Abstract
Benchmarks and metrics related to laboratory test utilization are based on evidence-based medical literature that may suffer from a positive publication bias. Guidelines are only as good as the data reviewed to create them. Disruptive technologies require time for appropriate use to be established before utilization review will be meaningful. Metrics include monitoring the use of obsolete tests and the inappropriate use of lab tests. Test utilization by clients in a hospital outreach program can be used to monitor the impact of new clients on lab workload. A multi-disciplinary laboratory utilization committee is the most effective tool for modifying bad habits, and reviewing and approving new tests for the lab formulary or by sending them out to a reference lab.
Keywords: Test utilization, Obsolete tests, Inappropriate test, Disruptive technology, Outreach, Metrics
Highlights
-
•
Laboratory test overutilization is 2.9% to 56% of all lab tests ordered.
-
•
Guidelines may have a positive publication bias.
-
•
Choosing Wisely is 35 medical subspecialty groups targeting overused tests or therapies.
-
•
Utilization of disruptive technologies requires time for appropriate use to be established.
-
•
Client test utilization by speciality can help predict needs to meet future outreach growth.
1. Introduction
Laboratory test overutilization is estimated to represent 2.9% to 56% of all laboratory tests internationally. Efforts have been made to reduce the demand for or utilization of these over utilized tests [1], [2], [3], [4], [5], [6]. The most efficient outcomes have involved the formation of a laboratory utilization committee [2], [6] or a laboratory formulary committee [5] based on the hospital pharmacy and therapeutics committee’s organizational structure. This committee evaluates the clinical value of laboratory tests using an evidence-based review of the appropriate medical literature. This same literature is reviewed by numerous professional specialty medical organizations as well as healthcare insurance carriers to determine what tests or procedures should be performed and reimbursed. The conclusions based on these reviews need to be updated on a regular basis.
“The quality of guidelines is only as good as the published studies on which they are based” [7]. Often relevant studies evaluating laboratory tests demonstrate negative findings and are not published [7], [8]. This phenomenon is referred to as positive publication bias or publication bias. Tzoulaki et al. [8] demonstrated publication bias during a review of reports evaluating emerging cardiovascular biomarkers. Therefore, misinterpretation is a potential impact of failing to publish studies with negative results during a review of evidence-based literature. Readers beware.
Tests may be obsolete and should be retired from clinical use, while others may be inappropriately used for specific disease categories. The playing field is not level. There are at least six newer game-changing disruptive technologies being evaluated [9], [10], [11] which will result in modifications of clinical practice and laboratory testing modalities. These newer disruptive technologies may replace obsolete or inappropriate tests. Lab utilization benchmarks and metrics are under continuous flux as a consequence. In the case of evolving newer technology, it is imperative to explore their impact early in their development to anticipate and monitor their impact on laboratory testing.
2. Approach
The three authors have reviewed the current literature related to laboratory test utilization with an emphasis on where do the definitions of obsolete or inappropriate test utilization originate. We evaluated whole genome sequencing, next generation sequencing and proteomics as examples of high impact disruptive technologies that generate large quantities of data that need software to reduce to clinically useful results. Practical examples of obsolete and inappropriate tests are reviewed as potential metrics to monitor improvement in test utilization. Another useful metric is test utilization by clients in a hospital outreach program which can be used to monitor the impact of new clients on laboratory workload. Finally, the result of published data from the work of laboratory utilization committees is summarized.
3. Benchmarks and metrics for laboratory utilization
Benchmarks and metrics for laboratory utilization will be reviewed for three disruptive technologies as well as obsolete and inappropriately used tests.
3.1. Disruptive technologies
Medical practice as well as pathology is in the midst of the rapid development of at least six major game-changing disruptive technologies. They include genetics, proteomics, digital pathology, informatics, therapeutic pathology and in vivo diagnostics [9], [10], [11]. All six of these disruptive technologies share similar issues like resolution of best applications for routine clinical use, paucity of evidence-based outcome literature for review, education of practitioners and physician users of the clinical information generated and software to convert big databases the method generates into clinical useful information [9], [10], [11]. The utilization of these techniques will increase as these barriers or obstacles to clinical use are overcome.
3.1.1. Whole genome sequence
An example of a disruptive technology is next generation sequencing or massively parallel sequencing [12], [13], [14]. This technique is currently not cleared by the U.S. Food and Drug Administration [13]. It has been used to generate genome wide sequences and one of the authors (FLK) has had his genome sequenced at the CLIA approved laboratory at Illumina (San Diego, CA). The results revealed 3.23 million variants compared with the reference method and 20,426 of these variants were in the exome or in the coding elements.
The study interrogated 344 genes causally associated with 140 conditions as recommended by the American College of Medical Genetics. In that limited number of genes, 1,254 variants were detected and classified as clinically significant (0), carrier status (1), variants of unknown significance (255), likely benign variants (356) and benign variants (642). The definition of these variants calls and the failure of this technique to detect deletions, insertions, interspersed repeats and tandem repeats (repeats adjacent to each other like triplet repeats [15]) may lead to inappropriate interpretation of the results and expensive follow up clinical and laboratory evaluation. For example, a clinically significant pathogenic variant reported in at least 3 unrelated cases with control data may be found in additional genome studies in other populations [16] to be a benign variant that is also found with a new variant which contains the mutation that leads to the most significant deleterious effect on gene function. The software application for variant significance assignment, like DataGenno [17], will need to be up-to-date with the latest genotype/phenotype associations to prevent false positive findings and inappropriate follow-up testing.
