Abstract
The nucleolus is the largest nuclear organelle and is the primary site of ribosome subunit biogenesis in eukaryotic cells. It is assembled around arrays of ribosomal DNA genes, forming specific chromosomal features known as nucleolar organizing regions (NORs) which are the sites of ribosomal DNA transcription. While the nucleolus main activity involve different steps of ribosome biogenesis, the presence of proteins with no obvious relationship with ribosome subunit production suggests additional functions for the nucleolus, such as regulation of mitosis, cell cycle progression, stress response and biogenesis of multiple ribonucleoprotein complexes. The many novel factors and separate classes of proteins identified within the nucleolus support this view that the nucleolus may perform additional functions beyond its known role in ribosome subunit biogenesis. Here we review our knowledge of the nucleolar functions and will provide a detailed picture of how the nucleolus is involved in many cellular pathways.
Keywords: Nucleolus, Ribosome biogenesis, rRNA transcription
Introduction
Nucleoli are present in almost every eukaryotic cell type and represent the most prominent compartment of the cell nucleus. The primary function of the nucleolus consists in ribosomal RNA (rRNA) transcription, rRNA processing and ribosome subunit assembly (Hernandez-Verdun et al. 2010; Pederson 2011; Raska et al. 2006). Nucleoli assemble at the end of mitosis around the tandemly repeated clusters of rDNA genes forming a subnuclear compartment that locally recruits the specific transcription and processing machineries that are responsible for generating ribosome subunits (Hernandez-Verdun 2011; Raska et al. 2006). The process of assembling a ribosome subunit requires the initial transcription of the ribosomal DNA (rDNA) genes by the RNA polymerase I. Because these rDNA genes are arranged in arrays of tandem repeats, it results in local concentration of proteins involved in different aspect of transcription, processing and assembly of rRNA into ribosomes. In higher eukaryotes, three sub-nucleolar compartments can be distinguished by their distinct morphology using electron and light microscopy: The fibrillar centres (FC) are surrounded by the dense fibrillar component (DFC) and the granular component (GC), in which the FC and DFC are embedded. The composition of these sub-compartments is tightly linked to sequential steps in ribosome biogenesis [reviewed in (Olson and Dundr 2005)]. The FC contains unengaged RNA polymerase I transcription factors, whereas the DFC contains mostly pre-RNA processing factors, indicating specialization of these compartments. Transcription occurs at the boundary of the FC and DFC (Raska et al. 2006), and the transcribed rRNA is then moving to the GC compartment for further maturation and assembly into ribosomes (Figs. 1 and 2).
Fig. 1.
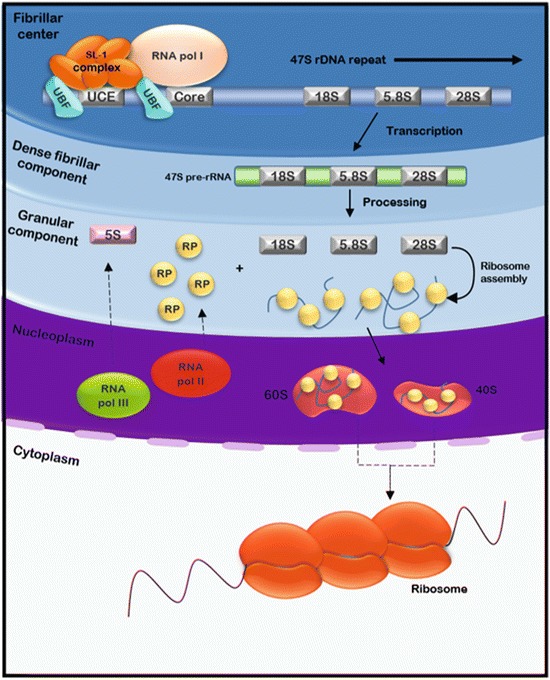
Ribosome biogenesis. Transcription of ribosomal DNA by RNA polymerase I occurs at the fibrillary centres or at the boundary with the dense fibrillary component. The 47S pre-ribosomal RNA transcripts are then further processed by specific cleavages and post-transcriptional modifications by small nucleolar ribonucleoproteins (snoRNPs) in the dense fibrillary component. Assembly of the rRNA with the ribosomal proteins then occurs in the granular component of the nucleolus prior to export of the 40 and 60S subunits to the cytoplasm for final assembly into the functional ribosome
Fig. 2.
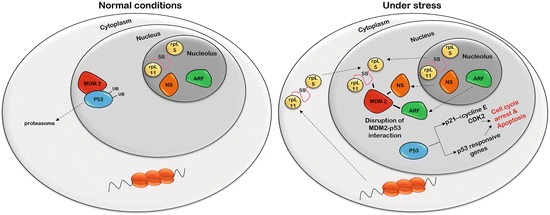
Nucleolus under stress. The nucleolus plays a central role during the cellular response to stress. Under normal conditions, p53 is kept at very low level by proteasomal mediated degradation through ubiquitination by MDM2. Following activation of stress responses, oncogene activation or DNA damage, the p14ARF tumor suppressor normally located in the nucleolus associates with HDM2 and sequesters it within the nucleolus. This segregation prevents the ubiquitination of p53 mediated by HDM2 resulting in increased levels of cellular p53 resulting in cell cycle arrest and apoptosis
In many cell types, only a subset of rDNA genes are transcriptionally active, even though inactive rDNA are still assembled into nucleoli. The initial 47S ribosomal RNA (rRNA) precursor transcript transcribed by RNA pol I is subsequently cleaved to form the mature 28S, 18S and 5.8S rRNAs which is then post-transcriptionally modified through interaction with small nucleolar ribonucleoproteins (snoRNPs) and additional protein processing factors (Henras et al. 2015). Finally, the processed and modified rRNAs are assembled with the many ribosomal proteins, prior to interaction with the export machinery and transport to the cytoplasm.
While the nucleolar function in ribosomal biogenesis is well characterized, many additional functions of the nucleolus besides ribosomal biogenesis have been uncovered, suggesting many important cellular functions for the nucleolus (Boisvert et al. 2007; Pederson and Powell 2015). This was made particularly evident following several reports of proteomic analyses to characterize the nucleolar proteome in human, mouse, as well as other organisms (Andersen et al. 2002, 2005; Kar et al. 2011; Pendle et al. 2005; Scherl et al. 2002). Nucleolar proteins identified in these studies have shown that over 70 % are not involved in the production of ribosome subunits, consistent with the nucleolus performing additional cellular functions (Boisvert et al. 2007; Lam and Trinkle-Mulcahy 2015; Pederson and Powell 2015). A further dimension to these studies has been added by recent studies that have characterized the dynamic protein composition of the nucleolus following treatment with actinomycin D (Andersen et al. 2005), etoposide (Boisvert et al. 2010) or through the cell cycle (Ly et al. 2014).
Additionally, the nucleolus has been linked to multiple forms of diseases involving a wide range of mechanisms including cancer (Tsai and Pederson 2014), viral infections (Hiscox 2007) and neurodegenerative diseases (Parlato and Kreiner 2013), affecting either ribosome biogenesis, nucleolar structure or other functions.
This chapter provides an overview of the characteristics of nucleolar organisation, discusses non-ribosomal functions of the nucleolus, and will present some of the latest findings regarding their regulation and dynamic behaviour, as well as its implications in cancers and diseases.
The Nucleolar Organisation
Nucleolar Organiser Regions (NORs)
The rDNA genes are arranged in arrays of head-to-tail tandem repeats, termed nucleolar organizer regions (NORs). In the human genome, approximately 400 copies of 43-kb repeat units are distributed along all acrocentric chromosomes (chromosomes 13, 14, 15, 21 and 22) to form NORs (Henderson et al. 1972), which are assumed to have resulted from interchromosomal recombination (Gonzalez and Sylvester 1997; Worton et al. 1988). While the rDNA repeats are mostly arranged into canonical head-to-tail repeats, around a third of them are arranged into palindromic or non-canonical repeats (Caburet et al. 2005). During metaphase, these clusters contain so called r-chromatin (for ribosomal genes complexed with proteins involved in rDNA transcription) and are constituted of 60–80 nm fibers showing a twisted loop organization as visualized by electron tomography (Heliot et al. 1997). These RNA Polymerase I associated transcription factors remain associated with NORs throughout mitosis as well (Roussel et al. 1996).
Each of the 43 kb repeats includes a 13–14 kb segment coding for the rRNA sequence and are separated from the next transcription unit by 30 kb intergenic spacers (IGS). These spacers contains regulatory elements such as the gene promoter, repetitive enhancer elements and terminator sequences. Polar replication fork barriers (RFBs) within the transcription termination element between the individual repeats ensure the stalling of the bidirectionally running replication fork opposite to the direction of transcription (Brewer and Fangman 1988; Gerber et al. 1997; Hernandez et al. 1993; Little et al. 1993; Wiesendanger et al. 1994). Additional elements located within the IGS including small, non-coding RNAs such promoter RNAs (pRNAs) have been recently identified. These 150–250 nucleotides long RNAs are transcribed from a promoter within the IGS and involved in epigenetic mechanisms acting on the rDNA locus (Mayer et al. 2006, 2008; Santoro et al. 2010).
