Abstract
Introduction
To consider alternative mechanisms that give rise to a refluxing ureterovesical junction (UVJ), we hypothesized that children with a common heritable urinary tract defect, vesicoureteric reflux (VUR), may have a defect in the extracellular matrix composition of the UVJ and other tissues that would be revealed by assessment of the peripheral joints. Hypermobile joints can arise from defects in the extracellular matrix within the joint capsule that affect proteins, including tenascin XB (TNXB).
Methods
We performed an observational study of children with familial and non-familial VUR to determine the prevalence of joint hypermobility, renal scarring, and DNA sequence variants in TNXB.
Results
Most children (27/44) exhibited joint hypermobility using the Beighton scoring system. This included 15/26 girls (57.7%) and 12/18 boys (66.7%), which is a significantly higher prevalence for both sexes when compared to population controls (p<0.005). We found no association between joint hypermobility and renal scarring. Seven of 49 children harbored rare pathogenic sequence variants in TNXB, and two also exhibited joint hypermobility. No sequence variants in TNXB were identified in 25/27 children with VUR and joint hypermobility. Due to the observational design of the study, there was missing data for joint hypermobility scores in six children and for dimercaptosuccinic acid (DMSA) scans in 17 children.
Conclusions
We observed a high prevalence of VUR and joint hypermobility in children followed within a tertiary care pediatric urology clinic. While mutations in TNXB have been reported in families with VUR and joint hypermobility, we identified only two children with these phenotypes and pathogenic variants in TNXB. We, therefore, speculate that VUR and joint hypermobility may be due to mutations in other extracellular matrix genes.
Introduction
Vesicoureteric reflux (VUR) is a congenital defect of the ureterovesical junction (UVJ) that results in the retrograde flow of urine from the bladder towards the kidneys. The ability of the UVJ to occlude the ureteral orifice requires adequate musculature within the intravesical portion of the ureter and sufficient collagen fibers to maintain its tensile strength.1,2 VUR affects a variable number of children ranging from 1–25% of the population and may result in recurrent pyelonephritis that can produce scars with loss of function.3–5 Renal scarring associated with VUR continues to be a relatively common cause of chronic kidney disease in children.
To consider alternative mechanisms that give rise to a refluxing UVJ, we hypothesized that children with a common heritable urinary tract defect, VUR, may have a defect in the extracellular matrix composition of the UVJ and other tissues that would be revealed by assessment of the peripheral joints. Several papers have reported that children with primary VUR are at higher risk for generalized joint hypermobility or “double jointedness,” which raises the intriguing possibility that there are common defects in the extracellular matrix of the joint capsules and the UVJ that result in hypermobile joints and a refluxing UVJ.6,7 Indeed, we discovered two families with dominantly inherited VUR and joint hypermobility and found that these phenotypes mapped to a locus on chromosome 6. Using whole exome sequencing, we determined that each family harbored a heterozygous mutation in a gene that encodes an extracellular matrix glycoprotein known as tenascin XB (TNXB).8,9 Tenascin XB is a large glycoprotein within the extracellular matrix that interacts with the fibrillary collagens, the fibril-associated collagens, and the proteoglycan, decorin.10 The protein consists of the N-terminal oligomerization domain, the epidermal growth factor (EGF)-like repeats, the fibronectin (FN) type III repeats, and a fibrinogen (FBG)-like domain at the C-terminus.10 Autosomal recessive mutations in TNXB have been reported in individuals with classical-like Ehlers-Danlos syndrome that encompasses skin hyperextensibility, skin ecchymoses, and joint hypermobility, typically from large deletions or truncating mutations.11 We tested children with both familial and non-familial primary VUR for joint hypermobility and demonstrate that there is a high prevalence of joint hypermobility that surpasses the population prevalence. We did not find that children with VUR and joint hypermobility were more likely to have recurrent urinary tract infections (UTIs), nor did they exhibit more renal scars. Children with VUR were sequenced to determine if they had mutations in TNXB. While we did identify pathogenic variants in 7/49 children with VUR, only two of them exhibited joint hypermobility. This suggests that the association between VUR and joint hypermobility may be due to mutations in other genes that are relevant for the formation of the extracellular matrix in these tissues.
Methods
Clinical phenotyping and joint hypermobility testing
We previously collected DNA samples from a large cohort of 250 children with primary VUR from pediatric urology clinics.12 Within the cohort, there were familial cases and the majority of these were identified by sibling screening. The children were recruited at the time of diagnosis and had a mean age of 18 months at the time of enrollment. The Beighton scoring system assesses joint hypermobility in children and is validated for use in children greater than or equal to six years of age.13 The scoring system uses a nine-point scale to assess the fifth fingers, the thumbs, the elbows, the knees, and the spine for joint hypermobility. Joint hypermobility is defined by a score ≥6/9 as per the guidelines established by the International Consortium of Ehlers-Danlos Syndrome.14 For this study, we re-approached children ≥6 years of age who presented for routine followups with their treating urologist and 50 of the 250 children were eligible by age for joint hypermobility testing. Of the 50 children, 44 consented to joint hypermobility testing, 49 agreed to submit DNA samples for sequencing, and 33 underwent dimercaptosuccinic acid (DMSA) scans as part of routine clinical care (Table 1). The children and their parents were recruited in accordance with the research ethics board at the McGill University Health Centre (protocol #1389) after obtaining informed consent. All children were tested for joint hypermobility by one investigator (IRG), who was blinded to their VUR status. The grade of VUR was defined by the initial voiding cystourethrogram (VCUG). In cases of bilateral VUR, grade was defined by the highest grade reported on either side. Nationality was determined by asking the parents their country of origin. Within the subset of 50 children, 33 underwent DMSA scans as requested by the treating pediatric urologist as part of routine clinical care. DMSA scans were performed at least six months after a UTI in the presence of recurrent febrile UTIs, severe VUR (grade IV or V) and/or renal abnormalities on ultrasound. Renal scintigraphy was performed using either Tc99m-DMSA or Tc99m-glucoheptonate and the presence of renal scarring was defined using RIVUR guidelines.15 Interpretation of the renal scan was performed by an experienced nuclear medicine physician (ST), who was blinded to the patient’s Beighton score and TNXB sequence, but not to the presence of VUR. Forty-nine patients consented to DNA sequencing and submitted saliva samples that were used for DNA extraction (Oragene DNA).
