Highlights
► Direct antiviral effect of purified Surface layer protein (S-layer) of Lactobacillus acidophilus. ► Inhibition of infection by Junin Virus that is mediated by DC-sign lectin. ► Basic nature of the S-layer is involved in the interaction with the viral receptor. ► Novel functionality for the S-layer not previously described including Carbohydrate recognition. ► Results support the probiotic status described for this lactobacilli strain.
Keywords: Surface layer, Lactobacillus, Antiviral
Abstract
It has been previously described that S-layer binds to the C-type lectin DC-specific ICAM-3-grabbing nonintegrin (DC-SIGN, CD209). It was also shown that DC-SIGN is a cell-surface adhesion factor that enhances viral entry of several virus families. Among those, Junin virus (JUNV) entry is enhanced in cells expressing DC-SIGN and for that reason surface-layer protein (S-layer) of Lactobacillus acidophilus ATCC 4365 was evaluated as a possible JUNV inhibitor. Experiments using 3T3 cells stably expressing DC-SIGN, showed an almost complete inhibition of JUNV infection when they were treated with S-layer in a similar extend as the inhibition shown by mannan. However no inhibition effect was observed in 3T3 wild type cells or in 3T3 cells expressing liver/lymph node-specific ICAM-3 grabbing nonintegrin (L-SIGN or DC-SIGNR or CD209L).
Treatments with S-layer during different times in the infection demonstrated that inhibition was only observed when S-layer was presented in early stages of the viral infection. This inhibition does not involve the classic recognition of mannose by this C-type lectin as the S-layer showed no evidence to be glycosylated. In fact, the highly basic nature of the S-layer (pI > 9.5) seems to be involved in electrostatic interactions between DC-SIGN and S-layer, since high pH abolished the inhibitory effect on infection cause by the S-layer. In silico analysis predicts a Ca2+-dependant carbohydrate recognition domain in the SlpA protein.
This novel characteristic of the S-layer, a GRAS status protein, contribute to the pathogen exclusion reported for this probiotic strain and may be applied as an antiviral agent to inhibit several kinds of viruses.
1. Introduction
Many species of the genus Lactobacillus possess surface layer (S-layer) proteins which are arrays of a single protein noncovalently bound that constitute the outermost cell envelope also found in several members of Bacteria and Archaea. One of the main characteristics of Lactobacillus S-layer proteins is their high isoelectric point (pI) showing them as highly basic [1]. Lactobacillus acidophilus is one of the major species found in human intestines and some strains have probiotic characteristics making them GRAS (generally recognized as safe). Probiotics are live microorganisms, usually contained in food, traditionally regarded as safe for human consume that, when ingested in sufficient number, play an important role in control of the host intestinal microbiota and in the modulation of the host immune response. L. acidophilus ATCC4356 has been described as a probiotic strain [2] with a 45 kDa S-layer protein highly basic with a calculated pI of 9.59, (Expasy) that was recently showed to have an antibacterial endopeptidase activity [3], [4]. S-layers have also been considered to function as protective coats, maintenance of cell shape and ion exchangers and be involved in adhesion to biotic and abiotic surfaces [5], [6]. Although many studies focus on the structure of the S-layer, their biological functions remains poorly understood. S-layer would account for the probiotic properties of some strains [7] being capable of influencing the immune response [8] and favoring cell adhesion [9], [10], [11], [12]. Several reports have involved the S-layer proteins in the enteric pathogen exclusion phenomena [13], [14].
In the gastro-intestinal tract (GIT) L. acidophilus regularly encounters many antigens presenting cell, such as dendritic cells (DCs) [15]. These cells express DC-specific ICAM-3-grabbing nonintegrin protein (DC-SIGN), which is a cell-surface receptor, that is express mainly in DCs and recognize mannose and fructose glycans that are present on microbial and viral surfaces. DCs play a very important role in the innate and adaptive immune response [16]. It has been shown that DC-SIGN can act enhancing viral entry of different viruses such as HIV type 1, hepatitis C, Ebola, Dengue and SARS [17], [18], [19]. It was also shown that DCs interact with L. acidophilus; this contact involve the DC-SIGN and the S-layer presented on the bacterial cell-envelope, and regulates the induction of a number of cytokines involved in cellular immune regulation [8], [20].