In many cell types, only a subset of rDNA genes are transcriptionally active, even though inactive rDNA are still assembled into nucleoli. (Akhmanova et al. 2000; Strohner et al. 2001; Sullivan et al. 2001). The difference results from different chromatin states of rDNA repeats that correspond with the state of the nucleosomes within the rDNA (Dammann et al. 1993; Langst et al. 1998; Li et al. 2006b; Sogo et al. 1984). Inactive rDNA has been shown to be inaccessible for psoralen crosslinking and exhibits regularly spaced nucleosomes, while active regions of rDNA is accessible for crosslinking and lack nucleosomes (Conconi et al. 1989; Dammann et al. 1993). These different chromatin states are usually stable throughout the cell cycle (Conconi et al. 1989). The rate of production of rDNA can therefore be regulated either by increasing the transcription rate of active genes and/or by activation of silent genes. In general, rapid changes in rRNA expression, i.e., in response to nutrient status or growth factor signalling, will result in a change in the transcription rate of rDNA genes that are already active (Grummt and Pikaard 2003; Russell and Zomerdijk 2005; Stefanovsky et al. 2006), whereas slower changes such as during development or differentiation results from a change in the number of genes that are actively transcribed (Haaf et al. 1991).
rDNA Transcription
The RNA Polymerase I does not require a TATA box sequence in the promoter, but instead relies on regulatory sequences that are divided in two functional elements: a core element (CORE) next to the transcription start site located between −45 to +20, and an upstream control element (UCE) located between −200 to −107 (Haltiner et al. 1986; Learned et al. 1986). Transcription of rRNA genes requires the formation of a preinitiation complex (PIC) that is composed of the RNA Polymerase I, the upstream binding factor (UBF) and the promoter selectivity factor (SL1) at the rDNA promoter [reviewed in (Grummt and Pikaard 2003; Moss et al. 2007; Russell and Zomerdijk 2005)]. Upon dimerization, the upstream binding factor UBF binds the UCE and the CORE element to recruit the SL1 protein complex composed of the TATA-binding protein (TBP) and TBP-associated factors (TAFs), TAFI 110/95; TAFI 68; TAFI 48 (Comai et al. 1992; Heix et al. 1997; Zomerdijk et al. 1994). The UBF dimer allows the UCE and CORE elements to come into contact by introducing loops in the upstream region into a structure called the enhanceosome to allow the binding of SL1 and formation of a stable PIC (Bazett-Jones et al. 1994; Bell et al. 1988). By interacting with UBF and the RNA Pol I component TIF-1A, TAFs recruit RNA Pol I to the rDNA promoter (Miller et al. 2001; Moorefield et al. 2000). UBF not only binds to the promoter region, but also throughout the whole rDNA repeat (Mais et al. 2005; O’Sullivan et al. 2002) where it acts as a transcriptional activator of RNA Pol I, modulator of elongation in response to growth factor signalling and as an anti-repressor by replacing histone H1 (Kermekchiev et al. 1997; Kuhn and Grummt 1992). Thus, regulation of UBF is central to the regulation of rDNA transcription by RNA Polymerase I. Important insight into the role of UBF in ribosomal chromatin remodelling and nucleolar formation has come from the studies of pseudoNORs [reviewed in (Prieto and McStay 2008)]. PseudoNORs are artificial, high-affinity UBF-binding arrays that are transcriptionally silent due to the lack of a promoter, but behave like active NORs. For example, they exhibit consistent under condensation throughout the cell cycle and are highly enriched in the RNA Pol I machinery through UBF (Mais et al. 2005; Prieto and McStay 2007) that stays associated with rDNA after nucleoli disassembly in mitosis (Roussel et al. 1993). Therefore, UBF is believed to establish a chromatin structure that allows rapid re-initiation of rRNA transcription after mitosis and thereby promotes nucleolar formation. This is also supported by studies that show rDNA gene silencing upon UBF deletion, suggesting a regulatory role for UBF in determining the number of active rDNA genes (Sanij and Hannan 2009; Sanij et al. 2008).
As RNA Polymerase I starts the elongation process of the rRNA, UBF and SL1 remain associated with the promoter region to allow recruitment of another RNA Polymerase I so that each rDNA gene can be transcribed multiple times simultaneously, contrary to RNA Polymerase II genes. This gives rise to the Christmas tree like structure as visualized by electron microscopy (Miller and Beatty 1969). Termination occurs through the binding of TTF-I at the 3′ end of the transcribed region, bending the termination site and, with the help of the transcript-releasing factor PTRF, will induce RNA Polymerase I to dissociate from the DNA and the new transcript.
Processing of rRNA and Ribosome Assembly
The production of human 40S and 60S ribosomal subunits prior to export into the cytoplasm is a complex mechanism that requires a large proportion of the cellular energy for production (Warner 1999). Indeed, the size and organisation of the nucleolus reflects the scale of this process (Sirri et al. 2008). The regulation of ribosome biogenesis is crucial for cellular growth and proliferation (Tschochner and Hurt 2003) and it is upregulated in the majority of cancers to accommodate the increase rate of cellular growth and metabolism (Ruggero and Pandolfi 2003). The two ribosomal subunits includes 79 major ribosomal proteins (RPs) as well as four mature ribosomal RNAs (Yusupova and Yusupov 2014). The smaller ribosomal subunit (40S) includes the 18S rRNA, whereas the large subunit (60S) includes the 28S, 5.8S as well as the RNA Polymerase III transcribed 5S rRNA (Fedoriw et al. 2012).
The rRNA precursor is transcribed by RNA Pol I at the FC/DFC border as one long 47S transcript (~13 kb in humans), which includes the sequences for the 18S, 5.8S and 28S rRNA in that order. The additional sequences are called external transcribed spacers called ETS at both ends of the rRNA (5′-ETS and 3′-ETS), as well as internal transcribed spacers (ITS1 and ITS2) within the rRNA precursor. Sequential cleavages at specific sites termed A′, A0, 1, 2 and 3 will generate the 18S rRNA and requires distinct small nucleolar RNAs (snoRNAs) (Henras et al. 2015). Cleavage at the T1 site by the U8 snoRNA will remove the 3′-ETS. Site 3 cleavage near the end of ITS1 between the 18S and 5.8S rRNAs will generate a 32S intermediate which will be further processed (Preti et al. 2013). Cleavage at the 4′ site will generate the 28S rRNA and will results in a 5.8S rRNA with a longer 3′-end that will be further processed. The maturing rRNAs will be assembled within a 90S pre-ribosome complex which accumulates in the DFC (Henras et al. 2015; Tschochner and Hurt 2003). The pre-ribosome contains the 47S rRNA, 5S rRNA, ribosomal proteins and ~150 non-ribosomal proteins, including factors involved in processing and maturation, for example endo- and exonucleases, pseudouridine synthases, methyltransferases, RNA chaperones, GTPases and AAA-ATPases helicases (Tschochner and Hurt 2003). The pre-ribosome is subsequently separated into pre-40S and pre-60S subunits in the GC. These subunits are exported to the cytoplasm, where they undergo further processing to form the mature small 40S and large 60S ribosome subunits. The 40S subunit contains 18S rRNA and ~33 ribosomal proteins, whereas the 60S subunit is composed of the 28S and 5.8S rRNAs, the RNA Pol III-transcribed 5S rRNA and ~49 ribosomal proteins. Interestingly, around two thirds of the ribosomal protein genes are duplicated in humans. Although it has been assumed that these copies are largely redundant, recent work has suggested that these copies exhibit functional specificity (Komili et al. 2007).
Other Functions for the Nucleolus
RNA Complexity in the Nucleolus
While the nucleolus is known to have a major role in coordinating the processing and maturation of rRNAs, there is now extensive literature demonstrating that the nucleolus is also involved in the processing and maturation of several different families of RNA. For example, the nucleolus has been suggested to be a site of covalent RNA modifications and protein assembly of multiple ribonucleoprotein complexes, such as the spliceosomal small nuclear RNPs, telomerase and several other small RNAs transcribed by RNA polymerase III, such as 5S rRNA, some tRNAs, RNAse P RNA, the signal recognition particle (SRP) RNA and now also miRNA (Gerbi et al. 2003; Lam and Trinkle-Mulcahy 2015).
The signal recognition particle (SRP) is a ribonucleoprotein complex responsible for the recognition of the N-terminal signal peptide sequence on nascent proteins and for proper targeting of proteins onto a receptor on the cytoplasmic face of the endoplasmic reticulum (Walter and Johnson 1994). This complex is formed by 6 proteins and a ~300 nucleotide RNA (Walter and Blobel 1982). Recent studies have shown that both the RNA and proteins from the SRP can be found to transit through the nucleolus of mammalian cells prior to SRP export to the cytoplasm (Jacobson and Pederson 1998). These results suggest that the nucleolus is the site of assembly and processing of the SRP complex prior to their cytoplasmic export, and that the RNA could be modified/matured within the nucleolus.
In addition to the above roles for the nucleolus in RNA modification and maturation, there are several other observations linking the nucleolus to RNA processing. The RNase P RNA, a component of the pre-tRNA processing enzyme RNase P, has been found in both the nucleolus and nucleoplasm (Jacobson et al. 1997), suggesting that some pre-tRNA processing happens within the nucleolus. An alternative possibility is that the nucleolus plays a role in the assembly of RNase P ribonucleoprotein complex. The nucleolus contains all the trans-acting factors that are responsible for the synthesis of the eight 2′-O-methylated nucleotides and three pseudouridine residues carried by the mammalian U6 spliceosomal small nuclear RNA (Ganot et al. 1999). These findings demonstrate a trafficking pathway in which the U6 spliceosomal RNA cycles through the nucleolus to undergo nucleolar RNA-directed processing. Interestingly, like 5S rRNA, these RNA (tRNA, RNase P RNA and U6 spliceosomal small nuclear RNA) are all transcribed outside the nucleolus by RNA polymerase III and subsequently transit through the nucleolus. This suggest a possible common maturation process shared by RNA pol III transcripts that could occur within the nucleolus. ADAR1 and ADAR2 are editing enzymes that deaminate adenosine to inosine in long double stranded RNA duplexes and specific pre-mRNA transcripts. Live microscopy experiments demonstrate that ADAR1 and ADAR2 are in constant flux in and out of the nucleolus (Desterro et al. 2003). Furthermore, it was shown that ADAR2- but not ADAR1-mediated RNA editing occurs within the nucleolus, indicating a role for the nucleolus in the regulation of RNA editing (Vitali et al. 2005).
