Abstract
The immense global burden of infectious disease outbreaks and the need to establish prediction and prevention systems have been recognized by the World Health Organization (WHO), the National Institutes of Health (NIH), the United States Agency of International Development (USAID), the Bill and Melinda Gates Foundation, and the international scientific community. Despite multiple efforts, this infectious burden is still increasing. For example, it has been reported that between 1.5 and 12 million people die each year from waterborne diseases and diarrheal diseases are listed within the top 15 leading causes of death worldwide. Rapid population growth, climate change, natural disasters, immigration, globalization, and the corresponding sanitation and waste management challenges are expected to intensify the problem in the years to come.
Introduction
The immense global burden of infectious disease outbreaks and the need to establish prediction and prevention systems have been recognized by the World Health Organization (WHO), the National Institutes of Health (NIH), the United States Agency of International Development (USAID), the Bill and Melinda Gates Foundation, and the international scientific community. Despite multiple efforts, this infectious burden is still increasing. For example, it has been reported that between 1.5 and 12 million people die each year from waterborne diseases [1, 2] and diarrheal diseases are listed within the top 15 leading causes of death worldwide [3]. Rapid population growth, climate change, natural disasters, immigration, globalization, and the corresponding sanitation and waste management challenges are expected to intensify the problem in the years to come.
Most infectious disease outbreaks in the United States have been related to microbial agents [4–7]. In the vast majority of cases, the infectious agents have not been identified. However, the Environmental Protection Agency (EPA) suggests that most outbreaks of unidentified etiology are caused by viruses [8]. Viruses have been cited as potentially the most important and hazardous pathogens found in wastewater [9] and are included in the EPA contaminant candidate list. Viruses can lead to serious health outcomes, especially for children, the elderly, and immunocompromised individuals, and are of great concern because of their low infectious dose, ability to mutate, inability to be treated by antibiotics, resistance to disinfection, small size that facilitates environmental transport, and high survivability in water and solids.
Infectious outbreaks can cause uncontrollable negative effects especially in dense urban areas. Traditional disease detection and management systems are based on diagnostic analyses of clinical samples. However, these systems fail to detect early warnings of public health threats at a wide population level and fail to predict outbreaks in a timely manner. Classic epidemiology observes disease outbreaks based on clinical symptoms and infection status but does not have the ability to predict “critical locations” and “critical moments” for viral disease onset. Recent research efforts in developing optimized detection systems focus on rapid methods for analyzing blood samples, but this approach assumes that patients are examined at a clinical setting after the outbreak has been established and recognized.
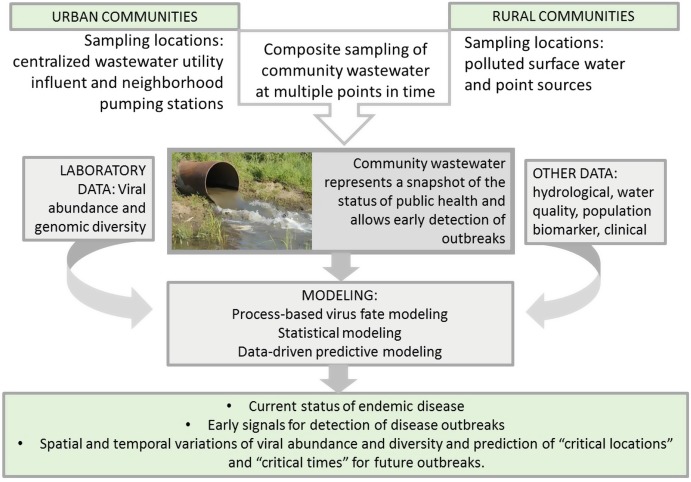
The central premise of the proposed approach is that community wastewater represents a snapshot of the status of public health. Wastewater analysis is equivalent to obtaining and analyzing a community-based urine and fecal sample. Monitoring temporal changes in virus concentration and diversity excreted in community wastewater, in combination with monitoring metabolites and biomarkers for population adjustments, allows early detection of outbreaks (critical moments for the onset of an outbreak). In addition, carefully designed spatial sampling will allow detection of locations where an outbreak may begin to develop and spread (critical locations for the onset of an outbreak) (Fig. 5.1).
Fig. 5.1.
Photomicrograph of adenovirus particles (left) and influenza virus particles (right). Adenovirus image from Dr. G. William Gary, Jr./CDC and influenza image from National Institute of Allergy and Infectious Diseases
Background
Similar detection systems have been used for the investigation of illicit drugs in various locations around the world [10–12]. The approach was first theorized in 2001 [10] and first implemented and reported for several illicit drugs in 2005 where the method was termed sewage epidemiology [11]. The methodology considers raw untreated wastewater as a reservoir of human excretion products; among these products are the parent compounds and metabolites of illicit drugs. If these excretion products are stable in wastewater as they travel through the sewage system, then the measured concentration from a wastewater treatment plant (WWTP) could correspond to the amount excreted by the serviced population. Table 5.1 presents a summary of prior studies utilizing the wastewater-based epidemiology methods to assess levels of various substances in a population.
Table 5.1.
Summary of substances investigated via wastewater-based epidemiology
| Substance | Country | References |
|---|---|---|
| Alcohol | Norway | [13] |
| Amphetamines | Australia, Belgium, Italy, Spain, South Korea, the United Kingdom, the United States | [14] |
| Cocaine | Australia, Belgium, Germany, Ireland, Italy, Spain, the United Kingdom, the United States | [14] |
| Counterfeit medicine | The Netherlands | [15] |
| Opiates | Germany, Italy, Spain, South Korea | [16] |
| Tobacco | Italy | [17] |
Any substance that is excreted by humans and is stable (or has known kinetic pathways) in wastewater can be back-calculated into an initial source concentration. An important step in the application of wastewater-based epidemiology is the estimation of the contributing population and its sampled wastewater. Both census and biomarker data can be used in this approach to estimate the number of individuals that contribute to the wastewater sample.
Occurrence of Viruses in Wastewater
Waterborne viruses comprise a significant component of wastewater microbiota and are known to be responsible for disease outbreaks. A critical characteristic of viruses is that they do not grow outside the host cells. Therefore, viral concentrations in the wastewater stream will represent the concentrations excreted by the corresponding human population. Table 5.2 summarizes studies that detected waterborne and non-waterborne viruses in wastewater and human excrement.
Table 5.2.
Summary of human viruses detected in wastewater or human excrement
| Virus | Detected in excrement | Detected in wastewater | Reported concentrations in wastewater (copies/L) | References |
|---|---|---|---|---|
| Adenoviruses | Yes | Yes | 6.0∗102 to 1.7∗108 | [21–36] |
| Astroviruses | Yes | Yes | 4.0∗104 to 4.1∗107 | [24, 29, 43, 44, 47, 50–54] |
| Enteroviruses | Yes | Yes | 6.9∗102 to 4.7∗106 | [24–26, 28, 29, 31, 33, 34, 36, 43, 53, 58–63] |
| Hepatitis A virus | Yes | Yes | 4.3∗103 to 8.9∗105 | [29, 30, 58, 67–72] |
| Hepatitis E virus | Yes | Yes | 7.8∗104 | [21, 34, 75–78] |
| Noroviruses | Yes | Yes | 4.9∗103 to 9.3∗106 | [24–26, 28–30, 32, 33, 43, 47, 53, 54, 59, 60, 79, 84–88] |
| Rotaviruses | Yes | Yes | 1.8∗103 to 8.7∗105 | [29–31, 36, 43–45, 47, 50, 53, 58, 59, 90–95] |
| Aichi virus | Yes | Yes | 9.7∗104 to 2.0∗106 | [36, 96, 114] |
| Polyomaviruses | Yes | Yes | 8.3∗101 to 5.7∗108 | [24, 35, 36, 85, 99, 115, 116] |
| Salivirus | Yes | Yes | 3.7∗105 to 9.7∗106 | [97, 114, 117] |
| Sapovirus | Yes | Yes | 1.0∗105 to 5.1∗105 | [24, 36, 98, 118] |
| Torque teno virus | Yes | Yes | 4.0∗104 to 5.0∗105 | [23, 24, 35, 119] |
| Coronaviruses | Yes | Yes | [120, 121] | |
| Influenza | Yes | Yes | [104–107, 122] | |
| Dengue virus | Yes | [111–113, 123] | ||
| West Nile virus | Yes | [109, 110, 124] | ||
| Zika virus | Yes | [108, 125] | ||
| Yellow fever virus | Yes | [126, 127] |
Note: The primary method of laboratory detection in the studies presented in this table is polymerase chain reaction (PCR), as well as real-time quantitative PCR (qPCR). PCR uses specific primers to replicate target sequences of nucleic acids; designing a primer to replicate a specific sequence in a given viral genome allows for the detection of that particular virus. qPCR can also determine the concentration of a virus in a sample by quantifying the number of copies of the target sequence
Waterborne Viruses
There are several groups of commonly detected and studied waterborne viruses, including adenoviruses, astroviruses, enteroviruses, hepatitis A and E viruses, noroviruses, and rotaviruses. Adenoviruses are known to cause gastroenteritis and respiratory disease [18] and have been linked to outbreaks of disease [19, 20]. Adenoviruses are a commonly studied group of viruses in water. They are commonly detected in raw wastewater [21–36] and have been cited as among the most significantly abundant human viruses in wastewater [24, 27, 28, 33, 37]. Adenoviruses have also been detected in human excrement of infected persons, including both feces and urine [38–47]. Studies have found the concentration of adenovirus in the stool of infected persons to range from 102 to 1011 copies per gram with an average concentration in the range from 105 to 106 copies per gram of stool [39, 41, 42, 46] as quantified by qPCR.
Astroviruses are a group of RNA viruses that have been linked to outbreaks of gastroenteritis [19, 48]. They have been cited as one of the more important viruses associated with gastroenteritis [49], but they have not been as commonly studied in wastewater compared to other groups of human enteric viruses. Nonetheless, they have been detected using standard PCR in wastewater in prior studies [24, 29, 50, 51]. They have also been detected in clinical samples of human excrement of infected people [43, 44, 47, 52–54], making them a viable candidate for wastewater epidemiology. While qPCR has been used as a detection method for astroviruses in human feces [44, 47, 54], and for quantification purposes in wastewater [51], no cited studies have reported quantitative values for astroviruses in human excrement.
