Abstract
Study objective
A key public health question is whether syndromic surveillance data provide early warning of infectious outbreaks. One cause for skepticism is that biological correlates of the administrative and clinical data used in these systems have not been rigorously assessed. This study measures the value of respiratory data currently used in syndromic surveillance systems to detect respiratory infections by comparing it against criterion standard viral testing within a pediatric population.
Methods
We conducted a longitudinal study with prospective validation in the emergency department (ED) of a tertiary care children’s hospital. Children aged 7 years or younger who presented with a respiratory syndrome or who were tested for respiratory syncytial virus (RSV), influenza virus, parainfluenza virus, adenovirus, or enterovirus between January 1993 and June 2004 were included. We assessed the predictive ability of the viral tests by fitting generalized linear models to respiratory syndrome counts.
Results
Of 582,635 patient visits, 89,432 (15.4%) were for respiratory syndromes, and of these, 7,206 (8.1%) patients were tested for the viruses of interest. RSV was significantly related to respiratory syndrome counts (adjusted rate ratio [RR] 1.33; 95% confidence interval [CI] 1.04 to 1.71). In multivariate models including all viruses tested, influenza virus was also a significant predictor of respiratory syndrome counts (RR 1.47; 95% CI 1.03 to 2.10). This model accounted for 81.6% of the observed variability in respiratory syndrome counts.
Conclusion
Respiratory syndromic surveillance data strongly correlate with virologic test results in a pediatric population, providing evidence of the biologic validity of such surveillance systems. Real-time outbreak detection systems relying on syndromic data may be an important adjunct to the current set of public health systems for the detection and surveillance of respiratory infections.
Introduction
Background
In recent years, syndromic surveillance has shown promise for identifying naturally occurring epidemics.1, 2, 3, 4 Nonetheless, uncertainty persists as to the meaningfulness of this methodology and the proportion of true biological events that it can detect.5, 6, 7, 8
Importance
The source of data for many syndromic surveillance systems consists of information routinely collected by health care personnel during emergency department (ED) and office-based visits.5, 9, 10 These types of data lend themselves well to use in surveillance because their acquisition requires minimal additional resources and they are frequently recorded electronically, enabling automated transmission and rapid availability.3, 10 Studies of the validity of information collected by current surveillance systems have primarily used clinical and public health data for validation, even though these sources have not been shown to provide an acceptable standard.11, 12 In addition, the greatest utility of surveillance systems would be their ability to detect and monitor infectious outbreaks, a capability that specifically requires further examination. To date, no study has assessed the ability of a syndromic surveillance system to detect and monitor infectious disease activity by measuring virologic disease within the surveyed population.13
Editor’s Capsule Summary.
What is already known on this topic
Syndromic surveillance systems have the potential for providing early warning of infectious outbreaks by identifying unusual patterns of rapidly available data such as emergency department chief complaints or diagnoses.
What question this study addressed
The authors sought to determine whether chief complaint data suggestive of respiratory syndromes correlate with virologic test results on the same pediatric patient population.
What this study adds to our knowledge
By demonstrating that variation in the frequency of respiratory-related chief complaints correlates with variation in positive viral tests, the study adds biological validity to the use of rapidly available chief complaint data for surveillance and detection purposes.
How this might change clinical practice
This study will not affect practice on individual patients but may hasten the development of accurate methods for the early detection of infectious epidemics.
Goals of This Investigation
To measure the value of current syndromic surveillance systems for monitoring infectious respiratory disease, we compared surveillance data on respiratory illnesses derived from an ED population to the results of viral tests from the same population.
