Abstract
Industrial enzymes have been produced from microorganisms for more than a century. Today, a large share of enzyme products is manufactured using recombinant microorganisms. This chapter focuses on major industrial fungal species belonging to the ascomycetes like Aspergillus niger, A. oryzae, and Trichoderma reesei. Many of the commercially available recombinant enzymes are manufactured using fungi. Examples of fungal enzymes used in food products are described. The enzyme industry is to a large extent cost-driven, so the enzyme product needs to meet strict COGS (cost of goods sold) targets. Therefore, the cell factory must be very efficient to produce the enzyme in high titers and efficiently utilize raw materials. Secondly, it must be designed for a robust and generic fermentation process. When developing fungal hosts for enzyme production, several properties of the system need to be considered relating to efficiency of the cell factory, purity of the product, and safety of both the cell factory and the product. Purity is secured by engineering of the cell factory, and properties related to safety must also be engineered into the fungal host. The methods used for strain improvement are continuously being developed to increase yields and are described herein. More automation using precision tools for modification of the genome (i.e., CRISPR) and low-cost sequencing have vastly expanded the possibilities and enabled fast strain development. Using systems biology approaches, better understanding of cellular processes is now possible enabling advanced engineering of fungal cell factories. Surprisingly, a survey of innovation in the field revealed a decrease in the number of patent applications in recent years. Finally, the requirements for enzyme approval, especially in food and feed, have increased significantly worldwide in the last few years. A description of the regulatory landscape and its challenges in food and feed is included.
The Industrial Enzyme Market
The global industrial enzyme market was 6 billion USD in 2017. The market is expected to grow up to 6% in emerging markets in the next years (Global Industrial Enzymes Markets, seventh Edition, Marketresearch.com). The largest segment of industrial enzyme application is food and beverage, amounting to approx. 2 billion USD in 2016, depending on the source (Patel et al. 2016; Chapman et al. 2018). Higher personal spending in developing countries will lead to growth in sales of higher-value products containing or manufactured with enzymes. Market trends also indicate growth in the use of technical enzymes (textile, paper, biodiesel, biofuel) where waste generation using chemicals will be penalized by environmental agencies (Chapman et al. 2018).
Enzymes are used today to make more than 700 commercial products. Enzyme-based products and solutions are used in over 40 industry sectors, from household care (e.g., detergents) and bioenergy to agriculture, animal health, and food. Food application of enzymes include baking, brewing, beverage, juice, wine, dairy, and oil/fats (Patel et al. 2016).
Using the current list of commercial enzymes for food, feed, and technical applications from the European industry association of manufacturers and formulators of enzyme products (AMFEP, list update of 2015), 243 enzymes manufactured by fermentation of microorganisms are commercially available. Of these, 114 (47%) are produced using recombinant host strains. Most of the commercial enzymes are used in food applications (225 or 93%). Currently, over 300 food enzyme dossiers are under evaluation by the European Food Safety Authority (EFSA) under the EU FIAP (Food Improvement Agents Package) regulatory frame. These applications include both the existing commercial enzymes and new enzymes. Approximately 50% of all submitted dossiers include enzymes manufactured from genetically modified microorganisms (GMM).
Interestingly, most of the commercial enzyme classes used in food are manufactured using fungal hosts (31 of 38 or 82%; see Table 7.1).
Table 7.1.
List of approved enzyme classes for use as a processing agent in food in Europe. Only one example of species of the production organism per enzyme is shown. For each product, the fungal or bacterial origin of the host organism is shown
| Enzyme activity | Production microorganism | Host organism |
|---|---|---|
| Aminopeptidase | Aspergillus niger | Fungal |
| Aminopeptidase (leucyl) | Aspergillus oryzae | Fungal |
| AMP deaminase | Aspergillus melleus | Fungal |
| Arabinofuranosidase | Aspergillus niger | Fungal |
| Catalase | Aspergillus niger | Fungal |
| Cellulase | Aspergillus niger | Fungal |
| Cellulase | Penicillium funiculosum | Fungal |
| Cellulase | Trichoderma reesei | Fungal |
| Dextranase | Chaetomium erraticum | Fungal |
| Glucanase (endo-beta) | Aspergillus niger | Fungal |
| Glucanase (exo-beta) | Aspergillus niger | Fungal |
| Glucanase (exo-beta) | Trichoderma reesei | Fungal |
| Glucanase (beta) | Aspergillus niger | Fungal |
| Glucanase (beta) | Talaromyces emersonii | Fungal |
| Glucoamylase | Aspergillus niger | Fungal |
| Glucose oxidase | Aspergillus niger | Fungal |
| Isoamylase | Pseudomonas amyloderamosa | Bacterial |
| Lactase or galactosidase (beta) | Aspergillus niger | Fungal |
| Lipase triacylglycerol | Candida rugosa | Fungal |
| Lipase triacylglycerol | Rhizopus niveus | Fungal |
| Lipase triacylglycerol | Rhizopus oryzae | Fungal |
| Lysophospholipase | Aspergillus niger | Fungal |
| Alpha amylase | Microbacterium imperial | Bacterial |
| Pectinase | Aspergillus niger | Fungal |
| Phospholipase A | Streptomyces violaceoruber | Bacterial |
| Protease (exopeptidase) | Aspergillus melleus | Fungal |
| Protease | Aspergillus oryzae | Fungal |
| Protease (mucorpepsin) | Rhizomucor miehei | Fungal |
| Protein glutaminase | Chryseobacterium proteolyticum | Bacterial |
| Ribonuclease | Penicillium citrinum | Fungal |
| Tannase | Aspergillus oryzae | Fungal |
| Transglutaminase | Streptoverticillium mobaraense | Bacterial |
| Xylanase | Aspergillus niger | Fungal |
| Glucanase (beta) | Humicola insolens | Fungal |
| Xylanase | Humicola insolens | Fungal |
| Pullulanase | Pullulanibacillus spp | Bacterial |
| Pullulanase | Klebsiella planticola | Bacterial |
| Xylanase | Trichoderma reesei | Fungal |
Enzymes Used in Food Products: New Solutions to Old and New Problems
Fungi, which consist of both yeasts (unicellular eukaryotes) and filamentous fungi (multicellular), are among the most widely used microorganisms in traditional and advanced food production. The yeast Saccharomyces cerevisiae has been extensively used traditionally for food production. In all cases (brewing, baking, wine making), the ability to ferment sugars into ethanol and carbon dioxide is the key feature.
Filamentous fungi have also been used for food. Traditional miso processes using Aspergillus oryzae and A. sake (the “koji” mold) for fermentation of soybean, barley, or rice have been used in Asia for more than 10,000 years in spite of the fact that for a very long time, it was not known that the “mold” was present in the inoculate used (Machida et al. 2008). In a few cases, the organism is a component of the final food product, e.g., Penicillium roqueforti strains used in the manufacture of blue cheese or “Quorn”, a product that has been marketed for decades by Marlow Foods in the UK and is made using Fusarium venenatum (Trinci 1992).
The original koji isolates were recognized as natural producers of enzyme “cocktails” useful for the food industry more than a century ago. In 1894, manufacturing of an enzyme complex from A. oryzae was established (Taka-diastase, Allen 1896; Stone and Wright 1898). A. oryzae was developed to produce an array of different enzyme products in submerged fermentation, still with emphasis on proteases and amylases. Other Aspergillus species like the black aspergilli, A. niger, A. awamori, A. foetidus, A. aculeatus, and A. japonicus were also used mainly for production of glucoamylase.
Development of commercial sources of glucoamylase was the basis for the enzyme revolution in the starch industry (Cairns et al. 2018). Traditionally, starch was hydrolyzed to glucose in an acid process using hydrochloric acid followed by neutralization. In this process, several by-products were formed, and heavy salt formation resulted from the neutralization step. By introducing enzymatic hydrolysis, by-product formation was avoided, and a product of a better quality could be manufactured at a lower price.
There is an increased attention on healthy food products where the use of new enzymes has shown a clear benefit. Among those, the use of enzymes for reduction of lactose content in food products derived from milk using a bacterial β-galactosidases is a great example (Ugidos-Rodriguez et al. 2018). Additionally, using enzymes to reduce the amount of substances with a demonstrated health risk or increasing properties involved in health like higher protein content or a lower salt concentration are worth mentioning. Below are two examples of enzymes manufactured using a fungal host for healthier food products.
Asparaginase for Acrylamide Reduction in Processed Food
Acrylamide is formed naturally in foods as a by-product during frying or baking at temperatures above 120 °C and at low moisture. These conditions are responsible for the flavor and color in toasted, fried, or baked starchy products. Acrylamide is known as a neurotoxin in humans, and it is classified as a probable human carcinogen by the International Agency for Research on Cancer, and its carcinogenic effect has been recently shown (Zhivagi et al. 2019). Furthermore, it is classified as mutagen by the European Union and added to the list of substances of very high concern by the European Chemical Agency (Palermo et al. 2016). It is formed mainly from free asparagine and reducing sugars during high-temperature processing of common foods. The main sources of dietary exposure are fried potato, bakery products, breakfast cereals, and coffee (Pedreschi et al. 2014). A very effective way to substantially reduce acrylamide formation is to convert asparagine to aspartic acid using asparaginase. The use of asparaginase has been suggested as the best approach to reduce acrylamide in food products (Palermo et al. 2016).
Different commercial sources of asparaginase are approved for use in food processing by, e.g., the US FDA (https://www.fda.gov/downloads/food/ucm374534.pdf). Importantly, this approach effectively reduces the acrylamide contents (Fig. 7.1a) while maintaining the organoleptic properties of the different food products (Fig. 7.1b) using a recombinant asparaginase manufactured using A. oryzae (Acrylaway®).
Fig. 7.1.
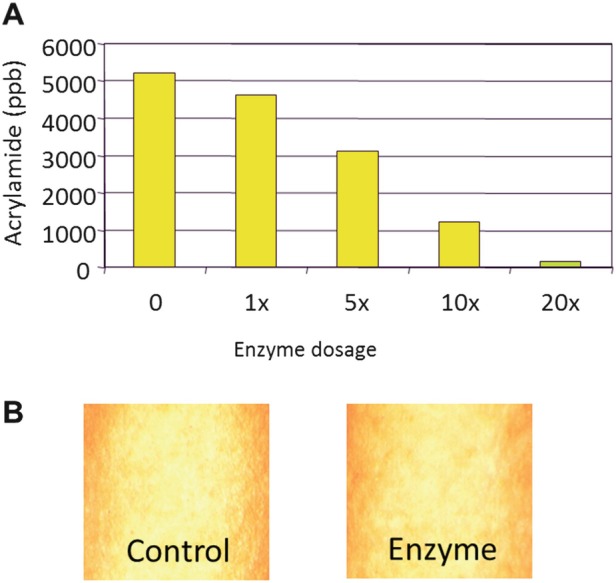
Use of commercial recombinant asparaginase (Acrylaway®) to reduce acrylamide content in potato chips. (a) Dose response test adding different amounts of the enzyme (0 to 20x). (b) Using the 10x dose (“Enzyme”), the potato chips display the same visual (and organoleptic) properties as the “Control.” More information available at https://www.novozymes.com/en/news/news-archive/2013/11/new-novozymes-solution-enables-acrylamide-mitigation-in-even-more-product-categories
Lipase for the Removal of Unsaturated Fatty Acids
Lipases are used as additives or biocatalysts to manufacture many food ingredients, including hydrolysis of milk fat, acceleration of cheese ripening, improvement of flavor, and extended freshness of bread among other applications (Ferreira-Dias et al. 2013).