Table 1.
Cohort of children with vesicoureteric reflux
| Patient ID | Population | Sex | Beighton score | UTI at VUR diagnosis | Recurrent UTI | VUR grade | Family history | Scarring |
|---|---|---|---|---|---|---|---|---|
| 1 | Other | F | 4 | No | No | 3 | VUR | NA |
| 2 | European (non-Finnish) | M | 6 | No | No | 3 | No | NA |
| 3 | South Asian | M | 2 | Yes | Yes | 5 | No | Yes |
| 4 | Other | F | 8 | Yes | Yes | 3 | HM | Yes |
| 5 | European (non-Finnish) | M | 5 | Yes | Yes | 2 | No | NA |
| 6 | Other | M | Not tested | Yes | Yes | 3 | No | Yes |
| 7 | Other | F | 4 | Yes | Yes | 5 | No | Yes |
| 8 | European (non-Finnish) | M | 2 | Yes | No | 2 | No | NA |
| 9 | Other | M | 6 | No | Yes | 3 | No | Yes |
| 10 | European (non-Finnish) | F | 6 | No | Yes | 4 | No | NA |
| 11 | European (non-Finnish) | F | 7 | No | No | 2 | VUR | NA |
| 12 | European (non-Finnish) | M | 5 | Yes | Yes | 5 | No | Yes |
| 13 | European (non-Finnish) | F | 9 | Yes | Yes | 4 | HM | Yes |
| 14 | European (non-Finnish) | F | 6 | No | Yes | 2 | HM | Yes |
| 15 | European (non-Finnish) | F | 8 | Yes | Yes | 4 | HM | Yes |
| 16 | European (non-Finnish) | F | 6 | Yes | Yes | 3 | No | Yes |
| 17 | European (non-Finnish) | F | 5 | Yes | Yes | 2 | VUR | Yes |
| 18 | European (non-Finnish) | F | 9 | Yes | No | 3 | VUR | NA |
| 19 | European (non-Finnish) | F | Not tested | Yes | No | 3 | No | NA |
| 20 | European (non-Finnish) | F | 8 | No | No | 2 | No | No |
| 21 | European (non-Finnish) | M | 3 | Yes | No | 4 | HM | Yes |
| 22 | European (non-Finnish) | F | 6 | Yes | Yes | 5 | VUR, HM | NA |
| 23 | European (non-Finnish) | M | 9 | Yes | Yes | 2 | No | NA |
| 24 | European (non-Finnish) | M | Not tested | Yes | Yes | 5 | No | Yes |
| 25 | South Asian | M | 7 | Yes | Yes | 4 | No | Yes |
| 26 | Other | F | Not tested | No | No | 5 | No | No |
| 27 | Other | M | 6 | No | No | 4 | No | Yes |
| 28 | European (non-Finnish) | M | 6 | Yes | No | 3 | No | Yes |
| 29 | European (non-Finnish) | F | 4 | No | No | 4 | No | NA |
| 30 | Other | M | 6 | Yes | Yes | 5 | No | Yes |
| 31 | European (non-Finnish) | F | 4 | No | Yes | 3 | No | NA |
| 32 | European (non-Finnish) | F | Not tested | Yes | No | 5 | VUR | Yes |
| 33 | European (non-Finnish) | F | 4 | Yes | Yes | 5 | HM | No |
| 34 | Other | M | 8 | Yes | Yes | 3 | No | Yes |
| 35 | European (non-Finnish) | F | 6 | No | Yes | 1 | No | NA |
| 36 | European (non-Finnish) | M | 6 | No | No | 5 | VUR | Yes |
| 37 | European (non-Finnish) | F | 8 | Yes | Yes | 3 | HM | No |
| 38 | European (non-Finnish) | F | 2 | Yes | Yes | 2 | No | Yes |
| 39 | European (non-Finnish) | F | 3 | Yes | Yes | 2 | No | NA |
| 40 | Other | M | Not tested | No | No | 4 | No | No |
| 41 | European (non-Finnish) | F | 4 | No | No | 3 | No | NA |
| 42 | European (non-Finnish) | M | 2 | No | No | 4 | VUR | Yes |
| 43 | European (non-Finnish) | F | 9 | Yes | No | 2 | HM | NA |
| 44 | European (non-Finnish) | M | 6 | No | No | 3 | No | NA |
| 45 | European (non-Finnish) | F | 9 | Yes | Yes | 2 | HM | Yes |
| 46 | South Asian | M | 7 | No | Yes | 3 | No | Yes |
| 47 | South Asian | F | 4 | Yes | Yes | 3 | No | Yes |
| 48 | European (non-Finnish) | F | 4 | Yes | Yes | 3 | No | Yes |
| 49 | European (non-Finnish) | M | 7 | No | Yes | 5 | No | Yes |
| 50 | European (non-Finnish) | F | 6 | Yes | Yes | 5 | HM | Yes |
F: female; HM: hypermobility; M: male: UTI: urinary tract infection; VUR: vesicoureteric reflux.
Sequencing studies of children with VUR
We used 41 primer pairs to sequence all exons,9,16 including a pair used for long-range polymerase chain reaction (PCR) to cover the duplicated region of TNXB (exons 32–44) in 49 children with VUR. Primer sequences are provided (Supplementary Table 1). Sequenced reads were mapped to the hg19 reference genome using the PHRAP algorithm of the PHRED/PHRAP/Consed software.17 Variants were determined using Atlas-SNP2 and the PHRED-base caller to detect single nucleotide variants. Variants were annotated using 1000 genome database,18 ExAC,19 dbSNP,20 RefSeq,21 HSF3.1,22 and dbNSFP.23 Variants not reported in these databases were considered novel. Variants with a minor allele frequency (MAF) of ≤1% when compared to the frequency in ethnically matched populations were considered as rare variants. The predicted effects of rare variants were determined using the PROVEAN,24 SIFT,25 and Polyphen226 algorithms. Clustal Omega alignment tool (The European Bioinformatics Institute, EMBL-EBI) was used for the phylogenetic analysis. Pathogenic coding variants were defined as such if two or more in silico prediction formulae reported they were deleterious.
