pteropus vampyrus bats, Malaysia. Emerg Infect Dis. 2010;16:1990–1993. doi: 10.3201/eid1612.091790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman MA, Hossain MJ, Sultana S, et al. Date palm sap linked to Nipah virus outbreak in Bangladesh, 2008. Vector Borne Zoonotic Dis. 2012;12:65–72. doi: 10.1089/vbz.2011.0656. [DOI] [PubMed] [Google Scholar]
- Rahman SA, Hassan L, Epstein JH, et al. Risk factors for Nipah virus infection among pteropid bats, peninsular Malaysia. Emerg Infect Dis. 2013;19:51–60. doi: 10.3201/eid1901.120221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravanini P, Huhtamo E, Ilaria V et al (2012) Japanese encephalitis virus RNA detected in Culex pipiens mosquitoes in Italy. Euro Surveill 17(28). pii: 20221 [DOI] [PubMed]
- Reusken CB, Ababneh M, Raj VS, et al. Middle East respiratory syndrome coronavirus (MERS-CoV) serology in major livestock species in an affected region in Jordan, June to September 2013. Euro Surveill. 2013;18:20662. doi: 10.2807/1560-7917.ES2013.18.50.20662. [DOI] [PubMed] [Google Scholar]
- Reusken CB, Haagmans BL, Muller MA, et al. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. Lancet Infect Dis. 2013;13:859–866. doi: 10.1016/S1473-3099(13)70164-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynes JM, Molia S, Audry L, et al. Serologic evidence of lyssavirus infection in bats, Cambodia. Emerg Infect Dis. 2004;10:2231–2234. doi: 10.3201/eid1012.040459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers RJ, Douglas IC, Baldock FC, et al. Investigation of a second focus of equine morbillivirus infection in coastal Queensland. Aust Vet J. 1996;74:243–244. doi: 10.1111/j.1751-0813.1996.tb15413.x. [DOI] [PubMed] [Google Scholar]
- Rohde RE, Mayes BC, Smith JS, et al. Bat rabies, Texas, 1996-2000. Emerg Infect Dis. 2004;10:948–952. doi: 10.3201/eid1005.030719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roine RO, Hillbom M, Valle M, et al. Fatal encephalitis caused by a bat-borne rabies-related virus. Clinical findings. Brain. 1988;111:1505–1516. doi: 10.1093/brain/111.6.1505. [DOI] [PubMed] [Google Scholar]
- Ronsholt L. A new case of European bat lyssavirus (EBL) infection in Danish sheep. Rabies Bull Eur. 2002;2:15. [Google Scholar]
- Ronsholt R, Sorensen KJ, Bruschke CJ, et al. Clinically silent rabies infection in (zoo) bats. Vet Rec. 1998;142:519–520. doi: 10.1136/vr.142.19.519. [DOI] [PubMed] [Google Scholar]
- Rosevear DR. The bats of West Africa. London: The British Museum (Natural History); 1965. [Google Scholar]
- Rupprecht CE, Gibbons RV. Clinical practice. Prophylaxis against rabies. N Engl J Med. 2004;351:2626–2635. doi: 10.1056/NEJMcp042140. [DOI] [PubMed] [Google Scholar]
- Sabeta Claude T., Markotter Wanda, Mohale Debrah K., Shumba Wonderful, Wandeler Alexander I., Nel Louis H. Mokola Virus in Domestic Mammals, South Africa. Emerging Infectious Diseases. 2007;13(9):1371–1373. doi: 10.3201/eid1309.070466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler WW, Enright JB. Effect of metabolic level of the host upon the pathogenesis of rabies in the bat. J Infect Dis. 1959;105:267–273. doi: 10.1093/infdis/105.3.267. [DOI] [PubMed] [Google Scholar]
- Sahu R, Verma R, Jain A, et al. Neurologic complications in dengue virus infection: a prospective cohort study. Neurology. 2014;83:1601–1609. doi: 10.1212/WNL.0000000000000935. [DOI] [PubMed] [Google Scholar]
- Samaratunga H, Searle JW, Hudson N. Non-rabies Lyssavirus human encephalitis from fruit bats: Australian bat Lyssavirus (pteropid Lyssavirus) infection. Neuropathol Appl Neurobiol. 1998;24:331–335. doi: 10.1046/j.1365-2990.1998.00129.x. [DOI] [PubMed] [Google Scholar]
- Sato G, Kobayashi Y, Shoji Y, et al. Molecular epidemiology of rabies from Maranhao and surrounding states in the northeastern region of Brazil. Arch Virol. 2006;151:2243–2251. doi: 10.1007/s00705-006-0770-7. [DOI] [PubMed] [Google Scholar]
