Abstract
Cleavage of the gasdermins to produce a pore-forming N-terminal fragment causes inflammatory death (pyroptosis)1. Caspase-3 cleaves gasdermin E (GSDME, also known as DFNA5), mutated in familial aging-related hearing loss2, which converts noninflammatory apoptosis to pyroptosis in GSDME-expressing cells3–5. GSDME expression is suppressed in many cancers and reduced GSDME is associated with decreased breast cancer survival2,6, suggesting GSDME might be a tumor suppressor. Here we show reduced GSDME function of 20 of 22 tested cancer-associated mutations. Gsdme knockout in GSDME-expressing tumors enhances, while ectopic expression in Gsdme-repressed tumors inhibits, tumor growth. Tumor suppression is mediated by cytotoxic lymphocyte killing since it is abrogated in perforin-deficient or killer lymphocyte-depleted mice. GSDME expression enhances tumor-associated macrophage phagocytosis and the number and functions of tumor-infiltrating NK and CD8+ T lymphocytes. Killer cell granzyme B also activates caspase-independent pyroptosis in target cells by directly cleaving GSDME at the same site as caspase-3. Non-cleavable or pore-defective GSDME are not tumor suppressive. Thus, tumor GSDME is a tumor suppressor by activating pyroptosis, which enhances anti-tumor immunity.
Evidence that GSDME might act as a tumor suppressor (epigenetic GSDME inactivation by promoter DNA methylation in many cancer lines and primary cancers2,6, GSDME suppression of colony formation and cell proliferation in gastric cancer, melanoma and colorectal cancer (CRC) and invasivity of breast cancer, worse 5-year survival and increased metastases in breast cancers that poorly express GSDME2,6) prompted us to probe whether and how GSDME might function as a tumor suppressor. Induction of inflammatory death in GSDME-expressing cancers subjected to intrinsic stresses (i.e., hypoxia, ER stress) or extrinsic challenges (i.e., chemotherapy, radiation, cytotoxic lymphocyte attack) that activate caspase-3 could have a profound effect on the tumor microenvironment (TME), immune cell recruitment and function, and tumor growth. Here we show that GSDME in tumors suppresses tumor growth by increasing anti-tumor functions of tumor-infiltrating NK and CD8+ T killer lymphocytes.
GSDME converts caspase-3-mediated apoptosis to pyroptosis
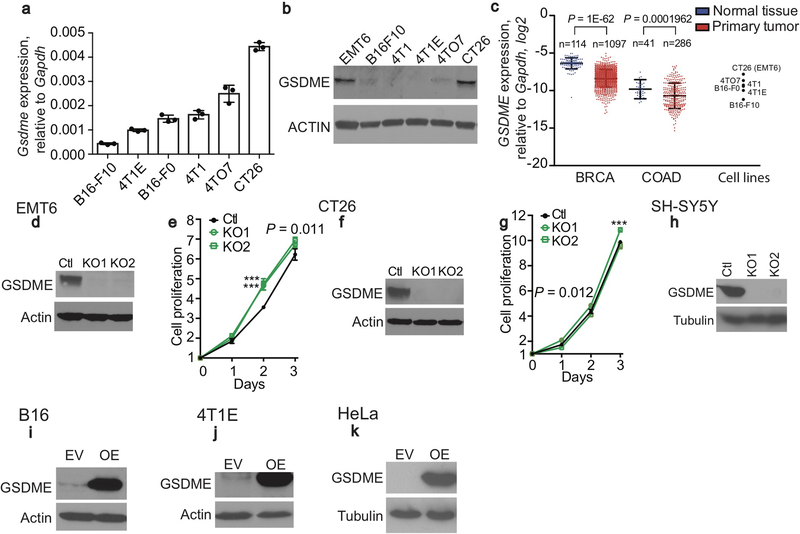
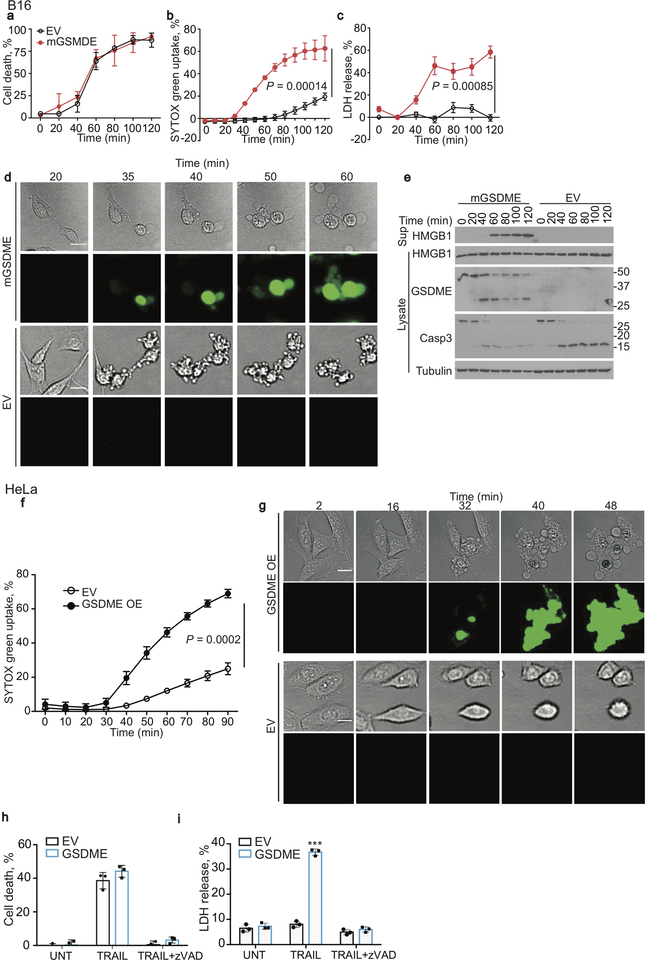
Gsdme mRNA and/or protein in 7 mouse tumor cell lines were within the range found in primary breast cancers and CRC in the TCGA database (https://www.cancer.gov/tcga, Extended Data Fig. 1a–c). Gsdme was knocked out in 2 highly expressing mouse lines, EMT6 triple negative breast cancer (TNBC) and CT26 CRC (Extended Data Fig. 1d–g) and 1 human neuroblastoma line (SH-SY5Y, Extended Data Fig. 1h), and Gsdme or GSDME was stably expressed in 2 poorly expressing mouse cancer cell lines, B16-F10 melanoma (hereafter referred to as B16) and 4T1E TNBC (Extended Data Fig. 1i,j), and in a human cervical carcinoma (HeLa) (Extended Data Fig. 1k). To examine GSDME’s effect on cell death, empty vector (EV) and mGSDME-overexpressing (OE) B16 were treated with raptinal, a rapid caspase-3 activator7. mGSDME did not alter cell death kinetics and extent, measured by annexin V/propidium iodide (PI) staining (Extended Data Fig. 2a). Raptinal did not trigger pyroptosis in EV B16, assessed by SYTOX green uptake and LDH release, but did so in mGSDME-OE B16 cells after ~40 min (Extended Data Fig. 2b,c). Pyroptotic ballooning cell membranes and SYTOX green uptake were detected by time-lapse fluorescence microscopy only in mGSDME-OE cells (Extended Data Fig. 2d, Supplementary Video 1,2). EV cells instead became apoptotic (detached, shrunken, with membrane blebbing). After adding raptinal, caspase-3 cleavage was detected independently of mGSDME-OE beginning within 20 min and mGSDME cleavage was detected coincidently if mGSDME was expressed (Extended Data Fig. 2e). Although raptinal did not appreciably change cellular HMGB1, only mGSDME-OE B16 released HMGB1. Similarly, raptinal or TRAIL converted apoptosis to pyroptosis only in hGSDME-OE HeLa (Extended Data Fig. 2f–i, Supplementary Video 3,4). Thus, GSDME is cleaved rapidly after caspase-3 activation to permeabilize the cell membrane, converting apoptosis to pyroptosis, as previously reported3,4.
Cancer-related GSDME mutations cause loss of function
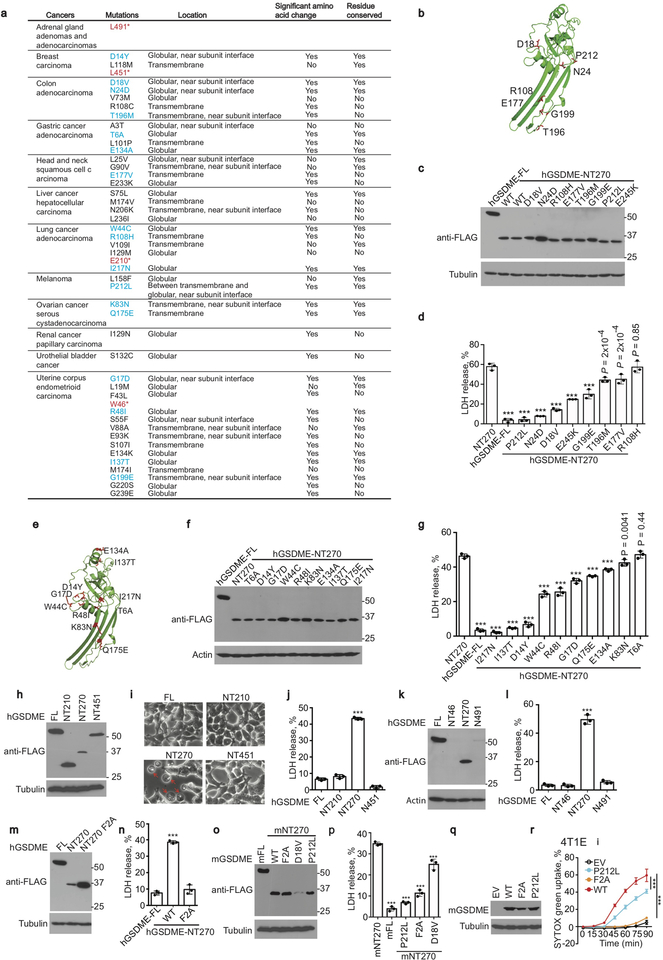
If GSDME is a tumor suppressor, some GSDME-expressing cancers might have loss of function (LOF) mutations. The TCGA of single nucleotide polymorphisms (SNP) in primary cancers was examined for GSDM mutations (Extended Data Fig. 3a). GSDME and GSDMC had the most mutations, and GSDME mutations were increased around the caspase-3 cleavage site. GSDME SNPs in the N-terminus (NT) were mapped onto the GSDME-NT pore, modeled based on mouse GSDMA3-NT8 (Extended Data Fig. 3b,e). Eighteen GSDME-NT conserved site mutants were expressed in HEK293T and tested for pyroptosis. All the mutant proteins were well expressed (Extended Data Fig. 4c,f). 16 of 18 cancer-associated SNPs significantly reduced LDH release compared to WT GSDME-NT (Extended Data Fig. 3d,g), suggesting some cancer-associated GSDME-NT mutations cause LOF. Globular domain mutations close to the oligomerization and cell membrane binding sites had the largest effect. Four premature stop mutants (GSDME 1–46, 1–210, 1–451 or 1–491) also did not cause pyroptosis in HEK293T cells, although some of the mutated proteins (NT451, NT491) may have been unstable (Extended Data Fig. 3h–l). A previously described3 LOF F2A mutation was verified (Extended Data Fig. 3m,n). F2, D18 and P212 are conserved between human and mouse GSDME. Expression of F2A, D18V or P212L mGSDME-NT in HEK293T or of F2A or P212L full-length mGSDME in 4T1E significantly reduced spontaneous or raptinal-induced pyroptosis, respectively, compared to unmutated mGSDME (Extended Data Fig. 3o–r). Thus, 20 of 22 (91%) studied cancer-related GSDME mutations cause LOF.