Statistical analysis
The R-squared statistic was used to assess the relationship between the grade of VUR and the Beighton score. Chi-squared analysis was used to compare the proportion of children with VUR and joint hypermobility who had renal scars on DMSA scans vs. those without joint hypermobility and/or recurrent UTIs.
Results
Children with VUR have joint hypermobility
We previously recruited a cohort of children with primary VUR12 and 50 were eligible for joint hypermobility testing using the Beighton scoring system. Of the 50 children, 44 consented to joint hypermobility testing, 49 agreed to submit DNA samples for sequencing, and 33 underwent DMSA scans as part of routine clinical care (Table 1). Fifteen of the 26 girls that were tested, 57.7%, exhibited joint hypermobility, while 12 of the 18 boys that were tested, 66.7%, exhibited joint hypermobility (Tables 1, 2) that was not significantly different (χ2=0.3613; p=0.54). This observation is of interest because in most studies, girls are more hypermobile than boys,27–29 but in our cohort, boys were especially hypermobile. There was a significantly higher proportion of girls and boys with VUR and joint hypermobility when compared to age-matched girls (91/516, 17.6%) and boys (67/530, 12.6%) in the general population (Z=3.45; p=0.0005 for girls; Z=5.04; p<0.00001 for boys using population proportions).29 Two children had bladder diverticula in addition to VUR: one had confirmed joint hypermobility with a Beighton score of 9, while the other did not. The parents self-reported their nationality and the majority of the children (36/50) were defined as European, non-Finnish using the categories as listed in the ExAc Browser.19 Within the cohort, 31 children were diagnosed with VUR because they presented with a UTI (19 females, 12 males), and most of these children (24/31) went on to experience recurrent UTIs (Tables 1, 2). The remaining 19 children that did not present with a UTI were diagnosed because of an abnormal antenatal ultrasound that prompted further postnatal evaluation, including a VCUG. Children with VUR and joint hypermobility were not more likely to present with a UTI or to experience recurrent UTIs compared to children without joint hypermobility (χ2=0.017; p=0.89). Similarly, there was no relationship between the grade of VUR and the Beighton score (r2=0.02; p=0.3511).
Table 2.
Summary of clinical phenotypes of children with vesicoureteric reflux
| Sex | |
| Male | 21 |
| Female | 29 |
| HM | |
| HM | 27 (15F, 12M) |
| No HM | 17 (11F, 6M) |
| Not tested | 6 (3F, 3M) |
| UTI at diagnosis | |
| Yes | 31 (12F, 19M) |
| No | 19 (10F, 9M) |
| VUR grade | |
| 1 | 1 |
| 2 | 11 |
| 3 | 17 |
| 4 | 9 |
| 5 | 12 |
| Family history (no. of index cases) | |
| VUR | 7 |
| HM | 10 |
| VUR and HM | 1 |
| Scarring | |
| Renal scarring | 28 |
| No scarring | 5 |
| No DMSA | 17 |
DMSA: dimercaptosuccinic acid; F: female; HM: hypermobility; M: male: UTI: urinary tract infection; VUR: vesicoureteric reflux.
Twenty-eight of the 44 children who underwent joint hypermobility testing had a DMSA scan as part of routine clinical care. While a large proportion (16/28) had VUR, joint hypermobility, and renal scars on DMSA scan, this was not statistically significant when compared to 9/28 children with VUR and renal scars who did not exhibit joint hypermobility (χ2= 0.0083; p=0.92).
TNXB sequence variants in children with VUR and joint hypermobility
We previously reported an association between heterozygous sequence variants in TNXB and familial VUR with joint hypermobility.8,9 We performed Sanger sequencing for this gene in 49 children with familial (8/49) and non-familial (41/49) VUR. Of the familial cases of VUR, in only one was there a family history of joint hypermobility, and this child did not harbor a pathogenic variant in TNXB (Patient 22, Table 1). Overall, a total of 100 sequencing variants in TNXB were identified in 49 children. Of the 100 variants, 66 were exonic variants: 25 synonymous and 41 non-synonymous. Among the non-synonymous variants, nine were defined as rare. The remaining 34 variants were intronic and 10 were splice variants. Two of the 10 splice variants were identified as rare. The eleven variants, nine exonic and two intronic, were inherited as heterozygous alleles in children with non-familial VUR. The variants were analyzed using prediction software (Table 3). None of the patients sequenced had more than one variant. Five patients harbor pathogenic exonic variants based on deleterious scores from two or more prediction software: G878D, G1163E, P2217T, R3721W, and R4173Q (Table 3). All variants, with the exception of the R4173Q variant, affect one of the FN type III (FNIII) repeat domains (Fig. 1). Among the five children, three are female and two are male, and two have VUR of grade 4 or higher (Patients 12, 29). Four of the five children are not hypermobile: Patients 5, 12, 29, and 48. Only one individual, Patient 20, is hypermobile: she has low-grade VUR (grade 2) and harbors a novel variant that is predicted to be deleterious, R3721W. This variant affects exon 33 that encodes a FNIII repeat (Fig. 1). Interestingly, among the five children, she is the only one with renal scars in spite of a mild grade of VUR. Two rare intronic variants were identified that are predicted to affect a splice acceptor site. One male patient harbors a rare two base pair intronic variant that is predicted to affect a splice acceptor site (Patient 27). He has high-grade VUR, joint hypermobility, and renal scars. A second patient, Patient 47, harbors a rare intronic variant, but she is not hypermobile (Tables 1, 3).
Table 3.
