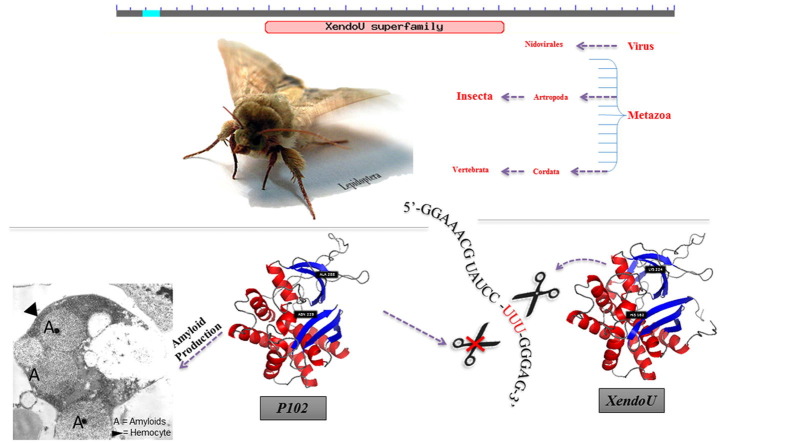
Graphical abstract
Keywords: Endoribonuclease-U, Lepidoptera, Amyloids, Heliothis virescens, Trichoplusia ni, Immunity
Highlights
-
•
The hemocytes of Heliothis virescens larvae produce a protein named P102.
-
•
P102 has a endoribonuclease-U (XendoU) domain but lacks essential catalytic residues.
-
•
P102 lost its original enzymatic activity to produce functional amyloids.
-
•
Amyloids formed by P102 are involved in Lepidopteran immune response.
-
•
Lepidoptera P102 orthologs form a XendoU subfamily with similar function.
Abstract
Hemocytes of Heliothis virescens (F.) (Lepidoptera, Noctuidae) larvae produce a protein, P102, with a putative endoribonuclease-U domain. In previous works we have shown that P102 is involved in Lepidopteran immune response by forming amyloid fibrils, which catalyze and localize melanin deposition around non-self intruders during encapsulation, preventing harmful systemic spreading. Here we demonstrate that P102 belongs to a new class of proteins that, at least in Lepidoptera, has a diminished endoribonuclease-U activity probably due to the lack of two out of five catalytically essential residues. We show that the P102 homolog from Trichoplusia ni (Lepidoptera, Noctuidae) displays catalytic site residues identical to P102, a residual endoribonuclease-U activity and the ability to form functional amyloids. On the basis of these results as well as sequence and structural analyses, we hypothesize that all the Lepidoptera endoribonuclease-U orthologs with catalytic site residues identical to P102 form a subfamily with similar function.
1. Introduction
P102, the amyloidogenic protein produced by hemocytes of Heliothis virescens (F.) (Lepidoptera, Noctuidae) larvae (Falabella et al., 2012) is a member of the novel XendoU protein family (Renzi et al., 2006) that includes several proteins annotated in distantly related organisms ranging from viruses to humans (Renzi et al., 2006, Snijder et al., 2003, Caffarelli et al., 1994). Members of the XendoU family are characterized by RNA binding and processing (hydrolytic) activity. They are Mn2+-dependent nucleases that cleave U-stretches of RNA and produce 2′, 3′-cyclic-phosphate termini products, which is a unique characteristic for this class of RNases (Laneve et al., 2008). The amphibian XendoU, the human PP11 and the genetic marker of Nidovirales NendoU (Nsp 15) are three well-characterized proteins belonging to this family. They all share conserved regions containing a proposed active site with five amino acid residues that are crucial for the catalytic activity (Renzi et al., 2006, Gioia et al., 2005). Despite their similarities these proteins are involved in distinct RNA processing pathways in different organisms (Bohn and Winckler, 1980, Bohn et al., 1981, Inaba et al., 1980a, Inaba et al., 1980b, Inaba et al., 1981, Inaba et al., 1982, Laneve et al., 2003, Laneve et al., 2008, Ivanov et al., 2004, Ulferts and Ziebuhr, 2011). Other XendoU family members were frequently, but erroneously annotated as serine protease-like enzymes on the basis of their homology with the human placental protein 11 (PP11), that previously was thought to be a protease (Grundmann et al., 1990, Laneve et al., 2008). However, the highly conserved active site found in a wide range of orthologs (Gioia et al., 2005, Renzi et al., 2006, Laneve et al., 2008) may suggest that they all have endoribonucleolytic activity, although they are involved in different RNA-processing pathways.
In previous work, (Falabella et al., 2012) we identified and characterized the 102 gene (EBL-Bank ID: FR751090) in larval hemocytes of the Lepidopteran H. virescens (F.) (Lepidoptera, Noctuidae), encoding a predicted protein (P102), that showed 86% sequence identity with a venom protein from Lonomia obliqua (Lepidoptera, Saturnidae) larvae (Veiga et al., 2005), and possesses a putative endoribonuclease-U (XendoU) domain. We demonstrated that P102 is involved in insect cellular defenses in hemocytes of H. virescens larvae by forming amyloid fibrils in the reticulum endoplasmic cisternae. Upon immune challenge, these amyloid fibrils are released and form a fibrillar scaffold around the non-self intruder, promoting melanin synthesis directly on this scaffold (Falabella et al., 2012, Grimaldi et al., 2012). To our knowledge this was the first model that describes the mechanism of melanin deposition during encapsulation in a Lepidopteran, shedding new light on an aspect of insect cellular immune response previously unknown.
Here, we investigate the putative enzymatic activity of P102, belonging to the novel poly(U) specific endoribonuclease protein family (Laneve et al., 2003, Renzi et al., 2006), trying to understand if it could have a catalytic function in RNA cleavage and fragmentation apart from its roles in amyloidogenesis (Falabella et al., 2012) and hemocytes capsule melanin deposition. The endoribonuclease assay shows only a slight residual activity for P102, which could be explained by the fact that two of the conserved residues crucial for catalysis (Gioia et al., 2005) are missing in P102. These results most likely rule out any direct involvement of P102 in RNA fragmentation. Sequence analysis of homologous proteins of P102 from other insects leads us to hypothesize that P102 belongs to a XendoU subfamily, characterized by a conserved set of alternative residues in the catalytic site, that is restricted to Lepidoptera. This protein subfamily is distinct from other insect XendoU sequences that generally belong to groups roughly according to species phylogeny. In addition, the in vitro functional characterization of the P102 homolog from Trichoplusia ni (Hübner) (Lepidoptera, Noctuidae) corroborates our hypothesis by demonstrating that the P102 characteristics extend to other members of this subfamily. Thus our results suggest that the amyloid-mediated melanin synthesis in the defense against intruders is a conserved mechanism within the order Lepidoptera, and that there could be an association between the loss of ancestral enzymatic RNA cleavage activity and the ability of the Lepidopteran alternative XendoU-like proteins to form amyloids.
2. Material and methods
2.1. Sequence analysis and molecular modeling
Database searches were performed using the nucleotide and the amino acid sequences of P102 (GenBank IDs: FR751090.1 and CBY85302.1, respectively) and the BLAST algorithm available at the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/). In particular, the complete P102 sequence was compared to the NCBI non-redundant protein database, the NCBI non-redundant nucleotide and EST databases and our extensive in-house transcriptome databases (H. Vogel, MPI Jena). The conserved domain architecture in protein sequences was identified searching the Conserved Domain Database (CDD) at NCBI. The translation into amino acid residue sequences and analysis of the physico-chemical parameters of proteins were carried out using on-line tools available at the Expasy SIB Bioinformatics Resource Portal (http://expasy.org/). Occurrence and position of signal peptides, glycosylation and phosphorylation sites in protein sequences were predicted using the CBS prediction server (http://www.cbs.dtu.dk/services/).
Multiple amino acid sequence alignments were performed using the default options of the Uniprot program (http://www.uniprot.org). Conserved residues in the alignments were highlighted with Multiple Align show (http://www.bioinformatics.org/sms/multi_align.html).
Phylogenetic analyses based on MAFFT (E-INS-I parameter set; Katoh et al., 2005) alignments were made using deduced amino acid sequences from insect endoribonuclease-U (XendoU) transcripts retrieved from NCBI and the other sources described above. The phylogenetic tree was inferred by the maximum likelihood (ML) method using PhyML (Dereeper et al., 2008) available at LIRMM (http://www.phylogeny.fr/) and displayed and edited with FigTree (http://tree.bio.ed.ac.uk/software/figtree). Additionally, a Bayesian analysis implemented in Mr. Bayes 3.2.2 (Ronquist and Huelsenbeck, 2003) was performed, using the Xenopus laevis XendoU sequence as outgroup. The Markov Chain Monte Carlo runs were carried out for 1,000,000 generations after which log likelihood values showed that equilibrium had been reached after the first 400 generations in all cases, and those data were discarded from each run and considered as ‘burnin’. Two runs were conducted per dataset showing agreement in topology and likelihood scores. The maximum likelihood and the Bayesian tree topologies including their general subfamily relationships were generally in good agreement.
Three-dimensional models of P102 and P102Tni were made with Modeller (Eswar et al., 2006) and analyzed with PyMol (http://www.pymol.org).
2.2. Production of recombinant proteins in Escherichia coli
The cDNA coding for the 102 ORF of H. virescens was cloned into pET32 Ek/LIC vector (Novagen, San Diego, California, USA) for bacterial expression, as previously described by Falabella et al., 2012. The cDNA coding for the mature T. ni P102 homolog lacking the signal peptide was amplified by PCR using the following primers:
Tn102 pET32 forw 5′ GACGACGACAAGATGGACAACCTAGCCAACGCA 3′
Tn102 pET32 rev 5′ GAGGAGAAGCCCGGTTAGGAGAAGGGGGTGGG 3′
The primers were designed by adding appropriate extensions (underlined) to allow direct cloning of the fragment into the expression vector pET32 Ek/LIC, in frame with Trx, His and S tags (a total of 17 kDa) according to the manufacturer’s instructions (Ek/LIC Cloning Kits Novagen, San Diego, California, USA). The obtained construct was sequenced and used to transform E. coli Rosetta-gami 2 (DE3) cells (Novagen, San Diego, California, USA) according to the manufacturer’s protocol.
