Abstract
Chameleon sequences can adopt either alpha helix sheet or a coil conformation. Defining chameleon sequences in PDB (Protein Data Bank) may yield to an insight on defining peptides and proteins responsible in neurodegeneration. In this research, we benefitted from the large PDB and performed a sequence analysis on Chameleons, where we developed an algorithm to extract peptide segments with identical sequences, but different structures. In order to find new chameleon sequences, we extracted a set of 8315 non-redundant protein sequences from the PDB with an identity less than 25%. Our data was classified to “helix to strand (HE)”, “helix to coil (HC)” and “strand to coil (CE)” alterations. We also analyzed the occurrence of singlet and doublet amino acids and the solvent accessibility in the chameleon sequences; we then sorted out the proteins with the most number of chameleon sequences and named them Chameleon Flexible Proteins (CFPs) in our dataset. Our data revealed that Gly, Val, Ile, Tyr and Phe, are the major amino acids in Chameleons. We also found that there are proteins such as Insulin Degrading Enzyme IDE and GTP-binding nuclear protein Ran (RAN) with the most number of chameleons (640 and 405 respectively). These proteins have known roles in neurodegenerative diseases. Therefore it can be inferred that other CFP's can serve as key proteins in neurodegeneration, and a study on them can shed light on curing and preventing neurodegenerative diseases.
Keywords: Protein secondary structure, Chameleon sequences, Neurodegenerative diseases, Sequence properties, Enrichment analysis
1. Introduction
Anfinsen proposed that the proteins are predisposed to fold into a unique three dimensional structure which is clearly specified by its amino acid sequence, according to Anfinsen's dogma, the amino acid sequence of a protein contains sufficient information to determine its three-dimensional structure [1]. This broadly accepted theory was used as the central dogma in predicting the secondary and tertiary structures from its sequence alone following the pioneer work of Chou and Fasman [2]. Anfinsen's theory was shaken by the discovery of the chameleon sequences which can fold as different secondary structures in proteins, these sequences are abundant in nature and play a crucial role in human diseases and are said to constitute as part of the proteome named ‘unfoldome’ [3]. These segments with ambivalent structures were first reported by Kabsch and Sander [4]. Examples of related studies are the involvement of chameleon sequences in the induction of misfolding diseases such as amyloid fibril formation of neurodegeneration [5], [6], [7], [8]. Neurodegenerative diseases including Alzheimer's, Parkinson's, Huntington's, Creutzfeldt-Jakob disease, etc. involve a series of brain proteins named amyloid proteins, which upon interaction of the neuronal membranes monomers of amyloid proteins undergo an alpha helix to sheet shift in their conformation. These sequences are also suggested to be one of the limiting factors for the accuracy of secondary structure prediction methods and one important reason for misprediction of programs designed for protein secondary structures is the structural diversity among the peptides with the same sequence i.e. the chameleons [9], [10], [11], [12], [13]. The relationship of an amino acid sequence to its eventual structures is important for the structural prediction and design purposes as well as for the comprehension of diseases caused by a protein conformation [10], [14], [15], and the identification of the propensity values would provide local sequence information for predicting secondary structures [16], [17].
Many studies showed that the sequence neighboring in secondary structure of proteins is important in forming these particular structures [18], [19]. It has been mentioned that the amino acid propensities for secondary structures can be still improved to obtain better predictive results and to reveal important structural information's [20]. Different amino acids have different preferences for their neighbors and that these local interactions are crucial for their structural conformation, and are also used in the secondary structure prediction methods [21], [22], [23], [24], [25]. Due to the importance of the chameleon sequences [5], [11], [13] and the involvement of the local amino acid interactions in secondary structure formation, we hereby present a comprehensive meta-analysis of single and double propensity of amino acids in chameleon sequences of the Protein databank (PDB), in order to find proteins with the most number of chameleons and name them Chameleon Flexible Proteins (CFPs). We have also built different chameleon groups corresponding to helix, strand and coil, along with the analysis of their solvent accessibility, presenting their singlet and doublet amino acid propensities.
2. Methods
2.1. Databases
A 8315 non-redundant protein chains in the PDB database was used for gathering the chameleon sequences. This set was generated from the current version of the PDB (Dec. 2014) [26] using the PISCES protein sequence algorithm [27] which provided the most up-to-date collection with the following criteria of non-redundant PDB chain database by the selection method of Hobohm et al. [28].
Experimental method = X-ray crystallography, maximum resolution 2.5 A°, maximum R-value 0.3, maximum sequence percentage identity = 25% or less. To avoid statistical bias caused by the large number of homologues proteins this dataset was used for our subsequent statistical analysis.
2.2. Chameleon sequence determination
Secondary structure assignments were made automatically using the DSSP [4] program. The 8 level secondary structural assignments in DSSP were reduced to the 3 classical states: helix including α, 310 and π-helices, strand the β -strand assignments, and coil which covered the rest of our assignments (γ-bridges, turns, bends and coils). For both datasets, three non-redundant sequence files were prepared based on DSSP (≥4 amino acids). Our first file was sequences with helix conformations only, our second file was β-strands and the third was restricted to the coils or unstructured sequences. The tool for extracting segments with identical sequences and complete different secondary structures was designed using our in house C-sharp program. In this process, we found the helix sequences (list H) with the strand (list E) and the coil sequences (list C) by sliding the helix sequence along the strand sequence, one residue at a time. In addition, sequences which corresponded to the entire strands were searched against the helix and the coil sequences, and sequences that correlated with the entire coils were searched against the helix and strand sequences. Finding the same sequences was performed as followed: first, we searched for all possible identical 4 residues (4-mer) in one list (e.g. H list) and another list (e.g. E list) using a matching matrix, wherever possible. These residues were then extended to identify longer identical sequence pairs (4–12-mer). Contiguous, overlapping 4-mers that could form higher order n-mers were not retained in the 4-mer dataset and were assigned as the appropriate n-mer while only the longest possible n-mer was considered. Where one sequence from one list exactly matched the target sequence from another list, it was designated as a “chameleon” sequence for the corresponding protein, identified by its PDB code (e.g. HHHHH in one protein and EEEEE in another protein) and then they were classified into 3 distinct groups namely, Helix-Strand (HE-Chameleons), Helix-Coil (HC-Chameleons) and Coil–Strand (CE-Chameleons). For each chameleon peptide, the peptide sequence length, the peptide sequence, the PDB code/chain, the protein name and the location of the chameleon sequence along the protein chain were recorded. Finally we sorted the proteins for their number of chameleons, in order to find the most flexible proteins in the protein databank.
2.3. Residue occurrence
In order to avoid biases in the statistical analyses, the following survey was accomplished on the chameleon sequences in our dataset (sequence identity less than 25%). To investigate the residue occurrences in the extracted chameleon peptides dataset, the amino acid frequencies were calculated from all n-mers in dataset that had undergone complete helix to strand (HE), helix to coil (HC) and strand to coil (CE) transitions. These values were normalized against the occurrence of the amino acid frequencies in the two types of structure involved in our dataset.
In order to calculate the amino acid neighboring preferences, we used the following methodology; for 20 amino acids, there were 400 possible amino acid doublets (i.e., neighbors). For amino acid i, all 20 nij values (with j = 1, 2,. . . 20, corresponding to the 20 amino acids), provided a profile of neighbor preference for amino acids found after amino acid i while all 20 nij values provided another profile for amino acids found before amino acid i along the amino acid sequence. Additionally, the situation of every doublet was analyzed. We assumed the neighbor-dependent propensity values as Ʃx (a ± 1) where the Ʃx (a ± 1) value of 1.0 means that the occurrence of the residue pair, ax (or xa), in the chameleon sequences is the same as its frequency of occurrence of the amino acid neighboring in the two types of structure involved in database. A value > 1.0 means that the pair has an occurrence in the chameleon sequences which is higher than its incident in the PDB, suggesting that the pair has a preference for adopting chameleon sequences. Furthermore, Ʃx (a ± 1) values lower than unity suggest less preference for the pair in the chameleon sequences, all of our singlet and doublet propensity calculations were mentioned in our previous studies [25], [29].