GSDME suppresses tumor growth and enhances functional properties of tumor-infiltrating immune cells
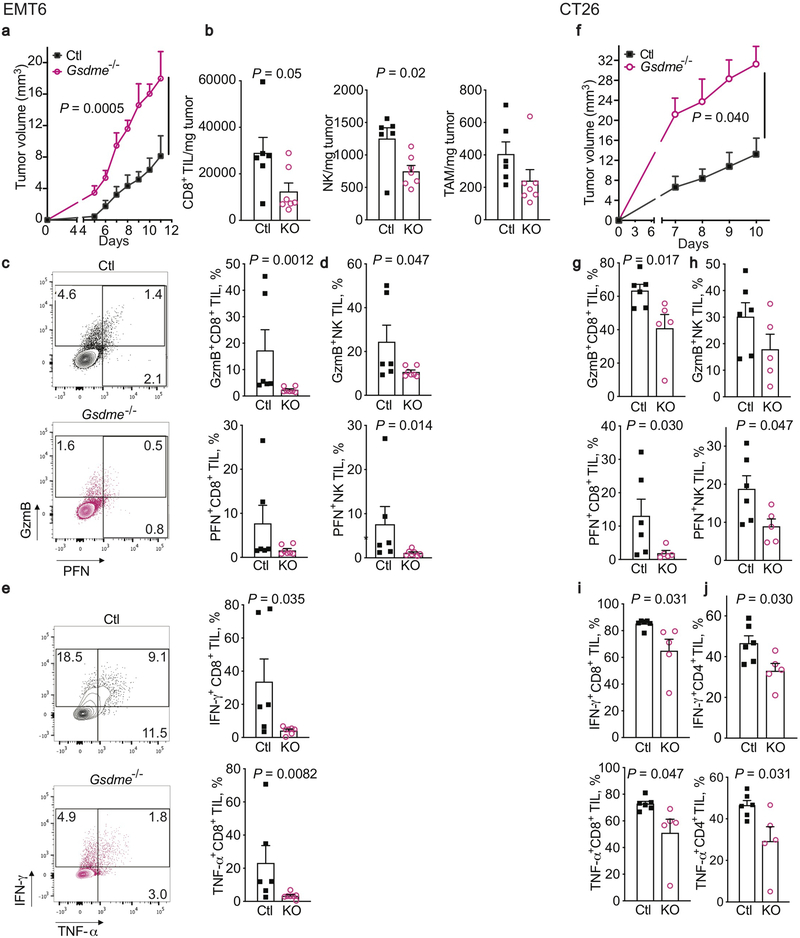
EMT6 (Extended Data Fig. 4a–e) and CT26 tumors (Extended Data Fig. 4f–j) knocked out for Gsdme grew significantly faster in immunocompetent mice than tumors expressing endogenous Gsdme (Extended Data Fig. 4a,f). The tumor microenvironment (TME) of Gsdme−/− EMT6 had fewer CD8+ T and NK and a trend towards fewer tumor-associated macrophages (TAM) (Extended Data Fig. 4b). Tumor-infiltrating lymphocytes (TIL, CD8+ T and NK) from Gsdme−/− EMT6 and CT26 also expressed less GzmB and/or perforin (PFN) (Extended Data Fig. 4c,d,g,h), and produced less IFNγ and TNFα after PMA and ionomycin stimulation (Extended Data Fig. 4e,i,j). Thus, endogenous GSDME suppresses tumor growth and promotes TIL function.
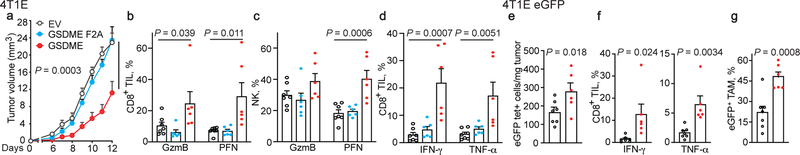
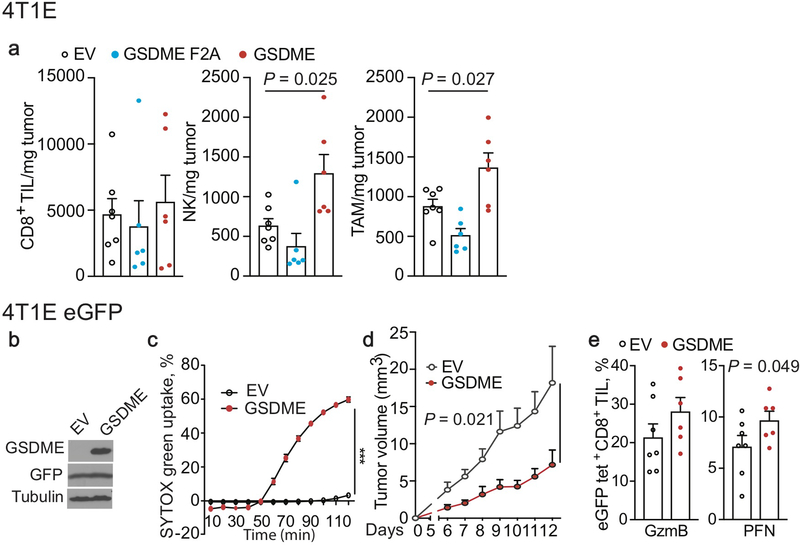
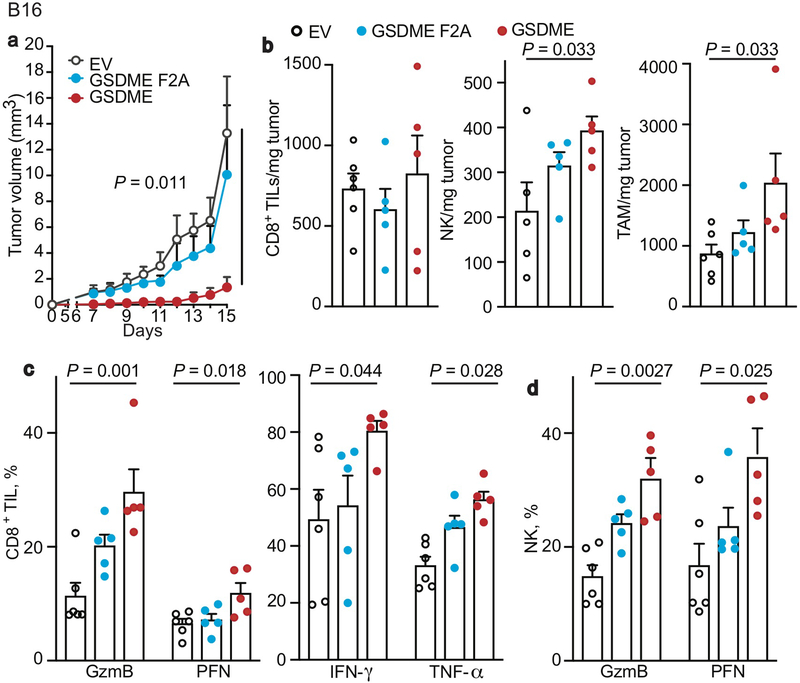
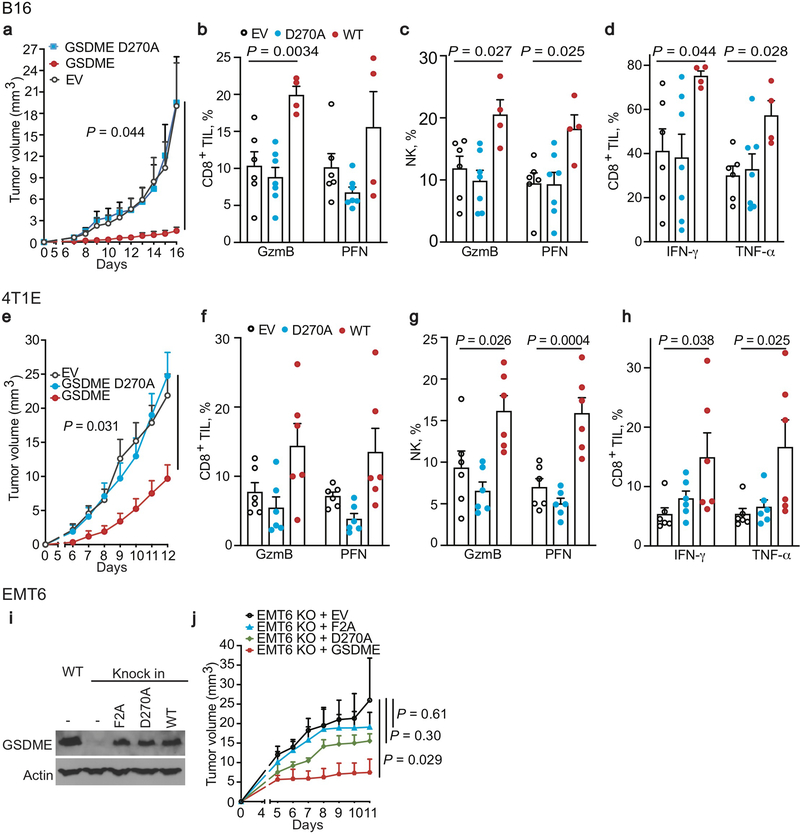
Conversely, ectopic mGSDME expression in 4T1E (Fig. 1, Extended Data Fig. 5) and B16 (Extended Data Fig. 6) significantly inhibited tumor growth (Fig. 1a, Extended Data Fig. 6a). GSDME-expressing 4T1E tumors had significantly more tumor-infiltrating NK and TAM, but not CD8+ TIL (Extended Data Fig. 5a). More NK and CD8+ TIL expressed GzmB and PFN (Fig. 1b,c) and more CD8+ TIL produced IFNγ and TNFα (Fig. 1d). 4T1E overexpressing F2A or P212L LOF mutants of GSDME did not have more functional TIL (Fig. 1a–d, Extended Data Fig. 5a, data not shown). Similarly, increased TIL numbers and function were observed in GSDME-OE, but not F2A GSDME-OE, B16 tumors (Extended Data Fig. 6b–d). Thus, GSDME pores were required for tumor suppressive immune enhancement.
