Abstract
Dressing the problem of virtual screening is a long-term goal in the drug discovery field, which if properly solved, can significantly shorten new drugs’ R&D cycle. The scoring functionality that evaluates the fitness of the docking result is one of the major challenges in virtual screening. In general, scoring functionality in docking requires a large amount of floating-point calculations, which usually takes several weeks or even months to be finished. This time-consuming procedure is unacceptable, especially when highly fatal and infectious virus arises such as SARS and H1N1, which forces the scoring task to be done in a limited time. This paper presents how to leverage the computational power of GPU to accelerate Dock6’s (http://dock.compbio.ucsf.edu/DOCK_6/) Amber (J. Comput. Chem. 25: 1157–1174, 2004) scoring with NVIDIA CUDA (NVIDIA Corporation Technical Staff, Compute Unified Device Architecture – Programming Guide, NVIDIA Corporation, 2008) (Compute Unified Device Architecture) platform. We also discuss many factors that will greatly influence the performance after porting the Amber scoring to GPU, including thread management, data transfer, and divergence hidden. Our experiments show that the GPU-accelerated Amber scoring achieves a 6.5× speedup with respect to the original version running on AMD dual-core CPU for the same problem size. This acceleration makes the Amber scoring more competitive and efficient for large-scale virtual screening problems.
Keywords: Virtual screen, Dock, Amber scoring, GPU, CUDA
Introduction
One early stage of new drug discovery is focused on finding pharmacologically active compounds from the vast number of chemical compounds. In order to reduce the amount of wet-lab experiments, virtual screening is developed to search chemical compounds database for potentially effective compounds. Computer-assisted virtual drug screening is a definite shortcut to develop new drugs. It can reduce the amount of candidate compounds for biological experiments by thousands of times and is very prospective in exploiting new drug candidates [4].
Identifying the interactions between molecules is critical both to understanding the structure of the proteins and to discovering new drugs. Small molecules or segments of proteins whose structures are already known and stored in database are called ligands, while macromolecules or proteins associated with a disease are called receptors [5]. The final goal is to find out whether the given ligand and receptor can form a favorable complex and how appropriate the complex is, which may inhibit a disease’s function and thus act as a drug.
Virtual screening can usually be roughly divided into two parts functionally:
Routines determining the orientation of a ligand relative to the receptor, which are known as docking;
Routines evaluating the orientation, which are known as scoring.
Docking is the first step in virtual screening that needs a potential site of interest on the receptor to be identified, which is also known as the active site. Within this site, points are identified where ligand atoms may be located. In dock6, a program routine called sphere center is to identify these points by generating spheres filling the site. To orient a ligand within the active site, some of the spheres are “paired” with ligand atoms, which are also called “matched” in docking.
Scoring is the step after docking, which is involved in evaluating the fitness for docked molecules and ranking them. A set of sphere-atom pairs will be on behalf of an orientation in receptor and evaluated with a scoring function on three-dimensional grids. At each grid point, interaction values are to be summed to form a final score. These processes need to be repeated for all possible translations and rotations. There are many kinds of existing scoring algorithms, while amber scoring is prevalent due to its fast speed and considerable high accuracy. The advantage of amber scoring is that both ligand and active sites of the receptor can be flexible during the evaluation, which allows small structural rearrangements to reproduce the so-called induced fit. While the disadvantage is also obvious, it brings tremendous intensive floating-point computations. When performing amber scoring, it calculates the internal energy of the ligand, receptor, and the complex, which can be broken down into three steps:
minimization,
molecule dynamics (MD) simulation,
more minimization using solvents.
The computational complexity of amber scoring is very huge, especially in the MD simulation stage. Three grids that individually have three dimension coordinates are used to represent the molecule during the orientation such as geometry, gradient, and velocity. In each grid, at least 128 elements are required to sustain the accuracy of the final score. During the simulation, scores are calculated in three nested loops, each of which walks through one of the three grids. Derived from vast practical experiences, the MD simulation is supposed to be performed 3,000 times until a preferable result is obtained. All above commonly means that the problem size can reach as large as  , which is computationally infeasible in one single computer.
, which is computationally infeasible in one single computer.