3.1.2. Tumor genome sequence
In 2009 the highest rate of reported cancers was prostate, lung and bronchus and colon and rectum for men with female breast replacing prostate for women in the U.S. [18]. The annual incidence rate was 459 cases per 100,000 individuals. Comprehensive sequencing of numerous human cancers have revealed driver genes, 2 to 8 such genes per tumor, which alter intracellular signal transduction pathways related to the cells future death or survival and/or genome maintenance [19], [20]. There are at least 10 FDA approved cancer therapies based on the inhibition of these tumor-activated intracellular pathways [19]. For example, the BRAF kinase inhibitor, Vemurafenib, has shown a response rate in 50% of patients with metastatic melanoma that have the BRAF valine to glutamic acid mutation at codon 600 (V600E) [21]. This V600E mutation is associated with aggressive clinical course in patients with thyroid papillary microcarcinoma [22]. In one study of a hybrid score composed of one molecular diagnostic (V600E) and 3 histopathologic parameters were used to predict this tumor's clinical course with a sensitivity of 96% and specificity of 80% [22].
The selection of the correct molecular diagnostic tests for specific tumors is aided by published guidelines. Immunohistochemistry detection of estrogen and progesterone receptors in breast cancer from American Society of Clinical Oncology and College of American Pathologists [23] and selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors from the International Association for Study of Lung Cancer, Association for Molecular Pathology and College of American Pathologists [24].
Whole genome sequencing of a tumor will provide access to all known and unknown variants related to the tumor's survival skills [25]. The development of software [26] which will convert the patient's raw genome sequence into a medically relevant assessment of therapeutic targets and drug metabolism based on the tumor's body site will be very useful. From this genome analysis, the clinician wants to know what anticancer drug or drugs will this patient respond to as well as the dose.
3.1.3. MALDI-TOF
MALDI-TOF (Matrix Assisted Laser Desorption Ionization-Time of Flight) spectroscopy is a relatively new technology to the Clinical Microbiology laboratory. Pathogen identification has always relied on visual and biochemical interrogation where the summary of results may point to a specific identification (genus and species) or sometimes to at least the genus level. Visual and biochemical results can sometimes yield variable results meaning in some cases the ID may change depending on the result. The use of MALDI-TOF allows the clinical microbiology laboratory to identify bacteria once an isolate has been cultured potentially without performing any biochemical testing [11], [27], [28]. The implications are quicker pathogen identifications to clinicians and the potential to affect antibiotic treatment before susceptibility results are available. The ability to obtain a quicker answer will disrupt the testing workflow and require a re-evaluation of that workflow to optimize the use of MALDI-TOF and antibiotic susceptibility testing [11], [27], [28].
3.2. Professional subspecialty medical organizations
Benchmarks and subsequent metrics for monitoring laboratory test utilization have been developed by professional subspecialty medical organizations in the format of recommendations and guidelines [29]. Examples include guidelines for hypothyroidism in adults from the American Association of Clinical Endocrinologists and the American Thyroid Association [30], definition of myocardial infarction from the American College of Cardiology Foundation and American Heart Association [31], definition of diabetes mellitus from the American Diabetes Association [32], pharmacogenetics as well as follow-up testing for metabolic diseases identified by expanded newborn screening using tandem mass spectrometry from the National Academy of Clinical Biochemistry [33], [34], and use of bone metabolic markers from the Japan Osteoporosis Society [35].
Thirty-five of these specialty societies have joined the Choosing Wisely project organized by the American Board of Internal Medicine. Societies are asked to provide five specific, evidence-based recommendations on when tests and procedures may be appropriate or inappropriate for patient care (www.choosingwisely.org).
A review of the lists from 26 specialty societies revealed 135 recommendations. Laboratory tests were referenced in 25 items or 18.5% of the total. Only one organization, American Society of Clinical Pathology, had a list of 5 laboratory test-related recommendations [36]. Kale et al. [37] reviewed the national annual savings if outpatient visits to the primary care physicians did not include unnecessary or inappropriate laboratory tests including CBC ($32.7 million), urinalysis ($3.3 million) and basic metabolic panel ($10.1 million). Those three procedures yield an annual cost savings of $46.1 million compared to the elimination of inappropriate Pap tests at an annual savings of $47.8 million. These figures illustrate the magnitude of healthcare savings by implementing simple laboratory test ordering practices which reduce duplication and/or inappropriate testing. Collaboration by subspecialty medical societies in disruptive technology development and improvements in routine clinical laboratory test utilization will be a fertile area for the development of benchmarks and metrics for future laboratory test utilization.
3.3. Metrics: obsolete tests
The appropriateness of laboratory tests and the appropriate utilization of laboratory tests are always important for patient care, but require increased scrutiny in the era of containment of healthcare costs. Objective criteria for the judging of appropriateness of tests and their utilization have not been universally developed or applied, so it is not always easy to define these terms [38]. Insurance companies are recognizing the medical and the financial burden of unnecessary testing and are taking action. Many companies have posted information on their websites defining obsolete and unreliable lab tests which are readily accessible on the internet, including Aetna [39], United Healthcare [40] and AmeriHealth [41].
One criterion for judging the appropriateness of a test is to determine if it is obsolete. The definition of “obsolete” as noted in the Merriam-Webster on-line dictionary at http://www.merriam-webster.com/dictionary/ is “no longer in use or no longer useful”. Synonyms include: antiquated, archaic, dated, démodé, demoded, fossilized, and kaput. Over time, with advances in medical technology, laboratory tests become outdated. Although it is difficult to remove a test from a laboratory's formulary, there are good reasons to do so. Reasons include the availability of a more sensitive, specific, or accurate test or new guidelines recommend the elimination of a test with the replacement of another.
There are tests in Clinical Pathology that must be considered for obsolescence. These include T3 uptake and lactic acid dehydrogenase (LDH) isoenzymes in the clinical chemistry lab and bleeding time in hematology. Obsolete tests in the microbiology laboratory include bacterial antigen detection tests, Group B Streptococcus antigen (GBS) testing and HIV-1 Western blot.