More recently, evidence has started to emerge demonstrating a role for the nucleolus in the regulation of small interfering RNA (siRNA). The finding that many proteins involved in siRNA processing, including RDR2, DCL3, AGO4, and NRPD1b (the largest subunit of RNA Pol IVb) were identified with siRNAs within the nucleolus in plant cells suggest that processing of endogenous nuclear siRNAs, and possibly RISC storage or sequestration, occurs within the nucleolus (Li et al. 2006a; Pontes et al. 2006). It was also reported in mammalian cells that a microRNA (miR-206) had been found to co-localize with the 28S rRNA in the granular component of the nucleolus, implying that this miRNA associates early with the ribosome subunits (Politz et al. 2006). Several other miRNAs have also been identified within the nucleolus, further supporting these observations (Bai et al. 2014a, b; Li et al. 2013). It will be interesting to determine whether multiple forms of miRNAs arise within the nucleolus and whether they either function in nucleolar processes or leave the nucleolus to regulate downstream cellular events, such as protein translation.
Mitosis and Cell Cycle Regulation
The nucleolus is a dynamic structure which is disassembled/re-assembly at each cell division. The first step in prophase is initiate by the phosphorylation of components of the rDNA-transcription machinery by cyclin B1 and CDK1 (Heix et al. 1998). The phosphorylation of these components triggers a repression of rRNA transcription but the machinery such as UBF remains localized to the NORs (Dundr et al. 2000). In contrast, during the disassembly of nucleolus, the rRNA-processing machinery does not remain associated with nucleolar regions and move to the cytoplasm or become attached to the surface of condensed chromosomes at the perichromosomal region (PR) (Gautier et al. 1992). During anaphase, the processing proteins remains attached to the PR while proteins within the cytoplasm become packaged into nucleolar-derived foci (NDF). Cyclin B1/CDK1 levels decrease during late anaphase and early telophase, which results in the reactivation of rRNA transcription by loss of hypherphosphorylation of the transcription machinery (Sirri et al. 2000). During the G1 phase, the rRNA-processing proteins are release in a specific order starting with proteins such as fibrillarin which plays a role early in the rRNA processing followed by proteins involved in late stage of processing (Leung et al. 2004). The dynamic of nucleoli during cell cycle involves several stages which are tightly regulated and still not fully understood. Interestingly, the rDNA-transcription and rRNA-processing proteins might be regulated independently during the cell cycle.
During the interphase, the nucleoli remains dynamic. The nucleolar proteome has been shown to contain approximately 4000 proteins which can change localization (Boisvert et al. 2010). This pool of proteins is not all involved in ribosome biogenesis and several proteins have been shown to interact with the nucleolus at different stages of the cell cycle, further underlining a potential role in cell cycle regulation. In fact, the nucleolus and cell cycle regulation are influenced mutually. The most striking situation where the cell cycle can alter the ribosome biogenesis is in response to DNA damage (Jordan and Carmo-Fonseca 1998). Two proteins involved in DNA damage response, DNA-PK and PARP1, have been identified to be responsible for this inhibition (Calkins et al. 2013). Additionally, the RNA Polymerase I can be inhibited by DNA lesions pathway dependent of ATM, NBS1 and MDC1 (Kruhlak et al. 2007). The nucleolus can also regulates key cell cycle checkpoints as well. One of the mechanisms is by modulation of post-translational modifications such as sumoylation and phosphorylation of proteins involved in the cell cycle. For example, SENP5, a SUMO-specific protease found within the nucleolus is involved in sumoylation of proteins that affect progression through cell division (Di Bacco et al. 2006). Another example is the sequestration in nucleoli of the telomerase reverse transcriptase, the RNP enzyme that adds telomeric sequences (Wong et al. 2002). The telomerase is retained in the nucleolus until the telomeres are replicated at stages of S phase.
Stress Sensor
The ribosome biogenesis is the mechanism that requires the largest amount of cell’s energy, consuming ~80 % of total cell’s energy (Schmidt 1999). Considering the large amount of effort needed for the biogenesis of ribosomes, it is surprising that one of the strategies used to preserve energy homeostasis is decreasing ribosome biosynthesis under stress conditions. In response to stress, such as nutrient deprivation, oxidative stress or drastic change in temperature, several steps in ribosome biogenesis is altered. These stresses can cause a change in the localization of nucleolar proteins, downregulation of RNA polymerase I activity, pre-rRNA processing and transport, or changes in chromatin structure (Grummt 2013). Many drug have also been shown to alter the integrity and the activities of the nucleolus. For example, UV irradiation leads to segregation of nucleolar components and ends with the complete disintegration of the nucleolus (Govoni et al. 1994).
The alteration of any of the steps in ribosome biogenesis, such as transcription, processing and assembly of the 40S and 60S subunits, results in the activation of nucleolar stress pathways leading to senescence or apoptosis. The activation of this pathway culminates in the stabilization of the p53 protein (Rubbi and Milner 2003) by disruption of its interaction with MDM2, which can no longer add ubiquitin on p53 and target it for proteasomal degradation. MDM2 can be targeted by three proteins in the nucleolus, alternative reading frame protein p14ARF (ARF), nucleostemin (NS) and ribosomal proteins (RPs), in response to nucleolar stresses. ARF is a tumor suppressor protein usually weakly expressed (Brady et al. 2004). Under different cellular stress conditions, an increase in the expression of ARF results in binding to MDM2 and disruption of the interaction between MDM2 and p53 (Weber et al. 1999). Nucleostemin is a nucleolar protein and its function in this structure is not well understood. However, following stress in the nucleolus, the protein delocalizes to the nucleoplasm and bind MDM2 to prevent the association with p53 (Tsai 2011). Finally, several ribosomal proteins have been found to be involved in p53 stabilization such as rpL11, rpL23, rpL5, rpL7 and rpL26. However recent study demonstrate that only rpL5 and rpL11 are essential for this function (Fumagalli et al. 2012). In addition, the 5s rRNA is also involved in the formation of the rpL5-rpL11-MDM2 complex (Donati et al. 2013). The change of localization of ribosomal proteins has long been considerate passive and a result of nucleolar disruption. However, the protein rpL11 can move to the nucleoplasm without loss of nucleolar integrity (Fumagalli et al. 2009), indicating that the relocalization of ribosomal proteins may be a regulated mechanism. For example, the localization of rpL11 is regulated by NEDDylation, a ubiquitin-like molecule (Sundqvist et al. 2009). Another method to retain the ribosomal proteins within the nucleolus is through their interaction with PICT1 who is degraded in response to nucleolar stress (Maehama et al. 2014; Sasaki et al. 2011). It is clear that cells react in response to environmental stress by decreasing ribosome biogenesis to decrease energy consumption and preserve homeostasis but the nucleolus is also directly involved in the regulation of cellular response to different type of stresses.
The Nucleolus and Diseases
Cancer and Genomic Instability
Several years before the discovery of the role of the nucleolus in the synthesis of ribosomes, cytological analysis had already made a connection between the size of nucleoli and cancer (Pinease et al. 1896). It was observed that tumor cells had an increase in the number and size of nucleoli. Thereafter, numerous studies confirms these observations and concluded that these abnormalities could be used as a marker for the aggressiveness of malignancies (Derenzini et al. 2009) and was directly related to cell grow rate (Derenzini et al. 1998, 2000). In addition, the biogenesis of ribosome is under control of key cellular growth and proliferation signaling pathways which are known to be frequently mutated in several types of cancer such MYC, RAS, phosphatidylinositol-3-kinase (PI3K), tumor suppressors including TP53 (p53), retinoblastoma protein (Rb) and PTEN (Moss 2004). Irregular shape and deregulation of ribosome synthesis rate has long been considered a consequence of cancer. However, new data suggest that dysfunction of rRNA synthesis is not only a consequence cellular transformation but is also required for the survival of tumor cells and can even initiate the tumor transformation. This is support by two evidences: first, perturbation in ribosome biogenesis activated stress pathways and second, the specific inhibition of RNA polymerase I transcription leads to tumor cell death.
The first evidence is that the cell were demonstrated to monitors closely ribosome synthesis and disruption of this process leads to the activation of a ribosomal surveillance pathway (Zhang et al. 2003). This stress pathway leads to the accumulation of p53 through a non-genotoxic activation, as described in the section stress sensor. The second evidence is based on the observation that deregulation of ribosome synthesis is essential for the survival of cancer and small molecule inhibitors of RNA Polymerase I transcription can, in tumors cells, selectively activate the nucleolar stress pathway (Bywater et al. 2012). Many drugs already used in cancer therapy affect, at least in part, ribosome biogenesis or rRNA processing such as doxorubicin, flavopiridol and roscovitine (Burger et al. 2010). However, these treatments induce a general nucleolar disruption and affect many other pathways resulting in several side effects. By contrast, a new generation of small molecules is being developed that selectively inhibit RNA Polymerase I transcription. The CX-3543 (first generation) and CX-5461 (second generation) bind GC-rich sequence, found in large proportion in rRNA and prevents RNA Polymerase I transcription (Balasubramanian et al. 2011). This treatment specifically induces p53 non-genotoxic stress pathway activation by release of ribosomal proteins from the nucleolus and only affect cancer cells and not in normal cells (Bywater et al. 2012). These targeted transcription therapies are less genotoxic for normal cells reducing risk of secondary cancer associated with classical chemotherapeutic agent (Godley and Larson 2008). Recent studies also show another possible pathway by which RNA Polymerase I transcription inhibitors are also effective independent of p53 activation (Peltonen et al. 2014). While the mechanisms involved remain unknown, these data underline the potential for selective RNA Polymerase I inhibition for cancer treatment whether p53 is present or mutated.