Baking fats and margarines have either 1) hardened fats with trans-fatty acids or 2) fatty acids removed through a chemical interesterification process. This results in oil of poor quality while also being problematic in terms of both safety and the environment. Current regulations prohibit manufacturers to use partially hydrogenated oils (PHOs) due to the trans-fat content. The World Health Organization launched a program to reduce PHOs and trans fats globally, as their intake is associated to risk for coronary heart disease (Shin et al. 2009). Trans fats are formed when manufacturers use a partial hydrogenation process to give margarines the correct melting properties and shelf stability. The same quality and avoidance of trans fats formation can be obtained using a different process, interesterification. Using enzymatic interesterification gives a more natural, better tasting product that contains healthier oils (Fig. 7.2; see below).
Fig. 7.2.

Color change and by-product formation using chemical esterification (“Chemical,” right flask) compared to the use of a lipase (Lipozyme TL®, a lipase from T. lanuginosus manufactured in A. oryzae; “Enzymatic,” middle flask) using plant oil (“Oil start,” left flask)
In interesterification, the fatty acids of the triglycerides are shifted around randomly—they exchange positions on the glycerol backbone molecule. The most common chemical catalyst is still either sodium methylate (methoxide) or sodium ethylate (ethoxylate). These chemicals are both highly toxic and explosive. The chemical reaction causes by-product formation (including color, Fig. 7.2). Therefore, the reaction is followed by washing, bleaching and deodorization. All these are unit operations, which result in yield loss during the process. By-products of chemical interesterification have been identified as dialkyl ketones (Santoro et al. 2018). Differences between the result of interesterification of a fat blend by chemical and enzymatic interesterification are also partly due to the higher level of diglycerides produced in the chemical process and partly because the enzymatic process does not produce full randomization (Holm et al. 2018).
The enzymatic process eliminates the need for chemicals, washing or post-bleaching, and it produces no wastewater. This process involves fewer steps and reduces energy costs. Several commercial enzymes are available for this application, e.g., Evonik Accurel MD® (non-recombinant Candida parapsilosis lipase) and Novozymes Lipozyme®. Lipozyme® is a series of lipase products (e.g., C. antarctica lipase B manufactured in A. niger or Thermomyces lanuginosus or Rhizomucor miehei lipase manufactured in A. oryzae) that are used as immobilized enzymes to reduce trans fats in, e.g., margarine.
Enzymatic interesterification provides a simple, efficient, and environmentally friendly way to produce margarines without the use of chemicals. The use of enzymes improves product quality and production yields (Ferreira-Dias et al. 2013).
A Survey of Innovation in the Field of Biotechnological Production
Since cost reduction is a major drive in enzyme manufacturing, we decided to look at the evolution of patenting in the field. The assumption was that since the market for industrial enzymes is growing, an underlying and growing technology development must occur to sustain this. The data presented here are not the result of a comprehensive survey and just serve the purpose of describing the trends in the field.
Looking at the number of patent families within the fields of biochemical production, recombinant DNA technology, expression systems, fermentation, production yield enhancement, and enzyme recovery and screening, 2873 patent applications were filed in 2012. These numbers decreased consistently every year (2832 applications in 2013, 2574 in 2014, and 2494 in 2015). In 2016, the total number of patent applications was just 2153.
Considering areas closely related to cell factories, numbers for recombinant DNA technology and expression systems have also consistently decreased in the period investigated (664 and 214 in 2012 to 358 and 160 in 2016 for recombinant DNA technology and expression system patents, respectively; Fig. 7.3). In contrast, gene editing patent numbers increased from 13 to 56 in the period 2013–2015 with a small decrease in 2016 (Fig. 7.3). This recent decrease may be due to the uncertainty on the outcome of the court disputes between Broad Institute and the University of Berkeley among others on CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats; see Sect. 4.4) rights. In a recent review, the total number of granted patents of CRISPR was described as over 90 (Ferreira et al. 2018). This number correlates with the aggregate numbers described here. However, the estimated number of filed applications under examination is 1300 ranging from CRISPR components to delivery systems and applications in medicine, agriculture, and biotechnology.
Fig. 7.3.
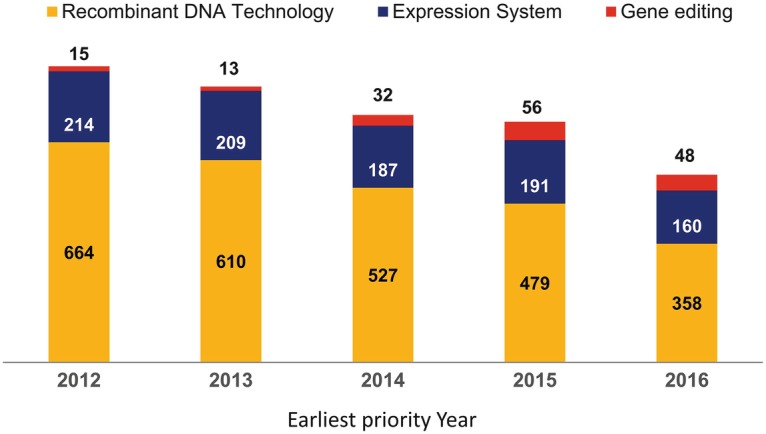
Number of patent families filed in the period 2012–2016 in Europe, USA, and World Intellectual Property Organization (WIPO). The numbers refer to the priority counts based on the earliest priority filing dates of the patent families in the dataset and separated by subject (“recombinant DNA technology,” “expression system,” and “gene editing”). The numbers depict the total number of patent families that originate from the identified priority filing dates. The analysis was performed using broad search terms and patent classification codes. The accuracy of this survey is only 60–70% as it was performed using semiautomated techniques and only limited manual review. Data for 2017 cannot be shown as there is an 18-month delay from application to publication date when searches apply
Except for CRISPR, the overall picture is that the pace at which patents are being filed on technology has slowed down although the number of strategies that can be used for optimization has significantly increased (see Sect. 4.3). It is possible to speculate that many of the existing systems for enzyme production include a mature technology and therefore the number of improvements might have become limiting. Alternative explanations for the steady decrease in the number of patents may include (1) consolidation of the industry, e.g., Danisco being acquired by DuPont in 2011 or BASF expanding its enzyme business in 2013, and (2) a US Supreme Court decision against Myriad Genetics in 2013 that “merely isolating genes that are found in nature does not make them patentable,” i.e., the fact that a DNA sequence alone can no longer be patented.
Fungal Expression Systems and Optimization for Enzyme Production
Since the development of the first recombinant commercial enzyme, research in fungal biotechnology has focused on increasing the yield of enzyme(s) produced in a given host species. The reduction in production costs is a main driver for the use of enzymes for, e.g., replacement of cheap chemicals in food applications. Excellent reviews on expression technology for production that describe the tools and the main approaches to achieve high yields of enzyme are available (Fleissner and Dersch 2010; Nevalainen and Peterson 2014; Davy et al. 2017; Xiao et al. 2014).
An industrially relevant, “basic” expression system consists of:
A host strain with a history of safe use that displays robust and efficient growth and protein production in a low-cost media at industrial (large) scale.
A selection system to ensure introduction of the enzyme gene into the host. Several nutritional marker genes (e.g., pyrG, amdS, and niaD) and antibiotic resistance markers (e.g., hygB) can be used. For industrial food enzyme production, the use of antibiotic resistance markers is not recommended due to regulatory requirements in the final strain.
A transformation procedure to introduce one or more copies of the enzyme expression cassette into the genome of the strain preferably at specific locations.
A strong promoter that enables expression of the enzyme gene under fermentation conditions. An overview of common fungal promoters and their use has been described (Fleissner and Dersch 2010). See also Sect. 4.3.1.
Once a strain producing the enzyme of interest is available, several approaches can be followed to increase yields to make it commercially feasible (see Sect. 4.3). Practically, using one of the generic two-way selection systems available in fungi (i.e., selection can be made for either the presence or the lack of the marker gene), it is possible to rationally improve the host strain in sequential steps of gene inactivation (Fig. 7.4a) or gene overexpression. A disruption in the endogenous pyrG gene encoding an orotidine-5′-phosphate decarboxylase results in the requirement of uridine for growth (Mattern et al. 1989). Lack of a functional pyrG can be selected directly by growth in a medium containing 5-fluoro-orotic acid (FOA). FOA is converted into a toxic intermediate only in a strain containing a wild-type pyrG gene. Therefore, selection of pyrG− transformants or spontaneous pyrG− strains in medium containing FOA allows the direct identification of strains bearing the intended modification.
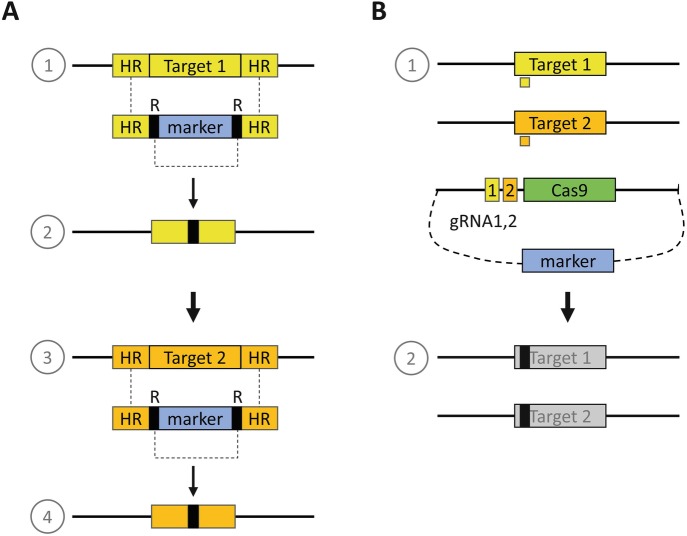
Fig. 7.4.
Traditional (a) and modern (genome editing, b) methods for construction of improved host strains. (a) Traditional methods can be based on the use of homologous recombination using upstream and downstream sequences (“HR”) of the target gene to be deleted (“Target 1”, “Target 2”) to flank a selective marker (“marker”) as indicated. For each target gene, two consecutive steps (1 and 2) are required for integration of the target site (1) and loop out of the marker (2) before a new target gene can be modified using the same selection (3 and 4). (b) Using CRISPR (illustrated with use of the most used Cas9 nuclease, green box), it is feasible to target modification of two or more genes in a single step using gRNAs directed to each target and a plasmid containing suitable expression cassettes for the Cas9 gene and the gRNAs. If the Cas9 expression cassette is introduced on a plasmid containing, e.g., pyrG as the selective marker, subsequent growth of the resulting strains in medium with FOA results in plasmid (Cas9) loss and enables identification of the desired strains. In the depicted example, two target genes are modified in a single step
Using a DNA fragment containing the wild-type pyrG gene as a marker flanked by DNA sequences of the target locus (to enable homologous recombination), disruption of the target gene can be achieved. Thus, using the wild-type pyrG and flanking regions of the target gene to be inactivated (e.g., an endogenous secreted protease), it is possible to introduce a deletion by selection in medium without uridine (requires a functional pyrG gene). Including a repeated DNA sequence at both ends of the marker, it is straightforward to loop out the marker by growing in medium containing FOA, thereby resulting in a pyrG− strain that can be used for the deletion of other target genes using the same marker (Fig. 7.4a) or to introduce the expression cassette for the enzyme gene.
