The proteome, phospho-proteome, and solubility-fractionated proteome was characterized using a fluorescent cell cycle reporter system (Fucci). Applying dimensionality-reduction, the data sets were systematically compared. This revealed novel patterns between mRNA and protein expression, cell-cycle related activity of kinases, and allowed for a search for proteins changing solubility throughout the cell cycle. The nuclear translocation of the S-adenosylmethionine synthase MAT2A during S-phase was confirmed.
Keywords: Cell cycle, phosphorylation, subcellular separation, computational biology, systems biology
Graphical Abstract
Highlights
A cell cycle resolved comparison between the proteome and the transcriptome.
Integrative analysis using phospho-proteome and dynamics of solubility.
Mechanistic validation of MAT2A as a nuclear translocator in S-phase.
Abstract
The cell cycle is a highly conserved process involving the coordinated separation of a single cell into two daughter cells. To relate transcriptional regulation across the cell cycle with oscillatory changes in protein abundance and activity, we carried out a proteome- and phospho-proteome-wide mass spectrometry profiling. We compared protein dynamics with gene transcription, revealing many transcriptionally regulated G2 mRNAs that only produce a protein shift after mitosis. Integration of CRISPR/Cas9 survivability studies further highlighted proteins essential for cell viability. Analyzing the dynamics of phosphorylation events and protein solubility dynamics over the cell cycle, we characterize predicted phospho-peptide motif distributions and predict cell cycle-dependent translocating proteins, as exemplified by the S-adenosylmethionine synthase MAT2A. Our study implicates this enzyme in translocating to the nucleus after the G1/S-checkpoint, which enables epigenetic histone methylation maintenance during DNA replication. Taken together, this data set provides a unique integrated resource with novel insights on cell cycle dynamics.
The mechanism of cell division has been extensively studied for many decades resulting in a very detailed picture of the genes and proteins involved and their temporal function within the dividing cell. Historically, the cell cycle is divided into a DNA synthesis phase (S-phase) and a cell division phase (Mitosis; M-phase), with these two phases separated by two gap phases, G1 and G2.
To date, many large-scale studies addressing the cell cycle focus on the transcriptional control of cell cycle regulated genes. Transcription can be used as a proxy for protein abundance and transcript dynamics translated to protein abundance on a larger scale for systems at the steady state (1), but extrapolation between different mRNA-protein pairs has very little explanatory power (2).
Aside from translational control, allowing for regulation of protein turnover that is not reflected in mRNA levels, regulation by the Ubiquitin-Proteasome-System (UPS)1 can also influence protein abundance, causing a rapid decline in protein levels through ubiquitin-mediated degradation, an effect that is not reflected by mRNA levels. In addition, many cellular functions are regulated not by protein abundance at all, but rather by protein activation through post-translational modifications (PTMs), such as phosphorylation, or differential localization within the cell (3).
Proteomic and transcriptomic analyses of cell cycle have previously been reported, often using chemical synchronization of cells. The disadvantage of synchronization-based cell-cycle analysis is the disruption of the natural cell cycle. Studies investigating cell cycle proteomics on asynchronous cells has been performed by Lamond and colleagues. In 2014 they used centrifugal elutriation to separate fractions enriched for cell cycle phases. They identified 358 cell-cycle dependent proteins, of which 31 also had cell-cycle dependent mRNA (4). In a 2017 follow-up study, they segmented cells based on DAPI- and phospho-histone-H3 antibody staining on paraformaldehyde-fixated cells. This study included a phospho-proteomic analysis, and a further focus on mitotic subphases (5).
We aimed to further dissect the complex and dynamic process of the cell cycle at the protein level in asynchronous cells. Unlike previous studies that relied on chemical synchronization of cells or centrifugal elutriation, (4, 6, 7) we used the Fucci system, a fluorescent cell cycle probe based on the ubiquitin-mediated degradation of CDT1 and Geminin (8), to sort HeLa cells into three cell cycle phases. CDT1 and Geminin are each degraded by the cell cycle regulating complexes APC/C and SCF, which exhibit oscillating activity during the cell cycle. The respective target sequences of CDT1 and Geminin are fused to a red and a green fluorescent protein, creating an oscillation of colors from red, to double-positive, to green, throughout the cell cycle. We combined this system with fluorescence-activated cell sorting (FACS), cellular fractionation, proteomics and phospho-proteomics analyses. Furthermore, we integrated our data set with a cell cycle transcriptomics expression profile (9) and in vitro essentiality data summarized from ten different CRISPR/Cas9 survivability cell line experiments (10).