- Sazzad HM, Hossain MJ, Gurley ES, et al. Nipah virus infection outbreak with nosocomial and corpse-to-human transmission, Bangladesh. Emerg Infect Dis. 2013;19:210–217. doi: 10.3201/eid1902.120971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scatterday JE. Bat rabies in Florida. J Am Vet Med Assoc. 1954;124:125. [PubMed] [Google Scholar]
- Schaefer R, Batista HB, Franco AC, et al. Studies on antigenic and genomic properties of Brazilian rabies virus isolates. Vet Microbiol. 2005;107:161–170. doi: 10.1016/j.vetmic.2005.01.023. [DOI] [PubMed] [Google Scholar]
- Schatz J, Fooks AR, McElhinney L, et al. Bat rabies surveillance in Europe. Zoonoses Public Health. 2013;60:22–34. doi: 10.1111/zph.12002. [DOI] [PubMed] [Google Scholar]
- Schatz J, Freuling CM, Auer E, et al. Enhanced passive bat rabies surveillance in indigenous bat species from Germany—a retrospective study. PLoS Negl Trop Dis. 2014;8(5):e2835. doi: 10.1371/journal.pntd.0002835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer W. F., Chia C. Wong, Terrian J., Dickerman R. W., La Fiandra R. P. Ecologic Studies of Venezuelan Encephalitis Virus in Southeastern México. The American Journal of Tropical Medicine and Hygiene. 1971;20(6):980–988. doi: 10.4269/ajtmh.1971.20.980. [DOI] [PubMed] [Google Scholar]
- Schneeberger K, Czirják GÁ, Voigt CC. Measures of the constitutive immune system are linked to diet and roosting habits of neotropical bats. PLoS One. 2013;8:e54023. doi: 10.1371/journal.pone.0054023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schountz T. Virology and immunobiology of bats. In: Adams RA, Pedersen SC, editors. Bat evolution, ecology, and conservation. New York: Springer; 2013. pp. 393–412. [Google Scholar]
- Selvey LA, Wells RM, McCormack JG, et al. Infection of humans and horses by a newly described morbillivirus. Med J Aust. 1995;162:642–645. doi: 10.5694/j.1326-5377.1995.tb126050.x. [DOI] [PubMed] [Google Scholar]
- Selvey LA, Taylor R, Arklay A, et al. Screening of bat carers for antibodies to equine morbillivirus. Commun Dis Intell. 1996;20:477–478. [Google Scholar]
- Sendow I, Field HE, Curran J, et al. Henipavirus in Pteropus vampyrus bats, Indonesia. Emerg Infect Dis. 2006;12:711–712. doi: 10.3201/eid1204.051181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra-Cobo J, Amengual B, Abellan C, et al. European bat lyssavirus infection in Spanish bat populations. Emerg Infect Dis. 2002;8:413–420. doi: 10.3201/eid0804.010263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seymour C, Dickerman RW, Martin MS. Venezuelan encephalitis virus infection in neotropical bats. I. Natural infection in a Guatemalan enzootic focus. Am J Trop Med Hyg. 1978;27:290–296. doi: 10.4269/ajtmh.1978.27.290. [DOI] [PubMed] [Google Scholar]
- Shi Z. Emerging infectious diseases associated with bat viruses. Sci China Life Sci. 2013;56:678–682. doi: 10.1007/s11427-013-4517-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z, Hu Z. A review of studies on animal reservoirs of the SARS coronavirus. Virus Res. 2008;133:74–87. doi: 10.1016/j.virusres.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shope RE, Murphy FA, Harrison AK, et al. Two African viruses serologically and morphologically related to rabies virus. J Virol. 1970;6:690–692. doi: 10.1128/jvi.6.5.690-692.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons NB, Conway TM. Evolution of ecological diversity of bats. In: Kunz TH, Fenton MB, editors. Bat ecology. Chicago: The University of Chicago Press; 2003. pp. 493–535. [Google Scholar]
- Simmons NB, Geisler JH. Phylogenetic relationships of Icaronycteris, Archaeonycteris, Hassianycteris, and Palaeochiropteryx to extant bat lineages, with comments on the evolution of echolocation and foraging strategies in Microchiroptera. New York: American Museum of Natural History; 1998. [Google Scholar]