The expression in bacterial cells and the purification of P102 and T. ni P102 (named P102Tni) recombinant proteins were performed as described by Falabella et al., 2012. Briefly, the recombinant proteins were expressed by growing the transformed Rosetta-gami 2 (DE3) cells at 37 °C and induced (OD600 0.6) with 1 mM isopropyl β-d-thiogalactoside (IPTG) for 2 h at room temperature. The recombinant proteins were subsequently purified by nickel-nitrilotriacetic acid-agarose affinity chromatography under native condition on Protino Ni-TED 1000 packed columns (Macherey–Nagel, Düren, Germany). The purified proteins were dialyzed to eliminate imidazole and digested with 1 U of enterokinase (Novagen, San Diego, California, USA) for 16 h at room temperature in order to eliminate the fusion tag. Purified proteins were quantified by using the Bio-Rad protein Assay kit (Bio-Rad, Hercules, California, USA).
2.3. Expression of P102, P102Tni and XendoU in insect cells
The expression of H. virescens cDNA 102 in Drosophila Schneider 2 (S2) cells (Life Technologies, Carlsbad, California, USA) was performed as described by Falabella et al., 2012. The cDNA coding for P102Tni and the cDNA coding for XendoU (kindly provided by Dr. P. Laneve) were used to amplify the coding region by PCR using specific primers containing restriction sites for Xba I and Sac II (underlined sequences), respectively:
Tn102 pIZT forw 5′ CTAGTCTAGAATGAAGATTGCCATTGTG 3′
Tn102 pIZT rev 5′ TCCCCGCGGGTATAGAGGGTAGG 3′
XendoU pIZT forw 5′ CTAGTCTAGAATGGCGAGTAACAGGGGGCA 3′
XendoU pIZT rev 5′ TCCCCGCGGCAATAACCCGGATCTGTAC 3′
The PCR products were both cloned into the pCR2.1 TOPO vector (Life Technologies, Carlsbad, California, USA) following the manufacturer’s protocol and the sequence identity was verified by sequencing. The inserts were then digested with Xba I and Sac II and cloned into the expression vector pIZT/V5-His (Life Technologies, Carlsbad, California, USA). The obtained constructs were sequenced and used to transfect S2 cells. The expression of P102Tni and XendoU in S2 cells and the selection of stable polyclonal cell lines were performed as described previously for P102 (Falabella et al., 2012). The production of the recombinant protein was verified by western blot analysis using the H. virescens P102 antibody for P102Tni with P102 as a positive control. The recombinant XendoU protein was verified using the anti-V5 antibody (Life Technologies, Carlsbad, California, USA).
2.4. Endoribonuclease-U specific activity
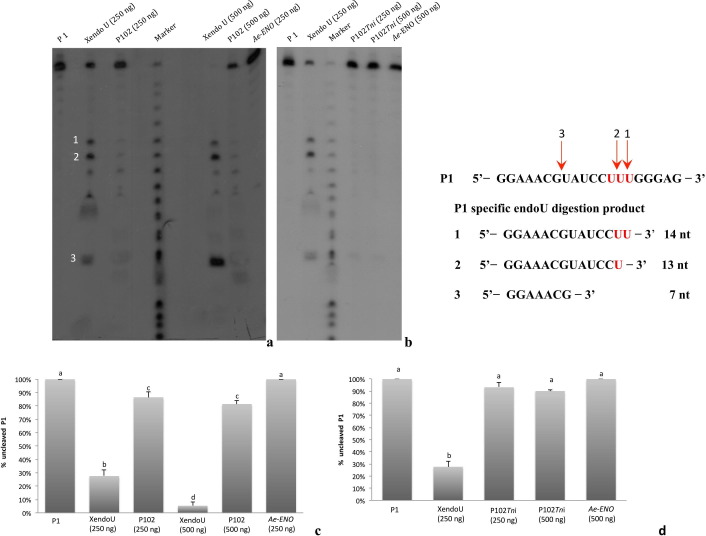
The enzymatic activity of the recombinant proteins, P102 and P102Tni, was assayed by electrophoretic analysis using conditions described by Laneve et al., 2003, Laneve et al., 2008. Briefly, the synthetic oligoribonucleotide P1 (5′ GGAAACGUAUCCUUUGGGAG 3′), containing a natural cleavage site for XendoU (Laneve et al., 2003), was purchased from Sigma and 5′ end-labeled with [γ-32P]ATP (Perkin Elmer Life Sciences, Waltham, Massachusetts, USA) by using T4 polynucleotide kinase (Promega, Fitchburg, Wisconsin, USA) according to the manufacturer’s instructions. The labeled RNA oligonucleotide was precipitated with ethanol and ammonium acetate following a standard protocol with minor modifications (Sambrook et al., 1989) and resuspended in sterile water. The activity, expressed in cpm/μl, was measured by using a scintillation counter (Beckman Coulter, Brea, California, USA).
The assay was carried out by incubating 3.5 × 104 cpm of 5′-32P-labeled oligoribonucleotide P1 with 250 and 500 ng of each protein in 5 mM MnCl2, 50 mM NaCl, 25 mM Hepes pH 7.5, 1 mM DTT, 10 μg of baking yeast tRNA (Roche, Penzberg, Germany), 20 units of RNase inhibitors (RNase OUT 40 U/μl, Life Technologies, Carlsbad, California, USA). The reactions were carried out at 24 °C for 30 min. 250 and 500 ng of the recombinant XendoU, kindly provided by Dr. E. Caffarelli (CNR, Rome, Italy), were used as a positive control, while 250 and 500 ng of an unrelated recombinant protein, the AeENO (Falabella et al., 2009) were used as negative control.
The products of the reactions were analyzed on 20% polyacrylamide gels containing 7 M urea by using the Sequi-Gen GT Nucleic Acid Electrophoresis Cell (Bio-Rad, Hercules, California, USA). Radioactive signals were revealed by exposing the gel to a Fuji RX film at room temperature for an appropriate time. The nucleotide RNA ladder was prepared by incubation of 250,000 cpm of 5’-32P-labeled oligoribonucleotide P1 in 500 mM NaHCO3 at 90 °C for 20 min. The experiment was replicated three times in the same experimental conditions.
The resulting gels were digitized as 300 dpi, 16 bit TIFF images using a desktop scanner and imported to GelCompar II (Applied Maths, Sint-Martens-Latem, Belgium) where background subtraction and detection for each lane of bands corresponding to 5′-32P-labeled oligoribonucleotide P1 and/or its digestion products were carried out. Band intensity was expressed as relative surface area considering to the sum of the intensities of all the bands of each lane corresponding to 100%. The measurement of enzyme activity was calculated as the percentage uncleaved RNA substrate (P1) compared to the input RNA (untreated P1). Data (mean ± SE) were analyzed by general linear model analysis of variance (ANOVA), with subsequent comparison between means using a post hoc Tukey test. All the analyses were carried out using R. 3.0.1. for windows (www.r-project.org).
2.5. Transmission electron microscopy and immunogold labeling
For routine TEM, collected S2 cells, stably expressing P102Tni, and XendoU, respectively, were fixed at 4 °C with 2% glutaraldehyde in 0.1 M Na-cacodylate buffer (pH 7.2) for 2 h. Cells were centrifuged at 400g for 7 min at 4 °C, the pellet washed in 0.1 M Na-cacodylate buffer (pH 7.2), and post-fixed at 4 °C for 2 h with 1% osmic acid in cacodylate buffer (pH 7.2). After standard dehydration in an ethanol series, samples were embedded in an Epon-Araldite 812 mixture (Sigma, St. Louis, Missouri, USA) and sectioned with a Reichert Ultracut S ultratome (Leica, Vienna, Austria). For morphological analysis, thin sections were stained by uranyl acetate and lead citrate. S2 cells were also processed for immunogold labeling according to published protocols (Donini et al., 1989). The used primary antibodies were: rabbit anti-P102 and rabbit anti-V5 (diluted 1:100 in saturation buffer). Primary antibodies were then visualized by immunochemical staining with secondary goat anti-rabbit IgG (H+L)-gold conjugate antibodies (GE Healthcare Amersham, Buckingamshire, UK) (particle size, 10 nm) diluted 1:40 (incubation 30 min at room temperature). Control sections were incubated with the secondary antibody alone. Samples were counterstained with uranyl acetate in water. Sections were observed with a Jeol 1010 EX electron microscope (Jeol). Data were recorded with a MORADA digital camera system (Olympus, Tokyo, Japan). S2 cells stably expressing the unrelated recombinant protein AeENO (Falabella et al., 2009), were used as control in both the TEM and the immunogold labeling experiments.