2.4. Solvent accessibility analysis
The solvent accessibility of each segment is the solvent accessibility value per residue as computed by the DSSP program averaged over the segment's length. The relative solvent accessibility of each residue was estimated by normalizing the absolute value by the maximum accessibility per residue. In this work, we assigned two values (i.e. buried (B) and exposed (E)), depending on the average accessibility value, for either being higher (or equal) and lower than the 16% threshold, respectively [30], [31].
2.5. Enrichment analysis of chameleon sequences
In order to investigate the human disease enrichment analysis of HE-, CE- and HC-Chameleon sequences were performed using interactive and collaborative gene list enrichment analysis tool (Enrichr: http://amp.pharm.mssm.edu/Enrichr/) [32]. Enrichr is an integrative web-based software application that includes 35 gene-set libraries, an approach to rank enriched terms. To find the disease categories, three gene set library databases such as OMIM (Online Mendelian Inheritance in man) [33], Disease Perturbations from GEO (Gene Expression Omnibus) up and Disease Perturbations from GEO down were used. In the results section of this tool, the computed p-value was combined using the Fisher exact test with the z-score of the deviation from the expected rank and produced a combined score rank. To gain insight into the potential map pathway of the proteins with chameleon sequences, KEGG (Kyoto of Encyclopedia of Genes and Genomes) enrichment analysis was performed.
3. Results
In order to get an insight on these chameleon sequences in nature, we surveyed the whole non redundant PDB, to find the so called chameleon sequences which can serve as the core of amyloid fibril formation. Subsequently, we analyzed the singlet and doublet propensity for different amino acids, and we extracted from our dataset the most abundant chameleon sequences in proteins and the proteins with the most chameleon sequences (CFPs).
3.1. Distribution of chameleon sequences
The distribution of chameleon sequences of ≥4 residues in length in our dataset were shown in Table 1 . We found 84266 HC-Chameleon sequences and 81921 HE- Chameleons. It is worth mentioning that the number of CE-Chameleons were considerably less than the other types (51795 peptides). The longest chameleon was 8-mer in our HC dataset. We found octapeptide chameleons, KKLREKVD (PDB ID: 4E4W, PDB ID: 1R1H) ENLYFQGQ (PDB ID: 4G3O, PDB ID: 4LQZ), GETNLYFQ (PDB ID: 4M7R, PDB ID: 4JG2), ELEHHHHH (PDB ID: 4F2L, PDB ID: 1WB4), SLLTEVET (PDB ID: 2Z16, PDB ID: 4N8C) and DEVKRNTE (PDB ID: 2F1F, PDB ID: 4GOU) which all of them were interestingly grouped in two enzymatic superfamilies: transferases and hydrolases.
Table 1.
Statistics of the chameleon sequences in the dataset.
| Length of chameleon | No of non-redundant HE-Chameleona (number of redundant)d | No of non-redundant HC-Chameleonb (number of redundant)d | No of non-redundant CE-Chameleonc (number of redundant)d |
|---|---|---|---|
| 4-mer | 55380(1125678) | 62129(984713) | 43645(347083) |
| 5-mer | 25056 (43550) | 21022(33288) | 7878(9670) |
| 6-mer | 1423(1501) | 1067(1268) | 267(283) |
| 7-mer | 62(63) | 42(57) | 4 (9) |
| 8-mer | 0 | 6(6) | 0 |
| Total | 81921(1170792) | 84266(1019332) | 51795(357045) |
Complete helix to strand.
Complete helix to coil.
Complete strand to coil.
The sequences were shown when chameleons are detected in more than one occasion.
We observed many specific highly repeated sequences in the chameleons (eg.: 55380 unique 4-mer HE-chameleons vs 1125678 repeated 4-mer HE-chameleons, Table 1 shows the most abundant naturally occurred chameleons in PDB and their corresponding codes. The highest repeated 4-mer chameleons were: AAVA, 927 times and AAAL, 922 times seen in HE, AAAL, 1123 and LEEL, 892 times seen in HC and LGAG, 250 and LGLP, 186 times in CE chameleons. The comparison of amino acid frequencies demonstrated that Leu, Val and Ala in HE and Leu, Ala, Glu in HC and Leu, Gly and Val in CE-chameleons were the most frequent residues in 4-mer. Furthermore, the study of the 5-mer chameleon sequences revealed that AALAA in HE and HHHHH in HC and CE chameleons were the sequences with the highest repeat.
It is obvious that the number of different n-mers detected in the protein structure dataset is declined with the growing length of the peptides. As it can be predicted, most chameleon segments in our datasets were 4 or 5 residues in length. In order to find the organisms with the most chameleon sequences, our chameleon proteins and their PDB ids were analyzed in UniProt [34] (http://www.uniprot.org/uploadlists/) and we derived the greatest chameleon sequences in Homo sapiens.
3.2. Amino acids distribution
3.2.1. Propensity of single amino acids
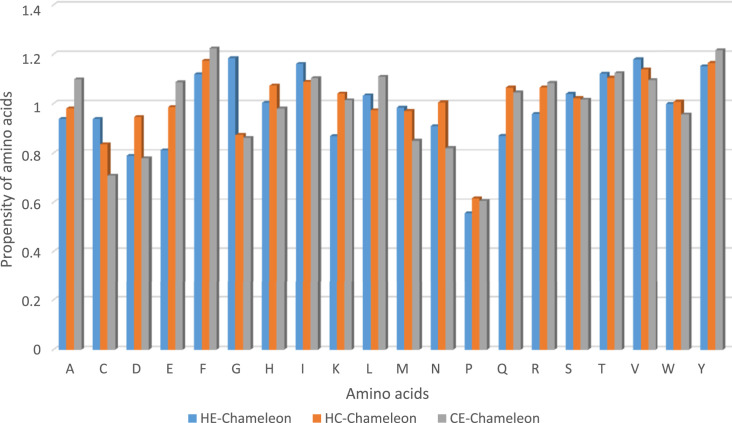
We calculated the propensity of the 20 amino acids in chameleon segments, as shown in Fig. 1 . The statistical analysis of protein residues in the chameleon sequences showed that Gly, Val, Ile, Tyr, and Thr in HE-Chameleons as well as Phe, Tyr, Val, Thr and Ile in HC-Chameleons along with Phe, Tyr, Thr, Leu and Ile in CE-Chameleons had the highest propensities in each type of the chameleons. Moreover, Pro, Asp and Glu in HE-Chameleons, Pro, Cys and Gly in HC-Chameleons as well as Pro, Cys and Asp in CE-chameleons residues were significantly less abundant in the chameleon segments. The amino acids were divided into 5 main groups, namely peptides with non–polar side chains (A, C, G, P, M, L, I, V), aromatic side chains (F, Y, W), positively-charged (H, K, R), negatively-charged (D, E) and non-charged polar residues (N, Q, T, S), were examined in the chameleon sequences. The aromatic residues had the highest frequencies in our chameleons. Accordingly, long aromatic amino acids can destabilize the secondary structure and its hydrophobic nature can help the aggregation responsibility.
Fig. 1.
A bar graph of normalized chameleon's propensity of amino acids in groups of different chameleon types (HE, HC and CE) in dataset1. The propensity values of amino acids in different groups are represented by different bars as indicated in the legend.
3.2.2. Propensity of doublet amino acids
In an attempt to evaluate how neighboring residues affect the chameleon conformation, the propensities of doublet amino acids were also determined. There were 400 possible pairs of amino acids which can occur in any doublet position of chameleon sequences. In Table 2, Table 3, Table 4 we calculated the neighbor-dependent propensities of 20 amino acids at the +1 and −1 situations of the chameleon residues [Ʃx (a ± 1)] in different groups, where the occurring dipeptide combinations was normalized by their natural frequencies in our dataset. The chameleons' neighbor-dependent propensities of amino acids often mirrored the individual chameleon propensities of the neighboring residues. In Table 2, Table 3, Table 4 we highlight the doublets with the highest possible propensities (at least 20% more than 1).
Table 2.