Enteroviruses comprise several types of human enteric viruses, including polioviruses, coxsackieviruses, and echoviruses [55, 56]. Enteroviruses can cause an array of afflictions depending on type, including common cold, meningitis, and poliomyelitis [57], and have been linked to outbreaks of these diseases [19]. Enteroviruses have been detected via PCR in raw wastewater by numerous studies [25, 26, 28, 29, 31, 33, 34, 58, 59], as well as detected in human feces [43, 53, 60–63]. qPCR has not as yet been extensively employed to quantify enteroviruses in stool samples, though one study determined the enterovirus load to be in the range of 1.4∗104 to 6.6∗109 copies per gram of stool [60].
Two species of hepatitis viruses, hepatitis A virus and hepatitis E virus, are considered to be waterborne viruses. Hepatitis is a liver disease that can cause numerous afflictions, including fever, nausea, and jaundice [64]. Hepatitis A virus has been linked to disease outbreaks [65], and it has been suggested that even low levels of viral water pollution can produce infection [66]. Hepatitis A virus is often detected via PCR in raw wastewater [29, 30, 58, 67, 68] and several studies have also detected the virus in human stool samples [69–72]. Like enteroviruses, there has not been significant investigation into the quantification of hepatitis A virus in stool, though one study reported values in the range of 3.6∗105 to 5.6∗109 copies per gram of stool [70].
Hepatitis E virus , meanwhile, has only recently begun to become a pathogen of interest compared to other waterborne human viruses [73]. Like hepatitis A, hepatitis E virus can cause liver disease with many of the same symptoms; in fact, hepatitis E is not clinically distinguishable from other types of viral hepatitis infection [74]. While not investigated to the extent of other human enteric viruses, hepatitis E virus has been detected via PCR in raw wastewater [21, 34, 75]. There have also been studies that have detected hepatitis E virus in human stool samples [76–78]. One such study also used RT-qPCR to quantify the concentration of hepatitis E virus in stool and reported values in the range of 101 to 106 copies per μL of stool [77].
Noroviruses , also known as Norwalk-like viruses, are a genus of viruses within the Caliciviridae family. They are one of the more significant gastroenteritis-causing viral agents, considered to be a leading cause of the disease [79–81], and are commonly associated with disease outbreaks [19, 82, 83]. Noroviruses are one of the more commonly investigated and detected viruses in wastewater [24–26, 28–30, 32, 33, 36, 59, 60, 84, 85]. A number of studies have also investigated the presence of noroviruses in human feces [43, 47, 53, 54, 79, 86–88]. One such cited study reported quantification values for norovirus in stool following qPCR, in the range of 9.7∗105 to 1.1∗1012 copies per gram, with a mean value of approximately 1011 copies per gram [87].
Rotaviruses are another primary cause of gastroenteritis with symptoms including diarrhea, vomiting, and fever, in accordance with other enteric viruses [89]. They are commonly detected via PCR in raw wastewater [29–31, 36, 50, 58, 59, 90–92] and are commonly investigated and detected in human feces [43–45, 47, 53, 93–95]. Like other waterborne viruses, though, only a handful of studies on rotaviruses have used qPCR as a detection tool, and none reported quantification values in terms of the number of copies.
In addition to the commonly investigated waterborne viruses described above, there are other human viruses that are commonly detected in wastewater and human stool but not as frequently studied, such as Aichi virus, polyomaviruses, salivirus, sapovirus, and torque teno virus. Aichi virus is a member of the Picornaviridae family, the same family as enteroviruses, and is believed to cause gastroenteritis [96]. Salivirus, another member of the Picornaviridae family, is also associated with gastroenteritis, as well as acute flaccid paralysis [97]. Sapovirus, like norovirus, is a member of the Caliciviridae family and like its relative is a common cause of gastroenteritis [98]. Polyomaviruses are associated with a variety of diseases in humans, including nephropathy, progressive multifocal leukoencephalopathy, and Mercel cell carcinoma [99]. Torque teno virus is commonly detected in humans, but the clinical consequences of infection are unclear [100]. These viruses are included in Table 5.2.
Non-waterborne Viruses
Non-waterborne viruses have also been detected in wastewater or human excrement (included in Table 5.2). While it is logical to investigate the applicability of waterborne viruses to wastewater-based epidemiology, it is also important to note the potential for other categories of viruses to fit into this methodology.
There exists a category of water-related viruses that are transmitted via insects (like mosquitos) that breed in water, such as Zika virus, West Nile virus, Rift Valley fever virus, yellow fever virus, dengue virus, and chikungunya virus, in addition to confirmed waterborne viruses. These viruses also fall into the category of zoonotic viruses, which are viruses that can be transmitted between humans and animals. Other zoonotic viruses include avian influenza virus, SARS (Severe Acute Respiratory Syndrome) coronavirus, Menangle virus, Tioman virus, Hendra virus, Australian bat lyssavirus, Nipah virus, and hantavirus. Specific animal species of concern that are vectors for these zoonotic viruses include avian species, bats, rodents, and mosquitos. While these zoonotic viruses are not classified as waterborne, they are associated with potential waterborne transmission, such as exposure to aerosolized wastewater, which can occur when wastewater undergoes turbulence, such as in flush toilets, converging sewer pipes, and aeration basins [101, 102] as well as irrigation and land application systems.
It has been shown that coronaviruses have been detected in wastewater [103] and SARS coronaviruses have been detected in stool and urine samples. Furthermore, detection in both human stool and urine [104–106] as well as wastewater [107] has been reported for influenza. Detection in urine has been reported for the mosquito-associated Zika virus [108], West Nile virus [109, 110], dengue virus [111, 112], and yellow fever virus [113]. These observations indicate that the concept of wastewater-based epidemiology could be applied to a wide range of viruses beyond the confirmed waterborne viruses.
Variations of Viruses in Wastewater
The quantity of human enteric viruses in wastewater has been shown to have seasonal variation, indicating that infection resulting from these viruses is more prevalent at certain times of the year. A study conducted in Japan by Katayama et al. (2008) found that norovirus concentrations in wastewater were highest during the months of November through April [26], while enterovirus and adenovirus concentrations were largely consistent throughout the year. A 9-year study in Milwaukee, Wisconsin, by Sedmak et al. (2005) found that concentrations of reoviruses, enteroviruses, and adenoviruses were highest during the months of July through December. This study also analyzed clinical specimens of enterovirus isolates and found the incidence of clinical enterovirus infection corresponded to the concentration of these viruses in wastewater during the same time periods [31]. Another study in Beijing, China, by Li et al. (2011) found that rotavirus concentrations were highest during the months of November through March [90] and that these findings also corresponded with clinical rotavirus data reported in China [128].
Additionally, variation in viral concentration in wastewater can occur on a smaller timescale. For example, tourist locations could experience higher wastewater loads, and consequently higher viral concentrations, on weekends where there is an influx of population. For example, Xagoraraki’s research group conducted a study which observed an increase in adenovirus concentration in wastewater following the July 4th holiday in Traverse City, Michigan, a popular vacation destination [27]. Likewise, urban centers may experience higher loads during the day on weekdays, while people are at work. Accounting for these population changes would be vital for understanding when viral outbreaks occur.
Wastewater has been used in the past as a tool to investigate viruses for other purposes as well, such as spatial surveillance and evaluation of immunization efficacy. Two particular studies were able to use wastewater to observe the spatial variation of particular viral strains; Bofill-Mas et al. observed that particular strains of polyomavirus were endemic to specific regions, while Clemente-Cesares et al. detected Hepatitis E virus in areas previously considered non-endemic for the virus [129, 130]. Lago et al. (2003) investigated the efficacy of a poliovirus (a type of enterovirus) immunization campaign in Havana, Cuba, by quantifying concentrations of the virus in wastewater [61]. Poliovirus was detected in 100% of wastewater samples prior to the start of the immunization campaign and dropped to a 0% detection rate in wastewater 15 weeks after the campaign, indicating the usefulness of wastewater surveillance. A study by Carducci et al. (2006) investigated the relationship between wastewater samples and clinical samples and found that the same viral strains could sometimes be detected between the two sets of samples [131].
Proposed Methodology
Waterborne and non-waterborne viruses have been detected in wastewater, variations of concentrations in time have been observed, and virus presence in wastewater has on occasion been correlated with occurrence of clinical disease. However, wastewater-based epidemiology methods have not yet been applied to assess and predict viral disease outbreaks in a systematic way. Wastewater-based epidemiology has the potential to predict “critical locations” and “critical moments” for viral disease onset. Designing spatial and temporal sampling appropriate to the area of concern, as well as modeling the fate of viruses, is critical for the effectiveness of the proposed method. This methodology is summarized in Fig. 5.2. In the following sections, critical factors for implementation are discussed.
Fig. 5.2.
Summary of the proposed wastewater-based epidemiology methodology
Sampling in Urban and Rural Locations
The most critical parameter for the effective application of wastewater-based epidemiology is the selection of a surveillance program, including spatial and temporal sampling. Considerations must be made in the differences between urban and rural wastewater systems. Urban sewage systems offer a convenient confluence of wastewater in the serviced population, as all wastewater will ultimately flow to a WWTP, providing a sampling point representing the entire community. Additionally, localized sampling can be performed in specific neighborhoods where access points are available. By surveying both the combined wastewater at the treatment plant and the localized samples from neighborhoods, viral outbreaks can be traced to a more specific location and the urban areas of concern can be identified. Xagoraraki’s research group is currently conducting an National Science Foundation-funded study of this nature in the city of Detroit, sampling at several interceptors at the Detroit wastewater treatment plant, as well as sampling from sewer lines in residential areas throughout the city.
More rural or underdeveloped areas that do not have sewage collection systems pose sampling problems. In these areas, wastewater is often disposed in open space, latrines, or septic tanks. As a result, for wastewater-based epidemiology sampling to be effectively applied to these areas, disposal, fate, and transport of wastewater in the environment must be taken into account. Watershed modeling would therefore become an integral component of the wastewater-based epidemiology methodology for rural locations. In a study performed by Xagoraraki’s research group, preliminary investigation into the wastewater epidemiology methodology was conducted [132]. Samples were collected from a wastewater treatment plant and surrounding surface waters in Kampala, Uganda. Three sampling events were conducted in 2-week intervals. Four human viruses (adenovirus, enterovirus, hepatitis A virus, and rotavirus) were quantified at each sampling location via qPCR. Concentrations of each virus at each location from each sampling event were compared to one another to determine if significant differences could be observed from one sampling event to the next. Results indicated that statistically significant differences in viral concentration were observed for the measured viruses at several sampling locations.
