Abstract
Despite many decades of multilateral global efforts, a significant portion of the world population continues to be plagued with one or more mosquito-vectored diseases. These include malaria and filariasis as well as numerous arboviral-associated illnesses including Dengue and Yellow fevers. The dynamics of disease transmission by mosquitoes is complex, and involves both vector competence and vectorial capacity. One area of intensive effort is the study of chemosensory-driven behaviours in the malaria vector mosquito Anopheles gambiae Giles, the modulation of which are likely to provide opportunities for disease reduction. In this context recent studies have characterized a large divergent family of An. gambiae odorant receptors (AgORs) that play critical roles in olfactory signal transduction. This work has facilitated high-throughput, cell-based calcium mobilization screens of AgOR-expressing HEK cells that have identified a large number of conventional AgOR ligands, as well as the first non-conventional Orco (olfactory receptor co-receptor) family agonist. As such, ligand-mediated modulation serves as a proof-of-concept demonstration that AgORs represent viable targets for high-throughput screening and for the eventual development of behaviour-modifying olfactory compounds. Such attractants or repellents could foster malaria reduction programmes.
Keywords: Active odorants, anopheline, control, host selection, mosquito, odour receptors, high-throughput screen
Introduction
A multitude of field and laboratory studies have linked female mosquito host preference to olfactory signals (Dekker et al., 1998, 2001; Mboera et al., 2000) and particularly human odours in the case of An. gambiae (For reviews see Bock & Cardew 1996; Takken & Knols, 1999 & 2009; Zwiebel & Takken, 2004; Besansky et al. 2004). Indeed, the propensity of female Anopheles gambiae mosquitoes to select humans for blood-feeding (anthropophily) is directly correlated to some of the highest-recorded Human Biting Indexes, (Tanga et al., 2011). The strong anthropophily in this system is furthermore demonstrated in two-choice studies in an olfactometer where An. gambiae moves significantly more towards human odour than does a sibling species An. quadriannulatus (Pates et al., 2001a). Moreover, An. gambiae shows a strong aversion to cow odour while An. quadriannulatus has no preference for either odour source (Pates et al., 2001a). In other field trials, An. gambiae has demonstrated a strong preference for human hosts from a distance, even in the absence of visual cues or CO2, while its sibling species An. arabiensis moves more towards animal odours (Costantini et al., 1996, 1998). Field collections of mosquitoes using tent traps containing sleeping humans, again without the possibility of visual contact between the mosquito and host, also catch large numbers of female An. gambiae mosquitoes. Odour-releasing traps routinely catch more Anophelines when human odours are used (Njiru et al., 2006), even when in direct competition with primate odours. Taken together these studies make a strong case for the use of novel olfactory-based interventions to reduce human-vector encounters.
The location of a blood-meal source involves a series of behaviours: activation of insect flight following stimulation with a host chemical odour (kairomone), upwind flight in the direction of the odour, and eventually alighting/probing on the host itself. In the context of human odour, approximately 350 different chemical compounds have been identified in sweat (Cork & Park, 1996). Attractive sweat compounds include carboxylic acids (Meijerink, 2001; Meijerink & van Loon, 1999), ammonia (Meijerink, 2001), lactic acid, and various other volatiles (Cork & Park, 1996; Meijerink et al., 2001; Healy & Copland 2001). Moreover, An. gambiae is more responsive to incubated human sweat than freshly-collected sweat (Braks et al., 2001). Skin microbes are responsible for the changes in chemical composition of sweat that increase its potency (Verhulst et al., 2009;). An. gambiae females also respond to human breath components, although though this response is sometimes based on repellent effects (Mukabana et al., 2004; Qiu et al., 2010). Another important component of human breath is CO2. While not a human-specific odour, CO2 has long been recognized as an important mosquito kairomone in both field and laboratory settings conditions (Costantini et al., 1996). Carbon dioxide acts synergistically to enhance the response to other volatiles and is particularly important for the activation phase of host seeking (Takken and Knols, 1999; Dekker et al., 2001; 2005; Lacey & Cardé, 2010). As is the case for CO2, ammonia also acts as a powerful synergist for human hosts (Smallegange et al., 2005).
The molecular basis of Anopheline chemosensation
At the molecular level, insect olfactory signal transduction results from the interaction of chemical odorants and several groups of proteins expressed on the peripheral dendrites of olfactory receptor neurones (ORNs) that typically are found on the antennae, maxillary palps and other adult head appendages. These include odorant binding proteins (OBPs), odorant degrading enzymes (ODEs), sensory neuron membrane proteins (SNMPs) as well as large families of variant ionotropic and odorant receptors (IRs and ORs, respectively) that have been extensively reviewed elsewhere (Rutzler & Zwiebel, 2005; Benton, 2009).
Because of their central role in olfactory signal transduction OR proteins have been the subject of extensive study. The first insect odorant receptors (Ors) were molecularly identified in Drosophila melanogaster by multiple groups using differing approaches (Clyne et al., 1999; Gao and Chess, 1999; Vosshall et al., 1999). As a result of genome sequencing projects, Or gene families have now been described in numerous insect species representing multiple orders (Krieger et al., 2003; Robertson et al., 2003; Robertson and Wanner, 2006; Smadja et al., 2009; Smith et al., 2011), including three mosquito species: An. gambiae (Hill et al., 2002), Ae. aegypti (Bohbot et al., 2007), and Culex quinquefasciatus (Pelletier et al., 2010). With one notable exception (see below) insect Ors are an extremely divergent gene superfamily, often sharing very low amino acid identities/similarities within the same species, and having few orthologs between species (Robertson et al., 2003; Ache & Young, 2005; Robertson, 2006).
The large family of 79 candidate AgOrs was identified using homology-based approaches (Fox et al., 2001, 2002; Hill et al., 2002). Of these 79, 75 appear to be transcriptionally active (Pitts et al., 2011). As is the case for other insect Ors, the majority of AgOr gene products share little identity at the primary amino acid level, usually less than 20%, although 14 pairs share >70% identity (Hill et al., 2002). In the olfactory tissues of insects, individual ORNs appear to express only one of these conventional ORs (also called “tuning” ORs), and it is members of this class of Or that then defines the unique odorant response profile of any given ORN (Wetzel et al., 2001; Sakurai et al., 2004; Neuhaus et al., 2005).
In stark contrast to the diversity and expression profiles of conventional, tuning Ors, one member of this gene family which is now uniformly known as Orco (olfactory receptor co-receptor) is both highly conserved across insect orders and is ubiquitously expressed in all ORNs. Orco is necessary and sufficient for the proper localization and retention of other conventional Ors at the dendritic membrane (Larsson et al., 2004, Benton et al., 2006), and is required for proper function of conventional Ors (Krieger et al., 2003; Pitts et al., 2004; Larsson et al., 2004; Jones et al., 2005; Xia & Zwiebel, 2006). This exceptional degree of sequence conservation and expression characteristics among insect OR gene families suggested that the Orco subfamily represents a non-conventional Or that is broadly required for olfactory signal transduction in all insects.