Normalized neighbor-dependent propensity in HE-Chameleons group.
| Ala | Arg | Asn | Asp | Cys | Glu | Gln | Gly | His | Ile | Leu | Lys | Met | Phe | Pro | Ser | Thr | Trp | Tyr | Val | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ala | 0.74 | 0.88 | 0.80 | 0.69 | 0.88 | 0.72 | 0.71 | 1.25 | 1.02 | 1.13 | 0.84 | 0.76 | 0.81 | 1.10 | 0.82 | 0.95 | 1.19 | 1.12 | 1.20 | 1.21 |
| Arg | 0.81 | 0.85 | 0.76 | 0.59 | 1.04 | 0.61 | 0.74 | 1.21 | 0.93 | 1.18 | 1.03 | 0.75 | 0.87 | 1.04 | 0.73 | 1.07 | 1.14 | 0.97 | 1.24 | 1.22 |
| Asn | 0.86 | 0.84 | 0.69 | 0.58 | 0.68 | 0.61 | 0.60 | 1.17 | 0.70 | 1.18 | 0.99 | 0.77 | 0.76 | 1.18 | 0.40 | 0.92 | 1.04 | 0.71 | 0.99 | 1.27 |
| Asp | 0.66 | 0.60 | 0.73 | 0.51 | 0.77 | 0.45 | 0.61 | 1.05 | 0.73 | 1.07 | 0.81 | 0.60 | 0.59 | 0.96 | 0.59 | 0.75 | 0.93 | 0.59 | 0.96 | 1.14 |
| Cys | 0.94 | 0.85 | 0.57 | 0.63 | 0.32 | 0.80 | 0.70 | 0.89 | 0.56 | 1.08 | 1.18 | 0.79 | 0.66 | 1.07 | 0.20 | 0.96 | 1.06 | 0.39 | 0.88 | 1.25 |
| Glu | 0.62 | 0.66 | 0.60 | 0.52 | 0.75 | 0.50 | 0.60 | 1.01 | 0.78 | 1.00 | 0.84 | 0.61 | 0.82 | 1.00 | 0.96 | 0.82 | 0.98 | 0.85 | 0.95 | 1.14 |
| Gln | 0.66 | 0.71 | 0.74 | 0.57 | 0.65 | 0.53 | 0.67 | 1.09 | 0.82 | 1.13 | 0.90 | 0.60 | 0.71 | 1.00 | 0.68 | 1.02 | 1.12 | 0.93 | 1.05 | 1.19 |
| Gly | 1.23 | 1.18 | 1.01 | 0.86 | 0.96 | 1.05 | 1.09 | 1.22 | 1.06 | 1.34 | 1.24 | 1.10 | 1.09 | 1.30 | 0.72 | 1.17 | 1.30 | 1.11 | 1.44 | 1.37 |
| His | 1.04 | 1.04 | 0.90 | 0.66 | 0.64 | 0.87 | 0.82 | 1.19 | 0.78 | 1.19 | 1.07 | 0.89 | 0.56 | 1.16 | 0.53 | 1.09 | 1.11 | 0.71 | 1.12 | 1.23 |
| Ile | 1.15 | 1.11 | 1.10 | 1.03 | 1.01 | 1.10 | 1.09 | 1.38 | 1.19 | 1.22 | 1.13 | 1.09 | 1.18 | 1.19 | 0.88 | 1.18 | 1.17 | 1.20 | 1.27 | 1.17 |
| Leu | 0.92 | 1.01 | 0.97 | 0.84 | 1.24 | 0.88 | 0.91 | 1.29 | 1.19 | 1.15 | 1.04 | 0.87 | 1.18 | 1.17 | 0.88 | 1.08 | 1.15 | 1.31 | 1.23 | 1.25 |
| Lys | 0.75 | 0.81 | 0.68 | 0.65 | 0.72 | 0.61 | 0.67 | 1.17 | 0.69 | 1.11 | 0.92 | 0.69 | 0.89 | 1.02 | 0.65 | 0.88 | 1.06 | 0.93 | 0.91 | 1.15 |
| Met | 0.90 | 0.90 | 0.77 | 0.68 | 0.53 | 0.78 | 0.75 | 1.25 | 0.94 | 1.18 | 1.03 | 0.87 | 0.73 | 0.99 | 0.38 | 1.04 | 1.13 | 0.61 | 1.04 | 1.30 |
| Phe | 1.14 | 1.10 | 1.07 | 0.98 | 0.96 | 1.06 | 1.01 | 1.35 | 1.16 | 1.26 | 1.04 | 1.08 | 1.13 | 1.24 | 0.78 | 1.15 | 1.21 | 1.10 | 1.28 | 1.24 |
| Pro | 0.55 | 0.46 | 0.38 | 0.27 | 0.45 | 0.19 | 0.30 | 0.53 | 0.49 | 1.03 | 0.85 | 0.42 | 0.62 | 0.78 | 0.32 | 0.36 | 0.65 | 0.35 | 0.74 | 1.00 |
| Ser | 1.12 | 1.04 | 0.91 | 0.70 | 0.98 | 0.88 | 0.88 | 1.26 | 0.99 | 1.24 | 1.09 | 0.82 | 1.01 | 1.22 | 0.74 | 1.07 | 1.20 | 1.17 | 1.20 | 1.21 |
| Thr | 1.16 | 1.14 | 1.16 | 0.97 | 0.85 | 1.04 | 1.11 | 1.31 | 1.16 | 1.15 | 1.16 | 1.13 | 1.14 | 1.17 | 0.78 | 1.16 | 1.17 | 1.14 | 1.26 | 1.17 |
| Trp | 1.06 | 1.00 | 0.81 | 0.70 | 0.57 | 0.85 | 0.88 | 1.04 | 0.77 | 1.07 | 1.08 | 0.95 | 0.69 | 1.06 | 0.32 | 1.01 | 1.08 | 0.61 | 1.14 | 1.19 |
| Tyr | 1.24 | 1.14 | 1.07 | 0.93 | 0.81 | 1.11 | 1.12 | 1.30 | 1.13 | 1.21 | 1.13 | 1.14 | 1.08 | 1.25 | 0.77 | 1.23 | 1.27 | 1.18 | 1.37 | 1.20 |
| Val | 1.20 | 1.20 | 1.12 | 1.07 | 1.12 | 1.14 | 1.20 | 1.34 | 1.28 | 1.21 | 1.23 | 1.13 | 1.31 | 1.23 | 0.86 | 1.22 | 1.17 | 1.27 | 1.19 | 1.14 |
* Amino acids in the columns precede amino acids in the rows. All the propensities >1.2 were bolded.
Table 3.
Normalized neighbor-dependent propensity in HC-Chameleons group.