Aside from identifying a great number of novel cell cycle regulated proteins, we employed novel methods to reduce dimensionality to be able to compare these different data sets and could further study the overlying patterns and dynamics between transcription and protein turnover. Using motif-recognition methods on the phospho-proteomic data set, we could characterize kinase activity patterns over the cell cycle. Finally, by identifying cell cycle changes in the proteomic data separated by solubility, and focusing on diametrically opposite patterns, we can present a shortlist of proteins that change solubility phase during the cell cycle, either through translocation between cellular compartments, or by changing their binding configuration. We furthermore uncovered a strong correlation between the level of regulation in a group of proteins (oscillation of abundance, phosphorylation, and translocation), to their essentiality score in CRISPR/Cas9 survivability assays.
Among the essential translocators, we identified the S-adenosylmethionine synthase MAT2A, which is involved in the synthesis of S-Adenosyl-Methionine, a major methyl donor. Methylation is a well-established epigenetic mark and suspected to be highly correlated to the etiology of hepatocellular carcinoma among other cancers (11). We show for the first time that nuclear localization of MAT2A is cell cycle dependent in proliferating cells.
Together, we provide an integrated resource database combining cell cycle dynamics of proteomics, transcriptomics, phospho-proteomics, and fractionation proteomics. We also integrate mathematical methods to compare the dynamics between the different modes of data and to essentiality data extracted from CRISPR/Cas9 survivability assays.
MATERIALS AND METHODS
Contact for Reagent and Resource sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Mikael Altun (mikael.altun@ki.se). For HeLa Fucci cells, we refer to the RIKEN Cell Bank. Supplemental Table S5: Key Resources Table contains details of all Antibodies, Cell Lines, Software, and data sets used in this study.
Cell Culture
HeLa-Fucci cells (obtained from Riken Cell Bank, Japan) were cultured in DMEM with 10% FBS, penicillin (100 μg/ml), streptomycin (100 μg/ml) at 37 °C containing 5% CO2, in a humidified incubator. Cells were regularly checked for Mycoplasma contamination (Lonza, MycoAlert, Lonza, Switzerland). This cell line was the same as used in Boström et al. to generate the transcriptomic data that we compared our data with and was thawed up from freezing vials from a similar passage number (9).
Western Blotting
Western blotting was carried out following standard protocols with Bio-Rad SDS gradient gels and the Trans-Blot Turbo transfer system (Bio-Rad, CA). Cells were lysed in RIPA buffer for 20 min on ice in presence of protease inhibitor mixture (Roche, Switzerland), followed by sonication with a needle sonicator (Hielscher UP100H, 70% amplitude, 0.7 cycle, 10 cycles, (Hielscher, Germany)). Protein concentration was measures according to BCA (Pierce, MA). Images were taken at a LI-COR Odyssey FC.
Cell Fractionation
The soluble fraction was isolated based on a protocol published by (12). Lysis buffer to isolate the cytosolic (soluble) fraction contained 42 μg/ml Digitonin, 2 mm DTT, 2 mm MgCl2, 150 mm NaCl, 0.2 mm EDTA, 20 mm Hepes-NaOH at pH 7.4. Protease and phosphatase inhibitors (including 1 mm sodium orthovanadate) were added fresh. The insoluble fraction was separated by centrifugation. The remaining insoluble fraction containing nucleus, chromatin, larger organelles, and membranes was isolated according to a protocol by (13). Lysis buffer contained 8 m Urea, 20 mm HEPES pH 7.5, 1 mm b-glycerophosphate, 2.5 mm Sodium pyrophosphate, 1 mm Na3VO4. Protease and phosphatase inhibitor were added fresh. Lysate was sonicated.