Viral Infections
Viruses are obligate intracellular parasites and used host cell for genome replication, protein expression and assembly of new virus particles. Several types of virus involved the nucleolus for effective infection. In fact, the RNA virus, retroviruses and DNA viruses interact with/or alter the nucleolus when they infect cells (Hiscox 2002). The infection results in change of morphology and in the proteome of the nucleolus (Dove et al. 2006). The association between viral proteins and the nucleolus results from three type of direct interaction : with rDNA, with nucleolar RNA (consisting mainly of rRNA) or with nucleolar protein components (Carmo-Fonseca et al. 2000). The viruses affect the nucleosome homeostasis at many level. First, the viral components can co-localize with the nucleolus. Second, the virus can use nucleolar proteins to allow his own proliferation and finally, viral infection can result in changes in the localization of nucleolar proteins.
Following infection with certain different viruses, such as coronavirus and arterivirus, some normally nuclear proteins are found within the nucleolus (Hiscox et al. 2001; Rowland et al. 1999). Changes in the protein distribution can be either through specific nucleolar-trafficking signal present in the viral proteins (Rowland and Yoo 2003) or viral proteins trafficking through the nucleolus can associate with cellular proteins and recruit them to a different localization such as the hepatitis delta antigen which requires nucleolin association for its nucleolar localization (Lee et al. 1998). The nucleolar localization of viral proteins is also important for an effective infection. For example the disruption of nucleolar localization of Semliki Forest virus non-structural protein nsP2 results in a reduction in neurovirulence (Fazakerley et al. 2002).
The three nucleolar proteins that have been most studied during viral infection are B23, fibrillarin and Nucleolin. The B23 protein acts in several functions associated with the nucleolus, such as ribosome assembly, nucleocytoplasmic shuttling, possibly regulating transcription of rDNA and recent study have indicated is implication in the p53 regulation (Boulon et al. 2010; Hiscox 2002). During infection with HIV, B23 facilitates the nuclear import of Rev proteins promoting virus mRNA trafficking (Szebeni et al. 1997). B23 protein can also stimulates the replication of adenovirus as well (Okuwaki et al. 2001). Fibrillarin is involved in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly (Tollervey et al. 1993). During adenovirus and coronavirus infection, fibrillarin redistribution leads to decrease in RNA Polymerase I transcription (Puvion-Dutilleul and Christensen 1993). Nucleolin has been involved in the regulation of gene expression, chromatin remodeling, DNA recombination and replication, RNA synthesis, rRNA processing, mRNA stabilization, cytokinesis and apoptosis (Mongelard and Bouvet 2007). During infection with HIV, nucleolin promotes DNA replication process and stimulates IRES-mediated translation of the poliovirus genome (Izumi et al. 2001). Nucleolin has also a membrane fraction which is involved in viral infection by facilitating the virus attachment (Nisole et al. 2002).
Finally, viral infection can results in relocalization of nucleolar protein to other cellular compartments. Nucleolin, B23 and fibrillarin change their localization after cell are infected with adenovirus or HIV (Matthews 2001). The mechanisms that are responsible for this delocalization are unknown, but the displacement of nucleolar proteins changes the nucleolar, nuclear or cytoplasmic pool of these proteins. The interaction between the nucleolus and viral components is thus an interesting potential therapeutic target. These associations can be used for the development of new therapies against viral infection. For example, the HIV Rev protein localizes in the nucleolus and is involved in viral mRNA trafficking (Dundr et al. 1995). A nucleolar localizing Rev response element (RRE) decoy in infected cells results in a significant inhibition of the replication of HIV-1 in cell culture (Michienzi et al. 2006).
Neurodegenerative Disorders
While the increased size of the nucleolus is associated with cell proliferation and a potential marker for cancer, the reduction in size of nucleoli is correlated with completely different disorders. It was reported patients affected by Alzheimer’s disease and Parkinson’s disease show a reduction in nucleolar size in different part of the brain (Mann et al. 1988; Mann and Yates 1982). In fact, the dysregulation of different nucleolar aspects is associated with several human neuropathological conditions (Kinderman and LaVelle 1976).
In Alzheimer’s disease, not only the size of the nucleolus is affected but the organization of NOR is also altered (Donmez-Altuntas et al. 2005). The aberrant structure is associated with a diminution of mature rRNA products and is found as an early event in patients affected (Ding et al. 2005). This is further confirmed by recent studies which demonstrate that rRNA promoter region is hypermethylated in early Alzheimer’s disease, leading to a diminution in transcription of rRNA (Pietrzak et al. 2011). Reducing nucleolar transcription may participate in the decline of the rRNA component of brain ribosomes contributing to Alzheimer associated synapse loss and dementia. In Parkinson’s disease, nucleolar integrity was also reported to be disrupted in the dopaminergic neurons of patients (Rieker et al. 2011). In support of this observation, two proteins which are associated with Parkinson’s disease, Alpha-synuclein and DJ-1, are capable of interacting with nucleolin (Jin et al. 2007), although the exact function of this interaction is not known and could also be a consequence of an accumulation of DNA damage and activation of oxidative stress (Markesbery and Lovell 2006; Rieker et al. 2011).
The Huntington’s disease and spinocerebellar ataxias belong to a group of poly-glutamine disease which are cause by additional repetition of CAG in particular genes. The repeat of these nucleotides alters the function of the affected proteins leading to toxicity (Orr and Zoghbi 2007). However, the toxicity is not only caused by the gain of function of the proteins. Recent study showed that mutant transcripts can also contribute to the adverse effect (Tsoi et al. 2012). The mutant transcripts can alter rDNA transcription and induced apoptosis by activating nucleolar stress pathways. In amyotrophic lateral sclerosis or frontotemporal dementia, an abortive transcripts of C9orf72 gene is produce with several repetitions of the hexanucleotide repeat region GGGGCC (Haeusler et al. 2014). This abortive transcript migrates to the nucleolus and binds Nucleolin. This association results in a mislocalized Nucleolin and likely contributes to the nucleolar stress activation observed (Kwon et al. 2014).
Summary
It has become increasingly apparent that the cellular function for the nucleolus goes well beyond ribosome subunit production. Multiple lines of investigation have demonstrated a wide range of function including cell cycle regulation, stress responses and maturation and biogenesis of a wide range of ribonucleoprotein complexes. Several recently developed technologies are driving forward our understanding of the extent and relevance of the nucleolus in these cellular processes, and the dynamic nature of the nucleoli underline the importance of studying the structure and function of this nuclear organelle under a wide range of conditions. Considering the large number of proteins identified in the nucleolus in proteomic experiments that have no known functions or that are still poorly characterized, there will be likely further functions that will be uncovered in the future associated with the nucleolus.
Cell growth and proliferation is critically dependent on an efficient supply of ribosomes to maintain protein synthesis levels. Therefore, the nucleolus is emerging as a key centre of cell growth regulation and it is not surprising that its activity is influenced by a wide range of signaling events that can modulate the efficiency of rRNA expression and ribosome subunit assembly and transport. Several examples where disruption of nucleolar components and activities result in human disease, including inherited genetic disorders and predisposition to cancer, directly reflecting the importance on cell function of disrupting mechanisms that occur in the nucleolus. The link between the nucleolus and regulation of such important cellular function demonstrate the potential as a therapeutic target for cancer treatment, viral infection and neurodegenerative diseases.
Acknowledgments
Work in the Boisvert laboratory is supported by the Canadian Institutes of Health Research (CIHR), and by the National Sciences and Engineering Research Council of Canada (NSERC).
Contributor Information
David P. Bazett-Jones, Phone: +11001(416) 813-2181, Email: David.Bazett-jones@sickkids.ca
Graham Dellaire, Phone: +11001(902) 494-4730, FAX: +11001(902) 494-2519, Email: dellaire@dal.ca.
François-Michel Boisvert, Email: fm.boisvert@usherbrooke.ca.