JUNV, a New World arenavirus, is the etiological agent of the Argentine Hemorrhagic Fever (AHF) [21], and endemo-epidemic disease that causes hemorrhagic and neurological complications. AHF presents mortality rates from 20–30% and mainly affects population on the fertile farming land of Argentina [22]. It has been shown that JUNV uses the human transferrin receptor 1 (TfR1) to infect human cells [23]. In addition to its primary receptor (TfR1), JUNV virus infection was shown to be enhanced by C-type lectins (Martinez & Candurra, unpublished results). Among the C-type lectins DC-SIGN and the Liver/lymph node–specific ICAM-3-grabbing nonintegrin (L-SIGN) were studied. DC-SIGN and L-SIGN are human homologs, share 77% of the amino-acid sequence and has function similarities. The role of these receptors (DC- and L-SIGN) in the infection of several major importance viruses placed them as a target for antiviral agents. The most common compounds used so far are carbohydrate-binding agents that can interact with the glycoprotein present on the virus surface and prevent the infection [24].
Lactobacilli have been described to have antiviral activity [25] however the nature of this effect has not been clearly established. In this work we evaluated the effect of the purified S-layer protein of L. acidophilus ATCC 4353 on JUNV infection to provide evidence on its role in pathogen exclusion and in the search for new antiviral agents with GRAS status. We used 3T3 cells stably expressing DC-SIGN, or its homolog L-SIGN or Vero cells for transient expression as in vitro models. Since 3T3 cells are poorly infect by JUNV in the absence of expression of C-type lectins this model provides a really strong tool to study the effect of S-layer protein on the infection enhanced by DC-SIGN or L-SIGN.
2. Materials and methods
2.1. Isolation of S-layer proteins
The S-layer proteins were extracted from overnight L. acidophilus ATCC 4356 cells grown in MRS medium at 37 °C, by using 6 M LiCl, extensively dialyzed against distilled water overnight at 4 °C and after centrifugation (10,000g 20 min) suspended in sterile PBS and store at −20 °C [3].
2.2. Cell culture
Monolayers of Vero cells (ATCC CCL 81) were grown in MEM containing 5% heat-inactivated fetal bovine serum (FBS) and supplemented with 50 μg/ml gentamycin. Maintenance medium (MM) consisted of MEM containing 1.5% fetal calf serum, and titration medium consisted of MEM 2X, 2% FBS and methyl cellulose (0.7%). 3T3 cells (ATCC CCL 1658) and 3T3-derived DC-SIGN and L-SIGN cells (NIAID AIDS Research and Reference Reagent Program) were grown in MEM supplemented with 10% FBS. In all cases, cultures were grown in 5% CO2 and were supplemented with HEPES (20 μM).
2.3. Viruses
The naturally attenuated Junin IV4454 strain was used in all experiments with live virus, and propagated in Vero cells. Virus yields were then determined by plaque formation (PFU) assays in Vero cells. The titers of the JUNV stock suspensions were 2–8 × 106 PFU/ml.
2.4. Reagents and antibodies
FITC-goat anti mouse and mannan were purchased from Sigma. Anti-JUNV monoclonal antibodies NA05-AG12 were kindly donated by Dr. A. Sanchez (Centers for Disease Control, Atlanta GA, USA). Anti-hDC-SIGN and anti-hL-SIGN MAbs (120526, 9E9A8, 14EG7) were obtained from the NIAID AIDS Research and Reference Reagent Program.
2.5. Inhibition of JUNV infectivity
Confluent 3T3 DC-SIGN cells grown in 24-well plates were treated with different concentrations of S-layer for 1 h. After the treatment period, the medium was removed and the cells were infected in the presence of the protein. After infection, the inoculum was removed and fresh MM was added. The viral supernatants were harvested after 24 h and JUNV yields were determined by PFU assays. Viability of cell cultures under each treatment condition was determined by the MTT assay.
2.6. Immunofluorescence assays
3T3 WT, DC-SIGN o L-SIGN expressing cells were grown on glass coverslips for 24 h. After this period cells were treated with the appropriate concentration of S-layer for 60 min and then infected with JUNV (m.o.i. = 1) or in other cases first infected with JUNV (m.o.i. = 1) and then treated with S-layer protein. After a 24 h incubation period, cells were rinsed three times with PBS, fixed with 4% formaldehyde, permeabilized with 0.2% Triton X-100 and incubated with a MAb specific for JUNV NP (NA05-AG12). After washing, anti-JUNV bound antibody was detected by incubation with FITC-goat anti-mouse antibody. Samples were examined using an Olympus BX51 microscope and the average number of positive and non infected cells on each coverslip was calculated as the average of 20 optical fields chosen randomly. Data is expressed as percentage of virus positive cells over the number of total cells.