With the advent of genome editing, multiple genetic modifications can be performed in a single step (see Sect. 4.3.3, Fig. 7.4b).
How to Choose the Best Fungal Host Species for Industrial Enzyme Production
One of the challenging tasks to produce a recombinant enzyme is the choice of fungal host. Although S. cerevisiae has been the subject of extensive research and it is currently one of the hosts of choice for production of biologicals, the yields typically obtained preclude its use for production of bulk enzymes (Bao et al. 2017).
Prediction of the most suitable host is still not possible, and rather a screening for multiple hosts is the standard approach whenever possible. As a rule of thumb, the host should be as taxonomically closely related to the donor organism (i.e., where the enzyme gene originates from) as possible. In this way, requirements for the cellular machinery (secretion, protein folding, etc.) should be somewhat similar in the recombinant host. Yields of 20–100 g/L have been published, but they are normally obtained for homologous or near-homologous enzymes (Meyer et al. 2015). The longer taxonomic distance between the donor organism of the enzyme gene and the host, the lower the likelihood of expression at a feasible level (Singh et al. 2015).
Not surprisingly though, there are exceptions to this rule as there are enzymes of bacterial origin that have been successfully produced in fungi, e.g., a Citrobacter braakii phytase used for feed applications that is manufactured in A. oryzae. A bacterial xylanase is produced at high yield in T. reesei (Paloheimo et al. 2003). This illustrates the lack of predictability of a suitable host.
Alternatively, a single host can be used in a high-throughput screening approach combining multiple expression tools, strain backgrounds, and growth conditions. Although the hope is to enable production of a wide variety of enzymes, the reality still suggests that only a limited share of the “protein universe” can be produced by a single host. As an example, the Dyadic International Inc. proprietary thermophilic fungal host C1 (Myceliophthora thermophila) discovered in 1992 as a natural neutral cellulase producer was developed extensively aiming at developing a universal system for recombinant protein production. It has shown great potential for cellulase production for biofuel and for the manufacture of pharmaceutical proteins and vaccines, which are proteins that require a much lower yield to be commercially feasible compared to bulk enzymes (Visser et al. 2011). For other enzymes, yields are typically lower than those from more conventionally used species (Berka et al. 2011).
In principle, there are additional reasons to choose a host among the most broadly used fungal species for enzyme production including A. niger, A. oryzae, and Trichoderma reesei. All three species have been used for decades and have an extensive history of safe use for enzyme production (Frisvad et al. 2018).
Another point of consideration when selecting a host species is the level of endogenous proteases as it can affect yield and stability of the product. Clearly, enzyme yields might not be feasible if the host strain secretes numerous proteases to the medium during fermentation. And protease activity varies depending on the strain, the medium, and the culture conditions. Choosing a host species with a low level of protease like A. vadensis has been proposed (Lubertozzi and Keasling 2009). Alternatively, deletion or reduction of expression of endogenous protease coding genes may result in higher enzyme yields (see Sect. 4.3.3).
Additionally, unwanted side activities (i.e., other secreted enzymes that are not suitable for the application) need to be removed. Examples of this include background glucoamylase and alpha-amylase in A. niger and A. oryzae and different cellulases and endoglucanases in T. reesei.
Finally, the selected host species should not produce any compound of concern during fermentation. As mentioned above, a safety history of use for the most frequently used species is an advantage since the use, e.g., as food additives of many different enzymes in different strain backgrounds, has been documented. As described below (Sect. 6), the major safety concern of a commercial enzyme is not the enzyme but the presence of known mycotoxins in the product. For A. niger, A. oryzae, and T. reesei, safety has been described in a recent review covering this aspect (Frisvad et al. 2018).
Scalability is also important. Growth of the host strains and production of the enzyme should be representative from small scale (preferably from a high-throughput screening format) to industrial production scale. This would enable the testing of several thousands of strains to select the most suitable candidates(s) for fermentation upscaling.
A Generic Fermentation Process for Enzyme Production in Fungal Hosts
During manufacturing at industrial scale, the enzyme should be efficiently secreted into the fermentation broth to ease recovery of the enzyme. A simple and robust process for submerged fermentation is typically used (Fig. 7.5). Shortly, a cell bank vial (typically a working cell bank, WCB) is used to inoculate a solid medium to generate sufficient spores to inoculate a medium-size seed tank (seed fermenter). This step enables the generation of suitable fungal biomass optimally without induction of enzyme production. The biomass is used as inoculum for the main tank (5000 L or larger) where induction of enzyme production occurs. Growth of filamentous fungi in a fermenter presents a number of challenges in terms of the morphology, pellet vs. dispersed growth, high viscosity, and transfer of oxygen to the cells compared to, e.g., unicellular, round yeast cells or bacteria. High viscosity typically limits oxygen transfer and result in loss of productivity (enzyme yield). Mycelial morphology during fermentation has an important effect on aeration and productivity (Amanullah et al. 2002). In submerged cultures, the observed macroscopic morphology of filamentous fungi varies from freely dispersed mycelium to dense pellets consisting of a more or less condensed and intertwined network of hyphae. Morphology and productivity are influenced by the environment and can be controlled by inoculum concentration and viability, pH value, temperature, dissolved oxygen concentration, medium composition, mechanical stress, or process mode as well as through the addition of inorganic salts (Walisko et al. 2015).
Fig. 7.5.
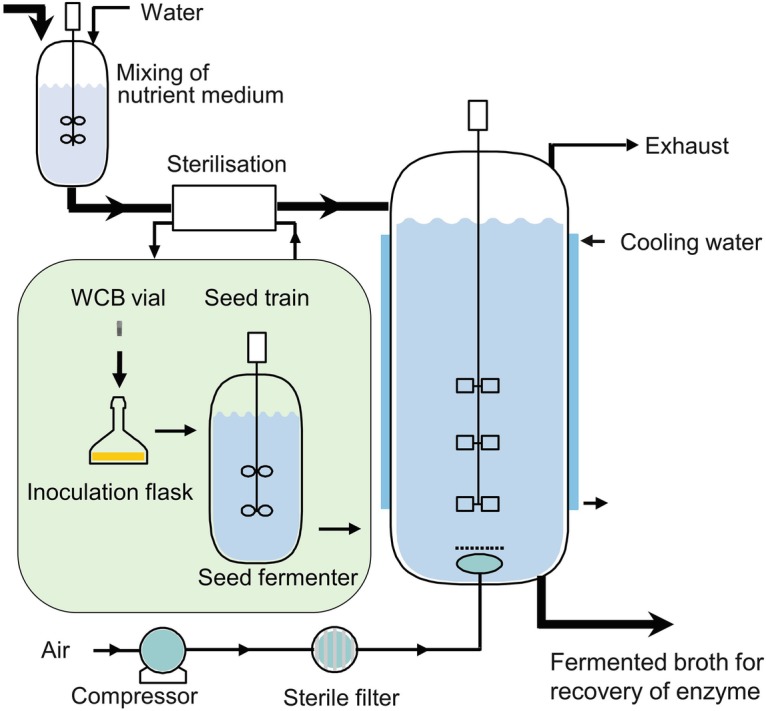
Overview of a generic fermentation process for enzyme production in a fungal host (see main text for explanation). Typically, a defined medium is used and sterilized. A vial of the production strain is used for cultivation in an inoculation flask. Spores or cells are harvested after 5–10 days depending on the host and used to inoculate a seed fermenter. After 1–4 days, the culture is used to inoculate a larger tank where induction of enzyme production occurs by, e.g., feeding with an inducer. Both medium pH and temperature are controlled, the latter by, e.g., cooling water
Depending on the promoter used to control expression of the enzyme gene, the feed rate can be adjusted to enable high productivity and low carbon catabolite repression by, e.g., glucose using the most common fungal-inducible promoters (see Sect. 4.3.1).
Production processes involving fungi are often highly aerobic in nature, which implies these cultures are routinely subject to oxidative stress (Li et al. 2011).
Most of the enzymes produced in fungal hosts and used commercially are secreted into the medium during fermentation. Once the fermentation is completed, the enzyme is recovered from the fermentation broth.
Recovery consists of a step for removal of the cellular debris and a step to concentrate the cell-free fraction, e.g., ultrafiltration, followed by stabilization, sterile filtration, and formulation (Fig. 7.6). Thus, contrary to production of biopharmaceuticals, the enzyme product is a concentrate of the culture supernatant with no physical/biochemical separation of molecules as in a chromatography column. This is a point of concern for the safety evaluation of food and feed enzymes by the authorities. Impurities and compounds of concern like mycotoxins should not be present in the enzyme product (see Sect. 6).
Fig. 7.6.
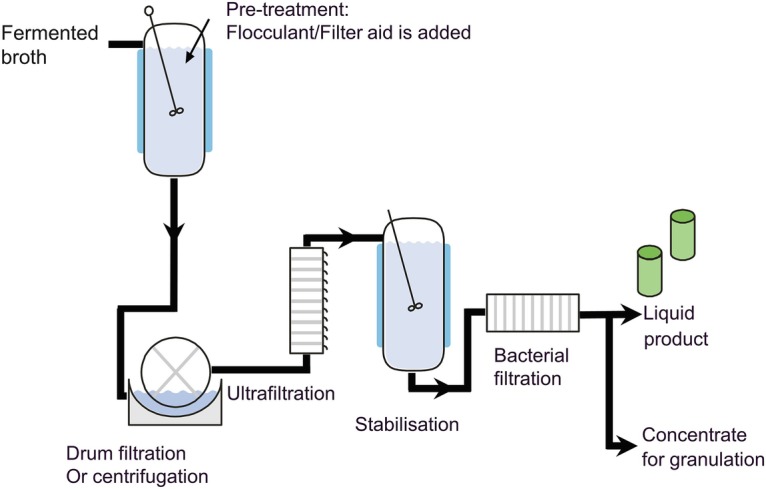
Overview of a generic recovery process for enzyme production. The fermented broth undergoes different steps to remove the cellular debris (flocculation, filtration, or centrifugation). Subsequently the product can be concentrated by ultrafiltration. Product stabilizers are added prior to filter sterilization (bacterial filtration). The enzyme preparation is used for formulation of a liquid or a solid (granulate) enzyme product
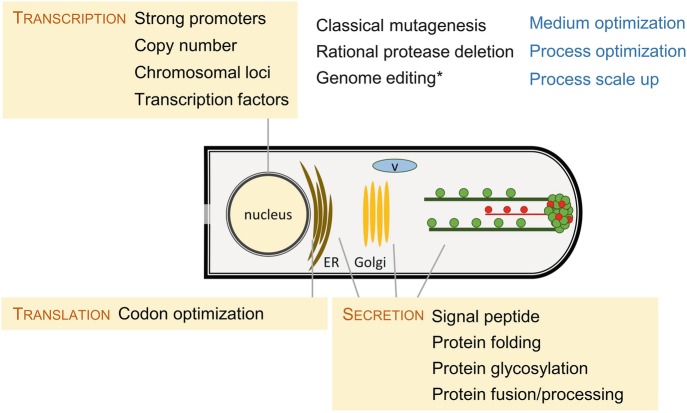
Significant work is devoted to optimizing the fermentation and recovery processes from media optimization, growth conditions, scale up from lab fermenter, etc. (Fig. 7.7).
Fig. 7.7.