- Simmons NB, Seymour KL, Habersetzer J, et al. Inferring echolocation in ancient bats. Nature. 2010;466(7309):E8. doi: 10.1038/nature09219. [DOI] [PubMed] [Google Scholar]
- Smith I, Wang LF. Bats and their virome: an important source of emerging viruses capable of infecting humans. Curr Opin Virol. 2013;3:84–91. doi: 10.1016/j.coviro.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JS, Reid-Sanden FL, Roumillat LF, et al. Demonstration of antigenic variation among rabies virus isolates by using monoclonal antibodies to nucleocapsid proteins. J Clin Microbiol. 1986;24:573–580. doi: 10.1128/jcm.24.4.573-580.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C, Skelly C, Kung N, et al. Flying-fox species diversity—a spatial risk factor for Hendra virus infection in horses in eastern Australia. PLoS One. 2014;9:e99965. doi: 10.1371/journal.pone.0099965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohaytati AR, Hassan L, Sharifah SH, et al. Evidence for Nipah virus recrudescence and serological patterns of captive Pteropus vampyrus. Epidemiol Infect. 2011;139:1570–1579. doi: 10.1017/S0950268811000550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon T, Dung NM, Vaughn DW, et al. Neurological manifestations of dengue infection. Lancet. 2000;355:1053–1059. doi: 10.1016/S0140-6736(00)02036-5. [DOI] [PubMed] [Google Scholar]
- Soria Baltazar R, Blancou J, Artois M. Etude de virus de la rage isolé d’une chauve-souris Européene Eptesicus serotinus: pouvoir pathogéne pour les ovins et le renard roux. Rev Méd Vét. 1988;139:621. [Google Scholar]
- Sotomayor-Bonilla J, Chaves A, Rico-Chávez O, et al. Dengue virus in bats from southeastern Mexico. Am J Trop Med Hyg. 2014;91:129–131. doi: 10.4269/ajtmh.13-0524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speakman JR, Thomas DW. Physiological ecology and energetics of bats. In: Kunz TH, Fenton MB, editors. Bat ecology. Chicago: The University of Chicago Press; 2003. pp. 430–490. [Google Scholar]
- Speare R, Skerratt L, Foster R, et al. Australian bat lyssavirus infection in three fruit bats from north Queensland. Commun Dis Intell. 1997;21:117–120. doi: 10.33321/cdi.1997.21.25. [DOI] [PubMed] [Google Scholar]
- Springer MS. Phylogenetics: bats united, microbats divided. Curr Biol. 2013;23:R999–R1001. doi: 10.1016/j.cub.2013.09.053. [DOI] [PubMed] [Google Scholar]
- Srinivasan A, Burton EC, Kuehnert MJ, et al. Transmission of rabies virus from an organ donor to four transplant recipients. N Engl J Med. 2005;352:1103–1111. doi: 10.1056/NEJMoa043018. [DOI] [PubMed] [Google Scholar]
- St George TD. Are there common features in lyssavirus diseases? In: Uren MF, Blok J, Manderson LH, editors. Arbovirus research in Australia: proceedings of the fifth symposium. Brisbane, Australia: Queensland Institute of Medical Research; 1989. pp. 264–267. [Google Scholar]
- Steece RS, Calisher CH. Evidence for prenatal transfer of rabies virus in the Mexican free-tailed bat (Tadarida brasiliensis Mexicana) J Wildl Dis. 1989;25:329–334. doi: 10.7589/0090-3558-25.3.329. [DOI] [PubMed] [Google Scholar]
- Streicker DG, Turmelle AS, Vonhof MJ, et al. Host phylogeny constrains cross-species emergence and establishment of rabies virus in bats. Science. 2010;329:676–679. doi: 10.1126/science.1188836. [DOI] [PubMed] [Google Scholar]
- Sulkin SE, Allen R. Virus infection in bats. Monogr Virol. 1974;8:1–103. doi: 10.1159/000395511. [DOI] [PubMed] [Google Scholar]
- Sulkin SE, Krutzsch PH, Allen R, et al. Studies on the pathogenesis of rabies in insectivorous bats. I. Role of brown adipose tissue. J Exp Med. 1959;110:369–388. doi: 10.1084/jem.110.3.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung JJ-Y (2006) Clinical features.In: SARS: how a global epidemic was stopped. World Health Organization Western Pacific Regional Office, Manila, pp 175–184