The selection of the sampling times and locations is of paramount importance to the methodology, regardless of whether sampling takes place in urban or rural areas. Sampling should be based upon expected critical pathways of viral transport and transmission. These critical pathways include environmental reservoirs for viruses and the timing and locations where viruses are most easily transported and transmitted between humans and the environment. By determining sampling times and locations based upon critical pathways, “critical locations,” and “critical moments,” areas and times most impactful to the spread of viral disease would be most readily and effectively identified.
Quantification of Viruses
Quantitative data of viruses of concern, such as those obtained with qPCR, are critical for the proposed methodology, as peaks in viral concentrations will indicate potential onset of disease outbreaks. While detection in human excrement or raw wastewater has not been reported for all viruses, it is possible that they have simply not been investigated in this context, as detection of viruses via conventional methods (cell culture, PCR, qPCR) is specific to the virus being investigated. Thus, while qPCR is important to detect and quantify common waterborne viruses, next-generation sequencing and metagenomic methods could also be performed to screen for the presence of other viruses. If genomic sequences of viruses of concern are found, then quantification with qPCR can follow.
Metagenomic methods have been applied to investigate viruses in wastewater and have been found to produce more conservative results of viral detection compared to conventional methods; viruses detected with metagenomic methods are typically also detected with conventional methods, whereas viruses detected via qPCR may not be detected with metagenomic methods. These metagenomic methods, however, can detect the presence of viruses not commonly quantified using qPCR [37, 133–136]. Xagoraraki’s research group’s studies have used metagenomic methods to identify human viruses of potential concern in wastewater. The first of these studies, conducted with samples from both Michigan and France, detected a comparatively high number of metagenomic hits for human herpesviruses and also detected human parvovirus and human polyomavirus in wastewater effluents [37]. Their other study, conducted in Uganda, detected human astroviruses, papillomaviruses, as well as a BLAST (Basic Local Alignment Search Tool) hit for Ebola virus [132]. While more research is still required to attain more robust genomic information and comparison databases, metagenomic methods can still be a useful tool for the identification of potential viruses that can then be monitored with qPCR methods. Table 5.3 presents a summary of studies that have used metagenomic methods to detect human viruses in wastewater and human excrement.
Table 5.3.
Summary of studies using metagenomic methods to detect viral sequences in wastewater and human excrement
| Detected in | Virus | References |
|---|---|---|
| Wastewater | Adenovirus, enterovirus, polyomavirus, papillomavirus | [135] |
| Adenovirus, Aichi virus, coronavirus, herpesvirus, torque teno virus | [137] | |
| Adenovirus, Aichi virus, astrovirus, coronavirus, enterovirus, herpesvirus, papillomavirus, parechovirus, parvovirus, rotavirus, salivirus, sapovirus, torque teno virus | [133] | |
| Adenovirus, Aichi virus, astrovirus, norovirus, papillomavirus, parechovirus, polyomavirus, salivirus, sapovirus | [136] | |
| Adenovirus, herpesvirus, parvovirus, polyomavirus | [37] | |
| Adenovirus, astrovirus, Ebola virus, enterovirus, papillomavirus, rotavirus, torque teno virus | [132] | |
| Human excrement | Adenovirus, astrovirus, enterovirus, norovirus, parvovirus, rotavirus, torque teno virus | [138] |
| Adenovirus, Aichi virus, enterovirus, parechovirus, rotavirus | [139] |
Population Normalization
Population normalization is also a critical factor for the application of wastewater-based epidemiology. Proper quantification of biomarkers in wastewater would allow for an appropriate estimation of serviced population via statistical modeling, which would provide context to measured viral concentrations and ensure that differences in viral concentration could not be attributed to changes in population. When observed viral concentrations are significantly high relative to the estimated population, a viral outbreak could be indicated.
Quantification of biomarkers (substances naturally excreted by humans) in wastewater can be used as a method of estimating population in an area. Governmental census information has been found to underestimate the population of a community compared to estimation using biomarkers [140], and certain substances detected in wastewater have been shown to correlate with census data [141]. Several substances have been proposed and investigated as population biomarkers (Table 5.4), including creatinine [142], cholesterol, coprostanol [143], nicotine [144], cortisol, androstenedione, and the serotonin metabolite 5-hydroxyindoleacetic acid (5-HIAA) [145]. Nutrients such as nitrogen, phosphorus, and oxygen [12], as well as ammonium [146], have also been proposed as population biomarkers, but these may more adequately reflect human activity and industry footprint rather than population [145, 147, 148].
Table 5.4.
Summary of biomarkers proposed for population adjustment
| Biomarker | Description | Excreted in | References |
|---|---|---|---|
| 5-HIAA | Metabolite of serotonin | Urine | [145] |
| Ammonium | Form of ammonia found in water | Urine | [146] |
| Androstenedione | Sex hormone precursor | Urine | [149] |
| Atenolol | Drug (beta blocker) used to treat hypertension | Urine | [140] |
| Cholesterol | Lipid molecule, key component of cell membranes | Feces | [143] |
| Coprostanol | Metabolite of cholesterol | Feces | [143] |
| Cortisol | Steroid hormone produced by adrenal glands | Urine | [150] |
| Cotinine | Metabolite of nicotine | Urine | [145] |
| Creatinine | Metabolite of creatine phosphate in muscle | Urine | [142] |
| Nicotine | Stimulant found in tobacco | Urine | [144] |
| Nutrients (N, P, BOD) | Water-quality parameters | n/a | [12] |
Estimation of Shedding Rates
The shedding rate (the rate with which viruses are released from the body in excrement) for each waterborne virus group encompasses a wide range, from 102 copies per gram at minimum to 1012 copies per gram at maximum. This variability is summarized for selected viruses in Table 5.5. For example, mean concentration values of adenoviruses in excrement ranged from 104 to 106 depending on the study and whether the virus is excreted in stool or urine, indicating a wide data variance [39, 41]. Many factors can impact the shedding rate of viruses in excrement, including viremia (the presence of the virus in the bloodstream) [40, 87, 151]. The duration of the presentation of a particular disease can also impact the shedding rate [105, 121].
Table 5.5.
Summary of reported shedding rates for viruses
| Virus | Range of shedding rate, copies/g stool | References |
|---|---|---|
| Adenoviruses | 1.0∗102 to 1.0∗1011 | [39, 41, 42, 46] |
| Enteroviruses | 1.4∗104 to 6.6∗109 | [60] |
| Hepatitis A virus | 3.6∗105 to 1.0∗1011 | [70] |
| Hepatitis E virus | 1.0∗101 to 1.0∗106 | [77] |
| Noroviruses | 1.1∗105 to 1.1∗1012 | [87] |
| Sapoviruses | 1.3∗105 to 2.5∗1011 | [98, 118] |
Transport of Viruses in the Environment
Waterborne viruses survive well in water, but all viruses are susceptible to natural degradation determined by factors such as temperature, exposure to UV light, and the microbial community [152, 153]. The kinetic decay rate of a virus would thereby be primarily dependent not only on the characteristics of the individual virus but also environmental conditions within the sewage system, which could vary from location to location. Moreover, the fate of viruses may be different between wastewater systems in urban areas which typically use enclosed underground sewer pipes and rural areas which may utilize septic tanks, catchments, and the open environment. Viruses can also adsorb to or be enveloped by particulate matter in wastewater which would lead to confounding factors in measurement of these viruses.
Correlation with Public Health Records and Unidentified Clinical Data
Comparison with clinical data is another key component of these methods. Correlations between measured viral concentrations in wastewater and reported clinical cases of disease could be established, strengthening the proposed methodology. The establishment of these correlations can serve as a validation for a prediction model that accounts for the factors discussed above, providing evidence for the notion that changes of viral concentrations in wastewater will indicate changes in viral disease cases in humans. Moreover, should preventative public health measures be implemented after the identification of an outbreak, the tracking of clinical data could provide a quantifiable indicator of the efficacy of these preventative measures.
Conclusions
Infectious viral outbreaks can cause uncontrollable negative effects especially in densely populated areas. Early detection is critical for effective management and prevention of outbreaks. Recent research efforts in developing optimized detection systems often focus on rapid methods for analyzing blood or excrement samples; however, these approaches require that individuals are examined in clinical settings, typically after an outbreak has been established. Wastewater-based epidemiology is a promising methodology for early detection of viral outbreaks at a population level. Analyzing wastewater is equivalent to obtaining and analyzing a community excrement sample. In the determination of whether an outbreak is imminent or already in progress, quantifying viral concentration in raw wastewater is a crucial first step in this process. Waterborne viruses appear to be prime candidates, as they are detectable and quantifiable in both wastewater and human excrement. Non-waterborne viruses have been shown to be detected in human excrement, and some have been reported to be detected in wastewater. Wastewater-based epidemiology therefore has the potential to expand beyond waterborne viruses.
Routine monitoring for temporal changes in virus concentration and diversity in community wastewater, in combination with monitoring metabolites and biomarkers for population adjustments, allows early detection of outbreaks (critical moments for the onset of an outbreak). In addition, carefully designed spatial sampling of wastewater will allow detection of locations where an outbreak may begin to develop and spread (critical locations for the onset of an outbreak). Considerations in sampling locations must be taken with regard to the area of investigation, as urban and rural areas may have differences in the respective wastewater systems that can affect viral transport in the water environment. Moreover, to obtain an accurate estimation of disease cases in a population, other factors must be considered such as viral shedding rates, environmental transport and degradation rates, and correlation with reported clinical disease data. Ultimately, there is great opportunity for the use of wastewater-based epidemiology to investigate viral outbreaks within a community. Comprehensive application of the various factors discussed above is crucial for the full potential of this methodology to be realized. Further research could clarify many of these issues and allow for the full development and application of this new epidemiological technique for studying, identifying, and predicting viral outbreaks.