Figure 1. Ectopic expression of pore-forming, but not inactive, GSDME reduces tumor growth and enhances tumor immune responses.
a-d, Orthotopically implanted 4T1E cells stably expressing WT mGSDME (n=6 mice/group), inactive F2A mGSDME (n=6 mice/group) or empty vector (EV, n=7 mice/group) were analyzed for tumor growth (a) and TIL function (b-d). Percentage of CD8+ (b) and NK (c) TIL expressing GzmB or PFN; CD8+ TIL producing IFN-γ or TNF-α induced by PMA and ionomycin (d). e-g, Anti-tumor immunity after orthotopic implantation of 4T1E, stably expressing eGFP and then stably transduced to express mGSDME or EV (n=7 mice/group). e, Mean numbers of tumor-specific GFP tetramer+ (tet+) CD8+ TIL/mg of tumor. f, Percentage of CD8+ TIL activated by eGFP peptide to produce IFN-γ or TNF-α. g, Percentage of GFP+ TAM that phagocytosed tumor cells. The area under the curve of tumor growth curves were compared by one-way ANOVA with Holm-Sidak correction for type I error (a). Comparisons in (b-d) were calculated by one-way ANOVA using the Holm-Sidak method for multiple comparisons; (e-g) were calculated by two-tailed Student’s t-test. Data shown are mean + SEM and are representative of two independent experiments.
Ectopic mGSDME expression in eGFP+ 4T1E (Fig. 1e–g, Extended Data Fig. 5b–e) also reduced tumor growth (Extended Data Fig. 5d). In this model tumor-specific CD8+ TIL could be examined using GFP tetramers. GSDME significantly increased GFP-tetramer+ CD8+ TIL (Fig. 1e). Tetramer+ TIL in GSDME-OE tumors expressed significantly more PFN (Extended Data Fig. 5e), and also produced more cytokines after GFP peptide stimulation (Fig. 1f). TAM in mGSDME-expressing tumors were twice as likely to be GFP+, indicating increased in vivo tumor cell phagocytosis (Fig. 1g).
Killer lymphocytes mediate tumor suppression
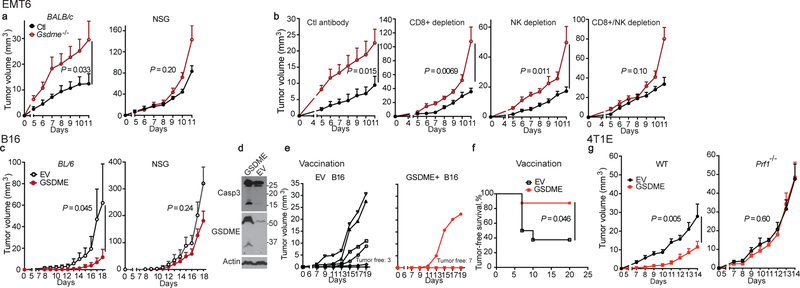
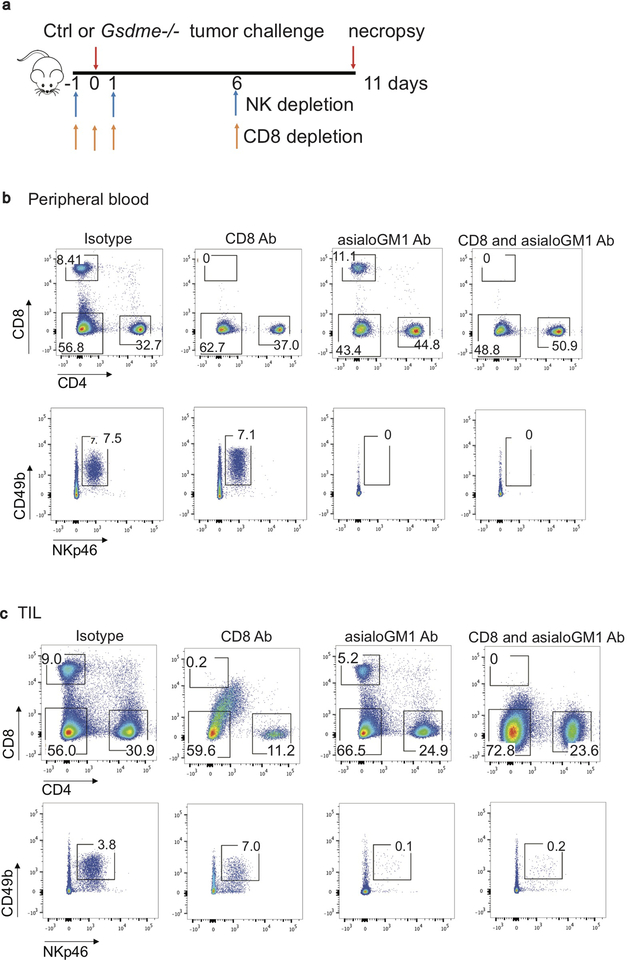
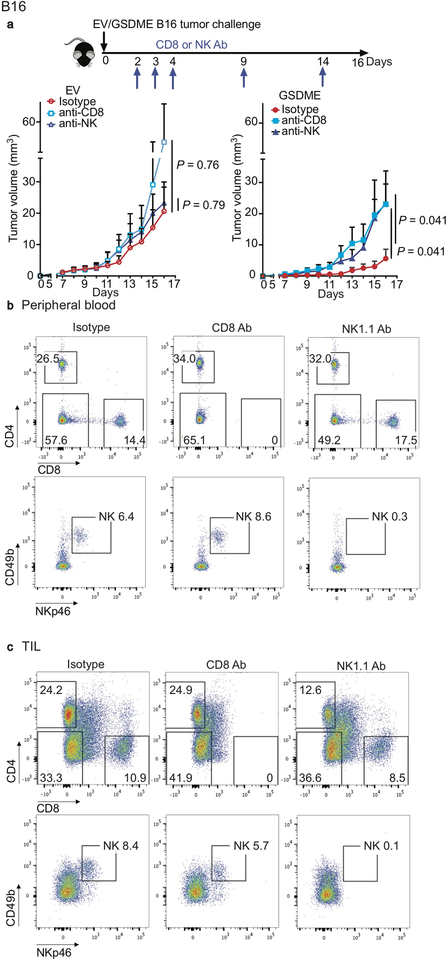
Enhanced immune function in mGSDME-expressing tumors suggests that tumor inhibition might be immune-mediated. To investigate this hypothesis, GSDME-expressing and Gsdme−/− EMT6 tumors were compared in WT BALB/c and NOD-scid IL2Rgnull (NSG) mice lacking mature lymphocytes (Fig. 2a). Both Gsdme+/+ and Gsdme−/− EMT6 grew more rapidly in NSG than WT mice, indicating immune protection even in the absence of GSDME. Gsdme deficiency did not significantly affect tumor growth in NSG mice, while Gsdme−/− EMT6 grew faster in immunocompetent mice. A requirement for lymphocytes in the anti-tumor effect of GSDME was also observed by comparing EV and mGSDME-OE B16 tumors (Fig. 2c). To determine whether killer cells are important for GSDME-mediated tumor inhibition, Gsdme+/+ and Gsdme−/− EMT6 were implanted in CD8+ T and/or NK-depleted or control antibody-treated mice (Fig. 2b, Extended Data Fig. 7). Depletion of either CD8+ T or NK modestly but significantly reduced GSDME-mediated tumor inhibition, but GSDME expression did not alter tumor growth significantly in mice depleted of both CD8+ T and NK, indicating that both types of killer cells are responsible for tumor suppression (Fig. 2b). Similarly, depletion of either killer cell subset only significantly increased the growth of GSDME-expressing but not EV B16 (Extended Data Fig. 8). Thus, both CD8+ T and NK mediate GSDME’s tumor suppression.
Figure 2. GSDME-mediated tumor inhibition depends on cytotoxic lymphocytes.
a,b, Growth of control (Ctl) or Gsdme−/− EMT6 in BALB/c (left, n=8 mice/group) and NSG mice (right, Ctl n=7 mice/group; Gsdme−/− n=8 mice/group) (a) or in BALB/c mice treated with an isotype control antibody (n=6 mice/group) or depleted of CD8+ T cells (n=8 mice/group), NK (n=7 mice/group) or both (n=8 mice/group) (b). Antibody depletion (schematic, Extended Data Fig. 7a) was verified on day 3 post tumor challenge and day 11 at necropsy (Extended Data Fig. 7b,c). c, Growth of EV or mGSDME-overexpressing B16 in C57BL/6 (left, EV n=5 mice/group, mGSDME n=8 mice/group) and NSG (right, n=6 mice/group). d-f, B16 vaccination model. C57BL/6 mice were vaccinated in the left flank with EV or GSDME+ B16 and challenged 10 days later with EV B16 in the right flank (n=8 mice/group). Shown are immunoblot of lysates of representative left flank tumors at necropsy probed for caspase-3, GSDME and actin loading control (d), right flank tumor growth (e) and tumor-free survival (f). g, Comparison of growth of orthotopic EV and mGSDME+ 4T1E tumors in WT (n=7 mice/group) and Prf1−/− (EV n=6 mice/group, mGSDME n=7 mice/group) BALB/c mice. The area under the growth curves was compared by two-tailed Student’s t-test. Log-rank test was used for survival analysis. Data are mean + SEM and are representative of two independent experiments.