While many virtual screening tools such as GasDock [6], FTDock [7], and Dock6 can utilize multi-CPUs to parallel the computations, the incapacity of CPU in processing floating-point computations still remains untouched. GPU has been widely used for general purpose computations because of its high floating-point computation capability [8]. Compared with CPU, GPU has the advantages of computational power and memory bandwidth. For example, a GeForce 9800 GT can reach 345 GFLOPS at peak rate and has an 86.4 GB/s memory bandwidth, whereas an Intel Core 2 Extreme quad core processor at 2.66 GHz has a theoretical 21.3 peak GFLOPS and 10.7 GB/s memory bandwidth. Another important factor why GPU is becoming widely used is that it is more cost effective than CPU.
Our contributions in this paper include porting the original Dock6 amber scoring to GPU using CUDA, which can archive a 6.5× speedup. We analyze the different memory access patterns in GPU which can lead to a significant divergence in performance. Discussions on how to hide the computation divergence on GPU are made. We also conduct experiments to see the performance improvement.
The rest of the paper is organized as follows. In Sect. 56.2, an overview of Dock6’s amber scoring and analysis of the bottleneck is given. In Sect. 56.3, we present the main idea and implementation of the amber scoring on GPU with CUDA, and details of considerations about performance are made. Then we give the results, including performance comparisons among various GPU versions. Finally, we conclude with discussion and future work.
Analysis of the Amber Scoring in DOCK6
Overview
A primary design philosophy of amber scoring is allowing both the atoms in the ligand and the spheres in the receptor to be flexible during the virtual screening process, generating small structural rearrangements, which is much like the actual situation and gives more accuracy. As a result, a large number of docked orientations need to be analyzed and ranked in order to determine the best fit set of the matched atom-sphere pairs.
In the subsection, we will describe the amber scoring program flow and profile the performance bottleneck of the original amber scoring, which can be perfectly accelerated on GPU.
Program Flow and Performance Analysis
Figure 56.1 shows the steps to score the fitness for possible ligand–receptor pairs in amber. The program first performs conjugate gradient minimization, MD simulation, and more minimization with solvation on the individual ligand, the individual receptor, and the ligand–receptor complex, and then calculates the score as follows:
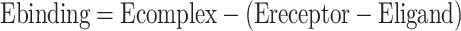 |
Fig. 56.1.
Program flow of amber scoring
The docked molecules are described using three-dimension intensive grids containing the geometry, gradient, or velocity coordinate’s information. The order of magnitude of these grids is usually very large. Data in these grids are represented using floating-point, which has little or no interactions during the computation. In order to archive higher accuracy, the scoring operation will be performed repeatedly, perhaps hundreds or thousands times.
Due to the characteristics of the amber scoring such as data independency and high arithmetic intensity, which are exactly the sweet spots of computing on GPU, it can be perfectly paralleled to leverage the computing power of GPU and gain preferable speedup.
Porting Amber Scoring to GPU
Overview
To determine the critical path of amber scoring, we conduct an experiment to make statistics about the cost of each step as shown in Table 56.1. We see that the time spent in processing a ligand is negligible, because ligand in docking always refers to small molecules or segments of protein whose information grids are small and can be calculated quickly. We also observe, however, that MD simulation on receptor and complex is the most time-consuming part, which takes up to 96.25% of the total time. Either on receptor or on complex, MD simulation performs the same functionality. Therefore, in our GPU-accelerated version, we focus on how to port the MD simulation to GPU, which could accelerate the bulk of the work.
Table 56.1.
Runtime statistics for each step of amber scoring 100 cycles are performed for minimization steps and 3,000 cycles for md simulation step
| Stage | Run time (s) | Ratio of total (%) | |
|---|---|---|---|
| Receptor protocol | Gradient minimization | 1.62 | 0.33 |
| MD simulation | 226.41 | 45.49 | |
| Minimization solvation | 0.83 | 0.17 | |
| Energy calculation | 2.22 | 0.45 | |
| Ligand protocol | Gradient minimization | ≈0 | 0 |
| MD simulation | 0.31 | 0.06 | |
| Minimization solvation | ≈0 | 0 | |
| Energy calculation | ≈0 | 0 | |
| Complex protocol | Gradient minimization | 8.69 | 1.75 |
| MD simulation | 252.65 | 50.76 | |
| Minimization solvation | 2.69 | 0.54 | |
| Energy calculation | 2.22 | 0.45 | |
| Total | 497.64 | 100 | |
For simplicity and efficiency, we take advantage of the Compute Unified Device Architecture (CUDA), a new parallel programming model that leverages the computational power in NVIDIA G80-based GPUs. We find that the key issue to utilize GPU fully is the high ratio of arithmetic operations to memory operations, which can be achieved through refined utilization of memory model, data transfer pattern, parallel thread management, and branch hidden.