3.3.1. T3 uptake and free thyroxine Index (FTI)
T3 Uptake and Free Thyroxine Index (FTI) are still ordered by physicians, despite the fact that alternative tests have been available for many years. T3 Uptake is an old test designed with a purpose of indirectly measuring free thyroxine (T4). It was developed before the availability of direct assays able to accurately measure free T4 levels [42]. Standardization of free T4 assays has been reported using the time-consuming equilibrium dialysis in combination with isotope dilution-liquid chromatography/tandem mass spectrometry [43]. T3 Uptake is an assessment of unsaturated (unbound to thyroxine) thyroid binding proteins in serum and is used with total T4 to calculate FTI. The FTI is obtained by multiplying the (Total T4) times (T3 Update). There is no longer a need to estimate free T4 when there are assays for the direct measurement of free T4 in every laboratory. Supporting evidence for the obsolescence of T3 Update and FTI has been available for decades [44]. Current guidelines for the diagnosis and management of hypothyroidism [30], hyperthyroidism [45], and thyroid disease in pregnancy [46], no longer include the assessment of T3 Update or FTI.
3.3.2. Lactic acid dehydrogenase (LDH) isoenzymes
The analysis of lactic acid dehydrogenase (LDH) Isoenzymes by electrophoresis has been utilized as an aid in the diagnosis of myocardial infarction. With the development of a more specific test for myocardial damage, troponin, there is little use for this insensitive and time-consuming electrophoretic assay. Current guidelines clearly establish that the preferred marker for cardiac injury is troponin [31].
3.3.3. Bleeding time
Bleeding time is a crude test of hemostasis (the arrest or stoppage of bleeding). It is an indication of how well platelets interact with blood vessel walls to form blood clots. Indirectly assesses platelet function. It is performed by making a small incision on the skin and measuring, in seconds, the time taken for bleeding to stop. The test was designed to assess platelet function or exclude von Willebrand Disease. This test is labor intensive, invasive, poorly reproducible, and insensitive [36]. Historically it was performed because screening tests with a higher sensitivity for platelet dysfunction and von Willebrand disease (vWD) were unavailable. Bleeding time has been replaced by instrumentation that can assess platelet function in whole blood by aggregation studies [47], [48]. Available instrumentation includes the PFA-10 (Platelet Function Analyzer, Siemens USA), the VerifyNow (Accumetrics), the Plateletworks (Helena), the IMPACT (Diamed) and the thromboelastograph (TEG) (Haemonetics). Initial tests for a bleeding disorder rule out more common causes of bleeding. These tests include complete blood and platelet counts, PTT, PT, and possibly fibrinogen level or thrombin time. Additional tests for von Willebrand Disease (vWF: Antigen, ristocetin cofactor activity. Factor VIII clotting activity) can confirm the disease [48].
3.3.4. Bacterial antigen detection
Bacterial antigen detection tests should be considered obsolete. They have historically been used as an adjunct to other laboratory tests for the diagnosis of bacterial meningitis. The test's purported advantages were the rapid detection of H. influenza, N. meningitides, S. pneumoniae, and S. agalactiae. Overall, the sensitivity is essentially the same as that of a Gram-stained smear of a cyto-centrifuged CSF specimen [49], [50]. With the advent of vaccines to H. influenza type b and N. meningitides (A, C, Y, and W-135) the antigen testing is even less useful. The literature confirms that the use of direct antigen testing from the CSF is neither sensitive nor specific [49], [50]. More importantly, the Gram stain and cultures still need to be performed regardless of the initial antigen test result. Based on the data reviewed, our laboratory has discontinued this testing in-house.
3.3.5. Group B Streptococcus antigen (GBS) testing
This is an example of testing that was removed from the market based on recommendations from the Centers for Disease Control (CDC) stating that the rapid antigen detection tests for GBS are not sensitive enough to replace the culture based prenatal screening or to use in place of the risk-based approach when culture results are unknown at the time of labor [51]. Because of the poor performance of the rapid antigen testing, CDC has recommended intrapartum chemoprophylaxis antibiotics be administered to women who have certain risk factors [51]. Our laboratory looked at internal data for our patient population and found the sensitivity of the rapid antigen test that was being used was 28%. Forty-one patients were missed on the rapid antigen test were detected only by culture. Data from the literature show an average sensitivity of 25.7% among various labs surveyed [52]. This is an example where CDC recommendations and assessment of testing performance within your laboratory supports moving a test into obsolescence.
3.3.6. HIV-1 Western Blot
The HIV-1 Western Blot (WB) has been a constant as one of the confirmatory tests for HIV antibody testing. However, the WB is moving its way into obsolescence as newer generation antigen/antibody and molecular assays become part of the new HIV testing algorithms. HIV-1 WBs have always had issues with indeterminate results due to a myriad of factors (false positives and/or lack of specificity, kit design, etc.), which required either re-testing at a later time and/or molecular testing for HIV-1 nucleic acid [53]. The CDC/APHL, WHO, and France each have different interpretation criteria for defining a positive confirmatory result which indicates a lack of standardization for defining a patient as positive for HIV-1 [54]. The immune response to HIV-2 is well known to produce antibodies that cross react on the HIV-1 WB which could lead to a false positive HIV-1 result [54]. The lack of improvements or advancements of the WB compared to other antibody based assays has been nicely shown by Masciotra et al. [55]. They demonstrate that the rapid HIV tests actually detect HIV-1 antibodies several days earlier before the WB becomes positive [55]. The introduction of 4th generation antigen/antibody testing has allowed clinicians to detect reactive patients earlier in their disease course compared to 3rd generation testing effectively narrowing the window of serological detection by approximately 4–8 days [56]. Because of these new developments, CLSI and APHL have recommended new algorithms that incorporate antigen/antibody combination tests, rapid tests, and molecular testing [57], [58]. The WB should be a test that will be obsolete as laboratories adopt the newer algorithms for HIV testing.
