Graphical abstract
Keywords: Porcine parvovirus, Sindbis virus, Water purification, Electrospinning, Pathogen removal, Microfiltration
Highlights
-
•
Quaternary amine (HTCC)/PVA nanofibers have been electrospun.
-
•
Glutaraldehyde crosslinking was used to impart water stability to the fibers.
-
•
Nanofibers had a maximum swelling of 30% after crosslinking.
-
•
Virus removal was achieved for an enveloped and nonenveloped virus.
Abstract
The burden of unsafe drinking water is responsible for millions of deaths each year. To relieve this burden, we are in search of an inexpensive material that can adsorb pathogens from drinking water. In this pursuit, we have studied the natural carbohydrate, chitosan. To impart virus removal features, chitosan has been functionalized with a quaternary amine to form quaternized chitosan N-[(2-hydroxyl-3-trimethylammonium) propyl] chitosan (HTCC). HTCC can be electrospun into nanofibers with the non-ionogenic polyvinyl alcohol (PVA), creating a high surface area mat. High surface area is a major requirement for effective adsorption processes. HTCC is antiviral and antimicrobial, making it a good material for water purification. However, HTCC dissolves in water. We have explored the parameters to crosslink the nanofibers with glutaraldehyde. We have imparted water stability so there is a maximum of 30% swelling of the fibers after 6 h in water. The water stable fibers retain their ability to adsorb virus, as shown for an enveloped and nonenveloped virus. HTCC now has the potential to be incorporated into a microfiltration membrane that can remove viruses. This could create an inexpensive, low pressure filtration membrane for drinking water purification.
1. Introduction
Drinking water contaminated with pathogens kills millions of people every year.1 The most common method to remove or inactivate pathogens is the use of chemical disinfectants.2, 3 However, the most common chemical disinfectant, chlorine, is suspected to create carcinogenic byproducts when natural organic matter is present.4 Newer methods include the use of ultrafiltration for bacteria and nanofiltration for viruses.5 However, small pore-sized nanofilters require high back pressures to function and are not commonly used in under developed countries. To help relieve the burden of unsafe drinking water, we are in search of an adsorption material that can remove pathogens from water. This material would ideally form a microfiltration membrane that would require low back pressures, but have the ability to clean non-potable water.
In our search for an inexpensive material that has the potential to adsorb pathogens, we have selected chitosan. Chitosan (chemical structure shown in Figure 1 ) is a polycationic polymer and the N-deacetylated derivative of the natural polymer chitin, the second most abundant polysaccharide found on earth next to cellulose.6, 7 Chitosan is insoluble in water and common organic solvents because of its rigid crystalline structure. It is soluble in acidic, aqueous solution if the pH value is less than 6.5.8
Figure 1.
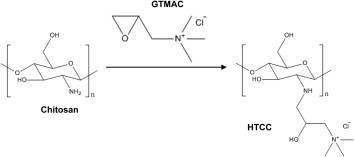
Formation of HTCC from chitosan.
Chitosan is well known to be non-toxic, biocompatible, biodegradable, biofunctional, and hydrophilic.9, 10, 11, 12 Chitosan has been used as an antimicrobial and antiviral material in the fields of biotechnology, pharmaceutics, wastewater treatment, cosmetics, agriculture, food science, and textiles because of its advantageous biological properties.12, 13, 14 However, this activity of chitosan against pathogens is limited to acidic conditions due to its poor solubility above pH value of 6.5, where chitosan starts to lose its polycationic nature.7, 12 The polycationic nature is what imparts most of chitosan’s antimicrobial and antiviral properties. Therefore, the preparation of chitosan derivatives with an improved cationic properties over a wide pH range was studied for their antimicrobial and antiviral activity.6, 7, 10
Among the various chitosan derivatives, the derivatives with quaternary ammonium groups have shown higher efficient activity against bacteria as compared to those of chitosan.6, 10, 12 Quaternized chitosan N-[(2-hydroxyl-3-trimethylammonium) propyl] chitosan (HTCC)7, 11, 14 (chemical structure shown in Figure 1), N–N-propyl-N, N-dimethyl chitosan15 and N-butyl-N,N-dimethyl chitosan iodide6, 10 showed enhanced antimicrobial activity compared with chitosan.14, 16 This may be due to the positively-charged quaternary amine group, known to target the negatively charged cytoplasmic membrane of microbes, altering membrane properties and impeding nutrients entering the cells.12, 16 HTCC and hydrophobically-modified HTCC compounds were shown to inhibit the human coronavirus NL63 and murine hepatitis virus.17 Double-stranded RNA formulated with quaternized chitosan derivative was shown to inhibit yellow head virus.18 HTCC blended with graphene can also reduce infectious porcine parvovirus (PPV) concentrations in high salt environments.19
To form HTCC fibers, we use the technique of electrospinning.19, 20 This method creates fibers that have a submicron diameter. An electric potential is applied to a polymer solution. When the electric potential overcomes the viscosity and surface tension, a jet is created from the polymer solution. The jet whips as the solvent dries, prior to the resting of polymer nanofibers on the collector. The use of a rotating drum collector helps to create more even surface coverage for filtration media. The high surface to volume ratio of nanofibers is sought to increase the adsorptive capacity and accessible surface area of the membrane.