CUDA Programming Model Highlights
At the core of CUDA programming model are three key abstractions – a hierarchy of thread groups, shared memories, and barrier synchronization, which provide fine-gained data parallelism, thread parallelism, and task parallelism. CUDA defines GPU as coprocessors to CPU that can run a large number of light-weight threads concurrently. The programming language of CUDA is a minimal set of C language extensions based on a few low learning carve abstractions for parallel computing. Threads are manipulated by kernels representing independent tasks mapped over each sub-problem. Kernels contain the processing assignments that each thread must carry out during the runtime. More specifically, same instruction sets are applied on different partitions of the original domain by the threads in SPMD fashion.
In order to process on the GPU, data should be prepared by copying it to the graphic board memory first. Actually, the problem domain is defined in the form of a 2D grid of 3D thread blocks. The significance of a thread block primitive is that it is the smallest granularity of work unit to be assigned to a single streaming multiprocessor (SM). Each SM is composed of eight scalar processors (SP) that indeed run threads on the hardware in a time-slice manner. Every 32 threads within a thread block are grouped into warps. At any time, there is only one warp active on the SM and it will proceed to run until it has to stop and wait for something to happen such as I/O operations. The hardware scheduler on the SM selects the next warp to execute. Within a warp the executions are in order, while beyond the warp the executions are out of order. Therefore, it does not matter if there are divergent threads among different warps. However, if threads within a warp follow divergent paths, only threads on the same path can be executed simultaneously.
In addition to global memory, each thread block has its own private shared memory that is only accessible to threads within that thread block. Threads within a thread block could cooperate by sharing data among shared memory with low latency. Among thread blocks, synchronization can be achieved by finishing a kernel and starting a new one. It is important to point out that the order of thread blocks assigned to the SMs is arbitrary and non-deterministic. Therefore, sequential semantics should not be fulfilled depending on the execution orders of the thread blocks, which may lead to race condition or deadlock.
Parallel Thread Management
To carry out the MD simulation on GPU, a kernel needs to be written, which is launched from the host (CPU) and executed on the device (GPU). A kernel is the same instruction set that will be performed by multiple threads simultaneously. This parallelism is implemented through the GPU hardware called Streaming Multi-processor (SM). By default, all the threads are distributed onto the same SM, which cannot fully explore the computational power of the GPU or may cause launch failure if the threads are more than what one SM can hold. In order to utilize the SMs more efficiently, thread management must be taken into account.
We divide the threads into multiple blocks, and each block can hold the same number of threads. These blocks will be distributed among SMs. There are two kinds of IDs in CUDA named blockId and threadId, which are used to simplify the memory addressing among threads. Blocks have their own blockId during the kernel lifetime, and threads within the same block can be identified by threadId. Therefore, we can take advantage of these two kinds of IDs to issue threads computing on different partitions of the grids concurrently.
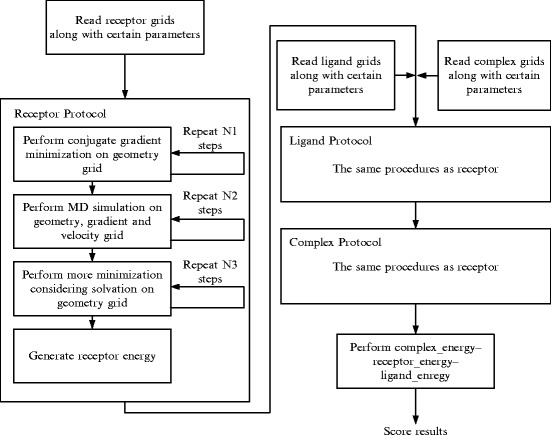
In the geometry, gradient, and velocity grids, 3D coordinates of atoms are stored sequentially and the size of the grid usually reaches as large as 7,000. Calculation works are assigned to blocks on different SMs; each thread within the blocks computes the energy of one atom, respectively, and is independent of the rest (see Fig. 56.2). We compose N threads into a block (N = 512 is the maximum number of threads per block in GeForce 9800 GT), which calculates N independent atoms in the grids. Assuming the grid size is M and M is divisible by N, there will happen to be M/N blocks.