Materials and methods
Study Design, Setting, and Selection of Participants
We conducted a longitudinal study in the ED of an urban, tertiary care children’s hospital. This study was conducted in 2 parts. In the first, retrospective data were used to determine whether variations in the frequency of positive viral cultures performed during routine clinical care correlated with variations in the frequency of cases meeting syndromic surveillance criteria for respiratory illness. All children aged 7 years or younger, who presented to the ED during an 11-year period between January 1, 1993, and July 9, 2004, and who had a presenting complaint related to a respiratory illness or who had viral testing performed as part of their routine clinical care were included in this analysis. In the second part, a prospective validation study was performed to validate the use of the clinically collected viral cultures in the retrospective analysis. We chose to perform this validation because the viral tests collected during routine care were not from a systematically selected population known to have an infectious respiratory illness. Subjects in the prospective study consisted of patients presenting to the ED with a respiratory illness between December 7, 2003, and June 19, 2004, who met a specific definition for an infectious respiratory illness. Nasopharyngeal aspirates were collected from these patients and tested for respiratory viruses. By comparing the results of these tests to the historical time series, we were able to measure whether the historical time series was a representative sample of the viral distribution among children with infectious respiratory illnesses.
The protocol was approved by the hospital institutional review board. For all patients prospectively enrolled, we obtained written informed consent from a parent and assent from the child when age appropriate.
Methods of Measurement and Data Collection and Processing
Patient visits for respiratory syndromes were identified based on presenting complaints, which are routinely elicited from all patients or parents on presentation to the ED and recorded by triage nurses. This free-text description is subsequently numerically encoded, using a constrained list of 181 possible codes. Seventeen of these describe respiratory problems and were used to define a group of patients as having a respiratory illness, using a previously validated classification.12
Viral tests for respiratory syncytial virus (RSV), influenza A and B viruses, parainfluenza virus types 1 through 3, adenovirus, and enterovirus performed by the hospital during the study period were identified. These tests were ordered by physicians during routine clinical care for patients treated in the ED. We included tests that were ordered up to 7 hours before the ED visit and 24 hours after the ED visit to capture tests performed on patients during clinic visits before referral to the ED and on patients admitted to the hospital from the ED with a respiratory illness and tested after admission. Results for influenza A and B viruses were grouped, as were results for parainfluenza virus types 1 through 3. If a test was positive for more than 1 virus, all viruses identified were included in the analysis.
For the validation group, we screened patients aged 7 years and younger and presenting to the ED with a respiratory problem, such as difficulty breathing, cough, wheezing, and shortness of breath. Patients presenting with fever were also screened because this is frequently the complaint reported by parents of young children with respiratory illnesses. Patients were then further screened to determine whether they met our definition of an infectious respiratory illness and were recruited for the study if they did. We defined an infectious respiratory illness as the presence of at least 2 of the following symptoms: fever, cough, sneezing, sore throat, runny nose, or congestion. This criterion was chosen because we sought to determine the viral distribution among patients with respiratory infections and wanted to examine the ability of the surveillance system to track these infectious respiratory illnesses.
A total of 81.6% of patients screened met enrollment criteria and participated in the study. The majority of patients who did not meet criteria had fever without other respiratory symptoms. Patients were recruited during a 6-month period from December 7, 2003, through June 19, 2004, for comparison with the patients who had routinely collected specimens during this time. Because of the large number of ED patients treated for respiratory illnesses during the winter, there was only a small overlap between the patients composing the validation group and those who were tested as part of their routine clinical care. The analysis was performed with these patients included and excluded from the study populations. Nasopharyngeal aspirates were obtained from all patients in the validation group and tested for RSV, influenza virus, parainfluenza virus, and adenovirus. A small number of specimens were not tested for all of the study viruses, because of insufficient volume and a brief shortage of cells for viral cultures in December. Rarely, an RSV test was read as indeterminate and was considered a negative test result in the analysis.14
Routine clinical testing on patients included direct immunofluorescent antibody stains and viral culture. Direct immunofluorescent antibody assays used pooled antibodies for influenza virus, parainfluenza virus, RSV, and adenovirus (SimulFluor Respiratory Screen, Light Diagnostics, Chemicon International, Temecula, CA), or single virus-specific antibodies (Imagen Influenza Virus A and B, DakoCytomation, Carpinteria, CA; Bartels RSV DFA Kit, Trinity Biotech, Carlsbad, CA; ViraStat Parainfluenza Test, ZymeTx, Inc., Oklahoma City, OK; Adenovirus Antibody FITC Reagent, Light Diagnostics, Chemicon International). Viral cultures were performed using RMK, Hep-2, MRC-5, and Hel cells. Cultures with cytopathic effect or hemagglutination were stained with immunofluorescent antibodies for identification of influenza, parainfluenza virus, RSV, or adenovirus. Enterovirus was identified by viral passage and characteristic cytopathic effect.