When pH treatment was performed 3T3 DC-SIGN or L-SIGN cells were treated with S-layer protein (400 μg/ml) during 60 min. After this period control cell were rinsed during one min with PBS or with basic pH medium. Then the cells were infected with JUNV (m.o.i. = 1). After 60 min of infection the virus inoculum was removed and fresh MM was added. 24 h after the infection cells were fixed with 4% formaldehyde and proceed for IF as described in the previous paragraph.
2.7. Transfection
Transfection was performed according to the Lipofectamine2000 manufacturer (Invitrogen).
2.8. Glycoprotein staining
Glycosylation detection was performed with periodic acid-Schiff’s reagent (PAS) according to the procedure of Segrest et al. [26] and Pro-Q_Emerald 300 Glycoprotein Gel Stain Kit (Invitrogen) on SDS–polyacrylamide gels. This fluorescence-based staining method allows the detection of less than 0.5 ng glycoprotein which is 50 times more sensitive than PAS staining.
2.9. Sequence-based analysis
Free access sites were used to predict protein structure and function for the S-layer protein SlpA CAA61560.1 [L. acidophilus ATCC 4356] and compared to possible orthologous proteins.
2.10. URL links
Protparam (http://web.expasy.org/cgi-bin/protparam)
CDART (http://www.ncbi.nlm.nih.gov/Structure/lexington/lexington.cgi)
Clustal-W (http://www.ch.embnet.org/software/ClustalW.html)
I-TASSER 3D structure and function prediction platform (http://zhanglab.ccmb.med.umich.edu/I-TASSER)
SMART, Simple modular architecture research tool (http://smart.embl-heidelberg.de/smart/do_annotation.pl?DOMAIN=CLECT)
3. Results
3.1. S-layer treatment blocks JUNV infection by C-type lectins
We infected 3T3 cells wt or stably expressing C-type lectins with JUNV. As it can be seen in Fig. 1 A 3T3 cells are poorly infected with JUNV in the absence of DC-sign or L-sign expression. To study the effect of S-layer protein on JUNV infection 3T3 cells and 3T3-derived stably expressing DC-SIGN and L-SIGN cells were incubated with different concentrations of S-layer protein, infected with JUNV and then cells expressing viral nucleoprotein (NP) were quantify. The results showed reduction in the infection of JUNV in 3T3DC-SIGN cells, but none inhibition was shown in 3T3 L-SIGN cell (Fig. 1A). When we performed a similar experiment but adding the S-layer protein after the infection with JUNV, there was almost no effect of this treatment in cell expressing JUNV NP. This suggests that the effect of the S-layer treatment in JUNV infection is only in an early step of the replication cycle, not affecting later stages (Fig. 1B).
Fig. 1.
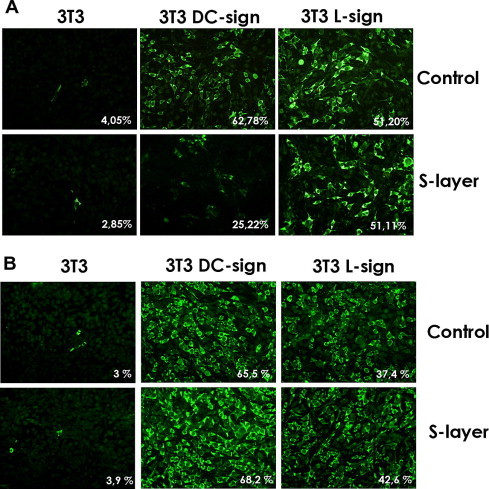
S-layer blocks virus infection at early stages. 3T3, DC-sign 3T3 or L-sign 3T3 cells treated with S-layer (400 μg/ml) for 60 min prior to infection (A) and then infected with JUNV at a m.o.i. = 1 or cells were infected (B) and then treated with S-layer. Fixed cells (24 h p.i.) were stained for IF microscopy. Images from three independent experiments were captured and quantified. Data are indicated in each figure as a percentage of virus positive cells over the number of total cells. Magnifications 400×.