Rare TNXB sequencing variants in children with vesicoureteric reflux and/or joint hypermobility
| Patient ID | Hypermobility status | Mutation | MAF | SIFT | Provean | Polyphen2 | Predicted effect |
|---|---|---|---|---|---|---|---|
| 3 | Not hypermobile | G3174R | 0.0031 | 0.059 | −4.12* | 0.033 | Neutral |
| 5 | Not hypermobile | R4171Q | Novel | 0.027* | −2.91* | 0.999* | Deleterious |
| 9 | Hypermobile | E2652K | 0.0011 | 0.206 | −1.98 | 0.926* | Neutral |
| 12 | Not hypermobile | G1163E | 0.0012 | 0.037* | −4.19* | 0.954* | Deleterious |
| 13 | Hypermobile | S1830R | 0.0009 | 0.071 | −1.66 | 0.039 | Neutral |
| 20 | Hypermobile | R3719W | Novel | 0.011* | −5.18* | 1.00* | Deleterious |
| 29 | Not hypermobile | G878D | 0.0017 | 0.151 | −3.07* | 1.00* | Deleterious |
| 46 | Hypermobile | G3161A | 0.0079 | 0.242 | −1.09 | 0.027 | Neutral |
| 48 | Not hypermobile | P2217T | 0.0023 | 0.004* | −2.78* | 0.204 | Deleterious |
| 27 | Hypermobile | Intronic (Chr6:32035762-3203576, CT>TG) | 0.004 | N/A | N/A | N/A | Possible effect on splicing |
| 47 | Not hypermobile | Intronic (Chr6:32015793, G>A) | 0.0018 | N/A | N/A | N/A | Possible effect on splicing |
Joint hypermobility status was determined using the Beighton scoring system. A score of ≥6/9 was defined as joint hypermobility. Exonic or coding variants were defined as pathogenic based on deleterious scores using two or more in silico prediction software and are indicated with a superscript asterisk symbol. SIFT scores range from 0–1.0, with 0 most damaging. Provean scores <−2.5 are deleterious and >−2.5 are considered neutral. Polyphen2 scores range from 0–1.0, with 1.0 most damaging. TNXB: tenascin XB.
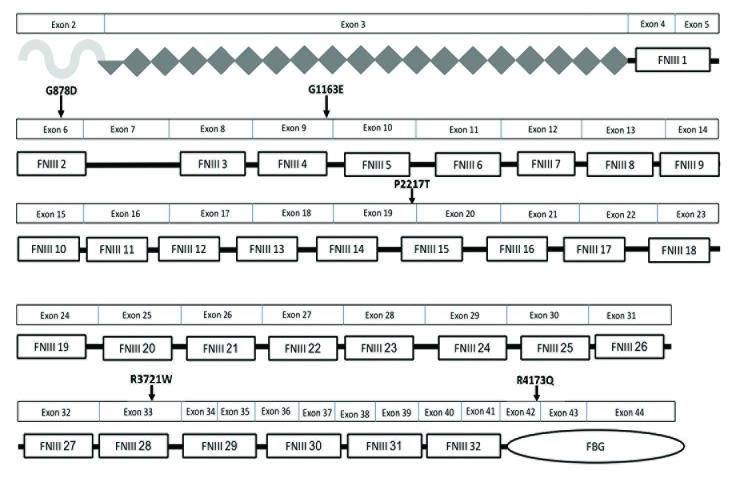
Fig. 1.
Location of pathogenic rare tenascin XB (TNXB) sequence variants within the TNXB protein domains. Five heterozygous TNXB missense variations were identified in the 49 children with vesicoureteric reflux who underwent sequencing and their corresponding amino acid changes and locations within the protein are shown. Most of the variants affect the fibronectin type III (FNIII) modules that are implicated in binding to decorin and in mediating extensibility of the TNX protein, which imparts elasticity to the tissue. One amino acid variant, R4173Q, affects the fibrinogen-like (FBG) domain, which directly binds to collagen fibrils and promotes the formation of collagen fibrils. The symbol under exon 2 depicts the N-terminal oligomerization domain. The diamonds depict the epidermal growth factor (EGF)-like repeats. The rectangular boxes depict the FNIII repeats, and the oval depicts the fibrinogen (FBG).
Discussion
While VUR remains a common pediatric disorder, the ability for clinicians to determine who will develop recurrent UTIs, who will develop renal scarring, and how these patients should be managed remains unclear. An additional challenge is the fact that VUR is both genetically30–34 and phenotypically heterogeneous. The purpose of this paper was to ascertain if VUR with joint hypermobility is a common phenotype and to determine if children with VUR and joint hypermobility might be at higher risk for recurrent UTIs and/or renal scars. We also sought to determine if children with VUR and joint hypermobility would be more likely to harbor coding DNA sequence variants in an extracellular matrix gene known as TNXB that has been reported in familial cases of VUR and joint hypermobility. We tested 44 children with VUR for joint hypermobility and determined that the majority exhibited joint hypermobility using the Beighton scoring system. We did not observe an association between VUR, joint hypermobility, and recurrent UTIs, nor did we observe an association between VUR, joint hypermobility, and renal scars, but this may be due to the sample sizes that were assessed. We identified rare heterozygous variants in TNXB in 7/49 children with VUR, of which five are predicted to be pathogenic exonic variants and two are predicted to affect a splice acceptor site. Among the seven children, two exhibit joint hypermobility and they both have renal scars. Importantly, 25/27 children who exhibited VUR and joint hypermobility did not have DNA sequence variants in TNXB, suggesting that either other genes implicated in the composition of the extracellular matrix or environmental factors might explain the co-occurrence of these phenotypes.
Our results support the findings of van Eerde et al,6 but demonstrate that an even higher percentage of children with VUR, 66.7 % of boys and 57.7 % of girls exhibit joint hypermobility using cutoffs of ≥6 for pre-pubertal children and ≥5 for pubertal adolescents, as reported by the International Consortium on the Ehlers-Danlos Syndromes.14 Our study did not include age and gender-matched controls without VUR, which ideally would have included children ≥6 years of age with a normal VCUG. Given the radiation exposure and the need for urethral catheterization, very few children undergo a VCUG unless there is a high suspicion of high-grade VUR. Therefore, we compared the prevalence of joint hypermobility in our VUR cohort to a population of Italian school-aged children with a mean age of 10.8 years in which the prevalence of joint hypermobility was reported to be 17.6% in girls and 12.6% in boys using the same cutoff criteria as our study.29 Other population estimates of joint hypermobility using the Beighton score and/or the Bulbena score report that 18–26% of girls and 12–18% of boys exhibit joint hypermobility.27,28,35,36 Taken together, it appears that joint hypermobility is more frequently observed in children with VUR.