| Ala | Arg | Asn | Asp | Cys | Glu | Gln | Gly | His | Ile | Leu | Lys | Met | Phe | Pro | Ser | Thr | Trp | Tyr | Val | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ala | 0.91 | 0.95 | 1.13 | 0.99 | 0.85 | 0.88 | 0.89 | 1.00 | 1.18 | 0.89 | 0.81 | 0.95 | 0.87 | 1.13 | 0.65 | 1.09 | 1.16 | 1.09 | 1.14 | 1.02 |
| Arg | 0.97 | 1.09 | 1.16 | 1.10 | 0.79 | 0.96 | 0.99 | 1.01 | 1.14 | 1.18 | 0.97 | 1.12 | 0.97 | 1.18 | 0.55 | 1.13 | 1.24 | 1.03 | 1.15 | 1.31 |
| Asn | 1.13 | 1.21 | 0.91 | 0.93 | 0.79 | 1.06 | 1.23 | 0.57 | 1.01 | 1.29 | 1.20 | 1.10 | 0.99 | 1.22 | 0.20 | 0.97 | 1.13 | 0.83 | 1.24 | 1.25 |
| Asp | 1.04 | 1.12 | 0.87 | 0.86 | 0.96 | 0.92 | 1.09 | 0.61 | 1.06 | 1.19 | 1.14 | 0.97 | 1.11 | 1.20 | 0.17 | 0.91 | 1.05 | 1.08 | 1.20 | 1.15 |
| Cys | 0.95 | 0.95 | 0.73 | 0.66 | 0.32 | 0.97 | 0.95 | 0.92 | 0.73 | 0.72 | 1.03 | 0.83 | 0.34 | 0.83 | 0.15 | 0.91 | 0.94 | 0.25 | 0.63 | 1.01 |
| Glu | 0.89 | 0.98 | 0.99 | 0.98 | 0.89 | 0.93 | 0.96 | 1.03 | 1.14 | 1.00 | 0.85 | 0.99 | 0.87 | 1.18 | 0.60 | 1.05 | 1.13 | 1.04 | 1.07 | 1.17 |
| Gln | 0.96 | 1.11 | 1.12 | 1.20 | 0.70 | 1.03 | 0.93 | 1.10 | 1.09 | 1.06 | 0.94 | 1.09 | 0.92 | 1.17 | 0.57 | 1.21 | 1.25 | 0.90 | 1.15 | 1.27 |
| Gly | 1.06 | 0.93 | 0.77 | 0.72 | 0.90 | 0.95 | 1.02 | 0.66 | 1.00 | 1.18 | 1.14 | 0.87 | 1.27 | 1.14 | 0.56 | 0.80 | 0.87 | 1.17 | 1.18 | 1.09 |
| His | 1.24 | 1.30 | 1.09 | 1.05 | 0.61 | 1.17 | 0.97 | 0.88 | 0.91 | 1.35 | 1.22 | 1.22 | 0.85 | 1.23 | 0.21 | 1.10 | 1.27 | 0.91 | 1.30 | 1.25 |
| Ile | 0.88 | 0.97 | 1.30 | 1.14 | 0.93 | 1.00 | 1.11 | 1.18 | 1.23 | 0.97 | 0.85 | 1.15 | 0.88 | 1.22 | 0.77 | 1.27 | 1.31 | 0.83 | 1.15 | 1.17 |
| Leu | 0.83 | 0.95 | 1.09 | 1.02 | 1.11 | 0.92 | 0.98 | 1.02 | 1.17 | 0.91 | 0.79 | 0.98 | 0.83 | 1.00 | 0.79 | 1.02 | 1.10 | 0.86 | 1.01 | 0.98 |
| Lys | 0.97 | 1.23 | 1.07 | 1.02 | 0.87 | 0.99 | 1.09 | 0.90 | 1.07 | 1.22 | 0.98 | 1.05 | 1.01 | 1.22 | 0.54 | 1.05 | 1.17 | 0.97 | 1.11 | 1.27 |
| Met | 1.03 | 0.96 | 1.03 | 0.98 | 0.47 | 1.01 | 0.89 | 1.09 | 0.73 | 0.87 | 0.89 | 1.06 | 0.72 | 0.92 | 0.57 | 1.14 | 1.04 | 0.38 | 1.03 | 1.02 |
| Phe | 1.16 | 1.28 | 1.25 | 1.20 | 0.87 | 1.19 | 1.27 | 1.10 | 1.26 | 1.05 | 0.97 | 1.15 | 0.89 | 1.25 | 0.64 | 1.26 | 1.41 | 1.07 | 1.23 | 1.26 |
| Pro | 0.86 | 0.73 | 0.49 | 0.46 | 0.36 | 0.64 | 0.82 | 0.41 | 0.74 | 1.16 | 1.01 | 0.64 | 0.96 | 0.98 | 0.19 | 0.56 | 0.69 | 0.84 | 1.01 | 0.98 |
| Ser | 1.11 | 1.07 | 0.98 | 0.89 | 1.05 | 0.99 | 1.13 | 0.74 | 1.03 | 1.17 | 1.09 | 1.08 | 1.18 | 1.22 | 0.24 | 0.98 | 1.12 | 1.08 | 1.29 | 1.22 |
| Thr | 1.16 | 1.21 | 1.07 | 0.95 | 0.91 | 1.09 | 1.25 | 0.82 | 1.10 | 1.26 | 1.13 | 1.23 | 1.13 | 1.46 | 0.37 | 1.13 | 1.27 | 1.25 | 1.36 | 1.33 |
| Trp | 0.92 | 0.96 | 1.07 | 1.00 | 0.44 | 1.07 | 0.91 | 1.22 | 0.90 | 0.90 | 0.89 | 1.14 | 0.63 | 0.90 | 0.73 | 1.07 | 1.13 | 0.65 | 0.99 | 1.10 |
| Tyr | 1.12 | 1.28 | 1.28 | 1.16 | 0.87 | 1.18 | 1.24 | 1.07 | 1.13 | 1.25 | 0.97 | 1.22 | 0.99 | 1.32 | 0.54 | 1.28 | 1.40 | 1.12 | 1.25 | 1.31 |
| Val | 1.01 | 1.16 | 1.29 | 1.20 | 0.90 | 1.08 | 1.21 | 1.19 | 1.34 | 1.13 | 1.00 | 1.15 | 1.07 | 1.28 | 0.68 | 1.20 | 1.30 | 1.08 | 1.28 | 1.35 |
* Amino acids in the columns precede amino acids in the rows. All the propensities >1.2 were bolded.
Table 4.
Normalized neighbor-dependent propensity in CE-Chameleons group.
| Ala | Arg | Asn | Asp | Cys | Glu | Gln | Gly | His | Ile | Leu | Lys | Met | Phe | Pro | Ser | Thr | Trp | Tyr | Val | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ala | 1.29 | 1.25 | 0.90 | 0.79 | 0.78 | 1.28 | 1.16 | 0.94 | 1.27 | 1.13 | 1.15 | 1.17 | 1.03 | 1.40 | 0.56 | 1.11 | 1.22 | 1.34 | 1.38 | 1.11 |
| Arg | 1.28 | 1.13 | 0.87 | 0.72 | 0.74 | 1.10 | 1.00 | 0.90 | 1.03 | 1.15 | 1.18 | 1.08 | 0.87 | 1.28 | 0.69 | 1.20 | 1.31 | 0.86 | 1.35 | 1.16 |
| Asn | 1.01 | 0.90 | 0.55 | 0.48 | 0.67 | 0.80 | 0.71 | 0.57 | 0.78 | 1.30 | 1.14 | 0.74 | 0.73 | 1.32 | 0.35 | 0.75 | 0.89 | 0.83 | 1.12 | 1.22 |
| Asp | 0.86 | 0.71 | 0.52 | 0.47 | 0.69 | 0.65 | 0.79 | 0.53 | 0.71 | 1.24 | 1.07 | 0.68 | 0.66 | 1.28 | 0.36 | 0.68 | 0.92 | 0.92 | 1.15 | 1.14 |
| Cys | 0.78 | 0.69 | 0.50 | 0.47 | 0.15 | 0.76 | 0.66 | 0.81 | 0.40 | 0.80 | 0.94 | 0.57 | 0.29 | 0.53 | 0.44 | 0.89 | 0.97 | 0.32 | 0.57 | 0.81 |
| Glu | 1.29 | 1.05 | 0.72 | 0.76 | 0.70 | 1.16 | 1.16 | 0.79 | 1.13 | 1.21 | 1.29 | 1.04 | 1.11 | 1.52 | 0.74 | 1.03 | 1.20 | 0.98 | 1.32 | 1.26 |
| Gln | 1.25 | 1.02 | 0.77 | 0.70 | 0.52 | 1.08 | 1.04 | 0.82 | 0.84 | 1.25 | 1.25 | 0.96 | 0.80 | 1.26 | 0.58 | 1.15 | 1.18 | 0.94 | 1.31 | 1.26 |
| Gly | 0.89 | 0.84 | 0.67 | 0.54 | 0.82 | 0.83 | 0.80 | 0.78 | 0.88 | 1.09 | 1.00 | 0.68 | 1.00 | 1.14 | 0.63 | 0.86 | 0.95 | 1.05 | 1.16 | 1.04 |
| His | 1.24 | 1.04 | 0.74 | 0.77 | 0.35 | 1.08 | 0.62 | 0.86 | 0.69 | 1.15 | 1.36 | 0.94 | 0.47 | 1.22 | 0.53 | 1.15 | 1.21 | 0.73 | 1.24 | 1.17 |
| Ile | 1.09 | 1.16 | 1.13 | 1.09 | 0.63 | 1.28 | 1.38 | 1.16 | 1.24 | 0.83 | 0.91 | 1.29 | 0.84 | 1.13 | 0.80 | 1.19 | 1.16 | 0.76 | 1.06 | 0.84 |
| Leu | 1.30 | 1.31 | 1.12 | 1.00 | 1.02 | 1.33 | 1.33 | 1.12 | 1.31 | 1.00 | 1.09 | 1.25 | 1.01 | 1.18 | 0.64 | 1.11 | 1.10 | 0.94 | 1.20 | 0.93 |
| Lys | 1.26 | 1.11 | 0.69 | 0.72 | 0.71 | 1.14 | 1.10 | 0.83 | 0.89 | 1.16 | 1.18 | 0.90 | 1.08 | 1.30 | 0.63 | 0.96 | 1.14 | 0.98 | 1.24 | 1.10 |
| Met | 1.16 | 0.74 | 0.80 | 0.76 | 0.44 | 0.98 | 0.68 | 1.07 | 0.53 | 0.81 | 1.03 | 0.74 | 0.53 | 0.86 | 0.65 | 0.93 | 0.92 | 0.11 | 0.94 | 0.96 |
| Phe | 1.34 | 1.39 | 1.19 | 1.12 | 0.75 | 1.39 | 1.25 | 1.17 | 1.15 | 1.03 | 1.31 | 1.35 | 0.63 | 1.34 | 0.66 | 1.30 | 1.30 | 0.95 | 1.32 | 1.03 |
| Pro | 0.61 | 0.53 | 0.33 | 0.26 | 0.35 | 0.39 | 0.44 | 0.37 | 0.56 | 1.01 | 0.87 | 0.45 | 0.72 | 0.91 | 0.13 | 0.41 | 0.61 | 0.41 | 0.79 | 0.98 |