To provide further evidence that blocking in the infection with S-layer was due to the presence of DC-SIGN but not L-SIGN, we performed transient transfection of Vero cells with plasmids expressing hDC-SIGN or hL-SIGN. After 24 h of the transfection the cells were treated with the S-layer protein and then infected with JUNV (Table 1 ). This confirms that the S-layer inhibitory effect was exclusive to cells expressing DC-SIGN but not L-SIGN.
Table 1.
Transient expression of lectins on Vero cells. Vero cells were transfected with plasmids encoding hDC-SIGN or hL-SIGN. 24 h post-transfection, cells were treated with S-layer (400 μg/ml), mannan (100 μg/ml) or left untreated (control) and then infected with JUNV (m.o.i. = 1). Cells were fixed at 24 h p.i. and stained for IF microscopy. Lectin expression was verified using anti hDC-/hL-sign antibodies provided by NIH (11423). Data is indicated as a mean of the percentage of inhibition relative to each control. Results from three independent experiments were captured and quantified and are shown.
| % of Inhibition | ||
|---|---|---|
| L-sign | DC-sign | |
| Mannan | 10.9 ± 1.1 | 13.6 ± 1.2 |
| S-layer | 2.8 ± 0.2 | 26.3 ± 2.5 |
To further investigate the potential of the S-layer protein to act as an inhibitor for JUNV infection 3T3 DC-SIGN cells were incubated with different concentrations of S-layer for 1 h ant then infected with JUNV. Then, 24 h post infection we measure viral production by PFU assay and quantified viral NP by IF (Fig. 2 ). As it is shown in Fig. 2A when we increased the concentration of S-layer we observe less JUNV infection. However when we exceed 500 μg/ml, where it is likely to have auto-assembly of a crystalline S-layer array, the inhibitory effect was suppressed. Virucidal effect on JUNV was assayed by treatment with S-layer during 90 min at 37 °C. No difference with the untreated control was observed by infectivity assay (Fig. 2C). This indicates that the S-layer protein have no effect on the viral particles. Cytotoxicity effect of the protein S-layer protein in 3T3 cells was also studied: no toxicity was found in the concentration of S-layer needed for the inhibition (Fig. 2B).
Fig. 2.
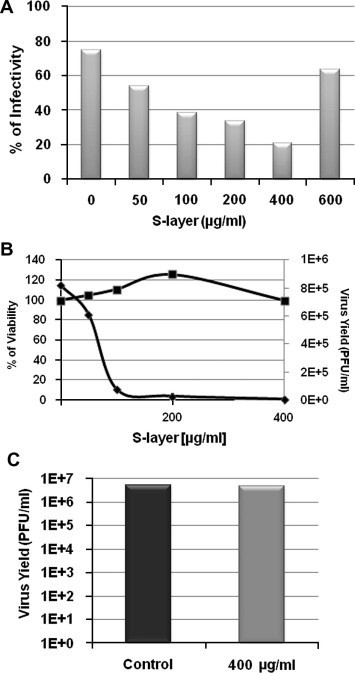
Inhibitory effect of S-layer on virus infection, dose response curves, cell viability analysis and virucidal activity. 3T3 DC-sign cells were treated with S-layer during 60 min and infected with JUNV (m.o.i. = 1). 24 h post infection supernatants were collected and cells were fixed for IF. (A) JUNV NP was detected by IF using anti-NP monoclonal antibodies and detected by IF. Data was expressed as percentage of positive cells in 3 independent experiments. (B) Cell viability was determined by MTT assay after a 24 h treatment. Supernatants from IF (A) were titrated by PFU assay. (C) Viral stocks were treated with S-layer protein, or only with medium (control), during 90 min at 37 °C. Then infectivity of the treated virus was studied by PFU assay in Vero cells.
3.2. Effect of the pH in the S-layer treatment to block JUNV infection cycle
Previous reports suggested that DC-SIGN and the S-layer protein interact [8]. To determine glycosylation state of the S-layer preparation, two assays were performed as shown in methods (Fig. 3 ). Results showed no glycosylation therefore S-layer protein of L. acidophilus recognition of DC-SIGN does not involve the classic mannose interaction.
Fig. 3.