Tuning Ors from multiple insect species have now been functionally characterized and greater than 60% of AgORs have been functionally characterized through various heterologous expression methods (Lu et al., 2007; Xia et al., 2008; Carey et al., 2010; Wang et al., 2010). In each of these systems a tuning OR is expressed along with Orco; signal is not observed in the absence of Orco. Although AgOrs expressed in heterologous systems are generally not as sensitive as the native neurones in their ability to detect nanomolar odorant concentrations, they nevertheless faithfully recapitulate the agonist rankings of AgOR-expressing neurones (Lu et al., 2007; Carey et al., 2010, Bohbot et al., 2011). This observation serves as a proof-of-concept that these expression systems are excellent substitutes for the native system, and provides a necessary rationale for extensive AgOR functional characterization in high throughput systems outside of the constraints of the mosquito.
Importantly, functional analyses using heterologous expression studies have revealed that on its own, Orco does not confer odorant sensitivity (Dobritsa et al., 2003; Elmore et. al., 2003; Benton et al., 2006; Sato et al., 2008; Wicher et al., 2008). Rather, Orco forms an essential part of a multimeric ion channel in cooperation with a tuning Or that is gated by its cognate odour ligand (Sato et al., 2008; Wicher et al., 2008; Smart et al., 2008) and elicits intracellular signals without GTP-binding proteins (Sato et al. 2009, Jones et al., 2011). Validation of these paradigms in vivo remains an important objective of current efforts.
Molecular targets of current insect repellents
The molecular mechanisms and targets of known insect repellents have been the subject of considerable study. While still somewhat controversial, several studies suggest that the most effective and wide used commercial repellent, N,N-Diethyl-meta-toluamide (DEET) triggers an aversive response through the bimodal activation of a subset of ORNs and specific Gustatory Receptor Neurones (GRNs; Syed & Leal, 2008; Xia et al., 2008; Liu et al., 2010). Citronella, the only other insect repellent with at least a partially-defined molecular target, activates a Transient Receptor Potential (TRPA1) channel that is normally responsible for heat detection in dipterans to trigger an avoidance response (Kwon et al., 2010). In addition, citronella also targets an Orco dependent pathway, although the conventional OR targets have yet to be defined (Kwon et al., 2010). However, despite its widespread use, DEET is somewhat toxic and hardly affects some vector mosquito species; factors that decrease consumer acceptance (reviewed in Paluch et al., 2010). For these reasons, there is widespread interest in developing next generation insect repellents, with increased efficacy, longer half-lives, and lower toxicity.
High throughput approach to insect control by modifying behaviours
Until recently, the development of new attractants and repellents for insects has largely focused on screening compounds using electrophysiology and whole-organism behavioural responses. Such approaches are resource-intensive, require large amounts of compound, and are not amenable to high-throughput approaches (Paluch, et al., 2010). In an effort to circumvent this bottleneck, the feasibility of employing high throughput, cell-screening technologies that have been principally focused upon pharmacologic drug discovery has been investigated. A critical component of such an undertaking is the ability to faithfully express screening targets in one or more heterologous systems in order to facilitate rapid and sensitive screens of small molecule libraries. As AgORs are key mediators of various mosquito behaviours and have been shown to express well in various heterologous systems, AgORs have been selected to be expressed in HEK293 cells (always in conjunction with the co-receptor, AgOrco) for the purposes of high throughput screening. These screens are designed to identify novel AgOR modulators that could potentially affect Anopheline behaviour (Lu et al., 2007; Carey et al., 2010, Wang et al., 2010; Bohbot et al., 2011).
In selecting the AgOR targets of these screens, specific AgORs are chosen from among the nearly 60 tuning ORs expressed in the antennal tissues of adult An. gambiae. Among these receptors, AgOR10 is seen as the prime candidate for extensive screening for two principal reasons. First, AgOR10 is one of only two AgORs that shows a greater than 69% conservation of peptide sequence between the blood-feeding mosquito subfamilies of Anophelinae and Culicinae (Bohbot et al., 2011; LJZ unpublished data). Secondly, when expressed heterologously, AgOR10 demonstrates a robust responsiveness to known, attractive semiochemicals, thus suggesting that AgOR10 plays an active role in olfactory-mediated behaviours of hematophagous mosquitoes (Carey et al., 2010, Wang et al., 2010; Bohbot et al., 2011). Consequently, a high-throughput screening protocol has been optimized around AgOrco + AgOR10 expressing HEK293 cells.
Materials and methods
Calcium fluorometry
The creation and validation of the AgOrco +AgOR10 cell line used in these screens has been previously described (Bohbot et al., 2011). All assays were conducted using the instrumentation available through the high-throughput screening Facility that is part of the Institute for Chemical Biology at Vanderbilt University. Twenty four hours prior to each assay, cells were plated at a concentration of 20000 cells/well in a black-walled, clear-bottomed 384-well plate (Greiner Bio-One, Longwood, Florida) and incubated for 8 h at 37 °C with 5% CO2 to facilitate adherence. Cells were then treated with 0.3 µg µL−1 tetracycline (Sigma-Aldrich) overnight to induce expression of the AgOR complexes. 12–16 h post-induction, cells were incubated for 45 min at 37 °C with 20 µL of 3 µM Fluo-4/acetoxymethyl ester (Fluo-4AM), prepared as a 2.3 mM stock in DMSO (Sigma-Aldrich), mixed in a 1:1 ratio with 10% (w/v) pluronic acid F-127 (Invitrogen), and diluted in assay buffer (Hanks' balanced salt solution, 20 mM HEPES, and 2.5 mM probenecid). Dye-buffer was then replaced with 20 µL of calcium assay buffer.
Vanderbilt Molecular Screening Library test compounds were prepared from 10 mM DMSO stocks, and diluted in assay buffer to 20 µM (i.e., 2× 10 µM) in the central 320 wells of a 384-well, 15mm poly-propylene plates using an Echo 555 acoustic liquid handler (Labcyte, Sunnyvale, California) and a Multidrop Combi (Themo Scientific). For the AgOR10 odour-control plates, EC20, EC80, and EC100 concentrations of 2-ethylphenol (Sigma Aldrich) were experimentally determined by half log-molar concentration response curves generated in the assay format employed. The odour-control compound was prepared for assay in each of either of two separate 384-well, deep well (22 mm) poly-propylene plates (Greiner Bio-One, Longwood, Florida). The first control plate contains a 5× EC20 (5 µM) concentration of 2-ethyl phenol (2-EP) in each of the 356 central wells (i.e., all wells exclusive of the outer columns on east/west edges). The wells of the east/west edge outer columns then received either a 5× EC100 concentration of 2-EP (500 µM) or a DMSO-only negative control (EC0). The second control plate contains a 5× EC80 (50 µM) concentration of 2-EP in each of the 356 central wells and assay buffer only in the east/west edge outer columns.