| Ser | 1.29 | 1.21 | 0.80 | 0.64 | 0.80 | 1.08 | 1.08 | 0.94 | 1.03 | 1.27 | 1.30 | 1.00 | 1.01 | 1.42 | 0.51 | 0.99 | 1.18 | 1.19 | 1.52 | 1.31 |
| Thr | 1.34 | 1.31 | 0.95 | 0.82 | 0.87 | 1.25 | 1.35 | 1.11 | 1.20 | 1.13 | 1.23 | 1.29 | 1.03 | 1.37 | 0.63 | 1.24 | 1.20 | 1.22 | 1.50 | 1.15 |
| Trp | 1.10 | 1.07 | 0.88 | 0.90 | 0.42 | 1.19 | 0.84 | 1.05 | 0.70 | 0.77 | 1.01 | 1.03 | 0.53 | 0.74 | 0.52 | 1.07 | 0.95 | 0.63 | 0.94 | 0.98 |
| Tyr | 1.33 | 1.40 | 1.11 | 1.06 | 0.82 | 1.44 | 1.27 | 1.26 | 1.12 | 1.09 | 1.24 | 1.32 | 1.00 | 1.27 | 0.67 | 1.25 | 1.41 | 0.97 | 1.28 | 1.04 |
| Val | 1.13 | 1.21 | 1.11 | 1.07 | 0.71 | 1.29 | 1.33 | 1.22 | 1.15 | 0.79 | 0.96 | 1.22 | 0.99 | 1.12 | 0.79 | 1.14 | 1.13 | 0.94 | 1.06 | 0.84 |
* Amino acids in the columns precede amino acids in the rows. All the propensities >1.2 were bolded.
As shown in Table 2, Table 3, Table 4, ten of the most frequently-detected dipeptides were GY, IG, YY, GV, FG, VG, GI, VM, LW and TG in HE-Chameleon sequences. The appearances of dipeptides SY, EF, TV, YE, SF, YT, AF, YR, FE and FR in CE-Chameleons and TF, FT, YT, TY, HI, VV, VH, TV, YF and RV in HC-Chameleons were much higher than others residues.
All the propensities >1.2 are emboldened in Table 2, Table 3, Table 4. In the above-mentioned tables, doublets with very low propensities were also seen, and there were many low propensity values when an amino acid was neighbored with Pro (a weak chameleon conformer amino acid).
3.3. Calculation of solvent accessibility in chameleon sequences
The analysis of solvent accessibility indicated that the majority of HE-chameleons and CE-chameleons in all n-mers were mixed with one segment exposed and the other one buried. Our results showed that the greater part of the couples with HC-Chameleons in all n-mers consisted of exposed solvent accessibility (Table 5 ).
Table 5.
Solvent accessibility of chameleon pairs.
| Segments | No | No (%) (both exposed) | No (%) (mixed) | No (%) (both buried) |
|---|---|---|---|---|
| 4-mers | ||||
| HE | 53235 | 14647(27.5) | 24502(46) | 14086(26.5) |
| HC | 58633 | 33562(57.2) | 21368(36.4) | 3703(6.3) |
| CE | 41402 | 4308(10.4) | 20397(49.3) | 166974(40.3) |
| 5-mers | ||||
| HE | 24133 | 5843(24.2) | 10857(45) | 7433(30.8) |
| HC | 19940 | 12079(60.6) | 6897(34.6) | 964(4.8) |
| CE | 7444 | 3069(41.2) | 3720(50) | 655(8.8) |
| 6,7,8-mers | ||||
| HE | 1429 | 366(25.6) | 657(46) | 406(28.4) |
| HC | 1052 | 661(62.8) | 341(32.4) | 50(4.8) |
| CE | 252 | 114(45.2) | 119(47.2) | 19(7.5) |
3.4. Enrichment analysis of chameleon sequences
In order to further evaluate the biological mechanisms led by chameleon sequences we performed the disease enrichment analysis. We performed an enrichment analysis for all different chameleon sequences with a threshold of P < 0.05, and we tabulated the top significant diseases in OMIM, up and down regulated GEO on all groups in Table 6 . We noticed that the most diseases found related to HE-and HC-Chameleons in the OMIM database was Alzheimer's (Pvalue = 0.002, combined score: 8.6) and colorectal cancer (Pvalue = 0.005, combined score: 7.48) respectively. In this study a total of 507, 305, 10 Disease Perturbations from GEO UP and 305, 319 and 1 Disease Perturbations from GEO Down in HE-chameleons, HC-Chameleons and CE-chameleons (7-mer) were identified respectively. Interestingly, data analyses on the 4-mer chameleon sequences between all groups illustrated that the same type of significant disease pattern could be seen in all databases. The immunodeficiency, colorectal cancer, cardiomyopathy, anemia and leukemia, were seen in all three chameleon groups in the OMIM database. The highest combined score in disease perturbation from GEO up regulated enrichment in chameleon groups belonged to lung disease, ALS (Amyotrophic Lateral Sclerosis), autism spectrum disorder, Purpura and juvenile dermatomyositis. Additionally, the collected data from disease perturbation from GEO down regulated enrichment showed that Cardiac Hypertrophy, diffuse large B-cell lymphoma (DLBCL), severe acute respiratory syndrome (SARS), multiple sclerosis (MS) and Familial combined hyperlipidaemia were the most significant diseases associated with all groups of chameleon proteins.
Table 6.
Disease enrichment analysis of chameleon sequences in seven residues in length.
| Gene-set library | Type of chameleons |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Type of disease | |||||||||||
| Neurodegenerative diseasea | Blood disorder | Colorectal cancer | Eye disorderb | Mental disorderc | Lung disease | Liver disease | Artritis | IBDd | Type 2 diabetes mellitus | CFe | |
| HE-Chameleons | |||||||||||
| OMIM | √ | √ | – | – | – | – | – | – | – | – | – |
| Disease Perturbations from GEO UP | √ | – | √ | √ | √ | √ | – | √ | √ | – | – |
| Disease Perturbations from GEO Down | √ | – | – | – | √ | – | – | – | – | – | – |
| HC-Chameleons | |||||||||||
| OMIM | – | √ | √ | – | – | – | – | – | – | – | – |
| Disease Perturbations from GEO UP | – | √ | – | – | – | √ | √ | √ | √ | – | – |
| Disease Perturbations from GEO Down | √ | √ | – | – | – | – | – | – | – | √ | √ |
| CE-Chameleons | |||||||||||
| OMIM | – | – | – | – | – | – | – | – | – | – | – |
| Disease Perturbations from GEO UP | – | – | – | – | – | – | √ | – | √ | – | – |
| Disease Perturbations from GEO Down | – | – | – | – | – | – | – | – | – | – | √ |
Neurodegenerative disease including Parkinson's disease, Alzheimer's disease, Huntington's disease, Amyotrophic lateral sclerosis (ALS), Alexander disease.