3. Results
3.1. P102 and P102Tni sequence analysis and molecular modeling
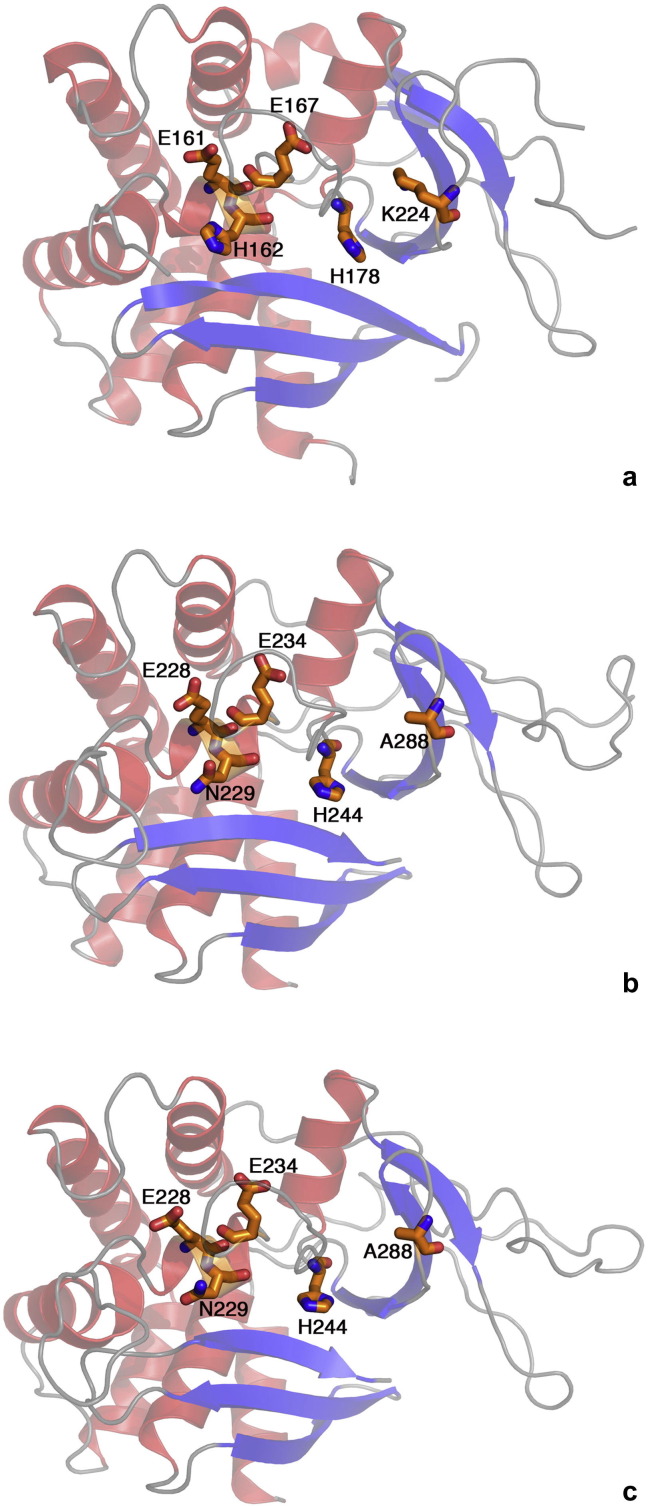
P102 (GenBank ID: CBY85302.1) consists of 349 amino acids with a molecular mass of 39.3 kDa, a theoretical isoelectric point of 6.02 and a predicted N-terminal signal peptide of 16 amino acids. One N-glycosylation site, two O-GlnNAc glycosylation sites and nineteen phosphorylation sites were predicted in silico as possible sites for post-translation modifications. A putative endoribonuclease-U (XendoU) domain was identified by Conserved Domain Database (CDD) analysis. To further support this prediction, a 3D homology model of P102 was generated in Modeller by using the closest sequence homologue in the Protein Data Bank (PDB) – the experimental 3D structure of XendoU (PDB ID: 2c1wB) (Renzi et al., 2006) – as a template (Fig. 1 a). The two sequences share 28% identical (45% similar) residues. The overall structural organization of the P102 model (Fig. 1b) is similar to that of the XendoU structure with a superimposed Root-Mean-Square Deviations (RMSD) of 0.443 Å for 252 paired α-carbon atoms. Also the secondary structure elements of the P102 model are similar to those of XendoU with the exception of a region of residues 99–136 of P102 due to low sequence similarity and a missing loop (residues 44–54) in the XendoU structure. These observations suggest that P102 could lack a small β-sheet consisting of two antiparallel β-strands and that this specific region is likely to be less ordered.
Fig.1.
Homology models of P102 and P102Tni. The structure of XendoU (a) as well as the homology models of P102 (b) and P102Tni (c) are visualized as ribbons by PyMOL. The α-helices are depicted in red, β-strands in blue and random coil in grey. The five residues involved in the enzymatic activity are highlighted in orange. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The 1050 nucleotide coding region of the homologous gene of T. ni encodes a putative protein (P102Tni) of 349 amino acids with a theoretical molecular weight of 39.3 kDa and an isoelectric point of 5.9, and a predicted N-terminal signal peptide of 16 amino acids. Two O-GlnNAc glycosylation sites and twenty-one phosphorylation sites were identified as predicted post-translation modifications. P102Tni is 89% identical to P102 and therefore CDD and PDB searches predicted that also P102Tni is a structural homologue of XendoU with 23% identical (40% similar) residues. The XendoU structure (Renzi et al., 2006) was used as a template in Modeller to create a homology model of P102Tni (Fig. 1c) that had an RMSD of 0.469 Å for 265 paired α-carbon atoms when superimposed with its template. The P102Tni 3D model has a similar overall structure to that of XendoU and similar secondary structural elements to that of the P102 model (Fig. 1a–c).
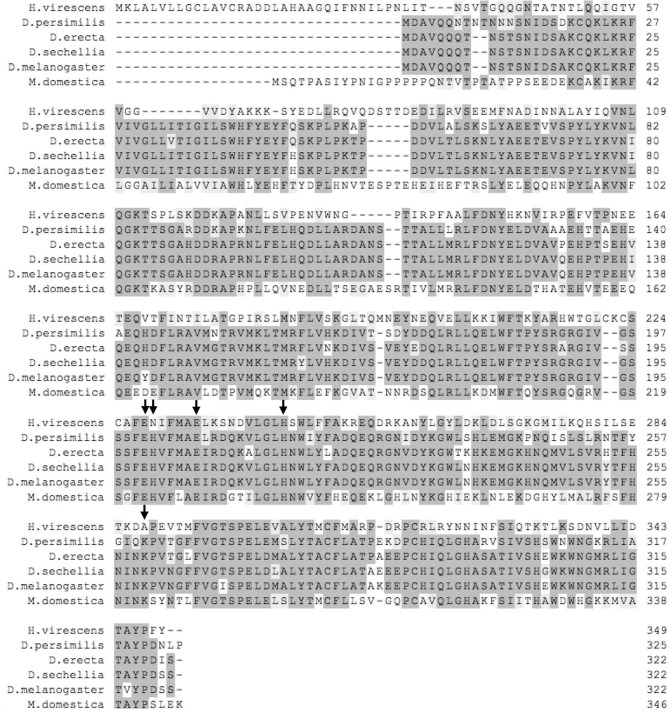
The alignment of P102 and P102Tni with XendoU showed two out of five catalytically essential residues (Gioia et al., 2005) lacked in these Lepidopteran homologues; the asparagine N229 and alanine A288, corresponding to H162 and K224, respectively, in XendoU, are different while E228, E234 and H244, corresponding to E161, E167 and H178 in XendoU, are conserved (Figs. 1 and 2 ).
Fig. 2.
Partial alignment of P102 and P102Tni sequences (residues 172–348) with XendoU. The number of the last amino acid of the aligned region is indicated. Identical and conserved amino acids are shaded in light gray and very light gray, respectively. Residues involved in the enzymatic activity are highlighted: arrows mark the conserved residues of the catalytic site of P102 and P102Tni compared to XendoU while arrowheads indicate alternative ones.
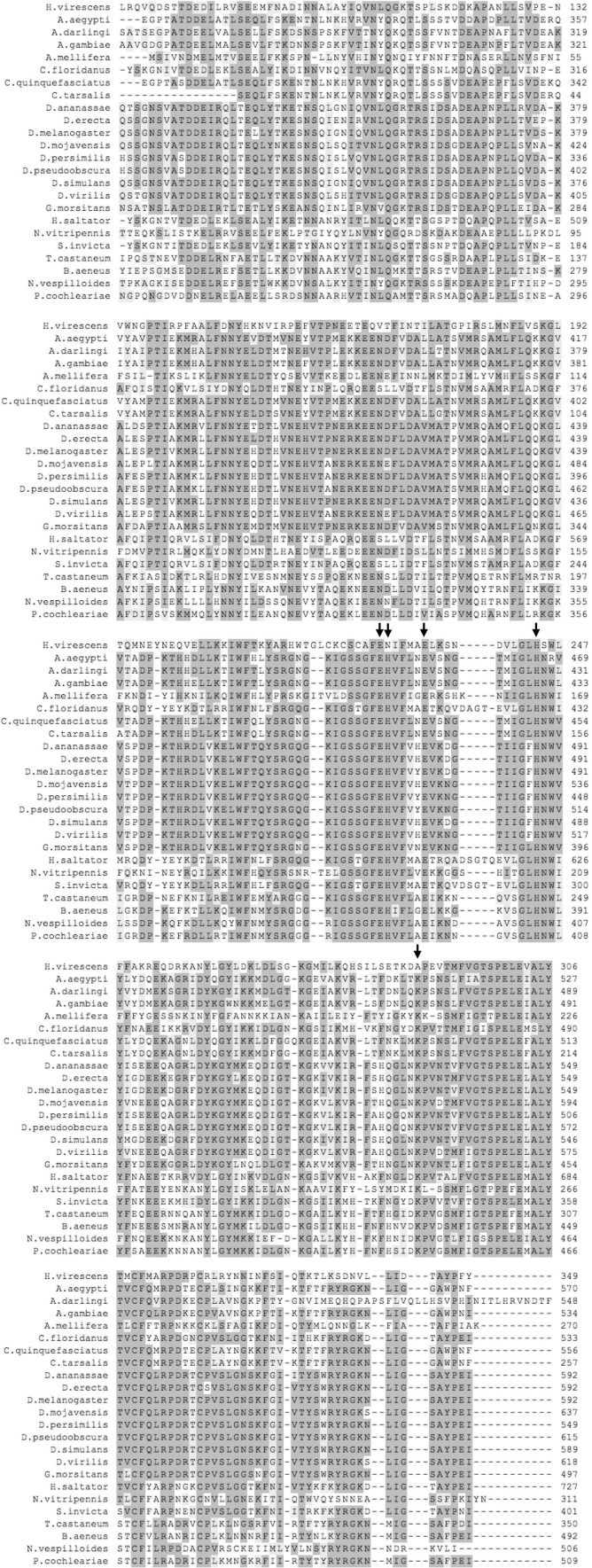
Searching the NCBI nr protein database and in-house transcriptome databases (Supplementary Fig. 1) (consisting of species from the orders Lepidoptera, Coleoptera and Hymenoptera) using the P102 amino acid sequence as query revealed two distinct groups of proteins which differed in their level of sequence conservation to the P102 protein. The proteins forming a group with very high (>75%) sequence identity are all from species of the order Lepidoptera. These include a XendoU-like protein from Papilio xuthus (GenBank ID: BAM17994.1) with 88% sequence identity and a XendoU-like protein from P. polytes (GenBank ID: BAM20530.1) with 86% sequence identity to P102. The other group of proteins displaying 42–43% sequence identity to P102 was found in Lepidoptera, such as from P. xuthus (GenBank ID: BAM19803.1), P. polytes (GenBank ID: BAM20523.1) and D. plexippus (GenBank ID: EHJ66341.1), and with a lower amino acid identity of around 40%, with characterized or predicted proteins from other insect orders, such as Diptera, Hymenoptera and Coleoptera.