The killer cell dependence of protection suggests that pyroptosis causes immunogenic cell death (ICD)9. The gold standard criterion of ICD is protection from secondary tumor challenge after vaccination with tumor cells undergoing ICD. To determine whether pyroptosis causes ICD, mice were vaccinated with either WT or GSDME-OE B16 subcutaneously in the left flank and challenged on the right flank 10 day later with WT B16 (Fig. 2d–f). At the vaccination site, GSDME and caspase-3 cleavage were easily detected by immunoblot only in GSDME-OE tumors (Fig. 2d), indicating that pyroptosis occurred spontaneously in vivo and cell death greatly increased in GSDME-OE tumors. Moreover, vaccination with GSDME-OE, compared to WT, B16 significantly reduced challenge WT B16 tumor growth (Fig. 2e) and improved tumor-free survival (Fig. 2f). 5/8 mice vaccinated with WT B16 developed palpable tumors, while only 1/8 mice vaccinated with GSDME-OE B16 did (P=0.039, χ2 test). Thus, pyroptosis is a form of ICD spontaneously occurring in GSDME-OE tumors.
CD8+ T and NK recognition triggers both cytokine secretion and cytotoxic granule-mediated, PFN-dependent killing, but the latter is generally considered key for anti-tumor immunity. To determine if killing mediates tumor suppression, EV and mGSDME-OE 4T1E tumor growth was compared in WT and PFN-deficient Prf1−/− mice (Fig. 2g). Both tumors grew significantly faster in PFN-deficient than WT mice. Although GSDME significantly reduced 4T1E growth in WT mice, GSDME conferred no significant advantage in PFN-deficient mice, indicating that granule-dependent cytotoxicity was responsible for GSDME’s tumor suppression. Since GSDME had no significant effect on tumor growth in mice lacking killer lymphocytes or PFN, GSDME’s tumor suppression was primarily mediated by killer lymphocytes. GSDME- tumors tended to be larger than GSDME+ tumors at later timepoints even in NSG and lymphocyte-depleted mice, suggesting cell-intrinsic mechanisms of GSDME tumor suppression may exist.
Killer cells induce pyroptosis in GSDME-expressing tumors
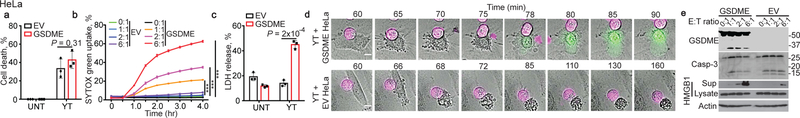
The strong dependence of GSDME-mediated tumor suppression on cytotoxicity suggested that killer lymphocytes might cause pyroptosis in GSDME-expressing targets. To determine if killer cells induce pyroptosis, the human NK line YT was incubated with EV and hGSDME-OE HeLa. Although cell death was comparable (Fig. 3a), pyroptosis only occurred in GSDME+ HeLa and increased with more NK (Fig. 3b,c). By time-lapse microscopy, both EV and GSDME+ HeLa began to detach about an hour after adding YT (Fig. 3d, Supplementary Video 5,6). EV HeLa showed progressive apoptotic morphology and did not take up SYTOX green over 160 min, while GSDME+ HeLa began to take up SYTOX green after 15–20 min and underwent increasing pyroptotic membrane ballooning. Caspase-3 was cleaved in YT-cocultured EV and GSDME+ HeLa harvested 4 hr after adding YT, but GSDME was cleaved only in GSDME+ tumors, which released much more HMGB1 (Fig. 3e).
Figure 3. Killer cells cleave GSDME and induce GSDME-dependent pyroptosis in target cells.
Cell death induced by YT NK in EV and hGSDME-expressing HeLa, assessed by CellTiter-Glo (a, E:T ratio=2:1, 4 h), SYTOX green uptake (b, E:T ratios indicated) and LDH release (c, E:T ratio=2:1, 4 h). d, Time-lapse confocal microscopy images of co-cultures of Vybrant DiD-labeled YT (magenta) with EV and hGSDME+ HeLa in SYTOX green containing medium. e, Caspase-3 and GSDME cleavage and HMGB1 release in HeLa incubated with YT at indicated E:T ratios for 4 h, assessed by immunoblot. Comparisons were calculated by two-tailed Student’s t-test (a,c) or by one-way ANOVA with Holm-Sidak method for multiple comparisons (b). Data are mean ± SD of biological triplicates and are representative of three independent experiments. *** P<0.0001. Scale bar, 10 μm.
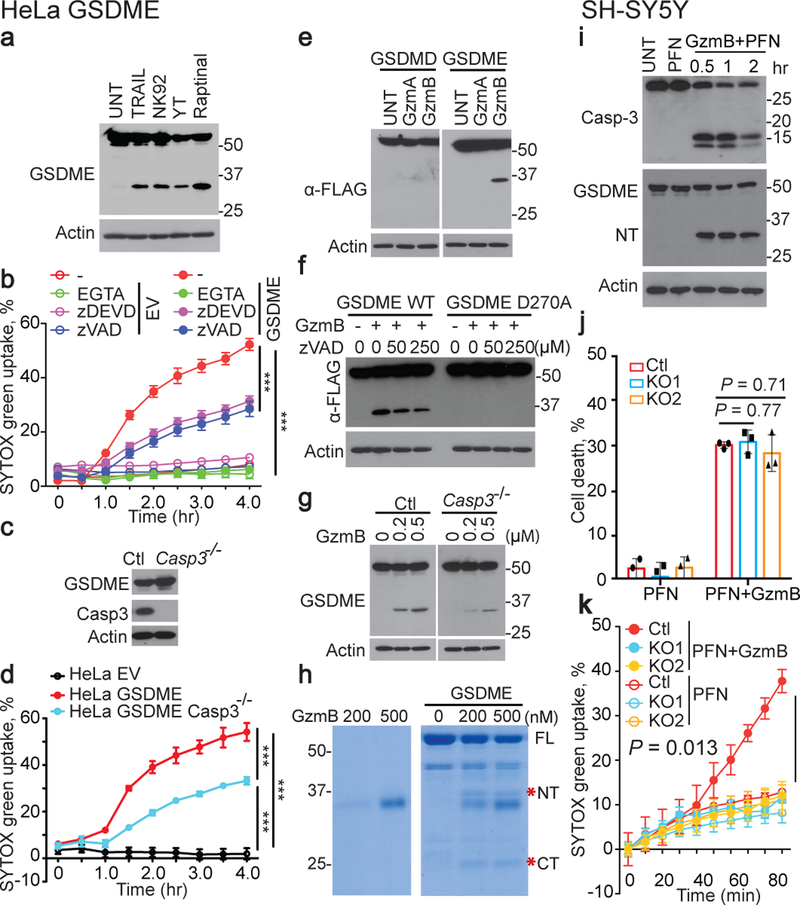
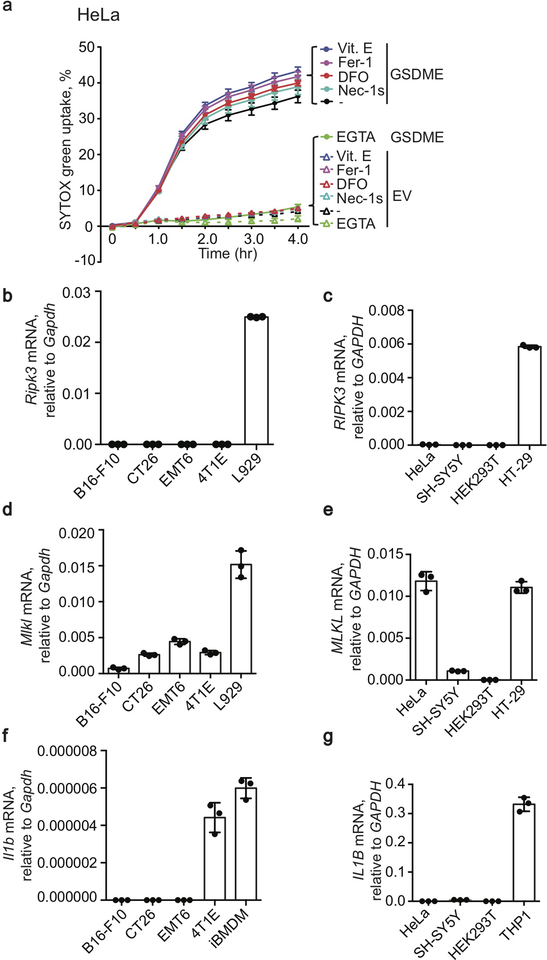
Treatment of GSDME+ HeLa with YT or another human NK line NK-92 or with raptinal or TRAIL produced a GSDME fragment of the same size (Fig. 4a), suggesting that NK cleaved GSDME at the caspase-3 site. To determine whether NK induced pyroptosis depends on cytotoxic granule release, pyroptosis was measured in the presence or absence of the Ca++ chelator EGTA, which inhibits cytotoxic granule release and PFN (Fig. 4b). EGTA completely blocked pyroptosis, suggesting that degranulation was required. Surprisingly, the caspase-3 inhibitor zDEVD-fmk or pan-caspase inhibitor zVAD-fmk only partially blocked YT-induced pyroptosis. To confirm caspase-3-independent pyroptosis, YT killing of Casp3+/+ and Casp3−/− hGSDME-OE HeLa were compared (Fig. 4c,d). Casp3-deficiency only partially reduced YT-induced pyroptosis, suggesting that NK activated both caspase-dependent and independent pyroptosis. To test if necroptosis or ferroptosis, other caspase-independent inflammatory death pathways, contribute, a necroptosis inhibitor Necrostatin-1s10 or three ferroptosis inhibitors (ferrostatin-1, α-tocopherol, desferoxamine11) were added to YT cocultures with EV and GSDME-OE HeLa (Extended Data Fig. 9a). None of these inhibitors suppressed YT-induced pyroptosis of GSDME-OE HeLa, suggesting that necroptosis and ferroptosis are not involved. Moreover, RIPK3 was not detected by qRT-PCR in any of the cell lines studied (Extended Data Fig. 9b–e). Thus, cytotoxic granule release induces partly caspase-independent pyroptosis in GSDME+ tumors.