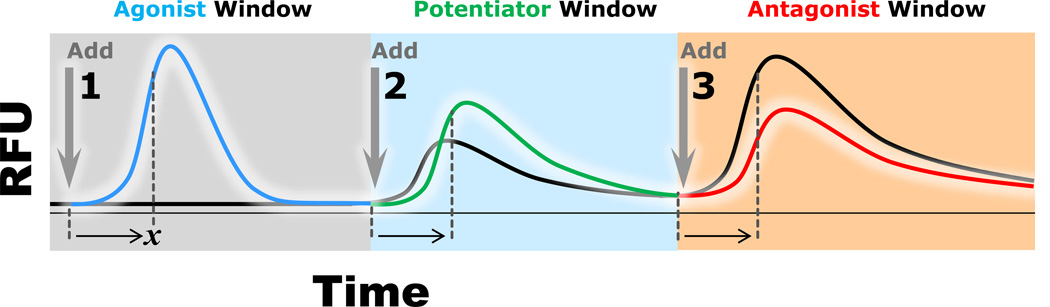
Fluorescence readings and integrated liquid handling were conducted in a Hamamatsu FDSS6000 plate reader (Excitation: 470 ± 20 nm; Emission: 540 ± 30 nm; Hamamatsu Corporation, Bridgewater, New Jersey). Each cell plate was assayed over the course of five min, and was subjected to of three separate compound additions interspersed with latency periods to allow for a return to baseline (Fig. 1). Compound additions were performed at assay time-points of 5, 120 and 220 s, from the test compound plate, the first control plate and the second control plate respectively. The volumes of each addition were 2l, 10, and 12 µL from the test compound plate (2× final concentration), the first control plate (5× final concentration) and second control plate (5× final concentration), respectively. Fluorescent readings were taken at 1-s intervals over the course of the entire assay. Additional, 0.5-s fluorescent readings were taken during the 30 s subsequent to each addition to better quantify response kinetics.
Figure 1.
Triple-add screen design. Well-by-well relative fluorescence units (RFU) are measured over a time course of 5.5 min. Compound activity is gauged against the RFU produced by control wells (black line). The first addition (Add 1) contains a 10 µM concentration of test compound and no control compound. The second addition (Add 2) is of an EC20 concentration of control compound and the third addition (Add 3) is one of an EC80 concentration of control compound. Agonist activity of a given test compound (blue line) is ascertainable following Add 1; potentiator activity of the compound (green line) is ascertainable following Add 2; antagonist activity of the compound (red line) is ascertainable following Add 3. Hit discrimination is algorithmically determined by comparing RFU intensities in each window following a set interval of time (x).
Hit identification
The following criteria and related algorithm were used to automatically identify (call) hits. Agonists were called if and only if: [A1+x]>(1.05*[avgEC0+x]) AND [A3+x]<[EC80+x] Where [A1+x] is the RFU (Relative Fluorescent Unit) taken at x sampling intervals after the first compound addition (A1; Fig. 1). The agonist criteria requires both a fluorescent increase of 5% over background (RFU of the avgEC0 at time x) to demonstrate efficacy, as well as a cumulative, time-dependent diminution in RFU following the third addition (A3). This latter criteria was employed to filter out both auto-fluorescing compounds as well as compounds that may have off-target effects
Potentiators (compounds that have no intrinsic activity, but that can amplify the activity of other agonists) were called if and only if: [A2+x]−[A2−3]>(1.2*[avgEC20+x]−[avgEC20-3]). Criteria for a potentiator required an RFU increase of greater than 20% (over control) in wells receiving an EC20 concentration of agonist control.
Antagonists were called if and only if [A3+x]<(0.7*[EC80+x]) AND [A1+x]=[A1+x+10] where A3 represents the third add, containing an EC80 concentration of control compound. The antagonist criteria requires that a given test compound be able to diminish the signal of an EC80 concentration of control compound by 30-percent or more. Additionally, it was that the test compound itself have no intrinsic efficacy.
Hit confirmation
To confirm potentially-active small-molecules identified in the primary screen, cells were plated and assayed as they were in the primary screen. Furthermore, to discriminate OR agonists from compounds capable of generating non-specific responses from HEK cells, all agonists were tested against uninduced HEK cells. Compounds were confirmed in three or more replicate assays in both tetracycline-induced cells and in uninduced cells. Only those small molecules that showed activity in the induced line and no activity in the uninduced line were classified as “confirmed hits”.
Results and Discussion
Z-factor determination
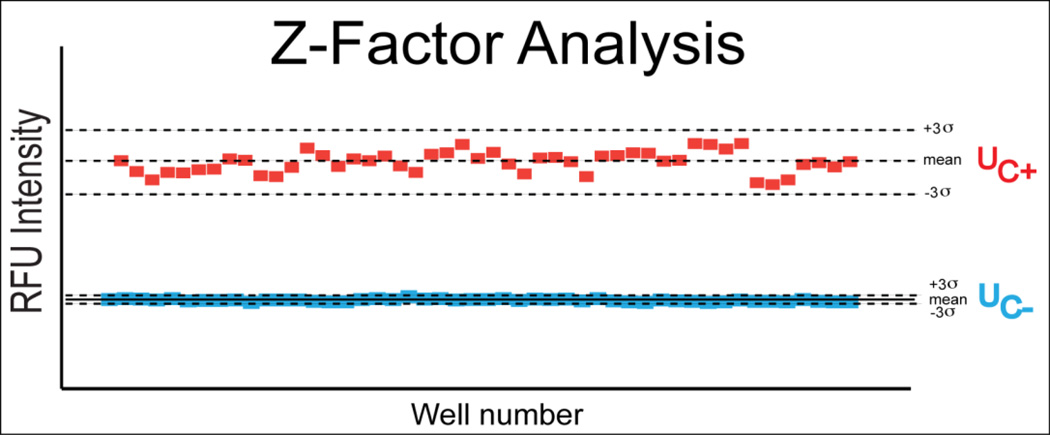
To determine if the resolution of this assay was suitable for high-throughput screening, the Z-factor was calculated within pilot experiments (Zhang et.al.,1999). A Z-factor was determined between the EC0 (agonist) and EC80 (antagonist) window. A Z-factor of 0.79 was found for the EC80 positive control (Figure 2), confirming that this assay has powerful hit discrimination capabilities
Figure 2.