3.4. Metrics: inappropriate laboratory tests
In addition to obsolete tests, tests may be inappropriately used. Inappropriate utilization includes the failure to follow current practice guidelines for the diagnosis and management of disease, thus ordering the incorrect test, panel of tests, or algorithm of tests; ordering tests too frequently or lack of medical rationale for the test. The problem and the resolution need to be reshaped as a way to “improve patient outcomes and lower costs” [59].
The acknowledgement of the laboratory test utilization problem has been known and published for more than 2 decades [1], [2], [3], [4], [5], [6], [60] Studies have been done to estimate and document the percentage of unnecessary tests [37], [61].
3.4.1. Thyroid tests
A typical scenario for inappropriate test ordering occurs with thyroid function testing in the diagnosis and management of hypothyroidism. The guidelines are clear on the appropriate tests [30]. Thyroid stimulating hormone (TSH) is regarded as the best screening test, followed by free thyroxine (free T4) if the TSH is abnormal. Additional tests are often ordered including total T4. There is no need for a total T4 measurement if a free T4 is provided. Furthermore, adding a total T4 level may confuse the diagnosis if changes in binding proteins via disease or drug therapy result in a total T4 that is inconsistent with other test results.
3.4.2. Vitamin D
Tests may be ordered inappropriately or at other times, the wrong test is ordered. Vitamin D testing is known to result in both inappropriate and erroneous ordering [62]. The correct test for the routine assessment of vitamin D status or deficiency is 25-hydroxy vitamin D. The test 1,25-dihydroxy vitamin D is often mistakenly ordered. The dihydroxy form of vitamin D is occasionally ordered in patients with kidney disease (decreased levels are one of the earliest changes to occur in persons with early kidney failure). However, most of the orders for 1,25-dihydroxy vitamin D are simply erroneous. Tests for both vitamin D2 and D3 are unnecessary for the assessment of vitamin D status. There is no need to differentiate between the D2 and D3 forms other than in the research setting.
3.4.3. Viral cultures
Viral cultures have traditionally been the gold standard for virological detection in the clinical microbiology laboratory. Direct viral detection direct from patient samples have also been utilized with monoclonal antibodies to detect viral antigen(s) with the intent of obtaining a result in a timelier manner than viral cultures. Molecular technologies have now become established in the clinical microbiology laboratory. With the increased sensitivity and specificity [63] and almost always a shorter turn-around-time, it is reasonable to ask “Why do viral cultures?” [63], [64].
One example of molecular testing that helps to answer this question are panels developed for respiratory viruses. There are several commercial companies that offer their version of an RVP (Respiratory Virus Panel). Our laboratory offers a RVP assay that detects 20 viral targets. Not only are results obtained sooner than traditional virology testing, but our molecular RVP offers greater sensitivity and specificity than our prior Direct Fluorescence Assay (DFA) that was sent out to a reference laboratory. Because of the increased sensitivity, the RVPs have allowed us to document co-viral infections, which have not been fully appreciated before with DFA or culture based testing [64]. There have been reports of increased severity of disease, as well as reports of decreased severity of disease in the setting of co-viral infections [65]. Much more research must be done to understand the potential interactions of different respiratory viruses in the setting of a respiratory infection. Our laboratory publishes a “Virogram” during the respiratory virus season and related viral coinfections (Fig. 1, Fig. 2 and Table 1 ). The “Virogram” charts the respiratory virus prevalence among the patient populations tested to give an idea what is circulating in the community.
Fig. 1.
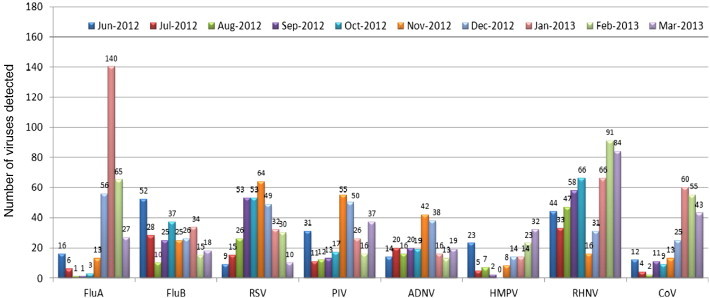
This bar graph shows the respiratory virus prevalence in our healthcare institution for all patients tested from June 2012 through March 2013. This is valuable information for clinicians to know what is circulating amongst the patient population. There are implications for those viruses, especially Influenza A and B, which have treatment options available. It would be important for clinicians to know when to most likely treat based on symptoms relative to the time of year. FluA (Influenza A). FluB (Influenza B). RSV (Respiratory Syncytial Virus). PIV (Parainfluenza virus subtypes 1, 2, 3, or 4). ADNV (Adenovirus subtypes B, C, or E). HMPV (Human Metapneumovirus). RHNV (Rhinovirus). CoV (Coronavirus subtypes 229E, HKU1, NL63, or OC43).
Fig. 2.
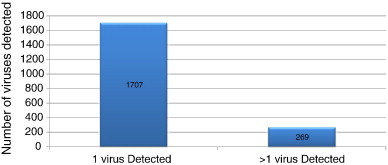
The use of molecular methods for respiratory virus testing has allowed us to detect patients with more than 1 virus present. The presence of multiple viruses within a patient sample is currently underappreciated and the effect on the overall disease presentation is currently unknown.
Table 1.