Figure 4. GzmB directly cleaves GSDME to cause pyroptosis.
a, Immunoblot of hGSDME+ HeLa after no treatment (UNT) or treatment with YT or NK92 or TRAIL for 4 h or raptinal for 1 h. b, Effect of EGTA, zVAD-fmk and zDEVD-fmk on YT-induced SYTOX green uptake in EV and hGSDME+ HeLa. c,d, Expression of GSDME and caspase-3 (c) and YT-induced SYTOX green uptake (d) in hGSDME+ and hGSDME- Casp3−/− HeLa. e,f, Immunoblots, probed for FLAG-GSDMD (e, left) or FLAG-GSDME (e, right; f), of cell lysates of HEK293T expressing FLAG-tagged WT or D270A (f, right lanes) hGSDME after 1 h incubation with PBS (UNT) or recombinant GzmA or GzmB (800 nM). g, Immunoblots, probed for GSDME, of cell lysates of hGSDME+ HeLa, knocked out or not for Casp3, after 1 h incubation with GzmB. h, Coomassie-stained SDS-PAGE gel of in vitro reaction of recombinant GzmB incubated with recombinant hGSDME for 1 h. *, N-terminal (NT) and C-terminal (CT) GSDME cleavage products. i, SH-SY5Y treated with PFN ± GzmB or medium (UNT) for 2 h or indicated times were analyzed by immunoblot of cell lysates probed for caspase-3, GSDME or tubulin. j,k, Effects of GSDME knockout (KO) on PFN ± GzmB-induced SH-SY5Y cell death and pyroptosis assessed after 1 h by CellTiter-Glo (j), SYTOX green uptake (k). Differences among multiple groups in (b,d,j,k) were analyzed by one-way ANOVA using the Holm-Sidak method for multiple comparisons. Data are mean ± SD of biological triplicates and are representative of three (a-h) or two (i-k) independent experiments. *** P<0.0001.
Granzyme B cleaves GSDME after D270 like caspase-3
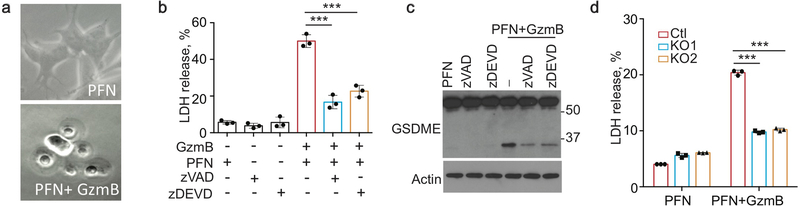
Because NK-induced pyroptosis in GSDME-expressing cells does not require caspases, we speculated that Gzm proteases might cleave GSDME. Gzm-mediated cell death is caspase-independent, but amplified by the caspases because GzmB cuts and activates caspase-312. To test if Gzms cleave GSDME, lysates from hGSDME-OE or hGSDMD-OE HEK293T were incubated with purified human GzmA or GzmB, the most abundant Gzms. GzmB, but not GzmA, led to GSDME, but not GSDMD, cleavage (Fig. 4e). Since caspase- and killer cell-activated GSDME-NT were of similar size (Fig. 4a), GzmB either activates caspase-3 to cleave GSDME and/or directly cleaves GSDME at the same residue. In fact, GzmB did not activate mutant D270A GSDME cleavage, but WT GSDME was still cleaved when all caspases were inhibited (Fig. 4f) or in lysates of Casp3−/− GSDME-OE HeLa (Fig. 4g), suggesting that GzmB directly cleaves GSDME after D270. Direct cleavage was verified by showing that GzmB cleaved recombinant GSDME (Fig. 4h). To determine whether GzmB cleaves GSDME in cells, SH-SY5Y, a GSDME+ human neuroblastoma, was treated with PFN ± GzmB (Fig. 4i, Extended Data Fig. 10a–c). The combination, but not PFN alone, caused pyroptotic morphology (Extended Data Fig. 10a). Within 30 min of adding GzmB and PFN, GSDME and caspase-3 cleavage were detected by immunoblot (Fig. 4i). GSDME cleavage and LDH release from SH-SY5Y required GzmB and PFN and were partially inhibited by inhibiting caspase-3 or all caspases (Extended Data Fig. 10b,c). CRISPR/Cas9 KO of GSDME in SH-SY5Y (Extended Data Fig. 1h) did not change overall GzmB and PFN-mediated cell death (Fig. 4j), but blocked pyroptosis (Fig. 4k, Extended Data Fig. 10d). Thus, GzmB induces GSDME-dependent pyroptosis in tumor targets by both directly cleaving GSDME and indirectly by activating caspase-3.
GSDME cleavage at D270 mediates its tumor suppressive activity
Since both GzmB and caspase-3 use D270 to activate GSDME, mutation of this residue in tumors should abrogate tumor suppression. To test this hypothesis, growth of B16 (Extended Data Fig. 11a–d) and 4T1E (Extended Data Fig. 11e–h) stably over-expressing WT or D270A mGSDME or EV were compared. Although WT GSDME reduced tumor growth as before (Fig. 1, Extended Data Fig.6), D270A GSDME OE and EV tumors grew indistinguishably (Extended Data Fig. 11a,e). WT GSDME OE enhanced CD8+ and NK TIL functionality, but OE of D270A GSDME did not alter GzmB, PFN or cytokine expression in CD8+ or NK TIL (Extended Data Fig. 11b–d,f–h). When Gsdme−/− EMT6 were knocked in to express EV or WT, F2A nonfunctional or D270A noncleavable GSDME at levels comparable to endogenous GSDME in WT EMT6, only WT GSDME significantly reduced tumor growth (Extended Data Fig. 11i,j), providing additional evidence that GSDME cleavage at D270 and pore formation are required for tumor suppression.
Discussion
Here we show in melanoma, TNBC and CRC tumors that GSDME expression acts as a tumor suppressor by inducing pyroptosis, which enhances anti-tumor killer cell cytotoxicity. GSDME’s anti-tumor activity was abrogated in mice lacking killer lymphocytes or PFN. Mutations abolishing GSDME pore formation or GzmB/Caspase-3 cleavage also blocked tumor suppression. Tumor suppression occurred without any extrinsic treatment. What initially triggers in vivo pyroptosis? It may have been initiated by spontaneous apoptosis of hypoxic or stressed regions of the tumor or by immune-mediated killing. Our working model is that GSDME expression in spontaneously dying tumor cells provides inflammatory danger signals that recruit immune cells to the TME and promote their functionality. GSDME expression not only increased the number and function of TIL, but also increased macrophage phagocytosis, which is predicted to enhance anti-tumor adaptive immunity. Some tumors evade immunity by resisting phagocytosis13,14; tumor GSDME may help overcome this immune evasion strategy.
Killer cell death was previously thought of as non-inflammatory. Here we show killer lymphocytes activate pyroptosis when GzmB cleaves GSDME at the caspase-3 site. Caspase-resistant cancer cells should be susceptible to both killer cell-mediated apoptosis (since Gzm-activated death is mostly caspase-independent) and pyroptosis, provided they express GSDME. Pyroptosis may augment killer cell immunity by providing adjuvant-like danger signals. Our results indicate that pyroptosis, like necroptosis15, is a form of ICD9. Implanted GSDME+ B16 melanoma, but not GSDME- B16, spontaneously underwent pyroptosis and protected mice from challenge with WT B16. Protection by vaccination, the gold standard for ICD9, did not require chemotherapy or radiation, suggesting that GSDME-expressing tumors, even otherwise “immunologically cold” tumors like B16, are spontaneously undergoing pyroptotic ICD in vivo. It is worth noting that GSDME+ tumors studied here do not release IL-1β or require it for immune protection since Il1b was only expressed by one of the mouse cell lines studied (Extended Data Fig. 9f,g) and IL-1β was not detected in sera of GSDME+ tumor-bearing mice (data not shown).
Direct Gzm induction of pyroptosis provides a simple mechanism for triggering inflammatory death, much simpler than canonical inflammasome activation that requires at least 4 molecules (a sensor, adaptor, inflammatory caspase and GSDMD) or even the noncanonical pathway which requires an inflammatory caspase and GSDMD. GSDME and other GSDMs may sense other mislocalized cytosolic proteases as danger signals. The GSDM linker region is an unstructured loop, making it a good protease substrate. Consistently, neutrophil serine proteases, which are homologous to Gzms, can cleave GSDMD to induce neutrophil netosis16,17.
Cancer cells have developed two strategies, epigenetic suppression of GSDME expression and LOF mutations, to avoid GSDME-mediated tumor suppression. Epigenetic suppression of GSDME is more common than GSDME mutation2,4,6. We show here that many cancer-related GSDME mutations reduce pyroptosis and mutations of D270, the shared GzmB/Caspase-3 cleavage site and a prominent cancer mutation, enabled tumors to evade tumor suppression by GSDME. Therapeutic strategies to induce GSDME, such as by using the DNA methylation inhibitor decitabine4, an approved leukemia and myelodysplasia drug, are worth exploring.
Online Methods
Data reporting.
No statistical methods were used to predetermine sample size. The experiments were not randomized and the investigators were not blinded to outcome assessment.□
Cell lines and cell culture conditions.
HeLa, HEK293T and EMT6 were purchased from ATCC. 4T1E and 4T1E-eGFP cells were generated in our lab by sorting 4T1 for high E-cadherin expression, as previously reported18. 4T1 and 4TO7 cells were kind gifts of Dr. Fred Miller (Wayne State University). B16F10 and CT26 were kindly provided by Dr. Gordon J. Freeman (Dana-Farber Cancer Institute). SH-SY5Y cell was a generous gift of Dr. Yang Shi (Boston Children’s Hospital). The YT-Indy NK cell line was a gift of Dr. Zacharie Brahmi (Indiana University). HeLa, HEK293T, 4T1, 4TO7, 4T1E, 4T1E GFP and B16F10 were cultured in DMEM medium supplemented with 10% heat-inactivated fetal bovine serum (FBS), 6 mM HEPES, 1.6 mM L-glutamine, 50 μM 2-mercaptoethonal, 100 U/ml Penicillin G and 100 μg/ml streptomycin sulphate. EMT6, CT26, SH-SY5Y and YT-Indy NK were cultured in RPMI medium with 10% FBS and the same supplements. All cells were verified to be free of mycoplasma by PCR and were only authenticated by morphology.
Plasmids.
Full-length human GSDMD and GSDME and mouse Gsdme were cloned into pFLAG-CMV4. GSDME and Gsdme point mutation plasmids were generated by quick-change PCR (Stratagene). GSDME truncations were amplified by PCR from pFLAG-CMV4-GSDME. pLVX-Puro empty vector was a kind gift of Dr. Christian Bowman-Colin (Dana-Farber Cancer Institute). Full-length wild type GSDME and mutant GSDMEs were cloned into pLVX-Puro using Hifi 1-step kit (Gibson Assembly). LentiCRISPR-v2 puro and LentiCRISPR-v2 hygro vector were obtained from Addgene and gRNAs were cloned into the vectors as previously described19,20. All plasmids were verified by sequencing.
Stable cell lines.
To generate lentiviruses, pLVX-puro Gsdme plasmid was transfected into HEK293T cells with pSPAX2 and pCMV-VSV-G at an 1:1:2 ratio. Supernatants collected 2 d later were used to transduce B16 and 4T1E for 48 hr. Puromycin (Sigma, 3 μg/ml) was then added to select GSDME-expressing cells. pLVX-puro GSDME was used to generate HeLa stably expressing GSDME and pLVX-puro empty vector was used to generate control cells.