Z- factor analysis of pilot experiments for EC80 concentrations of 2-ethyphenol and DMSO only control. Each data point represents the response to 2-EP (Uc+) or DMSO only (Uc−). Central dashed line for each group represents the mean, while upper and lower dashed lines represent (+/−) 3 standard deviations from the mean, respectively.
Screen design
The high throughput screening project was designed to identify novel modulators of mosquito ORs, which could potentially translate to behavioural-control agents for disease reduction programmes. AgOR10 was chosen both for its high signal levels in the presence of behaviourally-associated odorant stimuli and for its demonstrated functional conservation across mosquito species. In these assays, AgOR10 was co-expressed with AgOrco in HEK cells under a tetracycline inducible system and was validated as described (Bohbot et al., 2011).
The format of the screen was designed as a “triple-add” assay, which takes maximal advantage of the automated liquid handling and plate-reading capabilities of the FDSS instrumentation to examine a given compound from the small-molecule library for intrinsic, AgOR10 agonism or its ability to potentiate or antagonize the receptor’s response to a positive control in a single well over a 5-min time course visualized in real-time (Fig. 1). This was an efficient method to identify as many modulators as possible without losing resolution. In the first add, library compound was added to the experimental well to a final concentration of 10 uM and the library compound’s agonist capacity was examined. Subsequently, in the second add, an [EC20] of 2-EP was added to each well to determine the compounds’ ability to potentiate the ORs response to cognate ligand. Lastly, in the third add, an [EC80] of 2-EP was added to each test well to test for antagonism.
Odorant-based variation in the context of high-throughput screening
Initial pilot experiments involving various AgORs and their associated, odorants, revealed several phenomena that seemed to directly relate to the intrinsic volatility of the low-molecular weight odorants being employed as positive controls. Of particular note, it was observed that control plates containing the EC20 and EC80 concentrations of some odorants gradually lose their ability to elicit an equally efficacious response across successive cell plates, and that this decline in observed efficacy was odour-dependent. While some odours displayed remarkably stable efficacies across multiple, successive cell plates, the efficacy of other odours showed a marked, time-dependent decrease, even over the course of 2 or 3 plates. In the case of AgOR10, the 2-EP odour control displayed stable efficacies across multiple cell plates and allowed for very time-efficient screening. Conversely, in the case of more-volatile odour controls, it was necessary to replace the control plates much more frequently. In addition to the odorant-dependent phenomena of decreased efficacy, some highly-volatile odorants also displayed a tendency to “bleed over” into neighbouring wells such buffer-only control additions positioned adjacent to EC100 control wells could sometimes elicit cell responses. Numerous tests were conducted to rule out the possibility of technically-induced odour contamination and to rule out possibility that high signal levels in the EC100 control wells was causing spurious readings in the adjacent wells. This phenomenon has been observed for a number of cell lines and odorants and disappears when ligands of higher molecular weight are included. AgOR10 and 2-EP did not display this bleed-over effect.
Agonist hit identification and analysis
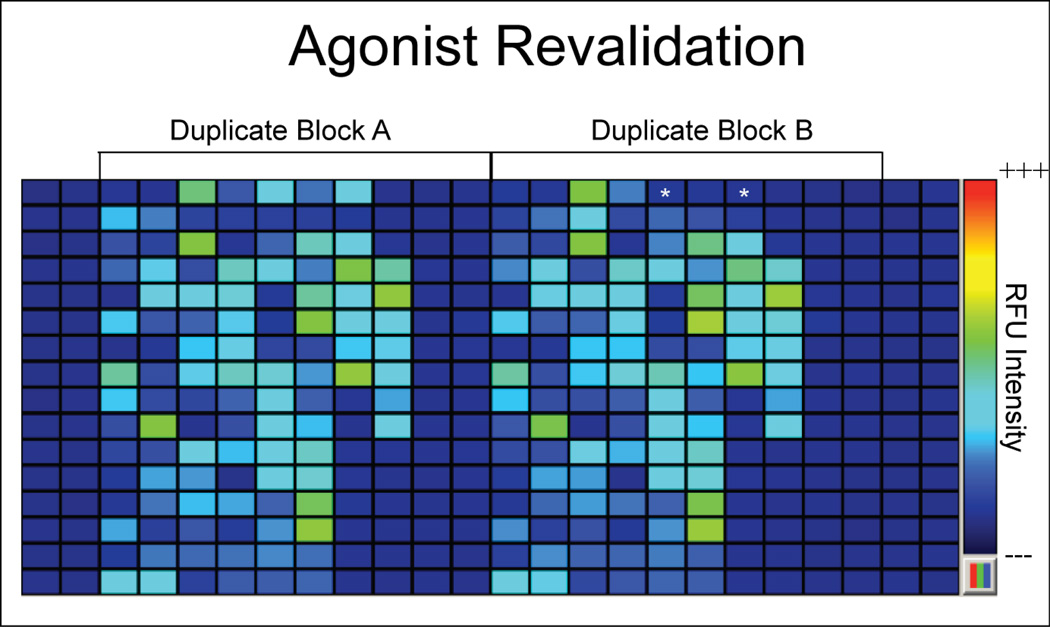
Agonists were determined by an algorithm (defined in methods), which identified compounds with a response that was significantly greater than the baseline control and that also removed auto-fluorescent compounds. Two hundred and sixty-six agonists were identified in the initial primary screen. All hits were retested in duplicate against AgOR10+AgOrco cells as well as against un-induced cells to test for responses specific to the OR complex (Fig. 3). Revalidation screening confirmed that 68 of the original 266 agonists (~25%) maintained activity that was contingent upon AgOR10 expression. This is in keeping with typical high throughput screening efforts and most likely reflects errors in compound loading and/or identification.
Figure 3.
A stylized schematic of a 384-well plate and the relative fluorescence unit (RFU) intensity for a representative agonist hit revalidation. Candidate hits were revalidated in duplicate blocks on the same 384-well plate to control for regional differences in RFU intensity scale as indicated using pseudo-colorization heat map from high (+++) to low (---) as depicted on extreme right. (*) indicates an example where a hit revalidated in one block, but failed to revalidate in another block.
AgOR10 tuning curves demonstrate that AgOR10 is strongly activated by a series of substituted aromatics (Bohbot et al., 2011). The high-throughput screening agonist hit results were no different, as AgOR10 demonstrated strong responses to a number of complex aromatic compounds. Eight tractable hit scaffolds were identified which accounted for more than 70% of all confirmed hits. One of the largest hit series centred upon an imine moiety that encompassed 43 compounds. The true activity of these imine hits will need further verification as this class of compound is generally unstable in solution; acid hydrolysis of imines generally leads to the production of aldehyde and aniline degradation products, classes of compounds that may have intrinsic activity of their own. It is therefore possible that the degradation products of this one particular hit series would yield a diverse set of compounds that could only be further investigated using mass spectrometry analysis to confirm hit identity.