Distribution of 269 patient samples with > 1 virus present.
| 2 Viral Co-detections | 248 |
| 3 Viral Co-detections | 20 |
| 4 Viral Co-detections | 1 |
Another example of molecular testing replacing viral cultures is the detection of Enterovirus (EV) from CSF. EV PCR has been shown to have greater sensitivity over culture [66]. A study (unpublished) performed within our institution looked at the utility of an in-house EV PCR and its affect on length of stay (LOS) and cost on 20 EV PCR negative and 20 EV PCR positive patients. Those with an EV PCR negative result had an average LOS of 2.1 days greater than those that were EV PCR positive and had an estimated $187,992 of additional cost related to in-patient care.
Viral cultures still have importance in growing the virus for the purposes of subtyping, identifying new strains, or antiviral testing. However, these should remain in specialty or research labs and not be routine for clinical diagnostic testing.
3.4.4. Borrelia burgdorferi (Lyme Disease) PCR
PCR testing for the causative agent of Lyme Disease may initially make sense, but the life cycle of B. burgdorferi is such that PCR detects Borrelia DNA in the blood in less than half of patients that are in the early acute stage of disease when the characteristic erythema migrans is present [67]. Therefore, PCR is not recommended as a first line test for making the initial diagnosis of Lyme disease. A review article showed that the median percent sensitivities of PCR testing from blood, skin biopsy, CSF, and synovial fluid are 10–48%, 64–76%, 23–73%, and 66–83%, respectively [67]. The current recommendation for diagnostic testing involves a two-tiered algorithmic approach involving antibody testing [68]. There may be clinical utility for PCR, but only if the serology testing is negative or inconclusive and clinical history and symptoms strongly suggest Lyme disease.
4. Outreach client test utilization by speciality
Hospitals have introduced outreach programs that market laboratory services to physician offices, nursing homes, and other hospitals [69], [70] to increase test volumes and reduce unit costs per test. This effort generates increased laboratory test utilization which should be monitored by average number of specific tests ordered per subspeciality physician per month ([69], Table 40.10). The data is collected from patient requisitions that are processed each day in the accessioning area of the outreach specimen receiving area. The sales force will introduce outreach administration to the projected number of new client accounts to be opened each month. The impact of the laboratory can be estimated by multiplying the volume of a specific test per physician for an office practice of multiple physicians. These volumes will estimate the utilization of tests per laboratory section and predict the future need for additional analyzers and/or personnel to perform the laboratory test procedures as the outreach client number increase.
Utilization data varies by medical subspeciality. For example, urology had 94 requisitions/physician, 1.4 procedures/requisition and 132 procedures/physician with the PSA being the highest test volume. Compare that to the nursing homes with 189 requisitions/physician, 3.6 procedures/requisition and 688 procedures/physician with electrolytes being the highest test volume [69]. This data is used primarily for planning the best strategies to absorb the increased workload new clients will bring to the outreach business. It can also be used to monitor the use of obsolete and inappropriate lab tests to develop educational efforts to improve test utilization practices by subspeciality among outreach clients.
5. Conclusions
An excellent review of laboratory test utilization, understanding the many factors involved, and steps to implement changes are provided in a recent publication [4]. The author provides insight and guidelines to assist the laboratory in initiating improvements in laboratory utilization. Ultimate goals include: developing and adopt more-effective testing algorithms, reducing testing costs, use new technologies cost-effectively, and shorten the time to diagnosis.
Interventions for hospitals and laboratories focus on changing physicians' test ordering behavior and include:
-
1.
Eliminating obsolete tests and modifying requisition forms. The laboratory can alter test-requisition forms to steer clinicians in the right direction. One such option is an "out of sight, out of mind" approach in which certain tests simply don't appear on the menu.
-
2.
Assisting in the education to promote appropriate lab testing can be part of a hospitals continuing medical education (CME) program for clinicians through grand rounds, newsletters, and CME lectures.
-
3.
Reinforcing positive changes by auditing clinicians’ use of new protocols and offering feedback.
-
4.
A two tier review process for molecular send out tests is a useful tool for pathologists to learn and advice on the wisdom of molecular assays [71].
Finally, all hospitals should implement a laboratory utilization or formulary committee to help in overseeing testing and promoting good testing practices, similar to pharmacy and therapeutics committees. This approach has been met with great success [2], [5], [6].