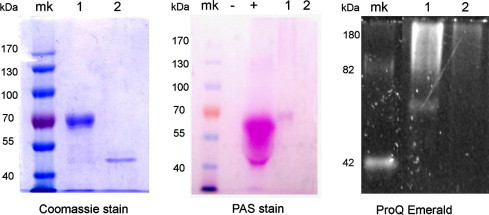
Absence of glycosylation in Lactobacillus acidophilus ATCC 4356 S-layer protein. Periodic acid-Schiff’detection (PAS) and ProQ Emerald 330 fluorescent detection were performed to detect glycosylation. For PAS assay Bovine serum albumin served as negative control (−) and TNFα was used as positive control (+). In ProQ Emerald the molecular weight standard CandyCane was used both as molecular marker and glycosylation control. S-layer protein from Lactobacillus kefir strains JCM 5818 is in Lane 1. S-layer protein from Lactobacillus acidophilus ATCC 4356 is in Lane 2.
To verify if the highly basic nature of the S-layer (pI > 9.5) was responsible for electrostatic interactions that could be responsible for the binding with DC-SIGN, interfering with JUNV interaction with the lectin, basic pH treatments were assayed. Increasing the pH in the medium reduced the inhibition of infection by JUNV in DC-SIGN expressing cells, due to a change in S-layer net charge (pI 9.59) (Fig. 4 A). This effect was not observed in treatments of L-SIGN expressing cells, where no virus inhibition was observed (Fig. 4B), indicating that there was not modification of the infectivity due to basic pH. Chelating effects were also verified to influence the interaction, reducing the inhibitory effect on virus infection (not shown).
Fig. 4.
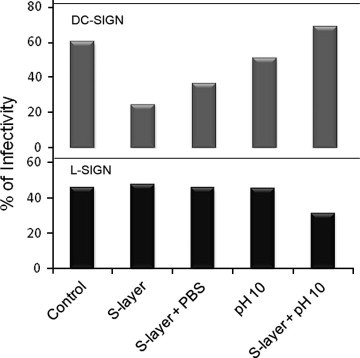
3T3 DC-SIGN (A) and 3T3 L-SIGN (B) cells were treated with S-layer (400 μg/ml) for 60 min. After treatment cells were rinsed during 1 min with PBS as control or with a basic pH buffer (pH 10). As control we also used non S-layer treated cells without pH treatment and with the pH treatment. Then cell cultures were infected with JUNV at a m.o.i. of 1. After 60 min of infection the virus was removed and cells were fixed at 24 h postinfection. Fixed cells were stained for IF microscopy with the anti-JUNV NP (NA05AG12). Images from three independent experiments were captured and quantified. Data is indicated as a percentage of virus positive cells over the number of total cells.
3.3. Bioinformatics analysis
We presumed that the C-terminal portion of the S-layer would be responsible of the interaction with the glycan strand of DC-SIGN, since we have already shown it to interact with cell wall peptidoglycan [3] and it has been reported, that two repeats sequences in the C-terminal portion of the protein showed homology to carbohydrate binding domains (CBD) of Clostridium difficile S-layer/toxin [27], [28]. The repeated sequences are a general theme for protein-carbohydrate interaction described for extracellular virulence factors of Gram positive bacteria [29]. SlpA protein sequence launched in CDART showed that the C-terminal part, Domain SLAP pfam03217, aligned with structural homology with putative CBD of a subgroup of bacterial protein homologous C-type lectin-like (CLECT or CTLD domain), protein domains homologous to the carbohydrate-recognition domains (CRDs) of the C-type lectins (CLECT cd03603 and CLECT_VCBS cd00037) however bacterial CTLDs within this group are functionally uncharacterized. When only the amino-acid sequence of SLAP pfam03217 domain (127 aa C-terminal portion) was used as query for SMART analysis two internal repeats not previously revealed with the whole protein sequence, because they overlap with pfam domain, were actually found with good accuracy (E-value 3.43e-05). The two internal repeats showed 33% identity (Table 2 ). Total number of positively charged residues (Arg + Lys) within the repeats gives a predicted pI of 9.26 and 9.90 for each sequence and supports the carbohydrate binding capacity suggested (Table 2). When using I-TASSER for 3D structure and function prediction, result obtained showed similarity to Ca2+ binding protein. The Consensus Prediction of Gene Ontology terms obtain include functions of cell adhesion, protein binding and surface expression (http://zhanglab.ccmb.med.umich.edu/I-TASSER/output/S97651). In summary these analyses results suggest that the S-layer of L. acidophilus is predicted to have a carbohydrate binding capacity.
Table 2.