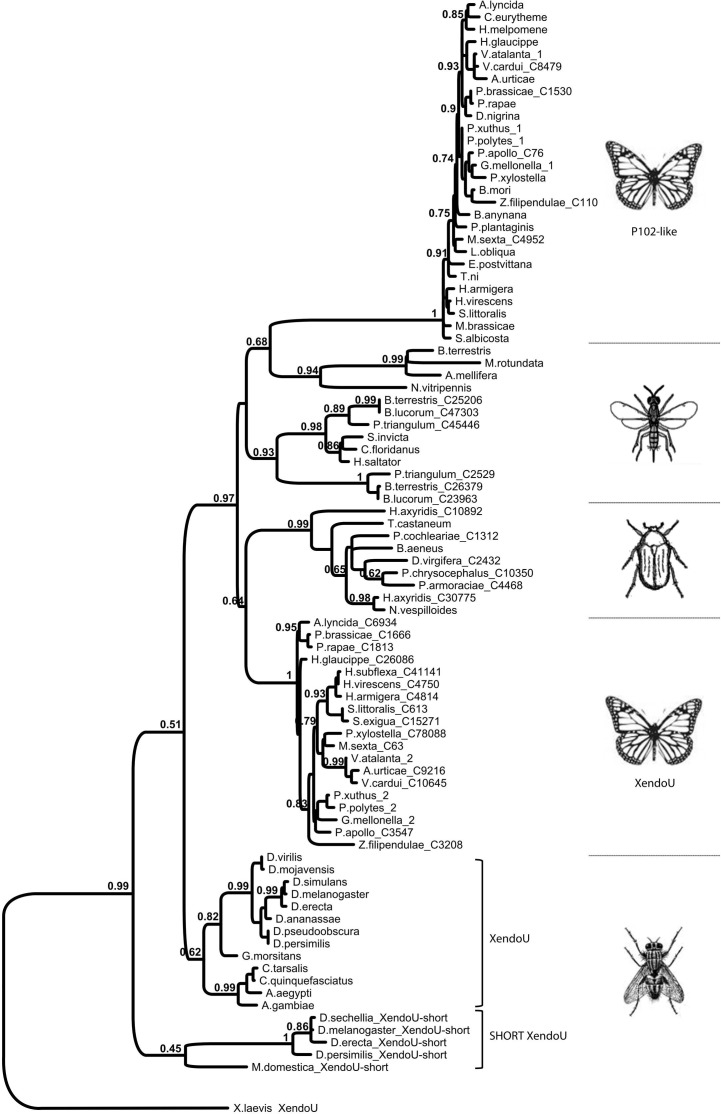
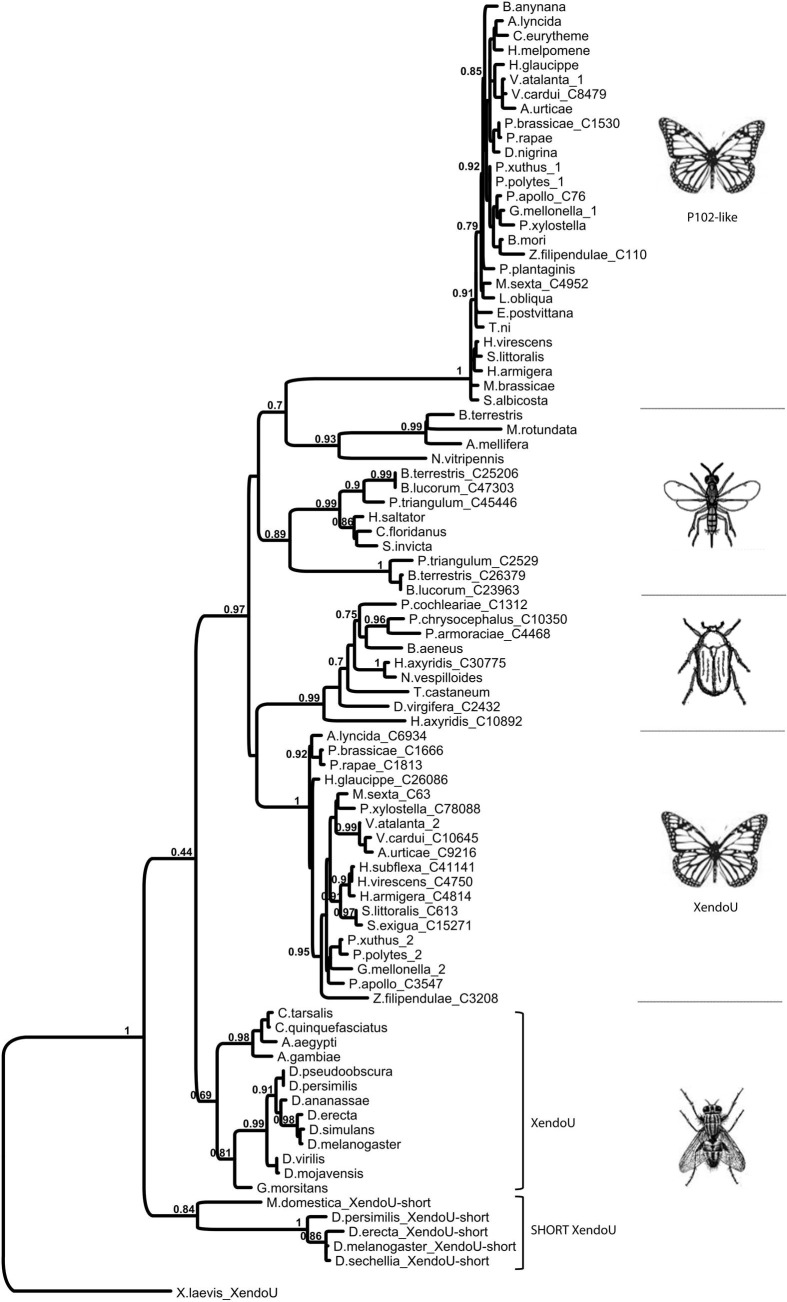
All nucleotide sequences were translated into the respective predicted amino acid sequences and the molecular parameters estimated using the on-line tools at Expasy SIB Bioinformatics Resource Portal. Comparison of the amino acid sequence of P102 with Lepidopteran homologs (identity > 75%), including P102Tni, revealed a high degree of sequence conservation along the complete amino acid sequence. Multiple alignments showed the occurrence of the same set of residues, as in the endoribonuclease-U catalytic site of P102, in all of the Lepidopteran sequences with very high sequence identity (Supplementary Fig. 2). In contrast to this, the other group of Lepidopteran P102-like homologs (identity around 40%) did not have this set of residues conserved in the catalytic site but the XendoU residues. The multiple amino acid alignment of P102 with homologous sequences from other insect orders displayed high sequence conservation only for the XendoU domain (Supplementary Fig. 3), but had the catalytic residues of XendoU. Furthermore, XendoU-like proteins generally have a rather variable N-terminal region, both in terms of length and amino acid identity, that precedes the XendoU domain. In contrast, P102-like proteins from Lepidoptera (Supplementary Fig. 2) as well as the short XendoU-like proteins from some Diptera (Supplementary Figs. 1 and 4), displayed a higher level of conservation in sequence and overall protein length. All of the available insect XendoU-like protein sequences were used to perform a phylogenetic analysis (Fig. 3 ). The Lepidopteran P102-like proteins form a highly supported clade, and include P102 of H. virescens and P102Tni, all of which have the same residues in the catalytic site. The clade of Lepidopteran P102 is clearly separated both from the other XendoU-like proteins from other insect orders and from the XendoU-like proteins of Lepidopteran origin with the XendoU set of residues in the active site (Fig. 3). This observation was further supported by the strength of the nodes. Short XendoU-like sequences that lack the variable N-terminal region were identified in several Dipteran species. These sequences have the XendoU set of residues in the active site and do not cluster together with the more highly conserved Lepidopteran P102-like proteins (Fig. 3). Importantly, the overall tree topology and the node support values for the major clades are almost identical for two different phylogenetic trees based on either the full-length sequence alignment (Fig. 3) or on the amino acid sequences trimmed to the shorter P102-like protein length (Supplementary Fig. 5).
Fig. 3.
Maximum-likelihood tree of the P102 sequence, indicated as H. virescens, and full-length sequence of homologs from Lepidoptera and other insect species. Mutated and conserved XendoU domain homologs from Lepidoptera are designated as 1 and 2, respectively. Names of insects are indicated for each branch. The accession numbers for mutated homologs from Lepidoptera and conserved homologs from other insects have been listed in Supplementary Figs. 2 and 3, respectively. The accession numbers for conserved homologs from Lepidoptera retrieved from NCBI are as follows: Papilio polytes (GenBank ID: BAM20523.1), P. xuthus (GenBank ID: BAM19803.1) and Danaus plexippus (GenBank ID: EHJ66341.1). Additional conserved sequences from Lepidoptera (Galleria mellonella, Parasemia plantaginis, Vanessa atalanta) were retrieved from in-house transcriptome databases. Numbers above branches are bootstrap support values shown as percentages of a total of 100 bootstrap replications. Branch lengths are proportional to evolutionary distance according to the provided scale.
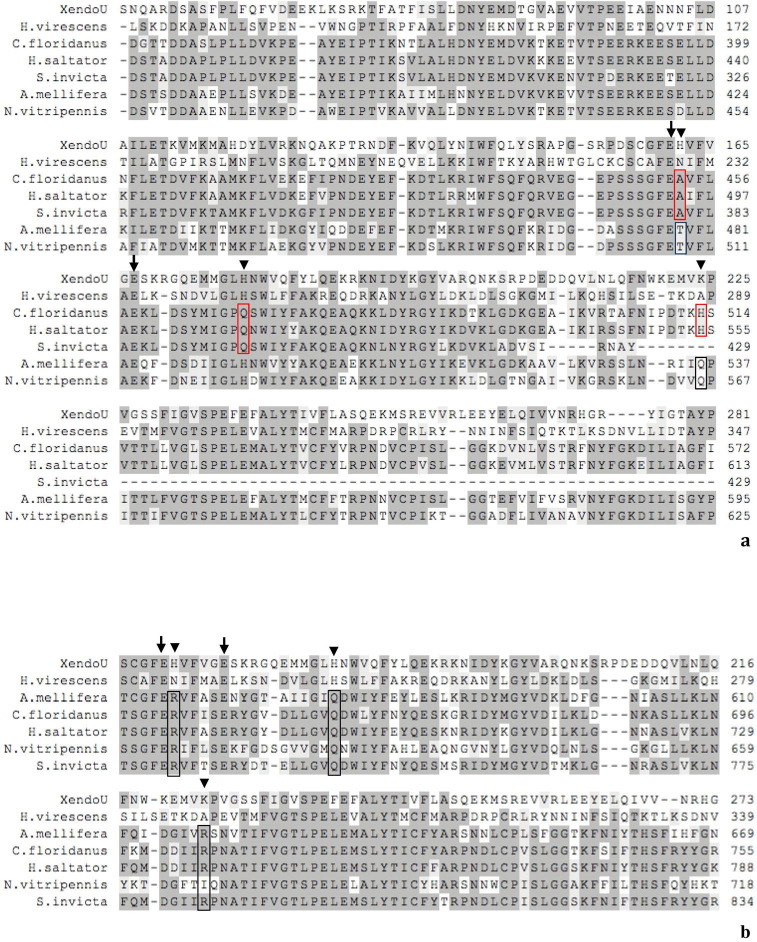
Moreover, we also found homologous sequences in several Hymenoptera sharing a sequence identity of about 40% with P102 that did not contain the canonical XendoU residues in the catalytic site (not included in the phylogenetic tree) but instead three different sets of the five catalytic residues (Supplementary Fig. 6).