For HTCC nanofibers to become an adsorptive membrane material, the fibers must not dissolve in water. Crosslinking of polymer structures is a common method to impart membrane stability in water soluble polymers.21 PVA fibers were crosslinked with maleic anhydride to create a filtration membrane.22 HTCC blended with polyvinyl alcohol (PVA) was photo-crosslinked to impart antimicrobial resistance to the fibers.10 Glutaraldehyde vapor was used to crosslink HTCC–PVA fibers to create an antimicrobial surface.7
Here, we demonstrate that HTCC nanofibers can be crosslinked while retaining their nonwoven structure and their ability to bind to negatively-charged viruses. We have explored the crosslinking conditions that give the greatest water stability. These conditions allowed for high virus removal. Our model viruses include the non-enveloped porcine parvovirus (PPV), one of the smallest known mammalian viruses and the enveloped virus, Sindbis virus. This nanofiber material has the potential to become an inexpensive defense against water-borne diseases.
2. Results and discussion
2.1. Characterization of HTCC
HTCC, was prepared by the reaction of chitosan with GTMAC in an aqueous solution, as shown in Figure 1. To confirm the successful synthesis of HTCC, FTIR spectra and NMR spectra of chitosan and HTCC are shown in Figure S1. Both the results of FTIR and NMR spectra are in agreement with previous reports.11, 14, 19, 23
The degree of quaternization (DQ) of HTCC was determined to be 76.4 ± 4.3%. This DQ value demonstrates that the amino groups on chitosan were substituted by quaternary ammonium salt groups. The remaining groups were likely acetylated since the chitosan used in this work was 75–85% deacetylated.
2.2. Electrospinning of HTCC blends
PVA has been shown to be a non-ionogenic partner for the formation of HTCC nanofibers.7 We explored how the ratio of HTCC to PVA affected the fiber formation. This has been explored in a wide range of values,7 and we chose a more narrow range. In Table 1 , it shows that the conductivity and absolute viscosity increase with an increase in HTCC content. Fiber diameter decreases with increasing HTCC content. Our results are consistent for conductivity and fiber diameter to those of Alipour et al.7 however, our increase in viscosity is not consistent with the reported decrease in viscosity.7 This may be due to the different molecular weights of chitosan and PVA used in each work. From SEM images, shown in Figure S2, the density of fibers formed from a 10 w/v% polymer blend greatly decreased once the HTCC content reached 50% of the total polymer content. This is consistent with other’s findings for 12 w/v% HTCC–PVA solutions.7 We desired the highest amount of HTCC as could be electrospun for water purification applications. For this reason, we chose to continue our studies with 4:6 HTCC–PVA blends.
Table 1.
Viscosity, conductivity, and fiber diameter of electrospun HTCC–PVA
| HTCC–PVA | Absolute viscosity (mPa s) | Conductivity (mS/cm) | Nanofiber diameter (nm) |
|---|---|---|---|
| 3:7 | 2000 ± 244 | 4.4 ± 0.7 | 119 ± 8 |
| 4:6 | 2409 ± 141 | 6.1 ± 0.5 | 102 ± 1 |
| 5:5 | 3023 ± 289 | 7.3 ± 0.8 | 97 ± 1 |
| 6:4 | 3536 ± 257 | 9.1 ± 0.8 | 84 ± 1 |
| 7:3 | 3805 ± 71 | 10.0 ± 0.5 | na |
na–not available.