Fig. 56.2.
Threads and blocks management about processing molecule grids on GPU: (a) blocks whose threads in the last block may calculate two atoms each (b) blocks whose threads in the last block may have nothing to do
While in most cases the grid size M is not divisible by N, we designed two schemes dealing with this situation. In the first scheme, there will be M/N blocks. Since there is M%N atoms left without threads to calculate, we will rearrange the atoms evenly to the threads in the last block. One more atom will be added to the threads in the last block until no atoms are left, which is ordered by ascending thread ID. The second scheme is to construct M/N + 1 blocks. Each thread in the blocks still calculates one atom; however, the last block may contain threads with nothing to do.
Our experiment proves that the former scheme obtained better performance. This is caused by underutilized SM resources and branch cost in the second scheme. When there is a branch divergence, all the threads must wait until they reach the same instructions again. Synchronization instructions are generated by the CUDA complier automatically, which is time consuming. Furthermore, the redundant threads have to wait for all the calculations to be done, while they take up the SM computing cycles which cannot be utilized by other working threads and which cause a waste of resources.
Memory Model and Data Transfer Pattern
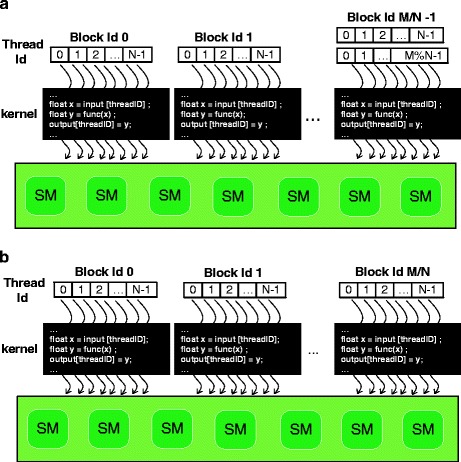
The first step to perform GPU computations is to transfer data from host (CPU) memory to device (GPU) memory since the receptor, ligand, and complex grids need to be accessible by all SMs during the calculations. There are two kinds of memory that can be used to hold these grids. One is the constant memory, which can be read and written by the host but can only be read by the device. The other is the global memory, which can be read and written by both the host and device. One important distinction between the two memories is the access latency. SMs can get access to the constant memory magnitude order faster than to the global memory. While the disadvantage of the constant memory is also obvious, it is much smaller, which is usually 64 KB compared to 512 MB global memory. Thus, a trade-off has to be made on how to store these grids.
During each MD simulation cycle, the gradient and velocity grids are read and updated. Therefore, they should be stored in global memory. While once entered in the MD simulation process, the geometry grids are never changed by the kernel. Hence, they can be stored in constant memory (see Fig. 56.3). Considering the out-bound danger due to the limited capacity of the constant memory, we observed the size of each geometry grid. The receptor and complex geometry grids usually contain no more than 2,000 atoms each, while the ligand geometry grid contains 700 atoms, which totally requires 2,000 × 3 × 4 × 2 + 700 × 3 × 4 bytes (56.4 KB) memory to store them. Since it is smaller than 64 KB, the geometry grids shall never go out-bound of the constant memory.
Fig. 56.3.
Memory model and data transfer pattern during the MD simulation cycles. Grids are transferred only once before the simulation. Atom results are first accumulated in the shared memory within the block. Then the accumulations per block are transferred into the host memory and summed up
The time to transfer molecule grids from the host to their corresponding GPU memory is likewise a critical issue, which may degrade the benefit archived from the parallel execution if not considered carefully [9]. For each MD simulation cycle, we could transfer one single atom 3D coordinates in the geometry, gradient, and velocity grids to device memory when they are required by the SMs. The other solution is to transfer the entire grids into the GPU memory before the MD simulation stage. When the simulation starts, these grids are already stored in device memory, which can be accessed by simulation cycles performed on SMs.
Based on our experiment, we noticed that there was a significant performance divergence between the two schemes. The former version turned out to be not speedup but obvious slowdown. Before data transformation, a certain time is required to get the PCIE bus and device ready. The data representing one atom only takes up 12 bytes, which is very tiny compared to the 8-GB bandwidth of the PCIE bus. Thus the time spent on real data transformation can be neglected, and most of the time is wasted in frequent bus communications.