3.2. Endoribonuclease-U specific activity of P102 and P102Tni
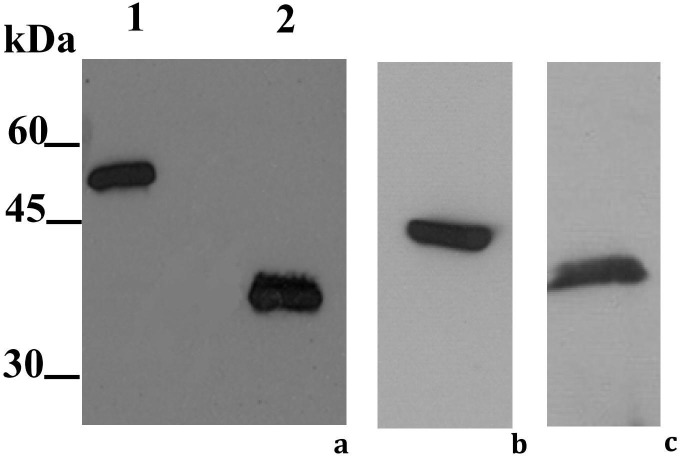
To investigate the endoribonuclease-U activity of P102 and P102Tni, we heterologously expressed the recombinant proteins in E. coli. Western blot analysis of the purified protein demonstrated that the anti P102 antibody specifically recognized P102Tni protein (Fig. 4 a), showing an analogous antigenicity already predicted on the basis of the strong sequence identity of 89% with P102. P102 was used as positive control (data not shown).
Fig. 4.
Western blot analysis of P102Tni. Positive signals to anti-P102 antibody both of the purified recombinant P102Tni protein before (1) and after (2) digestion by enterokinase (a) and of the medium collected from stably transformed Drosophila S2 cells (b), confirmed the strong identity of P102Tni with P102. Positive signal to anti-V5 antibody of purified recombinant XendoU protein recovered from S2 cytoplasm (c).
As a substrate for testing enzymatic activity, we employed the synthetic oligoribonucleotide P1 that contains the upstream distal cleavage site for XendoU, UUU (Laneve et al., 2003, Laneve et al., 2008). The analysis of the reaction products displayed only a slight residual activity for P102 (Fig. 5 a) and P102Tni (Fig. 5b), compared to XendoU activity (Fig. 5a, b). Processing experiments carried out in the presence of a greater quantity of enzymes (500 ng) did not show any significant differences in the catalytic activity of P102 and P102Tni (Fig. 5a, b), while excess of XendoU caused the cleavage at minor sites (Laneve et al., 2003). No activity was detected in absence of manganese, further confirming the specific ion requirement of this class of enzymes (data not shown). No activity was detected for both the used concentrations of the recombinant unrelated protein AeENO.
Fig. 5.
Analysis of the endoribonuclease-U activity of P102 (a) and P102Tni (b). Assay performed with 250 ng or 500 ng of each protein (P102 and P102Tni) with 5′-32P-labeled oligoribonucleotide P1, displayed only a slight residual activity. The greater quantity of enzymes (500 ng) did not show any significant differences in the catalytic activity of P102 and P102Tni. Labeled oligoribonucleotide P1 incubated in absence of enzymes was loaded in the first lanes (a, b). The XendoU enzyme (250 ng and 500 ng) and an unrelated protein, the Ae-ENO from Aphidius ervi (250 ng and 500 ng) were used as positive and negative controls, respectively. Marker: nucleotide ladder, generated by alkaline digestion of P1 oligoribonucleotide (Laneve et al., 2003). Densitometric analysis of P102 (c), P102Tni (d) and controls: enzymatic activity is expressed as relative surface area corresponding to band intensity of the uncleaved RNA (oligoribonucleotide P1 substrate) in each lane. Error bars represent the standard error of the mean for three independent experiments. Significant differences are denoted by different letters (Tukey’s test, p < 0.05).
The endoribonuclease-U activity of P102, P102Tni, XendoU and Ae-ENO was quantified and compared by densitometric analysis of band intensities of each lane. In Fig. 5c and d the relative surface area corresponding to band intensity of the uncleaved RNA (oligoribonucleotide P1 substrate) is reported. These data demonstrate that P102 and P102Tni have a statistically significant reduced RNase-like activity, leaving 86.3% ± 4 (SE) and 93.2% ± 3.6 oligoribonucleotide P1 substrate uncleaved, respectively, compared to XendoU that leaves 27.6% ± 4.4 uncleaved substrate at optimal enzyme concentration (250 ng).
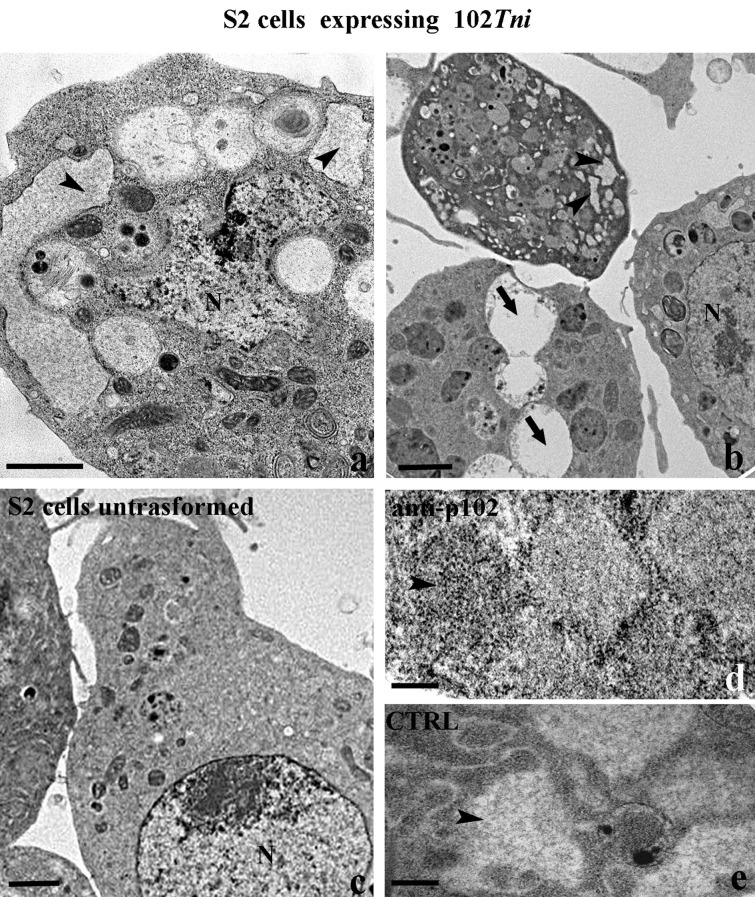
3.3. Production of fibrillar material in stable polyclonal insect cell line of P102Tni
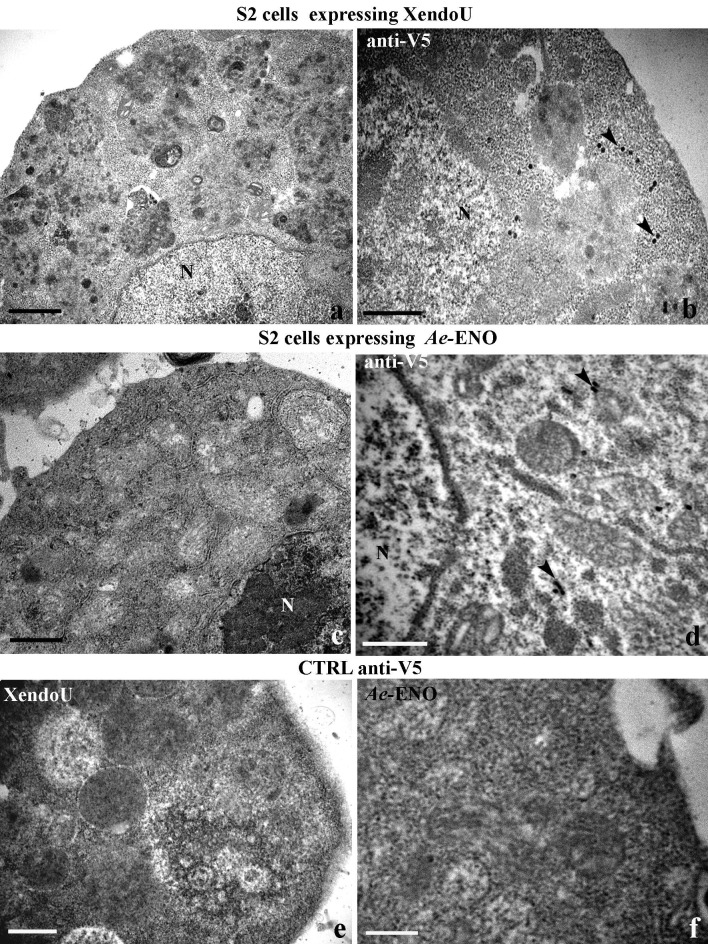
The complete coding region of cDNA 102Tni and XendoU were cloned into the pIZT/His-V5 vector and the constructs were transfected into S2 cells in order to produce polyclonal cell lines expressing these proteins. The recombinant protein, fused with a C-terminal tag containing the V5-epitope and the hexahistidine tail, was secreted into the medium in case of P102Tni and recovered from S2 cytoplasm in case of XendoU as demonstrated by western blot analysis (Fig. 4b, c).