Most of the children (31/50) reported at least one UTI, while 24/31 experienced recurrent UTIs. While we did not formally assess for bowel and bladder dysfunction, it is possible that in addition to VUR, these children had concurrent bowel and bladder dysfunction as a risk factor for UTI. This is of interest because de Kort et al have reported that children with generalized joint hypermobility are more likely to have lower urinary tract dysfunction, including chronic constipation, urinary incontinence, and UTIs.37 Therefore, we cannot exclude the possibility that children with VUR and joint hypermobility may also be at risk for bladder sphincter dysfunction, possibly due to a defect in the composition of the extracellular matrix within the bladder sphincter.
Fibrillary collagens, fibrillins, elastins, glycoproteins, and proteoglycans form the extracellular matrix within the joint capsule and the surrounding ligaments, as well as within the UVJ and bladder. Indeed, mutations in the genes that encode these proteins result in joint hypermobility in hereditary connective tissue disorders, including Cutis laxa and Ehlers-Danlos syndrome. VUR and bladder diverticula are reported in these disorders, although the penetrance of these urinary tract phenotypes has not been systematically examined.38 This suggests that a portion of children with VUR may have a global defect in the composition of the extracellular matrix that perturbs joint capsules and surrounding ligaments, as well as the UVJ and the integrity of the bladder wall.
To pursue the association of VUR with joint hypermobility, we performed candidate gene sequencing for TNXB, which is a large glycoprotein within the extracellular matrix that interacts with the fibrillary collagens, the fibril-associated collagens, and the proteoglycan, decorin.10 Autosomal recessive mutations in TNXB have been reported in individuals with classical-like Ehlers-Danlos syndrome that encompasses skin hyperextensibility, skin ecchymoses, and joint hypermobility, typically from large deletions or truncating mutations.11,16 While the genetic basis of autosomal-dominant joint hypermobility is defined as unknown in the most recent Ehlers-Danlos syndrome guidelines,14 we previously identified heterozygous mutations in TNXB in five families with VUR and joint hypermobility.8,9 Within the TNX protein, there are at least 32 FNIII repeats that are postulated to be important for facilitating the extensibility of the TNX protein itself, which imparts elasticity to the extracellular matrix.39 Most of the missense mutations identified to date in patients with VUR and joint hypermobility, including our study, localize to the FNIII repeats.8,9
In this current study, we sequenced 49 children for TNXB in which the majority (41) were sporadic cases of VUR, while the other eight cases had a positive family history of VUR. While none of the familial cases had pathogenic variants in TNXB, seven children with sporadic VUR did harbor pathogenic variants. One child with sporadic VUR and joint hypermobility harbored a novel variant in TNXB, R3721W, which is evolutionarily conserved and affects the FNIII repeat. Another child with VUR and joint hypermobility had a rare intronic variant that is predicted to affect a splice site. There were five other children with VUR and putative pathogenic variants in TNXB, but none of these children fulfilled criteria for joint hypermobility. Combining all of the TNXB sequencing studies,8,9 a total of 5/63 families (7.9%) exhibit a mutation in TNXB and two families have both VUR and joint hypermobility. Among the cases of sporadic or non-familial VUR, 9/96 (9.4%) harbor a mutation in TNXB, and of these, only three have both joint hypermobility and VUR. The TNXB protein is strongly expressed in the uroepithelium of the UVJ in children with VUR and without VUR.9 We speculate that heterozygous mutations in TNXB affect collagen deposition within the uroepithelial layer and may reduce the tensile strength of the UVJ and the joint capsule, predisposing to VUR and to joint hypermobility.
There are limitations to this study. It was an observational study of children seen in a tertiary care center in pediatric urology clinics. This factor explains the large number of severe cases of VUR within the cohort. We do not know if children with mild forms of VUR also exhibit a high prevalence of joint hypermobility. Another limitation of the study is the fact that the diagnosis of VUR is usually made at the age of two years, while the Beighton scoring system has been validated for use in children ≥6 years. Therefore, some children submitted DNA samples at the time of diagnosis but did not return for their followup appointment at the age of six years when joint hypermobility testing was performed. This accounts for the six children who did not undergo joint hypermobility testing. Another limitation is the fact that DMSA scans were requested as per clinical indications, so there was missing data on a total of 17/50 children. It would have been informative to perform DMSA scans on all of the children to ascertain if there is a relationship between VUR and renal scarring, but this would require a different study design in which we would need to convince parents to permit their children to undergo a DMSA scan even when not deemed to be clinically indicated.
Conclusions
In summary, children with VUR have a higher prevalence of joint hypermobility than age-matched population controls, suggesting there is a common mechanism that gives rise to both defects. From sequencing studies, it appears that mutations in TNXB do not explain most cases of VUR and joint hypermobility. This suggests that the association between VUR and joint hypermobility may be due to mutations in other genes that are relevant for the formation of the extracellular matrix in these tissues.
Supplementary Information
Supplementary Table 1.