FACS
HeLa-Fucci cells were seeded on 10 cm2 dishes and grown to 70–80% confluency. Cells were washed in 10 ml 37 °C PBS. PBS was removed and replaced with 1 ml Trypsin for 4 min until cells were in single cell suspension. Cells were then collected in 10 ml warm DMEM and counted. Cells were spun down for 3 min at 300 g at 4 °C. Media was removed and cells were washed in 10 ml of cold PBS. Cells were spun again for 3 min at 300 × g at 4 °C. The cell pellet was diluted in cold PBS with 5 mm EDTA to 3 million/ml and strained through a cell strainer FACS tube. Cells were sorted on a BD Influx. After every 30 min the sorted cells were snap frozen and pellets were stored at −80 °C.
Immunocytochemistry and Image Analysis
Cells were grown on 96 well plates, washed in PBS and fixed in 4% PFA in PBS for 20 min. Samples were permeabilized for 10 min with 0.3% Triton X-100 in PBS. Samples were blocked in 3% BSA in PBS for 40 min and incubated with primary antibody (1:500 in PBS 3% BSA) over night at 4 degrees. After 4 × 10 min washes with PBS samples were incubated with fluorescent secondary antibody for 1 h at RT. After another 4 × 10 min washes with PBS samples were stained with DAPI for 5 min at RT. After another 4 × 10 min washes with PBS samples were imaged. High-content imaging was performed with an ImageXpress XLS (Molecular Devices, CA) and data was analyzed with CellProfiler (v2.1.1) Cell cycle classifications were performed using R and nuclear localization classifications were performed using the Classifier tool in CellProfiler Analyst (v2.2.1) using information only measured in the MAT2A antibody channel, trained on a separate manually classified training set.
For the analysis in supplemental Fig. S2D–S2F, cells were incubated with 10 μm EdU 30 min prior to fixation, and stained with 2 μm Alexa FluorTM 647 Azide (Thermo Fisher, MA), 4 mm Copper(II)Sulfate, 10 mm ascorbic acid in PBS for 10 min. Fixation and analysis were done as described above except prior to DAPI incubation, cells were treated with 10 μg/ml RNase A in PBS for 1h at 4 °C in order to enhance DAPI quantification, and images were acquired on a Evos M5000 Imaging System (Thermo Fisher).
Proteomic Analyses
For the unfractionated proteome and phosphoproteome analyses of each of the three FACS sorted phases (G1, S, G2/M), cell pellets were harvested and resuspended in lysis buffer (1% Sodium deoxycholate, 100 mm Hepes pH 8, 1 mm sodium orthovanadate, 1 tablet of Complete mini EDTA-free mixture (Roche Applied Science) and one tablet of PhosSTOP phosphatase inhibitor mixture per 10 ml of lysis buffer (Roche Applied Science). Cells were then lysed by 10 rapid passages through a 23-gauge hypodermic syringe needle and by sonication on ice. After centrifugation (20,000 × g 30 min at 4 °C), the protein concentration was determined by Bradford assay (Pierce). The protein lysates of the three different phases (G1, S, G2/M) (total/soluble/insoluble fractions) were reduced by 2 mm DTT at room temperature for 1 h and alkylated by 4 mm chloroacetamide for 30 min at room temperature in the dark. A first enzymatic digestion was performed using Lys-C (1:75 w/w) at 37 °C overnight; a second enzymatic digestion was performed using Trypsin (1:75 w/w) at 37 °C overnight. Fifty micrograms of each sample was labeled by TMT10plex according to the manufacturer instructions (Thermo Fisher Scientific, Germany): G1 replicate 1 (126), G1 replicate 2 (127N), G1 replicate 3 (127C), S replicate 1 (128N), S replicate 2 (128C), S replicate 3 (129N), G2/M replicate 1 (129C), G2/M replicate 2 (130N), G2/M replicate 3 (130C), Pool of all sample (131) (Fig. 1A).