Internal repeats obtained with SMART within SLAP domain (pfam03217) and Clustal W alignment with Consensus Von Eichel-Streiber Carbohydrate binding sites [29]. pI was predicted with Protparam (Expasy). Consensuses are in italic or bold letters. (K + R) indicate positively charged residues (Arg + Lys), N° for number of residues and position within SlpA protein. Symbols:”|” identical aminoacid, “:” indicates group similarity, “.” indicates low group similarity. Amino acid notation according to IUPAC-IUB-CBN.
 |
4. Discussion
Cells stably expressing DC-SIGN can be infected by JUNV, but an almost complete inhibition of JUNV infection was shown when they were treated with purified S-layer protein from L. acidophilus ATCC 4365 prior to infection (Fig. 1). This effect was not observed in cell lines that do not express this C-type lectin. Post infection treatments with S-layer did not have an effect in viral infection, leading to the idea that the inhibition caused by S-layer was due to an early effect during viral replication cycle. Since no virucidal effect was observed for the S-layer, it seems that the target is not the viral particle but the direct interaction with the DC-SIGN. This inhibitory effect is a novel characteristic of the S-layer protein of L. acidophilus, which could account for the pathogen exclusion effect described for this probiotic bacterium. The S-layer is a highly stable protein with no toxicity to cells as cell viability was higher than 95% (Fig. 2).
Differences in binding properties between DC-SIGN and L-SIGN to its target have been reported [30]. Therefore it is not surprising the lack of an inhibitory effect in cell expressing L-SIGN (Fig. 1 and Table 1). The mechanism of interaction involved it is not expected to be the classic mannose recognition, known for this kind of lectins. In fact no glycosylation was observed in the S-layer preparation (Fig. 3). It has been previously described that DC-SIGN forms a tetramer that enables multivalent interaction with pathogens. DC-SIGN-pathogen complex can be internalized into compartments with acid pH, and it was shown that there is a pH sensitive control of the oligomerization state of this complex [31]. Therefore we believe that the highly basic nature (pI > 9.5) of this particular S-layer protein might be responsible of interfering with the calcium-dependent viral interaction by structural and electrostatic forces (Fig. 4).
The interaction of the S-layer protein that lacks glycan strands with the DC-SIGN was predicted by bioinformatics methods. It is interesting to remark that this novel mechanism of interaction between SlpA that lacks glycan strands and the DC-SIGN resembles the interaction between non glycosylated C-type animal lectins with their ligands, for example galectin-1 protein [32]. This family of soluble proteins displays conserved CRD that recognizes glycans in its glycoprotein ligand. The repeating sequences described in SlpA (Table 2) are a general theme for protein–carbohydrate interaction [29] and the S-layer protein is proposed to interact with carbohydrate moieties in a calcium dependant manner.
DC-SIGN has been shown to determine susceptibility to HIV type 1, hepatitis C virus, Ebola virus, cytomegalovirus, Dengue virus and SARS coronavirus [17], [18], [19]. The results obtained in this in vitro model, suggest that S-layer could also inhibit these viruses that use DC-SIGN for viral entry.
Although a great deal of work needs to be done to address the type of interaction predicted between the S-layer protein with the DC-SIGN, this is the first report of a direct viral inhibition by the purified S-layer protein that support the probiotic status of the strain.
Acknowledgments
This work was supported by research Grants from the Consejo Nacional de Investigaciones Científicas y Técnicas from Argentina (CONICET) and Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT-MINCyT).