To investigate if P102Tni is involved in formation of amyloid fibrils similar to P102, we observed stably transformed S2 cells expressing P102Tni by TEM. Several cells showed dilated reticulum cisternae filled with fibrillar material (Fig 6 a, b) while others were characterized by completely empty cisternae due to the exocytosis of fibrillar material (Fig. 6b) that was not visible in untransformed cells (Fig. 6c). Immunogold labeling experiments with the anti P102 antibody displayed positive signals in transfected S2 cells, confirming the role of P102Tni in the formation of fibrillar material (Fig. 6d, e). The same experiment conducted on S2 cells stably expressing XendoU (Fig. 7 a, b) or Ae-ENO (Fig. 7c, d) confirmed the expression of both these proteins by using the anti-V5 antibody but fibrillar material could not be observed by ultrastructural analysis. Control experiments performed in the absence of primary antibodies were negative for all the samples (Fig 7e, f).
Fig. 6.
In vivo detection of P102Tni (TEM). Dilated reticulum cisternae filled with fibrillar material (arrowheads in a, b) and completely empty cisternae (arrows in b) are recognizable in stably transformed Drosophila S2 cells expressing the cDNA 102Tni. Dilated reticulum cisternae are not present in untransformed Drosophila S2 cells (c). Immunogold staining using the antibody anti-P102 confirms the role of P102Tni in the production of fibrillar material (arrowhead) (d). Negative control, no signal was detectable in control experiment performed in the absence of the primary antibodies anti-P102 (e). N: nucleus. Bars in (a–c): 1 µm; bar in (d, e): 500 nm.
Fig. 7.
In vivo detection of XendoU and Ae-ENO (TEM). Dilated reticulum cisternae are not present in stably transformed Drosophila S2 cells expressing XendoU (a) or Ae-ENO (c). Immunogold staining, performed by using the anti-V5 antibody, shows the expression of XendoU protein (arrowheads in b) and of Ae-ENO protein (arrowheads in d) in the cytoplasm of cells. No signal was detectable in control experiments performed in the absence of the primary antibody anti-V5 (e, f). N: nucleus. Bars in (a–c): 1 µm; bars in (b, d, e, f): 500 nm.
4. Discussion
Very little is known about the role of the endoribonuclease-U like proteins in vertebrate and invertebrate organisms, and no biological function has been assigned to these proteins in insects. At the moment, our knowledge is restricted to the role of the Lepidopteran endoribonuclease P102, which was shown to form amyloid fibrils supporting melanogenesis and capsule formation, providing a physiological role of amyloidogenesis in insect immunity, at least in the order Lepidoptera (Falabella et al., 2012, Grimaldi et al., 2012).
The XendoU family is a novel protein class and includes enzymes which differ from all other known endoribonucleases. Proteins belonging to the XendoU family are highly conserved both in vertebrates and invertebrates (Laneve et al., 2003, Renzi et al., 2006). The founding member, XendoU (Swiss-Prot ID: B1H3D5.1), is involved in small RNA biogenesis in Xenopus laevis (Caffarelli et al., 1997, Gioia et al., 2005, Laneve et al., 2003). Five amino acids of the XendoU catalytic domain (E161, H162, E167, H178, K224) were identified as being essential for its Mn2+-dependent enzymatic activity by site-directed mutagenesis analysis (Gioia et al., 2005). Protein sequence homology analysis of P102 from Lepidopteran H. virescens revealed that it contains an endoribonuclease-U domain (Falabella et al., 2012) lacking two out of the five catalytically essential residues identified in XendoU from X. laevis (Gioia et al., 2005) and human PP11 (Swiss-Prot ID: P21128) (Laneve et al., 2008). The two glutamic acids E228 and E234, and the histidine H244 are conserved while the non-conserved N229 and A288 correspond to H162 and K224 in XendoU. Most of the other less-conserved residues, that have been suggested to form substrate binding pocket in the XendoU structure, such as R149, G176, H272, T278 and Y280 (Renzi et al., 2006), are all conserved in P102. The endoribonuclease assay showed only a residual XendoU-like activity for P102; this result is perfectly consistent with biochemical features of the phosphate-binding site of well-known RNases, that requires two histidines (Kawata et al., 1990, Nishikawa et al., 1987, Saïda et al., 2003, Thompson and Raines, 1994) and one basic amino acid, either an arginine (Zegers et al., 1994a) or a lysine (Sorrentino, 2010, Zegers et al., 1994b) that constitute the catalytic triad His-His-Lys/Arg for enzymatic activity. The phosphate-binding site has been reported as crucial for the catalytic activity of endoribonuclease-U described above (Renzi et al., 2006). The catalytic triad is also implicated in the enzymatic activity of NendoU (RefSeq ID: YP_460022.1), a homolog of XendoU found in coronavirus (Snijder et al., 2003). Although NendoU has aspartic acid instead of glutamate in its active site the acidic nature of these residues is conserved. The set of residues in the corresponding active site in the XendoU domain of P102 lacks the catalytic triad His-His-Lys (it has Asn-His-Ala) and the protein displays a strongly impaired RNase activity. It has been demonstrated that the K224A mutation in XendoU, i.e. alanine in the same position as in P102, causes loss of RNA processing activity (Gioia et al., 2005). Substitution of the corresponding lysine in PP11 (Laneve et al., 2008) and in NendoU (Ivanov et al., 2004) diminish or completely abolish enzymatic activity.
XendoU, PP11 and NendoU are all enzymes involved in RNA processing. XendoU and PP11 cleave single-strand RNA at uridylates, in the presence of Mn2+ ions as cofactor, and release 2′–3′ cyclic phosphodiester ends (Laneve et al., 2003, Laneve et al., 2008). Instead, NendoU displays a preference for cleavage of double-strand RNA (Ivanov et al., 2004, but also see Bhardwaj et al., 2004). Although the general enzymatic activity of these enzymes has been well defined, their physiological functions are not completely clarified. It has been shown that XendoU is involved in the nuclear biosynthesis of small non-coding RNAs in X. laevis (Caffarelli et al., 1997, Gioia et al., 2005, Laneve et al., 2003), whereas PP11 seems to participate in RNA degradation in the final stage of apoptosis cascade during syncytialization. In addition, PP11 may also be involved in the degradation of viral RNAs, as part of defense mechanisms that preserve the fetus during pregnancy (Laneve et al., 2008). Moreover, PP11 is highly expressed in malignant cells and may be associated with oncogenesis (Laneve et al., 2008). NendoU, instead, is a component of the replicase-transcriptase complex in coronavirus (Ivanov et al., 2004). The depleted residual activity of P102 seems not to agree with an active role in RNA processing. However, it is reasonable to speculate that during evolution the loss of RNA processing activity could be associated with capacity of P102 to form amyloid fibrils, which has allowed P102 subfamily to acquire a specific biological function in immune response in insects or at least in Lepidoptera (Falabella et al., 2012). However, it cannot be excluded that the other differences in the sequence of P102, such as in the N-terminal region containing the predicted signal peptide or in other parts of the protein, also have contributed to the transformation of the original U-specific RNase into a protein with less structured regions, that could take part in triggering the aggregation process and formation of amyloid fibers (Chiti and Dobson, 2006, Otzen et al., 2000, Schwartz et al., 2001). This essential function in insect immunity may have been the evolutionary selective force driving the separation of the P102-like proteins of Lepidoptera and the conserved endoribonuclease U-like proteins of Lepidoptera and of other insect orders. The functional characterization of the homolog from the Lepidopteran T. ni, which shares both a high sequence identity and the same set of residues in the active site with P102, as well as low residual enzymatic activity and the ability to produce fibrillar material in vitro, provide evidence for a similar function of P102 and P102Tni. TEM observations of the stably transformed Drosophila S2 cells, expressing P102Tni, confirmed that the fibrillar material is accumulated in large reticulum cisternae, as previously shown for P102 (Falabella et al., 2012), while S2 cells expressing XendoU do not produce any fibrillar material. Thus, it is reasonable to extend this finding to all P102-like proteins from Lepidoptera, as long as they share a high sequence homology and an identical set of residues in catalytic site. This finding is further supported by the recent characterization of the 102 gene homologue in the noctuid moth Spodoptera littoralis, showing its function in capsule formation and melanization when subjected to immune challenge (Di Lelio et al., 2014).
Examination of the available sequences of P102 homologs in insects revealed two main protein clusters. The first one is composed of proteins having a conserved and putatively active XendoU domain and about 40% of sequence identity with P102; these proteins belong to Hymenoptera, Diptera, Coleoptera and Lepidoptera orders. The second group of proteins is only found in Lepidoptera and is characterized by the different set of residues in catalytic site and a sequence identity of more than 80%. This suggests that although all of these proteins are likely to have 3D structures similar to XendoU they are divided into two functional subfamilies. Moreover, the hymenopteran XendoU-domain-like proteins could be grouped in three additional main classes, according to the three alternative combinations of the five amino acid residues in their catalytic domain. While Hymenoptera display a long evolutionary history and recent phylogenomic studies have placed them at the base of the radiation of holometabolous insects (Ishiwata et al., 2011, Savard et al., 2006), the functional role of these XendoU-domain-like proteins is yet unknown. The phylogenetic analysis of the highly conserved homologs from Lepidoptera and the conserved endoribonucleases from other insect orders confirmed the separation of the Lepidopteran P102 subfamily in a separate cluster. Based on both the phylogenetic analysis and the identification of similar set of residues in the catalytic site of the primary protein sequences, it seems likely that P102-like proteins are restricted to Lepidoptera and that the other insect XendoU sequences form groups roughly according to species phylogeny. As almost all of the XendoU proteins are much longer than the P102-like sequences, it might be conceivable to propose a gene duplication event at the origin/base of the Lepidoptera, which then lead to the formation of the P102-like gene, after loss of a larger part of the non-XndoU-like variable N-terminus and loss of the original U-specific RNase activity, rather than a loss of P102-like genes in all of the other insect orders.
In conclusion, our data support the idea of a functional role of Lepidopteran P102 in innate immunity. We show that the P102 subfamily proteins have almost completely lost the enzymatic RNase activity and gained the ability to form functional amyloid fibrils, that are involved in localizing melanin deposition for encapsulation and preventing its harmful diffusion outside the capsule. This mechanism sheds light on a crucial step of insect immunity, of which many other processes of humoral and cellular responses remain to be further clarified. Future work on enzymatic activities and biological roles of proteins from the P102 subfamily and the XendoU main family across different insect orders will ultimately lead to a better understanding of the evolutionary history of XendoU-like proteins.
5. Conflict of interest
All authors declare no conflict of interest.