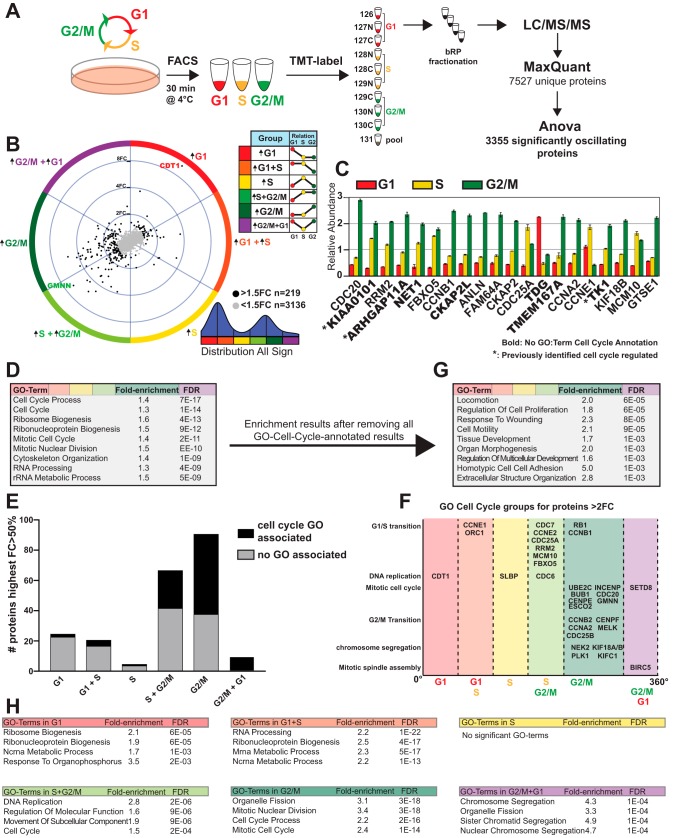
Fig. 1.
Characterization of Protein Oscillations over the Cell Cycle. A, Illustration of the workflow of cell cycle sorting and proteomic analysis. B, Distribution of all significantly changed proteins visualized with TriComp. Direction denotes the relationship between the three cell cycle phases, whereas distance from center denotes the intensity of the relationship. C, Bar graphs of relative abundance of the top 20 significantly oscillating proteins. Error bars denote the S.E. between the triplicate samples. Proteins without cell cycle GO terms are highlighted in bold. D, GSEA of all significantly oscillating proteins. E, Distribution of GO-Cell-Cycle-annotated proteins. F, Distribution of proteins annotated with GO-terms related to cell cycle. G, GSEA of all significantly oscillating proteins not annotated with a GO-term related to cell cycle. H, GSEA analysis on each of the six broad groups of oscillation patterns.
Two hundred μg of peptide mixture were fractionated using a Waters XBridge BEH300 C18 3.5 μm 2.1 × 250 mm column on an Agilent 1200 series operating at 200 μl/min. Buffer A consisted of 20 mm NH3, whereas buffer B of 80% ACN/20 mm NH3. The fractionation gradient was: 3–88% B in 63 min; 88% B for 15 min; and ramped to 100% B in 2.5 min; 100% B for 13.5 min. Fractions were collected into a polypropylene V-96 well microtiterplates (Microplate, 96 well PP, V-Bottom, IO-ONE, Grainer, Austria). At 97 min, fraction collection was halted, and the gradient was held at 3% B for 20 min. The concatenated fractions were collected in a plate, dried at room temperature using a SpeedVac (SPD 111V, Thermo), and stored at −20 °C until LC-MS/MS analyses.
Phosphoproteomics Analysis
For phosphoproteomics analyses, one hundred μg of tryptic digest of each cell sorted population and their respective replicates were labeled by TMT10plex as described above. Each sample underwent TiO2 phosphopeptide enrichment step as previously described (14); mixed 1:1; desalted by reverse phase using Waters Pak 1 cc (50 mg) cartridges (WAT054960; Waters, Milford, MA); and analyzed by LC-MS/MS using a Fusion OrbiTrap (Thermo Fisher, Germany). The mass spectrometer was connected to a Dionex UHPLC system (Thermo Fisher Scientific, Germany).