CRISPR/Cas9 knockout cells.
Gsdme gRNAs (5’-TTTCTTAAAGAAGTTGATGC-3’ and 5’-CAAGCTGCAACTTCTAAGTC-3’) and GSDME gRNAs (5’-TAAGTTACAGCTTCTAAGTC-3’ and 5’-TGACAAAAAAGAAGAGATTC-3’) were cloned into LentiCRISPR-v2 puro, and Casp3 gRNA (5’-GGAGAACACTGAAAACTCAG-3’) was cloned into LentiCRISPR-v2 hygro, as previously described19,20. The resulting plasmids were transfected into HEK293T with pSPAX2 and pCMV-VSV-G at an 1:1:2 ratio. Supernatants collected 2 days later were used to transduce mouse EMT6, CT26 or human SH-SY5Y and GSDME+ HeLa cells, respectively. Two days later, 3 μg/ml (8 μg/ml for CT26) puromycin or 200 μg/ml hygromycin was added to select for positive cells for 5 or 14 d, respectively. Cells were then subcloned in 96-well plates and screened for GSDME expression by immunoblot. Selected knockout clones were analyzed and verified by DNA sequencing. LentiCRISPR-v2 empty vector was used to generate control cells.
Reagents.
Raptinal, EGTA, necrostatin-1s (Nec-1s), ferrostatin-1 (Fer-1), α-tocopherol (Vit. E), and desferoxamine (DFO) were from Sigma. TRIzol was from Life Technologies. TRAIL was from Enzo Life Sciences. zVAD-fmk and zDEVD-fmk were from BD Biosciences. SYTOX green was from Invitrogen. Vybrant DiD dye was from ThermoFisher Scientific. CellTiter 96 kit (Promega) was used to measure cell proliferation.
Gene expression assays.
RNA was extracted using TRIzol reagent according to the manufacturer’s instructions and was subject to reverse transcription using SuperScript III system (Invitrogen). Gsdme expression was assayed by qRT-PCT using SsoFast Supermix (Bio-Rad). Breast cancer (BRCA) and colon cancer (COAD) RNA-seq expression data were obtained from The Cancer Genome Atlas (TCGA) using the University of California Santa Cruz (UCSC) Xena bioinformatic tool21. The log2 difference between GSDME and GAPDH expression was calculated for both tumor and normal tissue and plotted using Prism software.
Immunoblot.
Cells were lysed in lysis buffer (50 mM Tris-Cl pH7.4, 150 mM NaCl, 1% Triton-X100 supplemented with 1 mM PMSF before use). Cell lysate in SDS loading buffer (50 mM Tris-HCl pH 6.8, 10% glycerol, 2% SDS, 1% 2-mercaptoethanol, 0.1% Bromophenol blue) was boiled and analyzed by SDS-PAGE (Bio-Rad) and transfer to PVDF membranes (MilliporeSigma), which were then blocked with 2% milk in TBST buffer (150 mM NaCl, 20 mM Tris-HCl pH 7.6, 0.1% Tween-20) for 30 min before incubation with primary antibodies for 2 hr at room temperature or 4 °C overnight. After washing with TBST, membranes were incubated with secondary antibody for 40 min and then washed 3 times with TBST before adding the West Femto substrate (ThermoFisher Scientific) for detection. Antibodies were anti-GSDME (ab215191, Abcam), anti-FLAG (F1804, Sigma), anti-caspase-3 (9662S, Cell Signaling), anti-HMGB1 (ab18256, Abcam), anti-α-tubulin (T5168, Sigma), and anti-β-actin (JLA20, Developmental Studies Hybridoma Bank).
Cell death assays.
To measure membrane lysis, culture medium was collected and LDH release was measured using CytoTox 96 cytotoxicity assay (Promega) according to the manufacturer’s instructions. For GSDME-NT induced pyroptosis in HEK293T cells, HEK293T transfection was performed by the calcium-phosphate method and LDH release was measured 20 hours after transfection. Pyroptotic cells were also imaged using an Olympus IX70 inverted microscope and protein expression was analyzed by immunoblot. For raptinal-induced pyroptosis in B16 GSDME cells, 10 μM raptinal was used to treat B16 cells for 2 h and LDH release was measured at indicated timepoints. For TRAIL-induced pyroptosis in HeLa GSDME cells, LDH release was measured 16 h after cells were treated with 100 ng/ml TRAIL. For YT-induced pyroptosis in HeLa GSDME, LDH release was measured 4 h after treatment (E:T ratio = 2:1). For PFN/GzmB-induced pyroptosis in SH-SY5Y cells, LDH was measured 1 h after treatment in GSDME KO cells or 2 h post treatment in the presence of caspase inhibitors.
Overall cell death due to pyroptosis or apoptosis was measured at the same time as LDH release. To measure overall cell death in raptinal-treated B16 cells, samples were stained with APC-conjugated Annexin V (Invitrogen) and PI (Sigma) according to the manufacturer’s instructions and analyzed by BD FACSCanto II (BD Biosciences) using FlowJo V.10 (TreeStar) software. Cell death was determined by counting annexin V and/or PI positive cells. To measure overall cell death in HeLa or SH-SY5Y, cell viability was assessed by measuring ATP levels using CellTiter-Glo kit (Promega). The untreated cells were considered as an 100% viability control and cell death was inferred as the reduction in viable cells.
SYTOX green uptake and time-lapse microscopy.
For raptinal-induced cell death, cells were seeded in 96-well plates overnight and treated with 10 μM raptinal for 2 h in the presence of 2.5 μM SYTOX green. Fluorescence at 528 nM after excitation at 485 nM was continually recorded every 10 minutes using a Biotek Synergy plate reader. For time-lapse microscopy, cells seeded in glass bottom 35 mm dishes (MatTek) overnight were treated with 10 μM raptinal in complete RPMI medium containing 2.5 μM SYTOX green and imaged using a Zeiss 880 laser scanning confocal microscope within an environmental chamber maintained at 37 °C and 5% CO2.
For YT-induced cell death, HeLa cells were seeded in 96-well plates overnight and pretreated with EGTA (2 mM), zVAD-fmk (30 μM), zDEVD-fmk (30 μM), Nec-1s (20 μM), α-tocopherol (Vitamin E, 100 μM), Fer-1 (2 μM) or DFO (100 μM) as indicated for 0.5 h before YT cells at indicated E:T ratios and 2.5 μM SYTOX green were added. Fluorescence at 528 nM after excitation at 485 nM was continually recorded every 30 min using a Biotek Synergy plate reader. Readings were normalized to control wells containing only YT. For time-lapse microscopy, HeLa were seeded in glass bottom 35 mm dishes overnight. YT cells, stained with Vybrant DiD dye according to the manufacturer’s instructions, were added at an E:T ratio of 2:1 together with 2.5 μM SYTOX green. Beginning 1 h later, cells were visualized over 90 min in an environmental chamber using a Zeiss 880 laser scanning confocal microscope.
Animal studies.
All procedures were conducted in compliance with all the relevant ethical regulations and were approved by the Harvard Medical School IACUC. Female C57BL/6, BALB/c and NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NSG) mice (6–8 weeks) were purchased from Jackson Laboratories. Prf1−/− mice (Jackson Laboratories) in the BALB/c background were bred on site. All mice were housed in the Harvard Medical School Animal Facility. For tumor challenge experiments, B16 EV, B16 mGSDME and B16 mGSDME F2A cells (~1.5×105 cells/mouse) or CT26 control and Gsdme−/− cells (~2×106 cells/mouse) were injected subcutaneously (s.c.) into the right flank of C57BL/6 and BALB/c mice, respectively. 4T1E (EV, mGSDME, mGSDME F2A, mGSDME P212L or mGSDME D270A) cells (~5×104 cells/mouse); 4T1E-eGFP EV and GSDME OE cells (~1.5×105 cells/mouse); or EMT6 (control, Gsdme−/−, Gsdme−/− knocked in to express EV, GSDME F2A, GSDME D270A, or WT GSDME) (~3×105 cells/mouse) were injected into the 4th mammary fat pad of BALB/c mice. For the vaccine/challenge experiment, C57BL/6 mice were vaccinated with 1.5×105 B16 EV or B16 mGSDME cells in the left flank and challenged 10 d later with 2×105 B16 EV cells in the right flank. Tumor growth was monitored by measuring the perpendicular diameters of tumors every other day. When control tumors grew to ~3–4 mm in diameter (10–20 d after implantation), all mice in an experiment were sacrificed and tumors were harvested for analysis. For cell depletion in the B16 tumor model, CD8 Ab (clone 2.43), NK1.1 Ab (clone PK136) or the isotype control Ab (all from BioXCell) were injected intraperitoneally (300 μg/mouse) starting on day 2 after tumor challenge for three consecutive d, and every five d thereafter. For cell depletion in the EMT6 tumor model, NK were depleted with anti-asialo-GM1 Ab (30 μl per mouse, clone poly21460, BioLegend) on d −1, +1 and +6 of tumor challenge. Specific cell depletion was verified by staining for CD4, CD8, CD49b and NKp46 and flow cytometry of peripheral blood mononuclear cells and tumor-infiltrating lymphocytes obtained on day 3 or 7 after tumor challenge and at the time of necropsy (Extended Data Fig. 7a), respectively.
Isolation of tumor-infiltrating immune cells.
Tumors were harvested, cut into small pieces and treated with 2 mg/ml Collagenase D, 100 μg/ml DNase I (both from Sigma) and 2% FBS in RPMI with agitation for 30 min. Tumor fragments were homogenized, filtered through 70 μm strainers and immune cells were purified by Percoll-gradient centrifugation and washed with Leibovitz’s L-15 medium.
Antibody staining and flow cytometry.