2.3. Crosslinking of HTCC nanofibers
In order to stabilize the HTCC–PVA nanofibers before contacting the fibers with water, glutaraldehyde vapor7 was used. Different crosslinker concentration and crosslinking time were explored and the result is shown in Figure 2 . To determine the stability of crosslinked HTCC–PVA nanofibers against water, the nanofibers were filtered with water or immersed in water for 10 min.
Figure 2.
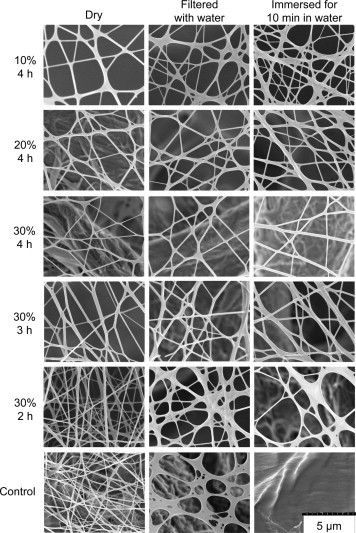
Images of glutaraldehyde crosslinked HTCC–PVA nanofibers. Electrospun nanofibers were crosslinked with different percentage of glutaraldehyde and for different times, as shown on the left. They were then tested for water stability by filtering with water or immersing in water for 10 min, as shown on the top.
The control samples in Figure 2 show the fresh electrospun HTCC–PVA nanofibers dissolving in water. For the uncrosslinked membrane, both the polymer of HTCC and PVA swells and the distance between any two polymer chains increases as the adsorbed water molecules increase.24 Once the attractive forces between these two polymers chains are not sufficient to keep them within a critical distance, the polymer dissolves.24
To stabilize the fibers, glutaraldehyde vapor was used to crosslink the fibers. This gave the fibers stability in water since the aldehyde group of glutaraldehyde will react with the unbound hydroxyl groups of both the HTCC and PVA to form crystallizations zones.25 Water caused fiber swelling to different degrees for all crosslinking conditions. Even the glutaraldehyde vapor caused some swelling of the fibers, since it was in an aqueous solution. As shown in Figure 2, nanofibers with a crosslinking time of 4 h in 30% glutaraldehyde had the least swelling and were the most water stable. This can also be confirmed in Figure 3 , which graphically shows the diameter of the fibers. The diameter of 30% glutaraldehyde, 4 h crosslinked nanofibers (dry, filtered and immersed for 10 min in water) were the closest to the diameter of fresh electrospun nanofibers. There was a statistically significant swelling of all crosslinked samples, as compared to the dry control without crosslinking. Both 10% glutaraldehyde, 4 h and 30% glutaraldehyde, 2 h crosslinked fibers started dissolving when contacted with water for 10 min and some coalescence of fibers was observed as shown in Figure 2. From Figure 2, Figure 3, it could be noted that the fiber stability increased with the increase of crosslinker concentration and crosslinking time, as demonstrated by a lack of fiber diameter increase.
Figure 3.
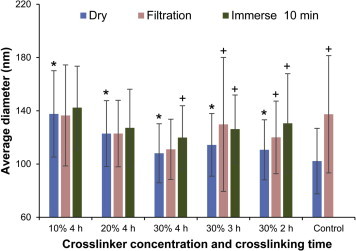
Change in fiber diameter. The fiber diameter of the crosslinking conditions found in Figure 2 were measured. Error bars are the standard deviation of 150 fibers that were measured from two independent experiments. ∗p <0.01 for Student’s t-test compared to the dry control fibers. +p <0.01 for Student’s t-test compared to the dry fibers within that condition.