LC-MS/MS Analyses
Each sample was analyzed on a HF Q-Exactive Orbitrap (Thermo Fisher) (full proteome samples) or a Fusion Orbitrap (Thermo Fisher) (soluble and insoluble fraction samples) using a data dependent acquisition mode and in both cases the instruments were connected to a Dionex UHPLC system (Thermo Fisher Scientific). The UHPLC was equipped with a trap column (Acclaim PepMap 100, 75 μm × 2 cm, nanoviper, C18, 3 μm, 100 Å; Thermo Fisher Scientific) and an analytical column (PepMap RSLC C18, 2 μm, 100 Å, 75 μm × 50 cm; Thermo Fisher Scientific). Mobile-phase buffers for nLC separation consisted of 0.1% FA in water (solvent A) and 80% ACN/0.1% FA (solvent B). The peptides were eluted during a 2 h gradient and directly sprayed into the mass spectrometer. The flow rate was set at 250 nl/min, and the LC gradient was as follows: 3–6% solvent B within 3 min, 6–35% solvent B within 117 min, 35–47% solvent B within 5 min, 47–100% solvent B within 5 min and 100% B for 8 min and 1% solvent B for 5 min. Nano spray was achieved with an applied voltage of 1.8 kV. For the analyses on HF Q-Exactive Orbitrap the mass spectrometer was programmed in a data-dependent acquisition mode (top 10 most intense peaks) and was configured to perform a Fourier transform survey scan from 370 to 1600 m/z (resolution 60,000), AGC target 3 e6, maximum injection time 250 ms. MS2 scans were acquired on the 10 most-abundant MS1 ions of charge state 2–7 using a Quadrupole isolation window of 1 m/z for HCD fragmentation. Collision energy was set at 34%; resolution = 30 000; AGC target 2e5, maximum injection time 200 ms; dynamic exclusion 15 s. For the analyses on Fusion Orbitrap the mass spectrometer was programmed as follows: Fourier transform survey scan was set from 350 to 1550 m/z (resolution 120,000), AGC target 2e5, maximum injection time 50 ms; MS2 scans were acquired in data-dependent mode. For the MS1 analysis ions of charge state 2–7 were isolated using a Quadrupole isolation window of 1.4 m/z for HCD fragmentation; collision energy was set at 35%; resolution = 30 000; AGC target 1e5, maximum injection time 100 ms; dynamic exclusion 30 s.
Proteomic and Phosphoproteomic Data Analyses
The raw data were analyzed using MaxQuant 1.5.3.30 (15) and Andromeda (16) was used to search the MS/MS data against the UniProt Homo sapiens database (containing canonical and isoforms_42145 entries downloaded on 15th September 2016) complemented with a list of common contaminants, the two fluorescent proteins used in the Fucci system, and concatenated with the reversed version of all sequences. In total the searched database contained 42,168 entries. TMT10plex was chosen as quantification platform. Trypsin/P was chosen as cleavage specificity allowing for two missed cleavages. Carbamidomethylation (C) was set as a fixed modification, whereas oxidation (M) (and phosphorylation of STY in the case of the phosphoproteome analysis) were used as variable modifications. The database search was performed with a mass deviation of the precursor ion of up to 4.5 ppm (main search). The mass tolerance for fragment ions was 0.5 Da. Data filtering was carried out using the following parameters: peptide and protein FDRs were set to 1%, minimum peptide length was set to 7 and Andromeda minimum score for modified peptides was set to 40, min reporter precursor ion fraction = 0.75. The reverse and common contaminant hits were removed from the output as well as those with localization probability <0.75. Phosphopeptide motif prediction was performed using Perseus (1.5.3.2) with Perseus' integrated motif list, additionally provided in supplemental Table S3. Motif enrichment analysis was performed with R using Fisher's exact test or a simulated Fisher's test.
To account for changes in total cell content during the cell cycle, normalization was deemed very important. Aside from using equal weight amounts of tryptic digest for TMT10plex-labeling, logarithmic reporter intensity values were normalized by subtracting the median logarithmic intensity for each sample prior to further analysis.
Experimental Design and Statistical Rationale
All experiments were performed on three replicates from each of three cell cycle stages. This was deemed adequate to identify the magnitude of differences intended in this study.
In the fractionation proteomic experiment, two insoluble fraction samples were excluded between peptide mapping and ANOVA analysis. After mapping, all proteomic samples (from full proteomics, insoluble fractions, soluble fractions) were hierarchically clustered in a heatmap, which independently clustered all triplicates of samples together except for two individual samples, one replicate for G-phase-Insoluble and one for S-phase-Insoluble. Therefore, the ANOVA for the insoluble data was performed on 8 total samples (3vs2vs2).