Overview of generic approaches used for enzyme yield increase in fungal cell factories. A simplified model of a fungal tip is depicted showing the apical cell, the nucleus, the endoplasmic reticulum (ER), the Golgi apparatus, the secretory vesicles containing proteins to be secreted (green circles) that are transported to the hyphal tip through tubulin filaments (green lines), the chitosomes (red circles) containing enzymes involved in the synthesis of cell wall that are directed through actin filaments (red lines), and the Spitzenkörper, where secretory vesicles accumulate before exocytosis into the extracellular environment. Secretory vesicles can also be directed to vacuoles (v) for recycling. ∗Genome editing can be used both as a rational and high-throughput method for functional genomics analysis
Strategies for the Optimization of Enzyme Yields
Product development in industrial biotechnology includes a continuous challenge to increase enzyme titers at large scale to reduce costs. Two major approaches have been used for this purpose in the last decades to attempt yield increase. The first one is based on classical mutagenesis and screening. Here, the specific genetic modification is not pre-defined. The main requirement is a screening assay that has the sensitivity and range of detection to allow the identification of mutants producing higher enzyme titers. Classical mutagenesis and screening strategies have been used for decades to improve small molecule (Adrio and Demain 2006) and enzyme production. The original Penicillium notatum strain isolated by Fleming contained a single copy of the penicillin biosynthetic genes, and the published yield using optimized strains that contain multiple copies is 70 g/L (Adrio and Demain 2006). Over several decades, a pedigree of A. niger strains with increased production of glucoamylase (GlaA) was developed by classical mutagenesis. Analysis of this pedigree identified both deletions and amplifications including repeated amplification of a 216 kb region that includes the glaA gene. The highest producing strain contained ten copies of glaA compared to two copies in the original parent strain (Cherry et al. 2010). High-throughput screening enables large numbers of mutants to be screened in search for the desired phenotype, i.e., higher enzyme yields. The increase in copy number typically leads to an increase in product formation, but it also reaches a maximum when transcription is not limiting and other cellular processes like secretion become a bottleneck (Gressler et al. 2015).
The second approach based on rational design includes numerous strategies including the use of stronger promoters and multicopy strains to ensure high expression of the gene of interest; codon-optimized gene sequences to improve translation; deletion of genes encoding endogenous proteases; gene fusions for enhanced secretion; and overexpression of accessory cell machinery to alleviate the burden of producing high amounts of protein (chaperones, protein fusions, etc.). Excellent reviews have been published on strategies for cell factory engineering (Fleissner and Dersch 2010; Nevalainen and Peterson 2014; Davy et al. 2017; Xiao et al. 2014).
Rational strategies also include the removal of unwanted side activities that could lead to a negative impact in the purity or the application (i.e., endogenous lipase activity in enzymes used for cheese manufacturing). Other approaches are directed to reduce endogenous protease levels in the host strain for yield and stability of the produced enzyme (Laustsen and Nielsson 1995). Altering the amino acid sequence of the enzyme for increased secretion and/or yield may also be a valid approach (Katakura et al. 1999; Held et al. 2018).
In industrial production strains, high-level production of a protein may trigger several bottlenecks in the cellular machinery for secretion of the enzyme of interest into the medium. Assuming high levels of transcription (result of strong promoters, high gene copy numbers, and sufficient mRNA stability) and translation (result of optimized codon usage, etc.), the next obvious bottlenecks might be encountered in the secretory pathway (translocation into the ER, protein folding, glycosylation, protein degradation, etc.; Fig. 7.7).
More targeted strategies to engineer fungi for high-level enzyme production can be hypothesized using omics tools like transcriptomics, proteomics, and metabolomics to identify cellular bottlenecks and suitable targets for yield optimization. However, multiple candidate target genes are normally identified as being upregulated or downregulated at specific growth conditions or by comparing strains overproducing different enzymes (see Sect. 4.3.2; Zubieta et al. 2018). Gene editing can be used to functionally test several to hundreds of targets. Most of this work is performed in model strains and at laboratory scale. Therefore, careful experimental design and correlation to large-scale production need to be taken into consideration.
Maximizing Transcription of the Enzyme Gene of Interest
A strong and regulated promoter is an important prerequisite for high yield. The stronger the promoter, the lower the requirement for copy number of the expression cassette to be inserted into the production strain to reach optimal enzyme titers. The advantages of using a regulated promoter include the possibility to design a fermentation process composed of a biomass production phase followed by induction of enzyme production. Strong and regulated promoters can be identified, e.g., looking at relatively abundant enzymes or mRNAs. The great majority of highly expressed genes include those encoding secreted enzymes that are an essential part of the life strategy of fungi, i.e., for degradation of polymers such as cellulose, starch, etc. (Wang et al. 2018; Yoder and Lehmbeck 2004). Examples of promoters that are used in industrial enzyme production include the Trichoderma reesei cbhI promoter (PcbhI), the A. oryzae TAKA amylase (PTAKA; Christensen et al. 1988) and the A. niger neutral amylase, and glucoamylase promoters (PamyB and PglaA). All the above promoters are strong and regulated. Induction is obtained in medium containing cellulose-like substrates like sophorose (PcbhI) or starch, maltose, and maltodextrin (PTAKA, PglaA, or PamyB). They are all subjected to carbon catabolite repression (CCR), and their use may negatively affect productivity as glucose is produced during growth. Many industrial fungal hosts have therefore been mutated in functions regulating CCR. One example is the mutation identified in the cre1 gene in T. reesei strains that produce cellulase during growth in glucose. Cre1 is the major CCR transcription factor that binds to a specific motif present in promoter of genes involved in glucose repression. Introducing the mutation in a production strain increases the enzyme yield without affecting growth (Nakari-Setälä et al. 2009).
Various non-native promoters have been engineered to control gene expression in fungi. Synthetic promoter systems (SES) have been developed that are useful for several yeast and fungal species and that show yields comparable to PcbhI for cellulase production. SES consists of a promoter with a core sequence and of a heterologous transcription factor which binds to repeated activating sequences placed in the promoter (Rantasalo et al. 2018). Other promoters like the Aspergillus tet-on/tet-off system are useful for research purposes rather than for industrial production (Wanka et al. 2016; Kluge et al. 2018). As mentioned above, a source of novel promoters may be obtained investigating mRNA levels and choosing the promoter of the highest expressed genes under relevant growth conditions. One challenge of this approach is the fact that unforeseen bottlenecks (titration of accessory functions like transcription factors) may appear when using these promoters at high copy number in a production strain background.
Another strategy to increase transcription from strong native promoters is artificial transcription factors that can be engineered to improve production of secreted heterologous enzymes. A recent paper highlighted improving cellulase expression in Trichoderma reesei by introducing artificial transcription activators (ATAs). The ATAs were constructed by linking the C-terminus of XYR1, ACE1, and ACE2 with an activation domain of herpes simplex virus protein VP16. Strains engineered with one of the three artificial transcription activators displayed different phenotypes of improved cellulase production (Zhang et al. 2018). Yet another approach is the use of polycistronic RNAs under the control of a regulated promoter for secondary metabolite production in A. niger (Geib and Brock 2017).
Long noncoding RNAs that are involved in gene regulation have been identified in yeast and filamentous fungi (Till et al. 2018a). Identification and manipulation of long noncoding RNAs represent another means to engineer filamentous fungi for improved heterologous protein production. Recently a long noncoding RNA, Hax1, that enhances cellulase expression was discovered in T. reesei (Till et al. 2018b). With low-cost sequencing, it is possible to sequence RNA to identify possible long noncoding RNAs present under relevant fermentation conditions for production followed by functional testing to determine if they have an impact on protein production.
The choice of promoter influences the fermentation process that can be developed, e.g., a process based on glucose as carbon source may not be suited if expression of the enzyme gene is under control of a promoter that is repressed totally or partially by glucose.
Optimization of Protein Secretion: Still a Black Box
Secretion is a complex cellular process that involves many steps, proteins and organelles in the cell. Shortly, secretion consists of the transfer of a nascent protein to the endoplasmic reticulum (ER), its appropriate folding, posttranslational modification and maturation (Golgi), sorting of vesicles to either early endosomes or secretory vesicles, directional transport of secretory vesicles to the growing hyphal tip, and fusion of the vesicles with the plasma membrane to release the enzyme to the extracellular environment (Fig. 7.7). Early endosomes might also be involved in the delivery of secretory proteins to the vacuole for degradation and recycling of nutrients (Hernandez-Gonzalez et al. 2018).
In 1994 a review was published in which protein secretion in filamentous fungi was called a “highly productive black box” because despite the use of these organisms to produce high yields of commercial enzymes applied in many industrial applications, very little was understood about the cellular mechanism of secretion (Peberdy 1994). Since this publication, research has broadened the understanding of secretion in filamentous fungi; however, many reports on attempts to improve “secretion” of a protein have been published, and no broadly successful strategy to manipulate the secretion pathway exists. Therefore, secretion optimization is probably one of the most challenging tasks to increase enzyme yields. Fungi like A. niger and A. oryzae adapt the protein load in the secretory pathway, probably as an evolutionary advantage of their lifestyle in nature. In addition, we continue to learn unique aspects of secretion in fungi as exemplified by recent insights into protein trafficking in A. nidulans. A genetic dissection of the exocytic pathway in A. nidulans has been described. It reveals numerous and yet unexplored cellular functions like Rab1and Sed5 (early Golgi), Sec7 and Trs120 (trans Golgi network), and Rab5 (early endosome) among others (Hernandez-Gonzalez et al. 2018).
To efficiently direct a protein for secretion via the ER, it is normally necessary to use of an optimal signal peptide (SP). A comprehensive review on the structure, features, and application of SPs has been published (Owji et al. 2018). In principle, SPs derived from efficiently secreted enzymes of the host should be suitable. Screening of different known signal peptides to find the most suitable sequence can be performed as described for the yeast Pichia pastoris. Although some signal peptides (e.g., the sequence derived from the yeast mating factor) result in an overall higher yield in yeast, no secretion was obtained using this sequence for other proteins (Obst et al. 2017). Suitable SPs may be obtained by identifying the secreted proteins of a fungal host and selecting the SP of the most abundant ones (Wang et al. 2018), although in vivo screening is needed as theoretical values generated by SignalP (a prediction software for SPs) are not associated with high levels of secretion in yeast (Mori et al. 2015).
Overexpression of secretory enzyme genes causes an overload of the secretory route. Excellent reviews have been published on the subject (Aviram and Schuldiner 2017; Meyer et al. 2015). To restore homeostasis, both ER-associated protein degradation and the unfolded protein response (UPR) aid in removing or folding the accumulated protein. The UPR is activated when misfolded proteins accumulate in the ER. It leads to the alternative splicing of the HAC1 gene product and the activation of UPR-regulated genes. In yeast, the secretion of α-amylase was significantly enhanced through the overexpression of HAC1 (Valkonen et al. 2003).
In addition, many genes become transcriptionally downregulated, maybe because they are less important for growth and survival under the specific growth condition. This phenomenon is called repression under secretion stress (RESS) and was first observed in cellulase production strains of T. reesei (Pakula et al. 2003).
To illustrate the complexity of enzyme secretion, it was shown that glucoamylase overproduction in A. niger is associated with a change in the expression of 1500 genes, including upregulation of genes that are involved in ER translocation, protein folding and glycosylation, and vesicle transport (Kwon et al. 2012).