References
- Akhmanova A, Verkerk T, Langeveld A, Grosveld F, Galjart N. Characterisation of transcriptionally active and inactive chromatin domains in neurons. J Cell Sci. 2000;113(Pt 24):4463–4474. doi: 10.1242/jcs.113.24.4463. [DOI] [PubMed] [Google Scholar]
- Andersen JS, Lam YW, Leung AK, Ong SE, Lyon CE, Lamond AI, Mann M. Nucleolar proteome dynamics. Nature. 2005;433:77–83. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]
- Andersen JS, Lyon CE, Fox AH, Leung AK, Lam YW, Steen H, Mann M, Lamond AI. Directed proteomic analysis of the human nucleolus. Curr Biol. 2002;12:1–11. doi: 10.1016/S0960-9822(01)00650-9. [DOI] [PubMed] [Google Scholar]
- Bai B, Liu H, Laiho M. Small RNA expression and deep sequencing analyses of the nucleolus reveal the presence of nucleolus-associated microRNAs. FEBS Open Biol. 2014;4:441–449. doi: 10.1016/j.fob.2014.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai B, Yegnasubramanian S, Wheelan SJ, Laiho M. RNA-Seq of the nucleolus reveals abundant SNORD44-derived small RNAs. PLoS One. 2014;9:e107519. doi: 10.1371/journal.pone.0107519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S, Hurley LH, Neidle S. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy? Nat Rev Drug Discov. 2011;10:261–275. doi: 10.1038/nrd3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazett-Jones DP, Leblanc B, Herfort M, Moss T. Short-range DNA looping by the Xenopus HMG-box transcription factor, xUBF. Science. 1994;264:1134–1137. doi: 10.1126/science.8178172. [DOI] [PubMed] [Google Scholar]
- Bell SP, Learned RM, Jantzen HM, Tjian R. Functional cooperativity between transcription factors UBF1 and SL1 mediates human ribosomal RNA synthesis. Science (New York, NY) 1988;241:1192–1197. doi: 10.1126/science.3413483. [DOI] [PubMed] [Google Scholar]
- Boisvert FM, Lam YW, Lamont D, Lamond AI. A quantitative proteomics analysis of subcellular proteome localization and changes induced by DNA damage. Mol Cell Proteomics. 2010;9:457–470. doi: 10.1074/mcp.M900429-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI. The multifunctional nucleolus. Nat Rev Mol Cell Biol. 2007;8:574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- Boulon S, Westman BJ, Hutten S, Boisvert FM, Lamond AI. The nucleolus under stress. Mol Cell. 2010;40:216–227. doi: 10.1016/j.molcel.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady SN, Yu Y, Maggi LB, Jr, Weber JD. ARF impedes NPM/B23 shuttling in an Mdm2-sensitive tumor suppressor pathway. Mol Cell Biol. 2004;24:9327–9338. doi: 10.1128/MCB.24.21.9327-9338.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer BJ, Fangman WL. A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell. 1988;55:637–643. doi: 10.1016/0092-8674(88)90222-X. [DOI] [PubMed] [Google Scholar]
- Burger K, Muhl B, Harasim T, Rohrmoser M, Malamoussi A, Orban M, Kellner M, Gruber-Eber A, Kremmer E, Holzel M, Eick D. Chemotherapeutic drugs inhibit ribosome biogenesis at various levels. J Biol Chem. 2010;285:12416–12425. doi: 10.1074/jbc.M109.074211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bywater MJ, Poortinga G, Sanij E, Hein N, Peck A, Cullinane C, Wall M, Cluse L, Drygin D, Anderes K, Huser N, Proffitt C, Bliesath J, Haddach M, Schwaebe MK, Ryckman DM, Rice WG, Schmitt C, Lowe SW, Johnstone RW, Pearson RB, McArthur GA, Hannan RD. Inhibition of RNA polymerase I as a therapeutic strategy to promote cancer-specific activation of p53. Cancer Cell. 2012;22:51–65. doi: 10.1016/j.ccr.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caburet S, Conti C, Schurra C, Lebofsky R, Edelstein SJ, Bensimon A. Human ribosomal RNA gene arrays display a broad range of palindromic structures. Genome Res. 2005;15:1079–1085. doi: 10.1101/gr.3970105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calkins AS, Iglehart JD, Lazaro JB. DNA damage-induced inhibition of rRNA synthesis by DNA-PK and PARP-1. Nucleic Acids Res. 2013;41:7378–7386. doi: 10.1093/nar/gkt502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca M, Mendes-Soares L, Campos I. To be or not to be in the nucleolus. Nat Cell Biol. 2000;2:E107–112. doi: 10.1038/35014078. [DOI] [PubMed] [Google Scholar]
- Comai L, Tanese N, Tjian R. The TATA-binding protein and associated factors are integral components of the RNA polymerase I transcription factor, SL1. Cell. 1992;68:965–976. doi: 10.1016/0092-8674(92)90039-F. [DOI] [PubMed] [Google Scholar]
- Conconi A, Widmer RM, Koller T, Sogo JM. Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell. 1989;57:753–761. doi: 10.1016/0092-8674(89)90790-3. [DOI] [PubMed] [Google Scholar]
- Dammann R, Lucchini R, Koller T, Sogo JM. Chromatin structures and transcription of rDNA in yeast Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:2331–2338. doi: 10.1093/nar/21.10.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derenzini M, Montanaro L, Trere D. What the nucleolus says to a tumour pathologist. Histopathology. 2009;54:753–762. doi: 10.1111/j.1365-2559.2008.03168.x. [DOI] [PubMed] [Google Scholar]
- Derenzini M, Trere D, Pession A, Govoni M, Sirri V, Chieco P. Nucleolar size indicates the rapidity of cell proliferation in cancer tissues. J Pathol. 2000;191:181–186. doi: 10.1002/(SICI)1096-9896(200006)191:2<181::AID-PATH607>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Derenzini M, Trere D, Pession A, Montanaro L, Sirri V, Ochs RL. Nucleolar function and size in cancer cells. Am J Pathol. 1998;152:1291–1297. [PMC free article] [PubMed] [Google Scholar]
- Desterro JM, Keegan LP, Lafarga M, Berciano MT, O’Connell M, Carmo-Fonseca M. Dynamic association of RNA-editing enzymes with the nucleolus. J Cell Sci. 2003;116:1805–1818. doi: 10.1242/jcs.00371. [DOI] [PubMed] [Google Scholar]
- Di Bacco A, Ouyang J, Lee HY, Catic A, Ploegh H, Gill G. The SUMO-specific protease SENP5 is required for cell division. Mol Cell Biol. 2006;26:4489–4498. doi: 10.1128/MCB.02301-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, Markesbery WR, Chen Q, Li F, Keller JN. Ribosome dysfunction is an early event in Alzheimer’s disease. J Neurosci Off J Soc Neurosci. 2005;25:9171–9175. doi: 10.1523/JNEUROSCI.3040-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donati G, Peddigari S, Mercer CA, Thomas G. 5S ribosomal RNA is an essential component of a nascent ribosomal precursor complex that regulates the Hdm2-p53 checkpoint. Cell Rep. 2013;4:87–98. doi: 10.1016/j.celrep.2013.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donmez-Altuntas H, Akalin H, Karaman Y, Demirtas H, Imamoglu N, Ozkul Y. Evaluation of the nucleolar organizer regions in Alzheimer’s disease. Gerontology. 2005;51:297–301. doi: 10.1159/000086365. [DOI] [PubMed] [Google Scholar]
- Dove BK, You JH, Reed ML, Emmett SR, Brooks G, Hiscox JA. Changes in nucleolar morphology and proteins during infection with the coronavirus infectious bronchitis virus. Cell Microbiol. 2006;8:1147–1157. doi: 10.1111/j.1462-5822.2006.00698.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M, Leno GH, Hammarskjold ML, Rekosh D, Helga-Maria C, Olson MO. The roles of nucleolar structure and function in the subcellular location of the HIV-1 Rev protein. J Cell Sci. 1995;108(Pt 8):2811–2823. doi: 10.1242/jcs.108.8.2811. [DOI] [PubMed] [Google Scholar]
- Dundr M, Misteli T, Olson MO. The dynamics of postmitotic reassembly of the nucleolus. J Cell Biol. 2000;150:433–446. doi: 10.1083/jcb.150.3.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazakerley JK, Boyd A, Mikkola ML, Kaariainen L. A single amino acid change in the nuclear localization sequence of the nsP2 protein affects the neurovirulence of Semliki Forest virus. J Virol. 2002;76:392–396. doi: 10.1128/JVI.76.1.392-396.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoriw AM, Starmer J, Yee D, Magnuson T. Nucleolar association and transcriptional inhibition through 5S rDNA in mammals. PLoS Genet. 2012;8:e1002468. doi: 10.1371/journal.pgen.1002468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli S, Di Cara A, Neb-Gulati A, Natt F, Schwemberger S, Hall J, Babcock GF, Bernardi R, Pandolfi PP, Thomas G. Absence of nucleolar disruption after impairment of 40S ribosome biogenesis reveals an rpL11-translation-dependent mechanism of p53 induction. Nat Cell Biol. 2009;11:501–508. doi: 10.1038/ncb1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli S, Ivanenkov VV, Teng T, Thomas G. Suprainduction of p53 by disruption of 40S and 60S ribosome biogenesis leads to the activation of a novel G2/M checkpoint. Genes Dev. 2012;26:1028–1040. doi: 10.1101/gad.189951.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganot P, Jady BE, Bortolin ML, Darzacq X, Kiss T. Nucleolar factors direct the 2′-O-ribose methylation and pseudouridylation of U6 spliceosomal RNA. Mol Cell Biol. 1999;19:6906–6917. doi: 10.1128/MCB.19.10.6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier T, Robert-Nicoud M, Guilly MN, Hernandez-Verdun D. Relocation of nucleolar proteins around chromosomes at mitosis. A study by confocal laser scanning microscopy. J Cell Sci. 1992;102(Pt 4):729–737. doi: 10.1242/jcs.102.4.729. [DOI] [PubMed] [Google Scholar]