Potentiator and antagonist hit identification and analysis
There were a smaller number of identified potentiators, which had the capacity to increase the response of the control ligand, 2-EP in the second assay window. To be classified as a potentiator, a library compound needed to increase the response of the EC20 control by greater than 20%. Overall, 4 small molecules were identified and confirmed as potentiators of 2-EP in revalidation experiments. Surprisingly, no compounds were found that were capable of significantly inhibiting 2-EP-mediated activation.
A novel allosteric agonist
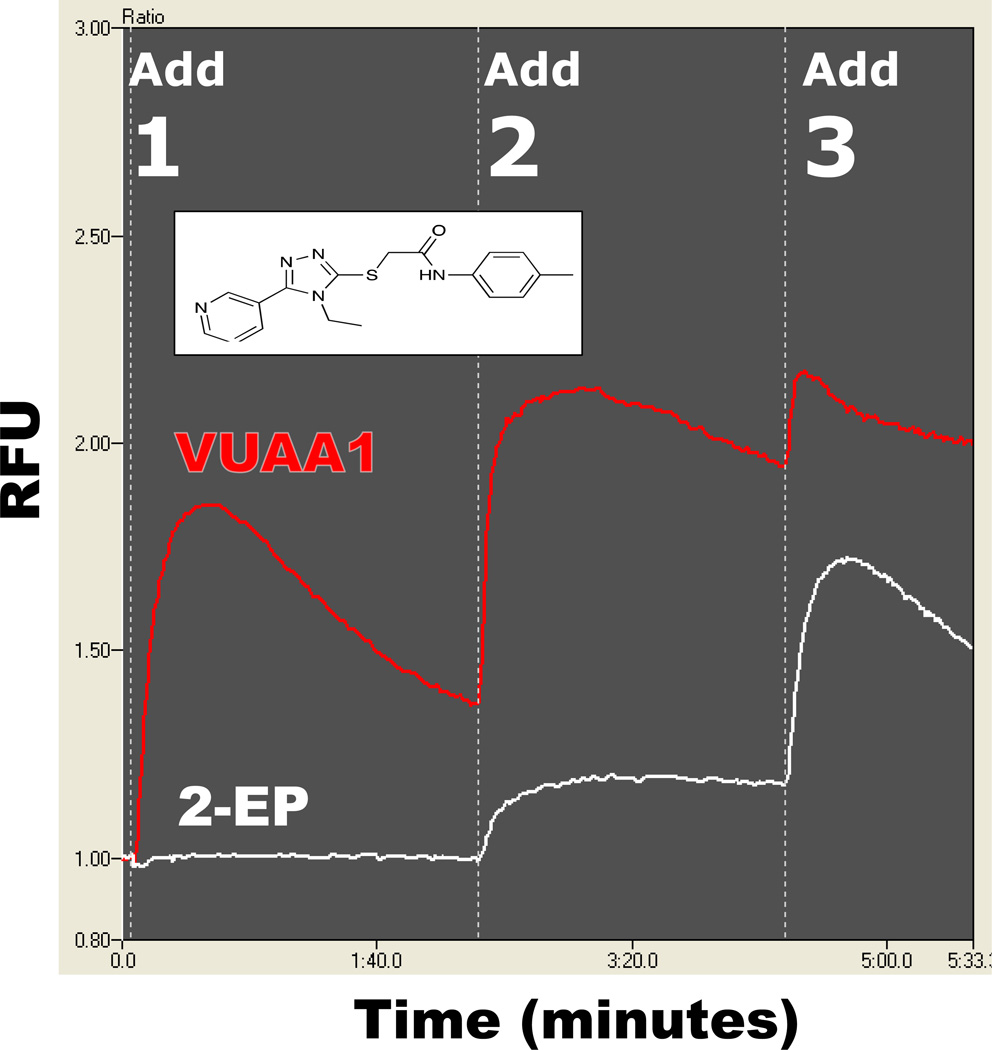
As an unintended consequence of the high-throughput screen for agents that could modify the activity of conventional mosquito AgORs, the first Orco agonist was discovered. This compound 2-(4-ethyl-5-(pyridin-3-yl)-4H-1,2,4-triazol-3-ylthio)-N-(4-ethylphenyl)acetamide was originally thought to be an allosteric agonist of AgOR10 and was designated as such (VUAA1, Vanderbilt University Allosteric Agonist 1; Jones et al., 2011). Indeed, of the more than 150000 compounds that have been screened against AgOR10+AgOrco from the Vanderbilt Small Molecule Library, VUAA1 is the only compound that displayed the hallmarks of allosteric agonism in that it possesses intrinsic agonism and it potentiates the response of 2-EP (Fig. 4). Interestingly, follow-up assays have demonstrated that VUAA1 activated multiple AgOR expressing cell lines, including AgOR8, AgOR10, and AgOR65 (L.J. Zwiebel, unpublished observation). As all of these cell lines also express AgOrco, it was possible that VUAA1 was acting directly on AgOrco. To test this hypothesis, cells expressing AgOrco and other Orco family members alone were treated with VUAA1; these cells display robust responses thereby confirming this molecule as the first-in-class ligand capable of directly gating AgOrco and other insect Orco ion channels (Jones et al., 2011). In addition, these experiments also demonstrated that Orco can form stand-alone ion channels which are themselves capable of being activated directly. Furthermore, VUAA1 provides a powerful positive control that allows for a better examination of the results of previous studies which suggest that Orco is also a cyclic nucleotide gated channel (Sato et al., 2008; Wicher et al., 2008; Smart et al., 2008); no evidence is observed for cyclic nucleotide gating of Orco. This analysis further redefines the model for insect odorant receptor signal transduction.
Figure 4.
Identification of a novel allosteric agonist. Activity of a novel compound is shown (red) and is compared to the control-only response (white). The compound (VUAA1, structure provided in inset) demonstrates the unique characteristics of both a stand-alone agonist and strong potentiating activator (Jones et al., 2011).
Conclusions
A calcium mobility assay that expresses and targets mosquito ORs has been developed in order to facilitate a novel high throughput screening-based approach to identify novel modulators of Anopheline ORs. Because advantage is taken of the pre-existing Institute for Chemical Biology at Vanderbilt University chemical library this initial effort has been biased towards molecules exhibiting known drug-like properties. These compounds are generally of a higher molecular weight than those chemical odorants thought to elicit responses from ORs in natural settings (Wang et al., 2010; Carey et al., 2010). These high molecular weights are generally indicative of low volatility, and thus the hits generated by these high throughput screening efforts are unlikely to be considered as a classical “odorant”. Therefore, lead characterization will likely involve further structure-activity relationship efforts to deconstruct these hits into lower molecular weight structures to increase volatility while maintaining efficacy as well as develop precise formulations to achieve this end. The identification and characterization of VUAA1 as an Orco family agonist raises the possibility of engineering broadly applicable formulations around this activity to hyper-stimulate the olfactory systems of most, if not all, insects. Efforts are currently engaged with behavioural validations of VUAA1 and other HTS hits against pre-adult and adult stage An. gambiae.