Primers used for sequencing of tenascin XB
| Exon | Forward primer (5′–3′) | Reverse primer (5′–3′) |
|---|---|---|
| 3 | ATGCCACAGTCGTCACCA | AGAGCAGAGCTGGGCTACAT |
| 3 | GCAATCGGTTCCAGTGTACC | GGTCGTTGCGTGTGCTTT |
| 3 | GCAGTCTTCCCCTGAGTAGC | GAATGCATTTGCGACACG |
| 3 | AGGCACACTCCTTGCACAC | GAGAACGGCGTGTGTGTTT |
| 3 | CCCTCTACACACACACACTGG | GGAAGGCTACGTGAGTGAGG |
| 3 | CATGCTCTCCCTCCACTCTT | GTGCAAGGAGTGTGCCTGT |
| 4 | GCCATCTGGACTCAACCAAT | CTGAGTAAAAGGGGCTGTGG |
| 5 | GGCAGATTCCCTCTCTAGTCC | GAGATAAGGGGGATTGAGCA |
| 6 | CCAGAAGCATTCAGAGGAGTC | TGGACTAGAGAGGGAATCTGC |
| 7 | CCAATAACCCCAGCTCCTC | GGACTGGGGATTCCTTTCTAGT |
| 8 | CCCAAAGCACTGAGAAAACC | ATCCAGGATGGAGTGAGGTG |
| 9 | CTGACACAGCCAGGGTATGA | CCTATGTGGGATTTGGCTTC |
| 10 | GGCAAAATGAGCTGAGAAGG | TGTCAGGCTTCCCAGAAGTT |
| 11 | CTGGAGCAAGGAGAGCAACT | TTTCCATGGCTGTCATCTGT |
| 12 | GGAGGAGTAAAGGGGTCAGG | GGTGACAGCGAGACTCCATC |
| 13 | CAGGTGGACAAAGGGAAGAC | CCCCATCTCAGTTCACAGC |
| 14 | CTGGGGCCAAATAATGGTAA | GCAGTTCTGGGTTTTTCCAG |
| 15 | AAAGGGGCACAAGGAAACTT | CCCAGTCTTCCAGAAACAGC |
| 16 | TTCTGAAGGCTTCTCCTCCTC | TTTCGATTGCTGACTGCTTG |
| 17 | ACCAAAGAGCAAGAGGGTGA | CTTTCAGATGGCTGGGAGAG |
| 18 | AGGAGATGCTGGAGGCTGTA | CCAGTCATAGCCTTGGCTTC |
| 19 | AGTGAAGGCACCAGCAGAA | CCTCAACACCTCCTTGCAG |
| 20 | ACCAAAGAGCAAGAGGGTGA | GCACCAGCATCCAGACTGT |
| 21 | GGTACCCATGAGGGAAAGGT | CCACGACGTAAGCACATCC |
| 22 | ACTGTGAGCCCCATCAAGAC | AGCAAAGCAAGTTGCCCTTA |
| 23 | ACCAAAGAGCAAGAGGGTGA | GGGCACTTTGTGTTTTGTGA |
| 24 | CATGGAAACGTGCAAAAGAA | CTTGAAGACCTGAGCACATCC |
| 25 | GTCAGTCCTCAGGGAAGTGG | AACAAAAGATGGCGAGGAGA |
| 26 | CGAAGACTGGAGAGACAGCA | CCTTCCTCACAAGACCCAAG |
| 27 | CCTGTTCTTGGGCACTTTGT | CCTCTGCAGTGGAGAAGGAG |
| 28 | AAGAGGTGCCAAGATCCAAA | CCAGTCATAGCCTTGGCTTC |
| 29 | ATCAGTGGGTGCTGAGGACT | GCCGCTAAGAAATGCTCACT |
| 30 | GAGGGACTCACTTTCGGAGTT | ATAGCAGCCCAGGAAGCTC |
| 31 | TTGTCTTCAGCCCAAATGC | CTCGATCACAGCAGGGAAG |
| 32 (start of duplicated region) | GGCCAAGCCTGGAAGATAAA | GATTGGAGACAGAAGCACAC |
| 33 | CCAGGGAGAGAGGATGGAT | GTCCCCAGGAATGGAAGT |
| 34, 35 | GACCTAGTGCCTCAGCCA | GGCTCTCTCTACTCCGTG |
| 36, 37, 38 | ATGGGTGGGAGTTGAGAG | TGGAAGCTGAGCAGGTAG |
| 38, 39, 40 | TCTCCTCTTCCTGCTTTCCC | CCCCATCAGTCTCCATGTC |
| 40, 41, 42, 43 | CAGGACCAGCACCATCTT | TTGAGGTTGGCGTAGTGG |
| 43, 44 | GCTGTCTCCTACCGAGGG | GCAGAGAAGGCTTCCTCC |
| Long-range primers for duplicated region | GTCTCTGCCCTGGGAATGA | TGTAAACACAGTGCTGCGA |
Acknowledgements
The authors would like to thank the children and the families that participated in this study.
Footnotes
Competing interests: The authors report no competing personal or financial interests related to this work.
Funding: This study was supported by the Canadian Institute of Health Research operating grant and Fondation des Etoiles grant to Dr. Gupta.