Eye disease including Leber congenital amaurosis and Retinoschisis.
Mental disorders including schizophrenia, bipolar disorder, Anxiety disorders.
IBD, inflammatory bowel disease.
CF, cystic fibrosis.
36 pathways in the genes of 7-mer HE-chameleons enriched in KEGG, of which neurodegenerative diseases (including APP and A2M genes) (Pvalue = 0.003) and focal adhesion (including COMP, MAP2K1 and TLN1 genes) (Pvalue = 0.01) were also the most significant cases. Proteins with 7-mer HC-Chameleons were significantly involved in 22 pathways of KEGG such as glycerolipid metabolism (including GK and LIPA genes) (Pvalue = 0.003) and tight junction (including PRKCI and PARD3 genes) (Pvalue = 0.04).
3.5. Chameleon Flexible Proteins (CFP) in human
We hereby present a new term named Chameleon Flexible Proteins or CFPs, as special proteins with a high number of chameleon sequences in our chameleon dataset. Table 7 shows 14 proteins with the most number of chameleon sequences. Insulin-degrading enzyme (IDE and GTP-binding nuclear protein Ran (RAN) has the most number of chameleon sequences in which we named CFP's.
Table 7.
Human flexible proteins.
| Name of proteins (gene names) | PDB ID | No of chameleon sequences in protein |
|||
|---|---|---|---|---|---|
| HE | HC | CE | Total | ||
| Insulin-degrading enzyme(IDE) | 3CWW | 257 | 261 | 122 | 640 |
| GTP-binding nuclear protein Ran(RAN) | 4HAT | 185 | 189 | 31 | 405 |
| Cytoplasmic FMR1-interacting protein 1 (CYFP1) | 3P8C | 157 | 181 | 24 | 379 |
| DNA damage-binding protein 1 (DDB1) | 3EI3 | 157 | 73 | 136 | 366 |
| SAM domain and HD domain-containing protein 1(SAMHD1) | 4MZ7 | 114 | 128 | 48 | 290 |
| pyruvate kinase muscle isozyme (PKM) | 3GR4 | 101 | 124 | 43 | 268 |
| Xaa-Pro aminopeptidase 1(,d.d24”>XPNPEP1) | 3ctz | 87 | 104 | 67 | 258 |
| SOSS complex subunit B1(SOSB1) | 4owt | 106 | 110 | 25 | 241 |
| Serum albumin(ALBU) | 1N5U | 86 | 130 | 21 | 237 |
| Presequence protease, mitochondrial(PREP) | 4L3T | 106 | 70 | 61 | 237 |
| 6-phosphogluconate dehydrogenase, decarboxylating(PGD) | 4GWG | 91 | 114 | 31 | 236 |
| Gamma-tubulin complex component 4 (,d.d24”>TUBGCP4) | 3RIP | 98 | 123 | 15 | 236 |
| Apoptosis inhibitor 5(API5) | 30UR | 102 | 116 | 16 | 234 |
| Beta adrenergic receptor kinase (,d.d24”>ADRBK1) | 4MK0 | 85 | 94 | 55 | 234 |
Proteins with a large number of chameleon sequences.
4. Discussion
Due to the lack of our molecular mechanism knowledge underlying neurodgeneration in neurodegenerative disease pathogenesis, current therapies can only somewhat relieve the symptoms and there is therefore no proven medication to cure or prevent the disease. Various neurodegenerative disorders show that there should be a misfolding and aggregation-prone mechanism underlying neurodegeneration, upon interaction with neuronal membranes. Monomers of amyloidoenic sequences undergo a shift in their conformation yielding to an aggregation. Amyloidogenic sequences are misfolded peptides or proteins that represent a supersecondary structure which are somewhat insoluble, fibrous like and proteolysis resistant. As mentioned earlier, amyloid proteins are mostly chameleon sequences involved in the widely accepted hypothesis in this amyloidogenesis. Chameleon sequences are therefore the basic known sequences responsible for the disease initiation. Although these sequences lack a well-defined conformation they may be involved in various biological functions [14] in which their structural instability and mobile flexibility is encoded by their amino acid sequences. Therefore in this study, we performed an extensive survey of peptide sequences of varying lengths (≥4 residues) with ≤25% sequence identity which could adopt either an α-helix structure in one protein and a β-strand or coil structures in another protein or a coil structure in one protein and an α-helix or β-strand structure in another protein (HE, HC and CE chameleons). Our investigation is the first report in which all possible types of conformational pairs (HE, HC, CE-Chameleons) have been classified and the neighboring residues and their propensities were carefully analyzed. We found an extensive number of chameleon sequences, which each can be used as a base for further related studies on chameleon sequences and their role in biological functions, particularly amyloidogenesis and neurodegeneration. It is worth mentioning that these sequences are recently widely recognized, due to their widely biological importance [26], [35], and their significant role in disease-associated proteins [9], [35], [36]. Many studies showed that the sequence neighboring in secondary structure of proteins is crucial in forming these particular structures [7], [18], [19]. To our knowledge, our work is the first comprehensive research on the occurrences of neighboring amino acid propensities (doublets) and disease enrichment analysis for all kinds of chameleon peptides. Our research showed that in general, the CE-Chameleon sequences are less abundant than the HE and HC Chameleons, but HC and HE show at a relatively comparable frequency in this study. This may be due to the more number of helices compared to the beta sheets in the PDB.
Our algorithm to find chameleon sequences was different from other groups and relatively more stringent [26], [37]. Other groups work in searching for identical sequences that adopt an H conformation in one protein and an E conformation in another protein, but were not as stringent as ours. In their studies, at least one of the middle two residues in the identical sequences may include both ‘E’ and ‘H’ (such as ‘CCCEEECC’ vs ‘CCHHHHHH’), while the criteria of our study was finding complete chameleon sequences and to find identical sequences with quite a different secondary structure (such as “EEEEEEE” vs “HHHHHHH” or “CCCCCCC”) compared to others.
In accordance to the earlier observations [9], [11], we noticed in this study that there are many more 4 or 5 residue length chameleons. This implies the flexibility of short peptide regions in proteins compared to longer ambivalent peptides in naturally occurring proteins.
The profiles of single amino acid propensity value for transition of helix to sheet (HE), helix to coil (HC) and sheet to coil (CE) chameleons is presented in Fig. 1, where we clearly witness the amino acids Phe and Tyr were more frequently seen in chameleons, although the most frequent amino acids in proteins are Leu and Ala as of very recent SwissProt database information [38], [39], [40]. This may indicate that aliphatic side-chained amino acids are preferred in chameleon sequences. The major challenge for neurodegenerative disease peptides is their propensity for aggregation which clearly stems from their hydrophobic residues [31]. Therefore our results show that not only hydrophobic residues are important, but close attention should be paid on Phe and Tyr as specific amino acids in chameleon sequences. The abundance of these two hydrophobic residues Phe and Tyr can clearly indicate these peptide properties for aggregation. Pro is the least amino acid present at these flexible regions. In other words, Pro residue is less abundant in all categories of the chameleons. This amino acid is unique among protein residues as it is a cyclic amino acid with no amide hydrogen to contribute in hydrogen bonding. Consequently, it cannot fit into the regular structure of either α-helix or β-sheet and is a common ‘breaker’ of secondary structure [41]. Although it is said to be a helix initiator as well [25], but as to our knowledge it has never been reported to be abundant in the middle of a helix or any other secondary structure. Therefore, the disruptive nature of Pro in the context of a helix or a sheet structure may be the reason for its low frequency in the chameleon peptides. We can also categorize Pro as a low flexible amino acid to form a chameleon.