Significant performance improvements are obtained from the second scheme since the molecule grids are transferred only once for all before the MD simulation. Therefore, the SMs donot have to halt and wait for the grids to be prepared. Generally, at least 3,000 MD simulation cycles are required for each molecular stage to maintain accuracy, which means highly intensive floating-point calculations are performed on the same molecule grids with updated values in each atom. Thus, the ratio of memory access and floating-point calculations should be pretty high, which obviously speeds up the parallel execution of the MD simulations by fully utilizing the SMs.
The MD simulations are executed parallelly on different SMs, and threads within the different blocks are responsible for the calculation of their assigned atoms of the grids. The traditional approach is to transfer all the atom results back to the host memory where the accumulation is performed. In practice, this may be inefficient since shared memory in GPU can be utilized to cut down the communication cost between the device and host. However, the limitation is that synchronization can only be applied within the block (see Fig. 56.3). Our solution is to synchronize threads within the blocks, which generates atom results separately. Then a transformation is performed to store the atom results from shared memory to host memory in a result array. The molecule result shall be achieved by adding up all the elements in the array without synchronization. The experiment has showed that this solution has a significant impact on performance improvement as the simulation size scales compared to the original approach without shared memory synchronization.
Divergence Hidden
Another important factor that dramatically impacts the benefits achieved by performing MD simulation on GPU is the branches. Original MD simulation procedure involves a bunch of nested control logics such as bonds of Van der Waals force and constrains of molecule energy. When the parallel threads computing on different atoms in the grids come to a divergence, a barrier will be generated and all the threads will wait until they reach the same instruction set again. The above situation can be time consuming and outweigh the benefits of parallel execution; thus divergence must be hidden to the minimum.
We extract the calculations out of the control logic. Each branch result of the atom calculation is stored in a register variable. Inside the nested control logic, only value assignments are performed, which means the divergence among all the threads will be much smaller; thus the same instruction sets can be reached with no extra calculation latency. Although this scheme will waste some computational power of the SMs since only a few branch results are useful in the end, it brings tremendous improvements in performance. These improvements can be attributed to that, in most cases, the computational power we required during the MD simulation is much less than the maximum capacity of the SMs. Hence, the extra calculations only consume vacant resources, which in turn speed up the executions. The feasibility and efficiency of our scheme have been demonstrated in our experiment.
Results
The performance of our acceleration result is evaluated for two configurations:
Two cores of a dual core CPU
GPU accelerated.
The base system is a 2.7-GHz dual core AMD Athlon processor. GPU results were generated using an NVIDIA GeForce 9800 GT GPU card.
We referred to the Dockv6.2 as the original code, which was somewhat optimized in amber scoring. We also used the CUDAv2.1, whose specifications support 512 threads per block, 64 KB constant memory, 16 KB shared memory, and 512 MB global memory. Since double precision floating point was not supported in our GPU card, transformation to single precision floating point was performed before the kernel was launched. With small precision losses, the amber scoring results were slightly different between CPU version and GPU version, which can be acceptable.
Table 56.2 compares the original CPU version with the GPU-accelerated version in runtime for various stages. The MD simulation performed comprises 3,000 cycles in each molecular stage, which clearly afford very high speedups due to the utilizations of multi-blocks, one time data transfer pattern, shared memory and divergence hidden. The overall speedup achieved for the entire amber scoring is over 6.5×. One interesting phenomenon we noticed is not all the minimizations are speeded up but ligand is slowed down. We find a reasonable explanation that the ligand is generally a small molecule requiring a negligible amount of floating-point calculations. When mapped on GPU, these calculations are insufficient to hide the latency of data transformation and the time consumed to initial the device.
Table 56.2.