To study the stability of the 30% glutaraldehyde, 4 h crosslinked HTCC–PVA nanofibers against water, the membrane was immersed in water for different lengths of time. The morphology of the fibers is shown in Figure 4 . The corresponding average fiber diameter distribution and the change of swelling degree calculated from the fiber diameter are shown in Figure 5 . It was found that 30% glutaraldehyde, 4 h crosslinked fibers kept their morphology and did not dissolve within 6 h of contact with water. The fibers swelled in water and the equilibrium swelling degree was found to be 30%.
Figure 4.
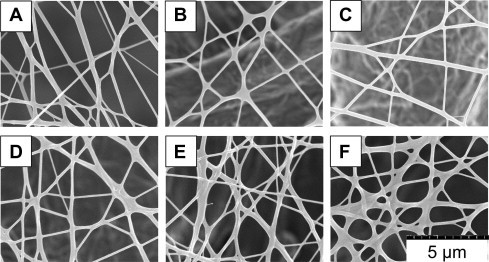
Effect of water immersion on fiber morphology. Crosslinked nanofibers were filtered or immersed in water for various times. (A) Dry crosslinked nanofibers, (B) nanofibers filtered with water, (C) nanofibers immersed for 10 min in water, (D) nanofibers immersed for 30 min in water, (E) nanofibers immersed for 60 min in water, and (F) nanofibers immersed for 360 min in water.
Figure 5.
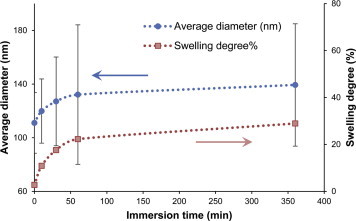
Diameter and swelling degree of fibers with different water immersion times. Error bars are the standard deviation of fibers found on three different images.
2.4. Electrospinning of HTCC fibers
The effect of two key electrospinning parameters, the applied voltage and the feed rate, on fiber morphology was studied with the water-stable 30% glutaraldehyde, 4 h crosslinked nanofibers. The fiber morphology is shown in Figure 6 and the fiber diameter distribution is shown in Figure 7 . The average nanofiber diameter significantly decreased with an increase in the applied voltage, which is similar to the data found previously.7, 10 This is likely due to an increased charge density on the polymer solution as the voltage increases. The greater Coulombic repulsive forces cause the polymer jet to accelerate faster and to have an increased stretch.26 In addition, the average diameter of nanofibers increased with an increase in the feed rate. This is likely due to insufficient time available for the electrospinning solution to evaporate, resulting in fusing of the fibers together at the higher feed rate.27 This can be seen in Figure 6. This demonstrates that we can control the fiber diameter. In the future, we will explore how fiber diameter effects virus removal.
Figure 6.
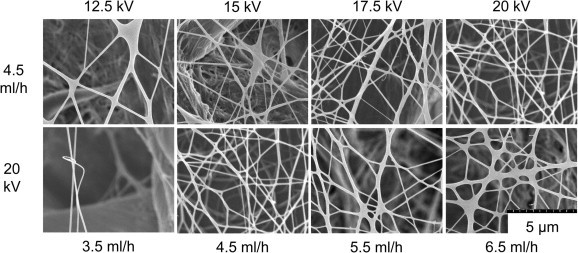
Effect of electrospinning conditions on fiber morphology. HTCC–PVA fibers were electrospun at different voltages and pump feed rates and then crosslinked. The top row was electrospun at 4.5 mL/h at different voltages. The bottom row was electrospun at 20 kV and different feed rates.
Figure 7.
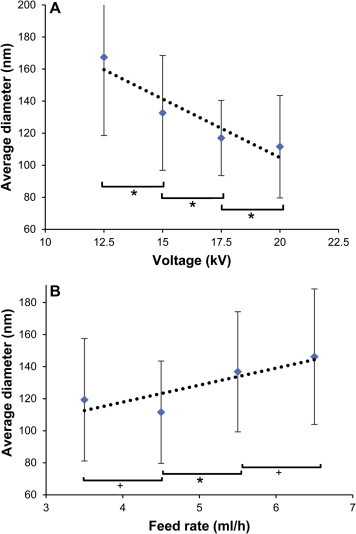
Diameter at different electrospinning conditions. (A) Effect of voltage and (B) effect of flow rate on crosslinked nanofibers. Error bars are the standard deviation of 150 fibers that were measured from two independent experiments. ∗p <0.01 and +p <0.05 for Student’s t-test.