This paper has been peer-reviewed
References
- 1.Lee BR, Silver RI, Partin AW, et al. A quantitative histologic analysis of collagen subtypes: The primary obstructed and refluxing megaureter of childhood. Urology. 1998;51:820–3. doi: 10.1016/S0090-4295(98)00013-2. [DOI] [PubMed] [Google Scholar]
- 2.Arena S, Fazzari C, Arena F, et al. Altered “active” anti-reflux mechanism in primary vesico-ureteric reflux: A morphological and manometric study. BJU Int. 2007;100:407–12. doi: 10.1111/j.1464-410X.2007.06921.x. [DOI] [PubMed] [Google Scholar]
- 3.Tullus K. Vesicoureteric reflux in children. Lancet. 2015;385:371–9. doi: 10.1016/S0140-6736(14)60383-4. [DOI] [PubMed] [Google Scholar]
- 4.Mattoo TK, Chesney RW, Greenfield SP, et al. Renal scarring in the randomized intervention for children with vesicoureteral rflux (RIVUR) trial. Clin J Am Soc Nephrol. 2016;11:54–61. doi: 10.2215/CJN.05210515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.The RIVUR Trial Investigators. Antimicrobial prophylaxis for children with vesicoureteral reflux. N Engl J Med. 2014;370:2367–76. doi: 10.1056/NEJMoa1401811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Eerde AM, Verhoeven VJ, de Jong TP, et al. Is joint hypermobility associated with vesicoureteral reflux? An assessment of 50 patients. BJU Int. 2012;109:1243–8. doi: 10.1111/j.1464-410X.2011.10469.x. [DOI] [PubMed] [Google Scholar]
- 7.Beiraghdar F, Rostami Z, Panahi Y, et al. Vesicourethral reflux in pediatrics with hypermobility syndrome. Nephrourol Mon. 2013;5:924–7. doi: 10.5812/numonthly.10770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elahi S, Homstad A, Vaidya H, et al. Rare variants in tenascin genes in a cohort of children with primary vesicoureteric reflux. Pediatr Nephrol. 2016;31:247–53. doi: 10.1007/s00467-015-3203-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gbadegesin RA, Brophy PD, Adeyemo A, et al. TNXB mutations can cause vesicoureteral reflux. J Am Soc Nephrol. 2013;24:1313–22. doi: 10.1681/ASN.2012121148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valcourt U, Alcaraz LB, Exposito JY, et al. Tenascin-X: Beyond the architectural function. Cell Adh Migr. 2015;9:154–65. doi: 10.4161/19336918.2014.994893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schalkwijk J, Zweers MC, Steijlen PM, et al. A recessive form of the Ehlers-Danlos syndrome caused by tenascin-X deficiency. N Engl J Med. 2001;345:1167–75. doi: 10.1056/NEJMoa002939. [DOI] [PubMed] [Google Scholar]
- 12.El Andalousi J, Murawski IJ, Capolicchio JP, et al. A single-center cohort of Canadian children with VUR reveals renal phenotypes important for genetic studies. Pediatr Nephrol. 2013;28:1813–9. doi: 10.1007/s00467-013-2440-9. [DOI] [PubMed] [Google Scholar]
- 13.Smits-Engelsman B, Klerks M, Kirby A. Beighton score: A valid measure for generalized hypermobility in children. J Pediatr. 2011;158:119–23. 123E1–4. doi: 10.1016/j.jpeds.2010.07.021. [DOI] [PubMed] [Google Scholar]
- 14.Malfait F, Francomano C, Byers P, et al. The 2017 international classification of the Ehlers-Danlos syndromes. Am J Med Genet C Semin Med Genet. 2017;175:8–26. doi: 10.1002/ajmg.c.31552. [DOI] [PubMed] [Google Scholar]
- 15.Ziessman HA, Majd M. Importance of methodology on (99m) technetium dmercapto-succinic acid scintigraphic image quality: Imaging pilot study for RIVUR (Randomized Intervention for Children with Vesicoureteral Reflux) multicenter investigation. J Urol. 2009;182:272–9. doi: 10.1016/j.juro.2009.02.144. [DOI] [PubMed] [Google Scholar]
- 16.Demirdas S, Dulfer E, Robert L, et al. Recognizing the tenascin-X deficient type of Ehlers-Danlos syndrome: A cross-sectional study in 17 patients. Clin Genet. 2017;91:411–25. doi: 10.1111/cge.12853. [DOI] [PubMed] [Google Scholar]
- 17.Ewing B, Green P. Base-calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res. 1998;8:186–94. doi: 10.1101/gr.8.3.186. [DOI] [PubMed] [Google Scholar]
- 18.Auton A, Abecasis G, Altshuler D, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60 706 humans. Nature. 2016;536:285–91. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sherry ST, Ward MH, Kholodov M, et al. DbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–11. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Leary NA, Wright MW, Brister JR, et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44:D733–45. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Desmet FO, Hamroun D, Lalande M, et al. Human splicing finder: An online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:E67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jian X, Boerwinkle E. DbNSFP: A lightweight database of human non-synonymous SNPs and their functional predictions. Hum Mutat. 2011;32:894–9. doi: 10.1002/humu.21517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi Y, Sims GE, Murphy S, et al. Predicting the functional effect of amino acid substitutions and indels. PLoS One. 2012;7:E46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–81. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 26.Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using polyphen-2. Curr Protoc Hum Genet. 2013;Chapter 7:20. doi: 10.1002/0471142905.hg0720s76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rikken-Bultman DG, Wellink L, van Dongen PW. Hypermobility in two Dutch school populations. Eur J Obstet Gynecol Reprod Biol. 1997;73:189–92. doi: 10.1016/S0301-2115(97)02745-0. [DOI] [PubMed] [Google Scholar]
- 28.El-Garf AK, Mahmoud GA, Mahgoub EH. Hypermobility among Egyptian children: Prevalence and features. J Rheumatol. 1998;25:1003–5. [PubMed] [Google Scholar]
- 29.Leone V, Tornese G, Zerial M, et al. Joint hypermobility and its relationship to musculoskeletal pain in schoolchildren: A cross-sectional study. Arch Dis Child. 2009;94:627–32. doi: 10.1136/adc.2008.150839. [DOI] [PubMed] [Google Scholar]
- 30.Darlow JM, Darlay R, Dobson MG, et al. Genome-wide linkage and association study implicates the 10q26 region as a major genetic contributor to primary non-syndromic vesicoureteric reflux. Sci Rep. 2017;7:14595. doi: 10.1038/s41598-017-15062-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feather SA, Malcolm S, Woolf AS, et al. Primary, non-syndromic vesicoureteric reflux and its nephropathy is genetically heterogeneous, with a locus on chromosome 1. Am J Hum Genet. 2000;66:1420–5. doi: 10.1086/302864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Eerde AM, Koeleman BP, van de Kamp JM, et al. Linkage study of 14 candidate genes and loci in four large Dutch families with vesicoureteral reflux. Pediatr Nephrol. 2007;22:1129–33. doi: 10.1007/s00467-007-0492-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weng PL, Sanna-Cherchi S, Hensle T, et al. A recessive gene for primary vesicoureteral reflux maps to chromosome 12p11-Q13. J Am Soc Nephrol. 2009;20:1633–40. doi: 10.1681/ASN.2008111199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Briggs CE, Guo CY, Schoettler C, et al. A genome scan in affected sib-pairs with familial vesicoureteral reflux identifies a locus on chromosome 5. Eur J Hum Genet. 2010;18:245–50. doi: 10.1038/ejhg.2009.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morris SL, O’Sullivan PB, Murray KJ, et al. Hypermobility and musculoskeletal pain in adolescents. J Pediatr. 2017;181:213–21. doi: 10.1016/j.jpeds.2016.09.060. [DOI] [PubMed] [Google Scholar]
- 36.Clinch J, Deere K, Sayers A, et al. Epidemiology of generalized joint laxity (hypermobility) in fourteen-year-old children from the U.K.: A population-based evaluation. Arthritis Rheum. 2011;63:2819–27. doi: 10.1002/art.30435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Kort LM, Verhulst JA, Engelbert RH, et al. Lower urinary tract dysfunction in children with generalized hypermobility of joints. J Urol. 2003;170:1971–4. doi: 10.1097/01.ju.0000091643.35118.d3. [DOI] [PubMed] [Google Scholar]
- 38.Tokhmafshan F, Brophy PD, Gbadegesin RA, et al. Vesicoureteral reflux and the extracellular matrix connection. Pediatr Nephrol. 2017;32:565–76. doi: 10.1007/s00467-016-3386-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oberhauser AF, Marszalek PE, Erickson HP, et al. The molecular elasticity of the extracellular matrix protein tenascin. Nature. 1998;393:181–50. doi: 10.1038/30270. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1.