Our data on single amino acid frequency of chameleon functions as a complement to previous related studies [9], [12]. Cys residues have also a low prevalence of occurrence in CE and HC-Chameleons. Since Cys tends to construct disulfide bonds which can impart higher stability to the sequence segment, therefore they could clearly inhibit the flexibility of sequences and hence it is rational not to be presented in these flexible regions. Our study revealed that Asp and Glu residues were found with a lower frequency in HE-Chameleon sequences. Our results confirm El Amiris studies [13]. But in contrast to Bhattacharjee's [10], the lack of Asp and Glu may be due to the acidic nature of these two amino acids which play an essential role in protein structures, and not to be restructured for their function. This is also in a very close agreement to the nature of neurodegenerative disease peptides with an aggregation nature in which these acidic amino acids give a more hydrophilic taste to the peptide, which prevents the required attachment for neurodegeneration.
Gly was seen as a major amino acid in HE-chameleons. This is apparently due to its nature being a flexible amino acid which can be sometimes seen in all four regions of ramachandran plot [42], which is widely accepted that the conformational freedom of Gly is remarkably vast.
The differential propensity values of amino acids in different groups were also reflected in the neighbor-dependent sequence analysis, where our highlighted doublets may be used as doublet words for pinpointing chameleons in any given sequence dataset.
Our solvent accessibility data clearly suggests that beta structured chameleons are more buried, which is in close agreement with recent works of Li and collaborators [26] indicating that the majority of sheet conformations were buried, or in other words tend to form aggregates in a hydrophilic environment and support the major role of chameleons in neurodegeneration.
The results of the disease enrichment analysis of HE-chameleon proteins in OMIM, Disease Perturbations from GEO up and down, indicated that the significant proteins including chameleon sequences were mostly a protein candidate for neurodegenerative diseases. In addition, our investigation for significant disease perturbations from GEO up enrichment, revealed that IBD disease which is a class of autoimmune diseases, was observed in all chameleon groups (HE, HC and CE chameleons) and genes such as NAT1; PTGDS; PRSS2; RAN, C1QBP; CIAPIN1; TLN1 and SERPINA7 were significantly involved in this disease.
Disease enrichment analysis may provide significant insights into the diagnosis and defining therapeutic targets for the disease. Conformational plasticity of chameleon sequences make them the prime candidates for amyloidogenic segments associated with neurodegenerative diseases which are identified by a conformational change from an α-helix to a β-sheet conformation. Better understanding of chameleon sequences are hence essential to examine the structure of amyloidogenic molecules for potential therapeutic intervention.
To our knowledge, there have been no such study for a comprehensive single and doublet amino acid propensity calculation on chameleons. Although there has been a high desire to find such important sequences, other works were first of all not as stringent as ours for finding chameleon sequences and not as comprehensive as ours for finding all three kinds of chameleons [13], [26]. Sorting out our whole dataset in order to find the most flexible proteins based on the number of chameleons presented in the protein, we found PDB ID: 3CWW the Insulin Degrading Enzyme (IDE) which can be mentioned as the most flexible human protein in PDB. Although the most flexible human protein was calculated based on its number of chameleon sequences, interestingly this protein turned out to be a link between Alzheimer's disease and Type II diabetes [43]. The next major flexible protein with the highest number of chameleon was RAN, PDB ID: 4HAT this newly deposited transport protein has been mentioned to be responsible for nuclear localization of Huntington's protein. This may be due to the disruption of nucleus cytoplasmic transport which has said to impair neuronal function, and literature supports that this mechanism is involved in the pathogenesis of neurodegenerative disorders [44]. We can therefore conclude that extracting the chameleon sequences and finding proteins with high number of chameleons can lead us to major proteins responsible in neurodegenerative disorders, although their role might have not yet been proved in lab experiments, investigations can be designed based on our Chameleon Flexible Proteins (CFP's).
In conclusion, our study aimed to investigate and present a novel dataset of chameleon sequences for neurodegenerative diseases. This study sets the foundation for a subsequent investigation on each sequence in native proteins, which along with presenting the mostly flexible proteins in our dataset, investigates the possibility to find which diseases are mostly affected by these sequences. The findings presented here confirm the existence of chameleon segments in their pathogenic conformational role in neurodegenerative diseases [8], [14]. Our study presents a new insight in defining new key proteins in neurodegeneration, where we hope our results would open the way to planning new strategies to overcome neurodegeneration and hence lead us to the cure and prevention of neurodegenerative diseases.
Acknowledgment
The authors would like to thank Dr. Pardis Minuchehr (George Washington University) and Professor Mihaly Mezei (Icahn School of medicine New York) for their valuable advice on the work. The work was supported by the National Institute of Genetic Engineering and Biotechnology grant no 303 and the Bioinformatics Lab.
Footnotes
Transparency document related to this article can be found online at http://dx.doi.org/10.1016/j.bbrc.2016.01.187.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.bbrc.2016.01.187.
Contributor Information
Bahram Goliaei, Email: goliaei@ut.ac.ir.
Zarrin Minuchehr, Email: minuchehr@nigeb.ac.ir.
Transparency document
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Anfinsen C.B. Principles that govern the folding of protein chains. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 2.Chou P.Y., Fasman G.D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv. Enzymol. Relat. Areas Mol. Biol. 1978;47 doi: 10.1002/9780470122921.ch2. 45–148. [DOI] [PubMed] [Google Scholar]
- 3.Uversky V.N. The mysterious unfoldome: structureless, underappreciated, yet vital part of any given proteome. J. Biomed. Biotechnol. 2010;2010:568068. doi: 10.1155/2010/568068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kabsch W., Sander C. On the use of sequence homologies to predict protein structure: identical pentapeptides can have completely different conformations. Proc. Natl. Acad. Sci. U. S. A. 1984;81:1075–1078. doi: 10.1073/pnas.81.4.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krishna N., Guruprasad K. Certain heptapeptide and large sequences representing an entire helix, strand or coil conformation in proteins are associated as chameleon sequences. Int. J. Biol. Macromol. 2011;49:218–222. doi: 10.1016/j.ijbiomac.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tidow H., Lauber T., Vitzithum K., Sommerhoff C.P., Rosch P., Marx U.C. The solution structure of a chimeric LEKTI domain reveals a chameleon sequence. Biochemistry. 2004;43:11238–11247. doi: 10.1021/bi0492399. [DOI] [PubMed] [Google Scholar]
- 7.Takano K., Katagiri Y., Mukaiyama A., Chon H., Matsumura H., Koga Y., Kanaya S. Conformational contagion in a protein: structural properties of a chameleon sequence. Proteins. 2007;68:617–625. doi: 10.1002/prot.21451. [DOI] [PubMed] [Google Scholar]