- Gerber JK, Gogel E, Berger C, Wallisch M, Muller F, Grummt I, Grummt F. Termination of mammalian rDNA replication: polar arrest of replication fork movement by transcription termination factor TTF-I. Cell. 1997;90:559–567. doi: 10.1016/S0092-8674(00)80515-2. [DOI] [PubMed] [Google Scholar]
- Gerbi SA, Borovjagin AV, Lange TS. The nucleolus: a site of ribonucleoprotein maturation. Curr Opin Cell Biol. 2003;15:318–325. doi: 10.1016/S0955-0674(03)00049-8. [DOI] [PubMed] [Google Scholar]
- Godley LA, Larson RA. Therapy-related myeloid leukemia. Semin Oncol. 2008;35:418–429. doi: 10.1053/j.seminoncol.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez IL, Sylvester JE. Beyond ribosomal DNA: on towards the telomere. Chromosoma. 1997;105:431–437. doi: 10.1007/BF02510479. [DOI] [PubMed] [Google Scholar]
- Govoni M, Farabegoli F, Pession A, Novello F. Inhibition of topoisomerase II activity and its effect on nucleolar structure and function. Exp Cell Res. 1994;211:36–41. doi: 10.1006/excr.1994.1055. [DOI] [PubMed] [Google Scholar]
- Grummt I. The nucleolus-guardian of cellular homeostasis and genome integrity. Chromosoma. 2013;122:487–497. doi: 10.1007/s00412-013-0430-0. [DOI] [PubMed] [Google Scholar]
- Grummt I, Pikaard CS. Epigenetic silencing of RNA polymerase I transcription. Nat Rev. 2003;4:641–649. doi: 10.1038/nrm1171. [DOI] [PubMed] [Google Scholar]
- Haaf T, Hayman DL, Schmid M. Quantitative determination of rDNA transcription units in vertebrate cells. Exp Cell Res. 1991;193:78–86. doi: 10.1016/0014-4827(91)90540-B. [DOI] [PubMed] [Google Scholar]
- Haeusler AR, Donnelly CJ, Periz G, Simko EA, Shaw PG, Kim MS, Maragakis NJ, Troncoso JC, Pandey A, Sattler R, Rothstein JD, Wang J. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 2014;507:195–200. doi: 10.1038/nature13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haltiner MM, Smale ST, Tjian R. Two distinct promoter elements in the human rRNA gene identified by linker scanning mutagenesis. Mol Cell Biol. 1986;6:227–235. doi: 10.1128/MCB.6.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heix J, Vente A, Voit R, Budde A, Michaelidis TM, Grummt I. Mitotic silencing of human rRNA synthesis: inactivation of the promoter selectivity factor SL1 by cdc2/cyclin B-mediated phosphorylation. EMBO J. 1998;17:7373–7381. doi: 10.1093/emboj/17.24.7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heix J, Zomerdijk JC, Ravanpay A, Tjian R, Grummt I. Cloning of murine RNA polymerase I-specific TAF factors: conserved interactions between the subunits of the species-specific transcription initiation factor TIF-IB/SL1. Proc Natl Acad Sci U S A. 1997;94:1733–1738. doi: 10.1073/pnas.94.5.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heliot L, Kaplan H, Lucas L, Klein C, Beorchia A, Doco-Fenzy M, Menager M, Thiry M, O’Donohue MF, Ploton D. Electron tomography of metaphase nucleolar organizer regions: evidence for a twisted-loop organization. Mol Biol Cell. 1997;8:2199–2216. doi: 10.1091/mbc.8.11.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson AS, Warburton D, Atwood KC. Location of ribosomal DNA in the human chromosome complement. Proc Natl Acad Sci U S A. 1972;69:3394–3398. doi: 10.1073/pnas.69.11.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras AK, Plisson-Chastang C, O’Donohue MF, Chakraborty A, Gleizes PE. An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip Rev RNA. 2015;6:225–242. doi: 10.1002/wrna.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Verdun D. Assembly and disassembly of the nucleolus during the cell cycle. Nucleus. 2011;2:189–194. doi: 10.4161/nucl.2.3.16246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Verdun D, Roussel P, Thiry M, Sirri V, Lafontaine DL. The nucleolus: structure/function relationship in RNA metabolism. Wiley Interdiscip Rev RNA. 2010;1:415–431. doi: 10.1002/wrna.39. [DOI] [PubMed] [Google Scholar]
- Hernandez P, Martin-Parras L, Martinez-Robles ML, Schvartzman JB. Conserved features in the mode of replication of eukaryotic ribosomal RNA genes. EMBO J. 1993;12:1475–1485. doi: 10.1002/j.1460-2075.1993.tb05791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscox JA. The nucleolus—a gateway to viral infection? Arch Virol. 2002;147:1077–1089. doi: 10.1007/s00705-001-0792-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscox JA. RNA viruses: hijacking the dynamic nucleolus. Nat Rev Microbiol. 2007;5:119–127. doi: 10.1038/nrmicro1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscox JA, Wurm T, Wilson L, Britton P, Cavanagh D, Brooks G. The coronavirus infectious bronchitis virus nucleoprotein localizes to the nucleolus. J Virol. 2001;75:506–512. doi: 10.1128/JVI.75.1.506-512.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi RE, Valdez B, Banerjee R, Srivastava M, Dasgupta A. Nucleolin stimulates viral internal ribosome entry site-mediated translation. Virus Res. 2001;76:17–29. doi: 10.1016/S0168-1702(01)00240-4. [DOI] [PubMed] [Google Scholar]
- Jacobson MR, Cao LG, Taneja K, Singer RH, Wang YL, Pederson T. Nuclear domains of the RNA subunit of RNase P. J Cell Sci. 1997;110(Pt 7):829–837. doi: 10.1242/jcs.110.7.829. [DOI] [PubMed] [Google Scholar]
- Jacobson MR, Pederson T. Localization of signal recognition particle RNA in the nucleolus of mammalian cells. Proc Natl Acad Sci U S A. 1998;95:7981–7986. doi: 10.1073/pnas.95.14.7981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Li GJ, Davis J, Zhu D, Wang Y, Pan C, Zhang J. Identification of novel proteins associated with both alpha-synuclein and DJ-1. Mol Cell Proteomics. 2007;6:845–859. doi: 10.1074/mcp.M600182-MCP200. [DOI] [PubMed] [Google Scholar]
- Jordan P, Carmo-Fonseca M. Cisplatin inhibits synthesis of ribosomal RNA in vivo. Nucleic Acids Res. 1998;26:2831–2836. doi: 10.1093/nar/26.12.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar B, Liu B, Zhou Z, Lam YW. Quantitative nucleolar proteomics reveals nuclear re-organization during stress-induced senescence in mouse fibroblast. BMC Cell Biol. 2011;12:33. doi: 10.1186/1471-2121-12-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermekchiev M, Workman JL, Pikaard CS. Nucleosome binding by the polymerase I transactivator upstream binding factor displaces linker histone H1. Mol Cell Biol. 1997;17:5833–5842. doi: 10.1128/MCB.17.10.5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinderman NB, LaVelle A. Ultrastructural changes in the developing nucleolus following axotomy. Brain Res. 1976;108:237–247. doi: 10.1016/0006-8993(76)90183-9. [DOI] [PubMed] [Google Scholar]
- Komili S, Farny NG, Roth FP, Silver PA. Functional specificity among ribosomal proteins regulates gene expression. Cell. 2007;131:557–571. doi: 10.1016/j.cell.2007.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruhlak M, Crouch EE, Orlov M, Montano C, Gorski SA, Nussenzweig A, Misteli T, Phair RD, Casellas R. The ATM repair pathway inhibits RNA polymerase I transcription in response to chromosome breaks. Nature. 2007;447:730–734. doi: 10.1038/nature05842. [DOI] [PubMed] [Google Scholar]
- Kuhn A, Grummt I. Dual role of the nucleolar transcription factor UBF: trans-activator and antirepressor. Proc Natl Acad Sci U S A. 1992;89:7340–7344. doi: 10.1073/pnas.89.16.7340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon I, Xiang S, Kato M, Wu L, Theodoropoulos P, Wang T, Kim J, Yun J, Xie Y, McKnight SL. Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science. 2014;345:1139–1145. doi: 10.1126/science.1254917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam YW, Trinkle-Mulcahy L (2015) New insights into nucleolar structure and function. F1000Prime Rep 7:48 [DOI] [PMC free article] [PubMed]
- Langst G, Becker PB, Grummt I. TTF-I determines the chromatin architecture of the active rDNA promoter. EMBO J. 1998;17:3135–3145. doi: 10.1093/emboj/17.11.3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Learned RM, Learned TK, Haltiner MM, Tjian RT. Human rRNA transcription is modulated by the coordinate binding of two factors to an upstream control element. Cell. 1986;45:847–857. doi: 10.1016/0092-8674(86)90559-3. [DOI] [PubMed] [Google Scholar]
- Lee CH, Chang SC, Chen CJ, Chang MF. The nucleolin binding activity of hepatitis delta antigen is associated with nucleolus targeting. J Biol Chem. 1998;273:7650–7656. doi: 10.1074/jbc.273.13.7650. [DOI] [PubMed] [Google Scholar]