Primers used for sequencing of tenascin XB
| Exon | Forward primer (5′–3′) | Reverse primer (5′–3′) |
|---|---|---|
| 3 | ATGCCACAGTCGTCACCA | AGAGCAGAGCTGGGCTACAT |
| 3 | GCAATCGGTTCCAGTGTACC | GGTCGTTGCGTGTGCTTT |
| 3 | GCAGTCTTCCCCTGAGTAGC | GAATGCATTTGCGACACG |
| 3 | AGGCACACTCCTTGCACAC | GAGAACGGCGTGTGTGTTT |
| 3 | CCCTCTACACACACACACTGG | GGAAGGCTACGTGAGTGAGG |
| 3 | CATGCTCTCCCTCCACTCTT | GTGCAAGGAGTGTGCCTGT |
| 4 | GCCATCTGGACTCAACCAAT | CTGAGTAAAAGGGGCTGTGG |
| 5 | GGCAGATTCCCTCTCTAGTCC | GAGATAAGGGGGATTGAGCA |
| 6 | CCAGAAGCATTCAGAGGAGTC | TGGACTAGAGAGGGAATCTGC |
| 7 | CCAATAACCCCAGCTCCTC | GGACTGGGGATTCCTTTCTAGT |
| 8 | CCCAAAGCACTGAGAAAACC | ATCCAGGATGGAGTGAGGTG |
| 9 | CTGACACAGCCAGGGTATGA | CCTATGTGGGATTTGGCTTC |
| 10 | GGCAAAATGAGCTGAGAAGG | TGTCAGGCTTCCCAGAAGTT |
| 11 | CTGGAGCAAGGAGAGCAACT | TTTCCATGGCTGTCATCTGT |
| 12 | GGAGGAGTAAAGGGGTCAGG | GGTGACAGCGAGACTCCATC |
| 13 | CAGGTGGACAAAGGGAAGAC | CCCCATCTCAGTTCACAGC |
| 14 | CTGGGGCCAAATAATGGTAA | GCAGTTCTGGGTTTTTCCAG |
| 15 | AAAGGGGCACAAGGAAACTT | CCCAGTCTTCCAGAAACAGC |
| 16 | TTCTGAAGGCTTCTCCTCCTC | TTTCGATTGCTGACTGCTTG |
| 17 | ACCAAAGAGCAAGAGGGTGA | CTTTCAGATGGCTGGGAGAG |
| 18 | AGGAGATGCTGGAGGCTGTA | CCAGTCATAGCCTTGGCTTC |
| 19 | AGTGAAGGCACCAGCAGAA | CCTCAACACCTCCTTGCAG |
| 20 | ACCAAAGAGCAAGAGGGTGA | GCACCAGCATCCAGACTGT |
| 21 | GGTACCCATGAGGGAAAGGT | CCACGACGTAAGCACATCC |
| 22 | ACTGTGAGCCCCATCAAGAC | AGCAAAGCAAGTTGCCCTTA |
| 23 | ACCAAAGAGCAAGAGGGTGA | GGGCACTTTGTGTTTTGTGA |
| 24 | CATGGAAACGTGCAAAAGAA | CTTGAAGACCTGAGCACATCC |
| 25 | GTCAGTCCTCAGGGAAGTGG | AACAAAAGATGGCGAGGAGA |
| 26 | CGAAGACTGGAGAGACAGCA | CCTTCCTCACAAGACCCAAG |
| 27 | CCTGTTCTTGGGCACTTTGT | CCTCTGCAGTGGAGAAGGAG |
| 28 | AAGAGGTGCCAAGATCCAAA | CCAGTCATAGCCTTGGCTTC |
| 29 | ATCAGTGGGTGCTGAGGACT | GCCGCTAAGAAATGCTCACT |
| 30 | GAGGGACTCACTTTCGGAGTT | ATAGCAGCCCAGGAAGCTC |
| 31 | TTGTCTTCAGCCCAAATGC | CTCGATCACAGCAGGGAAG |
| 32 (start of duplicated region) | GGCCAAGCCTGGAAGATAAA | GATTGGAGACAGAAGCACAC |
| 33 | CCAGGGAGAGAGGATGGAT | GTCCCCAGGAATGGAAGT |
| 34, 35 | GACCTAGTGCCTCAGCCA | GGCTCTCTCTACTCCGTG |
| 36, 37, 38 | ATGGGTGGGAGTTGAGAG | TGGAAGCTGAGCAGGTAG |
| 38, 39, 40 | TCTCCTCTTCCTGCTTTCCC | CCCCATCAGTCTCCATGTC |
| 40, 41, 42, 43 | CAGGACCAGCACCATCTT | TTGAGGTTGGCGTAGTGG |
| 43, 44 | GCTGTCTCCTACCGAGGG | GCAGAGAAGGCTTCCTCC |
| Long-range primers for duplicated region | GTCTCTGCCCTGGGAATGA | TGTAAACACAGTGCTGCGA |