Samples from the validation set were tested by culture with RMK cells for influenza virus, adenovirus, and parainfluenza virus and by direct immunofluorescent antibody for RSV. To increase the sensitivity for influenza virus, these specimens were also assayed by reverse transcriptase polymerase chain reaction. Ribonucleic acid was purified using the QIAamp Viral RNA Mini Kit (Qiagen, Valencia, CA) and amplified with the Access RT-PCR System (Promega, Madison, WI). Influenza A and B viruses were detected using previously published primers.15
Primary Data Analysis
Details of the data analysis are available in an online appendix (Appendix E1, available at http://www.annemergmed.com).16, 17, 18
Results
During the study period, there were a total of 582,635 visits to the ED. Presenting complaints were available for 99.6% of these visits, and 15.4% (89,432) were classified as having a respiratory syndrome.
Viral tests for RSV, influenza virus, parainfluenza virus, adenovirus, and enterovirus were performed, under routine care, for 7,206 ED visits. Our analysis included 7,046 tests for RSV, 3,671 tests for influenza virus, 3,494 tests for adenovirus, 7,035 tests for parainfluenza virus, and 3,322 tests for enterovirus. The overall rate of tests positive for at least 1 viral agent was 32.7%, with 24.8% positive for RSV, 5.3% positive for influenza virus, 3.4% positive for adenovirus, 3.3% positive for parainfluenza virus, and 2.6% positive for enterovirus. A total of 24 (0.3%) patient visits were associated with a test result positive for 2 viruses.
Three hundred sixty-one specimens were collected during the 6-month period of prospective recruitment. A total of 39.3% (142/361) of tests were positive for one of the viruses tested. The positivity rates for individual viruses were 21.1% (75/356) for RSV, 11.7% (41/351) for influenza virus, 7.2% (25/347) for parainfluenza virus, and 2.0% (7/347) for adenovirus. During the same period, 46.6% of the routine viral tests collected on patients presenting to the ED were positive, with 28.6% (165/576) positive for RSV, 16.2% (58/358) positive for influenza virus, 6.7% (38/570) positive for parainfluenza virus, and 8.6% (23/269) positive for adenovirus. There were 41 subjects who were recruited prospectively and subsequently also tested during routine clinical care.
A strong correlation was found between these 2 test groups, with a Pearson correlation coefficient of 0.914 (P<.0001) (Figure 1). A linear regression model also revealed a powerful association (χ2=60.64; P<.0001). Similar results were found excluding patients who were in both groups (Pearson correlation coefficient of 0.889, P<.0001; linear regression model χ2=24.31, P<.0001). This association strongly suggests that the results from viral tests obtained during routine clinical care are an appropriate representation of viral activity among children who are treated in the ED for infectious respiratory illnesses, supporting the use of the historical test results in the analysis.
Figure 1.
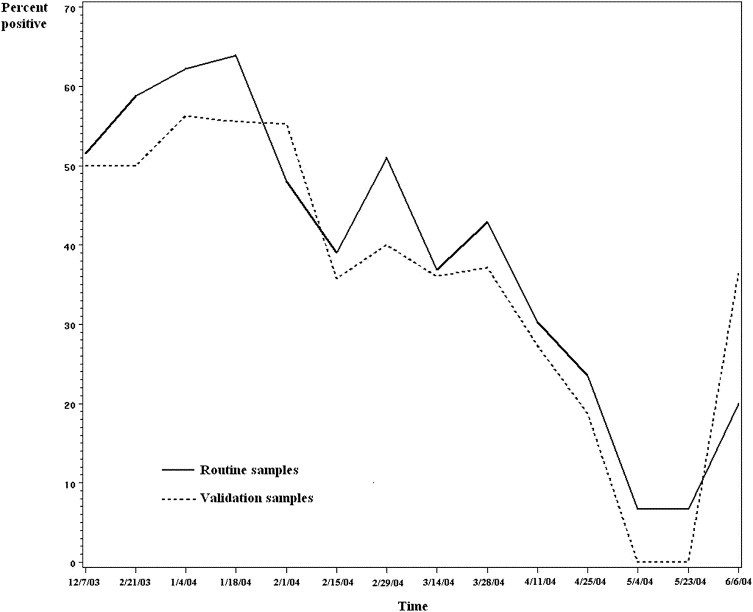
Positivity for routinely obtained viral tests (solid line) and validation sample (dotted line), December 2003 to June 2004. The proportion of positive viral test results for all children ≤7 years of age who were tested as part of their routine clinical care in the ED is compared to the results of tests collected prospectively from a validation group. A linear regression model revealed a significant association between the routine clinical tests and the validation sample (χ2=60.64; P<.0001).