If folding is a bottleneck in enzyme overproducing strains, an obvious target for optimizing enzyme yield is overexpression of ER-resident chaperones (Lubertozzi and Keasling 2009; Davy et al. 2017). Significant upregulation of bipA, clxA, and pdiA among others was reported for glucoamylase production in A. niger (Fleissner and Dersch 2010; Kwon et al. 2012). The challenge is the choice of chaperone and the appropriate level of overexpression that may be necessary to boost yields since different enzymes or different sequence variants of the same enzyme may require different cellular functions. In S. cerevisiae, moderate expression of SEC16 has been shown to increase secretion of two heterologous enzymes (Bao et al. 2017) adding extra complexity to the process of optimization of enzyme secretion.
Another approach to improve secretion of a desired protein is a fusion of the protein to a well-expressed, secreted protein from the host. The rationale for this approach is that the homologous protein may facilitate passage through the secretory pathway, although the precise mechanism is not understood (Yoder and Lehmbeck 2004). For production of calf chymosin in A. awamori, a protein fusion to the endogenous glucoamylase was used to increase yields (Ward et al. 1990).
Given that there are no clear strategies to engineer filamentous fungi for increased secretion of a protein of interest, the ability to build and identify recombinant strains with improved secretion of a given enzyme requires methods to be able to construct and test many different combinations of transcriptional control elements, signal peptides, and genome modifications. Automation and high-throughput screening that correlate with production scale are required.
Improving Protein Quality and Stability
The involvement of endogenous proteases on the yield is suspected whenever degradation of the enzyme of interest is observed during fermentation or during storage. Production and secretion of endogenous proteases and peptidases is rather widespread among fungal species. Filamentous fungi have the potential to produce numerous endogenous protein-degrading enzymes.
To identify the proteases responsible for product degradation, investigation of the proteins secreted into the medium can be performed. This may also be a substantial task since the number of secreted proteins in fungal cultivations may vary from 300 to 700 or more, depending on the growth conditions and the species (Jun et al. 2013; Schmoll et al. 2016; Terfrüchte et al. 2018). In A. oryzae, more than 130 exo- and endopeptidase genes have been identified (Kobayashi et al. 2007). In Ustilago maydis, the use of proteomics identified 477 proteins in submerged cultivation of which 21 were annotated as proteases or peptidases. Surprisingly, engineered strains in which deletion of major protease genes has been introduced and not displaying detectable protease activity in casein-indicator plates still contained a significant protease activity (Terfrüchte et al. 2018). As in many other fungi, the arsenal of proteases present in wild-type strains ensures that available protein sources can be degraded under different growth conditions even if loss of function mutations in major protease genes occur. In T. reesei, strains engineered for production of enzymes for biomass degradation, protease levels (e.g., PepM and Asp1) vary depending on growth conditions (Jun et al. 2013; Stappler et al. 2017). Additionally, a strain containing deletions in the seven genes encoding the more predominant proteases was developed, using a combination of inhibitor studies and protease purification, to enable production of biopharmaceuticals in T. reesei (Landowski et al. 2015).
Identifying major secreted proteases has resulted in the development of improved industrial strains carrying deletions in one or more protease genes (Laustsen and Nielsson 1995; Wang et al. 2018). An alternative approach is to introduce a deletion of the gene encoding a transcription factor, PrtT, that controls the expression of multiple protease genes (Hjort et al. 2001; Punt et al. 2008). A protease regulator, Pea1, has been identified in T. reesei and Fusarium sp. that is required for expression of several proteases. Interestingly, Pea1 does not share similarity to other known regulatory proteins (Paloheimo et al. 2018).
Modification of Other Important Features of the Host
Filamentous fungi have the potential to produce a large number of secondary metabolites (SM). Some SM display antimicrobial or antioxidant activity and are used as a defense mechanism in nature. These compounds are normally not produced during growth in the laboratory or in large-scale growth in a fermenter. Mycotoxins are a small group of SM representing a health risk for animal and humans. Mycotoxins are acutely or chronically toxic and pose health hazards or death in humans and other vertebrates when acquired in small amounts via a natural route, i.e., orally, by inhalation, or via the skin (Frisvad et al. 2018). Genes responsible for mycotoxin biosynthesis are normally grouped in gene clusters. Although mycotoxin production is not normally associated to submerged growth of pure cultures, the potential for production of these compounds can be eliminated by deletion of the responsible gene cluster resulting in strains with a reduced toxigenic potential. To do this, the genes responsible for production of the mycotoxin need to be identified. In A. oryzae, two neighboring gene clusters involved in the synthesis of cyclopiazonic acid and aflatoxin, respectively, have been removed from the genome in production strains (Christensen et al. 1999; Olempska-Beer et al. 2006). Similarly, genes responsible for production of most relevant fungal mycotoxins like fumonisin and ochratoxin (Khaldi and Wolfe 2011; Gill-Serna et al. 2018) are known, and strains lacking the potential to produce them can be engineered.
An alternative to this approach, e.g., in cases where the genes responsible for production of a mycotoxin are not known, is to monitor the presence of relevant mycotoxin in production and in the enzyme product.
As mentioned earlier, to keep COGs low, production processes do not include purification steps. Purity of the product is solely controlled by the level of production of the enzyme product and the concentration of other secreted proteins in the recovered and formulated product. Many of the industrial fungal host species produce one or more secreted enzymes at reasonable titers, i.e., amylase, cellulases, and glucoamylase by A. niger, A. oryzae, and T. reesei. This ability to produce high levels of a secreted enzyme(s) has been exploited both for production of these native enzymes as well as for heterologous enzymes. In strains producing a heterologous enzyme, genes coding for abundant native secreted enzymes are deleted or classical mutants that are unable to produce the native enzymes have been isolated. T. reesei produces a complex set of cellulases and hemicellulases that provides the capability to degrade lignocellulose. Deletion of the most abundant enzyme cellobiohydrolase I (cbh1) gene leads to an increase in production of cellobiohydrolase II (cbh2), whereas deletion of endoglucanase I or endoglucanase II increased levels of both cbh1 and cbh2 (Seiboth et al. 1997).
Genome Editing as a Revolutionizing Tool for Production Strain Development
Genome editing has become a true game changer in research, medicine, and biotechnology. CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) is an acronym whose impact has reached even outside the scientific community through headlines in everyday news as a tool to combat diseases and provide food for a growing population.
CRISPR is a natural mechanism that allows bacterial cells to detect and destroy the viruses that attack them. Since the publication describing how CRISPR and the Cas9 nuclease can be used to edit DNA sequences (Jinek et al. 2012), the possibilities for application of genome editing have extended to medicine and agriculture. In industrial biotechnology, the use of CRISPR has enabled a faster method for strain construction. A recent review was published highlighting the recent applications of CRISPR/Cas9 in industrial biotechnology as well as the patent landscape (Ferreira et al. 2018).
A review on CRISPR systems and tools for genome editing in filamentous fungi has recently been published (Deng et al. 2017). RNA-guided Cas9 endonuclease can be used to cleave any DNA sequence in a fungal host with high specificity, and repair of the double-strand breaks can be mediated by error-prone, nonhomologous end joining (NHEJ) or by the introduction of specific changes via homologous recombination using donor DNA. In strains deficient in nonhomologous end joining (NHEJ), highly efficient marker-free gene targeting can be performed. Even single-stranded oligonucleotides work efficiently as repair templates for Cas9-induced DNA double-strand breaks in different Aspergillus species (Nødvig et al. 2018). This indicates that this type of repair may be widespread in filamentous fungi. Using oligonucleotides for CRISPR-Cas9-mediated gene editing, it is now possible to introduce specific point mutations as well as gene deletions at efficiencies approaching 100%. In A. nidulans, up to three editing events can be achieved in one transformation (Nødvig et al. 2018).
For research purposes in, e.g., functional genomics, CRISPR has already shown a tremendous impact. CRISPR-Cas9 and the nuclease inactivated variant of Cas9 (Cas9dead or Cas9d) can be used for studies of gene regulation (activation and repression), to reconstruct and modify metabolic pathways among numerous other approaches (Deng et al. 2017). The ability to perform multiple modifications in a single step using CRISPR (Fig. 7.4b) will enable faster construction of yield-optimized strains and will fuel its use for rational design in strain construction as well as for more genome-wide searches for gene modifications that improve production of an enzyme. Genome editing in the thermophilic fungus Myceliophthora was used to increase cellulase production by 13-fold by editing up to four genes at one time (Liu et al. 2017).
Having the possibility to modify a target sequence regardless of the number of copies present in the genome using CRISPR, yet intractable but important microorganisms should now be amenable for strain improvement (e.g., diploid fungi).
Automated Strain Construction, Systems Biology, and Synthetic Biology Approaches
The emergence of inexpensive, base-perfect genome editing along with low-cost sequencing is revolutionizing biology. Modern industrial biotechnology exploits the advances in genome editing in combination with plug and play strain construction systems, automation, analytics, and data integration to build high-throughput automated strain engineering pipelines (Marcellin and Nielsen 2018). In this way, strain construction can be accelerated and expression results from a large number of unique strains can be used to begin to develop machine learning models to predict optimal expression cassette elements and genome modifications.
Systems biology provides rational approaches and analytical tools to understand complex biological processes like manufacture of industrial enzymes using fungal cell factories and to enable production of novel molecules (Martins-Santana et al. 2018; Hammer and Avalos 2017). Mathematical modeling is central in systems biology to integrate information and quantitatively analyze phenotypes to enable predictions about the behavior of the organism or elucidating mechanisms underlying experimental observations. For this purpose, a genome model of the host species provides the “scaffold” for the integration of the large datasets that are generated and that enable computational predictions (Campbell et al. 2017). These approaches address a key issue in the construction of efficient cell factories, i.e., the ability to control where and when chemical reactions take place in the cell (Jakobson et al. 2018).
An overview of systems biology research conducted in Aspergillus has recently been published (Brandl and Andersen 2018). A review on the impact of systems biology on cell factories including those producing heterologous proteins in different bacterial and fungal hosts like A. niger highlights possible applications of this approach for improving protein production (Campbell et al. 2017).
Synthetic biology approaches can also be used to engineer strains for improved yield and productivity; it provides an opportunity to engineer biology in a more standardized and rational fashion, and several examples using a synthetic yeast genome have been described (Liu et al. 2018). One of these approaches is the rewiring of metabolic flux by deleting genes that negatively affect enzyme production. Using protein localization tags, beneficial modifications (by engineering the cellular location of the reaction) of the metabolic flux can be obtained (Martins-Santana et al. 2018). In yeast, several organelles (mitochondria, peroxisomes, and even the cell wall) have been redesigned to improve processes such as bioethanol production (Hammer and Avalos 2017).
The Evolution of Methods for Construction of Production Strains to Adapt to Regulatory Requirements
In the early years of modern biotechnology, production strains were constructed using free replicating plasmids and antibiotic resistance as a dominant selective marker on the plasmid containing the expression cassette for the gene of interest. Although not a strict requirement for technical enzymes, the presence of genes encoding antibiotic resistance imposes additional requirements from regulatory authorities on food and feed enzymes (see Sect. 6). Thus, auxotrophic markers or markers that enable growth in specific nitrogen or carbon sources have been introduced, e.g., the amdS gene (Christensen et al. 1988). Genetic stability issues have also resulted in the scarcity of strains constructed using free replicating plasmid based on, e.g., the Aspergillus AMA1 fragment (Aleksensko and Clutterbuck 1997). Even though transformation frequencies are much lower than for free replicating plasmids, current production strains are therefore constructed using chromosomal integration. Because NHEJ is the dominant mechanism for DNA integration in industrial fungal strains, initially, production strains were the result of ectopic (nonhomologous) integration. The regulatory requirement—determination of the site of integration of the expression cassette—has driven the construction of strains through homologous recombination at one of more defined chromosomal locations. A suitable host strain should harbor multiple and reusable integration loci (i.e., a “plug and play” system) in such a way that newly identified yield improvements can be readily incorporated to produce different enzymes (Udagawa 2017).