Biographies
Irene Xagoraraki
is an Associate Professor of Environmental Engineering in the Department of Civil and Environmental Engineering at Michigan State University. Her bachelor’s degree (1993) in environmental science is from the University of the Aegean in Greece. Her bachelor’s thesis focused on landfill biogas production and recovery, under the guidance of Prof. Halvadakis. She earned her MS (1995) and PhD (2001) degrees in Civil and Environmental Engineering from the University of Wisconsin-Madison. Her MS degree, under the guidance of Prof. Berthouex, focused on pumping station reliability analysis for the Madison metropolitan sewerage district. Her PhD advisor was Prof. Harrington, and her dissertation focused on coagulation and sedimentation of Cryptosporidium parvum in water systems. Between 2001 and 2005, she held a postdoctoral position at the University of Wisconsin-Madison where she worked on multiple pathogen removal research projects for drinking water treatment systems. She joined the faculty of Michigan State University in 2006. The focus of her teaching is on environmental engineering, water quality and public health.
Dr. Xagoraraki’s research program at Michigan State University is focused on water-quality engineering, emphasizing protection of public health and prevention of waterborne disease. In particular, she is interested in microbial contaminants (such as human viruses, zoonotic viruses, antibiotic-resistant bacteria) and their detection, occurrence, fate, removal, inactivation, and associated risk, in water systems. Her research projects are funded by National Science Foundation, US Environmental Protection Agency, US Geological Survey, US Department of Homeland Security, Water Research Foundation, and Water Environmental Research Foundation. Her latest research project, funded by the National Science Foundation, explores wastewater-based epidemiology methods for early detection and prediction of water-related viral outbreaks. The basic principles of this methodology are described in the article presented in this book. Dr. Xagoraraki is also involved in multiple international projects in Uganda, Mexico, France, and the Republic of Georgia. Dr. Xagoraraki loves to be surrounded by water. She enjoys the beautiful shores of the Great Lakes in Michigan, but to find inspiration and reconnect with her origins, she visits the incredible Aegean Sea in the island of Crete, Greece, where she was born and raised.
Evan O’Brien
earned his PhD in Environmental Engineering at Michigan State University in 2018. He earned his MS degree (2014) in Environmental Engineering from Michigan State University and his BS degree (2011) in Nuclear Engineering with a minor in Environmental Engineering from The Pennsylvania State University. His research work involves the investigation and characterization of viral diversity in wastewater and surface waters. His first project utilized next-generation sequencing and metagenomic analyses to compare viral diversity in wastewater effluents between conventional activated sludge and membrane bioreactor wastewater treatment plants. His subsequent project employed these methods in addition to RT-qPCR to investigate viral abundance and diversity in wastewater and its impact on surface water in Uganda. His interest in the field of water resources arises from a lifelong appreciation of wilderness and the outdoors, cultivated while working as a camp counselor in northern Wisconsin. He hopes that his future career will work to ensure public and environmental health, so that these natural resources can be enjoyed for generations to come.
Contributor Information
Deborah Jean O’Bannon, Phone: +118162351287, Email: obannond@umkc.edu.
Irene Xagoraraki, Email: xagorara@egr.msu.edu.
References
- 1.Gleick PH. Dirty-water: estimated deaths from water-related diseases 2000–2020. Oakland, CA: Pacific Institute for Studies in Development, Environment, and Security; 2002. [Google Scholar]
- 2.Prüss-Üstün A, Bos R, Gore F, Bartram J, World Health Organization. 2008. Safer water, better health: costs, benefits and sustainability of interventions to protect and promote health. World Health Organization, Geneva.
- 3.Mathers CD, Loncar D. Projections of global mortality and burden of disease from 2002 to 2030. PLoS Med. 2006;3:e442. doi: 10.1371/journal.pmed.0030442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barwick RS, Levy DA, Craun GF, Beach MJ, Calderon RL. Surveillance for waterborne-disease outbreaks—United States, 1997–1998. MMWR CDC Surveill Summ. 2000;49:1–21. [PubMed] [Google Scholar]
- 5.Kramer MH, Herwaldt BL, Craun GF, Calderon RL, Juranek DD (1996) Surveillance for waterborne-disease outbreaks–United States, 1993–1994. MMWR CDC Surveill Summ 45:1–33 [PubMed]
- 6.Levy DA, Bens MS, Craun GF, Calderon RL, Herwaldt BL (1998) Surveillance for waterborne-disease outbreaks—United States, 1995–1996. MMWR CDC Surveill Summ: 47:1–34 [PubMed]
- 7.Liang JL, Dziuban EJ, Craun GF, Hill V, Moore MR, Gelting RJ, Calderon RL, Beach MJ, Roy SL. Surveillance for waterborne disease and outbreaks associated with drinking water and water not intended for drinking – United States, 2003–2004. Morb Mortal Wkly Rep. 2006;55:31–65. [PubMed] [Google Scholar]
- 8.US EPA National primary drinking water regulations: ground water rule; final rule. Fed Regist. 2006;71:65574–65660. [PubMed] [Google Scholar]
- 9.Toze S. Microbial pathogens in wastewater: literature review for urban water systems multi-divisional research program. Floreat Park: CSIRO Land and Water; 1997. [Google Scholar]
- 10.Daughton CG, Jones-Lepp TL. Pharmaceuticals and personal care products in the environment: scientific and regulatory issues. Washington, DC: American Chemical Society; 2001. [Google Scholar]
- 11.Zuccato E, Chiabrando C, Castiglioni S, Bagnati R, Fanelli R. Estimating community drug abuse by wastewater analysis. Environ Health Perspect. 2008;116:1027–1032. doi: 10.1289/ehp.11022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Nuijs ALN, Castiglioni S, Tarcomnicu I, Postigo C, de Alda ML, Neels H, Zuccato E, Barcelo D, Covaci A. Illicit drug consumption estimations derived from wastewater analysis: a critical review. Sci Total Environ. 2011;409:3564–3577. doi: 10.1016/j.scitotenv.2010.05.030. [DOI] [PubMed] [Google Scholar]
- 13.Reid MJ, Langford KH, Mørland J, Thomas KV. Analysis and interpretation of specific ethanol metabolites, ethyl sulfate, and ethyl glucuronide in sewage effluent for the quantitative measurement of regional alcohol consumption. Alcohol Clin Exp Res. 2011;35:1593–1599. doi: 10.1111/j.1530-0277.2011.01505.x. [DOI] [PubMed] [Google Scholar]
- 14.Irvine RJ, Kostakis C, Felgate PD, Jaehne EJ, Chen C, White JM. Population drug use in Australia: a wastewater analysis. Forensic Sci Int. 2011;210:69–73. doi: 10.1016/j.forsciint.2011.01.037. [DOI] [PubMed] [Google Scholar]
- 15.Venhuis BJ, Voogt P, Emke E, Causanilles A, Keizers PHJ. Success of rogue online pharmacies: sewage study of sildenafil in the Netherlands. BMJ. 2014;349:g4317. doi: 10.1136/bmj.g4317. [DOI] [PubMed] [Google Scholar]
- 16.Hummel D, Löffler D, Fink G, Ternes TA. Simultaneous determination of psychoactive drugs and their metabolites in aqueous matrices by liquid chromatography mass spectrometry. Environ Sci Technol. 2006;40:7321–7328. doi: 10.1021/es061740w. [DOI] [PubMed] [Google Scholar]
- 17.Castiglioni S, Senta I, Borsotti A, Davoli E, Zuccato E. A novel approach for monitoring tobacco use in local communities by wastewater analysis. Tob Control. 2015;24:38–42. doi: 10.1136/tobaccocontrol-2014-051553. [DOI] [PubMed] [Google Scholar]
- 18.Ganesh A, Lin J. Waterborne human pathogenic viruses of public health concern. Int J Environ Health Res. 2013;23:544–564. doi: 10.1080/09603123.2013.769205. [DOI] [PubMed] [Google Scholar]
- 19.Maunula L, Klemola P, Kauppinen A, Söderberg K, Nguyen T, Pitkänen T, Kaijalainen S, Simonen ML, Miettinen IT, Lappalainen M, Laine J, Vuento R, Kuusi M, Roivainen M. Enteric viruses in a large waterborne outbreak of acute gastroenteritis in finland. Food Environ Virol. 2008;1:31–36. doi: 10.1007/s12560-008-9004-3. [DOI] [Google Scholar]
- 20.Papapetropoulou M, Vantarakis AC. Detection of adenovirus outbreak at a municipal swimming pool by nested PCR amplification. J Infect. 1998;36:101–103. doi: 10.1016/S0163-4453(98)93414-4. [DOI] [PubMed] [Google Scholar]
- 21.Bofill-Mas S, Albinana-Gimenez N, Clemente-Casares P, Hundesa A, Rodriguez-Manzano J, Allard A, Calvo M, Girones R. Quantification and stability of human adenoviruses and polyomavirus JCPyV in wastewater matrices. Appl Environ Microbiol. 2006;72:7894–7896. doi: 10.1128/AEM.00965-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bofill-Mas S, Rodriguez-Manzano J, Calgua B, Carratala A, Girones R. Newly described human polyomaviruses Merkel Cell, KI and WU are present in urban sewage and may represent potential environmental contaminants. Virol J. 2010;7:141. doi: 10.1186/1743-422X-7-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carducci A, Morici P, Pizzi F, Battistini R, Rovini E, Verani M. Study of the viral removal efficiency in a urban wastewater treatment plant. Water Sci Technol. 2008;58:893–897. doi: 10.2166/wst.2008.437. [DOI] [PubMed] [Google Scholar]