All enrichment analyses were performed using either Fisher's exact test for comparing two different distributions (simulated if too large to calculate) using the fisher.test function in base R, or Log-Likelihood-Ratio analysis for comparing a distribution relative to its parental distribution, using the xmulti (xmonte for simulation) function in the XNomial package in R.
All data sets with three groups underwent ANOVA analysis in R (3.4.2) to generate p values for each protein/phosphopeptide. For a few proteins with one or two replicates missing data (Overall proteomics: 5/7527 proteins, Soluble Fraction: 6/4609, Insoluble Fraction: 2/4466), were exchanged for the minimal measured value. FDR-values were calculated in R using the “qvalue” package. All cutoffs of significance were based at the point where = 0.05, i.e. where the next hit would have more than a 5% chance of being a false positive, contrary to classical FDR-cutoff strategies (FDR< 0.05) where the entire data set contains 5% false hits. This method yielded classical FDR-value cutoffs ranging from FDR = 0.0073 to FDR = 0.017. The regression model used to generate the model determining the cutoffs was 8-polynomial and was performed in R using the “polynom” package. All final regression models are shown in supplemental Fig. S1E, supplemental Fig. S2C, and supplemental Fig. S4C respectively.
Data Analysis
As the cells studied are unsynchronized and constantly dividing, any significant difference in protein or phosphopeptide amount between the cell cycle phases in the study indicates a cell-cycle-dependent change in abundance. We thus classify these as exhibiting oscillatory behavior. For visualization of oscillatory patterns and to be able to compare patterns of oscillation between experiments, the algorithm TriComp was used according to (9). A generalized version of the TriComp code in R script can be found in supplemental File S1.
TriComp first calculates the plotting coordinates using Eq. (1) and (2)
| (Eq. 1) |
| (Eq. 2) |
Then these coordinates are converted from Euclidean coordinates into polar coordinates using Eq. (3) and (4).
| (Eq. 3) |
| (Eq. 4) |
Finally, a summarizing group variable is calculated by checking which hexagonal section the data point belongs to using Eq. (5).
| (Eq. 5) |
The TriComp algorithm generates two polar coordinates from the relative logarithmic relationship between three groups, in our study the three cell cycle phases, without losing any information. The output degree coordinate corresponds to the pattern of relationship, and the radius coordinate corresponds to the quantification/intensity of the relationship. By filtering on statistics and radius, the degree coordinate was used throughout the study as a quantitative variable describing the kind of cell cycle oscillation pattern a protein or phosphopeptide exhibited.
Preparation of figures were performed in R (3.4.2) using the libraries ggplot and extrafont
Annotation and Enrichment Analysis
GO-terms associated with cell cycle were classified as being annotated with GO:7049 or a child GO-term of GO:7049 (17). A grouping of these terms into five major cell cycle processes was done using the same grouping as (9). Kinase classifications were retrieved from the Kinome Database, accessed 181210 (18). Phosphatase classification was retrieved from DGIdb 3.0, accessed 181210 (19). Enrichment analysis was performed using hypergeometrical test, with a significance cutoff of FDR<0.01 based on biological function-ontologies from GSEA C5 v6.1 (Broad Institute) after re-annotation of Uniprot IDs to entrez IDs using BioMart version: 0.7. For all enrichment tests, relevant subsets of all detected protientes with unambiguous ID were used as background. Enrichment analysis of comparing Insoluble-specific proteins to Soluble-specific proteins was performed with DAVID after specifying hit lists using multiple T-tests corrected for multiplicity with the qvalue module in R. Uniprot IDs were provided to DAVID. Fraction-specific proteins were specified as more than 2-fold higher in soluble than insoluble and vice-versa, with a significance cutoff of FDR<0.001. Only proteins with available data in both unsorted Soluble and unsorted insoluble fractions were used.
For the literature-based kinase enrichment analysis, the Ma'ayan Lab Kinase Enrichment Analysis 2 tool was used, and each phase group of significantly fluctuating phosphopeptides was submitted, with the results presented as a network analysis.