Using the results from routine viral tests for the study period from 1993 to 2004, we measured the relationship between the prevalence of winter respiratory viruses and respiratory syndrome counts. The frequency of patients presenting with respiratory syndromes over time is compared to the viral test positivity rate for all viruses in Figure 2. Univariate Poisson regression showed varying association between respiratory illness counts and specific viral tests. The model for the rate of positive RSV tests revealed a significant positive relationship with respiratory illnesses (rate ratio [RR] 1.33; 95% confidence interval [CI] 1.04 to 1.71). Influenza virus (RR 1.15; 95% CI 0.83 to 1.60), parainfluenza virus (RR 1.03; 95% CI 0.66 to 1.57), adenovirus (RR 0.83; 95% CI 0.52 to 1.30), or enterovirus (RR 0.79; 95% CI 0.48 to 1.28) alone did not explain variation in respiratory syndrome frequencies.
Figure 2.
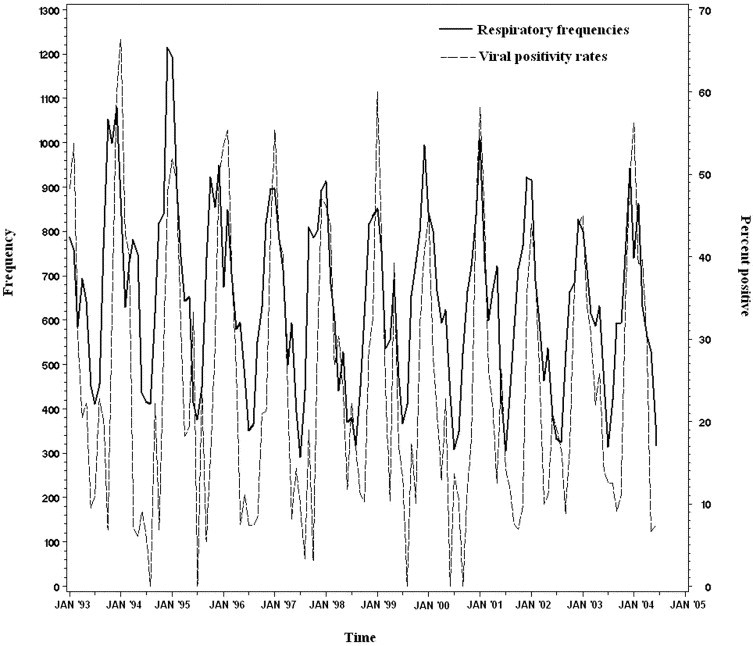
Respiratory syndrome frequencies (solid line) and viral positivity rates (dashed line), 1993 to 2004. The frequency of patients presenting with a respiratory illness syndrome is plotted against the viral test positivity rate for all respiratory viruses among patients tested in the ED.
However, when all viruses were included as covariates, influenza virus became a significant predictor (adjusted RR 1.47; 95% CI 1.03 to 2.10). Stepwise removal of the variables revealed that the effect of influenza virus depended on inclusion of RSV in the model. Thus, the effect of influenza is dominated by RSV and can only be uncovered when accounting for the variability of RSV in the model. The model with all covariates explains 81.6% of the deviance in the frequency of presentations for respiratory syndromes. We also examined different combinations of viral tests into a single predictor. Combining test results for RSV and influenza produced the best-fitting effect (adjusted RR 1.51; 95% CI 1.10 to 2.07). Figure 3 demonstrates the correlation of respiratory syndrome frequencies with RSV and influenza.
