Abstract
RNA Polymerase II (RNAPII) is responsible for transcribing multiple RNA species throughout eukaryotes. A variety of protein-protein interactions occur throughout the transcription cycle for coordinated regulation of transcription initiation, elongation, and/or termination. Taking a proteomics approach to study RNAPII transcription thereby offers a comprehensive view of both RNAPII biology and the variety of proteins that regulate the process itself. This review will focus on how mass spectrometry (MS) methods have expanded understanding of RNAPII and its transcription-regulatory interaction partners. The application of affinity purification mass spectrometry has led to the discovery of a number of novel groups of proteins that regulate an array of RNAPII biology ranging from nuclear import to regulation of phosphorylation state. Additionally, a number of methods have been developed using mass spectrometry to measure protein subunit stoichiometry within and across protein complexes and to perform various types of architectural analysis using structural proteomics approaches. The key methods that we will focus on related to RNAPII mass spectrometry analyses include: affinity purification mass spectrometry, protein post-translational modification analysis, crosslinking mass spectrometry, and native mass spectrometry.
Keywords: RNA Polymerase II, C-terminal domain, CTD, transcription, mass spectrometry, proteomics, nano-electrospray ionization, affinity purification, kinase, phosphorylation, non-covalent interaction networks, crosslinking mass spectrometry, structural proteomics
1. Introduction
RNA Polymerase II (RNAPII) is responsible for transcribing multiple RNA species, including messenger RNA (mRNA), noncoding RNAs (ncRNAs), and small nuclear/nucleolar RNAs (sn/snoRNAs) [1, 2]. In order to ensure proper transcription of these RNA species regulatory proteins are dynamically recruited to RNAPII. Transcription regulatory proteins may be recruited through interactions with the template DNA, the nascent RNA, or through protein-protein interactions (PPIs) within the transcription complex. The C-terminal domain (CTD) of the largest of the 12 RNAPII subunits, Rpb1, is a critical regulator of these PPIs through its dynamic phosphorylation.
Achieving a comprehensive understanding of transcription on the levels of DNA, RNA and protein requires diverse methodology. High-throughput sequencing method development has revolutionized the analysis of DNA and RNA and has greatly benefited the transcription field. These efforts have been driven by a wide variety of high-throughput nucleic acid sequencing approaches. Proteins are challenging to interrogate using similar high-throughput approaches as a consequence of their diversity in structure, size, and amino acid composition (including post-translational modifications). In recent years, proteomics and complementary structural mass spectrometry approaches have continued to develop at a rapid rate spurred on by developments in mass spectrometry instrumentation, analysis software, and various chemical tools.
This review will focus on the application of mass spectrometry to the study of RNA Polymerase II (RNAPII) transcription. Mass spectrometry-based approaches to study RNAPII transcription offers a wholistic view of both the core RNAPII machinery and also the proteins that regulate overall transcription including RNA processing and chromatin biology. Therein, this review will discuss how mass spectrometry (MS) methods have expanded understanding of RNAPII and its transcription-regulatory interaction partners.
2. Overview of Methods for MS Analysis of RNAPII Complexes
2.1. Analysis of RNAPII Interactors by MS based methods
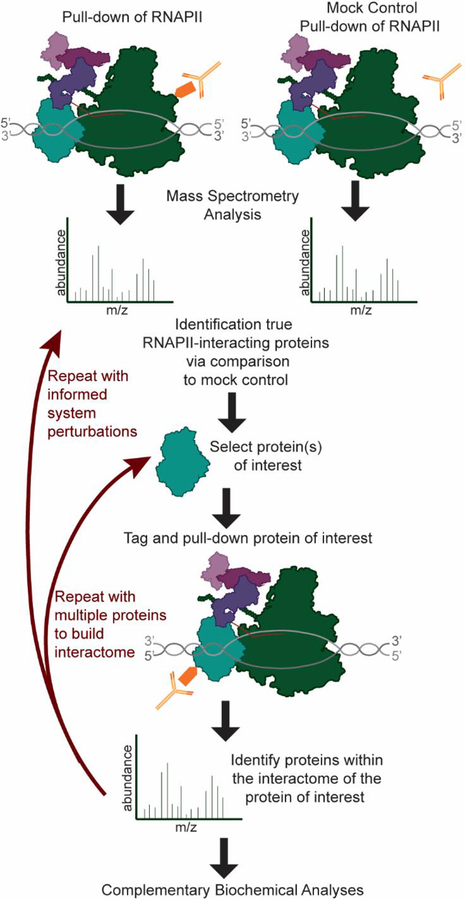
Identification of protein-protein interaction partners of the RNAPII machinery has been a longstanding interest of the molecular biology community. Many of the known RNAPII interacting proteins were isolated based on their affinity for RNAPII, with subsequent identification of the interacting proteins performed using approaches such as cDNA library screening [3]. However, mass spectrometry-based methods rapidly gained popularity after FASTA-based protein sequence database searching was enabled by the development of SEQUEST, and other algorithms, allowing researchers to establish a rapid and direct link between protein and gene sequences [4]. Using database sequence-based identification, subunits of large protein complexes, such as the chromatin/transcription related Spt-Ada-Gcn5-acetyltransferase (SAGA) complex, could be rapidly identified after biochemical purification [5]. Many of the proteins within the basic transcription machinery have now been identified as a consequence of wide-spread application of mass spectrometry to RNAPII transcription; however, adoption of new MS techniques and improved instrumentation continues to further the transcription field. MS approaches have not only confirmed and detailed suspected interactions between RNAPII and regulatory proteins, but have also continued to identify novel interactions. Specifically, affinity purification-mass spectrometry (AP-MS) has been a key approach in analyzing and mapping RNAPII protein-protein interactions (PPIs) (Figure 1).
Figure 1: AP-MS of RNAPII and regulators to build the RNAPII interactome.
MS/MSanalysis of RNAPII purifications allows identification of RNAPII-interactors. Subsequent purifications of proteins of interest enable interaction mapping.
The yeast transcription elongation complex has particularly benefitted from the application of AP-MS to studying RNAPII PPIs. Multiple groups used AP-MS pull-downs to identify that Set2, a methyl transferase, is a direct interactor of RNAPII [6–9]. Additional experiments showed that the Set2-RNAPII interaction is dependent upon a serine 2 phosphorylated (Ser2P) CTD. The discovery of this interaction was critical for understanding the coregulation of histone methylation and transcription elongation [6–9]. Similarly, Asr1, a yeast RING finger protein, was found to interact with RNAPII through AP-MS analysis, in which Asr1 was used as the bait protein (Asr1-TAP). Asr1 has some homology to human rA9, which was identified as a candidate RNAPII CTD binding protein through yeast two-hybrid studies prompting the analysis of Asr1 interactors in yeast [10]. Asr1 has been shown to function as a novel E3 ligase for RNAPII and functions through a unique mechanism that is not fully resolved in which two RNAPII subunits, Rpb4 and Rpb7 (hence Rpb4/7), are ejected from the RNAPII complex [10]. Interestingly, AP-MS studies have found that Rpb4/7 levels are reduced when RNAPII complexes are purified using a number of different elongation factors as bait [11]. Another prime example of the impact of AP-MS has had on the discovery of novel transcription regulatory proteins is the CTD phosphatase Rtr1. Mosley et al. [12] identified Rtr1 as a bona fide interactor of RNAPII through reciprocal TAP purifications and Multidimensional Protein Identification Technology (MudPIT) MS. Rtr1 was also isolated from human cells through analysis of the RNAPII interactome and named RPAP2 [13]. Additional experiments confirmed Rtr1 as a CTD Ser5 phosphatase and identified changes in RNAPII phosphorylation upon its deletion. Further studies on RPAP2/Rtr1 in yeast and human cells has confirmed its role as a CTD phosphatase that has now also been implicated in the regulation of the unfolded protein response [14–20]. These discoveries are highlighted here as key examples of the utility of APMS as a method to identify new interactors and potentially new biology (Figure 4A).
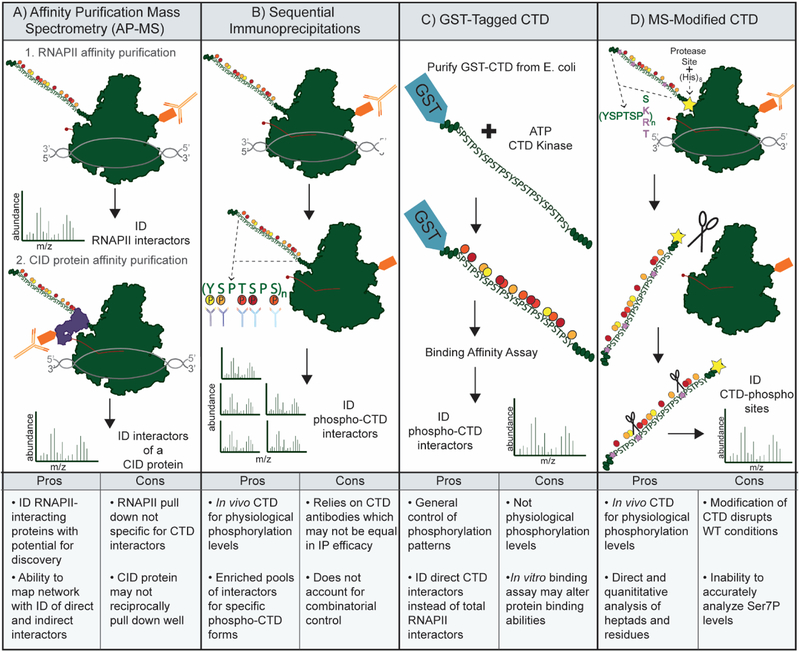
Figure 4: Strategies for Analysis of the CTD and CTD interactors.