To detect eGFP tetramer-specific CD8+ TIL, cells were stained with PE-labeled eGFP-specific MHC class I (H-2Kd) tetramer carrying the peptide HYLSTQSAL22 (NIAID Tetramer Facility). Immune cells isolated from tumors were stained with CD45-PerCPCy5.5 (clone 30-F11), CD8-PacBlue, -PerCPCy5.5, -Alexa700, -FITC or -APC (clone 53–6.7), CD4-PE-Cy7, -APC or -PerCPCy5.5 (clone GK 1.5), CD49b-FITC, -PacBlue or -PerCPCy5.5 (clone DX5), NKp46-APC (clone 29A1.4), CD11b-Alexa700 (clone M1170), F4/80-PE-Cy7 (clone BM8) and CD44- PerCPCy5.5 (clone IM7) (BioLegend). Dead cells were excluded using Aqua dead cell stain kit (ThermoFisher Scientific) added with cell surface antibodies. For intracellular staining, cells were first stained with antibodies to cell surface markers for 30 min, then fixed and permeabilized with fixation/permeabilization buffer (BD Pharmingen) and stained with granzyme B-PacBlue (clone GB11, ThermoFisher Scientific) and Perforin-PE (clone S16009B, BioLegend). For intracellular cytokine staining (ICS), ~106 cells/sample were cultured in RPMI medium containing 2% FBS and stimulated with either PMA (50 ng/ml, Sigma), ionomycin (2 μg/ml, Sigma) and Golgiplug (1.5 μl/ml, ThermoFisher Scientific) for 4 h, or with eGFP200–208: HYLSTQSAL peptide (10 μg/ml, Genscript) and Golgiplug for 6 h. Cells were then stained with IFN-γ-PacBlue (clone XMG1.2) and TNF-α-PE-Cy7 (clone MP6-XT22) after fixation/permeabilization.
Protein expression and purification.
The full-length coding sequence of human GSDME was cloned into the pDB.His.MBP vector to generate a recombinant construct with an N-terminal His6-MBP tag followed by a tobacco etch virus protease (TEV) cleavage site. The plasmid was verified by DNA sequencing and transformed into E. coli BL21 (DE3) cells. Successful transformants were selected on an LB plate supplemented with 50 μg/mL kanamycin, transferred to LB medium with the same antibiotic, and grown at 37 °C with vigorous shaking until the OD reached 1.0. Protein expression was then induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 26 °C overnight. Cells were collected by centrifugation and frozen in liquid nitrogen for long-term storage at −80 °C. To purify human GSDME, thawed E. coli pellets were resuspended in Buffer A (50 mM Tris-HCl at pH 8.0, 150 mM NaCl) and sonicated for cell lysis. The recombinant protein was captured on Ni-NTA beads (Qiagen) using a gravity-flow column and eluted with Buffer A supplemented with 500 mM imidazole. The His6-MBP tag was removed by overnight incubation with TEV at 4 °C followed by Ni-NTA affinity chromatography. The flow-through containing GSDME was concentrated and further fractionated using a Superdex 200 gel filtration column (GE Healthcare Life Sciences) equilibrated with Buffer A. Monomer fractions of GSDME were pooled and frozen at −80 °C for further use.
Recombinant GzmA and GzmB were purified from HEK293T cells and PFN was purified from YT-Indy NK cells as described23,24.
PFN/GzmB killing assay.
PFN/GzmB killing assay in SH-SY5Y cells was performed as described24. Briefly, 500 nM GzmB and/or sublytic PFN in buffer P (10 mM HEPES pH 7.5 in Hanks’ balanced salt solution (HBSS)) were added to SH-SY5Y cells in buffer C (10 mM HEPES pH 7.5, 4 mM CaCl2, 0.4% bovine serum albumin (BSA) in HBSS). The sublytic concentration of PFN was determined as a concentration that caused 5–15% cytolysis of the target cell on its own. After 2 h incubation, LDH release, cell death, SYTOX green uptake and GSDME cleavage were assayed as described above.
In vitro cleavage assay.
For in vitro cleavage in cell lysates, HEK293T cells transiently expressing FLAG-hGSDMD or FLAG-hGSDME for 48 h were lysed in lysis buffer containing 50 mM Tris-HCl pH 7.4, 150 mM NaCl and 1% Triton X100 (2×106 cells/ml) and cell lysates (20 μl) were incubated with GzmA or GzmB at 37 °C for 1 h. Cleavage products were detected by anti-FLAG immunoblot. Cleavage of recombinant GSDME protein by recombinant GzmB was analyzed by SDS-PAGE and Coomassie staining after incubation in buffer A at 37 °C for 1 h.
Statistics.
Student’s t-test (two-tailed) or Mann-Whitney test were used to determine differences between 2 groups. Multiple comparisons between two groups were performed by multiple t-test with type I error correction. One- or two-way ANOVA were used to calculate differences among multiple populations. Differences between tumor growth curves and SYTOX green uptake curves were compared by first calculating the area under curve values for each sample and comparing different groups using the Student’s t-test or one-way ANOVA. Type I errors were corrected by the Holm-Sidak method. P values<0.05 were considered significant.
Data Availability.
All relevant data are available in the Source Data or Supplementary Information of the manuscript.
Extended Data
Extended Data Figure 1. GSDME expression in human and mouse tumor cell lines.
a,b, Gsdme mRNA (a) and protein (b) levels of indicated mouse cancer cell lines assessed by qPCR, relative to Gapdh, and by immunoblot, respectively. c, GSDME expression relative to GAPDH in normal tissue and tumors from breast invasive carcinoma (BRCA) and colon adenocarcinoma (COAD) patients from The Cancer Genome Atlas (TCGA) compared to the qRT-PCR values for mouse cancer cell lines used in this study. P-values comparing normal tissue and cancer tissues were calculated using unpaired two-tailed Student’s t-test. d,f, Expression of mGSDME in EMT6 (d), CT26 (f) clones knocked out (KO) for Gsdme or control (Ctl) cells treated with nontargeting vector, assessed by immunoblot for GSDME. Actin serves as loading control. e,g Cell proliferation determined by CellTiter 96 in EMT6 (e) or CT26 (g) ctl and Gsdme KO cells (n=6 samples/group). h, Expression of hGSDME in SH-SY5Y clones knocked out for GSDME or control (Ctl) cells treated with nontargeting vector, assessed by immunoblot for GSDME. i, j, Expression of mGSDME in GSDME OE and empty vector (EV)-transduced B16 (i) or 4T1E (j) by immunoblot. k, Expression of hGSDME in GSDME OE and empty vector (EV)-transduced HeLa by immunoblot. Differences among multiple groups in (e,g) were analyzed by one-way ANOVA, using the Holm-Sidak method for multiple comparisons. P-values in (e,g) compare KO and Ctl cells. *** P<0.0001. Data are mean ± SD of three technical (a) or six biological (e,g) replicates. Data are representative of at least two independent experiments.
Extended Data Figure 2. Raptinal and/or TRAIL induce pyroptosis in B16 and HeLa overexpressing GSDME.
a-e, Comparison of cell death after adding raptinal to EV and mGSDME OE B16. a, Kinetics of overall cell death assayed by counting annexin V+ and/or PI+ cells by flow cytometry. b,c, Pyroptosis assessed by SYTOX green uptake (b) and LDH release (c). d, Time-lapse microscopy images showing morphological changes and SYTOX green uptake. e, Kinetics of caspase-3 and GSDME cleavage and HMGB1 release by immunoblot of cell lysates and culture supernatants. f,g, SYTOX green uptake by plate reader (f) and time-lapse confocal microscopy (g) after raptinal treatment of EV and hGSDME OE HeLa. h,i, TRAIL induction of cell death 16 h post treatment, assessed by CellTiter-Glo (h), and LDH release (i) in HeLa cells transduced with an EV or in hGSDME OE HeLa in the presence or absence of the pan-caspase inhibitor zVAD-fmk. UNT, untreated. The area under the curve in (a-c,f) and data in (h,i) were compared by two-tailed Student’s t-test. Data are mean ± SD of biological triplicate wells. *** P<0.0001. Scale bar, 20 μm. Data are representative of two independent experiments.
Extended Data Figure 3. GSDME mutations in tumors are mostly loss of function.
a, GSDME-NT mutations in primary cancer cells in The Cancer Genome Atlas (TCGA) (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga). *, stop codon. Red, truncations. Blue, mutants analyzed in this study. b-g, 18 GSDME-NT cancer mutations mapped onto GSDME-NT (amino acids 1–241), modeled based on the cryo-EM structure of the pore conformation of GSDMA3-NT (PDB ID: 6CB8)8 (b,e) were analyzed after transient expression of WT FL GSDME or WT or mutated GSDME-NT (amino acids 1–270) in HEK293T cells for expression by immunoblot (c,f) and LDH release by CytoTox 96 assay (d,g). h-l, Effect of four cancer-related truncation mutations (amino acid 1–46, 1–210, 1–451 and 1–491) on GSDME function. GSDME expression of truncated proteins or FL or GSDME-NT (1–270) in HEK293T (h,k) and their effect on HEK293T cell death, assessed by morphology using microscopy (i) and LDH release (j,l). The NT46 truncated protein is too small to be detected in (k). Red arrows indicate ballooning pyroptotic cells. m,n, Effect of F2A mutation on GSDME-NT-induced pyroptosis in HEK293T. Protein expression (m) detected by anti-FLAG immunoblot and pyroptosis (n) assessed by LDH release. o,p Effect of transient expression of mouse WT FL GSDME or WT or mutated GSDME-NT (mNT270, amino acids 1–270) in HEK293T (o) on LDH release (p). q,r, Effect in 4T1E of expression of WT or mutated FL mGSDME (g) on raptinal-induced SYTOX green uptake (r). Differences among multiple groups in (d,g,j,l,n,p,r) were analyzed by one-way ANOVA, using the Holm-Sidak method for multiple comparisons. Data are mean ± SD of 3 technical (d,g,j,l,n,p) or biological (r) replicates. P-values compare unmutated to mutated constructs. *** P<0.0001. Data are representative of three independent experiments.
Extended Data Figure 4. Gsdme knockout in EMT6 and CT26 increases tumor growth and reduces immune responses within tumors.
Comparison of control (Ctl) and Gsdme−/− orthotopic EMT6 (Ctl n=6, Gsdme−/− n=7) (a-e) and subcutaneous CT26 (Ctl n=6, Gsdme−/− n=5) (f-j) tumors in BALB/c mice. (Knockout (KO) is demonstrated in Extended Data Fig. 1.) Shown are tumor growth (a,f), numbers of CD8+ TIL (left), NK (middle) and TAM (right), normalized to tumor weight (b), representative flow plots (left) and mean percentage of CD8+ TILs expressing GzmB or PFN (right) (c,g), mean percentage of NK expressing GzmB or PFN (d,h), and representative flow plots (left) and mean percentage of CD8+TIL (right e,i) and CD4 TIL (j) producing IFN-γ or TNF-α after PMA and ionomycin stimulation. The area under the curve in (a,f) and differences between two groups in (b-e, g-j) were compared by two-tailed Student’s t-test. Data are mean + SEM. Data are representative of at least two independent experiments.