- 24.Hata A, Kitajima M, Katayama H. Occurrence and reduction of human viruses, F-specific RNA coliphage genogroups and microbial indicators at a full-scale wastewater treatment plant in Japan. J Appl Microbiol. 2013;114:545–554. doi: 10.1111/jam.12051. [DOI] [PubMed] [Google Scholar]
- 25.Hewitt J, Leonard M, Greening GE, Lewis GD. Influence of wastewater treatment process and the population size on human virus profiles in wastewater. Water Res. 2011;45:6267–6276. doi: 10.1016/j.watres.2011.09.029. [DOI] [PubMed] [Google Scholar]
- 26.Katayama H, Haramoto E, Oguma K, Yamashita H, Tajima A, Nakajima H, Ohgaki S. One-year monthly quantitative survey of noroviruses, enteroviruses, and adenoviruses in wastewater collected from six plants in Japan. Water Res. 2008;42:1441–1448. doi: 10.1016/j.watres.2007.10.029. [DOI] [PubMed] [Google Scholar]
- 27.Kuo DH-W, Simmons FJ, Blair S, Hart E, Rose JB, Xagoraraki I. Assessment of human adenovirus removal in a full-scale membrane bioreactor treating municipal wastewater. Water Res. 2010;44:1520–1530. doi: 10.1016/j.watres.2009.10.039. [DOI] [PubMed] [Google Scholar]
- 28.La Rosa G, Pourshaban M, Iaconelli M, Muscillo M. Quantitative real-time PCR of enteric viruses in influent and effluent samples from wastewater treatment plants in Italy. Ann Ist Super Sanità. 2010;46:266–273. doi: 10.4415/ANN_10_03_07. [DOI] [PubMed] [Google Scholar]
- 29.Petrinca AR, Donia D, Pierangeli A, Gabrieli R, Degener AM, Bonanni E, Diaco L, Cecchini G, Anastasi P, Divizia M. Presence and environmental circulation of enteric viruses in three different wastewater treatment plants. J Appl Microbiol. 2009;106:1608–1617. doi: 10.1111/j.1365-2672.2008.04128.x. [DOI] [PubMed] [Google Scholar]
- 30.Prado T, Silva DM, Guilayn WC, Rose TL, Gaspar AMC, Miagostovich MP. Quantification and molecular characterization of enteric viruses detected in effluents from two hospital wastewater treatment plants. Water Res. 2011;45:1287–1297. doi: 10.1016/j.watres.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 31.Sedmak G, Bina D, MacDonald J, Couillard L. Nine-year study of the occurrence of culturable viruses in source water for two drinking water treatment plants and the influent and effluent of a wastewater treatment plant in Milwaukee, Wisconsin (August 1994 through July 2003) Appl Environ Microbiol. 2005;71:1042–1050. doi: 10.1128/AEM.71.2.1042-1050.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sima LC, Schaeffer J, Saux J-CL, Parnaudeau S, Elimelech M, Guyader FSL. Calicivirus removal in a membrane bioreactor wastewater treatment plant. Appl Environ Microbiol. 2011;77:5170–5177. doi: 10.1128/AEM.00583-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simmons FJ, Kuo DH-W, Xagoraraki I. Removal of human enteric viruses by a full-scale membrane bioreactor during municipal wastewater processing. Water Res. 2011;45:2739–2750. doi: 10.1016/j.watres.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 34.Masclaux FG, Hotz P, Friedli D, Savova-Bianchi D, Oppliger A. High occurrence of hepatitis E virus in samples from wastewater treatment plants in Switzerland and comparison with other enteric viruses. Water Res. 2013;47:5101–5109. doi: 10.1016/j.watres.2013.05.050. [DOI] [PubMed] [Google Scholar]
- 35.Hamza IA, Jurzik L, Überla K, Wilhelm M. Evaluation of pepper mild mottle virus, human picobirnavirus and Torque teno virus as indicators of fecal contamination in river water. Water Res. 2011;45:1358–1368. doi: 10.1016/j.watres.2010.10.021. [DOI] [PubMed] [Google Scholar]
- 36.Kitajima M, Iker BC, Pepper IL, Gerba CP. Relative abundance and treatment reduction of viruses during wastewater treatment processes—Identification of potential viral indicators. Sci Total Environ. 2014;488–489:290–296. doi: 10.1016/j.scitotenv.2014.04.087. [DOI] [PubMed] [Google Scholar]
- 37.O’Brien E, Munir M, Marsh T, Heran M, Lesage G, Tarabara VV, Xagoraraki I. Diversity of DNA viruses in effluents of membrane bioreactors in Traverse City, MI (USA) and La Grande Motte (France) Water Res. 2017;111:338–345. doi: 10.1016/j.watres.2017.01.014. [DOI] [PubMed] [Google Scholar]
- 38.Allard A, Albinsson B, Wadell G. Detection of adenoviruses in stools from healthy persons and patients with diarrhea by two-step polymerase chain reaction. J Med Virol. 1992;37:149–157. doi: 10.1002/jmv.1890370214. [DOI] [PubMed] [Google Scholar]
- 39.Berciaud S, Rayne F, Kassab S, Jubert C, Faure-Della Corte M, Salin F, Wodrich H, Lafon ME. Adenovirus infections in Bordeaux University Hospital 2008–2010: clinical and virological features. J Clin Virol. 2012;54:302–307. doi: 10.1016/j.jcv.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 40.Heim A, Ebnet C, Harste G, Pring-Åkerblom P. Rapid and quantitative detection of human adenovirus DNA by real-time PCR. J Med Virol. 2003;70:228–239. doi: 10.1002/jmv.10382. [DOI] [PubMed] [Google Scholar]
- 41.Jeulin H, Salmon A, Bordigoni P, Venard V. Diagnostic value of quantitative PCR for adenovirus detection in stool samples as compared with antigen detection and cell culture in haematopoietic stem cell transplant recipients. Clin Microbiol Infect. 2011;17:1674–1680. doi: 10.1111/j.1469-0691.2011.03488.x. [DOI] [PubMed] [Google Scholar]
- 42.Lion T, Kosulin K, Landlinger C, Rauch M, Preuner S, Jugovic D, Pötschger U, Lawitschka A, Peters C, Fritsch G, Matthes-Martin S. Monitoring of adenovirus load in stool by real-time PCR permits early detection of impending invasive infection in patients after allogeneic stem cell transplantation. Leukemia. 2010;24:706–714. doi: 10.1038/leu.2010.4. [DOI] [PubMed] [Google Scholar]
- 43.Martínez MA, de Soto-Del Río MLD, Gutiérrez RM, Chiu CY, Greninger AL, Contreras JF, López S, Arias CF, Isa P. DNA microarray for detection of gastrointestinal viruses. J Clin Microbiol. 2015;53:136–145. doi: 10.1128/JCM.01317-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mori K, Hayashi Y, Akiba T, Nagano M, Tanaka T, Hosaka M, Nakama A, Kai A, Saito K, Shirasawa H. Multiplex real-time PCR assays for the detection of group C rotavirus, astrovirus, and Subgenus F adenovirus in stool specimens. J Virol Methods. 2013;191:141–147. doi: 10.1016/j.jviromet.2012.10.019. [DOI] [PubMed] [Google Scholar]
- 45.Ribeiro A, Ramalheira E, Cunha Â, Gomes N, Almeida A. Incidence of rotavirus and adenovirus: detection by molecular and immunological methods in human faeces. J PURE Appl Microbiol. 2013;7:1505–1513. [Google Scholar]
- 46.Takayama R, Hatakeyama N, Suzuki N, Yamamoto M, Hayashi T, Ikeda Y, Ikeda H, Nagano H, Ishida T, Tsutsumi H. Quantification of adenovirus species B and C viremia by real-time PCR in adults and children undergoing stem cell transplantation. J Med Virol. 2007;79:278–284. doi: 10.1002/jmv.20796. [DOI] [PubMed] [Google Scholar]
- 47.van Maarseveen NM, Wessels E, de Brouwer CS, Vossen ACTM, Claas ECJ. Diagnosis of viral gastroenteritis by simultaneous detection of Adenovirus group F, Astrovirus, Rotavirus group A, Norovirus genogroups I and II, and Sapovirus in two internally controlled multiplex real-time PCR assays. J Clin Virol. 2010;49:205–210. doi: 10.1016/j.jcv.2010.07.019. [DOI] [PubMed] [Google Scholar]
- 48.Oishi I, Yamazaki K, Kimoto T, Minekawa Y, Utagawa E, Yamazaki S, Inouye S, Grohmann G, Monroe S, Stine S, Carcamo C, Ando T, Glass R. A large outbreak of acute gastroenteritis associated with astrovirus among students and teachers in Osaka, Japan. J Infect Dis. 1994;170:439–443. doi: 10.1093/infdis/170.2.439. [DOI] [PubMed] [Google Scholar]
- 49.Clark B, McKendrick M. A review of viral gastroenteritis. Curr Opin Infect Dis. 2004;17:461–469. doi: 10.1097/00001432-200410000-00011. [DOI] [PubMed] [Google Scholar]
- 50.Arraj A, Bohatier J, Aumeran C, Bailly JL, Laveran H, Traoré O. An epidemiological study of enteric viruses in sewage with molecular characterization by RT-PCR and sequence analysis. J Water Health. 2008;6:351–358. doi: 10.2166/wh.2008.053. [DOI] [PubMed] [Google Scholar]
- 51.Le Cann P, Ranarijaona S, Monpoeho S, Le Guyader F, Ferré V. Quantification of human astroviruses in sewage using real-time RT-PCR. Res Microbiol. 2004;155:11–15. doi: 10.1016/j.resmic.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 52.Ashley CR, Caul EO, Paver WK. Astrovirus-associated gastroenteritis in children. J Clin Pathol. 1978;31:939–943. doi: 10.1136/jcp.31.10.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guyader FSL, Saux J-CL, Ambert-Balay K, Krol J, Serais O, Parnaudeau S, Giraudon H, Delmas G, Pommepuy M, Pothier P, Atmar RL. Aichi virus, norovirus, astrovirus, enterovirus, and rotavirus involved in clinical cases from a French oyster-related gastroenteritis outbreak. J Clin Microbiol. 2008;46:4011–4017. doi: 10.1128/JCM.01044-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Logan C, O’Leary JJ, O’Sullivan N. Real-time reverse transcription PCR detection of norovirus, sapovirus and astrovirus as causative agents of acute viral gastroenteritis. J Virol Methods. 2007;146:36–44. doi: 10.1016/j.jviromet.2007.05.031. [DOI] [PubMed] [Google Scholar]