CPU times, GPU times, and speedups with respect to 3,000 MD simulation cycles per molecule protocol
| Stage | CPU | GPU | Speedup | |
|---|---|---|---|---|
| Receptor protocol | Gradient minimization | 1.62 | 0.89 | 1.82 |
| MD simulation | 226.41 | 31.32 | 7.23 | |
| Minimization solvation | 0.83 | 0.15 | 5.53 | |
| Energy calculation | 2.22 | 1.21 | 1.83 | |
| Ligand protocol | Gradient minimization | ≈0 | 0.02 | – |
| MD simulation | 0.31 | 0.60 | – | |
| Minimization solvation | ≈0 | 0.03 | – | |
| Energy calculation | ≈0 | ≈0 | 0 | |
| Complex protocol | Gradient minimization | 8.69 | 2.88 | 3.02 |
| MD simulation | 252.65 | 34.79 | 7.26 | |
| Minimization solvation | 2.69 | 2.05 | 1.31 | |
| Energy calculation | 2.22 | 1.47 | 1.51 | |
| Total | 497.64 | 75.41 | 6.5 | |
The CPU version was performed using dual core, while GPU version with all superior scheme
Figure 56.4 depicts the total speedups of different GPU schemes with respect to the range of increasing MD simulation cycles. As mentioned in Sect. 56.3.3, the GPU version with only one block did not speedup the process of MD simulation but slowed down, which should be attributed to the poor management of threads since each block has a boundary of maximum active threads. The most significant performance improvements are achieved by transferring the molecule grids only once during the MD simulation in addition to the usage of multi-blocks. This scheme can greatly diminish the overhead produced by duplicate transferring the molecule grids from CPU to GPU, which dominates the time consumed during the MD simulation.
Fig. 56.4.
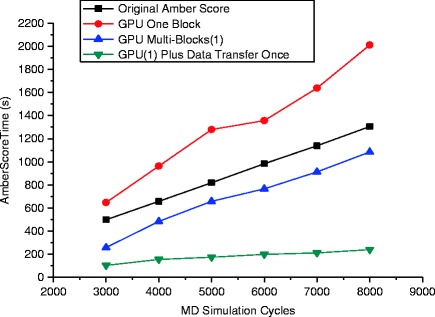
Shown is a comparison of amber scoring time between original amber and different GPU versions whose speedup varies significantly as the MD simulation cycles increase from 3,000 to 8,000
Figure 56.5 depicts the second speedup in performance obtained from the utilizations of divergence hidden and synchronization on shared memory. Since the branch calculations are extracted from the control logic and stored in temporary variables, only one single instruction will be performed which assigns corresponding values into the final result when divergences occur. While threads within a block will accumulate atom simulation values into a partial result of a molecule on shared memory, the result array transferred back to the host is very small. Performance improvements are obtained when summing up the elements in the array to form the molecule simulation result. We also notice that as the MD simulation cycles scale up, the speedup becomes more considerable in our best GPU version.
Fig. 56.5.
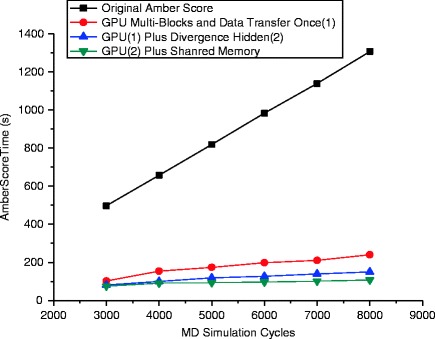
Comparison of speedups among different GPU versions based on Fig. 56.4 in addition to divergence hidden and shared memory
Related Work
Studies on utilizing GPU to accelerate molecule docking and scoring problems are rare, the only work that we find more related to our concern is in the paper of Bharat Sukhwani [10]. The author described a GPU-accelerated production docking code, PIPER [11], which achieves an end-to-end speedup of at least 17.7× with respect to a single core. Our contribution is different from the former study in two aspects. First, we focus our energy on flexible docking such as amber scoring, while the previous study mainly focuses on rigid docking using FFT. Thus our work is more complex and competitive in the real world. Second, we noticed that the logic branches in the parallel threads on GPU degraded the entire performance sharply. We also described the divergence hidden scheme and represented the comparison on speedup with and without our scheme.
Another attractive work that needs to be mentioned is that by Michael Showerman and Jeremy Enos [12] in which they developed and deployed a heterogeneous multi-accelerator cluster at NCSA. They also migrated some existing legacy codes to this system and measured the performance speedup, such as the famous molecular dynamics code called NAMD [13, 14]. However, the overall speedup they achieved was limited to 5.5×.