2.5. Virus removal by incubation of nanofibers
The virus binding ability of crosslinked HTCC–PVA nanofibers was tested and is shown in Figure 8 . The log removal value (LRV) was used to describe the virus removal and the equation can be found in Section 4.2.5 of the Methods section. The fibers were tested for their ability to remove two different viruses, porcine parvovirus (PPV) and Sindbis virus. PPV is a nonenveloped virus and Sindbis is an enveloped virus. These viruses represent two distinct classes of viruses that could contaminate water. A range of enveloped and non-enveloped viruses are typically used to evaluate generic virus removal techniques.28 The water-stable nanofibers achieved a 3.3 LRV for PPV and a 4.2 LRV for Sindbis. No virus was removed when only blank filter paper with no crosslinking was contacted with the virus. Crosslinking of blank filter paper (the control) did remove some virus. This demonstrates that the glutaraldehyde, even after water washing, has some virus adsorption properties. Nevertheless, the addition of nanofibers greatly increased the virus removal as compared to glutaraldehyde exposed filter paper (the control). The maximum theoretical removal was 4.4 LRV. These values come close to or exceed the EPA regulation of a 4 LRV for virus removal processes.29 This is a clear demonstration that these stable nanofibers have the ability to bind to two different viruses and could become a new material for virus removal in drinking water applications.
Figure 8.
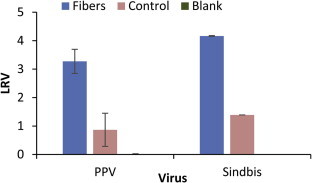
Virus removal of crosslinked nanofibers. PPV and Sindbis virus were incubated with crosslinked HTCC–PVA nanofibers for 10 min. Blank filter paper, control crosslinked filtered paper and crosslinked nanofibers on filter paper were compared. Error bars are the standard deviation of three separate experiments.
3. Conclusions
The high charge density on the quaternary amine functionalized chitosan, HTCC, has been demonstrated to have antimicrobial7, 11 and antiviral17, 19 properties. We pursued this polymer for the creation of electrospun membranes that remove pathogens from drinking water. To realize this goal, we have examined the crosslinking of glutaraldehyde vapor to impart water stability to HTCC–PVA nanofibers. It was found that using high glutaraldehyde concentration and crosslinking time created a membrane that was stable for a minimum of 6 h in water with a maximum swelling degree of 30%. The water-stable nanofibers had a high virus removal, with an LRV of 3.3 and 4.2 for a non-enveloped and an enveloped virus, respectively. This is approaching or exceeding the EPA regulation of 4 LRV.29 With careful design of a filtration apparatus, along with other polymer modifications, this system has high potential to become a future pathogen removal system for water purification.
4. Materials and methods
4.1. Materials
Chitosan (75–85% deacetylated, MW = 190,000–310,000 Da), glycidyltrimethylammonium chloride (GTMAC) (⩾90%), PVA (99% hydrolyzed, MW = 89,000–98,000 Da), silver nitrate (⩾99.0%), potassium chromate (⩾99.0%), deuterium oxide and trifluoroacetic acid-d were all purchased from Sigma–Aldrich (St. Louis, MO). To crosslink HTCC nanofibers, glutaraldehyde solution was purchased from Sigma–Aldrich (St. Louis, MO).
Minimum essential medium (MEM), penicillin–streptomycin, trypsin and phosphate buffered saline (PBS) (pH = 7.2) were purchased from Life Technologies (Grand Island, NY). Fetal bovine serum (FBS) was purchased from Atlanta Biologicals (Grand Island, NY). MTT agent, thiazolyl blue tetrazolium bromide (⩾97.5%) was purchased from Sigma–Aldrich (St. Louis, MO). Hydrochloric acid (HCl) (12.1 M) was purchased from VWR (Radnor, PA). All aqueous solutions were prepared using purified water with a resistivity of ⩾18 MΩ cm from a Nanopure filtration system (Fisher Scientific, Pittsburgh, PA).