- 55.Kuan MM. Detection and rapid differentiation of human enteroviruses following genomic amplification. J Clin Microbiol. 1997;35:2598–2601. doi: 10.1128/jcm.35.10.2598-2601.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Muir P, Kämmerer U, Korn K, Mulders MN, Pöyry T, Weissbrich B, Kandolf R, Cleator GM, van LAM. Molecular typing of enteroviruses: current status and future requirements. Clin Microbiol Rev. 1998;11:202–227. doi: 10.1128/CMR.11.1.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.CDC. Non-Polio Enterovirus | Home | Picornavirus. Available at: http://www.cdc.gov/non-polio-enterovirus/index.html. Accessed 24 May 2016
- 58.Tsai YL, Tran B, Sangermano LR, Palmer CJ. Detection of poliovirus, hepatitis A virus, and rotavirus from sewage and ocean water by triplex reverse transcriptase PCR. Appl Environ Microbiol. 1994;60:2400–2407. doi: 10.1128/aem.60.7.2400-2407.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou J, Wang XC, Ji Z, Xu L, Yu Z. Source identification of bacterial and viral pathogens and their survival/fading in the process of wastewater treatment, reclamation, and environmental reuse. World J Microbiol Biotechnol. 2015;31:109–120. doi: 10.1007/s11274-014-1770-5. [DOI] [PubMed] [Google Scholar]
- 60.Kitajima M, Hata A, Yamashita T, Haramoto E, Minagawa H, Katayama H. Development of a reverse transcription-quantitative PCR system for detection and genotyping of Aichi viruses in clinical and environmental samples. Appl Environ Microbiol. 2013;79:3952–3958. doi: 10.1128/AEM.00820-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lago PM, Gary HE, Pérez LS, Cáceres V, Olivera JB, Puentes RP, Corredor MB, Jímenez P, Pallansch MA, Cruz RG. Poliovirus detection in wastewater and stools following an immunization campaign in Havana, Cuba. Int J Epidemiol. 2003;32:772–777. doi: 10.1093/ije/dyg185. [DOI] [PubMed] [Google Scholar]
- 62.Nijhuis M, van Maarseveen N, Schuurman R, Verkuijlen S, de Vos M, Hendriksen K, van Loon AM. Rapid and sensitive routine detection of all members of the genus enterovirus in different clinical specimens by real-time PCR. J Clin Microbiol. 2002;40:3666–3670. doi: 10.1128/JCM.40.10.3666-3670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xiao X-L, He Y-Q, Yu Y-G, Yang H, Chen G, Li H-F, Zhang J-W, Liu D-M, Li X-F, Yang X-Q, Wu H. Simultaneous detection of human enterovirus 71 and coxsackievirus A16 in clinical specimens by multiplex real-time PCR with an internal amplification control. Arch Virol. 2008;154:121–125. doi: 10.1007/s00705-008-0266-8. [DOI] [PubMed] [Google Scholar]
- 64.CDC. Hepatitis A Information | Division of Viral Hepatitis. Available at: http://www.cdc.gov/hepatitis/hav/. Accessed 24 May 2016
- 65.Wheeler C, Vogt TM, Armstrong GL, Vaughan G, Weltman A, Nainan OV, Dato V, Xia G, Waller K, Amon J, Lee TM, Highbaugh-Battle A, Hembree C, Evenson S, Ruta MA, Williams IT, Fiore AE, Bell BP. An outbreak of hepatitis a associated with green onions. N Engl J Med. 2005;353:890–897. doi: 10.1056/NEJMoa050855. [DOI] [PubMed] [Google Scholar]
- 66.Grabow WOK. Hepatitis viruses in water: update on risk and control. Water SA. 1997;23:379–386. [Google Scholar]
- 67.Morace G, Aulicino FA, Angelozzi C, Costanzo L, Donadio F, Rapicetta M. Microbial quality of wastewater: detection of hepatitis A virus by reverse transcriptase-polymerase chain reaction. J Appl Microbiol. 2002;92:828–836. doi: 10.1046/j.1365-2672.2002.01587.x. [DOI] [PubMed] [Google Scholar]
- 68.Villar LM, De Paula VS, Diniz-Mendes L, Guimarães FR, Ferreira FFM, Shubo TC, Miagostovich MP, Lampe E, Gaspar AMC. Molecular detection of hepatitis A virus in urban sewage in Rio de Janeiro, Brazil. Lett Appl Microbiol. 2007;45:168–173. doi: 10.1111/j.1472-765X.2007.02164.x. [DOI] [PubMed] [Google Scholar]
- 69.Chitambar SD, Joshi MS, Sreenivasan MA, Arankalle VA. Fecal shedding of hepatitis A virus in Indian patients with hepatitis A and in experimentally infected Rhesus monkey. Hepatol Res. 2001;19:237–246. doi: 10.1016/S1386-6346(00)00104-2. [DOI] [PubMed] [Google Scholar]
- 70.Costafreda MI, Bosch A, Pintó RM. Development, evaluation, and standardization of a real-time TaqMan reverse transcription-PCR assay for quantification of hepatitis A virus in clinical and shellfish samples. Appl Environ Microbiol. 2006;72:3846–3855. doi: 10.1128/AEM.02660-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sjogren MH, Tanno H, Fay O, Sileoni S, Cohen BD, Burke DS, Feighny RJ. Hepatitis A virus in stool during clinical relapse. Ann Intern Med. 1987;106:221–226. doi: 10.7326/0003-4819-106-2-221. [DOI] [PubMed] [Google Scholar]
- 72.Yotsuyanagi H, Koike K, Yasuda K, Moriya K, Shintani Y, Fujie H, Kurokawa K, Iino S. Prolonged fecal excretion of hepatitis A virus in adult patients with hepatitis A as determined by polymerase chain reaction. Hepatology. 1996;24:10–13. doi: 10.1002/hep.510240103. [DOI] [PubMed] [Google Scholar]
- 73.Bonnet D, Kamar N, Izopet J, Alric L. L’hépatite virale E: une maladie émergente. Rev Médecine Interne. 2012;33:328–334. doi: 10.1016/j.revmed.2012.01.017. [DOI] [PubMed] [Google Scholar]
- 74.CDC. Hepatitis E Information | Division of Viral Hepatitis. Available at: http://www.cdc.gov/hepatitis/hev/. Accessed 24 May 2016
- 75.Jothikumar N, Aparna K, Kamatchiammal S, Paulmurugan R, Saravanadevi S, Khanna P. Detection of hepatitis E virus in raw and treated wastewater with the polymerase chain reaction. Appl Environ Microbiol. 1993;59:2558–2562. doi: 10.1128/aem.59.8.2558-2562.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Clayson ET, Myint KSA, Snitbhan R, Vaughn DW, Innis BL, Chan L, Cheung P, Shrestha MP. Viremia, Fecal Shedding, and IgM and IgG responses in patients with hepatitis E. J Infect Dis. 1995;172:927–933. doi: 10.1093/infdis/172.4.927. [DOI] [PubMed] [Google Scholar]
- 77.Orrù G, Masia G, Orrù G, Romanò L, Piras V, Coppola RC. Detection and quantitation of hepatitis E virus in human faeces by real-time quantitative PCR. J Virol Methods. 2004;118:77–82. doi: 10.1016/j.jviromet.2004.01.025. [DOI] [PubMed] [Google Scholar]
- 78.Turkoglu S, Lazizi Y, Meng H, Kordosi A, Dubreuil P, Crescenzo B, Benjelloun S, Nordmann P, Pillot J. Detection of hepatitis E virus RNA in stools and serum by reverse transcription-PCR. J Clin Microbiol. 1996;34:1568–1571. doi: 10.1128/jcm.34.6.1568-1571.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fankhauser RL, Noel JS, Monroe SS, Ando T, Glass RI. Molecular epidemiology of “Norwalk-like viruses” in outbreaks of gastroenteritis in the United States. J Infect Dis. 1998;178:1571–1578. doi: 10.1086/314525. [DOI] [PubMed] [Google Scholar]
- 80.Fankhauser RL, Monroe SS, Noel JS, Humphrey CD, Bresee JS, Parashar UD, Ando T, Glass RI. Epidemiologic and molecular trends of “Norwalk-like viruses” associated with outbreaks of gastroenteritis in the United States. J Infect Dis. 2002;186:1–7. doi: 10.1086/341085. [DOI] [PubMed] [Google Scholar]
- 81.Mead PS, Slutsker L, Dietz V, McCaig LF, Bresee JS, Shapiro C, Griffin PM, Tauxe RV. Food-related illness and death in the United States. Emerg Infect Dis. 1999;5:607–625. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.de Andrade Jda SR, Rocha MS, Carvalho-Costa FA, Fioretti JM, Xavier Mda PTP, Nunes ZMA, Cardoso J, Fialho AM, Leite JPG, Miagostovich MP. Noroviruses associated with outbreaks of acute gastroenteritis in the State of Rio Grande do Sul, Brazil, 2004–2011. J Clin Virol. 2014;61:345–352. doi: 10.1016/j.jcv.2014.08.024. [DOI] [PubMed] [Google Scholar]
- 83.Kukkula M, Maunula L, Silvennoinen E, von Bonsdorff C-H. Outbreak of viral gastroenteritis due to drinking water contaminated by Norwalk-like viruses. J Infect Dis. 1999;180:1771–1776. doi: 10.1086/315145. [DOI] [PubMed] [Google Scholar]
- 84.da Silva AK, Saux J-CL, Parnaudeau S, Pommepuy M, Elimelech M, Guyader FSL. Evaluation of removal of noroviruses during wastewater treatment, using real-time reverse transcription-PCR: different behaviors of genogroups I and II. Appl Environ Microbiol. 2007;73:7891–7897. doi: 10.1128/AEM.01428-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hewitt J, Greening GE, Leonard M, Lewis GD. Evaluation of human adenovirus and human polyomavirus as indicators of human sewage contamination in the aquatic environment. Water Res. 2013;47:6750–6761. doi: 10.1016/j.watres.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 86.Atmar RL, Opekun AR, Gilger MA, Estes MK, Crawford SE, Neill FH, Graham DY. Norwalk virus shedding after experimental human infection. Emerg Infect Dis. 2008;14:1553–1557. doi: 10.3201/eid1410.080117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fumian TM, Justino MCA, Mascarenhas JDP, Reymão TKA, Abreu E, Soares L, Linhares AC, Gabbay YB. Quantitative and molecular analysis of noroviruses RNA in blood from children hospitalized for acute gastroenteritis in Belém, Brazil. J Clin Virol. 2013;58:31–35. doi: 10.1016/j.jcv.2013.06.043. [DOI] [PubMed] [Google Scholar]