Figure 3.
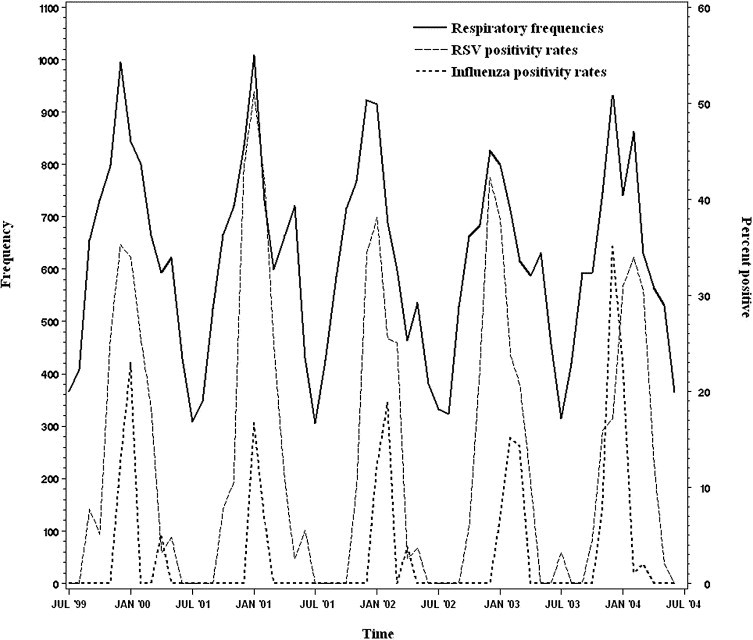
Respiratory syndrome frequencies (solid line) and rates of positive tests for RSV (dashed line) and influenza virus (dotted line), 1999 to 2004. The frequency of patients presenting with a respiratory illness syndrome is plotted against the positivity rate for RSV and influenza virus among patients tested in the ED.
The effect of RSV and influenza virus on respiratory illnesses is shown graphically for the respiratory virus season of 2003-04 in Figure 4. As in the previous figures, the test results shown are for the routinely collected specimens among ED patients. The earlier peak in respiratory presentations observed in December corresponds to a peak in influenza virus activity while the second peak in February represents RSV activity.
Figure 4.
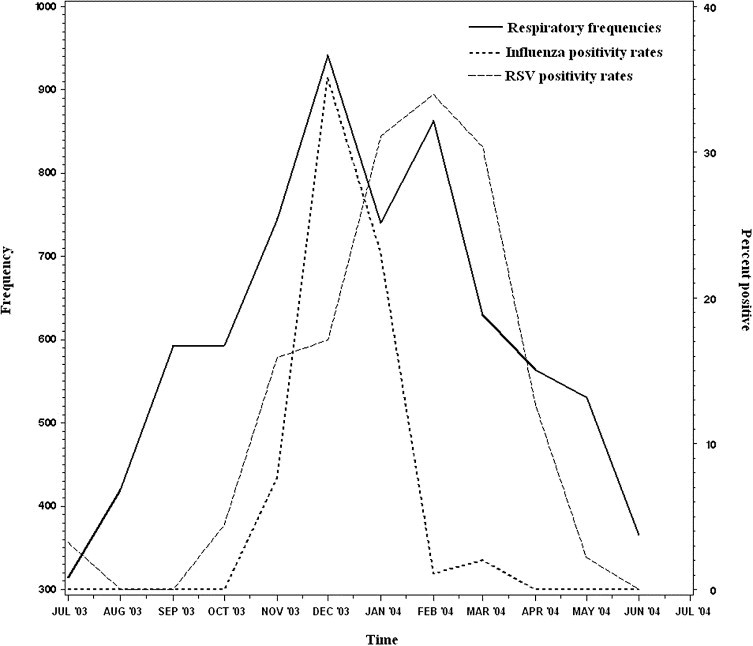
Respiratory syndrome frequencies (solid line) and rates of positive tests for RSV (dashed line) and influenza virus (dotted line), July 2003 to June 2004. The frequency of patients presenting with a respiratory illness syndrome is plotted against the positivity rate for RSV and influenza virus among patients tested in the ED.