Extended Data Figure 5. The effects of GSDME expression on tumor growth and immune responses within 4T1E tumors.
a, Mammary fat pad-implanted 4T1E cells stably expressing WT mGSDME (n=6 mice/group), the inactive F2A mutant of mGSDME (n=6 mice/group) or an empty vector (EV, n=7 mice/group) were analyzed for tumor-infiltrating immune cell numbers. b-e, 4T1E, stably expressing eGFP and then stably transduced to express mGSDME or EV (b), were compared for raptinal-induced SYTOX green uptake in vitro (c), tumor growth after orthotopic implantation (n=7 mice/group) (d), and percentage of CD8+ TIL expressing GzmB or PFN (e); Comparisons in (a) were calculated by one-way ANOVA using the Holm-Sidak method for multiple comparisons; (e) were calculated by two-tailed Student’s t-test. Comparisons in (c,d) were calculated by comparing the difference between the area under the curve by two-tailed Student’s t-test. Data shown are mean + SEM. *** P<0.0001. All data are representative of two independent experiments.
Extended Data Figure 6. GSDME overexpression in B16 reduces tumor growth and increases immune responses within tumors.
a, Tumor growth in C57BL/6 mice that were implanted subcutaneously with B16 stably transduced with empty vector (EV) (n=6 mice/group) or to express mGSDME (n=8 mice/group) or inactive F2A GSDME (n=5 mice/group). b, Numbers of CD8+ (left) and NK (middle) TIL and TAM (right) in tumors, normalized to tumor weight. c, Percentage of CD8+ TIL expressing GzmB or PFN (left), and IFN-γ or TNF-α after PMA and ionomycin stimulation (right). d, Percentage of NK expressing Gzm B or PFN. Area under the curve of tumor growth curves in (a) was compared by one-way ANOVA with Holm-Sidak correction for type I error. Comparisons in (b-d) were calculated by one-way ANOVA using the Holm-Sidak method for multiple comparisons. Data are mean + SEM. All data are representative of at least two independent experiments.
Extended Data Figure 7. CD8+ T and NK depletion in mice bearing EMT6 tumors.
Experimental scheme (a) and representative flow plots of CD4+ and CD8+ T cell and NK in the peripheral blood (b) and tumors (c) of mice bearing EMT6 tumors treated with isotype control antibody or anti-CD8 and/or anti-asialo GM1 antibodies in Fig. 2b. Samples were obtained on day 3 (blood) and day 11 (tumors) post tumor challenge. Data are representative of at least two independent experiments.
Extended Figure 8. B16 tumor growth in mice depleted of CD8+ or NK.
a, Experimental scheme and growth of EV or mGSDME+ B16 tumors in mice depleted of CD8+ T cells (n=6 mice/group) or NK (n=6 mice/group) or treated with an isotype control antibody (EV n=6 mice/group, mGSDME+ n=7 mice/group). b,c, Antibody depletion was verified by flow cytometry using PBMC on day 7 post tumor challenge or using TILs at the time of necropsy. Representative flow plots of CD8+ T cell (upper) or NK cell (lower) depletion in the peripheral blood (b) and tumors (c) of mice bearing B16 EV or B16 GSDME tumors treated with isotype control, anti-CD8 or anti-NK1.1 antibodies. Area under the curve of tumor growth curves was compared by one-way ANOVA with Holm-Sidak correction for type I error. Data are mean + SEM. All data are representative of two independent experiments.
Extended Data Figure 9. Necroptosis and ferroptosis are not involved in GSDME-mediated pyroptosis.
a, Effect of EGTA, necrostatin-1s (Nec-1s), α-tocopherol (Vit. E), ferrostatin-1 (Fer-1) and desferoxamine (DFO) on YT-induced SYTOX green uptake in EV and hGSDME+ HeLa. b-g, Cell death-related gene expression in mouse and human cancer cell lines. Ripk3 (b), RIPK3 (c), Mlkl (d), MLKL (e), Il1b (f) and IL1B (g) mRNA levels of indicated mouse (b,d,f) or human (c,e,g) cancer cell lines assayed by qRT-PCR, relative to Gapdh or GAPDH, respectively. Data are mean ± SD of biological (a) or technical (b-g) triplicates and are representative of two independent experiments.
Extended Data Figure 10. PFN plus GzmB induce pyroptosis in SH-SY5Y cells.
SH-SY5Y treated with PFN ± GzmB or medium (UNT) for 2 hr or indicated time were analyzed by microscopy (a), immunoblot of cell lysates probed for GSDME and tubulin (c) or LDH release (b). In (b,c), treatment was in the presence of 30 μM caspase inhibitors as indicated. d, Effects of GSDME knockout on PFN ± GzmB-induced pyroptosis assessed after 1 hr treatment by LDH release. Differences among multiple groups in (b,d) were analyzed by one-way ANOVA using the Holm-Sidak method for multiple comparisons. Data are mean ± SD of biological triplicate wells. *** P<0.0001. Data are representative of two independent experiments.
Extended Data Figure 11. Noncleavable D270A mutation blocks GSDME-mediated tumor protection and induction of anti-tumor immunity.
a-d, B16 stably expressing EV (n=7 mice/group), WT (n=8 mice/group) or D270A non-cleavable GSDME (n=7 mice/group) were implanted in C57BL/6 mice and followed for tumor growth (a) and the functional phenotype of CD8+ and NK TIL (b-d). Shown are the percentage of CD8+ or NK TIL expressing GzmB or PFN (b, c) or CD8+ TIL expressing IFN-γ or TNF-α induced by PMA and ionomycin (d). e-h, 4T1E stably expressing EV, WT or D270A non-cleavable GSDME were implanted in syngeneic (BALB/c, n=6 mice/group) mice and followed for tumor growth (e) and the functional phenotype of CD8+ and NK TIL (f-h). Shown are the percentage of CD8+ TIL expressing GzmB or PFN (f) or induced by PMA and ionomycin to express IFN-γ or TNF-α (h) and of NK TIL expressing GzmB or PFN (g). i, j, Gsdme−/− EMT6 rescued by transduction with lentiviruses expressing EV, WT, F2A or D270A non-cleavable GSDME were examined by immunoblot for GSDME expression (i) and for tumor growth after orthotopic implantation in BALB/c mice (n=8 mice/group) (j). Area under the curve of tumor growth curves or differences among multiple groups were compared by one-way ANOVA with Holm-Sidak correction for type I error. Data are mean + SEM. All data are representative of two independent experiments.
Supplementary Material
Acknowledgements
We thank the NIH Tetramer Core Facility for providing the eGFP tetramer. This work was supported by NIH R01 AI139914 (H.W., J.L.), Charles A. King Trust Fellowship (Z.Z.) and Department of Defense Breast Cancer Breakthrough Fellowship Award (Y.Z.). The mutations in GSDME primary tumors and primary human breast cancer and CRC expression analyses in this study are based upon data generated by The Cancer Genome Atlas (TCGA) Research Network: https://www.cancer.gov/tcga.
Footnotes
Competing interests The authors declare no competing interests.
References
- 1.Liu X & Lieberman J A Mechanistic Understanding of Pyroptosis: The Fiery Death Triggered by Invasive Infection. Adv. Immunol 135, 81–117 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Beeck KO, Van Laer L & Van Camp G DFNA5, a gene involved in hearing loss and cancer: a review. Ann. Otol. Rhinol. Laryngol 121, 197–207 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Rogers C et al. Cleavage of DFNA5 by caspase-3 during apoptosis mediates progression to secondary necrotic/pyroptotic cell death. Nature Commun. 8, 14128 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y et al. Chemotherapy drugs induce pyroptosis through caspase-3 cleavage of a gasdermin. Nature 547, 99–103 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Rogers C et al. Gasdermin pores permeabilize mitochondria to augment caspase-3 activation during apoptosis and inflammasome activation. Nature Commun. 10, 1689 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xia X et al. The role of pyroptosis in cancer: pro-cancer or pro-”host”? Cell death dis. 10, 650 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Palchaudhuri R et al. A Small Molecule that Induces Intrinsic Pathway Apoptosis with Unparalleled Speed. Cell Rep. 13, 2027–2036 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruan J, Xia S, Liu X, Lieberman J & Wu H Cryo-EM structure of the gasdermin A3 membrane pore. Nature 557, 62–67 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galluzzi L, Buque A, Kepp O, Zitvogel L & Kroemer G Immunogenic cell death in cancer and infectious disease. Nat. Rev. Immunol 17, 97–111 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Teng X et al. Structure-activity relationship study of novel necroptosis inhibitors. Bioorg. Med. Chem. Lett 15, 5039–5044 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Wenzel SE et al. PEBP1 Wardens Ferroptosis by Enabling Lipoxygenase Generation of Lipid Death Signals. Cell 171, 628–641 e626 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chowdhury D & Lieberman J Death by a thousand cuts: granzyme pathways of programmed cell death. Annu. Rev. Immunol 26, 389–420 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagata S Apoptosis and Clearance of Apoptotic Cells. Annu. Rev. Immunol 36, 489–517 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Werfel TA & Cook RS Efferocytosis in the tumor microenvironment. Semin. immunopathol 40, 545–554 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aaes TL et al. Vaccination with Necroptotic Cancer Cells Induces Efficient Anti-tumor Immunity. Cell Rep. 15, 274–287 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Sollberger G et al. Gasdermin D plays a vital role in the generation of neutrophil extracellular traps. Sci. Immunol 3, eaar6689 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Kambara H et al. Gasdermin D Exerts Anti-inflammatory Effects by Promoting Neutrophil Death. Cell Rep. 22, 2924–2936 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petrocca F et al. A genome-wide siRNA screen identifies proteasome addiction as a vulnerability of basal-like triple-negative breast cancer cells. Cancer Cell 24, 182–196 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shalem O et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanjana NE, Shalem O & Zhang F Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldman M et al. The UCSC Xena platform for public and private cancer genomics data visualization and interpretation. bioRxiv, 326470 (2019). [Google Scholar]
- 22.Gambotto A et al. Immunogenicity of enhanced green fluorescent protein (EGFP) in BALB/c mice: identification of an H2-Kd-restricted CTL epitope. Gene Ther. 7, 2036–2040 (2000). [DOI] [PubMed] [Google Scholar]
- 23.Dotiwala F et al. A High Yield and Cost-efficient Expression System of Human Granzymes in Mammalian Cells. Journal of visualized experiments. JoVE e52911 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thiery J, Walch M, Jensen DK, Martinvalet D & Lieberman J Isolation of cytotoxic T cell and NK granules and purification of their effector proteins. Curr. Protoc. Cell Biol Chapter 3, Unit3 37 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are available in the Source Data or Supplementary Information of the manuscript.