A) Affinity Purification Mass Spectrometry (AP-MS) of RNAPII complexes allows identification (ID) of RNAPII interactors. Interactors of interest with CTD Interaction Domains (CIDs) may then be purified to analyze the CTD interactome. B) Purification of total RNAPII followed by IPs using antibodies specific for CTD phospho-sites. MS analysis to identify protein interactome for CTD phospho-forms. C) Purification of GST-tagged CTD peptides to be phosphorylated by CTD kinases and used in binding assays. MS analysis of bound proteins to ID CTD-interacting proteins. D) Modification of the CTD in cells to increase suitability for MS analysis. Mutation of select Ser7 residues to lysine to provide trypsin cleavage sites, as well as additional mutations to create unique mass peptides. MS analysis of these msCTDs allows mapping of specific post-translational modifications.
Furthermore, Krogan et al. [21] demonstrated the usefulness of such an AP-MS approach with an early effort in characterizing the components of RNAPII transcription elongation complexes. Suspected elongation factors (DSIF, FACT, Spt6, TFIIF, Rtf1 and Elongator) were TAP-tagged (tandem affinity purification) and purified from Saccharomyces cerevisiae yeast. Different biochemical conditions (e.g. salt concentration) in the purification will determine the degree of interacting proteins that co-purify with the tagged proteins, referred to in AP-MS as baits. The pull-down products, referred to in AP-MS as prey proteins, were then subjected to MALDI-TOF MS for protein identification. These data were able to show the interaction of Spt5 with Spt4 (both members of DSIF) and also with Spt6. Interestingly, Spt6 itself was found to interact with the then uncharacterized protein, Iws1/Spn1, an interaction that is involved in a number of transcription-coupled processes [22–25]. Post-translational modification (PTM) searches of the MS analyses also revealed two phosphorylated casein kinase II (CKII) consensus sites within Spt6 [21]. In fact, PTM mapping depth is enabled through coupling it with AP-MS. Recent papers, utilizing quantitative AP-MS approaches have further detailed the phosphorylation of Spt6 by CKII and how this modification regulates the interaction with Spn1[22, 23]. Additionally, purification of the FACT subunits by Krogan et al. revealed novel interactions of FACT with CKII, both individually and in a complex with the chromatin remodeler Chd1, as well as with all the subunits of the complex now known as the RNAPII associated PAF Complex (PAFC). Bedard et al. further characterized these findings using MS methods to illuminate that FACT interacts with PAFC to facilitate CKII phosphorylation of PAFC [26]. Of note, the two largest subunits of RNAPII were reproducibly identified in purifications of PAFC and FACT; however, the identification of the remaining 12-subunits of RNAPII was stochastic across replicates. These data suggest that the majority of purified PAF-C, for instance, does not associate with RNAPII but rather interacts in a transient fashion during different stages of transcription.
It is important to note that the impact of AP-MS on the characterization of protein-protein interactions within the transcription complex has not been limited to the yeast basal elongation complex. These contributions include the identification of a number of uncharacterized RNA Polymerase Associated Proteins (RPAPs) in tandem affinity purifications (TAP) of human RNAPII from HEK293 cells[13, 27]. Follow up studies on XAB1/Npa3 and the RPAPs have revealed that they play diverse roles in the regulation of RNAPII biology that has been uncovered as consequence of AP-MS experiments. XAB1/Npa3 is a conserved GPN-loop GTPase that has been implicated in RNAPII nuclear import as an RNAPII chaperone [28–31]. RPAP3 has been characterized as a R2TP-like co-chaperone that may regulate RNAPII stability as well as that of other proteins/protein complexes[32–34]. RPAP1 has been characterized to play a role in the interaction between RNAPII and Mediator [35].
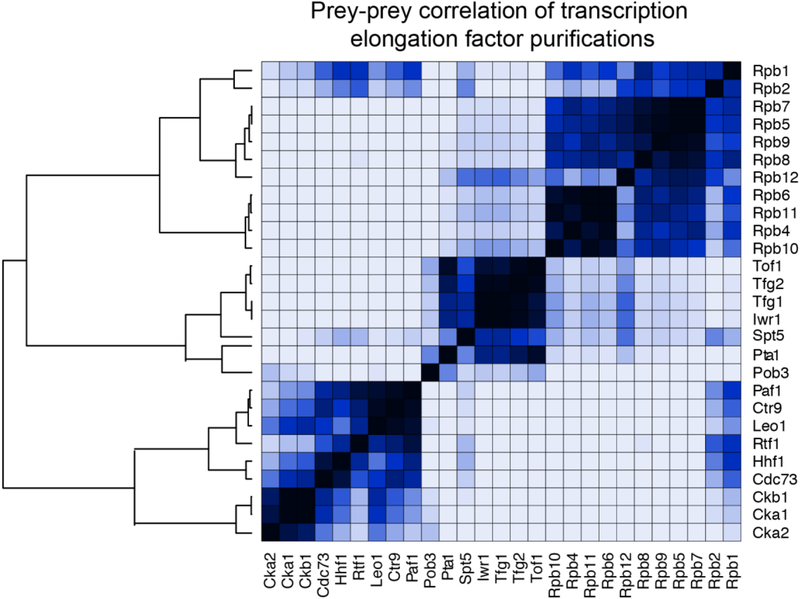
However, while AP-MS of RNAPII and/or interacting, proteins has the ability to both identify and aid in the characterization of interactions within the transcription complex. The dynamic nature of these PPIs during transcription can make the determination of specific bait-prey interactions, and any changes in them, hard to distinguish from non-specific or random preys. An early approach to solve this problem was pioneered by Ranish et al. [36] that provided benefits to both the proteomics and transcription fields. Isotope-coded affinity tag (ICAT) reagents were used in both a purification of the pre-initiation complex (PIC) [37] and a control purification. The use of ICAT reagents coupled with MudPIT MS/MS improved the quantitation of relative abundance measurements for prey proteins, allowing the identification of specific interactions over background noise. Thus, the true components of the PIC were able to be identified and comprehensively analyzed [36]. More recent development of database and computational tools such as the CRAPome [38], significant analysis of interactome (SAINT) [39] and the Coon OMSSA Proteomic Analysis Software Suite (COMPASS) [40] have aided in the ability to distinguish real bait-prey interactions from contaminants, and score these interactions, both with probability and fold change values. For example, using SAINT analysis followed by data visualization by ProHits-viz [41] of the dataset from Bedard et. al, we performed prey-prey correlation analysis. Using these approaches, the high degree of copurification dynamics of the two largest RNAPII subunits can be seen across various baits (Figure 2). As mentioned above, Rpb1 and Rpb2 are readily identified in transcription-related AP-MS studies due to their large size. The reproducible identification of the other subunits of RNAPII is also readily seen when a core RNAPII subunit is used as the bait protein (Figure 2, [42]). Along these same lines, a high degree of correlation is observed between subunits of individual protein complexes such as PAFC (subunits: Ctr9, Paf1, Cdc73, Leo1, and Rtf1). With the proper controls and statistical tools, AP-MS can provide insightful data on interaction dynamics within and between complexes regulating transcription.
Figure 2: Prey-prey correlation analysis of RNAPII, FACT, and PAFC purifications.
Following significance analysis of interactome (SAINT), the degree of correlation between the quantity of various co-purifying proteins was analyzed using ProHits-viz (Nat Methods. 2017 Jun 29;14(7):645–646.). Quantitative data was reanalyzed from Bedard LG, Dronamraju R, et al. J Biol Chem. 2016 Jun 24;291(26):13410–20.
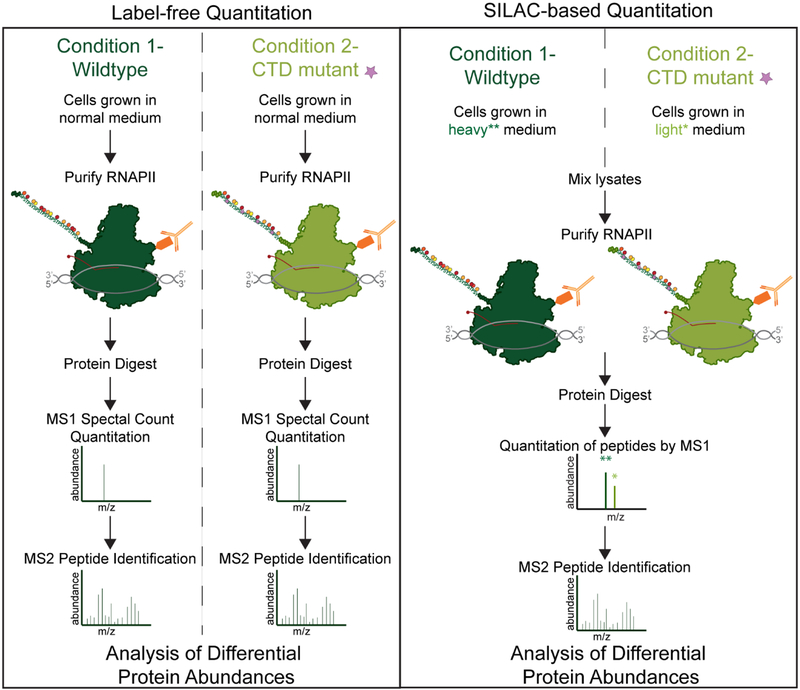
Assuredly, the quality of data AP-MS will continue to improve with the development of new affinity purification mass spectrometry approaches. Emerging methods include, proximity biotin-labeling of interacting proteins through approaches including APEX [43], BioID [44], and antibody recognition [45]. These methods [43–45] have been shown to further improve the sorting true interactors from contaminants, a fundamental problem for AP-MS, as discussed previously. Improvements in MS-based quantitation between samples through isotopic labeling-based multiplexing techniques [46–48] also have the potential to benefit the accuracy and sensitivity of AP-MS. Multiplexing through isotopic labeling allows multiple samples to be analyzed within the same MS run, thereby reducing batch effect and improving quantitation. These approaches have not been broadly applied to the RNAPII interactome at this point in time, and so are not discussed at length in this review. However, Figure 3 depicts an example RNAPII experiment comparing MS quantitation using a label-free method versus using stable isotope labeling in cell (SILAC)-based multiplexing.