The ultimate aim of this work is to utilize state of the art, high-throughput screening-based approaches to develop a rapid method to begin to identify next generation insect excito-repellents that could be used for a number of applications including agriculture, personal protection and public health. While there is, as yet, no comprehensive study that examines the feasibility of a repellent-based disease reduction programme, there is reason to posit that because known repellents have demonstrated remarkable protection against mosquito bites (approximately 96% in numerous field trials), an efficacious repellent would reduce disease burdens (Moore et al., 2002; Costantini et al., 2004). The few studies that have been performed suggest that with high compliance and repellent efficacy, malaria levels would drop below those achieved with insecticide treated nets (Moore, et al., 2007). However, such studies have drawn on the benchmark mosquito repellent DEET, which has a number of limitations. In addition, in areas of high compliance, those users who do not apply repellent are put at greater risk because mosquitoes are diverted to non-compliant individuals; this raises a number of ethical problems (Moore et al., 2007). The use of high-throughput screening-based approaches described here represents a unique opportunity to revisit the search for next-generation insect excito-repellents that may be used for multiple purposes against a wide range of economically and medically important insects.
Acknowledgments
The authors wish to thank staff members at The Vanderbilt Institute of Chemical Biology High-throughput Screening Facility for instrumentation support and members of the Zwiebel laboratory for helpful discussions. This work was supported by Vanderbilt University and a grant from the Foundation for the National Institutes of Health to L.J. Z through the Grand Challenges in Global Health Initiative.
References
- Ache BW, Young JM. Olfaction: Diverse species, conserved principles. Neuron. 2005;48:417–430. doi: 10.1016/j.neuron.2005.10.022. [DOI] [PubMed] [Google Scholar]
- Benton R, Sachse S, Michnick SW, et al. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biology. 2006;4:e20. doi: 10.1371/journal.pbio.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton R, Vannice KS, Gomez-Diaz C, et al. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell. 2009;136:149–162. doi: 10.1016/j.cell.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besansky NJ, Hill CA, Costantini C. No accounting for taste: host preference in malaria vectors. Trends in Parasitology. 2004;20:249–251. doi: 10.1016/j.pt.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Bock GR, Cardew G. Olfaction in Mosquito-Host Interactions. John Wiley and Sons; 1996. [Google Scholar]
- Bohbot JD, Fu L, Le TC, et al. Multiple activities of insect repellents on odorant receptors in mosquitoes. Medical and Veterinary Entomology. 2011 Dec;25(4):436–444. doi: 10.1111/j.1365-2915.2011.00949.x. [DOI] [PubMed] [Google Scholar]
- Bohbot J, Pitts RJ, Kwon HW, et al. Molecular characterization of the Aedesaegypti odorant receptor gene family. Insect Molecular Biology. 2007;16:525–537. doi: 10.1111/j.1365-2583.2007.00748.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey AF, Wang G, Su CY, et al. Odorant reception in the malaria mosquito Anopheles gambiae. Nature. 2010;464:66–71. doi: 10.1038/nature08834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clyne PJ, Warr CG, Freeman MR, et al. A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron. 1999;22:327–338. doi: 10.1016/s0896-6273(00)81093-4. [DOI] [PubMed] [Google Scholar]
- Cork A, Park KC. Identification of electrophysiologically-active compounds for the malaria mosquito, Anopheles gambiae, in human sweat extracts. Medical and Veterinary Entomology. 1996;10:269–276. doi: 10.1111/j.1365-2915.1996.tb00742.x. [DOI] [PubMed] [Google Scholar]
- Costantini C, Badolo A, Ilboudo-Sanogo E. Field evaluation of the efficacy and persistence of insect repellents DEET, IR3535, and KBR 3023 against Anopheles gambiae complex and other Afrotropical vector mosquitoes. Transactions of the Royal Society of Tropical Medicine and Hygiene. 2004 Nov;98(11):644–652. doi: 10.1016/j.trstmh.2003.12.015. [DOI] [PubMed] [Google Scholar]
- Costantini C, Gibson G, Sagnon N, et al. Mosquito responses to carbon dioxide in a west African Sudan savanna village. Medical and Veterinary Entomology. 1996;10:220–227. doi: 10.1111/j.1365-2915.1996.tb00734.x. [DOI] [PubMed] [Google Scholar]
- Costantini C, Sagnon NF, della Torre A, et al. Odor-mediated host preferences of West African mosquitoes, with particular reference to malaria vectors. American Journal of Tropical Medicine and Hygiene. 1998;58:56–63. doi: 10.4269/ajtmh.1998.58.56. [DOI] [PubMed] [Google Scholar]
- Dekker T, Takken W. Differential responses of mosquito sibling species Anopheles arabiensis and An. quadriannulatus to carbon dioxide, a man or a calf. Medical and Veterinary Entomology. 1998;12:136–140. doi: 10.1046/j.1365-2915.1998.00073.x. [DOI] [PubMed] [Google Scholar]
- Dekker T, Takken W, Braks MAH. Innate preference for host-odor blends modulates degree of anthropophagy of Anopheles gambiae sensu lato (Diptera: Culicidae) Journal of Medical Entomology. 2001;38:868–871. doi: 10.1603/0022-2585-38.6.868. [DOI] [PubMed] [Google Scholar]
- Dekker T, Geier M, Cardé RT. Carbon dioxide instantly sensitizes female yellow fever mosquitoes to human skin odours. Journal of Experimental Biology. 2005 doi: 10.1242/jeb.01736. [DOI] [PubMed] [Google Scholar]
- Dobritsa AA, van der Goes van Naters W, et al. Integrating the molecular and cellular basis of odor coding in the Drosophila antenna. Neuron. 2003;37:827–841. doi: 10.1016/s0896-6273(03)00094-1. [DOI] [PubMed] [Google Scholar]
- Elmore T, Ingell R, Carlson JR, et al. Targeted mutation of a Drosophila odor receptor defines receptor requirement in a novel class of sensillum. Journal of Neuroscience. 2003;23(30):9906–9912. doi: 10.1523/JNEUROSCI.23-30-09906.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox AN, Pitts RJ, Zwiebel LJ. A cluster of candidate odorant receptors from the malaria vector mosquito, Anopheles gambiae. Chemical Senses. 2002;27:453–459. doi: 10.1093/chemse/27.5.453. [DOI] [PubMed] [Google Scholar]