Limitations
There are some potential limitations to this study. The first is that tests were not performed to identify several respiratory viruses that may have contributed to the observed burden of respiratory illness, including rhinovirus, coronavirus, and human metapneumovirus.19, 20 Although adding 1 or more of these viruses to our study might have changed the attributed variability, our findings of the influence of RSV and influenza virus are quite robust, and it is unlikely that their effect would be negated by the inclusion of the additional viruses in the model. Another potential concern is that biases in test ordering might make the routine viral test results unrepresentative of the patients who presented to the ED with respiratory syndromes. However, the results of prospective testing among ED patients with respiratory problems are closely correlated with the results obtained among the study population, strongly suggesting that the patients tested are in fact representative of the patients presenting with respiratory syndromes. Finally, our analytic methods may be limited by the presence of yearly seasonality in overall respiratory illness and viral prevalence. Although we have attempted to control for seasonal trends by including month as a categorical variable, it is difficult to know whether the correct adjustments were made for this effect.
Discussion
The variability over time in rates of positive laboratory tests for common respiratory viruses is largely predictive of the fluctuation in patients presenting with respiratory problems, suggesting that the seasonal trends in respiratory illnesses observed by syndromic surveillance systems are a direct result of respiratory virus activity. RSV and influenza virus appear to have the greatest impact on trends of respiratory illnesses observed in the clinical setting. This is best demonstrated during the most recent respiratory virus season of 2003 to 2004, during which influenza activity began unusually early in October, peaked in late November to December, and then rapidly declined in January and February.21 RSV followed a typical course with an increase in disease activity beginning in October and peaking in February before gradually declining.22 Laboratory tests at our institution showed peak influenza and RSV activity in December and February, respectively, and presenting complaint-based respiratory syndrome surveillance demonstrated a bifid peak corresponding to this viral activity.
Many of the current syndromic surveillance systems rely on data collected as part of the routine clinical workflow.5, 10 Examples of such data include symptoms recorded by telephone triage centers or nurse advice lines, patient complaints obtained on presentation to EDs, and diagnosis assigned by physicians at the end of ambulatory and emergency encounters.3, 10, 23, 24, 25, 26 The advantage of using these data is that they are readily available in a timely fashion, their collection does not require additional staff resources, and in many cases they are in electronic form.9 Several attempts have been made to measure the accuracy of these data sources, but none measured the relationship with laboratory-proven viral infections in the same population.
The emergence of West Nile virus, SARS, and avian influenza virus, among other infections, has demonstrated the need for effective surveillance systems. Rapid and accurate disease detection and monitoring are required to reduce the potential morbidity associated with such an outbreak. Our study validates the correlation between viral illness and respiratory syndromic data in a pediatric ED population. The presenting complaints of patients treated by physicians in EDs and office-based practices are likely to reflect the disease activity of winter respiratory viruses and could be a useful early indicator of viral activity for public health purposes. The information may also benefit physicians by alerting them to unusual patterns in illnesses commonly seen in communities and guiding diagnostic and therapeutic decisions.
Footnotes
Supervising editor: Jonathan M. Teich, MD, PhD
Author contributions: FTB and KDM conceived and designed the study and obtained research funding. FTB, AJM, and KDM supervised the conduct of the study and data collection. FTB and KLO were responsible for data acquisition and management. FTB, JSB, and KDM undertook interpretation and analysis of the data. The manuscript was drafted by FTB and KDM, and all authors contributed substantially to critical revision of the final document. FTB takes responsibility for the paper as a whole.
Funding and support: This work was supported by MO1 RR-02172 from the General Clinical Research Center at Children’s Hospital Boston, Contract No. 290-00-0020 from the Agency for Healthcare Research and Quality; by R01LM007970-01 from the National Library of Medicine, National Institutes of Health; and by National Research Service Awards 5 T32 HD40128-03 and 5 T32 HD40128-04.