6. Author contributions
P.F., M.P. and S.L. conceived the research, planned the molecular/functional experiments and analyzed the data. P.F., M.P., S.L., L.R and performed all molecular biology experiments, bioinformatic analyses and functional studies. A.G. and G.T. planned, performed and analyzed all electron microscopy experiments, amyloid staining and immunogold labeling. H.V. provided the homologous gene of cDNA 102 from Trichoplusia ni, made available the Max Planck Institute (Jena, Germany) private database and performed bioinformatic analyses. M.M. performed bioinformatic analyses. F.P., M.P. and H.V. wrote the paper, which was edited by H.V., M.M., and A.G.
Acknowledgments
This work was supported by the Italian Ministry of University and Research (MiUR), in the framework of a national research program (PRIN 2008FBJPR8). We would like to thank Prof. Angharad Gatehouse (Newcastle University, UK) and Prof. Magda de Eguileor (University of Insubria, Varese, Italy) for their critical reading of the manuscript, Dr. Elisa Caffarelli (CNR, Rome, Italy) for providing the recombinant XendoU used in enzymatic activity assays, Dr. Pietro Laneve for providing the cDNA coding for Xendo U Dr. Gennaro Sansone for realizing the graphical abstract, Dr. Teresa Zotta (University of Basilicata, Potenza, Italy) for providing the GelCompar II software and Luca Trotti for Heliothis virescens picture.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.dci.2014.07.009.
Appendix A. Supplementary data
Aminoacidic sequences used in alignments and phylogenetic trees. Signal peptide is highlighted in red. Where possible, sequence accession numbers are shown.
Supplementary Fig. 2.
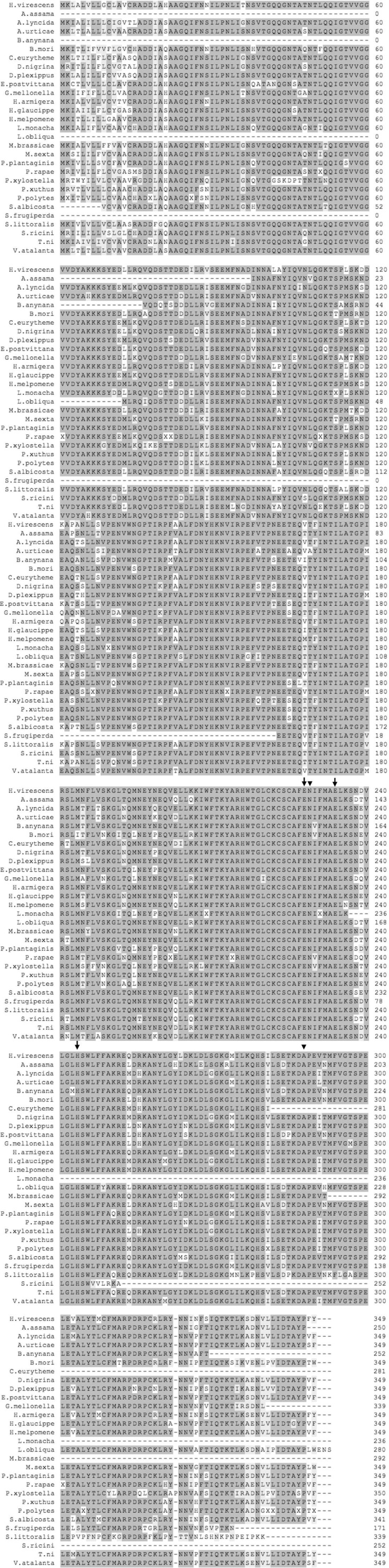
Multiple alignment of P102 sequence with homologs from Lepidoptera (identity > 75%). The name of the organism and the number of the first amino acid of the aligned region are indicated. The accession numbers for sequences retrieved from NCBI are the following: Antheraea assama (GenBank IDs: FG207602.1 and FG207602.1), Bicyclus anynana (GenBank ID: GE686877.1), Bombyx mori (GenBank ID: AK383576.1), Danaus plexippus (GenBank ID: EHJ69917.1), Epiphyas postvittana (GenBank ID: EV813526.1), Lonomia obliqua (GenBank ID: AAV91433.1), Papilio polytes (GenBank ID: BAM20530.1), P. xuthus (GenBank ID: BAM17994.1), Striacosta albicosta (GenBank ID: EZ591155.1), Spodoptera frugiperda (GenBank ID: FP366351.1), S. littoralis (GenBank ID: FQ014754.1), Samia cynthia ricini (GenBank IDs: DC860291.1 and DC864934.1). The following sequences were retrieved from in-house transcriptome databases: Appias lyncida, Aglais urticae, Colias eurytheme, Delias nigrina, Galleria mellonella, Helicoverpa armigera, Hebomoia glaucippe, Heliconius melpomene, Lymantria monacha, Mamestra brassicae, Manduca sexta, Parasemia plantaginis, Pieris rapae, Plutella xylostella, Trichoplusia ni, Vanessa atalanta. All nucleotide sequences were translated into proteins before alignment. Identical and conserved amino acids are shaded in light gray and very light gray, respectively. Residues involved in the enzymatic activity are highlighted: arrows mark the conserved residues of the catalytic site while arrowheads indicate the alternative ones.
Supplementary Fig. 3.
Partial multiple alignment of P102 sequence (residues 72-349) with homologs from insect orders except Lepidoptera. The name of the organism and the number of the first amino acid of the aligned region are indicated. The accession numbers for sequences retrieved from NCBI are the following: Aedes aegypti (RefSeq ID: XP_001660282.1), Anopheles darlingi (GenBank ID: EFR21399.1), A. gambiae (RefSeq ID: XP_311978.5), Apis mellifera (RefSeq ID: XP_003251941.1), Camponotus floridanus (GenBank ID: EFN64513.1), Culex quinquefasciatus (RefSeq ID: XP_001864562.1), C. tarsalis (GenBank ID: ACJ64345.1), Drosophila ananassae (RefSeq ID: XP_001963598.1), D. erecta (RefSeq ID: XP_001977186.1), D. melanogaster (RefSeq ID: NP_572668.1), D. mojavensis (RefSeq ID: XP_002010982.1), D. persimilis (RefSeq ID: XP_002025808.1), D. pseudoobscura pseudoobscura (RefSeq ID: XP_001354551.2), D. simulans (RefSeq ID: XP_002106640.1), D. virilis (RefSeq ID: XP_002056809.1), Glossina morsitans morsitans (GenBank ID: ADD19839.1), Harpegnathos saltator (GenBank ID: EFN80024.1), Nasonia vitripennis (RefSeq ID: XP_001603664.1), Solenopsis invicta (GenBank ID: EFZ16938.1), Tribolium castaneum (RefSeq ID: XP_968069.1). Three sequences of Coleoptera (Brassicogethes aeneus, Nicrophorus vespilloides and Phaedon cochleariae), were retrieved from in-house transcriptome databases Identical and conserved amino acids are shaded in light gray and very light gray, respectively. Residues involved in the enzymatic activity are indicated by arrows.
Supplementary Fig. 4.
Multiple alignment of P102 sequence with short XendoU-like sequences from Diptera. Short XendoU-like sequences lack the variable N-terminal region and have the XendoU canonical set of residues in the active site. The name of the organism and the number of the last amino acid of the aligned region are indicated. Identical and conserved amino acids are shaded in light gray and very light gray, respectively. Residues involved in the enzymatic activity are indicated by arrows.
Supplementary Fig. 5.
Maximum-likelihood tree of the P102 sequence, indicated as H. virescens, and sequences trimmed to the shorter P102-like protein length of homologs from Lepidoptera and other insect species. Mutated and conserved XendoU domain homologs from Lepidoptera are designated as 1 and 2, respectively. Names of insects are indicated for each branch. The accession numbers for mutated homologs from Lepidoptera and conserved homologs from other insects have been listed in Supplementary figures 2 and3, respectively. The accession numbers for conserved homologs from Lepidoptera retrieved from NCBI are as follows: Papilio polytes (GenBank ID: BAM20523.1), P. xuthus (GenBank ID: BAM19803.1) and Danaus plexippus (GenBank ID: EHJ66341.1). Additional conserved sequences from Lepidoptera (Galleria mellonella, Parasemia plantaginis, Vanessa atalanta) were retrieved from in-house transcriptome databases. Numbers above branches are bootstrap support values shown as percentages of a total of 100 bootstrap replications. Branch lengths are proportional to evolutionary distance according to the provided scale.
Supplementary Fig. 6.
Partial alignment of XendoU (residues 48-281 (a) residues 157-273 (b)) and P102 sequences with homologs from Hymenoptera (identity with P102 about 40%) not containing the canonical XendoU residues in the catalytic site. Three different classes of variants of the five catalytic residues compared to XendoU are shown. Simultaneous substitution of the histidines at positions H162 and H178 in alanine and glutamine respectively and the lisyne at position K224 in histidine (red box), discovered only in Hymenoptera Vespoidea (Camponotus floridanus, GenBank ID: EFN67408.1; Harpegnathos saltator, GenBank ID: EFN88107.1; Solenopsis invicta, GenBank ID: EFZ20078.1) and substitution of the histidine at position H162 in threonine and of lysine at position K224 in glutamine (blue box) in Hymenoptera Apoidea and Chalcidoidea (Apis mellifera, RefSeq ID: XP_625112.2; Nasonia vitripennis, RefSeq ID: XP_001606738.1)(a). In all three families the histidines were identified at positions H162 and H178 and of lysine at position K224 varied in arginine, glutamine and arginine or isoleucine, respectively (black box) (A. mellifera, RefSeq ID: XP_625106.3; C. floridanus, GenBank ID: EFN67407.1; H. saltator, GenBank ID: EFN88106.1; N. vitripennis, RefSeq ID: XP_003424318.1; S. invicta, GenBank ID: EFZ20057.1) (b). Residues involved in the enzymatic activity are highlighted: arrows mark the conserved residues of the catalytic site while arrowheads indicate the alternative ones.