References
- 1.Lewandrowski K. Managing utilization of new diagnostic tests. Clin Leadersh Manag Rev. 2003;17:318–324. [PubMed] [Google Scholar]
- 2.Kiechle F.L., Collins L. Clinical laboratory tests not performed in a central hospital laboratory. J Clin Ligand Assay. 2005;28:198–201. [Google Scholar]
- 3.Janssens P.M.W. Managing the demand for laboratory testing: options and opportunities. Clin Chim Acta. 2010;411:1596–1602. doi: 10.1016/j.cca.2010.07.022. [DOI] [PubMed] [Google Scholar]
- 4.Smellie W.S.A. Demand management and test request rationalization. Ann Clin Biochem. 2012;49:323–336. doi: 10.1258/acb.2011.011149. [DOI] [PubMed] [Google Scholar]
- 5.Warren J.S. Laboratory test utilization program. Structure and impact in a large academic medical center. Am J Clin Pathol. 2013;139:289–297. doi: 10.1309/AJCP4G6UAUXCFTQF. [DOI] [PubMed] [Google Scholar]
- 6.Lundberg G.D. Changing physician behavior in ordering diagnostic tests. JAMA. 1998;280:2036. doi: 10.1001/jama.280.23.2036. [DOI] [PubMed] [Google Scholar]
- 7.Nissen S.E. Biomarkers in cardiovascular medicine. The shame of publication bias, JAMA. Intern Med. 2013;173:671–672. doi: 10.1001/jamainternmed.2013.4074. [DOI] [PubMed] [Google Scholar]
- 8.Tzoulaki I., Siontis K.C., Evangelou E., Ioannidis J.P.A. Bias in association of emerging biomarkers with cardiovascular disease. JAMA Intern Med. 2013;173:664–671. doi: 10.1001/jamainternmed.2013.3018. [DOI] [PubMed] [Google Scholar]
- 9.Ziai J.M., Smith B.R. Pathology resident and fellow education in a time of disruptive technologies. Clin Lab Med. 2012;32:623–638. doi: 10.1016/j.cll.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 10.Tonellato P.J., Crawford J.M., Boguski M.S., Jaffitz J.E. A national agenda for the future of pathology in personalized medicine. Am J Clin Pathol. 2011;135:668–672. doi: 10.1309/AJCP9GDNLWB4GACI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kok J., Chen S.C., Dwyer D.E., Iredell J.R. Current status of matrix-assisted laser desorption ionisation-time of light mass spectrometry in the clinical microbiology laboratory. Pathology. 2013;45:4–17. doi: 10.1097/PAT.0b013e32835be408. [DOI] [PubMed] [Google Scholar]
- 12.Green E.D., Guyer M.S. National Human Research Institute, charting a course for genomic medicine from base pairs to bedside. Nature. 2011;470:204–213. doi: 10.1038/nature09764. [DOI] [PubMed] [Google Scholar]
- 13.Gargis A.S., Kalman L., Berry M.W., Blick D.P., Dimmock D.P., Hambuch T. Assuring the quality of next-generation sequencing in clinical laboratory practice. Nat Biotechnol. 2012;30:1033–1035. doi: 10.1038/nbt.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coonrod E.M., Durtschi J.D., Margrof R.L., Voelberding K.V. Developing genome and exome sequencing for candidate gene identification in inherited disorders. An integrated technical and bioinformatics approach. Arch Pathol Lab Med. 2013;137:415–433. doi: 10.5858/arpa.2012-0107-RA. [DOI] [PubMed] [Google Scholar]
- 15.McMurray C.T. Mechanisms of trinucleotide repeat instability during human development. Nat Rev Genet. 2010;11:786–799. doi: 10.1038/nrg2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.The 1000 Genomes Project Consortium. Nature. 2012;491:56–65. [Google Scholar]
- 17.Costa F.F., Foly L.S., Coutinho M.P. DataGenno: building a new tool to bridge molecular and clinical genetics. Appl Clin Genet. 2011;4:45–54. doi: 10.2147/TACG.S17597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Centers for Disease Control and Prevention Invasive cancer incidence – United States, 2009. MMWR. 2013;7:113–118. [PMC free article] [PubMed] [Google Scholar]
- 19.Idris S.F., Ahmad S.S., Scott M.A., Vassileou G.S., Hadfield J. The role of high-throughput technologies in clinical cancer genomics. Expert Rev Mol Diagn. 2013;13:167–181. doi: 10.1586/erm.13.1. [DOI] [PubMed] [Google Scholar]
- 20.Vogelslein B., Papadopoulos N., Velculescu V.E., Zhou S., Diaz L.A., Jr., Wikinzler K. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chapman P.B., Houschild A., Robert C., Haanen J.B., Ascierto P., Larkin J. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neimeier L.A., Akatsu H.K., Song C., Carter S.E., Hodak S.P., Yip L. A combined molecular-pathologic score improves risk stratification of thyroid papillary microcarcinoma. Cancer. 2012;118:2069–2077. doi: 10.1002/cncr.26425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hammond M.E., Hayes D.F., Dowsett M., Allred D.C., Hagerty K.L., Badve S. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunochemical testing of estrogen and progesterone receptors in breast cancer (unabridged version) Arch Pathol Lab Med. Jul 2010;134:e48–e72. doi: 10.5858/134.7.e48. [DOI] [PubMed] [Google Scholar]
- 24.Lindeman N.I., Cagle P.T., Beasley M.B., Chitale D.A., Dacic S., Giaccone G. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors. Arch Pathol Lab Med. 2013;137:828–860. doi: 10.5858/arpa.2012-0720-OA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsimberidou A.M., Iskander N.G., Hong D.S., Wheeler J.J., Fu S., Piha-paul S. Personalized medicine in a phase 1 clinical trials program: the MD Anderson Cancer Center initiative. Clin Cancer Res. 2012;18:6373–6383. doi: 10.1158/1078-0432.CCR-12-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang W., Hu W., Hou F., Hu P., Wei Z. SNVer GUI: a desktop tool for variant analysis of next-generation sequencing data. J Med Genet. 2012;49:753–755. doi: 10.1136/jmedgenet-2012-101001. [DOI] [PubMed] [Google Scholar]