Reprints not available from the authors.
Appendix E1
Primary Data Analysis
Our objective was to identify the extent to which respiratory syndromes are representative of respiratory virus activity, that is, the degree to which respiratory viruses determine the frequency of visits for respiratory syndromes. Thus, to assess the predictive ability of these patterns of viral illness, we fit generalized linear models to the respiratory presenting complaints.16 Because of frequent skewing in count data, a Poisson distribution was assumed instead of normality for modeling the respiratory count data. Overdispersion in the count data (extra variability not accounted for by Poisson regression) was also accounted for.17
Because of the relatively low counts in positive viral tests, aggregation of data to 2-week intervals was used to reduce random noise in the response variable while maintaining a reasonable sample size for time-series analysis. The aggregated data were fit against the proportion of positive viral samples. The proportions for each virus were included in the Poisson models as covariates separately and in aggregate. Confounding by seasonal trends in respiratory illness was considered by including months as indicator variables. Positive serial correlation in time-series data (lack of independence between observations over time) may result in underestimation of the standard errors of regression coefficients. Thus, to control for the effect of time, we also included the counts from the previous time point as a predictor in the model. The model framework is as follows:
where R i is the respiratory illness count for a 2-week time point i, V i is the proportion of positive samples for a given virus, M i are the dummy variables for month of year, and R i–1 is the respiratory illness count from the previous 2 weeks.
Overall model fit for each of the Poisson regression models was calculated using an alternative R 2 calculation because traditional linear regression statistics are inappropriate for Poisson regression models. This method calculates the proportion of the deviance explained by the model.18 Parameter estimates were transformed to rate ratios. A significant contribution of each effect to model fit was assessed by using 2-tailed χ2 tests and α<0.05 for rejecting the null hypothesis of no effect. All analyses were carried out using SAS statistical software (version 9.0, SAS Institute, Inc., Cary, NC).
Six months of viral test data from routinely collected specimens were validated against the prospectively collected data. Correlation was measured using the Pearson correlation coefficient on a pairwise comparison of percent viral test positivity over 2-week periods between the two datasets. To account for temporal autocorrelation, linear models were used to quantify the relationship between the 2 datasets where the historical time series was treated as the response variable. Seasonality was modeled through the inclusion of indicator variables for season. A normal distribution was assumed for the response variable after arcsine transformation of the proportional data to normality.
References
- 1.Reis B.Y., Pagano M., Mandl K.D. Using temporal context to improve biosurveillance. Proc Natl Acad Sci U S A. 2003;100:1961–1965. doi: 10.1073/pnas.0335026100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wagner M.M., Robinson J.M., Tsui F.C. Design of a national retail data monitor for public health surveillance. J Am Med Inform Assoc. 2003;10:409–418. doi: 10.1197/jamia.M1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lazarus R., Kleinman K.P., Dashevsky I. Using automated medical records for rapid identification of illness syndromes (syndromic surveillance): the example of lower respiratory infection. BMC Public Health. 2001;1:9. doi: 10.1186/1471-2458-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buehler J.W. Syndromic surveillance and bioterrorism-related epidemics. Emerg Infect Dis. 2003;9:1197–1204. doi: 10.3201/eid0910.030231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bravata D.M., McDonald K.M., Smith W.M. Systematic review: surveillance systems for early detection of bioterrorism-related diseases. Ann Intern Med. 2004;140:910–922. doi: 10.7326/0003-4819-140-11-200406010-00013. [DOI] [PubMed] [Google Scholar]