References
- 1.Deepika G., Charalampopoulos D. Surface and adhesion properties of lactobacilli. Adv. Appl. Microbiol. 2010;70:127–152. doi: 10.1016/S0065-2164(10)70004-6. [DOI] [PubMed] [Google Scholar]
- 2.Resta-Lenert S., Barrett K.E. Live probiotics protect intestinal epithelial cells from the effects of infection with enteroinvasive Escherichia coli (EIEC) Gut. 2003;52:988–997. doi: 10.1136/gut.52.7.988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prado Acosta M., Palomino M.M., Allievi M.C., Sanchez Rivas C., Ruzal S.M. Murein hydrolase activity in the surface layer of Lactobacillus acidophilus ATCC 4356. Appl. Environ. Microbiol. 2008;74:7824–7827. doi: 10.1128/AEM.01712-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prado-Acosta M., Ruzal S.M., Allievi M.C., Palomino M.M., Sanchez Rivas C. Synergistic effects of the Lactobacillus acidophilus surface layer and nisin on bacterial growth. Appl. Environ. Microbiol. 2010;76:974–977. doi: 10.1128/AEM.01427-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sleytr U.B., Huber C., Ilk N., Pum D., Schuster B., Egelseer E.M. S-layers as a tool kit for nanobiotechnological applications. FEMS Microbiol. Lett. 2007;267:131–144. doi: 10.1111/j.1574-6968.2006.00573.x. [DOI] [PubMed] [Google Scholar]
- 6.Allievi M.C., Sabbione F., Prado-Acosta M., Palomino M.M., Ruzal S.M., Sanchez-Rivas C. Metal biosorption by surface-layer proteins from Bacillus species. J. Microbiol. Biotechnol. 2011;21:147–153. doi: 10.4014/jmb.1009.09046. [DOI] [PubMed] [Google Scholar]
- 7.Beganović J., Frece J., Kos B. Functionality of the S-layer protein from the probiotic strain Lactobacillus helveticus M92. Antonie Van Leeuwenhoek. 2011 doi: 10.1007/s10482-011-9563-4. [DOI] [PubMed] [Google Scholar]
- 8.Konstantinov S.R., Smidt H., de Vos W.M. S layer protein A of Lactobacillus acidophilus NCFM regulates immature dendritic cell and T cell functions. Proc. Natl. Acad. Sci. USA. 2008;105:19474–19479. doi: 10.1073/pnas.0810305105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frece J., Kos B., Svetec I.K. Importance of S-layer proteins in probiotic activity of Lactobacillus acidophilus M92. J. Appl. Microbiol. 2005;98:285–292. doi: 10.1111/j.1365-2672.2004.02473.x. [DOI] [PubMed] [Google Scholar]
- 10.Buck B.L., Altermann E., Svingerud T., Klaenhammer T.R. Functional analysis of putative adhesion factors in Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 2005;71:8344–8351. doi: 10.1128/AEM.71.12.8344-8351.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jakava-Viljanen M., Palva A. Isolation of surface (S) layer protein carrying Lactobacillus species from porcine intestine and faeces and characterization of their adhesion properties to different host tissues. Vet. Microbiol. 2007;124:264–273. doi: 10.1016/j.vetmic.2007.04.029. [DOI] [PubMed] [Google Scholar]
- 12.Liu Z., Shen T., Zhang P., Ma Y., Qin H. Lactobacillus plantarum surface layer adhesive protein protects intestinal epithelial cells against tight junction injury induced by enteropathogenic Escherichia coli. Mol. Biol. Rep. 2011;38:3471–3480. doi: 10.1007/s11033-010-0457-8. [DOI] [PubMed] [Google Scholar]
- 13.Johnson-Henry K.C., Hagen K.E., Gordonpour M. Surface-layer protein extracts from Lactobacillus helveticus inhibit enterohaemorrhagic Escherichia coli O157 H7 adhesion to epithelial cells. Cell. Microbiol. 2007;9:356–367. doi: 10.1111/j.1462-5822.2006.00791.x. [DOI] [PubMed] [Google Scholar]
- 14.Li P., Yin Y., Yu Q., Yang Q. Lactobacillus acidophilus S-layer protein-mediated inhibition of Salmonella-induced apoptosis in Caco-2 cells. Biochem. Biophys. Res. Commun. 2011 doi: 10.1016/j.bbrc.2011.04.131. [DOI] [PubMed] [Google Scholar]
- 15.Penders J., Thijs C., Mommers M. Intestinal Lactobacilli and the DC-SIGN gene for their recognition by dendritic cells play a role in the aetiology of allergic manifestations. Microbiology. 2010;156:3298–3305. doi: 10.1099/mic.0.042069-0. [DOI] [PubMed] [Google Scholar]