- 27.Intelicato-Young J., Fox A. Mass spectrometry and tandem mass spectrometry characterization of protein patterns, protein markers and whole proteomes for pathogenic bacteria. J Microbiol Methods. 2013;92:381–386. doi: 10.1016/j.mimet.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 28.Bader O. MALDI-TOF-MS-based species identification and typing approaches in medical mycology. Proteomics. 2013;13:788–799. doi: 10.1002/pmic.201200468. [DOI] [PubMed] [Google Scholar]
- 29.Margolis C.Z., Cretin S. AHA Press; Chicago, IL: 1999. Implementing clinical practice guidelines. [Google Scholar]
- 30.Garber J.R., Cobin R.H., Gharib H., Hennessey J.V., Klein I., Mechanick J.I. American Association of Clinical Endocrinologists and American Thyroid Association Taskforce on Hypothyroidism in Adults, Clinical practice guidelines for hypothyroidism in adults. Thyroid. 2012;12:1200–1235. doi: 10.1089/thy.2012.0205. [DOI] [PubMed] [Google Scholar]
- 31.Newby L.K., Jesse R.L., Babb J.D., Christenson R.H., De Fer T.M., Diamond G.A. ACCF 2012 expert consensus document on practical considerations in the interpretation of troponin elevations. A report of the American College of Cardiology Foundation Task Force on clinical expert consensus documents. J Am Coll Cardiol. 2012;60:2427–2463. doi: 10.1016/j.jacc.2012.08.969. [DOI] [PubMed] [Google Scholar]
- 32.American Diabetes Association Diagnosis and classification of diabetes mellitus. Diabetes Care. 2013;36:567–574. [Suppl.] [Google Scholar]
- 33.Valdes R., Jr., Payne D.A., Linder M.W., Burckhart G., Farkas D.H., Fureh Fl. National Academy of Clinical Biochemistry; Washington, DC: 2010. Laboratory analysis and application of pharmacogenetics to clinical practice. [Google Scholar]
- 34.Bennett M.J., Rinaldo P., Whitley R.J., Rhead W.J., Hannon W.H., Dietzen D.J. National Academy of Clinical Biochemistry; Washington, DC: 2008. Follow up testing for metabolic diseases identified by expanded newborn screening using tandem mass spectrometry. [DOI] [PubMed] [Google Scholar]
- 35.Nishizawa Y., Ohta H., Mura M., Inaba M., Ichimura S., Shiraki M. Guidelines for the use of bone metabolic markers in the diagnosis and treatment of osteoporosis (2012 edition) J Bone Miner Metab. 2013 doi: 10.1007/s00774-012-0392-y. [DOI] [PubMed] [Google Scholar]
- 36.Hilborne L.H. When less is more for patients in laboratory testing. Am J Clin Pathol. 2013;139:271–272. doi: 10.1309/AJCPNZ41OFBRFVWD. [DOI] [PubMed] [Google Scholar]
- 37.Kale M.S., Bishop T.F., Federman A.D., Keyhani S. “Top 5” lists top $5 billion. Arch Intern Med. 2011;171:1856–1857. doi: 10.1001/archinternmed.2011.501. [DOI] [PubMed] [Google Scholar]
- 38.van Walraven C., Naylor C.D. Do we know what inappropriate laboratory utilization is? A systematic review of laboratory clinical audit. JAMA. 1998;280:550–558. doi: 10.1001/jama.280.6.550. [DOI] [PubMed] [Google Scholar]
- 39.Aetna clinical policy bulletin: obsolete and unreliable tests and procedures. August 2012. http://www.aetna.com/cpb/medical/data/400_499/0438.html
- 40.United Healthcare's Policy Number 300.1 Obsolete or Unreliable Tests. August 2012. https://www.unitedhealthcareonline.com/ccmcontent/ProviderII/UHC/en-US/Main%20Menu/Tools%20&%20Resources/Policies%20and%20Protocols/Medicare%20Advantage%20Reimbursement%20Policies/O/ObsoleteUnreliableDxTests_NCD300-1_10192012.pdf
- 41.AmeriHealth's Medical Policy Bulletin #00.01.24D Obsolete or unreliable diagnostic tests and medical services. August 2012. http://medpolicy.amerihealth.com/policies/mpi.nsf/e94faffabc7b0da6852569e0068df65/85256aa800623d7a852579d50069fe18!opendocument
- 42.Filee C., Cumps J., Ketelslegers J.M. Comparison of three free T4 (FT$) and free T3 (FT#) immunoassays in healthy subjects and patients with thyroid diseases and severe non-thryoid illnesses. Clin Lab. 2012;58:725–736. [PubMed] [Google Scholar]
- 43.Van Uytfanghe K., Stockl D., Ross H.A., Thienpont L.M. Use of frozen sera for FT4 standardization: investigation by equilibrium dialysis combined with isotope dilution-mass spectrometry and immunoassay. Clin Chem. 2006;52:1817–1821. doi: 10.1373/clinchem.2006.070425. [DOI] [PubMed] [Google Scholar]
- 44.Goldie D.J., Jones S.R., Thomas M.J. A reappraisal of the free thyroxine index. Lancet. 1981;318:572–573. doi: 10.1016/s0140-6736(81)90951-x. [DOI] [PubMed] [Google Scholar]
- 45.Bahn R.S., Burch H.B., Cooper D.S., Garber J.R., Greenlee M.C., Klein I. Hyperthyroidism and other causes of thyrotoxicosis: Management Guidelines of the American Thyroid Association and American Association of Clinical Endocrinologists. Thyroid. 2011;21:593–646. doi: 10.1089/thy.2010.0417. [DOI] [PubMed] [Google Scholar]
- 46.Stagnaro-Green A., Abalovich M., Alexander E., Azizi F., Mestman J., Negro R. Guidelines of the American Thyroid Association for the diagnosis and management of thyroid disease during pregnancy and postpartum. Thyroid. 2011;21:1081–1125. doi: 10.1089/thy.2011.0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Standardization of platelet function testing by aggregometry through new CLSI guideline. J Lab Med. 2009;40:269–270. [Google Scholar]
- 48.Yawn B., Nichols W.L., Rick M.E. Diagnosis and management of von Willebrand disease: guidelines for primary care. Am Fam Physician. 2009;80:1261–1268. [PubMed] [Google Scholar]