4.2. Methods
4.2.1. Synthesis and characterization of HTCC polymer
HTCC was synthesized according to a known method,19 with minor modifications. The scheme for the synthesis of HTCC is shown in Figure 1. First, chitosan (6 g, 37.0 mmol) was dispersed in Nanopure water (240 mL) in a 500 mL three-neck round-bottom flask. GTMAC (16.82 g, 21.3 mL, 111 mmol) was added in three portions (7.1 mL each) at 2 h intervals, and the mixture was stirred at 85 °C for 10 h. Then, the reaction solution was dialyzed for 3 days with Fisherbrand 3.5 kDa MWCO regenerated cellulose dialysis tubing (Fisher Scientific, Pittsburg, PA) against water to remove any unreacted GTMAC. Unreacted chitosan was removed by vacuum filtered with a Buchner funnel containing a fine porosity fritted disc. The clear solution was concentrated under vacuum with a Buchi R-200 rotavapor (New Castle, DE) at 70 °C. The concentrated solution was precipitated in cold acetone (250 mL) using an ice bath and washed twice with acetone. The precipitate was dried at 110 °C for 12 h to obtain the final product.
The degree of quaternization (DQ) was measured by the titration of chloride using silver nitrate, as has been described.19
4.2.2. Electrospinning of HTCC blends
HTCC at 10 w/v% was dissolved in Nanopure water at room temperature with gentle stirring for a day in order to prepare a homogeneous solution. PVA at 10 w/v% was dissolved in Nanopure water at 85 °C with gentle stirring for 5 h. HTCC–PVA ratios ranging from 3:7 to 7:3 were explored and the total polymer concentration of the electrospinning solution was kept constant at 10 w/v%.
Prior to electrospinning, the viscosity of the electrospinning solution was measured using a SV-10 viscometer (Malvern, United Kingdom). The conductivity of the electrospinning solution was measured using an AB-30 conductivity meter (Fisher Scientific, Pittsburg, PA).
The description of the home-made electrospinning apparatus can be found elsewhere.19 The electrospinning solutions were placed into a 3 mL, disposal, plastic syringe with a detachable needle (0.6 mm × 40 mm) (Fisher Scientific, Pittsburgh, PA). The needle was connected to a Glassman positive DC high voltage power supply (High Bridge, NJ), capable of generating voltages in the range of 0–30 kV, while the ground was connected to a rotating drum collector covered with aluminum foil run by an Electro Craft Torque power pump (Gallipolis, OH). The electrospun nanofibers were collected on Whatman filter papers circles, which were taped on the aluminum foil and used to support the nanofibers. A multi speed syringe pump (Braintree Scientific Inc., Braintree, MA) was used to feed the solution at a constant speed.
The applied voltage was 20 kV, the tip-to-collector distance was 5 cm and the feed rate of solution was 4.5 mL/h, unless otherwise stated. The rotational speed of the drum collector was set at 1500 rpm.
4.2.3. Crosslinking of HTCC nanofibers
The electrospun HTCC–PVA nanofibers were crosslinked with glutaraldehyde vapor,7 at 37 °C for various times. The crosslinked electrospun mats were washed with water for 20 min. All samples were dried in a Gold Series DP-32 vacuum drying oven (Ontario, Canada) at 120 °C for 1 h.
4.2.4. Characterization of HTCC nanofibers
The morphology of HTCC nanofibers was observed with a Hitachi S-4700 cold-field emission scanning electron microscope (FE-SEM) (Tustin, CA). The accelerating voltage for the FE-SEM was 5 kV, and the magnification was from 1000× to 80,000×. To increase the SEM sensitivity to polymer nanofibers, the fibers were coated with 5 nm of platinum/palladium with a sputter coater (Hummer Sputtering System, Union City, CA) at a rate of 0.1 nm/min.
To determine the stability of crosslinked electrospun HTCC–PVA nanofibers against water, the nanofibers were filtered with water and immersed in water. All samples were dried at 120 °C for 1 h.