Figure 3: Strategies for Quantitative Analysis of RNAPII interactors.
A) Label-free quantitation of RNAPII associated proteins can be done from multiple conditions. Individual mass spectrometry is performed and then quantitation is performed by either MS1 precursor quantitation of ion intensity and/or MS2 fragment ion count based quantitation (referred to as spectral counting or peptide-spectrum match (PSM) counting). B) SILAC based quantitation can be performed using cells grown in various combinations of light and heavy amino acids typically focused on lysine and arginine. Samples can be mixed together based on equal cell numbers or by equal protein concentration. Quantitation is then performed by analysis of the heavy isotope containing or light isotope containing MS1 ion intensities. MS2 based fragment ion analysis is still used for peptide fingerprinting but is not used for quantitation in SILAC based approaches.
A new MS-based method aimed at achieving deeper mechanistic insights from RNAPII AP-MS, by specifically looking at 5’ and 3’ RNAPII complexes, has recently been developed by Harlen and Churchman [49]. RNA stem-loop sequences were inserted into the untranslated regions (UTRs) of a single gene with 2 PP7 sequences in the 5’ UTR and 2 MS2 sequences in the 3’ UTR. Plasmids containing either GFP-PP7 coat binding protein or RFP-MS2 coat binding protein are expressed in these same cells. After total RNAPII is purified via an epitope tag on Rpb3, antibodies against the RNA stem loop coat proteins are used to isolate RNAPII complexes that were specifically enriched at either the 5’ or 3’ end of the gene. The purifications were then analyzed via MS to precisely determine the proteins present at early (5’) and late (3’) transcription. The results for the single gene locus used in these experiments implicate a novel, uncharacterized role for the exonuclease complex Rat1 and Rai1—typically associated with termination—during the earlier phases of transcription [49–52]. Additional results from these experiments, in conjunction with other techniques, support a role for the ubiquitin ligase Bre1 later in transcription than previously thought, through regulation of RNAPII pausing. This method provides an avenue for analysis of RNAPII interactors at specific stages of transcription, although it does require multiple genetic changes to the target gene of interest. Defining the stages of transcription that PPIs occur during will help to elucidate the mechanisms and functions of RNAPII-interacting proteins. The specificity of this approach has unique advantages when compared to purifications based on CTD phosphorylation marks, as these marks are not clear-cut between the different stages of transcription and may be heterogeneous across different gene types. In contrast, a disadvantage of analyzing only the 5’ and 3’ ends of a gene, compared to phospho-CTD pulldowns, is the limitation of single gene loci, and the loss of elongation dynamics that may occur in the gene body. For projects interested in looking at the RNAPII interactome at the start and ends of genes of interest, this approach holds the potential to increase the specificity of RNAPII interactome, increasing the amount of PPI mechanism and function data garnered from AP-MS.
Finally, an important application of AP-MS to the study of RNAPII is the analysis of posttranslational modification that may regulate transcription dynamics. Although, the CTD of Rpb1 receives a majority of the focus in the context of post-translational modifications, it is not the only region of RNAPII that is modified. Mohammed et al. [53] digested RNAPII with multiple proteases before multiplexing the digested peptides to be analyzed via MS, using both collision-induced dissociation (CID) and electron-transfer dissociation (ETD) fragmentation techniques. These methods allowed the detection of 19 phosphorylation-sites outside of the CTD, 12 of these being novel, with possible effects on the conformation of the clamp region. The biological pathways that may be regulated by these phosphorylation events could regulate RNAPII PPIs, which could be characterized through a variety of biochemical studies, including AP-MS. Thus, AP-MS has the potential to not only identify interactors of RNAPII, but also provide essential information regarding RNAPII PTM dynamics that may regulate those interactions. For both interactor identification and PTM analysis, AP-MS offers decreased sample population and increased coverage depth compared to global proteomics. The benefits of AP-MS studies may also be complemented by other methods. For example, structural studies on the RNAPII interacting protein Spt6 have found that previously uncharacterized phosphorylation sites in the linker region of RNAPII (S1493, T1471, and/or Y1473) stabilize the interaction between RNAPII and Spt6, playing an important role in the recruitment of Spt6 throughout the genome [54]. Phosphorylation of RNAPII at T1471 was initially reported by a large scale phosphoproteomics analysis using immobilized metal affinity chromatography (IMAC) in yeast [55] as well as in a focused proteomics study on RNAPII [53]. Further proteomics-based exploration of RNAPII PTMs outside of the CTD will undoubtedly benefit understanding of the complex regulation of the dynamic PPI network that regulates RNAPII transcription.
2.2. Analysis of CTD Interactome Dynamics
The CTD of the largest subunit of RNAPII is known to be an essential regulator of transcription, as its deletion is not compatible with life [56, 57]. Although its composition is simple, consisting of repeats (number varies with organism) of the amino acid consensus sequence Tyr1Ser2Pro3Thr4Ser5Pro6Ser7, its role in the mechanism of transcription has proved to be dynamic and intricate. The hypothesis of a CTD Code has been proposed, wherein the CTD is differentially phosphorylated at each stage of transcription, and this differential phosphorylation is responsible for recruiting the appropriate transcription factors to RNAPII [58]. Decoding the full range of PTM states found within the native CTD in eukaryotic systems is important to understand how RNAPII PPIs are regulated and has been an intense area of focus in the transcription field. Characterization of both CTD interacting proteins and CTD PTM states has been driven by a variety of MS-based methods (Figure 4).
One popular approach to identify candidate CTD interactors is to use a purified GSTCTD for binding assays with lysate from cells of interest (Figure 4C). Carty and Greenleaf [59] used MALDI-MS to identify phospho-CTD associated proteins (PCAPs) after performing an in vitro binding assay using a kinase (CTK1) modified CTD as bait and HeLa cell extract as a source for interacting partners (preys). The novel PCAPs identified in this study suggested that the phospho-CTD was responsible for recruiting more than just RNA-processing proteins, and that CTD phosphorylation played a role in the regulation of a variety of nuclear processes. A recent publication from Ebmeier et al. [60] used a similar approach with GST-purified CTD in a binding assay with HeLa nuclear extract. By first phosphorylating GST-CTD in a kinase assay with either Cdk7 (TFIIH) or Cdk9 (P-TEFb) before the binding assay, Orbitrap MS of pCTD-bound proteins was able to identify overlapping, yet somewhat distinct interactomes for the two different pCTD isoforms. Main findings were that 5’ mRNA capping enzymes and SETD1A/B were identified in both interactomes, suggesting they bind to a phosphorylated CTD. However, SETD2 bound specifically to a P-TEFb phosphorylated CTD. This type of approach can help better understand how differential phosphorylation of the CTD recruits specific proteins to the site of transcription. A GST-CTD approach has also aided in the characterization of an understudied CTD PTM, O-linked N-acetylglucosamine (O-GlcNAc) [61]. While it was shown over 25 years ago that the RNAPII CTD is modified by O-GlcNAc [62], the function this modification has during transcription has remained elusive. Recent ETD MS analysis of GSTCTD after incubation with O-GlcNAc-transferase (OGT) was able to detect O-GlcNAc modifications on Ser2 and Ser5 residues [61]. Further experiments demonstrated that O-GlcNAc exists on RNAPII at promoters, and inhibition of O-GlcNAcylation prevented RNAPII progression [61]. These data suggest an important role for O-GlcNAc in regulating transcription initiation and early elongation, possibly even through a reciprocal relationship with Ser2 and Ser5 phosphorylation. The interplay between the different CTD PTMs and the proteins that regulate them is an exciting and ongoing area of study in the transcription field that has been aided by MS analysis.
Mass spectrometry has helped fine tune the details of how phosphorylation sites of the CTD are regulated by the interplay between protein kinases and phosphatases, using both in vitro and in vivo approaches. Smith-Kinnaman et al. [63] used and AP-MS approach to focus on investigating the role of phosphorylation on the recruitment of the CTD phosphatase Rtr1, which is enriched in early elongation by ChIP-qPCR. AP-MS was able to identify 20 proteins within the interactome of an epitope tagged version of the Rtr1 phosphatase, including 9 subunits of RNAPII. However, when Rtr1 was affinity purified from CTK1 deletion yeast (CTDK-1 Ser2 kinase), the interaction probability of Rtr1 with RNAPII was decreased. These results, along with other experiments, suggest that the interaction of Rtr1 and RNAPII is regulated by CTDK-1 and that hyperphosphorylation of the CTD is required for Rtr1 recruitment. Synthetic peptides have also been a valuable MS application to study CTD kinases and phosphatases considering that it is a highly quantitative approach. Czudnochowski [64] et al. were able to use synthetic CTD peptides that were either hyperphosphorylated, Ser2P, Ser5P or Ser7P in a time-course experiment with P-TEFb, a well described Ser2 kinase in vivo. The phosphorylation status of the CTD after incubation with P-TEFb, was analyzed by ESI-MS and it was observed that only the Ser7P CTD peptide was further phosphorylated, and that this phosphorylation occurred on Ser5. These results indicate a Ser7P CTD as the preferred substrate for the P-TEFb kinase to phosphorylate Ser5, challenging the dogma of P-TEFb as a Ser2 kinase. Using the Drosophila melanogaster CTD (DmCTD) as a substrate, Gibbs et. al similarly found that DmP-TEFb heavily modifies Ser5P in vitro in 12/42 DmCTD repeats [65]. These studies suggest that phosphorylation of the CTD at Ser2P in vivo may require additional factors which contribute to the change in P-TEFb specificity from Ser5P in vitro. Luo et al. [66] investigated whether Thr4P affects Ssu72 activity, as the phosphatase seems to require a very specific CTD substrate with Pro6 in a cis-confirmation [67]. In their experiments, Ssu72 activity was measured by MS, and it was observed that the presence of Thr4P on the CTD lowered Ssu72 activity by approximately four-fold, but did not completely abolish it. Ssu72 also did not remove Thr4P itself. The authors propose that this activity decrease could be a mechanism of fine-tuning phosphatase activity.