- 49.Karre T., Vetter E.A., Mandrekar J.N., Patel R. Comparison of bacterial antigen test and gram stain for detecting classic meningitis bacteria in cerebrospinal fluid. J Clin Microbiol Rev. 2008;21:716–747. doi: 10.1128/JCM.00153-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perkins M.D., Mirrett S., Reller L.B. Rapid bacterial antigen detection is not clinically useful. J Clin Microbiol. 1995;33:1486–1491. doi: 10.1128/jcm.33.6.1486-1491.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Centers for Disease Control Prevention of perinatal group B streptococcal disease. MMWR. 2002;51 [Google Scholar]
- 52.Thinkkhamrop J., Lumpongsanurak S., Festin M.R., Daly S., Schuchat A., Lumbiganon P. Infections in international pregnancy study: performance of the optical immunoassay test for detection of group B streptococcus. J Clin Microbiol. 2003;41:5288–5290. doi: 10.1128/JCM.41.11.5288-5290.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guan M. Frequency, causes, and new challenges of indeterminate results in Western blot confirmatory testing for antibodies to human immunodeficiency virus. Clin Vaccine Immunol. 2007;14:649–659. doi: 10.1128/CVI.00393-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nasrullaha M., Ethridge S.F., Delaney K.P., Wesolowski L.G., Granade T.C., Schwendemann J. Comparison of alternative interpretive criteria for the HIV-1 and HIV-2 infections. J Clin Virol. 2011;52S:S23–S27. doi: 10.1016/j.jcv.2011.09.020. [DOI] [PubMed] [Google Scholar]
- 55.Masciotra S., McDougal J.S., Feldman J., Sprinkle P., Wesolowski L., Owen S.M. Evaluation of an alternative HIV diagnostic algorithm using specimens from seroconversion panels and persons with established HIV infections. J Clin Virol. 2011;52S:S17–S22. doi: 10.1016/j.jcv.2011.09.011. [DOI] [PubMed] [Google Scholar]
- 56.Chavez P., Wesolowski L., Patel P., Delaney K., Owen S.M. Evaluation of the performance of the Abbott ARCHITECT HIV Ag/Ab Combo Assay. J Clin Virol. 2011;52S:S51–S55. doi: 10.1016/j.jcv.2011.09.010. [DOI] [PubMed] [Google Scholar]
- 57.Clinical Laboratory Standards Institute (CLSI) CLSI; Wayne, Pennsylvania: 2011. Criteria for laboratory testing and diagnosis of human immunodeficiency virus infection; approved guideline, CLSI document M53-a. [Google Scholar]
- 58.Bennett B., Branson B., Delaney K., Owen S.M., Pentella M., Werner B. The Association of Public Health Laboratories; Silver Spring, Maryland: April 2009. HIV testing algorithms: a status report. [Google Scholar]
- 59.Lusky K. Order patrol – how to rein in test requests. CAP Today. 2006;20(1):52–58. [Google Scholar]
- 60.McConnell T.S., Berger P.R., Dayton H.H., Umland B.E., Skipper B.E. Professional review of laboratory utilization. Hum Pathol. 1982;13:399–403. doi: 10.1016/s0046-8177(82)80229-3. [DOI] [PubMed] [Google Scholar]
- 61.Wong E.T., Lincoln T.L. Ready, fire! Aim! An enquiry into laboratory test ordering. JAMA. 1983;250:2510–2513. doi: 10.1001/jama.250.18.2510. [DOI] [PubMed] [Google Scholar]
- 62.Rollins G. Vitamin D testing—What's the right answer? Clin Lab News. 2009;35 [ http://www.aacc.org/publications/cln/2009/july/Pages/CovStory1July09.aspx#] [Google Scholar]
- 63.Leland D.S., Ginocchio C.S. Role of cell culture for virus detection in the age of technology. Clin Microbiol Rev. 2009;20:49–78. doi: 10.1128/CMR.00002-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mahony J.B. Detection of respiratory viruses by molecular methods. Clin Microbiol Rev. 2008;21:716–747. doi: 10.1128/CMR.00037-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Popowitch E.B. Clinical virology symposium poster. April 2012. An analytical comparison of four commercial respiratory virus panels. [Google Scholar]
- 66.Kupila L., Vuorinen T., Vainionpää R., Marttila R.J., Kotilainen P. Diagnosis of enteroviral meningitis by use of polymerase chain reaction of cerebrospinal fluid, stool and serum specimens. Clin Infect Dis. 2005;40:982–987. doi: 10.1086/428581. [DOI] [PubMed] [Google Scholar]
- 67.Aguero-Rosenfeld M.E., Wang G., Schwartz I., Wormser G.P. Diagnosis of lyme borreliosis. Clin Microbiol Rev. 2005;18:484–509. doi: 10.1128/CMR.18.3.484-509.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Center for Disease Control Notice to readers: Recommendations for test performance and interpretation from the Second National Conference on Serological Diagnosis of Lyme Disease. MMWR. 1995;44 [PubMed] [Google Scholar]
- 69.Kiechle F.L., Skrisson J.E. Outreach implementation requirements: a case study. In: Garcia L.S., editor. Clinical laboratory management. ASM Press; Washington, D.C.: 2004. pp. 654–677. [Google Scholar]
- 70.Anderson V. Hospital laboratory outreach: benefits and planning. Clin Lab Med. 2007;27:791–805. doi: 10.1016/j.cll.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 71.Carter A.B. Clinical requests for molecular tests. The 3-step evidence check. Arch Pathol Lab Med. 2012;136:1585–1592. doi: 10.5858/arpa.2011-0691-SA. [DOI] [PubMed] [Google Scholar]
Three Potential Referees
-
[1]
James Faix, MD
Stanford Clinical Labs of Hillview 3375 Hillview Avenue #5627 Palo Alto, CA 94304-1204 Tel: + 001-650-736-1857 E-mail: jfaix@standfordmed.org
-
[2]
George S. Cembrowski, MD, PhD
University of Alberta Hospital 481.24 Walter Mackenzie Centre 8440 112th Street Edmonton AB T6G 2B7 Canada Tel: + 001-780-407-31-85 E-mail: cembr001@cha.ab.ca
-
[3]
Tar Choon Aw, MB Ch
Changi General Hospital 2 Simei Street 3 Singapore 529889 Singapore Tel: + 0065-1-6850-4927 E-mail: tarchoon@gmail.com