- Sureau P, Germain M, Herve JP, et al. Isolation of the Lagos-bat virus in the Central African Republic [Article in French] Bull Soc Pathol Exot Filiales. 1977;70:467–470. [PubMed] [Google Scholar]
- Swanepoel R, Barnard BJ, Meredith CD, et al. Rabies in southern Africa. Onderstepoort J Vet Res. 1993;60:325–346. [PubMed] [Google Scholar]
- Tan CT, Goh KJ, Wong KT, et al. Relapsed and late-onset Nipah encephalitis. Ann Neurol. 2002;51:703–708. doi: 10.1002/ana.10212. [DOI] [PubMed] [Google Scholar]
- Tan V, Thai le H, Phu NH, et al. Viral aetiology of central nervous system infections in adults admitted to a tertiary referral hospital in southern Vietnam over 12 years. PLoS Negl Trop Dis. 2014;8(8):e3127. doi: 10.1371/journal.pntd.0003127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeling EC, Scally M, Kao DJ, et al. Molecular evidence regarding the origin of echolocation and flight in bats. Nature. 2000;403:188–192. doi: 10.1038/35003188. [DOI] [PubMed] [Google Scholar]
- Teeling EC, Madsen O, Van Den Bussche RA, et al. Microbat paraphyly and the convergent evolution of a key innovation in Old World rhinolophoid microbats. Proc Natl Acad Sci U S A. 2002;99:1431–1436. doi: 10.1073/pnas.022477199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeling EC, Springer MS, Madsen O, et al. A molecular phylogeny for bats illuminates biogeography and the fossil record. Science. 2005;307:580–584. doi: 10.1126/science.1105113. [DOI] [PubMed] [Google Scholar]
- Thompson NN, Auguste AJ, Travassos da Rosa AP, et al. Seroepidemiology of selected alphaviruses and flaviviruses in bats in Trinidad. Zoonoses Public Health. 2015;62:53–60. doi: 10.1111/zph.12118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tignor GH, Murphy FA, Clark HF, et al. Duvenhage virus: morphological, biochemical, histopathological and antigenic relationships to the rabies serogroup. J Gen Virol. 1977;37:595–611. doi: 10.1099/0022-1317-37-3-595. [DOI] [Google Scholar]
- Tjornehoj K, Fooks AR, Agerholm JS, et al. Natural and experimental infection of sheep with European bat lyssavirus type-1 of Danish bat origin. J Comp Pathol. 2006;134:190–201. doi: 10.1016/j.jcpa.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Towner JS, Pourrut X, Albariño CG, et al. Marburg virus infection detected in a common African bat. PLoS One. 2007;2(8):e764. doi: 10.1371/journal.pone.0000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towner JS, Amman BR, Sealy TK, et al. Isolation of genetically diverse Marburg viruses from Egyptian fruit bats. PLoS Pathog. 2009;5(7):e1000536. doi: 10.1371/journal.ppat.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimarchi CV, Debbie JG. Naturally occurring rabies virus and neutralizing antibody in two species of insectivorous bats of New York State. J Wildl Dis. 1977;13:366–369. doi: 10.7589/0090-3558-13.4.366. [DOI] [PubMed] [Google Scholar]
- Tu C, Crameri G, Kong X, et al. Antibodies to SARS coronavirus in civets. Emerg Infect Dis. 2004;10:2244–2248. doi: 10.3201/eid1012.040520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuttle MD. Population ecology of the gray bat (Myotis grisescens): philopatry, timing and patterns of movement, weight loss during migration, and seasonal adaptive strategies. Lawrence: Museum of Natural History, University of Kansas; 1976. pp. 1–38. [Google Scholar]
- Ubico SR, McLean RG. Serologic survey of neotropical bats in Guatemala for virus antibodies. J Wildl Dis. 1995;31:1–9. doi: 10.7589/0090-3558-31.1.1. [DOI] [PubMed] [Google Scholar]
- Van den Hurk AF, Smith CS, Field HE, et al. Transmission of Japanese encephalitis virus from the black flying fox, Pteropus Alecto, to Culex annulirostris mosquitoes, despite the absence of detectable viremia. Am J Trop Med Hyg. 2009;81:457–462. [PubMed] [Google Scholar]