Thr4P has also recently been implicated in the recruitment of additional transcription regulators using an AP-MS approach with mutant CTD constructs in yeast. Nemec et al. [68] affinity purified RNAPII with either WT or T4A (Thr4->Ala) CTDs. Label-free MS-based quantitation was then used to identify significant interacting proteins (Figure 4A). These studies found that termination factors, including Rtt103, were significantly decreased in the T4A mutant. These data, along with other experiments showing termination defects in the presence of T4A, and the direct binding of Rtt103 to a Thr4P CTD, implicate a role for Thr4P in the regulation of Rtt103 termination at specific genes. Harlen and Churchman [69] have also identified roles for Thr4P in the regulation of global transcription. In an effort to analyze phospho-specific CTD interactomes, sequential immunoprecipitations (IP) with phospho-site specific monoclonal antibodies were used (Figure 4B). First total RNAPII was purified via epitope tag and then phospho-site CTD antibodies were used to isolate the specific populations containing the phospho-CTD of interest for MS. The IPs were then analyzed via label-free MS to identify prey proteins and the data showed distinct interactomes for each specific phospho-isoform of RNAPII. In agreement with the Nemec, et al. study, Rtt103 was found to be enriched in the Thr4P interactome although a different AP-MS approach was used. Additionally, spliceosome proteins were depleted in the RNAPII Thr4P interactome, but were enriched in the Ser5P interactome. Along with other experiments in the study, this data implicates Thr4 in a recruitment and release mechanism for the spliceosome machinery; unphosphorylated Thr4 may allow its recruitment, while Thr4P stimulates its release. The data discussed in this section of the review presents roles for CTD phosphorylation sites other than Ser2 and Ser5 in global regulation of transcription.
Additionally, it is critical to note that in higher eukaryotes, there are heptads within the CTD that vary from the consensus repeat sequence. A common variant in the 7th position, normally a serine residue, is lysine. The presence of Lys7 opens up additional possible modifications, such as acetylation and methylation (discussed below). However, it is possible that the lysine residue in the 7th position may also be important to mediate specific metazoan specific PPIs. Recent work has characterized proteins with altered RNAPII interaction following mutation of eight Lys7 residues to arginine using culture SILAC and AP-MS (Figure 4B). Lys7 to Arg mutations will retain a positive charge but can no longer be modified by enzymes who target primary amines for modification. Using this approach, it was discovered that the RPRD family of proteins (RPRD1A, RPRD1B, RPRD2) are significantly reduced in RNAPII purifications from Lys7 to Arg expressing HEK293T cells[70]. Additionally, proteins that have previously been shown to interact with the RPRDs were also reduced including: RPAP2, RPAP3, MCM7 and RUVB1. The RPRD family of proteins had previously been shown to have an increased affinity for doubly phosphorylated CTD peptides[71]. Interestingly, the combination of Ser2P and Lys7Ac CTD modification also shows an increased affinity for RPRD1B.
The methods described in this section include both in vitro and in vivo modified CTD approaches. Both approaches have advantages and disadvantages that are important to consider when deciding how to best experimentally answer a question. A major drawback of in vitro modified CTD substrates (either GST-CTD or synthetic peptides) is the inability to fully recapitulate the endogenous heterogeneity of the phosphorylated CTD due to contributions in vivo by a variety of kinases [72], removal of an unknown portion of phosphorylation sites by phosphatases, and sequential modification/modification removal cycles. For instance, studies on the CTD phosphatase Ssu72 have shown that Ssu72 dephosphorylation may serve as an upstream regulator of another CTD phosphatase, Fcp1 [73, 74]. However, a major advantage of in vitro approaches, as described earlier in this section by the data gained, is the ability to control how the CTD is modified and analysis of CTD interactors by MS is thereby more likely to be precise and quantitative. In contrast, in vivo modified CTDs are more likely to retain their endogenous, and thereby biologically relevant, modification states. However, the difficulty is that endogenous CTD is highly heterogeneously, and thereby it becomes very difficult to analyze specific CTD PTM interactomes quantitatively. Both approaches have also suffered from the lack of a technique to map individual PTM sites on the CTD, limiting the ability to detail exactly where and how much the CTD is modified at any one time, which will be critical to further characterizing the dynamics of CTD PTMs regulate PPIs at the site of transcription. Recently developed methods that aim to directly analyze the post-translational modification sites of the CTD are discussed in the next section (See Figure 4 for a comparison of CTD analysis methods).
2.3. Direct Analysis of CTD Post-translational Modification Site Composition
As discussed in the above section, mass spectrometry has been used to great effect for the identification and characterization of CTD protein interactors, as well as the dynamics of these interactions based on general phosphorylation patterns. However, direct MS analysis of the CTD itself has posed a challenge. Due to the lack of tryptic cleavage sites among consensus heptads and the repetitive nature of the CTD, the mapping of individual modification sites has been problematic. However, recent method development provided an avenue for such analyses. Lys7 residues also provide tryptic cleavage sites at the end of variant heptads. While coverage is still not ideal, there is some opportunity for modification mapping. Voss et al. [75] used mass spectrometry to identify acetylation as well as mono-, di-, or tri-methylation of the human CTD. These findings were supported by another group which identified the same modifications using an antibody-based approach [76]. In all, these data found that acetylation or di-/tri-methylation only occurred on phosphorylated CTD peptides and that specific Lys7 residues could be alternatively acetylated and methylated. In studies such as these, mass spectrometry provides an advantage over western blot analysis because the degree of cooccurrence of PTMs on RNAPII CTD peptides can be directly assessed using MS. Voss et al. found acetylation in combination with mono-repeat phosphorylation (within two CTD consensus repeats) at Tyr1, Ser2, Thr4, or Ser5 [75]. In contrast, mono-methylation could occur on either mono-repeat phosphorylated or hypo-phosphorylated CTD di-heptad peptides. The combined data shows modification of variant Lys7 residues to be as dynamic as that of consensus residues. As Lys7 is not essential for life, but conserved in vertebrates, the authors speculate that Lys7 modification could regulate the transcription of a subset of vertebrate genes.
Recent method development has taken further advantage of Lys7 residues. These variants do not occur naturally in yeast, and their presence in higher eukaryotes does not occur until well toward the distal end of the CTD, thus limiting tryptic cleavage over the full length of the CTD. To address the compatibility of the CTD for MS analysis, both Suh et al. [77] and Shuller et al. [78] endogenously modified the CTD of Rpb1 to be more suitable for MS analysis, termed the msCTD (Figure 4D). Within a subset of heptads, the 7th position residue, normally serine, is replaced with either lysine or arginine to facilitate tryptic cleavage. The msCTDs also contain subsets of heptads with additional residue substitutions in order to give cleaved fragments unique precursor masses, thus enabling modification mapping to individual residues within the natively repetitive CTD sequence. Suh et al. [77] used yeast CTD while Shuller et al. [78] focused mainly on mammalian CTD; despite the difference in model systems the MS data show striking similarities. Both studies confirm the presence of phosphorylation on Tyr1, Ser2, Thr4, Ser5, and Ser7, some of which had only previously been identified by antibody-based approaches. One major conclusion from both data sets is that on average, Ser2P and Ser5P levels are fold-change levels higher than Tyr1P, Thr4P, and Ser7P. Authors used this result to caution against direct comparison of different antibodies for relative quantitation of CTD phosphorylation marks. To confirm that MS analysis was able to measure phosphorylation changes at a physiologically relevant level, both studies employed Ser2 kinase inhibition, whether through mutation or drug treatment. Positively, both data sets showed marked decrease in Ser2P levels upon inhibition, while there was very little change in phosphorylation of the other CTD residues.
In both msCTD studies, the total phosphorylation level of the CTD was found to be much lower than expected. Multiply-phosphorylated heptads were found to be rare in both yeast and mammalian cells. With this initial data observing low total phosphorylation dominated by Ser2P and Ser5P, authors speculate that the CTD code might end up being simpler than expected. This idea is somewhat contrary to data presented in the above section, which suggest important roles for Ser7P, Thr4P and Lys7 modifications in metazoans. One caveat for these msCTDs is that Ser7 phosphorylation levels may be measured as low due to the mutations made to make the CTD suitable for mass spectrometry (although this possibility was considered in the published works). A potential drawback to using a modified msCTD is that the CTD is no longer wildtype. Both studies confirmed that cells carrying only msCTD grew in a manner approximate to wildtype. However, it remains to be seen how the mutations and trypsin cleavage may alter the ability to detect wildtype phosphorylation marks. Finally, it is also possible that heavily phosphorylated CTD peptides were refractory to reversed-phase column elution and/or electrospray ionization possibly losing the net positive charge required for guidance of the peptides through the mass spectrometer. Challenges with multiply-phosphorylated peptides in reversed phase chromatography have been reported previously [79] as have ionization issues for heavily phosphorylated sequences [80], although the latter issue has been reported to have a high-degree of peptide sequence dependence. Nevertheless, msCTDs are an exciting new tool for analysis of quantitative PTM mapping of an in vivo modified CTD.