- 6.Fee E., Brown T.M. Preemptive biopreparedness: can we learn anything from history? Am J Public Health. 2001;91:721–726. doi: 10.2105/ajph.91.5.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stoto M., Matthias S., Mariano L. Syndromic surveillance: is it worth the effort? Chance. 2004;17:19–24. [Google Scholar]
- 8.Reingold A. If syndromic surveillance is the answer, what is the question? Biosecur Bioterror. 2003;1:77–81. doi: 10.1089/153871303766275745. [DOI] [PubMed] [Google Scholar]
- 9.Mandl K.D., Overhage J.M., Wagner M.M. Implementing syndromic surveillance: a practical guide informed by the early experience. J Am Med Inform Assoc. 2004;11:141–150. doi: 10.1197/jamia.M1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lober W.B., Karras B.T., Wagner M.M. Roundtable on bioterrorism detection: information system-based surveillance. J Am Med Inform Assoc. 2002;9:105–115. doi: 10.1197/jamia.M1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsui F.C., Wagner M.M., Dato V. Value of ICD-9 coded chief complaints for detection of epidemics. Proc AMIA Symp. 2001:711–715. [PMC free article] [PubMed] [Google Scholar]
- 12.Beitel A.J., Olson K.L., Reis B.Y. Use of emergency department chief complaint and diagnostic codes for identifying respiratory illness in a pediatric population. Pediatr Emerg Care. 2004;20:355–360. doi: 10.1097/01.pec.0000133608.96957.b9. [DOI] [PubMed] [Google Scholar]
- 13.Buehler J.W., Hopkins R.S., Overhage J.M. Framework for evaluating public health surveillance systems for early detection of outbreaks: recommendations from the CDC Working Group. MMWR Morb Mortal Wkly Rep. 2004;53:1–11. [PubMed] [Google Scholar]
- 14.Levine D.A., Platt S.L., Dayan P.S. Risk of serious bacterial infection in young febrile infants with respiratory syncytial virus infections. Pediatrics. 2004;113:1728–1734. doi: 10.1542/peds.113.6.1728. [DOI] [PubMed] [Google Scholar]
- 15.Wright K.E., Wilson G.A., Novosad D. Typing and subtyping of influenza viruses in clinical samples by PCR. J Clin Microbiol. 1995;33:1180–1184. doi: 10.1128/jcm.33.5.1180-1184.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nelder J.A., Wedderburn R.W. Generalized linear models. J R Stat Soc Series A. 1972;175:370–384. [Google Scholar]
- 17.McCullagh P., Nelder J.A. Generalized Linear Models. Chapman and Hall; London: 1989. [Google Scholar]
- 18.Williams D.A. Generalized linear model diagnostics using the deviance and single case deletions. Appl Stat. 1987;36:181–191. [Google Scholar]
- 19.Michelow I.C., Olsen K., Lozano J. Epidemiology and clinical characteristics of community-acquired pneumonia in hospitalized children. Pediatrics. 2004;113:701–707. doi: 10.1542/peds.113.4.701. [DOI] [PubMed] [Google Scholar]
- 20.Jartti T., Lehtinen P., Vuorinen T. Respiratory picornaviruses and respiratory syncytial virus as causative agents of acute expiratory wheezing in children. Emerg Infect Dis. 2004;10:1095–1101. doi: 10.3201/eid1006.030629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Update: Influenza activity: United States, 2003-2004 season. MMWR Morb Mortal Wkly Rep. 2004;53:284–287. [PubMed] [Google Scholar]
- 22.Centers for Disease Control and Prevention. Respiratory syncytial virus trends [Centers for Disease Control and Prevention Web site]. Available at: http://www.cdc.gov/ncidod/dvrd/revb/nrevss/rsvtre1.htm. Accessed January 27, 2005.
- 23.Lombardo J., Burkom H., Elbert E. A systems overview of the Electronic Surveillance System for the Early Notification of Community-Based Epidemics (ESSENCE II) J Urban Health. 2003;80:i32–i42. doi: 10.1007/PL00022313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Espino J.U., Hogan W.R., Wagner M.M. Telephone triage: a timely data source for surveillance of influenza-like diseases. Proc AMIA Symp. 2003:215–219. [PMC free article] [PubMed] [Google Scholar]
- 25.Rodman J.S., Frost F., Jakubowski W. Using nurse hot line calls for disease surveillance. Emerg Infect Dis. 1998;4:329–332. doi: 10.3201/eid0402.980226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harcourt S.E., Smith G.E., Hollyoak V. Can calls to NHS Direct be used for syndromic surveillance? Commun Dis Public Health. 2001;4:178–182. [PubMed] [Google Scholar]