Environmental Aspects of the Yield Improvement in Industrial Recombinant Enzyme Production
Enzymes enable the increase in quality, speed, and yield of numerous processes and reduce both energy consumption and the use of chemicals. Life cycle assessment of industrial enzyme production has been performed. The fermentation step accounts for half of the environmental impact of enzyme manufacturing (energy consumption), while recovery and waste management represent 1/8 of the total contribution in the commercial setting at Novozymes A/S in Denmark (Nielsen et al. 2007). The environmental impact resulting from the optimization of enzyme yields from using recombinant technologies has been compared to non-recombinant enzyme production. Considerable and consistent improvements (e.g., energy consumption, global warming, nutrient enrichment, ozone formation) have been clearly demonstrated for the use of optimized recombinant strains (Nielsen et al. 2007).
The environmental impact of enzymes in some applications is quite striking. The textile industry has a reputation as one of the most polluting industries in the world. The problem is the traditional, extensive use of chemicals, energy, and water to meet the needs of the fashion industry. For instance, it takes about 150,000 L of clean water to make one ton of knitwear. The estimated savings if the world’s textile industry implements the whole range of available enzymatic solutions in the production of cotton textiles include 28% of water consumption which is 1250 billion liters of water. Additionally, the use of textile enzymes will save 80% of chemical consumption—that is, 10 mn tons of chemicals—and up to 25% of the energy consumption. Numbers are based on a world production of 25 million tons of cotton textiles per year which is a very conservative estimate and LCA studies made by Novozymes.
Approval of Enzymes for Use as Food Processing Aids and in Feed Applications
In principle, the same safety considerations apply to enzymes derived from native or recombinant microorganisms. The main aspect of the safety of food enzymes is the production strain and the potential risk of producing compounds with a toxicogenic or pathogenic effect (e.g., mycotoxins) during manufacturing, i.e., mainly during fermentation (Pariza and Johnson 2001). Contrary to biopharmaceuticals, industrial enzymes are typically not purified products but rather derive from a concentrated fermentation broth where the production organism has been removed. This principle of “contained use” where the enzyme product does not contain material from the production organism is important for the classification of the product and requirements from regulatory authorities. Furthermore, the use of antibiotic resistance genes as selective markers for strain construction should be avoided due to concerns of its transferability and effect on the spread of resistance to antibiotics normally used to treat infections in humans.
In this section, a short overview of the requirements for the description and characterization of the production organism used to manufacture enzymes for use as food processing aids and in feed applications is presented. A comprehensive review on the regulations, definitions, and approval processes in different regions of the world is available (Magnuson et al. 2013). Significantly lower requirements for the strain description applies for technical and detergent enzymes. However, the general trend is that the requirements for the characterization for the production strain used for food and feed applications increase steadily.
Regulatory Considerations for the Approval of Recombinant Food Aids and Feed Enzymes in the European Union
The European legislation for the use of additives, flavorings, and enzymes in foods is set out in the Food Improvement Agents Package (FIAP). A regulatory authority (European Commission, Directorate General for Health and Consumers) and an advisory scientific body responsible for the risk assessment [EFSA (European Food Safety Authority)] participate in the approval process to ensure, e.g., the safety of GM food products for animal and human consumption (Aguilera et al. 2013). These regulations were introduced in 2009 and reached a key milestone on March 11, 2015, the first deadline for food enzyme manufacturers to submit dossiers on all food enzymes currently sold or used in Europe under the new regulation. More than 300 enzyme dossiers were submitted. These submissions are required to ensure compliance of the enzyme industry and its customers when FIAP gets fully implemented in 2021. An update of the requirements for the characterization of the production strain has recently been implemented (EFSA 2018).
For enzymes manufactured using a recombinant strain, a comprehensive description of the genetic elements and methods used to construct the strain from a publicly available parental strain is required. One major aspect is the requirement to demonstrate the absence of the production organism and DNA in the final enzyme product to enable the approval as a Category 2 product (“Complex products in which both GMMs and newly introduced genes are no longer present”) as opposed to the use of, e.g., a recombinant organism directly in the food. To issue an opinion on the safety of the enzyme, EFSA requires—among many other aspects relating to, e.g., the technical need or calculation of the margin of safety for intake—a description of each modification step from the parental to the actual production strain, in addition to data on the taxonomical identification, safety, pathogenicity, toxicological studies, history of safe use, genetic stability and the absence of vector or any other sequence used during construction steps that is not intended for insertion into the genome, etc. (Aguilera et al. 2013).
In the latest update, and although not yet a requirement for fungal hosts, the genome sequence (i.e., a comparison between the parental and production strain) can be used as the basis for the description of the changes introduced in the production strain. Additionally, further constrains have been adopted for the PCR method used for demonstration of the absence of DNA in the product (EFSA 2018).
In the current stage, where EFSA has a backlog of submitted dossiers in 2015 and the expected publication of a positive enzyme list in 2021, it is difficult to predict the time that will be required to obtain approval of a new food or feed enzyme in Europe.
Regulatory Considerations for the Approval of Recombinant Food Aids and Feed Enzymes in the US
In the US, FDA and the GRAS (generally regarded as safe) notification system provides a relatively short standard review and approval time. GRAS food ingredients are generally recognized, among qualified experts, to be safe under the conditions for its intended use (GRAS final rule 21CFR170.250(b)). In the past, FDA reviewed GRAS affirmation petitions for enzymes. This process has been replaced by a voluntary notification program under the FDA’s proposed regulation. A successful notification process leads to an FDA letter to the manufacturer stating the agency has no questions on the manufacturer’s conclusion that the use of the enzyme for the described application is safe (Olempska-Beer et al. 2006).
There is no specific regulation governing enzymes. Depending on the intended use and the method, enzymes are regulated as direct (i.e., added to the food product) or—most common—secondary additives (i.e., processing aids in food manufacturing to fulfil a technological purpose) or GRAS substances (Magnuson et al. 2013).
In 2017, the accumulated number of submitted GRAS notifications in a recent pilot program (1997–2015) was 678, including enzymes, fats and oils, probiotics, sweeteners, and carotenoids. With a total of 70 enzyme notifications, the average review time by the FDA was 168 days. In 68 of the notifications, the application did not trigger questions whereas two applications were withdrawn (Hanlon et al. 2017).
The science-based process used by FDA for risk assessment is more of a product-centric approach where safety is assessed on the product and not on the methodology used to bring about the product (i.e., GM or not, CRISPR or not, etc.). Requirements for strain description and characterization are not as comprehensive as in other geographies. The data included in the enzyme dossier can be accessed by third parties following the freedom of information act (FOIA, a federal law that allows for the full or partial disclosure of previously unreleased information and documents controlled by agencies like FDA). This has direct consequences on the extent of the information that enzyme manufactures can disclose on, e.g., strain construction and gene expression technology in a GRAS notification.
Evolution of Regulatory Requirements in the Rest of the World
In the rest of the world, the trend is also a steady increase in the requirements for the characterization of the production strain fuelled by the development of new technologies and methods. Currently, data on the absence of DNA in the enzyme product that has been a requirement in Europe and Japan for several years, are also requested in China. Although FIAP has created a comprehensive, high-level standard for the characterization of production strains, countries like China have additional requirements compared to, e.g., FIAP that include a comprehensive genetic and expression stability study that spans to five consecutive cultivations of the production strain. Similarly, a requirement to identify the chromosomal location of the inserted DNA exists in Japan. Although technically feasible with novel sequencing technology, this can be rather challenging if the production strain is constructed using ectopic integration in multiple copies at single or multiple locations, considering the relatively high level of repeated DNA sequences in a fungal genome.
Finally, alignment of requirements between regulatory authorities would enable the characterization of the production strain and the submission of dossiers according to one standard. As an example, the modification of the host strain can be described with less detail in a GRAS notification than in a FIAP dossier where each step needs to be described in detail and supported with data confirming that the modification has happened as intended. Alignment of requirements for strain characterization in enzyme dossiers would represent a tremendous advantage for industrial biotechnology companies.
The Future of Industrial Enzyme Production in Fungal Hosts
Although many commercial enzymes have been successfully produced in filamentous fungi, it is still not possible to predict the necessary combination of engineered cellular functions and expression cassette elements required for optimal production of a novel enzyme in a fungal host.
Increasing numbers of fungal genome sequences are available, but gene annotation is still a limitation. A large number of putative genes still have an unknown function. Assigning functions to these genes using the appropriate method (transcriptomics, proteomics, etc.) will improve the knowledge about the biology and physiology of the host strains and contribute to our understanding of protein production and secretion in filamentous fungi.
With increasing accessibility to CRISPR and “omics” technologies, the expected growth of genomic and experimental datasets is of paramount importance to accelerate knowledge about complex cellular processes related to enzyme production and the development of new and improved fungal hosts.
Being able to construct many strains using targeted integration should enable genome-wide screening in fungi as it has been performed in yeast to identify optimization candidate genes (e.g., via knockout libraries). Novel technologies allowing for even larger number of strains to be tested (e.g., ultra-HTS) will contribute to this. Combining these approaches with emerging genome models and available “omics” will accelerate the capability of fungal cell factories to produce more diverse classes of enzymes at higher yields. This will enable production of many novel enzyme classes for existing and novel applications.
The development of novel fungal species as hosts may also facilitate production of difficult to express enzymes using traditional systems.
Increasing knowledge of SM pathways and genes involved will enable the construction of safer strains containing deletions in one or more SM gene clusters, especially for novel host species. The continuous improvement of technologies like genomics to characterize the production strains will impact the requirements from regulatory authorities for the characterization of the strains used for food and feed enzyme production.
The promise of industrial biotechnology becomes more relevant as we face the need of new biological solutions to replace chemicals and to support sustainability in a growing world.
Acknowledgments
We thank Amitabh Gupta (Novozymes) for the support in the patent survey.
Contributor Information
Helena Nevalainen, Email: helena.nevalainen@mq.edu.au.
Debbie Yaver, Email: dsy@novozymes.com.