- 16.Khoo U.S., Chan K.Y., Chan V.S., Lin C.L. DC-SIGN and L-SIGN the SIGNs for infection. J. Mol. Med. (Berl.) 2008;86:861–874. doi: 10.1007/s00109-008-0350-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoorelbeke B., Van Damme E.J., Rougé P. Differences in the mannose oligomer specificities of the closely related lectins from Galanthus nivalis and Zea mays strongly determine their eventual anti-HIV activity. Retrovirology. 2011;8:10. doi: 10.1186/1742-4690-8-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bertaux C., Daelemans D., Meertens L. Entry of hepatitis C virus and human immunodeficiency virus is selectively inhibited by carbohydrate-binding agents but not by polyanions. Virology. 2007;366:40–50. doi: 10.1016/j.virol.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 19.Alen M.M., Kaptein S.J., De Burghgraeve T. Antiviral activity of carbohydrate-binding agents and the role of DC-SIGN in dengue virus infection. Virology. 2009;387:67–75. doi: 10.1016/j.virol.2009.01.043. [DOI] [PubMed] [Google Scholar]
- 20.Weiss G., Rasmussen S., Zeuthen L.H. Lactobacillus acidophilus induces virus immune defence genes in murine dendritic cells by a Toll-like receptor-2-dependent mechanism. Immunology. 2010;131:268–281. doi: 10.1111/j.1365-2567.2010.03301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parodi S., Greenway D.J., Rugiero H.R., Frigerio M., De La Barrera J.M., Mettler N., Garzon F., Boxaca M., Guerrero L., Nota N. Concerning the epidemic outbreak in Junin. Dia. Med. 1958;30:2300–2301. [PubMed] [Google Scholar]
- 22.Weissenbacher M.C., Laguens R.P., Coto C.E. Argentine hemorrhagic fever. Curr. Top. Microbiol. Immunol. 1987;134:79–116. doi: 10.1007/978-3-642-71726-0_4. [DOI] [PubMed] [Google Scholar]
- 23.Radoshitzky S.R., Abraham J., Spiropoulou C.F. Transferrin receptor 1 is a cellular receptor for New World haemorrhagic fever arenaviruses. Nature. 2007;446:92–96. doi: 10.1038/nature05539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balzarini J. Targeting the glycans of glycoproteins a novel paradigm for antiviral therapy. Nat. Rev. Microbiol. 2007;5:583–597. doi: 10.1038/nrmicro1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Conti C., Malacrino C., Mastromarino P. Inhibition of herpes simplex virus type 2 by vaginal lactobacilli. J. Physiol. Pharmacol. 2009;60:19–26. [PubMed] [Google Scholar]
- 26.Segrest J.P., Jackson R.L., Marchesi V.T. Red cell membrane glycoprotein amino acid sequence of an intramembranous region. Biochem. Biophys. Res. Commun. 1972;49:964–969. doi: 10.1016/0006-291x(72)90306-3. [DOI] [PubMed] [Google Scholar]
- 27.Smit E., Oling F., Demel R., Martinez B., Pouwels P.H. The S-layer protein of Lactobacillus acidophilus ATCC 4356: identification and characterisation of domains responsible for S-protein assembly and cell wall binding. J. Mol. Biol. 2001;305:245–257. doi: 10.1006/jmbi.2000.4258. [DOI] [PubMed] [Google Scholar]
- 28.Smit E., Pouwels P.H. One repeat of the cell wall binding domain is sufficient for anchoring the Lactobacillus acidophilus surface layer protein. J. Bacteriol. 2002;184:4617–4619. doi: 10.1128/JB.184.16.4617-4619.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.von Eichel-Streiber C., Sauerborn M., Kuramitsu H.K. Evidence for a modular structure of the homologous repetitive C-terminal carbohydrate-binding sites of Clostridium difficile toxins and Streptococcus mutans glucosyltransferases. J. Bacteriol. 1992;174:6707–6710. doi: 10.1128/jb.174.20.6707-6710.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van Liempt E., Imberty A., Bank C.M. Molecular basis of the differences in binding properties of the highly related C-type lectins DC-SIGN and L-SIGN to Lewis X trisaccharide and Schistosoma mansoni egg antigens. J. Biol. Chem. 2004;279:33161–33167. doi: 10.1074/jbc.M404988200. [DOI] [PubMed] [Google Scholar]
- 31.Tabarani G., Thépaut M., Stroebel D. DC-SIGN neck domain is a pH-sensor controlling oligomerization SAXS and hydrodynamic studies of extracellular domain. J. Biol. Chem. 2009;284:21229–21240. doi: 10.1074/jbc.M109.021204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Kooyk Y., Rabinovich G.A. Protein–glycan interactions in the control of innate and adaptive immune responses. Nat. Immunol. 2008;9:593–601. doi: 10.1038/ni.f.203. [DOI] [PubMed] [Google Scholar]