RESULTS
Characterizing Protein Oscillation Patterns Over The Cell Cycle
To analyze the dynamics of the proteome across the cell cycle without introducing potential artifacts from chemical synchronization, we combined fluorescence-based cell sorting with mass spectrometry-based proteomic analysis. The cell cycle reporter cell line HeLa-Fucci (8) was used throughout this study for precise separation of the cell cycle phases (Fig. 1A; supplemental Fig. S2A–S2C). Live HeLa-Fucci cells were sorted into three separate groups, G1, S and G2/M, based on expression of either or both of its two reporters. A microscopy-based characterization of the three groups was also performed using EdU-incorporation-labeling of actively replicating DNA and quantification of chromatin status (supplemental Fig. S2D–S2F). To be able to quantify the relative abundance of a given protein in G1, S or G2/M phases of the cell cycle we employed a TMT isobaric labeling approach (20) of three replicates for each cell cycle phase (Fig. 1A, supplemental Fig. S2G). Nearly 7500 proteins were identified per cell cycle phase (Fig. 1A, supplemental Table S1), and our ANOVA statistical analysis (corrected by FDR) revealed that 3317 proteins were significantly altered across the cell cycle (supplemental Fig. S2H), of which 219 undergo changes larger than 50%, and 87 more than 2-fold. We refer to these dynamic protein fluctuations between cell cycle phases as oscillatory behaviors. Visualization and comparison of the three groups was achieved with the use of the TriComp algorithm (9) (supplemental Fig. S1A–S1D). TriComp translates three quantitative measurements into two polar descriptive variables: one variable describes the relationship type (θ, degree), whereas the other reports the relative intensity of the relationship (radius). Thus, each angle in the polar coordinate system corresponds to a specific relationship between G1, S and G2/M phase, whereas the distance from origo is a quantifying descriptor of that relationship and can be used for filtering. The angle (degree) was also used to distinguish six separate groups, corresponding to an up-regulation in either one or two of the three cell cycle phases. We further categorized a group of high-oscillating proteins as having at least a 50% increase from one group to another, while also being statistically significant. The results show all possible cell-cycle oscillation patterns, with the high-oscillating group comprised of two prominent clusters: 21% (46/219) of proteins were found to be increased in G1 and G1+S; whereas 72% (158/219) of proteins increased in S+G2/M and G2/M (Fig. 1B, supplemental Table S1).
Among the 20 highest oscillating proteins—excluding the cell cycle probes CDT1 and Geminin, which rank number 1 and 2—we identified well-known cell cycle regulators, as well as proteins not previously associated with the cell cycle (Fig. 1C). The thymine DNA glycosylase TDG is high in G1 whereas the uracil DNA glycosylase UNG2 peaks during S-phase. TK1 (thymidine kinase), involved in the production of the nucleotide dTTP, is increased during S-phase carrying on through G2/M phase. Proteins involved in Rho signaling such as ARHGAP11A and NET1 as well as the mitotic spindle protein CKAP2L have increased abundance in S and G2/M, which is likely associated with mitosis. Some of the highest oscillating proteins that are not GO-annotated as related to the cell cycle have previously been identified to be cell cycle regulated, including ARHGAP11A (4) which has previously been identified to have a critical function in mitosis (21), and KIAA0101 and TK1 which have both been identified to undergo cell-cycle-regulated degradation (22, 23).
Performing gene-set enrichment analysis (GSEA) on the entire hit-list, we observed a major enrichment for cell-cycle-annotated proteins, as expected (Fig. 1D). A closer inspection of these oscillating proteins with cell cycle-related GO terms revealed a strong bias toward the GO annotation of S+G2/M-phase enriched proteins, whereas proteins with higher abundance in G1- or G1+S-phase were underrepresented (Fig. 1E). Confirming the quality of our cell cycle sorting we detected bona fide cell cycle regulators in their corresponding phases (Fig. 1F). After removing all non-cell-cycle-annotated proteins, GSEA reveals that GO cellular process terms such as Locomotion, Cell Motility, Adhesion, and Extracellular Structure Organization are still enriched, suggesting that many proteins involved in these processes are under regulation of the cell cycle machinery (Fig. 1G). We also performed GSEA on each individual group of cell cycle patterns, which revealed a plethora of cellular processes with different protein dynamics. For example, Ribosome Biogenesis was enriched in the G1 pattern, RNA Processing in the G1+S pattern, DNA Replication in the S+G2/M pattern, Organelle Fission in the G2/M pattern, and Chromosome Segregation in the G2/M+G1 pattern (Fig. 1H). We conclude that many vital cellular processes, even those not directly cell cycle-regulatory, are still affected by the cell cycle.