References
- Adrio JL, Demain AL. Genetic improvement of processes yielding microbial products. FEMS Microbiol Rev. 2006;30:187–214. doi: 10.1111/j.1574-6976.2005.00009.x. [DOI] [PubMed] [Google Scholar]
- Aguilera J, Gomes AR, Olaru I. Principles for the risk assessment of genetically modified microorganisms and their food products in the European Union. Int J Food Microbiol. 2013;167:2–7. doi: 10.1016/j.ijfoodmicro.2013.03.013. [DOI] [PubMed] [Google Scholar]
- Aleksensko A, Clutterbuck AJ. Autonomous plasmid replication in Aspergillus nidulans: AMA1 and MATE elements. Fungal Genet Biol. 1997;21:373–387. doi: 10.1006/fgbi.1997.0980. [DOI] [PubMed] [Google Scholar]
- Allen TH. Notes on Taka-diastase. JAMA. 1896;27:374–375. doi: 10.1001/jama.1896.02430850034001i. [DOI] [Google Scholar]
- Amanullah A, Chirstensen LH, Hansen K, et al. Dependence of morphology on agitation intensity in fed-batch cultures of Aspergillus oryzae and its implications for recombinant protein production. Biotechnol Bioeng. 2002;77:815. doi: 10.1002/bit.10181. [DOI] [PubMed] [Google Scholar]
- Aviram N, Schuldiner M. Targeting and translocation of proteins to the endoplasmic reticulum at a glance. J Cell Sci. 2017;130:4079–4085. doi: 10.1242/jcs.204396. [DOI] [PubMed] [Google Scholar]
- Bao J, Huang M, Petranovic D, et al. Moderate expression of SEC16 increases protein secretion by Saccharomyces cerevisiae. Appl Environ Microbiol. 2017;83:e03400–e03416. doi: 10.1128/AEM.03400-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berka RM, Grigoriev IV, Otillar R, et al. Comparative genomic analysis of the thermophilic biomass-degrading fungi Myceliophthora thermophila and Thielavia terrestris. Nat Biotechnol. 2011;29:922–927. doi: 10.1038/nbt.1976. [DOI] [PubMed] [Google Scholar]
- Brandl J, Andersen MR. Aspergilli: models for systems biology in filamentous fungi. Curr Opin Syst Biol. 2018;6:67–73. doi: 10.1016/j.coisb.2017.09.005. [DOI] [Google Scholar]
- Cairns TC, Nai C, Meyer V. How a fungus shapes biotechnology: 100 years of Aspergillus niger research. Fungal Biol Biotechnol. 2018;5:13. doi: 10.1186/s40694-018-0054-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell K, Xia J, Nielsen J. The impact of systems biology on bioprocessing. Trends Biotechnol. 2017;35:1156–1168. doi: 10.1016/j.tibtech.2017.08.011. [DOI] [PubMed] [Google Scholar]
- Chapman J, Ismail AE, Dinu CZ. Industrial applications of enzymes: recent advances, techniques and outlooks. Catalysis. 2018;8:230. doi: 10.3390/catal8060238. [DOI] [Google Scholar]
- Cherry B, Bashkirova E, Lopez de Leon A, et al. Analysis of an Aspergillus niger glucoamylase strain pedigree using comparative genome hybridization and real-time quantitative polymerase chain reaction. Ind Biotechnol. 2010;5:237–244. doi: 10.1089/ind.2009.5.237. [DOI] [Google Scholar]
- Christensen T, Woeldike H, Boel E, et al. High level expression of recombinant genes in Aspergillus oryzae. Nat Biotechnol. 1988;6:1419–1422. doi: 10.1038/nbt1288-1419. [DOI] [Google Scholar]
- Christensen BE, Mollgaard H, Kaasgaard S et al. (1999) Methods for producing polypeptides in Aspergillus mutant cells. US6383781
- Davy AM, Kildegaard HF, Andersen MR. Cell factory engineering. Cell Syst. 2017;4:262–275. doi: 10.1016/j.cels.2017.02.010. [DOI] [PubMed] [Google Scholar]
- Deng H, Gao R, Liao X, et al. CRISPR system in filamentous fungi: current achievements and future directions. Gene. 2017;627:212–221. doi: 10.1016/j.gene.2017.06.019. [DOI] [PubMed] [Google Scholar]
- EFSA Guidance on the characterisation of microorganisms used as feed additives or as production organisms. EFSA J. 2018;16(3):e05206. doi: 10.2903/j.efsa.2018.5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira R, David F, Nielsen J. Advancing biotechnology with CRISPR/Cas9: recent applications and patent landscape. J Ind Microbiol Biotechnol. 2018;45:467–480. doi: 10.1007/s10295-017-2000-6. [DOI] [PubMed] [Google Scholar]
- Ferreira-Dias S, Sandoval G, Plou F et al (2013) The potential use of lipases in the production of fatty acid derivatives for the food and nutraceutical industries. Electron J Biotechnol 16. 10.2225/vol16-issue3-fulltext-5
- Fleissner A, Dersch P. Expression and export: recombinant protein production systems for Aspergillus. Appl Microbiol Biotechnol. 2010;87:1255–1270. doi: 10.1007/s00253-010-2672-6. [DOI] [PubMed] [Google Scholar]
- Frisvad JC, Møller LLH, Larsen TO, et al. Safety of the fungal workhorses of industrial biotechnology: update on the mycotoxin and secondary metabolite potential of Aspergillus niger, Aspergillus oryzae, and Trichoderma reesei. Appl Microbiol Biotechnol. 2018;102:9481–9515. doi: 10.1007/s00253-018-9354-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geib E, Brock M. ATNT: an enhanced system for expression of polycistronic secondary metabolite gene clusters in Aspergillus niger. Fungal Biol Biotechnol. 2017;4:13. doi: 10.1186/s40694-017-0042-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill-Serna J, Garcia-Diaz M, Gonzalez-Jaen MT, et al. Description of an orthologous cluster of ochratoxin a biosynthetic genes in Aspergillus and Penicillium species. A comparative analysis. Int J Food Microbiol. 2018;268:35–43. doi: 10.1016/j.ijfoodmicro.2017.12.028. [DOI] [PubMed] [Google Scholar]
- Gressler M, Hortschansky P, Geib E, et al. A new high-performance heterologous fungal expression system based on regulatory elements from the Aspergillus terreus terrein gene cluster. Front Microbiol. 2015;6:184. doi: 10.3389/fmicb.2015.00184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer SK, Avalos JL. Harnessing yeast organelles for metabolic engineering. Nat Chem Biol. 2017;13:823–832. doi: 10.1038/nchembio.2429. [DOI] [PubMed] [Google Scholar]
- Hanlon PR, Frestedt J, Magurany K. GRAS from the ground up: review of the interim pilot program for GRAS notification. Food Chem Toxicol. 2017;105:140–150. doi: 10.1016/j.fct.2017.03.064. [DOI] [PubMed] [Google Scholar]
- Held D, Luttrignger S, Doty T (2018) Laccase variants and polynucleotides encoding same. US0282708
- Hernandez-Gonzalez M, Pantazopoulou A, Spanoudakis D, et al. Genetic dissection of the secretory route followed by a fungal extracellular glycosyl hydrolase. Mol Microbiol. 2018;109:781–800. doi: 10.1111/mmi.14073. [DOI] [PubMed] [Google Scholar]
- Hjort CM, van den Hondel CM, Punt PJ et al. (2001) Fungal transcriptional activator useful in methods for producing polypeptides. WO2001068864
- Holm HC, Nielsen PM, Longin F. Upscaling of enzymatic processes. In: Bornscheuer U, editor. Lipid modification by enzymes and engineered microbes. London: Elsevier; 2018. pp. 343–373. [Google Scholar]
- Jakobson CM, Tullman-Ercek D, Mangan NM. Spatially organizing biochemistry: choosing a strategy to translate synthetic biology to the factory. Sci Rep. 2018;8:8196. doi: 10.1038/s41598-018-26399-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, et al. A programmable dual-RNA-guided DNA endonuclease in adaptative bacterial immunity. Science. 2012;337:816–822. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun H, Guangye H, Daiwen C. Insights into enzyme secretion by filamentous fungi: comparative proteome analysis of Trichoderma reesei grown on different carbon sources. J Proteome. 2013;89:191–201. doi: 10.1016/j.jprot.2013.06.014. [DOI] [PubMed] [Google Scholar]
- Katakura Y, Ametani A, Totsuka M, et al. Accelerated secretion of mutant β-lactoglobulin in Saccharomyces cerevisiae resulting from a single amino acid substitution. Biochim Biophys Acta. 1999;1432:302–312. doi: 10.1016/S0167-4838(99)00099-0. [DOI] [PubMed] [Google Scholar]
- Khaldi N, Wolfe KH. Evolutionary origins of the fumonisin secondary metabolite gene cluster in Fusarium verticillioides and Aspergillus niger. Int J Evol Biol. 2011;2011:7. doi: 10.4061/2011/423821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluge J, Terfehr D, Kück U. Inducible promoters and functional genomic approaches for the genetic engineering of filamentous fungi. Appl Microbiol Biotech. 2018;102:6357–6372. doi: 10.1007/s00253-018-9115-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Abe K, Gomi K, et al. Genomics of Aspergillus oryzae. Biosci Biotechnol Biochem. 2007;71:646–670. doi: 10.1271/bbb.60550. [DOI] [PubMed] [Google Scholar]
- Kwon MJ, Jørgensen TR, Nitsche BM, et al. The transcriptomic fingerprint of glucoamylase over-expression in Aspergillus niger. BMC Genomics. 2012;13:701. doi: 10.1186/1471-2164-13-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landowski CP, Huuskonen A, Wahl R, et al. Enabling low cost biopharmaceuticals: a systematic approach to delete proteases from a well-known protein production host Trichoderma reesei. PLoS One. 2015;10(8):e0134723. doi: 10.1371/journal.pone.0134723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laustsen MA, Nielsson S (1995) Selective inactivation of Aspergillus proteases. US6080564
- Li Q, Bai Z, O’Donnell A, et al. Oxidative stress in fungal fermentation processes: the roles of alternative respiration. Biotechnol Lett. 2011;33:457–467. doi: 10.1007/s10529-010-0471-x. [DOI] [PubMed] [Google Scholar]
- Liu Q, Gau R, Li J et al (2017) Development of a genome-editing CRISPR/Cas9 system in thermophilic fungal Myceliophthora species and its application to hypercellulase production strain engineering. Biotechnol Biofuels 10:1. 10.1186/s13068-016-0693-9 [DOI] [PMC free article] [PubMed]
- Liu W, Luo Z, Wang Y, et al. Rapid pathway prototyping and engineering using in vitro and in vivo synthetic genome SCRaMbLE-in methods. Nat Commun. 2018;8:1926. doi: 10.1038/s41467-018-04254-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubertozzi D, Keasling JD. Developing Aspergillus as a host for heterologous expression. Biotechnol Adv. 2009;27:53–75. doi: 10.1016/j.biotechadv.2008.09.001. [DOI] [PubMed] [Google Scholar]
- Machida M, Yamada O, Gomi K. Genomics of Aspergillus oryzae: learning from the history of Koji Mold and exploration of its future. DNA Res. 2008;15:173–183. doi: 10.1093/dnares/dsn020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnuson B, Munro I, Abbot P, et al. Review of the regulation and safety assessment of food substances in various countries and jurisdictions. Food Addit Contam. 2013;30:1147–1220. doi: 10.1080/19440049.2013.795293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcellin E, Nielsen LK. Advances in analytical tools for high throughput strain engineering. Curr Opin Biotechnol. 2018;54:33–40. doi: 10.1016/j.copbio.2018.01.027. [DOI] [PubMed] [Google Scholar]