Conclusions and Future Works
In this paper, we present a GPU-accelerated amber score in Dock6.2, which achieves an end-to-end speedup of at least 6.5× with respect to 3,000 cycles during MD simulation compared to that of a dual core CPU. We find that thread management utilizing multiple blocks and single transferring of the molecule grids dominates the performance improvements on GPU. Furthermore, dealing with the latency attributed to thread synchronization, divergence hidden, and shared memory can lead to elegant solutions, which will additionally double the speedup of the MD simulation. Unfortunately, the speedup of Amber scoring cannot go much higher due to Amdahl’s law. The limitations are as follows:
With the kernel running faster because of GPU acceleration, the rest of the Amber scoring takes a higher percentage of the run time.
Partitioning the work among SMs will eventually decrease the individual job size to a point where the overhead of initializing an SP dominates the application execution time.
The work we presented in this paper only shows a kick-off stage of our exploration in GPGPU computation. We will proceed to use CUDA acceleration various applications with different data structures and memory access patterns, and hope to be able to work out general strategies about how to use GPU more efficiently. With greater divergences in architectural designs of CPU and GPU, our goal is to find a parallel programming model to leverage the computation power of CPU and GPU simultaneously.
Acknowledgment
Many thanks to Ting Chen for thoughtful discussions and comments about our implementation and paper work. This work was supported by the National High Technology Research and Development Program of China under the grant No. 2007AA01A127.
Contributor Information
Hamid R. Arabnia, Phone: 706542-3480, FAX: 706542-2966, Email: hra@cs.uga.edu
Hailong Yang, Email: hailong.yang@jsi.buaa.edu.cn.
References
- 1.Dock6: http://dock.compbio.ucsf.edu/DOCK_6/.
- 2.Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general Amber force field. Journal of Computational Chemistry. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 3.NVIDIA Corporation Technical Staff, Compute Unified Device Architecture – Programming Guide, NVIDIA Corporation, 2008.
- 4.Sukhwani, B. and Herbordt, M. Acceleration of a production rigid molecule docking code. In Proceedings of the IEEE Conference on Field Programmable Logic and Applications Pages: 341–346, 2008.
- 5.Kuntz I, Blaney J, Oatley S, Langridge R, Ferrin T. A geometric approach to macromolecule–ligand interactions. Journal of Molecular Biology. 1982;161:269–288. doi: 10.1016/0022-2836(82)90153-X. [DOI] [PubMed] [Google Scholar]
- 6.Honglin Lia, Chunlian Lia, Chunshan Guib, Xiaomin Luob and Hualiang Jiangb. GAsDock: a new approach for rapid flexible docking based on an improved multi-population genetic algorithm. Bioorganic & Medicinal Chemistry Letters 14(18), Pages: 4671–4676, 2004. [DOI] [PubMed]
- 7.Servat H., Gonzalez C., Aguilar X., Cabrera D., Jimenez D. Drug design on the cell broadband engine. Parallel Architecture and Compilation Techniques, Pages: 2007;16:425–425. [Google Scholar]
- 8.Krüger J, Westermann R. Linear algebra operators for GPU implementation of numerical algorithms. ACM Transactions on Graphics. 2003;22(3):908–916. doi: 10.1145/882262.882363. [DOI] [Google Scholar]
- 9.Govindaraju, N.K., Gray, J., Kumar, R. and Manocha, D. GPUTeraSort: High-performance graphics coprocessor sorting for large database management. Proceedings of the 2006 ACM SIGMOD International Conference on Management of Data.
- 10.Bharat Sukhwani and Martin C. Herbordt. GPU acceleration of a production molecular docking code. In Proceedings of 2nd Workshop on General Purpose Processing on GPUs, Pages: 19–27, 2009.
- 11.PIPER: http://structure.bu.edu/index.html
- 12.Michael Showerman, Wen-Mei Hwu, Jeremy Enos, Avneesh Pant, Volodymyr Kindratenko, Craig Steffen and Robert Pennington. QP: A Heterogeneous Multi-Accelerator Cluster. In 10th LCI International Conference on High-Performance Clustered Computing, 2009.
- 13.NAMD: http://www.ks.uiuc.edu/Research/namd/.
- 14.James C. Phillips, Gengbin Zheng, Sameer Kumar and Laxmikant V. Kalé. NAMD: Biomolecular Simulation on Thousands of Processors, Conference on High Performance Networking and Computing, Pages: 1–18, 2002.