- van der Poel WH, Lina PH, Kramps JA. Public health awareness of emerging zoonotic viruses of bats: a European perspective. Vector Borne Zoonotic Dis. 2006;6:315–324. doi: 10.1089/vbz.2006.6.315. [DOI] [PubMed] [Google Scholar]
- Van Gucht S, Verlinde R, Colyn J, et al. Favourable outcome in a patient bitten by a rabid bat infected with the European bat lyssavirus-1. Acta Clin Belg. 2013;68:54–58. doi: 10.2143/ACB.68.1.2062721. [DOI] [PubMed] [Google Scholar]
- van Thiel PP, de Bie RM, Eftimov F, et al. Fatal human rabies due to Duvenhage virus from a bat in Kenya: failure of treatment with coma-induction, ketamine, and antiviral drugs. PLoS Negl Trop Dis. 2009;3(7):e428. doi: 10.1371/journal.pntd.0000428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco-Villa A, Orciari LA, Juarez-Islas V, et al. Molecular diversity of rabies viruses associated with bats in Mexico and other countries of the Americas. J Clin Microbiol. 2006;44:1697–1710. doi: 10.1128/JCM.44.5.1697-1710.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Teichman BF, de Koker WC, Bosch SJ, et al. Mokola virus infection: description of recent South African cases and a review of the virus epidemiology. J S Afr Vet Assoc. 1998;69:169–171. doi: 10.4102/jsava.v69i4.847. [DOI] [PubMed] [Google Scholar]
- Vos A, Muller T, Neubert L, et al. Rabies in red foxes (Vulpes vulpes) experimentally infected with European bat lyssavirus type 1. J Vet Med B Infect Dis Vet Public Health. 2004;51:327–332. doi: 10.1111/j.1439-0450.2004.00793.x. [DOI] [PubMed] [Google Scholar]
- Wacharapluesadee S, Lumlertdacha B, Boongird K, et al. Bat Nipah virus, Thailand. Emerg Infect Dis. 2005;11:1949–1951. doi: 10.3201/eid1112.050613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacharapluesadee S, Boongird K, Wanghongsa S, et al. A longitudinal study of the prevalence of Nipah virus in Pteropus lylei bats in Thailand: evidence for seasonal preference in disease transmission. Vector Borne Zoonotic Dis. 2010;10:183–190. doi: 10.1089/vbz.2008.0105. [DOI] [PubMed] [Google Scholar]
- Wang LF, Crameri G. Emerging zoonotic viral diseases. Rev Sci Tech. 2014;33:569–581. doi: 10.20506/rst.33.2.2311. [DOI] [PubMed] [Google Scholar]
- Wang L, Yu M, Hansson E, et al. The exceptionally large genome of Hendra virus: support for creation of a new genus within the family Paramyxoviridae. J Virol. 2000;74:9972–9979. doi: 10.1128/JVI.74.21.9972-9979.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LF, Shi Z, Zhang S, et al. Review of bats and SARS. Emerg Infect Dis. 2006;12:1834–1840. doi: 10.3201/eid1212.060401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JL, Pan XL, Zhang HL, et al. Japanese encephalitis viruses from bats in Yunnan, China. Emerg Infect Dis. 2009;15:939–942. doi: 10.3201/eid1506.081525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LF, Walker PJ, Poon LLM. Mass extinctions, biodiversity and mitochondrial function: are bats ‘special’ as reservoirs for emerging viruses? Curr Opin Virol. 2011;1:649–657. doi: 10.1016/j.coviro.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LF, Collins PI, Fouchier RAM, et al. Paramyxoviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, et al., editors. Virus taxonomy: classification and nomenclature of viruses: ninth report of the International Committee for the Taxonomy of Viruses. San Diego: Elsevier; 2011. pp. 640–653. [Google Scholar]
- Ward MP, Black PF, Childs AJ, et al. Negative findings from serological studies of equine morbillivirus in the Queensland horse population. Aust Vet J. 1996;74:241–243. doi: 10.1111/j.1751-0813.1996.tb15412.x. [DOI] [PubMed] [Google Scholar]
- Warrell M. Rabies and African bat lyssavirus encephalitis and its prevention. Int J Antimicrob Agents. 2010;36(Suppl 1):S47–S52. doi: 10.1016/j.ijantimicag.2010.06.021. [DOI] [PubMed] [Google Scholar]