- 88.Schvoerer E, Bonnet F, Dubois V, Cazaux G, Serceau R, Fleury HJ, Lafon ME. PCR detection of human enteric viruses in bathing areas, waste waters and human stools in southwestern France. Res Microbiol. 2000;151:693–701. doi: 10.1016/S0923-2508(00)90132-3. [DOI] [PubMed] [Google Scholar]
- 89.CDC. Rotavirus | Home | Gastroenteritis. Available at: http://www.cdc.gov/rotavirus/. Accessed 24 May 2016
- 90.Li D, Gu AZ, Zeng S-Y, Yang W, He M, Shi H-C. Monitoring and evaluation of infectious rotaviruses in various wastewater effluents and receiving waters revealed correlation and seasonal pattern of occurrences. J Appl Microbiol. 2011;110:1129–1137. doi: 10.1111/j.1365-2672.2011.04954.x. [DOI] [PubMed] [Google Scholar]
- 91.Meleg E, Bányai K, Martella V, Jiang B, Kocsis B, Kisfali P, Melegh B, Szűcs G. Detection and quantification of group C rotaviruses in communal sewage. Appl Environ Microbiol. 2008;74:3394–3399. doi: 10.1128/AEM.02895-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fumian TM, Gagliardi Leite JP, Rose TL, Prado T, Miagostovich MP. One year environmental surveillance of rotavirus specie A (RVA) genotypes in circulation after the introduction of the Rotarix® vaccine in Rio de Janeiro, Brazil. Water Res. 2011;45:5755–5763. doi: 10.1016/j.watres.2011.08.039. [DOI] [PubMed] [Google Scholar]
- 93.Baggi F, Peduzzi R. Genotyping of rotaviruses in environmental water and stool samples in Southern Switzerland by nucleotide sequence analysis of 189 base pairs at the 5’ end of the VP7 gene. J Clin Microbiol. 2000;38:3681–3685. doi: 10.1128/jcm.38.10.3681-3685.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gouvea V, Glass RI, Woods P, Taniguchi K, Clark HF, Forrester B, Fang ZY. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J Clin Microbiol. 1990;28:276–282. doi: 10.1128/jcm.28.2.276-282.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mukhopadhya I, Sarkar R, Menon VK, Babji S, Paul A, Rajendran P, Sowmyanarayanan TV, Moses PD, Iturriza-Gomara M, Gray JJ, Kang G. Rotavirus shedding in symptomatic and asymptomatic children using reverse transcription-quantitative PCR. J Med Virol. 2013;85:1661–1668. doi: 10.1002/jmv.23641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ambert-Balay K, Lorrot M, Bon F, Giraudon H, Kaplon J, Wolfer M, Lebon P, Gendrel D, Pothier P. Prevalence and genetic diversity of Aichi virus strains in stool samples from community and hospitalized patients. J Clin Microbiol. 2008;46:1252–1258. doi: 10.1128/JCM.02140-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Haramoto E, Kitajima M, Otagiri M. Development of a reverse transcription-quantitative PCR assay for detection of salivirus/klassevirus. Appl Environ Microbiol. 2013;79:3529–3532. doi: 10.1128/AEM.00132-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Oka T, Katayama K, Hansman GS, Kageyama T, Ogawa S, Wu F, White PA, Takeda N. Detection of human sapovirus by real-time reverse transcription-polymerase chain reaction. J Med Virol. 2006;78:1347–1353. doi: 10.1002/jmv.20699. [DOI] [PubMed] [Google Scholar]
- 99.Siebrasse EA, Reyes A, Lim ES, Zhao G, Mkakosya RS, Manary MJ, Gordon JI, Wang D. Identification of MW polyomavirus, a novel polyomavirus in human stool. J Virol. 2012;86:10321–10326. doi: 10.1128/JVI.01210-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hino S, Miyata H. Torque teno virus (TTV): current status. Rev Med Virol. 2006;17:45–57. doi: 10.1002/rmv.524. [DOI] [PubMed] [Google Scholar]
- 101.Lin K, Marr LC. Aerosolization of Ebola virus surrogates in wastewater systems. Environ Sci Technol. 2017;51:2669–2675. doi: 10.1021/acs.est.6b04846. [DOI] [PubMed] [Google Scholar]
- 102.Cotruvo JA, Dufour A, Rees G, Bartram J, Carr R, Cliver DO, Craun GF, Fayer R, Gannon VP (2004) Waterborne zoonoses. IWA Publishing, London, UK
- 103.Gundy PM, Gerba CP, Pepper IL. Survival of coronaviruses in water and wastewater. Food Environ Virol. 2008;1:10–14. doi: 10.1007/s12560-008-9001-6. [DOI] [Google Scholar]
- 104.Hu Y, Lu S, Song Z, Wang W, Hao P, Li J, Zhang X, Yen H-L, Shi B, Li T, Guan W, Xu L, Liu Y, Wang S, Zhang X, Tian D, Zhu Z, He J, Huang K, Chen H, Zheng L, Li X, Ping J, Kang B, Xi X, Zha L, Li Y, Zhang Z, Peiris M, Yuan Z. Association between adverse clinical outcome in human disease caused by novel influenza A H7N9 virus and sustained viral shedding and emergence of antiviral resistance. The Lancet. 2013;381:2273–2279. doi: 10.1016/S0140-6736(13)61125-3. [DOI] [PubMed] [Google Scholar]
- 105.Lee N, Chan PK, Wong CK, Wong K-T, Choi K-W, Joynt GM, Lam P, Chan MC, Wong BC, Lui GC, Sin WW, Wong RY, Lam W-Y, Yeung AC, Leung T-F, So H-Y, Yu AW, Sung JJ, Hui DS. Viral clearance and inflammatory response patterns in adults hospitalized for pandemic 2009 influenza A(H1N1) virus pneumonia. Antivir Ther. 2011;16:237–247. doi: 10.3851/IMP1722. [DOI] [PubMed] [Google Scholar]
- 106.To KKW, Chan K-H, Li IWS, Tsang T-Y, Tse H, Chan JFW, Hung IFN, Lai S-T, Leung C-W, Kwan Y-W, Lau Y-L, Ng T-K, Cheng VCC, Peiris JSM, Yuen K-Y. Viral load in patients infected with pandemic H1N1 2009 influenza A virus. J Med Virol. 2010;82:1–7. doi: 10.1002/jmv.21664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Heijnen L, Medema G. Surveillance of influenza A and the pandemic influenza A (H1N1) 2009 in sewage and surface water in the Netherlands. J Water Health. 2011;9:434–442. doi: 10.2166/wh.2011.019. [DOI] [PubMed] [Google Scholar]
- 108.Gourinat A-C, O’Connor O, Calvez E, Goarant C, Dupont-Rouzeyrol M. Detection of Zika virus in urine. Emerg Infect Dis. 2015;21:84–86. doi: 10.3201/eid2101.140894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Barzon L, Pacenti M, Franchin E, Pagni S, Martello T, Cattai M, Cusinato R, Palù G. Excretion of West Nile virus in urine during acute infection. J Infect Dis. 2013;208:1086–1092. doi: 10.1093/infdis/jit290. [DOI] [PubMed] [Google Scholar]
- 110.Tonry JH, Brown CB, Cropp CB, Co JKG, Bennett SN, Nerurkar VR, Kuberski T, Gubler DJ. West Nile Virus detection in urine. Emerg Infect Dis. 2005;11:1294–1296. doi: 10.3201/eid1108.0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hirayama T, Mizuno Y, Takeshita N, Kotaki A, Tajima S, Omatsu T, Sano K, Kurane I, Takasaki T. Detection of dengue virus genome in urine by real-time reverse transcriptase PCR: a laboratory diagnostic method useful after disappearance of the genome in serum. J Clin Microbiol. 2012;50:2047–2052. doi: 10.1128/JCM.06557-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mizuno Y, Kotaki A, Harada F, Tajima S, Kurane I, Takasaki T. Confirmation of dengue virus infection by detection of dengue virus type 1 genome in urine and saliva but not in plasma. Trans R Soc Trop Med Hyg. 2007;101:738–739. doi: 10.1016/j.trstmh.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 113.Poloni TR, Oliveira AS, Alfonso HL, Galvao LR, Amarilla AA, Poloni DF, Figueiredo LT, Aquino VH. Detection of dengue virus in saliva and urine by real time RT-PCR. Virol J. 2010;7:22. doi: 10.1186/1743-422X-7-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Nielsen ACY, Gyhrs ML, Nielsen LP, Pedersen C, Böttiger B. Gastroenteritis and the novel picornaviruses Aichi virus, cosavirus, saffold virus, and salivirus in young children. J Clin Virol. 2013;57:239–242. doi: 10.1016/j.jcv.2013.03.015. [DOI] [PubMed] [Google Scholar]
- 115.Vanchiere JA, Nicome RK, Greer JM, Demmler GJ, Butel JS. Frequent detection of polyomaviruses in stool samples from hospitalized children. J Infect Dis. 2005;192:658–664. doi: 10.1086/432076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Vanchiere JA, Abudayyeh S, Copeland CM, Lu LB, Graham DY, Butel JS. Polyomavirus shedding in the stool of healthy adults. J Clin Microbiol. 2009;47:2388–2391. doi: 10.1128/JCM.02472-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Li L, Victoria J, Kapoor A, Blinkova O, Wang C, Babrzadeh F, Mason CJ, Pandey P, Triki H, Bahri O, Oderinde BS, Baba MM, Bukbuk DN, Besser JM, Bartkus JM, Delwart EL. A novel picornavirus associated with gastroenteritis. J Virol. 2009;83:12002–12006. doi: 10.1128/JVI.01241-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chan MCW, Sung JJY, Lam RKY, Chan PKS, Lai RWM, Leung WK. Sapovirus detection by quantitative real-time RT-PCR in clinical stool specimens. J Virol Methods. 2006;134:146–153. doi: 10.1016/j.jviromet.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 119.Ross RS, Viazov S, Runde V, Schaefer UW, Roggendorf M. Detection of TT virus DNA in specimens other than blood. J Clin Virol. 1999;13:181–184. doi: 10.1016/S1386-6532(99)00015-3. [DOI] [PubMed] [Google Scholar]