4.2.5. Virus removal assessment
PPV strain NADL-2 and porcine kidney-13 (PK-13) cells were a gift from Dr. Ruben Carbonell, North Carolina State University. Sindbis virus (heat resistant strain) and baby hamster kidney (BHK-21) cells were a gift from Dr. Raquel Hernandez, North Carolina State University. Cells and virus were propagated, as described earlier.30 The virus was titrated with an MTT assay.30, 31 The incubation time for PPV was 5 days and the incubation time for Sindbis virus was 2 days. The MTT assay is a colorimetric cell viability method, based on the reaction of MTT agent (thiazolyl blue tetrazolium bromide) with the mitochondria of metabolically active cells.30
The log removal value (LRV) was used to determine the amount of virus that adsorbed to the nanofibers,
| (1) |
where, cf is the pathogen concentration after water purification and ci is the initial pathogen concentration.
For virus removal studies, 1 layer of 0.5024 cm2 filter paper with HTCC–PVA nanofibers after crosslinking with 30% glutaraldehyde for 4 h at 37 °C was placed into a 1.5 mL micro-centrifuge tube with 500 μL of 6 logs (MTT/mL) of virus in water. One layer of the same size blank filter paper and control crosslinked filter paper without nanofibers were put into separate tubes. Tubes were rotated for 10 min (unless otherwise stated) on a Roto-shake Genie rocker (Scientific Industries Inc., Bohemia, NY). Then, the nanofibers were taken out of the tubes, and the virus solutions after incubation were centrifuged for 30 min at a speed of 14,000 rpm in a Sorvall ST16R Centrifuge (Thermo Scientific, Pittsburgh, PA) to remove any remaining fibers in the tubes. The supernatant was removed and tested with the MTT assay to determine the concentration of infectious virus and calculate the LRV.
Acknowledgements
The authors would like to thank Ching-An Peng for the use of his viscometer, Adrienne Minerick for the use of her conductivity meter, and Patricia Heiden for use of her electrospinning apparatus, as well as fruitful discussions. Funding was gratefully received from the Department of Chemical Engineering at Michigan Tech, the Research Enhancement Fund at Michigan Tech and NSF (CBET-1125585).
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.carres.2014.01.017.
Supplementary data
This document file contains supplementary data.
References
- 1.WHO, Water and Sanitation Facts and Figures, Vol. (accessed 22 Nov 2010).
- 2.Samson L., Czegeny I., Mezosi E., Erdei A., Bodor M., Cseke B., Burman K.D., Nagy E.V. J. Endocrinol. Invest. 2012;35:21–24. doi: 10.3275/7759. [DOI] [PubMed] [Google Scholar]
- 3.Schmidt W., Bohme U., Sacher F., Brauch H.J. Ozone Sci. Eng. 2000;22:215–226. [Google Scholar]
- 4.Komulainen H. Toxicology. 2004;198:239–248. doi: 10.1016/j.tox.2004.01.031. [DOI] [PubMed] [Google Scholar]
- 5.Antony A., Blackbeard J., Leslie G. Crit. Rev. Env. Sci. Technol. 2012;42:891–933. [Google Scholar]
- 6.Ignatova M., Manolova N., Rashkov I. Eur. Polym. J. 2007;43:1112–1122. [Google Scholar]
- 7.Alipour S.M., Nouri M., Mokhtari J., Bahrami S.H. Carbohydr. Res. 2009;344:2496–2501. doi: 10.1016/j.carres.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 8.No H.K., Meyers S.P., Prinyawiwatkul W., Xu Z. J. Food Sci. 2007;72:R87–R100. doi: 10.1111/j.1750-3841.2007.00383.x. [DOI] [PubMed] [Google Scholar]