- Warrilow D, Harrower B, Smith IL, et al. Public health surveillance for Australian bat lyssavirus, in Queensland, Australia, 2000-2001. Emerg Infect Dis. 2003;9:262–264. doi: 10.3201/eid0902.020264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S, Nowak K, Fahr J, et al. Henipavirus-related sequences in fruit bat bushmeat, Republic of Congo. Emerg Infect Dis. 2012;18:1536–1537. doi: 10.3201/eid1809.111607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellenberg GJ, Audry L, Ronsholt L, et al. Presence of European bat lyssavirus RNAs in apparently healthy Rousettus aegyptiacus bats. Arch Virol. 2002;147:349–361. doi: 10.1007/s705-002-8324-3. [DOI] [PubMed] [Google Scholar]
- WHO (2015) Surveillance and outbreak alert. Nipah virus outbreaks in the WHO South-Eastern Region. World Health Organization Regional Office for South-East Asia. http://www.searo.who.int/entity/emerging_diseases/links/nipah_virus_outbreaks_sear/en/. Accessed 4 June 2015
- Wiktor TJ, Macfarlan RI, Foggin CM, et al. Antigenic analysis of rabies and Mokola virus from Zimbabwe using monoclonal antibodies. Dev Biol Stand. 1984;57:199–211. [PubMed] [Google Scholar]
- Wilson DE, Reeder DM. Mammal species of the world. Baltimore: Johns Hopkins University Press; 2005. [Google Scholar]
- Winkler WG. Airborne rabies virus isolation. Wildl Dis. 1968;4:37–40. doi: 10.7589/0090-3558-4.2.37. [DOI] [PubMed] [Google Scholar]
- Wong S, Lau S, Woo P, et al. Bats as a continuing source of emerging infections in humans. Rev Med Virol. 2007;17:67–91. doi: 10.1002/rmv.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo PC, Wang M, Lau SK, et al. Comparative analysis of twelve genomes of three novel group 2c and group 2d coronaviruses reveals unique group and subgroup features. J Virol. 2007;81:1574–1585. doi: 10.1128/JVI.02182-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood JL, Leach M, Waldman L, et al. A framework for the study of zoonotic disease emergence and its drivers: spillover of bat pathogens as a case study. Philos Trans R Soc Lond B Biol Sci. 2012;367:2881–2892. doi: 10.1098/rstb.2012.0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Yang L, Yang F, et al. Novel henipa-like virus, Mojiang paramyxovirus, in rats, China, 2012. Emerg Infect Dis. 2014;20:1064–1066. doi: 10.3201/eid2006.131022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynne JW, Tachedjian M. Bat genomics. In: Wang L-F, Cowled C, editors. Bats and viruses. New Jersey: Wiley Blackwell; 2015. pp. 315–326. [Google Scholar]
- Wynne JW, Shiell BJ, Marsh GA, et al. Proteomics informed by transcriptomics reveals Hendra virus sensitizes bat cells to TRAIL-mediated apoptosis. Genome Biol. 2014;15(11):532. doi: 10.1186/s13059-014-0532-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yob JM, Field H, Rashdi AM, et al. Nipah virus infection in bats (order Chiroptera) in peninsular Malaysia. Emerg Infect Dis. 2001;7:439–441. doi: 10.3201/eid0703.017312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young PL, Halpin K, Selleck PW, et al. Serologic evidence for the presence in Pteropus bats of a paramyxovirus related to equine morbillivirus. Emerg Infect Dis. 1996;2:239–240. doi: 10.3201/eid0203.960315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacks MA, Paessler S. Encephalitic alphaviruses. Vet Microbiol. 2010;140:281–286. doi: 10.1016/j.vetmic.2009.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki M, van Boheemen S, Bestebroer TM, et al. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zhang G, Cowled C, Shi Z, et al. Comparative analysis of bat genomes provides insight into the evolution of flight and immunity. Science. 2013;339:456–460. doi: 10.1126/science.1230835. [DOI] [PMC free article] [PubMed] [Google Scholar]