- 120.Chan KH, Poon LL, Cheng VC, Guan Y, Hung IF, Kong J, Yam LY, Seto WH, Yuen KY, Peiris JS. Detection of SARS coronavirus in patients with suspected SARS. Emerg Infect Dis. 2004;10:294–299. doi: 10.3201/eid1002.030610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Poon LLM, Chan KH, Wong OK, Cheung TKW, Ng I, Zheng B, Seto WH, Yuen KY, Guan Y, Peiris JSM. Detection of SARS coronavirus in patients with severe acute respiratory syndrome by conventional and real-time quantitative reverse transcription-PCR assays. Clin Chem. 2004;50:67–72. doi: 10.1373/clinchem.2003.023663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Vester D, Lagoda A, Hoffmann D, Seitz C, Heldt S, Bettenbrock K, Genzel Y, Reichl U. Real-time RT-qPCR assay for the analysis of human influenza A virus transcription and replication dynamics. J Virol Methods. 2010;168:63–71. doi: 10.1016/j.jviromet.2010.04.017. [DOI] [PubMed] [Google Scholar]
- 123.dos Santos HWG, Poloni TRRS, Souza KP, Muller VDM, Tremeschin F, Nali LC, Fantinatti LR, Amarilla AA, Castro HLA, Nunes MR, Casseb SM, Vasconcelos PF, Badra SJ, Figueiredo LTM, Aquino VH. A simple one-step real-time RT-PCR for diagnosis of dengue virus infection. J Med Virol. 2008;80:1426–1433. doi: 10.1002/jmv.21203. [DOI] [PubMed] [Google Scholar]
- 124.Linke S, Ellerbrok H, Niedrig M, Nitsche A, Pauli G. Detection of West Nile virus lineages 1 and 2 by real-time PCR. J Virol Methods. 2007;146:355–358. doi: 10.1016/j.jviromet.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 125.Faye O, Faye O, Diallo D, Diallo M, Weidmann M, Sall A. Quantitative real-time PCR detection of Zika virus and evaluation with field-caught mosquitoes. Virol J. 2013;10:311. doi: 10.1186/1743-422X-10-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Domingo C, Yactayo S, Agbenu E, Demanou M, Schulz AR, Daskalow K, Niedrig M. Detection of yellow fever 17D genome in urine. J Clin Microbiol. 2011;49:760–762. doi: 10.1128/JCM.01775-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bae H-G, Nitsche A, Teichmann A, Biel SS, Niedrig M. Detection of yellow fever virus: a comparison of quantitative real-time PCR and plaque assay. J Virol Methods. 2003;110:185–191. doi: 10.1016/S0166-0934(03)00129-0. [DOI] [PubMed] [Google Scholar]
- 128.Orenstein EW, Fang ZY, Xu J, Liu C, Shen K, Qian Y, Jiang B, Kilgore PE, Glass RI. The epidemiology and burden of rotavirus in China: a review of the literature from 1983 to 2005. Vaccine. 2007;25:406–413. doi: 10.1016/j.vaccine.2006.07.054. [DOI] [PubMed] [Google Scholar]
- 129.Bofill-Mas S, Pina S, Girones R. Documenting the epidemiologic patterns of polyomaviruses in human populations by studying their presence in urban sewage. Appl Environ Microbiol. 2000;66:238–245. doi: 10.1128/AEM.66.1.238-245.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Clemente-Casares P, Pina S, Buti M, Jardi R, Martín M, Bofill-Mas S, Girones R. Hepatitis E virus epidemiology in industrialized countries. Emerg Infect Dis. 2003;9:448–454. doi: 10.3201/eid0904.020351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Carducci A, Verani M, Battistini R, Pizzi F, Rovini E, Andreoli E, Casini B. Epidemiological surveillance of human enteric viruses by monitoring of different environmental matrices. Water Sci Technol. 2006;54:239–244. doi: 10.2166/wst.2006.475. [DOI] [PubMed] [Google Scholar]
- 132.O’Brien E, Nakyazze J, Wu H, Kiwanuka N, Cunningham W, Kaneene JB, Xagoraraki I. Viral diversity and abundance in polluted waters in Kampala, Uganda. Water Res. 2017;127:41–49. doi: 10.1016/j.watres.2017.09.063. [DOI] [PubMed] [Google Scholar]
- 133.Bibby K, Peccia J. Identification of viral pathogen diversity in sewage sludge by metagenome analysis. Environ Sci Technol. 2013;47:1945–1951. doi: 10.1021/es305181x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Tamaki H, Zhang R, Angly FE, Nakamura S, Hong P-Y, Yasunaga T, Kamagata Y, Liu W-T. Metagenomic analysis of DNA viruses in a wastewater treatment plant in tropical climate. Environ Microbiol. 2012;14:441–452. doi: 10.1111/j.1462-2920.2011.02630.x. [DOI] [PubMed] [Google Scholar]
- 135.Aw TG, Howe A, Rose JB. Metagenomic approaches for direct and cell culture evaluation of the virological quality of wastewater. J Virol Methods. 2014;210:15–21. doi: 10.1016/j.jviromet.2014.09.017. [DOI] [PubMed] [Google Scholar]
- 136.Cantalupo PG, Calgua B, Zhao G, Hundesa A, Wier AD, Katz JP, Grabe M, Hendrix RW, Girones R, Wang D, Pipas JM. Raw sewage harbors diverse viral populations. MBio. 2011;2:e00180–e00111. doi: 10.1128/mBio.00180-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bibby K, Viau E, Peccia J. Viral metagenome analysis to guide human pathogen monitoring in environmental samples. Lett Appl Microbiol. 2011;52:386–392. doi: 10.1111/j.1472-765X.2011.03014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Finkbeiner SR, Allred AF, Tarr PI, Klein EJ, Kirkwood CD, Wang D. Metagenomic analysis of human diarrhea: viral detection and discovery. PLoS Pathog. 2008;4:e1000011. doi: 10.1371/journal.ppat.1000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Victoria JG, Kapoor A, Li L, Blinkova O, Slikas B, Wang C, Naeem A, Zaidi S, Delwart E. Metagenomic analyses of viruses in stool samples from children with acute flaccid paralysis. J Virol. 2009;83:4642–4651. doi: 10.1128/JVI.02301-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lai FY, Anuj S, Bruno R, Carter S, Gartner C, Hall W, Kirkbride KP, Mueller JF, O’Brien JW, Prichard J, Thai PK, Ort C. Systematic and day-to-day effects of chemical-derived population estimates on wastewater-based drug epidemiology. Environ Sci Technol. 2015;49:999–1008. doi: 10.1021/es503474d. [DOI] [PubMed] [Google Scholar]
- 141.O’Brien JW, Thai PK, Eaglesham G, Ort C, Scheidegger A, Carter S, Lai FY, Mueller JF. A model to estimate the population contributing to the wastewater using samples collected on census day. Environ Sci Technol. 2014;48:517–525. doi: 10.1021/es403251g. [DOI] [PubMed] [Google Scholar]
- 142.Chiaia AC, Banta-Green C, Field J. Eliminating solid phase extraction with large-volume injection LC/MS/MS: analysis of illicit and legal drugs and human urine indicators in US wastewaters. Environ Sci Technol. 2008;42:8841–8848. doi: 10.1021/es802309v. [DOI] [PubMed] [Google Scholar]
- 143.Daughton CG. Real-time estimation of small-area populations with human biomarkers in sewage. Sci Total Environ. 2012;414:6–21. doi: 10.1016/j.scitotenv.2011.11.015. [DOI] [PubMed] [Google Scholar]
- 144.Baker DR, Kasprzyk-Hordern B. Multi-residue analysis of drugs of abuse in wastewater and surface water by solid-phase extraction and liquid chromatography–positive electrospray ionisation tandem mass spectrometry. J Chromatogr A. 2011;1218:1620–1631. doi: 10.1016/j.chroma.2011.01.060. [DOI] [PubMed] [Google Scholar]
- 145.Chen C, Kostakis C, Gerber JP, Tscharke BJ, Irvine RJ, White JM. Towards finding a population biomarker for wastewater epidemiology studies. Sci Total Environ. 2014;487:621–628. doi: 10.1016/j.scitotenv.2013.11.075. [DOI] [PubMed] [Google Scholar]
- 146.Been F, Rossi L, Ort C, Rudaz S, Delémont O, Esseiva P. Population normalization with ammonium in wastewater-based epidemiology: application to illicit drug monitoring. Environ Sci Technol. 2014;48:8162–8169. doi: 10.1021/es5008388. [DOI] [PubMed] [Google Scholar]
- 147.Edwards AC, Withers PJA. Linking phosphorus sources to impacts in different types of water body. Soil Use Manag. 2007;23:133–143. doi: 10.1111/j.1475-2743.2007.00110.x. [DOI] [Google Scholar]
- 148.James E, Kleinman P, Veith T, Stedman R, Sharpley A. Phosphorus contributions from pastured dairy cattle to streams of the Cannonsville Watershed, New York. J Soil Water Conserv. 2007;62:40–47. [Google Scholar]
- 149.Chang H, Wan Y, Wu S, Fan Z, Hu J. Occurrence of androgens and progestogens in wastewater treatment plants and receiving river waters: comparison to estrogens. Water Res. 2011;45:732–740. doi: 10.1016/j.watres.2010.08.046. [DOI] [PubMed] [Google Scholar]
- 150.Chang H, Hu J, Shao B. Occurrence of natural and synthetic glucocorticoids in sewage treatment plants and receiving river waters. Environ Sci Technol. 2007;41:3462–3468. doi: 10.1021/es062746o. [DOI] [PubMed] [Google Scholar]
- 151.Cheng H-Y, Huang Y-C, Yen T-Y, Hsia S-H, Hsieh Y-C, Li C-C, Chang L-Y, Huang L-M. The correlation between the presence of viremia and clinical severity in patients with enterovirus 71 infection: a multi-center cohort study. BMC Infect Dis. 2014;14:417. doi: 10.1186/1471-2334-14-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Bosch A, Pinto RM, Abad FX (2006) Survival and transport of enteric viruses in the environment. In: Goyal S.M. (eds) Viruses in Foods. Food Microbiology and Food Safety. Springer, Boston, MA
- 153.Xagoraraki I, Yin Z, Svambayev Z. Fate of viruses in water systems. J Environ Eng. 2014;140:04014020. doi: 10.1061/(ASCE)EE.1943-7870.0000827. [DOI] [Google Scholar]