Beyond the addition of the phosphate group itself, phosphorylation has the potential to change the structure of the CTD. Recent work has focused on investigating how phosphorylation might alter the CTD and regulate its interactions beyond the addition of the mark itself. Gibbs et al. [65] used a combination of mass spectrometry and NMR spectroscopy to observe that hyper Ser5P alters the local CTD structure via cis-proline isomerization in a sequence-dependent manner. This proline isomerization was then shown to modulate the activity of Ssu72, which is known to be cis-proline specific CTD interactor. Specific mammalian CTD repeat variants were found to more frequently be in a cis-proline state upon Ser5P, suggesting a potential mechanism for increased Ssu72 recruitment to specific regions of the CTD.
2.4. Analysis of RNAPII by structural proteomics
Structural proteomics is a rapidly developing area of mass spectrometry-based method development (reviewed in [81]). Many methods that fall under the umbrella of structural proteomics have been used to analyze RNAPII and its interactions with accessory proteins to provide novel insights into RNAPII biology and to aid in MS method development efforts. Crosslinking-mass spectrometry (XL-MS), for instance, has been used to study a number of macromolecular complexes including TFIIH [82]. The cross-linker Bis (sulphosuccinimidyl) suberate (BS3) has been used to map interactions between RNAPII and TFIIF [83] as well as interactions between TFIIF and TFIIH [84] to assist in modeling protein complex positioning within a cryoEM structure of a 32-protein RNAPII pre-initiation complex (PIC). These structural studies were expanded to a full 52 protein PIC with Mediator using additional crosslinking experiments of Mediator-PIC and Mediator-RNAPII with the 1-hydroxy-7-azabenzotriazole analog of DSS (disuccinimidyl suberate), 1,1’-(suberoyldioxy)bisazabenzotriazole (SBAT) [85]. The data from the BS3 and SBAT XL-MS studies were later utilized in a cryoEM study of the yeast PIC and PIC-core Mediator complexes. This 2017 study additionally obtained novel crosslinks using the 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC) crosslinker and were able to retain structures cryoEM with resolutions of 4.7 Å and 5.8 Å, respectively [86].
BS3-based XL-MS has continued to couple well with cryo-electron microscopy based structural biology analysis of large transcription-related complexes and has been used to provide peptide level resolution of amino acid proximity for: RNAPII-PAFC-TFIIS [87], RNAPII–DSIF–NELF [88], and RNAPII-DSIF-PAFC-SPT6 complexes These approaches provide a high degree of analytical power for coupling with protein complexes used in structural biology studies.. An emerging approach for large-scale such as organelle scale XL-MS is using mass-spectrometry cleavable crosslinkers (reviewed in [89]). MS-cleavable crosslinkers have been applied to large-scale protein-protein interaction analysis [90] and could be used for both qualitative and quantitative studies. Although disuccinimidyl sulfoxide cleavable crosslinking has not been used to interrogate the RNAPII interactome directly at this point, recent studies on the histone interactions isolated from intact nuclei could provide novel insights into co-transcriptional histone biology. DSSO crosslinking of intact nuclei and subsequent analysis by a MS3-based method identified a large number of potential histone interacting proteins (n=778) as well as histone crosslinks with known interactors [91]. While organelle level analysis of protein-protein interactions with DSSO is a powerful approach, there are still challenges to overcome since the depth of coverage is limited by multiple factors including the longer cycle times needed to carry out MS3 analysis.
An additional emerging approach in structural proteomics for analysis of RNAPII complexes involves the application of native spray mass spectrometry. Advances in this area have been driven by changes in sample preparation approaches, as well as technological developments and optimizations that allow for large protein assemblies to both ionize and retain their native quaternary state in gas phase. These approaches can be applied to macromolecular complexes to obtain precise analytical data related to intact protein complex mass, which can reveal protein subunit stoichiometry, post-translational modifications, and protein complex modularity. For RNAPII complexes, native electrospray mass spectrometry has been used to monitor complex engagement of alpha-amanitin [92, 93], as well as to investigate the stoichiometry of the yeast capping enzyme alone and in complex with transcribing phosphorylated-RNAPII (with a 20.2 kDa DNA-RNA scaffold) [94]. These native-MS studies revealed that phosphorylated RNAPII can interact with either heterotrimeric (Cet1-Cet1-Ceg1) or heterotetrameric (Ceg1-Cet1-Cet1-Ceg1) forms of the capping enzyme. The capping enzyme-RNAPII complexes were also monitored in the presence or absence of either GTP or the GTP analog, GpCpp. Theremarkable utility of the native-MS approach was clearly illustrated by the authors’ use of negative ion mode (positive ion mode is typically used for protein analysis in MS) to quantitatively monitor the addition of the RNA modification (5′-triphosphate end) by capping enzyme. While, protein complex modularity of RNAPII has not been monitored by native spray mass spectrometry, this application of the technique is not necessary, due to the wealth of high resolution RNAPII structural data that has already been obtained. However, noncovalent nanoelectrospray ionization mass spectrometry-based investigations of the yeast RNAPII-associated cleavage and polyadenylation factor (CPF) complex has revealed a wealth of information regarding the modularity of the complex [95]. The native-MS studies of CPF were able to define polyA-polymerase, nuclease, and phosphatase modules, aidingd in the selection of subunits for cryoEM structural analysis of the polymerase module of CPF [95]. This study, along with the others highlighted in this section demonstrate the utility of MS-based structural proteomics approaches in gaining novel insights into the protein complexes that regulate RNAPII transcription. Native spray MS has also been applied to other transcription related complexes of interest including RNAPI, RNAPIII, and TFIID [96–98].
3. Conclusion
While there is valuable knowledge of transcription dynamics to be gained through high-throughput sequencing methods, mass spectrometry-based analysis of RNAPII and its associated proteins is critical to advance our discovery of new transcription-related biology and related mechanisms. Application of mass spectrometry to the study of RNAPII transcription has shown to be immensely useful in both the qualitative and quantitative analysis of protein-protein interactions that regulate the process. Advances in MS technologies in sample preparation, instrumentation, and data analysis will increasingly facilitate sensitive and accurate quantitation of protein-protein interactions, post-translational modifications, and enzymatic activity. The emerging area of structural proteomics has already greatly benefitted the transcription field and there is no doubt that technological advancements will continue to change the types of mechanistic questions that can be asked and answered about RNAPII transcription.
Highlights.
Mass spectrometry analysis has advanced our understanding of RNA Polymerase biology from both qualitative and quantitative perspectives.
The various applications of mass spectrometry to RNA Polymerase II complexes and relevant results are reviewed.