- Leung AK, Gerlich D, Miller G, Lyon C, Lam YW, Lleres D, Daigle N, Zomerdijk J, Ellenberg J, Lamond AI. Quantitative kinetic analysis of nucleolar breakdown and reassembly during mitosis in live human cells. J Cell Biol. 2004;166:787–800. doi: 10.1083/jcb.200405013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CF, Pontes O, El-Shami M, Henderson IR, Bernatavichute YV, Chan SW, Lagrange T, Pikaard CS, Jacobsen SE. An ARGONAUTE4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana. Cell. 2006;126:93–106. doi: 10.1016/j.cell.2006.05.032. [DOI] [PubMed] [Google Scholar]
- Li J, Langst G, Grummt I. NoRC-dependent nucleosome positioning silences rRNA genes. EMBO J. 2006;25:5735–5741. doi: 10.1038/sj.emboj.7601454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZF, Liang YM, Lau PN, Shen W, Wang DK, Cheung WT, Xue CJ, Poon LM, Lam YW. Dynamic localisation of mature microRNAs in Human nucleoli is influenced by exogenous genetic materials. PLoS One. 2013;8:e70869. doi: 10.1371/journal.pone.0070869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little RD, Platt TH, Schildkraut CL. Initiation and termination of DNA replication in human rRNA genes. Mol Cell Biol. 1993;13:6600–6613. doi: 10.1128/MCB.13.10.6600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly T, Ahmad Y, Shlien A, Soroka D, Mills A, Emanuele MJ, Stratton MR, Lamond AI. A proteomic chronology of gene expression through the cell cycle in human myeloid leukemia cells. Elife. 2014;3:e01630. doi: 10.7554/eLife.01630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maehama T, Kawahara K, Nishio M, Suzuki A, Hanada K. Nucleolar stress induces ubiquitination-independent proteasomal degradation of PICT1 protein. J Biol Chem. 2014;289:20802–20812. doi: 10.1074/jbc.M114.571893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mais C, Wright JE, Prieto JL, Raggett SL, McStay B. UBF-binding site arrays form pseudo-NORs and sequester the RNA polymerase I transcription machinery. Genes Dev. 2005;19:50–64. doi: 10.1101/gad.310705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann DM, Marcyniuk B, Yates PO, Neary D, Snowden JS. The progression of the pathological changes of Alzheimer’s disease in frontal and temporal neocortex examined both at biopsy and at autopsy. Neuropathol Appl Neurobiol. 1988;14:177–195. doi: 10.1111/j.1365-2990.1988.tb00880.x. [DOI] [PubMed] [Google Scholar]
- Mann DM, Yates PO. Pathogenesis of Parkinson’s disease. Arch Neurol. 1982;39:545–549. doi: 10.1001/archneur.1982.00510210015004. [DOI] [PubMed] [Google Scholar]
- Markesbery WR, Lovell MA. DNA oxidation in Alzheimer’s disease. Antioxid Redox Signal. 2006;8:2039–2045. doi: 10.1089/ars.2006.8.2039. [DOI] [PubMed] [Google Scholar]
- Matthews DA. Adenovirus protein V induces redistribution of nucleolin and B23 from nucleolus to cytoplasm. J Virol. 2001;75:1031–1038. doi: 10.1128/JVI.75.2.1031-1038.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer C, Neubert M, Grummt I. The structure of NoRC-associated RNA is crucial for targeting the chromatin remodelling complex NoRC to the nucleolus. EMBO Rep. 2008;9:774–780. doi: 10.1038/embor.2008.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer C, Schmitz KM, Li J, Grummt I, Santoro R. Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol Cell. 2006;22:351–361. doi: 10.1016/j.molcel.2006.03.028. [DOI] [PubMed] [Google Scholar]
- Michienzi A, De Angelis FG, Bozzoni I, Rossi JJ. A nucleolar localizing Rev binding element inhibits HIV replication. AIDS Res Ther. 2006;3:13. doi: 10.1186/1742-6405-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G, Panov KI, Friedrich JK, Trinkle-Mulcahy L, Lamond AI, Zomerdijk JC. hRRN3 is essential in the SL1-mediated recruitment of RNA Polymerase I to rRNA gene promoters. EMBO J. 2001;20:1373–1382. doi: 10.1093/emboj/20.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller OL, Jr, Beatty BR. Visualization of nucleolar genes. Science. 1969;164:955–957. doi: 10.1126/science.164.3882.955. [DOI] [PubMed] [Google Scholar]
- Mongelard F, Bouvet P. Nucleolin: a multiFACeTed protein. Trends Cell Biol. 2007;17:80–86. doi: 10.1016/j.tcb.2006.11.010. [DOI] [PubMed] [Google Scholar]
- Moorefield B, Greene EA, Reeder RH. RNA polymerase I transcription factor Rrn3 is functionally conserved between yeast and human. Proc Natl Acad Sci U S A. 2000;97:4724–4729. doi: 10.1073/pnas.080063997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss T. At the crossroads of growth control; making ribosomal RNA. Curr Opin Genet Dev. 2004;14:210–217. doi: 10.1016/j.gde.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Moss T, Langlois F, Gagnon-Kugler T, Stefanovsky V. A housekeeper with power of attorney: the rRNA genes in ribosome biogenesis. Cell Mol Life Sci. 2007;64:29–49. doi: 10.1007/s00018-006-6278-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisole S, Krust B, Hovanessian AG. Anchorage of HIV on permissive cells leads to coaggregation of viral particles with surface nucleolin at membrane raft microdomains. Exp Cell Res. 2002;276:155–173. doi: 10.1006/excr.2002.5522. [DOI] [PubMed] [Google Scholar]
- O’Sullivan AC, Sullivan GJ, McStay B. UBF binding in vivo is not restricted to regulatory sequences within the vertebrate ribosomal DNA repeat. Mol Cell Biol. 2002;22:657–668. doi: 10.1128/MCB.22.2.657-668.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuwaki M, Iwamatsu A, Tsujimoto M, Nagata K. Identification of nucleophosmin/B23, an acidic nucleolar protein, as a stimulatory factor for in vitro replication of adenovirus DNA complexed with viral basic core proteins. J Mol Biol. 2001;311:41–55. doi: 10.1006/jmbi.2001.4812. [DOI] [PubMed] [Google Scholar]
- Olson MO, Dundr M. The moving parts of the nucleolus. Histochem Cell Biol. 2005;123:203–216. doi: 10.1007/s00418-005-0754-9. [DOI] [PubMed] [Google Scholar]
- Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- Parlato R, Kreiner G. Nucleolar activity in neurodegenerative diseases: a missing piece of the puzzle? J Mol Med (Berl) 2013;91:541–547. doi: 10.1007/s00109-012-0981-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T (2011) The nucleolus. Cold Spring Harb Perspect Biol 3 [DOI] [PMC free article] [PubMed]
- Pederson T, Powell K. Thoru Pederson: spotting novel roles for the nucleolus. J Cell Biol. 2015;208:384–385. doi: 10.1083/jcb.2084pi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltonen K, Colis L, Liu H, Jaamaa S, Zhang Z, Af Hallstrom T, Moore HM, Sirajuddin P, Laiho M. Small molecule BMH-compounds that inhibit RNA polymerase I and cause nucleolar stress. Mol Cancer Ther. 2014;13:2537–2546. doi: 10.1158/1535-7163.MCT-14-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendle AF, Clark GP, Boon R, Lewandowska D, Lam YW, Andersen J, Mann M, Lamond AI, Brown JW, Shaw PJ. Proteomic analysis of the Arabidopsis nucleolus suggests novel nucleolar functions. Mol Biol Cell. 2005;16:260–269. doi: 10.1091/mbc.E04-09-0791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrzak M, Rempala G, Nelson PT, Zheng JJ, Hetman M. Epigenetic silencing of nucleolar rRNA genes in Alzheimer’s disease. PLoS One. 2011;6:e22585. doi: 10.1371/journal.pone.0022585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz JC, Zhang F, Pederson T. MicroRNA-206 colocalizes with ribosome-rich regions in both the nucleolus and cytoplasm of rat myogenic cells. Proc Natl Acad Sci U S A. 2006;103:18957–18962. doi: 10.1073/pnas.0609466103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes O, Li CF, Nunes PC, Haag J, Ream T, Vitins A, Jacobsen SE, Pikaard CS. The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell. 2006;126:79–92. doi: 10.1016/j.cell.2006.05.031. [DOI] [PubMed] [Google Scholar]
- Preti M, O’Donohue MF, Montel-Lehry N, Bortolin-Cavaille ML, Choesmel V, Gleizes PE. Gradual processing of the ITS1 from the nucleolus to the cytoplasm during synthesis of the human 18S rRNA. Nucleic Acids Res. 2013;41:4709–4723. doi: 10.1093/nar/gkt160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto JL, McStay B. Recruitment of factors linking transcription and processing of pre-rRNA to NOR chromatin is UBF-dependent and occurs independent of transcription in human cells. Genes Dev. 2007;21:2041–2054. doi: 10.1101/gad.436707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto JL, McStay B. Pseudo-NORs: a novel model for studying nucleoli. Biochim Biophys Acta. 2008;1783:2116–2123. doi: 10.1016/j.bbamcr.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Puvion-Dutilleul F, Christensen ME. Alterations of fibrillarin distribution and nucleolar ultrastructure induced by adenovirus infection. Eur J Cell Biol. 1993;61:168–176. [PubMed] [Google Scholar]