- Martins-Santana L, Nora LC, Sanches-Medeiros A et al (2018) Systems and synthetic biology approaches to engineer fungi for fine chemical production. Front Bioeng Biotechnol 6. 10.3389/fbioe.2018.00117 [DOI] [PMC free article] [PubMed]
- Mattern IE, Unkles S, Kinghorn JR, et al. Transformation of Aspergillus oryzae using the A. niger pyrG gene. Mol Gen Genet. 1989;210:460–461. doi: 10.1007/BF00327197. [DOI] [PubMed] [Google Scholar]
- Meyer V, Fiedler M, Nitsche B, et al. The cell factory Aspergillus enters the big data era: opportunities and challenges for optimising product formation. Adv Biochem Eng Biotechnol. 2015;149:91–132. doi: 10.1007/10_2014_297. [DOI] [PubMed] [Google Scholar]
- Mori A, Hara S, Sugahara T, et al. Signal peptide optimization tool for the secretion of recombinant protein from Saccharomyces cerevisiae. J Biosci Bioeng. 2015;120:518–525. doi: 10.1016/j.jbiosc.2015.03.003. [DOI] [PubMed] [Google Scholar]
- Nakari-Setälä T, Paloheimo M, Kallio J, et al. Genetic modification of carbon catabolite repression in Trichoderma reesei for improved protein production. Appl Environ Microbiol. 2009;75:4583–4860. doi: 10.1128/AEM.00282-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevalainen H, Peterson R. Making recombinant proteins in filamentous fungi-are we expecting too much? Front Microbiol. 2014;5:75. doi: 10.3389/fmicb.2014.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen PH, Oxenbøll KM, Wenzel H. Cradle-to-gate environmental assessment of enzyme products produced industrially in Denmark by Novozymes A/S. Int J Life Cycle Assess. 2007;12(6):432. doi: 10.1065/lca2006.08.265.1. [DOI] [Google Scholar]
- Nødvig CS, Hoof JB, Kogle ME, et al. Efficient oligo nucleotide mediated CRISPR-Cas9 gene editing in Aspergilli. Fungal Genet Biol. 2018;115:78–89. doi: 10.1016/j.fgb.2018.01.004. [DOI] [PubMed] [Google Scholar]
- Obst U, Lu TK, Sieber V. A modular toolkit for generating Pichia pastoris secretion libraries. ACS Synth Biol. 2017;6:1016–1025. doi: 10.1021/acssynbio.6b00337. [DOI] [PubMed] [Google Scholar]
- Olempska-Beer ZS, Merker RI, Ditto MD, et al. Food-processing enzymes from recombinant microorganisms – a review. Regul Toxicol Pharmacol. 2006;45:144–158. doi: 10.1016/j.yrtph.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Owji H, Nezafat N, Negahdaripour M, et al. A comprehensive review of signal peptides: structure, roles and applications. Eur J Cell Biol. 2018;97:422–441. doi: 10.1016/j.ejcb.2018.06.003. [DOI] [PubMed] [Google Scholar]
- Pakula TM, Laxell M, Huuskonen A, et al. The effects of drugs inhibiting protein secretion in the filamentous fungus Trichoderma reesei. Evidence for down-regulation of genes that encode secreted proteins in the stressed cells. J Biol Chem. 2003;278:45011–45020. doi: 10.1074/jbc.M302372200. [DOI] [PubMed] [Google Scholar]
- Palermo M, Gökmen V, De Melauner B, et al. Acrylamide mitigation strategies: critical appraisal of the FoodDrinkEurope toolbox. Food Funct. 2016;7:2516–2525. doi: 10.1039/c5fo00655d. [DOI] [PubMed] [Google Scholar]
- Paloheimo M, Mäntylä A, Kallio J, et al. High-yield production of a bacterial xylanase in the filamentous fungus Trichoderma reesei requires a carrier polypeptide with an intact domain structure. Appl Environ Microbiol. 2003;69(12):7073–7082. doi: 10.1128/AEM.69.12.7073-7082.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paloheimo M, Mäkinen S, Punt P et al. (2018) Methods for controlling protease production. US20180037920
- Pariza MW, Johnson EA. Evaluating the safety of microbial enzyme preparations used in food processing: update for a new century. Regul Toxicol Pharmacol. 2001;33:173–186. doi: 10.1006/rtph.2001.1466. [DOI] [PubMed] [Google Scholar]
- Patel AK, Singhania RR, Pandey A. Novel enzymatic processes applied to the food industry. Curr Opin Food Sci. 2016;7:64–72. doi: 10.1016/j.cofs.2015.12.002. [DOI] [Google Scholar]
- Peberdy JF. Protein secretion in filamentous fungi- trying to understand a highly productive black box. Trends Biotechnol. 1994;12:50–57. doi: 10.1016/0167-7799(94)90100-7. [DOI] [PubMed] [Google Scholar]
- Pedreschi F, Mariotti MS, Granby K. Current issues in dietary acrylamide: formation, mitigation and risk assessment. J Sci Food Agric. 2014;94:9–20. doi: 10.1002/jsfa.6349. [DOI] [PubMed] [Google Scholar]
- Punt PJ, Schuren FHJ, Lehmbeck J, et al. Characterization of the Aspergillus niger prtT, a unique regulator of extracellular protease encoding genes. Fungal Genet Biol. 2008;45:1591–1599. doi: 10.1016/j.fgb.2008.09.007. [DOI] [PubMed] [Google Scholar]
- Rantasalo A, Landowski CP, Kuivanen J, et al. A universal gene expression system for fungi. Nucl Acid Res. 2018;46(18):e111. doi: 10.1093/nar/gky558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro V, Baiocchi C, Dal Bello F, et al. Formation of by-products during chemical interesterification of lipids. Detection and characterization of dialkyl ketones by non-aqueous reversed-phase liquid chromatography-high resolution mass spectrometry and gas chromatography-mass spectrometry. J Chromatogr. 2018;1581–1582:63–70. doi: 10.1016/j.chroma.2018.11.001. [DOI] [PubMed] [Google Scholar]
- Schmoll M, Dattenbock C, Carreras-Vilasenor N, et al. The genomes of three uneven siblings: footprints of the lifestyles of three Trichoderma species. Microbiol Mol Biol Rev. 2016;80:205–327. doi: 10.1128/MMBR.00040-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiboth B, Hakola S, Mach RL, et al. Role of four major cellulases in triggering of cellulase gene expression by cellulose in Trichoderma reesei. J Bacteriol. 1997;179:5318–5320. doi: 10.1128/jb.179.17.5318-5320.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JA, Akoh CC, Lee KT. Production and physicochemical properties of functional-butterfat through enzymatic interesterification in a continuous reactor. J Agric Food Chem. 2009;57:888–900. doi: 10.1021/jf802678a. [DOI] [PubMed] [Google Scholar]
- Singh A, Taylor LE, Wall TAV, et al. Heterologous protein expression in Hypocrea jecorina: a historical perspective and new developments. Biotechnol Adv. 2015;33:142–154. doi: 10.1016/j.biotechadv.2014.11.009. [DOI] [PubMed] [Google Scholar]
- Stappler E, Walton JD, Beier S, et al. Abundance of secreted proteins of Trichoderma reesei is regulated by light of different intensities. Front Microbiol. 2017;6:2586. doi: 10.3389/fmicb.2017.02586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone WE, Wright HE. Notes on taka-diastase. J Am Chem Soc. 1898;20:639–647. doi: 10.1021/ja02071a001. [DOI] [Google Scholar]
- Terfrüchte M, Wewetzer S, Sarkari P, et al. Tackling destructive proteolysis of unconventionally secreted heterologous proteins in Ustilago maydis. J Biotechnol. 2018;284:37–51. doi: 10.1016/j.jbiotec.2018.07.035. [DOI] [PubMed] [Google Scholar]
- Till P, Mach RL, Mach-Aigner AR. A current view on long noncoding RNAs in yeast and filamentous fungi. Appl Microbiol Biotechnol. 2018;102:7319–7331. doi: 10.1007/s00253-018-9187-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till P, Pucher M, Mach RL, et al. A long noncoding RNA promotes cellulase expression in Trichoderma reesei. Biotechnol Biofuels. 2018;11:78. doi: 10.1186/s13068-018-1081-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinci APJ. Myco-protein: a twenty-year overnight success. Mycol Res. 1992;96:1–13. doi: 10.1016/S0953-7562(09)80989-1. [DOI] [Google Scholar]
- Udagawa H (2017) Simultaneous site-specific integrations of multiple gene-copies. US9850501
- Ugidos-Rodriguez S, Matallana-Gonzalez MC, Sanchez-Mata MC. Lactose malabsorption and intolerance: a review. Food Funct. 2018;9:4056. doi: 10.1039/c8fo00555a. [DOI] [PubMed] [Google Scholar]
- Valkonen M, Penttilä M, Saloheimo M. Effects of inactivation and constitutive expression of the unfolded-protein response pathway on protein production in the yeast Saccharomyces cerevisiae. Appl Environ Microbiol. 2003;69:2065–2072. doi: 10.1128/AEM.69.4.2065-2072.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser H, Joosten V, Punt PJ, et al. Development of a mature fungal technology and production platform for industrial enzymes based on a Myceliophthora thermophila isolate, previously known as Chrysosporium lucknowense C1. Ind Biotechnol. 2011;7:214–223. doi: 10.1089/ind.2011.7.214. [DOI] [Google Scholar]
- Walisko R, Moench-Tegeder J, Blotenberg J, et al. The taming of the shrew – controlling the morphology of filamentous eukaryotic and prokaryotic microorganisms. Adv Biochem Eng Biotechnol. 2015;149:1–27. doi: 10.1007/10_2015_322. [DOI] [PubMed] [Google Scholar]
- Wang L, Zhao S, Chen XX, et al. Secretory overproduction of a raw starch-degrading glucoamylase in Penicillium oxalicum using strong promoter and signal peptide. Appl Microbiol Biotechnol. 2018;102:9291–9301. doi: 10.1007/s00253-018-9307-8. [DOI] [PubMed] [Google Scholar]
- Wanka F, Cairns T, Boecker S, et al. Tet-on, or tet-off, that is the question: advanced conditional gene expression in Aspergillus. Fungal Genet Biol. 2016;89:72–83. doi: 10.1016/j.fgb.2015.11.003. [DOI] [PubMed] [Google Scholar]
- Ward M, Wilson LJ, Kodama KH, et al. Improved production of chymosin in Aspergillus by expression as a glucoamylase-chymosin fusion. Nat Biotechnol. 1990;8:435–440. doi: 10.1038/nbt0590-435. [DOI] [PubMed] [Google Scholar]
- Xiao S, Shiloach J, Betenbaugh M. Engineering cells to improve protein expression. Curr Opin Struct Biol. 2014;26:32–38. doi: 10.1016/j.sbi.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder WT, Lehmbeck J. Heterologous expression and protein secretion in filamentous fungi. In: Tkacz JS, Lange L, editors. Advances in fungal biotechnology for industry, agriculture, and medicine. New York: Kluwer Academic/Plenum Publishers; 2004. [Google Scholar]
- Zhang J, Wu C, Wang W, et al. Construction of enhanced transcriptional activators for improving cellulase production in Trichoderma reesei RUT C30. Bioresour Bioprocess. 2018;5:40. doi: 10.1186/s40643-018-0226-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhivagi M, Ng AWT, Audin M, et al. Experimental and pan-cancer genome analyses reveal widespread contribution of acrylamide exposure to carcinogenesis in humans. Genome Res. 2019;29:1–11. doi: 10.1101/gr.242453.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubieta MP, Contestini FJ, Rubio MV, et al. Protein profile in Aspergillus nidulans recombinant strains overproducing heterologous enzymes. Microb Biotechnol. 2018;11:346–358. doi: 10.1111/1751-7915.13027. [DOI] [PMC free article] [PubMed] [Google Scholar]