Affinity purification-mass spectrometry of RNAPII complexes has led to the discovery of novel transcription- and protein complex assembly-related biology
Analysis of C-terminal domain modifications and interactors by mass spectrometry continues to inform our understanding of the CTD code
Structural proteomics methods are rapidly advancing our understanding of the mechanisms of RNAPII transcription and its associated protein complexes
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tan-Wong SM, Zaugg JB, Camblong J, Xu Z, Zhang DW, Mischo HE, Ansari AZ, Luscombe NM, Steinmetz LM, and Proudfoot NJ, Gene Loops Enhance Transcriptional Directionality. Science, 2012. 338(6107): p. 671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wyers F, Rougemaille M, Badis G, Rousselle J-C, Dufour M-E, Boulay J, Régnault B, Devaux F, Namane A, Séraphin B, Libri D, and Jacquier A, Cryptic Pol II Transcripts Are Degraded by a Nuclear Quality Control Pathway Involving a New Poly(A) Polymerase. Cell, 2005. 121(5): p. 725–737. [DOI] [PubMed] [Google Scholar]
- 3.Sopta M, Burton ZF, and Greenblatt J, Structure and associated DNA-helicase activity of a general transcription initiation factor that binds to RNA polymerase II. Nature, 1989. 341(6241): p. 410–4. [DOI] [PubMed] [Google Scholar]
- 4.Eng JK, McCormack AL, and Yates JR, An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom, 1994. 5(11): p. 976–89. [DOI] [PubMed] [Google Scholar]
- 5.Grant PA, Schieltz D, Pray-Grant MG, Steger DJ, Reese JC, Yates JR, and Workman JL, A Subset of TAFIIs Are Integral Components of the SAGA Complex Required for Nucleosome Acetylation and Transcriptional Stimulation. Cell, 1998. 94(1): p. 45–53. [DOI] [PubMed] [Google Scholar]
- 6.Li J, Moazed D, and Gygi SP, Association of the histone methyltransferase Set2 with RNA polymerase II plays a role in transcription elongation. J Biol Chem, 2002. 277(51): p. 49383–8. [DOI] [PubMed] [Google Scholar]
- 7.Li B, Howe L, Anderson S, Yates JR 3rd, and Workman JL, The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem, 2003. 278(11): p. 8897–903. [DOI] [PubMed] [Google Scholar]
- 8.Xiao T, Hall H, Kizer KO, Shibata Y, Hall MC, Borchers CH, and Strahl BD, Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev, 2003. 17(5): p. 654–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krogan NJ, Kim M, Tong A, Golshani A, Cagney G, Canadien V, Richards DP, Beattie BK, Emili A, Boone C, Shilatifard A, Buratowski S, and Greenblatt J, Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol Cell Biol, 2003. 23(12): p. 4207–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daulny A, Geng F, Muratani M, Geisinger JM, Salghetti SE, and Tansey WP, Modulation of RNA polymerase II subunit composition by ubiquitylation. Proc Natl Acad Sci U S A, 2008. 105(50): p. 19649–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mosley AL, Hunter GO, Sardiu ME, Smolle M, Workman JL, Florens L, and Washburn MP, Quantitative proteomics demonstrates that the RNA polymerase II subunits Rpb4 and Rpb7 dissociate during transcriptional elongation. Mol Cell Proteomics, 2013. 12(6): p. 1530–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mosley AL, Pattenden SG, Carey M, Venkatesh S, Gilmore JM, Florens L, Workman JL, and Washburn MP, Rtr1 is a CTD phosphatase that regulates RNA polymerase II during the transition from serine 5 to serine 2 phosphorylation. Mol Cell, 2009. 34(2): p. 168–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeronimo C, Forget D, Bouchard A, Li Q, Chua G, Poitras C, Therien C, Bergeron D, Bourassa S, Greenblatt J, Chabot B, Poirier GG, Hughes TR, Blanchette M, Price DH, and Coulombe B, Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol Cell, 2007. 27(2): p. 262–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Irani S, Yogesha SD, Mayfield J, Zhang M, Zhang Y, Matthews WL, Nie G, Prescott NA, and Zhang YJ, Structure of Saccharomyces cerevisiae Rtr1 reveals an active site for an atypical phosphatase. Sci Signal, 2016. 9(417): p. ra24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsu PL, Yang F, Smith-Kinnaman W, Yang W, Song JE, Mosley AL, and Varani G, Rtr1 is a dual specificity phosphatase that dephosphorylates Tyr1 and Ser5 on the RNA polymerase II CTD. J Mol Biol, 2014. 426(16): p. 2970–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hunter GO, Fox MJ, Smith-Kinnaman WR, Gogol M, Fleharty B, and Mosley AL, Phosphatase Rtr1 Regulates Global Levels of Serine 5 RNA Polymerase II C-Terminal Domain Phosphorylation and Cotranscriptional Histone Methylation. Molecular and Cellular Biology, 2016. 36(17): p. 2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith-Kinnaman WR, Berna MJ, Hunter GO, True JD, Hsu P, Cabello GI, Fox MJ, Varani G, and Mosley AL, The interactome of the atypical phosphatase Rtr1 in Saccharomyces cerevisiae. Molecular bioSystems, 2014. 10(7): p. 1730–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ni Z, Xu C, Guo X, Hunter GO, Kuznetsova OV, Tempel W, Marcon E, Zhong G, Guo H, Kuo W-HW, Li J, Young P, Olsen JB, Wan C, Loppnau P, El Bakkouri M, Senisterra GA, He H, Huang H, Sidhu SS, Emili A, Murphy S, Mosley AL, Arrowsmith CH, Min J, and Greenblatt JF, RPRD1A and RPRD1B are human RNA polymerase II C-terminal domain scaffolds for Ser5 dephosphorylation. Nature structural & molecular biology, 2014. 21(8): p. 686–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Egloff S, Zaborowska J, Laitem C, Kiss T, and Murphy S, Ser7 phosphorylation of the CTD recruits the RPAP2 Ser5 phosphatase to snRNA genes. Mol Cell, 2012. 45(1): p. 111–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang TK, Lawrence DA, Lu M, Tan J, Harnoss JM, Marsters SA, Liu P, Sandoval W, Martin SE, and Ashkenazi A, Coordination between Two Branches of the Unfolded Protein Response Determines Apoptotic Cell Fate. Mol Cell, 2018. 71(4): p. 629–636.e5. [DOI] [PubMed] [Google Scholar]
- 21.Krogan NJ, Kim M, Ahn SH, Zhong G, Kobor MS, Cagney G, Emili A, Shilatifard A, Buratowski S, and Greenblatt JF, RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol Cell Biol, 2002. 22(20): p. 6979–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dronamraju R, Kerschner JL, Peck SA, Hepperla AJ, Adams AT, Hughes KD, Aslam S, Yoblinski AR, Davis IJ, Mosley AL, and Strahl BD, Casein Kinase II Phosphorylation of Spt6 Enforces Transcriptional Fidelity by Maintaining Spn1-Spt6 Interaction. Cell Rep, 2018. 25(12): p. 3476–3489 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gouot E, Bhat W, Rufiange A, Fournier E, Paquet E, and Nourani A, Casein kinase 2 mediated phosphorylation of Spt6 modulates histone dynamics and regulates spurious transcription. Nucleic Acids Research, 2018. 46(15): p. 7612–7630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li S, Almeida AR, Radebaugh CA, Zhang L, Chen X, Huang L, Thurston AK, Kalashnikova AA, Hansen JC, Luger K, and Stargell LA, The elongation factor Spn1 is a multi-functional chromatin binding protein. Nucleic Acids Research, 2018. 46(5): p. 2321–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thurston AK, Radebaugh CA, Almeida AR, Argueso JL, and Stargell LA, Genome Instability Is Promoted by the Chromatin-Binding Protein Spn1 in Saccharomyces cerevisiae. Genetics, 2018. 210(4): p. 1227–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bedard LG, Dronamraju R, Kerschner JL, Hunter GO, Axley ED, Boyd AK, Strahl BD, and Mosley AL, Quantitative Analysis of Dynamic Protein Interactions during Transcription Reveals a Role for Casein Kinase II in Polymerase-associated Factor (PAF) Complex Phosphorylation and Regulation of Histone H2B Monoubiquitylation. J Biol Chem, 2016. 291(26): p. 13410–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeronimo C, Langelier MF, Zeghouf M, Cojocaru M, Bergeron D, Baali D, Forget D, Mnaimneh S, Davierwala AP, Pootoolal J, Chandy M, Canadien V, Beattie BK, Richards DP, Workman JL, Hughes TR, Greenblatt J, and Coulombe B, RPAP1, a novel human RNA polymerase II-associated protein affinity purified with recombinant wild-type and mutated polymerase subunits. Mol Cell Biol, 2004. 24(16): p. 7043–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Minaker SW, Filiatrault MC, Ben-Aroya S, Hieter P, and Stirling PC, Biogenesis of RNA polymerases II and III requires the conserved GPN small GTPases in Saccharomyces cerevisiae. Genetics, 2013. 193(3): p. 853–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niesser J, Wagner FR, Kostrewa D, Mühlbacher W, and Cramer P, Structure of GPN-Loop GTPase Npa3 and Implications for RNA Polymerase II Assembly. Molecular and Cellular Biology, 2016. 36(5): p. 820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reyes-Pardo H, Barbosa-Camacho AA, Perez-Mejia AE, Lara-Chacon B, Salas-Estrada LA, Robledo-Rivera AY, Montero-Moran GM, Lara-Gonzalez S, Calera MR, and Sanchez-Olea R, A nuclear export sequence in GPN-loop GTPase 1, an essential protein for nuclear targeting of RNA polymerase II, is necessary and sufficient for nuclear export. Biochim Biophys Acta, 2012. 1823(10): p. 1756–66. [DOI] [PubMed] [Google Scholar]