- Fox AN, Pitts RJ, Robertson HM, et al. Candidate odorant receptors from the malaria vector mosquito Anopheles gambiae and evidence of down-regulation in response to blood feeding. Proceedings of the National Academy of Sciences U.S.A. 2001;98:14693–14697. doi: 10.1073/pnas.261432998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q, Chess A. Identification of candidate Drosophila olfactory receptors from genomic DNA sequence. Genomics. 1999;60:31–39. doi: 10.1006/geno.1999.5894. [DOI] [PubMed] [Google Scholar]
- Healy TP, Copland MJW. Human sweat and 2-oxopentanoic acid elicit a landing response from Anopheles gambiae. Med Vet Entomol. 2001;14:195–200. doi: 10.1046/j.1365-2915.2000.00238.x. [DOI] [PubMed] [Google Scholar]
- Hill CA, Fox AN, Pitts RJ, Kent LB, Tan PL, et al. G protein-coupled receptors in Anopheles gambiae. Science. 2002;298:176–178. doi: 10.1126/science.1076196. [DOI] [PubMed] [Google Scholar]
- Jones WD, Nguyen TA, Kloss B, et al. Functional conservation of an insect odorant receptor gene across 250 million years of evolution. Current Biology. 2005;15:R119–R121. doi: 10.1016/j.cub.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Jones* PL, Pask* GM, Rinker DC, Zwiebel LJ. Functional agonism of insect odorant receptor ion channels. Proceedings of the National Academy of Sciences U.S.A. 2011 May 24;108(21):8821–8255. doi: 10.1073/pnas.1102425108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger J, Klink O, Mohl C, et al. A candidate olfactory receptor subtype highly conserved across different insect orders. Journal of Comparative Physiology A. 2003 Jul;189(7):519–526. doi: 10.1007/s00359-003-0427-x. [DOI] [PubMed] [Google Scholar]
- Kwon Y, Kim SH, Ronderos DS, et al. Drosophila TRPA1 channel is required to avoid the naturally occurring insect repellent citronellal. Current Biology. 2010 Sep 28;20(18):1672–1678. doi: 10.1016/j.cub.2010.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacey ES, Cardé RT. Activation, orientation and landing of female Culex quinquefasciatus in response to carbon dioxide and odour from human feet: 3-D flight analysis in a wind tunnel. Medical and Veterinary Entomology. 2011 Mar;25(1):94–103. doi: 10.1111/j.1365-2915.2010.00921.x. 2011. [DOI] [PubMed] [Google Scholar]
- Larsson MC, Domingos AI, Jones WD, et al. Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron. 2004;43:703–714. doi: 10.1016/j.neuron.2004.08.019. [DOI] [PubMed] [Google Scholar]
- Liu Q, Ye W, Hu N, et al. Olfactory receptor cells respond to odors in a tissue and semiconductor hybrid neuron chip. Biosensor Bioelectronics. 2010 Dec 15;26(4):1672–1678. doi: 10.1016/j.bios.2010.09.019. [DOI] [PubMed] [Google Scholar]
- Lu T, Qiu YT, Wang G, Kwon JY, et al. Odor coding in the maxillary palp of the malaria vector mosquito Anopheles gambiae. Current Biology. 2007 Sep 18;17(18):1533–1544. doi: 10.1016/j.cub.2007.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mboera LEG, Knols BGJ, Takken W, della Torre A. The response of Anopheles gambiae s.I. and A. funestus (Diptera: Culicidae) to tents baited with human odour or carbon dioxide in Tanzania. Bulletin of Entomological Research. 1997;87:173–178. [Google Scholar]
- Mboera LE, Takken W, Sambu EZ. The response of Culex quinquefasciatus (Diptera: culicidae) to traps baited with carbon dioxide, 1-octen-3-ol, acetone, butyric acid and human foot odour in Tanzania. Bulletin of Entomological Research. 2000;90:155–159. doi: 10.1017/s0007485300000262. [DOI] [PubMed] [Google Scholar]
- Meijerink J, Braks MA, van Loon JJ. Olfactory receptors on the antennae of the malaria mosquito Anopheles gambiae are sensitive to ammonia and other sweat-borne components. Journal of Insect Physiology. 2001;47:455–464. doi: 10.1016/s0022-1910(00)00136-0. [DOI] [PubMed] [Google Scholar]
- Meijerink J, van Loon JJ. Sensitivities of antennal olfactory neurons of the malaria mosquito, Anopheles gambiae, to carboxylic acids. Journal of Insect Physiology. 1999;45:365–373. doi: 10.1016/s0022-1910(98)00135-8. [DOI] [PubMed] [Google Scholar]
- Moore SJ, Hill N, Ruiz C, Cameron MM. Field evaluation of traditionally used plant-based insect repellents and fumigants against the malaria vector Anopheles darlingi in Riberalta, Bolivian Amazon. Journal of Medical Entomology. 2007 Jul;44(4):624–630. doi: 10.1603/0022-2585(2007)44[624:feotup]2.0.co;2. 2007. [DOI] [PubMed] [Google Scholar]
- Moore SJ, Lenglet A, Hill N. Field evaluation of three plant-based insect repellents against malaria vectors in Vaca Diez Province, the Bolivian Amazon. Journal of the American Mosquito Control Association. 2002 Jun;18(2):107–110. [PubMed] [Google Scholar]
- Neuhaus EM, Gisselmann G, Zhang W, et al. Odorant receptor heterodimerization in the olfactory system of Drosophila melanogaster. Nature Neuroscience. 2005;8:15–17. doi: 10.1038/nn1371. [DOI] [PubMed] [Google Scholar]
- Njiru BN, Mukabana WR, Takken W, Knols BG. Trapping of the malaria vector Anopheles gambiae with odour-baited MM-X traps in semi-field conditions in western Kenya. Malaria Journal. 2006;5:39. doi: 10.1186/1475-2875-5-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paluch G, Bartholomay L, Coats J. Mosquito repellents: a review of chemical structure diversity and olfaction. Pest Management Science. 2010 Sep;66(9):925–935. doi: 10.1002/ps.1974. [DOI] [PubMed] [Google Scholar]
- Pates HV, Takken W, Stuke K, Curtis CF. Differential behaviour of Anopheles gambiae sensu stricto (Diptera: Culicidae) to human and cow odours in the laboratory. Bulletin of Entomological Research. 2001a;91:289–296. doi: 10.1079/ber200198. [DOI] [PubMed] [Google Scholar]
- Pelletier J, Hughes DT, Luetje CW, et al. An odorant receptor from the southern house mosquito Culex pipiens quinquefasciatus sensitive to oviposition attractants. PLoS One. 2010;5:e10090. doi: 10.1371/journal.pone.0010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitts RJ, Zwiebel LJ. Antennal sensilla of two female Anopheline sibling species with differing host ranges. Malaria Journal. 2006;5:26. doi: 10.1186/1475-2875-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitts RJ*, Rinker DC*, Jones PL*, Rokas A, Zwiebel LJ. Transcriptome profiling of chemosensory appendages in the malaria vector Anopheles gambiae reveals tissue and sex-specific signatures of odor coding. BMC Genomics. 2011;12:271. doi: 10.1186/1471-2164-12-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitts RJ, Fox AN, Zwiebel LJ. A highly conserved candidate chemoreceptor expressed in both olfactory and gustatory tissues in the malaria vector Anopheles gambiae. Proceedings of the National Academy of Sciences U.S.A. 2004;101:5058–5063. doi: 10.1073/pnas.0308146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu YT, Smallegange RC, VAN Loon JJ, Takken W. Behavioural responses of Anopheles gambiae sensu stricto to components of human breath, sweat and urine depend on mixture composition and concentration. Medical and Veterinary Entomology. 2010 Nov 25; doi: 10.1111/j.1365-2915.2010.00924.x. [DOI] [PubMed] [Google Scholar]