References
- Bhardwaj K., Guarino L., Kao C.C. The severe acute respiratory syndrome coronavirus Nsp15 protein is an endoribonuclease that prefers manganese as a cofactor. J. Virol. 2004;78:12218–12224. doi: 10.1128/JVI.78.22.12218-12224.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohn H., Winckler W. Isolation and characterization of a new placental tissue protein (PP11) Arch. Gynecol. 1980;229:293–301. doi: 10.1007/BF02108580. [DOI] [PubMed] [Google Scholar]
- Bohn H., Inaba N., Luben G. New placental proteins and their potential diagnostic significance as tumor markers. Oncodev. Biol. Med. 1981;2:141–153. [PubMed] [Google Scholar]
- Caffarelli E., Arese M., Santoro B., Fragapane P., Bozzoni I. In vitro study of processing of the intron-encoded U16 small nucleolar RNA in Xenopus laevis Mol. Cell. Biol. 1994;14:2966–2974. doi: 10.1128/mcb.14.5.2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffarelli E., Maggi L., Fatica A., Jiricny J., Bozzoni I. A novel Mn++-dependent ribonucleasi that functions in U16 SnoRNA processing in X. laevis. Biochem. Biophys. Res. Commun. 1997;233:514–517. doi: 10.1006/bbrc.1997.6487. [DOI] [PubMed] [Google Scholar]
- Chiti F., Dobson C.M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J.F., Guindon S., Lefort V., Lescot M., Claverie J.M., Gascuel O. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Lelio I., Varricchio P., Di Prisco G., Marinelli A., Lasco V., Caccia S., Casartelli M., Giordana B., Rao R., Gigliotti S., Pennacchio F. Functional analysis of an immune gene of Spodoptera littoralis by RNAi. J. Insect Physiol. 2014;64:90–97. doi: 10.1016/j.jinsphys.2014.03.008. [DOI] [PubMed] [Google Scholar]
- Donini U., Casanova S., Zucchelli P., Linke R.P. Immunoelectron microscopic classification of amyloid in renal biopsies. J. Histochem. Cytochem. 1989;37:1101–1106. doi: 10.1177/37.7.2732456. [DOI] [PubMed] [Google Scholar]
- Eswar N., Webb B., Marti-Renom M.A., Madhusudhan M., Eramian D., Shen M.-Y., Pieper U., Sali A. Comparative protein structure modeling using Modeller. Curr. Protoc. Bioinformatics. 2006;15:5.6.1–5.6.30. doi: 10.1002/0471250953.bi0506s15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falabella P., Riviello L., De Stradis M.L., Stigliano C., Varricchio P., Grimaldi A., de Eguileor M., Graziani F., Gigliotti S., Pennacchio F. Aphidius ervi teratocytes release an extracellular enolase. Insect Biochem. Mol. Biol. 2009;39:801–813. doi: 10.1016/j.ibmb.2009.09.005. [DOI] [PubMed] [Google Scholar]
- Falabella P., Riviello L., Pascale M., Di Lelio I., Tettamanti G., Grimaldi A., Iannone C., Monti M., Pucci P., Tamburro A.M., de Eguileor M., Gigliotti S., Pennacchio F. Functional amyloids in insect immune response. Insect Biochem. Mol. Biol. 2012;42:203–211. doi: 10.1016/j.ibmb.2011.11.011. [DOI] [PubMed] [Google Scholar]
- Gioia U., Laneve P., Dlakic M., Arceci M., Bozzoni I., Caffarelli E. Functional characterization of XendoU, the endoribonuclease involved in small nucleolar RNA biosynthesis. J. Biol. Chem. 2005;280:18996–19002. doi: 10.1074/jbc.M501160200. [DOI] [PubMed] [Google Scholar]
- Grimaldi A., Girardello R., Malagoli D., Falabella P., Tettamanti G., Valvassori R., Ottaviani E., de Eguileor M. Amyloid/Melanin distinctive mark in invertebrate immunity. Invertebrate Surviv. J. 2012;9:153–162. [Google Scholar]
- Grundmann U., Römisch J., Siebold B., Bohn H., Amann E. Cloning and expression of a cDNA encoding human placental protein 11, a putative serine protease with diagnostic significance as a tumor marker. DNA Cell Biol. 1990;9:243–250. doi: 10.1089/dna.1990.9.243. [DOI] [PubMed] [Google Scholar]
- Inaba N., Renk T., Wurster K., Rapp W., Bohn H. Ectopic synthesis of pregnancy specific beta 1-glycoprotein (SP1) and placental specific tissue proteins (PP5, PP10, PP11, PP12) in nontrophoblastic malignant tumours. Possible markers in oncology. Klin. Wochenschr. 1980;58:789–791. doi: 10.1007/BF01478287. [DOI] [PubMed] [Google Scholar]
- Inaba N., Renk E., Bohn H. Immunohistochemical location of placental proteins (PP8, 9, 10, 11, 12) in human term placentae. Arch. Gynecol. 1980;230:109–121. doi: 10.1007/BF02108266. [DOI] [PubMed] [Google Scholar]
- Inaba N., Renk T., Daume E., Bohn H. Ectopic production of placenta-“specific” tissue proteins (PP5 and PP11) by malignant breast tumors. Arch. Gynecol. 1981;231:87–90. doi: 10.1007/BF02110028. [DOI] [PubMed] [Google Scholar]
- Inaba N., Ishige H., Ijichi M., Satoh N., Ohkawa R., Sekiya S., Shirotake S., Takamizawa H., Renk T., Bohn H. Immunohistochemical detection of pregnancy-specific protein (SP1) and placenta-specific tissue proteins (PP5, PP10, PP11 and PP12) in ovarian adenocarcinomas. Oncodev. Biol. Med. 1982;3:379–389. [PubMed] [Google Scholar]
- Ishiwata K., Sasaki G., Ogawa J., Miyata T., Su Z.H. Phylogenetic relationships among insect orders based on three nuclear protein-coding gene sequences. Mol. Phylogenet. Evol. 2011;58:169–180. doi: 10.1016/j.ympev.2010.11.001. [DOI] [PubMed] [Google Scholar]
- Ivanov K.A., Hertzig T., Rozanov M., Bayer S., Thiel V., Gorbalenya A.E., Ziebuhr J. Major genetic marker of nidoviruses encodes a replicative endoribonuclease. Proc. Natl. Acad. Sci. U.S.A. 2004;101:12694–12699. doi: 10.1073/pnas.0403127101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Kuma K., Toh H., Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawata Y., Sakiyama F., Hayashi F., Kyogoku Y. Identification of two essential histidine residues of ribonuclease T2 from Aspergillus oryzae. Eur. J. Biochem. 1990;187:255–262. doi: 10.1111/j.1432-1033.1990.tb15303.x. [DOI] [PubMed] [Google Scholar]
- Laneve P., Altieri F., Fiori M.E., Scaloni A., Bozzoni I., Caffarelli E. Purification, cloning and characterization of XendoU, a novel endoribonuclease involved in processing of intron-encoded small nucleolar RNAs in Xenopus laevis. J. Biol. Chem. 2003;11:13026–13032. doi: 10.1074/jbc.M211937200. [DOI] [PubMed] [Google Scholar]
- Laneve P., Gioia U., Ragno R., Altieri F., Di Franco C., Santini T., Arceci M., Bozzoni I., Caffarelli E. The tumor marker Human Placental Protein 11 is an endoribonuclease. J. Biol. Chem. 2008;283:34712–34719. doi: 10.1074/jbc.M805759200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa S., Morioka H., Kim H.J., Fuchimura K., Tanaka T., Uesugi S., Hakoshima T., Tomita K., Ohtsuka E., Ikehara M. Two histidine residues are essential for ribonuclease T1 activity as is the case for ribonuclease A. Biochemistry. 1987;26:8620–8624. doi: 10.1021/bi00400a019. [DOI] [PubMed] [Google Scholar]
- Otzen D.E., Kristensen O., Oliveberg M. Designed protein tetramer zipped together with a hydrophobic Alzheimer homology: a structuralclue to amyloid assembly. Proc. Natl. Acad. Sci. U.S.A. 2000;97:9907–9912. doi: 10.1073/pnas.160086297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renzi F., Caffarelli E., Laneve P., Bozzoni I., Brunori M., Vallone B. The structure of the endoribonuclease XendoU: from small nucleolar RNA processing to severe acute respiratory syndrome coronavirus replication. Proc. Natl. Acad. Sci. U.S.A. 2006;103:12365–12370. doi: 10.1073/pnas.0602426103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Saïda F., Uzan M., Bontems F. The phage T4 restriction endoribonuclease RegB: a cyclizing enzyme that requires two histidines to be fully active. Nucleic Acids Res. 2003;31:2751–2758. doi: 10.1093/nar/gkg377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E.F., Maniatis T. Vol. 1–3. Cold Spring Harbor Laboratory Press; New York: 1989. (Molecular Cloning: A Laboratory Manual). [Google Scholar]
- Savard J., Tautz D., Richards S., Weinstock G.M., Gibbs R.A., Werren J.H., Tettelin H., Lercher M.J. Phylogenomic analysis reveals bees and wasps (Hymenoptera) at the base of the radiation of Holometabolous insects. Genome Res. 2006;16:1334–1338. doi: 10.1101/gr.5204306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz R., Istrail S., King J. Frequencies of amino acid strings in globular protein sequences indicate suppression of blocks of consecutive hydrophobic residues. Protein Sci. 2001;10:1023–1031. doi: 10.1110/ps.33201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J., Poon L.L., Guan Y., Rozanov M., Spaan W.J., Gorbalenya A.E. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrentino S. The eight human “canonical” ribonucleases: molecular diversity, catalytic properties, and special biological actions of the enzyme proteins. FEBS Lett. 2010;584:2194–2200. doi: 10.1016/j.febslet.2010.04.018. [DOI] [PubMed] [Google Scholar]
- Thompson J.E., Raines R.T. Value of general acid–base catalysis to ribonuclease A. J. Am. Chem. Soc. 1994;116:5467–5468. doi: 10.1021/ja00091a060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulferts R., Ziebuhr J. Nidovirus ribonucleases: structures and functions in viral replication. RNA Biol. 2011;8(2):295–304. doi: 10.4161/rna.8.2.15196. [DOI] [PubMed] [Google Scholar]
- Veiga A.B., Ribeiro J.M., Guimarães J.A., Francischetti I.M. A catalog for the transcripts from the venomous structures of the caterpillar Lonomia obliqua: identification of the proteins potentially involved in the coagulation disorder and hemorrhagic syndrome. Gene. 2005;355:11–27. doi: 10.1016/j.gene.2005.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegers I., Haikal A.F., Palmer R., Wyns L. Crystal structure of RNase T1 with 3′-guanylic acid and guanosine. J. Biol. Chem. 1994;269:127–133. [PubMed] [Google Scholar]
- Zegers I., Maes D., Dao-Thi M.H., Poortmans F., Palmer R., Wyns L. The structures of RNase A complexed with 3’-CMP and d(CpA): active site conformation and conserved water molecules. Protein Sci. 1994;3:2322–2339. doi: 10.1002/pro.5560031217. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Aminoacidic sequences used in alignments and phylogenetic trees. Signal peptide is highlighted in red. Where possible, sequence accession numbers are shown.