- 31.Staresincic L, Walker J, Dirac-Svejstrup AB, Mitter R, and Svejstrup JQ, GTP-dependent binding and nuclear transport of RNA polymerase II by Npa3 protein. J Biol Chem, 2011. 286(41): p. 35553–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maurizy C, Quinternet M, Abel Y, Verheggen C, Santo PE, Bourguet M, Paiva ACF, Bragantini B, Chagot M-E, Robert M-C, Abeza C, Fabre P, Fort P, Vandermoere F, Sousa PMF, Rain J-C, Charpentier B, Cianférani S, Bandeiras TM, Pradet-Balade B, Manival X, and Bertrand E, The RPAP3-Cterminal domain identifies R2TP-like quaternary chaperones. Nature Communications, 2018. 9(1): p. 2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Henri J, Chagot ME, Bourguet M, Abel Y, Terral G, Maurizy C, Aigueperse C, Georgescauld F, Vandermoere F, Saint-Fort R, Behm-Ansmant I, Charpentier B, Pradet-Balade B, Verheggen C, Bertrand E, Meyer P, Cianferani S, Manival X, and Quinternet M, Deep Structural Analysis of RPAP3 and PIH1D1, Two Components of the HSP90 Co-chaperone R2TP Complex. Structure, 2018. 26(9): p. 1196–1209.e8. [DOI] [PubMed] [Google Scholar]
- 34.Martino F, Pal M, Muñoz-Hernández H, Rodríguez CF, Núñez-Ramírez R, Gil-Carton D, Degliesposti G, Skehel JM, Roe SM, Prodromou C, Pearl LH, and Llorca O, RPAP3 provides a flexible scaffold for coupling HSP90 to the human R2TP co-chaperone complex. Nature Communications, 2018. 9(1): p. 1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lynch CJ, Bernad R, Calvo I, Nobrega-Pereira S, Ruiz S, Ibarz N, Martinez-Val A, Grana-Castro O, Gomez-Lopez G, Andres-Leon E, Espinosa Angarica V, Del Sol A, Ortega S, Fernandez-Capetillo O, Rojo E, Munoz J, and Serrano M, The RNA Polymerase II Factor RPAP1 Is Critical for Mediator-Driven Transcription and Cell Identity. Cell Rep, 2018. 22(2): p. 396–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ranish JA, Yi EC, Leslie DM, Purvine SO, Goodlett DR, Eng J, and Aebersold R, The study of macromolecular complexes by quantitative proteomics. Nat Genet, 2003. 33(3): p. 349–55. [DOI] [PubMed] [Google Scholar]
- 37.Ranish JA, Yudkovsky N, and Hahn S, Intermediates in formation and activity of the RNA polymerase II preinitiation complex: holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB. Genes Dev, 1999. 13(1): p. 49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mellacheruvu D, Wright Z, Couzens AL, Lambert J-P, St-Denis NA, Li T, Miteva YV, Hauri S, Sardiu ME, Low TY, Halim VA, Bagshaw RD, Hubner NC, al-Hakim A, Bouchard A, Faubert D, Fermin D, Dunham WH, Goudreault M, Lin Z-Y, Badillo BG, Pawson T, Durocher D, Coulombe B, Aebersold R, Superti-Furga G, Colinge J, Heck AJR, Choi H, Gstaiger M, Mohammed S, Cristea IM, Bennett KL, Washburn MP, Raught B, Ewing RM, Gingras A-C, and Nesvizhskii AI, The CRAPome: a contaminant repository for affinity purification–mass spectrometry data. Nature Methods, 2013. 10: p. 730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choi H, Larsen B, Lin Z-Y, Breitkreutz A, Mellacheruvu D, Fermin D, Qin ZS, Tyers M, Gingras A-C, and Nesvizhskii AI, SAINT: probabilistic scoring of affinity purification-mass spectrometry data. Nature methods, 2011. 8(1): p. 70–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wenger CD, Phanstiel DH, Lee MV, Bailey DJ, and Coon JJ, COMPASS: a suite of pre- and post-search proteomics software tools for OMSSA. Proteomics, 2011. 11(6): p. 1064–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Knight JDR, Choi H, Gupta GD, Pelletier L, Raught B, Nesvizhskii AI, and Gingras AC, ProHits-viz: a suite of web tools for visualizing interaction proteomics data. Nat Methods, 2017. 14(7): p. 645–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mosley AL, Sardiu ME, Pattenden SG, Workman JL, Florens L, and Washburn MP, Highly reproducible label free quantitative proteomic analysis of RNA polymerase complexes. Molecular & Cellular Proteomics , 2010: p. mcp.M110.000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Braisted JC, Kuntumalla S, Vogel C, Marcotte EM, Rodrigues AR, Wang R, Huang ST, Ferlanti ES, Saeed AI, Fleischmann RD, Peterson SN, and Pieper R, The APEX Quantitative Proteomics Tool: generating protein quantitation estimates from LCMS/MS proteomics results. BMC Bioinformatics, 2008. 9: p. 529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roux KJ, Kim DI, and Burke B, BioID: a screen for protein-protein interactions. Curr Protoc Protein Sci, 2013. 74: p. Unit 19.23. [DOI] [PubMed] [Google Scholar]
- 45.Bar DZ, Atkatsh K, Tavarez U, Erdos MR, Gruenbaum Y, and Collins FS, Biotinylation by antibody recognition-a method for proximity labeling. Nat Methods, 2018. 15(2): p. 127–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thompson A, Schäfer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, Neumann T, and Hamon C, Tandem Mass Tags: A Novel Quantification Strategy for Comparative Analysis of Complex Protein Mixtures by MS/MS. Analytical Chemistry, 2003. 75(8): p. 1895–1904. [DOI] [PubMed] [Google Scholar]
- 47.Wiese S, Reidegeld KA, Meyer HE, and Warscheid B, Protein labeling by iTRAQ: a new tool for quantitative mass spectrometry in proteome research. Proteomics, 2007. 7(3): p. 340–50. [DOI] [PubMed] [Google Scholar]
- 48.McAlister GC, Nusinow DP, Jedrychowski MP, Wuhr M, Huttlin EL, Erickson BK, Rad R, Haas W, and Gygi SP, MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal Chem, 2014. 86(14): p. 7150–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harlen KM and Churchman LS, Subgenic Pol II interactomes identify region-specific transcription elongation regulators. Mol Syst Biol, 2017. 13(1): p. 900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim M, Krogan NJ, Vasiljeva L, Rando OJ, Nedea E, Greenblatt JF, and Buratowski S, The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature, 2004. 432(7016): p. 517–22. [DOI] [PubMed] [Google Scholar]
- 51.Jimeno-Gonzalez S, Haaning LL, Malagon F, and Jensen TH, The yeast 5’–3’ exonuclease Rat1p functions during transcription elongation by RNA polymerase II. Mol Cell, 2010. 37(4): p. 580–7. [DOI] [PubMed] [Google Scholar]
- 52.Brannan K, Kim H, Erickson B, Glover-Cutter K, Kim S, Fong N, Kiemele L, Hansen K, Davis R, Lykke-Andersen J, and Bentley DL, mRNA decapping factors and the exonuclease Xrn2 function in widespread premature termination of RNA polymerase II transcription. Mol Cell, 2012. 46(3): p. 311–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mohammed S, Lorenzen K, Kerkhoven R, van Breukelen B, Vannini A, Cramer P, and Heck AJ, Multiplexed proteomics mapping of yeast RNA polymerase II and III allows near-complete sequence coverage and reveals several novel phosphorylation sites. Anal Chem, 2008. 80(10): p. 3584–92. [DOI] [PubMed] [Google Scholar]
- 54.Sdano MA, Fulcher JM, Palani S, Chandrasekharan MB, Parnell TJ, Whitby FG, Formosa T, and Hill CP, A novel SH2 recognition mechanism recruits Spt6 to the doubly phosphorylated RNA polymerase II linker at sites of transcription. Elife, 2017. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Albuquerque CP, Smolka MB, Payne SH, Bafna V, Eng J, and Zhou H, A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol Cell Proteomics, 2008. 7(7): p. 1389–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nonet M, Sweetser D, and Young RA, Functional redundancy and structural polymorphism in the large subunit of RNA polymerase II. Cell, 1987. 50(6): p. 909–915. [DOI] [PubMed] [Google Scholar]
- 57.Zehring WA, Lee JM, Weeks JR, Jokerst RS, and Greenleaf AL, The C-terminal repeat domain of RNA polymerase II largest subunit is essential in vivo but is not required for accurate transcription initiation in vitro. Proceedings of the National Academy of Sciences, 1988. 85(11): p. 3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Buratowski S, The CTD code. Nat Struct Biol, 2003. 10(9): p. 679–80. [DOI] [PubMed] [Google Scholar]
- 59.Carty SM and Greenleaf AL, Hyperphosphorylated C-terminal repeat domain-associating proteins in the nuclear proteome link transcription to DNA/chromatin modification and RNA processing. Mol Cell Proteomics, 2002. 1(8): p. 598–610. [DOI] [PubMed] [Google Scholar]
- 60.Ebmeier CC, Erickson B, Allen BL, Allen MA, Kim H, Fong N, Jacobsen JR, Liang K, Shilatifard A, Dowell RD, Old WM, Bentley DL, and Taatjes DJ, Human TFIIH Kinase CDK7 Regulates Transcription-Associated Chromatin Modifications. Cell Rep, 2017. 20(5): p. 1173–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lewis BA, Burlingame AL, and Myers SA, Human RNA Polymerase II Promoter Recruitment in Vitro Is Regulated by O-Linked N-Acetylglucosaminyltransferase (OGT). J Biol Chem, 2016. 291(27): p. 14056–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kelly WG, Dahmus ME, and Hart GW, RNA polymerase II is a glycoprotein. Modification of the COOH-terminal domain by O-GlcNAc. J Biol Chem, 1993. 268(14): p. 10416–24. [PubMed] [Google Scholar]
- 63.Smith-Kinnaman WR, Berna MJ, Hunter GO, True JD, Hsu P, Cabello GI, Fox MJ, Varani G, and Mosley AL, The interactome of the atypical phosphatase Rtr1 in Saccharomyces cerevisiae. Mol Biosyst, 2014. 10(7): p. 1730–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Czudnochowski N, Bosken CA, and Geyer M, Serine-7 but not serine-5 phosphorylation primes RNA polymerase II CTD for P-TEFb recognition. Nat Commun, 2012. 3: p. 842. [DOI] [PubMed] [Google Scholar]