- Robertson HM, Wanner KW. The chemoreceptor superfamily in the honey bee, Apis mellifera: expansion of the odorant, but not gustatory, receptor family. Genome Research. 2006;16:1395–1403. doi: 10.1101/gr.5057506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson HM, Warr CG, Carlson JR. Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster. Proceedings of the National Academy of Sciences U.S.A. 2003;100(Supplement 2):14537–14542. doi: 10.1073/pnas.2335847100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rützler M, Zwiebel LJ. Molecular biology of insect olfaction: recent progress and conceptual models. Journal of Comparative Physiology A. 2005;191(9):777–790. doi: 10.1007/s00359-005-0044-y. [DOI] [PubMed] [Google Scholar]
- Sakurai T, Nakagawa T, Mitsuno H, Mori H, et al. Identification and functional characterization of a sex pheromone receptor in the silkmoth Bombyx mori. Proceedings of the National Academy of Sciences U.S.A. 2004;101:16653–16658. doi: 10.1073/pnas.0407596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart R, Kiely A, Beale M, Vargas E, et al. Drosophila odorant receptors are novel seven transmembrane domain proteins that can signal independently of heterotrimeric G proteins. Insect Biochemistry and Molecular Biology. 2008 Aug;38(8):770–780. doi: 10.1016/j.ibmb.2008.05.002. 2008. [DOI] [PubMed] [Google Scholar]
- Sato K, Touhara K. Insect olfaction: receptors, signal transduction, and behavior. Results and Problems in Cell Differetiation. 2009;47:121–138. doi: 10.1007/400_2008_10. [DOI] [PubMed] [Google Scholar]
- Sato K, Pellegrino M, Nakagawa T, et al. Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature. 2008;452:1002–1006. doi: 10.1038/nature06850. [DOI] [PubMed] [Google Scholar]
- Smadja C, Shi P, Butlin RK, Robertson HM. Large gene family expansions and adaptive evolution for odorant and gustatory receptors in the pea aphid, Acyrthosiphon pisum. Molecular Biology. Evolution. 2009;26:2073–2086. doi: 10.1093/molbev/msp116. [DOI] [PubMed] [Google Scholar]
- Smallegange RC, Qiu YT, van Loon JJ, Takken W. Synergism between ammonia, lactic acid and carboxylic acids as kairomones in the host-seeking behaviour of the malaria mosquito Anopheles gambiae sensu stricto (Diptera: Culicidae) Chemical Senses. 2005;30:145–152. doi: 10.1093/chemse/bji010. [DOI] [PubMed] [Google Scholar]
- Smith CR, Smith CD, Robertson HM, et al. Draft genome of the red harvester ant Pogonomyrmex barbatus. Proceedings of the National Academy of Sciences U.S.A. 2011 Apr 5;108(14):5667–5672. doi: 10.1073/pnas.1007901108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed Z, Leal WS. Mosquitoes smell and avoid the insect repellent DEET. Proceedings of the National Academy of Sciences U.S.A. 2008 Sep 9;105(36):13598–13603. doi: 10.1073/pnas.0805312105. Epub 2008 Aug 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takken W, Knols BG. Odor-mediated behavior of Afrotropical malaria mosquitoes. Annual Review of Entomololgy. 1999;44:131–157. doi: 10.1146/annurev.ento.44.1.131. [DOI] [PubMed] [Google Scholar]
- Takken W, Knols BGJ, editors. Olfaction in Vector-Host Interactions: Ecology and Control of Vector Borne Diseases. Wageningen Academic; 2009. [Google Scholar]
- Tanga MC, Ngundu WI, Tchouassi PD. Daily survival and human blood index of major malaria vectors associated with oil palm cultivation in Cameroon and their role in malaria transmission. Tropical Medicine & International Health. 2011 Jan 19; doi: 10.1111/j.1365-3156.2011.02726.x. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Verhulst NO, Beijleveld H, Knols BG, et al. Cultured skin microbiota attracts malaria mosquitoes. Malaria Journal. 2009;8:302. doi: 10.1186/1475-2875-8-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosshall LB, Amrein H, Morozov PS, et al. A spatial map of olfactory receptor expression in the Drosophila antenna. Cell. 1999;96:725–736. doi: 10.1016/s0092-8674(00)80582-6. [DOI] [PubMed] [Google Scholar]
- Wang G, Carey AF, Carlson JR, et al. Molecular basis of odor coding in the malaria vector mosquito Anopheles gambiae. Proceedings of the National Academy of Sciences U.S.A. 2010;107:4418–4423. doi: 10.1073/pnas.0913392107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetzel CH, Behrendt H, Gisselmann G, et al. Functional expression and characterization of a Drosophila odorant receptor in a heterologous cell system. Proceedings of the National Academy of Sciences U.S.A. 2001;98:9377–9380. doi: 10.1073/pnas.151103998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicher D, Schafer R, Bauernfeind R, et al. Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide- activated cation channels. Nature. 2008;452:1007–1011. doi: 10.1038/nature06861. [DOI] [PubMed] [Google Scholar]
- Xia Y, Wang G, Buscariollo D, et al. The molecular basis of olfactory-based behavior in Anopheles gambiae larvae. Proceedings of the National Academy of Sciences U.S.A. 2008;105:6433–6438. doi: 10.1073/pnas.0801007105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, Zwiebel LJ. Identification and characterization of an odorant receptor from the West Nile virus mosquito, Culex quinquefasciatus. Insect Biochemistry and Mol ecular Biology. 2006;36:169–176. doi: 10.1016/j.ibmb.2005.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. Journal of Biomolecular Screening. 1999;4(2):67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- Zwiebel LJ, Takken W. Olfactory regulation of mosquito-host interactions. Insect Biochemistry and Molecular Biology. 2004;34:645–652. doi: 10.1016/j.ibmb.2004.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]