- Raska I, Shaw PJ, Cmarko D. Structure and function of the nucleolus in the spotlight. Curr Opin Cell Biol. 2006;18:325–334. doi: 10.1016/j.ceb.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Rieker C, Engblom D, Kreiner G, Domanskyi A, Schober A, Stotz S, Neumann M, Yuan X, Grummt I, Schutz G, Parlato R. Nucleolar disruption in dopaminergic neurons leads to oxidative damage and parkinsonism through repression of mammalian target of rapamycin signaling. J Neurosci Off J Soc Neurosci. 2011;31:453–460. doi: 10.1523/JNEUROSCI.0590-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussel P, Andre C, Comai L, Hernandez-Verdun D. The rDNA transcription machinery is assembled during mitosis in active NORs and absent in inactive NORs. J Cell Biol. 1996;133:235–246. doi: 10.1083/jcb.133.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussel P, Andre C, Masson C, Geraud G, Hernandez-Verdun D. Localization of the RNA polymerase I transcription factor hUBF during the cell cycle. J Cell Sci. 1993;104(Pt 2):327–337. doi: 10.1242/jcs.104.2.327. [DOI] [PubMed] [Google Scholar]
- Rowland RR, Kervin R, Kuckleburg C, Sperlich A, Benfield DA. The localization of porcine reproductive and respiratory syndrome virus nucleocapsid protein to the nucleolus of infected cells and identification of a potential nucleolar localization signal sequence. Virus Res. 1999;64:1–12. doi: 10.1016/S0168-1702(99)00048-9. [DOI] [PubMed] [Google Scholar]
- Rowland RR, Yoo D. Nucleolar-cytoplasmic shuttling of PRRSV nucleocapsid protein: a simple case of molecular mimicry or the complex regulation by nuclear import, nucleolar localization and nuclear export signal sequences. Virus Res. 2003;95:23–33. doi: 10.1016/S0168-1702(03)00161-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubbi CP, Milner J. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J. 2003;22:6068–6077. doi: 10.1093/emboj/cdg579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggero D, Pandolfi PP. Does the ribosome translate cancer? Nat Rev Cancer. 2003;3:179–192. doi: 10.1038/nrc1015. [DOI] [PubMed] [Google Scholar]
- Russell J, Zomerdijk JC. RNA-polymerase-I-directed rDNA transcription, life and works. Trends Biochem Sci. 2005;30:87–96. doi: 10.1016/j.tibs.2004.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanij E, Hannan RD. The role of UBF in regulating the structure and dynamics of transcriptionally active rDNA chromatin. Epigenetics. 2009;4:374–382. doi: 10.4161/epi.4.6.9449. [DOI] [PubMed] [Google Scholar]
- Sanij E, Poortinga G, Sharkey K, Hung S, Holloway TP, Quin J, Robb E, Wong LH, Thomas WG, Stefanovsky V, Moss T, Rothblum L, Hannan KM, McArthur GA, Pearson RB, Hannan RD. UBF levels determine the number of active ribosomal RNA genes in mammals. J Cell Biol. 2008;183:1259–1274. doi: 10.1083/jcb.200805146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro R, Schmitz KM, Sandoval J, Grummt I. Intergenic transcripts originating from a subclass of ribosomal DNA repeats silence ribosomal RNA genes in trans. EMBO Rep. 2010;11:52–58. doi: 10.1038/embor.2009.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M, Kawahara K, Nishio M, Mimori K, Kogo R, Hamada K, Itoh B, Wang J, Komatsu Y, Yang YR, Hikasa H, Horie Y, Yamashita T, Kamijo T, Zhang Y, Zhu Y, Prives C, Nakano T, Mak TW, Sasaki T, Maehama T, Mori M, Suzuki A. Regulation of the MDM2-P53 pathway and tumor growth by PICT1 via nucleolar RPL11. Nat Med. 2011;17:944–951. doi: 10.1038/nm.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherl A, Coute Y, Deon C, Calle A, Kindbeiter K, Sanchez JC, Greco A, Hochstrasser D, Diaz JJ. Functional proteomic analysis of human nucleolus. Mol Biol Cell. 2002;13:4100–4109. doi: 10.1091/mbc.E02-05-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt EV. The role of c-myc in cellular growth control. Oncogene. 1999;18:2988–2996. doi: 10.1038/sj.onc.1202751. [DOI] [PubMed] [Google Scholar]
- Sirri V, Roussel P, Hernandez-Verdun D. In vivo release of mitotic silencing of ribosomal gene transcription does not give rise to precursor ribosomal RNA processing. J Cell Biol. 2000;148:259–270. doi: 10.1083/jcb.148.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirri V, Urcuqui-Inchima S, Roussel P, Hernandez-Verdun D. Nucleolus: the fascinating nuclear body. Histochem Cell Biol. 2008;129:13–31. doi: 10.1007/s00418-007-0359-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo JM, Ness PJ, Widmer RM, Parish RW, Koller T. Psoralen-crosslinking of DNA as a probe for the structure of active nucleolar chromatin. J Mol Biol. 1984;178:897–919. doi: 10.1016/0022-2836(84)90318-8. [DOI] [PubMed] [Google Scholar]
- Stefanovsky V, Langlois F, Gagnon-Kugler T, Rothblum LI, Moss T. Growth factor signaling regulates elongation of RNA polymerase I transcription in mammals via UBF phosphorylation and r-chromatin remodeling. Mol Cell. 2006;21:629–639. doi: 10.1016/j.molcel.2006.01.023. [DOI] [PubMed] [Google Scholar]
- Strohner R, Nemeth A, Jansa P, Hofmann-Rohrer U, Santoro R, Langst G, Grummt I. NoRC--a novel member of mammalian ISWI-containing chromatin remodeling machines. EMBO J. 2001;20:4892–4900. doi: 10.1093/emboj/20.17.4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan GJ, Bridger JM, Cuthbert AP, Newbold RF, Bickmore WA, McStay B. Human acrocentric chromosomes with transcriptionally silent nucleolar organizer regions associate with nucleoli. EMBO J. 2001;20:2867–2874. doi: 10.1093/emboj/20.11.2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundqvist A, Liu G, Mirsaliotis A, Xirodimas DP. Regulation of nucleolar signalling to p53 through NEDDylation of L11. EMBO Rep. 2009;10:1132–1139. doi: 10.1038/embor.2009.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szebeni A, Mehrotra B, Baumann A, Adam SA, Wingfield PT, Olson MO. Nucleolar protein B23 stimulates nuclear import of the HIV-1 Rev protein and NLS-conjugated albumin. Biochemistry. 1997;36:3941–3949. doi: 10.1021/bi9627931. [DOI] [PubMed] [Google Scholar]
- Tollervey D, Lehtonen H, Jansen R, Kern H, Hurt EC. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell. 1993;72:443–457. doi: 10.1016/0092-8674(93)90120-F. [DOI] [PubMed] [Google Scholar]
- Tsai RY-L. New frontiers in nucleolar research: nucleostemin and related proteins. In: Olson MOJ, editor. The nucleolus, protein reviews. New York: Springer; 2011. pp. 301–320. [Google Scholar]
- Tsai RY, Pederson T. Connecting the nucleolus to the cell cycle and human disease. FASEB J. 2014;28:3290–3296. doi: 10.1096/fj.14-254680. [DOI] [PubMed] [Google Scholar]
- Tschochner H, Hurt E. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 2003;13:255–263. doi: 10.1016/S0962-8924(03)00054-0. [DOI] [PubMed] [Google Scholar]
- Tsoi H, Lau TC, Tsang SY, Lau KF, Chan HY. CAG expansion induces nucleolar stress in polyglutamine diseases. Proc Natl Acad Sci U S A. 2012;109:13428–13433. doi: 10.1073/pnas.1204089109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitali P, Basyuk E, Le Meur E, Bertrand E, Muscatelli F, Cavaille J, Huttenhofer A. ADAR2-mediated editing of RNA substrates in the nucleolus is inhibited by C/D small nucleolar RNAs. J Cell Biol. 2005;169:745–753. doi: 10.1083/jcb.200411129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Blobel G. Signal recognition particle contains a 7S RNA essential for protein translocation across the endoplasmic reticulum. Nature. 1982;299:691–698. doi: 10.1038/299691a0. [DOI] [PubMed] [Google Scholar]
- Walter P, Johnson AE. Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu Rev Cell Biol. 1994;10:87–119. doi: 10.1146/annurev.cb.10.110194.000511. [DOI] [PubMed] [Google Scholar]
- Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–440. doi: 10.1016/S0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- Weber JD, Taylor LJ, Roussel MF, Sherr CJ, Bar-Sagi D. Nucleolar Arf sequesters Mdm2 and activates p53. Nat Cell Biol. 1999;1:20–26. doi: 10.1038/8991. [DOI] [PubMed] [Google Scholar]
- Wiesendanger B, Lucchini R, Koller T, Sogo JM. Replication fork barriers in the Xenopus rDNA. Nucleic Acids Res. 1994;22:5038–5046. doi: 10.1093/nar/22.23.5038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JM, Kusdra L, Collins K. Subnuclear shuttling of human telomerase induced by transformation and DNA damage. Nat Cell Biol. 2002;4:731–736. doi: 10.1038/ncb846. [DOI] [PubMed] [Google Scholar]
- Worton RG, Sutherland J, Sylvester JE, Willard HF, Bodrug S, Dube I, Duff C, Kean V, Ray PN, Schmickel RD. Human ribosomal RNA genes: orientation of the tandem array and conservation of the 5′ end. Science (New York, NY) 1988;239:64–68. doi: 10.1126/science.3336775. [DOI] [PubMed] [Google Scholar]
- Yusupova G, Yusupov M. High-resolution structure of the eukaryotic 80S ribosome. Annu Rev Biochem. 2014;83:467–486. doi: 10.1146/annurev-biochem-060713-035445. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wolf GW, Bhat K, Jin A, Allio T, Burkhart WA, Xiong Y. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol Cell Biol. 2003;23:8902–8912. doi: 10.1128/MCB.23.23.8902-8912.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zomerdijk JC, Beckmann H, Comai L, Tjian R. Assembly of transcriptionally active RNA polymerase I initiation factor SL1 from recombinant subunits. Science (New York, NY) 1994;266:2015–2018. doi: 10.1126/science.7801130. [DOI] [PubMed] [Google Scholar]