- 65.Gibbs EB, Lu F, Portz B, Fisher MJ, Medellin BP, Laremore TN, Zhang YJ, Gilmour DS, and Showalter SA, Phosphorylation induces sequence-specific conformational switches in the RNA polymerase II C-terminal domain. Nat Commun, 2017. 8: p. 15233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Luo Y, Yogesha SD, Cannon JR, Yan W, Ellington AD, Brodbelt JS, and Zhang Y, novel modifications on C-terminal domain of RNA polymerase II can fine-tune the phosphatase activity of Ssu72. ACS Chem Biol, 2013. 8(9): p. 2042–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Werner-Allen JW, Lee CJ, Liu P, Nicely NI, Wang S, Greenleaf AL, and Zhou P, cis-Proline-mediated Ser(P)5 dephosphorylation by the RNA polymerase II C-terminal domain phosphatase Ssu72. J Biol Chem, 2011. 286(7): p. 5717–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nemec CM, Yang F, Gilmore JM, Hintermair C, Ho YH, Tseng SC, Heidemann M, Zhang Y, Florens L, Gasch AP, Eick D, Washburn MP, Varani G, and Ansari AZ, Different phosphoisoforms of RNA polymerase II engage the Rtt103 termination factor in a structurally analogous manner. Proc Natl Acad Sci U S A, 2017. 114(20): p. E3944–E3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Harlen KM, Trotta KL, Smith EE, Mosaheb MM, Fuchs SM, and Churchman LS, Comprehensive RNA Polymerase II Interactomes Reveal Distinct and Varied Roles for Each Phospho-CTD Residue. Cell Rep, 2016. 15(10): p. 2147–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ali I, Garrido-Ruiz D, Ni Z, Johnson JR, Zhang H, Li P-C, Conrad RJ, Guo X, Min J, Greenblatt J, Jacobson M, Krogan NJ, and Ott M, Crosstalk between RNA Pol II C-Terminal Domain Acetylation and Phosphorylation via RPRD Proteins. bioRxiv, 2018: p. 442491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ni Z, Xu C, Guo X, Hunter GO, Kuznetsova OV, Tempel W, Marcon E, Zhong G, Guo H, Kuo WH, Li J, Young P, Olsen JB, Wan C, Loppnau P, El Bakkouri M, Senisterra GA, He H, Huang H, Sidhu SS, Emili A, Murphy S, Mosley AL, Arrowsmith CH, Min J, and Greenblatt JF, RPRD1A and RPRD1B are human RNA polymerase II C-terminal domain scaffolds for Ser5 dephosphorylation. Nat Struct Mol Biol, 2014. 21(8): p. 686–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nemec CM, Singh AK, Ali A, Tseng SC, Syal K, Ringelberg KJ, Ho YH, Hintermair C, Ahmad MF, Kar RK, Gasch AP, Akhtar MS, Eick D, and Ansari AZ, Noncanonical CTD kinases regulate RNA polymerase II in a gene-class-specific manner. Nat Chem Biol, 2019. 15(2): p. 123–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang DW, Mosley AL, Ramisetty SR, Rodriguez-Molina JB, Washburn MP, and Ansari AZ, Ssu72 phosphatase-dependent erasure of phospho-Ser7 marks on the RNA polymerase II C-terminal domain is essential for viability and transcription termination. J Biol Chem, 2012. 287(11): p. 8541–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bataille AR, Jeronimo C, Jacques PE, Laramee L, Fortin ME, Forest A, Bergeron M, Hanes SD, and Robert F, A universal RNA polymerase II CTD cycle is orchestrated by complex interplays between kinase, phosphatase, and isomerase enzymes along genes. Mol Cell, 2012. 45(2): p. 158–70. [DOI] [PubMed] [Google Scholar]
- 75.Voss K, Forne I, Descostes N, Hintermair C, Schuller R, Maqbool MA, Heidemann M, Flatley A, Imhof A, Gut M, Gut I, Kremmer E, Andrau JC, and Eick D, Site-specific methylation and acetylation of lysine residues in the C-terminal domain (CTD) of RNA polymerase II. Transcription, 2015. 6(5): p. 91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dias JD, Rito T, Torlai Triglia E, Kukalev A, Ferrai C, Chotalia M, Brookes E, Kimura H, and Pombo A, Methylation of RNA polymerase II non-consensus Lysine residues marks early transcription in mammalian cells. Elife, 2015. 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Suh H, Ficarro SB, Kang UB, Chun Y, Marto JA, and Buratowski S, Direct Analysis of Phosphorylation Sites on the Rpb1 C-Terminal Domain of RNA Polymerase II. Mol Cell, 2016. 61(2): p. 297–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schuller R, Forne I, Straub T, Schreieck A, Texier Y, Shah N, Decker TM, Cramer P, Imhof A, and Eick D, Heptad-Specific Phosphorylation of RNA Polymerase II CTD. Mol Cell, 2016. 61(2): p. 305–14. [DOI] [PubMed] [Google Scholar]
- 79.Fleitz A, Nieves E, Madrid-Aliste C, Fentress SJ, Sibley LD, Weiss LM, Angeletti RH, and Che F-Y, Enhanced Detection of Multiply Phosphorylated Peptides and Identification of Their Sites of Modification. Analytical Chemistry, 2013. 85(18): p. 8566–8576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Steen H, Jebanathirajah JA, Rush J, Morrice N, and Kirschner MW, Phosphorylation analysis by mass spectrometry: myths, facts, and the consequences for qualitative and quantitative measurements. Mol Cell Proteomics, 2006. 5(1): p. 172–81. [DOI] [PubMed] [Google Scholar]
- 81.Kaur U, Johnson DT, Chea EE, Deredge D, Espino JA, and Jones LM, Evolution of Structural Biology Through the Lens of Mass Spectrometry. Anal Chem, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Luo J, Cimermancic P, Viswanath S, Ebmeier CC, Kim B, Dehecq M, Raman V, Greenberg CH, Pellarin R, Sali A, Taatjes DJ, Hahn S, and Ranish J, Architecture of the Human and Yeast General Transcription and DNA Repair Factor TFIIH. Mol Cell, 2015. 59(5): p. 794–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen ZA, Jawhari A, Fischer L, Buchen C, Tahir S, Kamenski T, Rasmussen M, Lariviere L, Bukowski-Wills JC, Nilges M, Cramer P, and Rappsilber J, Architecture of the RNA polymerase II-TFIIF complex revealed by cross-linking and mass spectrometry. EMBO J, 2010. 29(4): p. 717–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Murakami K, Elmlund H, Kalisman N, Bushnell DA, Adams CM, Azubel M, Elmlund D, Levi-Kalisman Y, Liu X, Gibbons BJ, Levitt M, and Kornberg RD, Architecture of an RNA Polymerase II Transcription Pre-Initiation Complex. Science, 2013. 342(6159): p. 1238724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Robinson PJ, Trnka MJ, Bushnell DA, Davis RE, Mattei PJ, Burlingame AL, and Kornberg RD, Structure of a Complete Mediator-RNA Polymerase II Pre-Initiation Complex. Cell, 2016. 166(6): p. 1411–1422.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schilbach S, Hantsche M, Tegunov D, Dienemann C, Wigge C, Urlaub H, and Cramer P, Structures of transcription pre-initiation complex with TFIIH and Mediator. Nature, 2017. 551: p. 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xu Y, Bernecky C, Lee CT, Maier KC, Schwalb B, Tegunov D, Plitzko JM, Urlaub H, and Cramer P, Architecture of the RNA polymerase II-Paf1C-TFIIS transcription elongation complex. Nat Commun, 2017. 8: p. 15741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vos SM, Farnung L, Urlaub H, and Cramer P, Structure of paused transcription complex Pol II-DSIF-NELF. Nature, 2018. 560(7720): p. 601–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yu C and Huang L, Cross-Linking Mass Spectrometry: An Emerging Technology for Interactomics and Structural Biology. Analytical Chemistry, 2018. 90(1): p. 144–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yu C, Huszagh A, Viner R, Novitsky EJ, Rychnovsky SD, and Huang L, Developing a Multiplexed Quantitative Cross-Linking Mass Spectrometry Platform for Comparative Structural Analysis of Protein Complexes. Analytical Chemistry, 2016. 88(20): p. 10301–10308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fasci D, van Ingen H, Scheltema RA, and Heck AJR, Histone interaction landscapes visualized by crosslinking mass spectrometry in intact cell nuclei. Molecular & Cellular Proteomics, 2018: p. mcp.RA118.000924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lu J, Trnka MJ, Roh S-H, Robinson PJJ, Shiau C, Fujimori DG, Chiu W, Burlingame AL, and Guan S, Improved Peak Detection and Deconvolution of Native Electrospray Mass Spectra from Large Protein Complexes. Journal of The American Society for Mass Spectrometry, 2015. 26(12): p. 2141–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Guan S, Trnka MJ, Bushnell DA, Robinson PJJ, Gestwicki JE, and Burlingame AL, Deconvolution Method for Specific and Nonspecific Binding of Ligand to Multiprotein Complex by Native Mass Spectrometry. Analytical Chemistry, 2015. 87(16): p. 8541–8546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Martinez-Rucobo Fuensanta W., Kohler R, van de Waterbeemd M, Albert J Heck R, Hemann M, Herzog F, Stark H, and Cramer P, Molecular Basis of Transcription-Coupled Pre-mRNA Capping. Molecular Cell, 2015. 58(6): p. 1079–1089. [DOI] [PubMed] [Google Scholar]
- 95.Casañal A, Kumar A, Hill CH, Easter AD, Emsley P, Degliesposti G, Gordiyenko Y, Santhanam B, Wolf J, Wiederhold K, Dornan GL, Skehel M, Robinson CV, and Passmore LA, Architecture of eukaryotic mRNA 3′-end processing machinery. 2017. 358(6366): p. 1056–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lorenzen K, Vannini A, Cramer P, and Heck AJ, Structural biology of RNA polymerase III: mass spectrometry elucidates subcomplex architecture. Structure, 2007. 15(10): p. 1237–45. [DOI] [PubMed] [Google Scholar]
- 97.Lane LA, Fernandez-Tornero C, Zhou M, Morgner N, Ptchelkine D, Steuerwald U, Politis A, Lindner D, Gvozdenovic J, Gavin AC, Muller CW, and Robinson CV, Mass spectrometry reveals stable modules in holo and apo RNA polymerases I and III. Structure, 2011. 19(1): p. 90–100. [DOI] [PubMed] [Google Scholar]
- 98.Trowitzsch S, Viola C, Scheer E, Conic S, Chavant V, Fournier M, Papai G, Ebong IO, Schaffitzel C, Zou J, Haffke M, Rappsilber J, Robinson CV, Schultz P, Tora L, and Berger I, Cytoplasmic TAF2-TAF8-TAF10 complex provides evidence for nuclear holo-TFIID assembly from preformed submodules. Nat Commun, 2015. 6: p. 6011. [DOI] [PMC free article] [PubMed] [Google Scholar]