- 8.Gendoo D.M., Harrison P.M. Discordant and chameleon sequences: their distribution and implications for amyloidogenicity. Protein Sci. 2011;20:567–579. doi: 10.1002/pro.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuznetsov I.B., Rackovsky S. On the properties and sequence context of structurally ambivalent fragments in proteins. Protein Sci. 2003;12:2420–2433. doi: 10.1110/ps.03209703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhattacharjee N., Biswas P. Statistical analysis and molecular dynamics simulations of ambivalent alpha-helices. BMC Bioinforma. 2010;11:519. doi: 10.1186/1471-2105-11-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo J.T., Jaromczyk J.W., Xu Y. Analysis of chameleon sequences and their implications in biological processes. Proteins. 2007;67:548–558. doi: 10.1002/prot.21285. [DOI] [PubMed] [Google Scholar]
- 12.Zhou X., Alber F., Folkers G., Gonnet G.H., Chelvanayagam G. An analysis of the helix-to-strand transition between peptides with identical sequence. Proteins. 2000;41:248–256. doi: 10.1002/1097-0134(20001101)41:2<248::aid-prot90>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 13.El Amri C., Nicolas P. Plasticins: membrane-damaging peptides with'chameleon-like' properties. Cell. Mol. Life Sci. 2008;65:895–909. doi: 10.1007/s00018-007-7445-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamamoto N. Hot spot of structural ambivalence in prion protein revealed by secondary structure principal component analysis. J. Phys. Chem. B. 2014;118:9826–9833. doi: 10.1021/jp5034245. [DOI] [PubMed] [Google Scholar]
- 15.Biran Y., Masters C.L., Barnham K.J., Bush A.I., Adlard P.A. Pharmacotherapeutic targets in Alzheimer's disease. J. Cell. Mol. Med. 2009;13:61–86. doi: 10.1111/j.1582-4934.2008.00595.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Minor D.L., Jr., Kim P.S. Measurement of the beta-sheet-forming propensities of amino acids. Nature. 1994;367:660–663. doi: 10.1038/367660a0. [DOI] [PubMed] [Google Scholar]
- 17.Creamer T.P., Rose G.D. Alpha-helix-forming propensities in peptides and proteins. Proteins. 1994;19:85–97. doi: 10.1002/prot.340190202. [DOI] [PubMed] [Google Scholar]
- 18.Chakrabartty A., Baldwin R.L. Stability of alpha-helices. Adv. Protein Chem. 1995;46:141–176. [PubMed] [Google Scholar]
- 19.Sagermann M., Baase W.A., Matthews B.W. Structural characterization of an engineered tandem repeat contrasts the importance of context and sequence in protein folding. Proc. Natl. Acad. Sci. U. S. A. 1999;96:6078–6083. doi: 10.1073/pnas.96.11.6078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Costantini S., Colonna G., Facchiano A.M. Amino acid propensities for secondary structures are influenced by the protein structural class. Biochem. Biophys. Res. Commun. 2006;342:441–451. doi: 10.1016/j.bbrc.2006.01.159. [DOI] [PubMed] [Google Scholar]
- 21.Wang J., Feng J.A. Exploring the sequence patterns in the alpha-helices of proteins. Protein Eng. 2003;16:799–807. doi: 10.1093/protein/gzg101. [DOI] [PubMed] [Google Scholar]
- 22.Xia X., Xie Z. Protein structure, neighbor effect, and a new index of amino acid dissimilarities. Mol. Biol. Evol. 2002;19:58–67. doi: 10.1093/oxfordjournals.molbev.a003982. [DOI] [PubMed] [Google Scholar]
- 23.Kim O.T., Yura K., Go N. Amino acid residue doublet propensity in the protein-RNA interface and its application to RNA interface prediction. Nucleic Acids Res. 2006;34:6450–6460. doi: 10.1093/nar/gkl819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu M., Huang Z., Mao Y., Tao S. Neighbor preferences of amino acids and context-dependent effects of amino acid substitutions in human, mouse, and dog. Int. J. Mol. Sci. 2014;15:15963–15980. doi: 10.3390/ijms150915963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goliaei B., Minuchehr Z. Exceptional pairs of amino acid neighbors in alpha-helices. FEBS Lett. 2003;537:121–127. doi: 10.1016/s0014-5793(03)00105-4. [DOI] [PubMed] [Google Scholar]
- 26.Li W., Kinch L.N., Karplus P.A., Grishin N.V. ChSeq: A database of chameleon sequences. Protein Sci. 2015;24:1075–1086. doi: 10.1002/pro.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang G., Dunbrack R.L., Jr. PISCES: a protein sequence culling server. Bioinformatics. 2003;19:1589–1591. doi: 10.1093/bioinformatics/btg224. [DOI] [PubMed] [Google Scholar]
- 28.Hobohm U., Sander C. Enlarged representative set of protein structures. Protein Sci. 1994;3:522–524. doi: 10.1002/pro.5560030317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Minuchehr Z., Goliaei B. Propensity of amino acids in loop regions connecting beta-strands. Protein Pept. Lett. 2005;12:379–382. doi: 10.2174/0929866053765617. [DOI] [PubMed] [Google Scholar]
- 30.Rost B., Sander C. Conservation and prediction of solvent accessibility in protein families. Proteins. 1994;20:216–226. doi: 10.1002/prot.340200303. [DOI] [PubMed] [Google Scholar]
- 31.Jacoboni I., Martelli P.L., Fariselli P., Compiani M., Casadio R. Predictions of protein segments with the same aminoacid sequence and different secondary structure: a benchmark for predictive methods. Proteins. 2000;41:535–544. doi: 10.1002/1097-0134(20001201)41:4<535::aid-prot100>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 32.Gu S., Chan W.Y. Flexible and versatile as a chameleon-sophisticated functions of microRNA-199a. Int. J. Mol. Sci. 2012;13:8449–8466. doi: 10.3390/ijms13078449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nemeth-Pongracz V., Barabas O., Fuxreiter M., Simon I., Pichova I., Rumlova M., Zabranska H., Svergun D., Petoukhov M., Harmat V., Klement E., Hunyadi-Gulyas E., Medzihradszky K.F., Konya E., Vertessy B.G. Flexible segments modulate co-folding of dUTPase and nucleocapsid proteins. Nucleic Acids Res. 2007;35:495–505. doi: 10.1093/nar/gkl1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruvinsky A.M., Vakser I.A. Sequence composition and environment effects on residue fluctuations in protein structures. J. Chem. Phys. 2010;133:155101. doi: 10.1063/1.3498743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oldfield C.J., Dunker A.K. Intrinsically disordered proteins and intrinsically disordered protein regions. Annu. Rev. Biochem. 2014;83:553–584. doi: 10.1146/annurev-biochem-072711-164947. [DOI] [PubMed] [Google Scholar]
- 36.Uversky V.N., Oldfield C.J., Dunker A.K. Intrinsically disordered proteins in human diseases: introducing the D2 concept. Annu. Rev. Biophys. 2008;37:215–246. doi: 10.1146/annurev.biophys.37.032807.125924. [DOI] [PubMed] [Google Scholar]
- 37.Yoon S., Jung H. Analysis of chameleon sequences by energy decomposition on a pairwise per-residue basis. Protein J. 2006;25:361–368. doi: 10.1007/s10930-006-9023-6. [DOI] [PubMed] [Google Scholar]
- 38.Apweiler R., Bairoch A., Wu C.H., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Magrane M., Martin M.J., Natale D.A., O'Donovan C., Redaschi N., Yeh L.S. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res. 2004;32:D115–D119. doi: 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holliday G.L., Bairoch A., Bagos P.G., Chatonnet A., Craik D.J., Finn R.D., Henrissat B., Landsman D., Manning G., Nagano N., O'Donovan C., Pruitt K.D., Rawlings N.D., Saier M., Sowdhamini R., Spedding M., Srinivasan N., Vriend G., Babbitt P.C., Bateman A. Key challenges for the creation and maintenance of specialist protein resources. Proteins. 2015;83:1005–1013. doi: 10.1002/prot.24803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bairoch A. SEQANALREF: a sequence analysis bibliographic reference databank. Comput. Appl. Biosci. 1991;7:268. doi: 10.1093/bioinformatics/7.2.268. [DOI] [PubMed] [Google Scholar]
- 41.George R.A., Heringa J. An analysis of protein domain linkers: their classification and role in protein folding. Protein Eng. 2002;15:871–879. doi: 10.1093/protein/15.11.871. [DOI] [PubMed] [Google Scholar]
- 42.Ramakrishnan C., Ramachandran G.N. Stereochemical criteria for polypeptide and protein chain conformations. II. Allowed conformations for a pair of peptide units. Biophys. J. 1965;5:909–933. doi: 10.1016/S0006-3495(65)86759-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qiu W.Q., Folstein M.F. Insulin, insulin-degrading enzyme and amyloid-beta peptide in Alzheimer's disease: review and hypothesis. Neurobiol. Aging. 2006;27:190–198. doi: 10.1016/j.neurobiolaging.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 44.Maiuri T., Woloshansky T., Xia J., Truant R. The huntingtin N17 domain is a multifunctional CRM1 and ran-dependent nuclear and cilial export signal. Hum. Mol. Genet. 2013;22:1383–1394. doi: 10.1093/hmg/dds554. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.