- 9.Huang X.J., Ge D., Xu Z.K. Eur. Polym. J. 2007;43:3710–3718. [Google Scholar]
- 10.Ignatova M., Starbova K., Markova N., Manolova N., Rashkov I. Carbohydr. Res. 2006;341:2098–2107. doi: 10.1016/j.carres.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 11.Deng H.B., Lin P.H., Xin S.J., Huang R., Li W., Du Y.M., Zhou X., Yang J.H. Carbohydr. Polym. 2012;89:307–313. doi: 10.1016/j.carbpol.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 12.Li Q.L., Mahendra S., Lyon D.Y., Brunet L., Liga M.V., Li D., Alvarez P.J.J. Water Res. 2008;42:4591–4602. doi: 10.1016/j.watres.2008.08.015. [DOI] [PubMed] [Google Scholar]
- 13.Lu B.G., Li T., Zhao H.T., Li X.D., Gao C.T., Zhang S.X., Xie E.Q. Nanoscale. 2012;4:2978–2982. doi: 10.1039/c2nr11958g. [DOI] [PubMed] [Google Scholar]
- 14.Lim S.H., Hudson S.M. Carbohydr. Res. 2004;339:313–319. doi: 10.1016/j.carres.2003.10.024. [DOI] [PubMed] [Google Scholar]
- 15.Jia Z.S., Shen D.F., Xu W.L. Carbohydr. Res. 2001;333:1–6. doi: 10.1016/s0008-6215(01)00112-4. [DOI] [PubMed] [Google Scholar]
- 16.Rabea E.I., Badawy M.E.T., Stevens C.V., Smagghe G., Steurbaut W. Biomacromolecules. 2003;4:1457–1465. doi: 10.1021/bm034130m. [DOI] [PubMed] [Google Scholar]
- 17.Milewska A., Ciejka J., Kaminski K., Karewicz A., Bielska D., Zeglen S., Karolak W., Nowakowska M., Potempa J., Bosch B.J., Pyrc K., Szczubialka K. Antiviral Res. 2013;97:112–121. doi: 10.1016/j.antiviral.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Theerawanitchpan G., Saengkrit N., Sajomsang W., Gonil P., Ruktanonchai U., Saesoo S., Flegel T.W., Saksmerprome V. J. Biotechnol. 2012;160:97–104. doi: 10.1016/j.jbiotec.2012.04.011. [DOI] [PubMed] [Google Scholar]
- 19.Bai B., Mi X., Xiang X., Heiden P.A., Heldt C.L. Carbohydr. Res. 2013;380:137–142. doi: 10.1016/j.carres.2013.08.020. [DOI] [PubMed] [Google Scholar]
- 20.Desai K., Kit K., Li J., Zivanovic S. Biomacromolecules. 2008;9:1000–1006. doi: 10.1021/bm701017z. [DOI] [PubMed] [Google Scholar]
- 21.Vondran J.L., Sun W., Schauer C.L. J. Appl. Polym. Sci. 2008;109:968–975. [Google Scholar]
- 22.Qin X.-H., Wang S.-Y. J. Appl. Polym. Sci. 2008;109:951–956. [Google Scholar]
- 23.Xiao B., Wan Y., Wang X.Y., Zha Q.C., Liu H.M., Qiu Z.Y., Zhang S.M. Colloid Surf., B. 2012;91:168–174. doi: 10.1016/j.colsurfb.2011.10.053. [DOI] [PubMed] [Google Scholar]
- 24.Han B., Li J., Chen C., Xu C., Wickramasinghe S.R. Chem. Eng. Res. Des. 2003;81:1385–1392. [Google Scholar]
- 25.Young R.J., Lovell P.A. 3rd ed. CRC Press; 2011. Introduction to Polymers. [Google Scholar]
- 26.Lee Y.J., Shin D.S., Kwon O.W., Park W.H., Choi H.G., Lee Y.R., Han S.S., Noh S.K., Lyoo W.S. J. Appl. Polym. Sci. 2007;106:1337–1342. [Google Scholar]
- 27.Han S.O., Son W.K., Youk J.H., Park W.H. J. Appl. Polym. Sci. 2008;107:1954–1959. [Google Scholar]
- 28.Kempf C., Stucki M., Boschetti N. Biologicals. 2007;35:35–42. doi: 10.1016/j.biologicals.2006.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.EPA, Safe Drinking Water Act, 1974.
- 30.Heldt C.L., Hernandez R., Mudiganti U., Gurgel P.V., Brown D.T., Carbonell R.G. J. Virol. Methods. 2006;135:56–65. doi: 10.1016/j.jviromet.2006.01.022. [DOI] [PubMed] [Google Scholar]
- 31.Tafur M.F., Vijayaragavan K.S., Heldt C.L. Antiviral Res. 2013;99:27–33. doi: 10.1016/j.antiviral.2013.04.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This document file contains supplementary data.