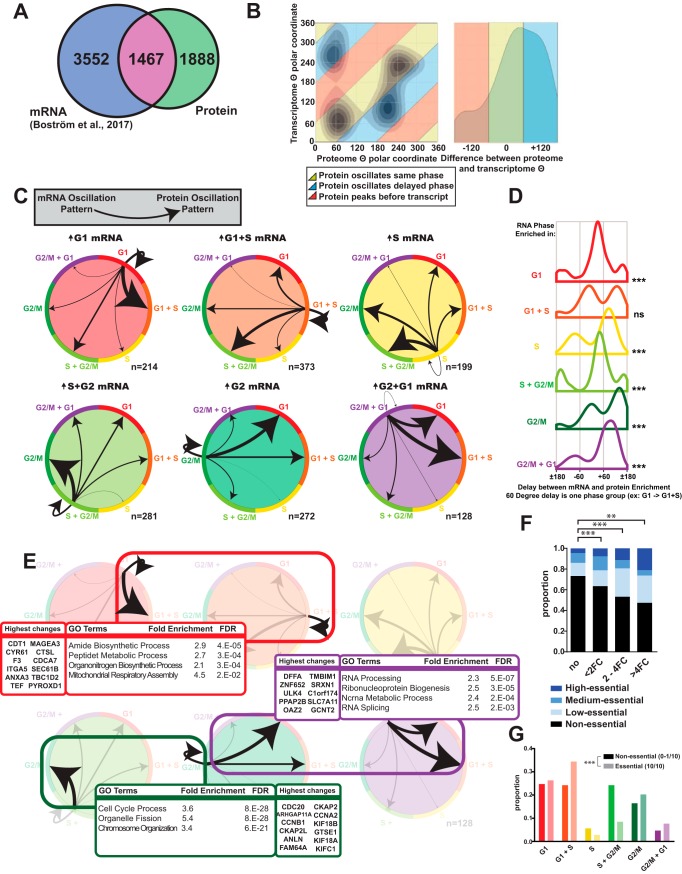
Substantiating the Relationship Between mRNA Expression and Protein Abundance
A comparison between our proteome data set with a recently published transcriptomic data set on cell cycle dynamics using the Fucci system as a biological model (9) revealed an overlap of 1467 transcripts that were significantly oscillating in both data sets. Interestingly, 56% of these showed a variation at the protein level but not at the transcript level, implying that a large extent of protein turnover is regulated via translational or post-translational mechanisms (Fig. 2A). Using the θ variable of both data sets on proteins significantly oscillating in both systems, we could compare variations at transcript and protein abundance, and these strongly correlated. The oscillation patterns for most proteins were either synchronized in the same cell cycle phase as the corresponding transcript or delayed into the next phase (Fig. 2B). By investigating and comparing the relationship between mRNA and protein in the six different cell cycle phase groups, we detected profound differences between mRNA categories. The percentage of proteins that are delayed differ depending on how the mRNA is regulated. Some mRNA-protein relationships are essentially self-supporting, such as G1-up-regulated mRNA leading to G1- or G1+S-up-regulated protein. Genes transcribed in G2/M have the highest percentage of delayed proteins, as a majority of corresponding oscillating proteins are enriched in the following G1 and G1+S groups (Fig. 2C, 2D), with similar magnitudes as the total hit list of oscillating proteins (supplemental Fig. S2I). These findings suggest that most of the transcriptional output during G2/M phase that can be seen as fluctuations on the proteome, only show enrichment in the following G1 phase. This could be part of a preparatory effort in G2/M for the future proteomic composition of the two daughter cells. GSEA analysis of some of the major groups of cross-regulated hits revealed that mRNA up-regulation starting in G1 phase (Group G1 or G1+S), with a protein up-regulation also starting in G1 phase, is enriched for multiple anabolic processes, such as amide biosynthesis and mitochondrial assembly. Proteins that are up-regulated solely in G2/M phase and which transcripts are also up-regulated in G2/M are enriched for chromosome organization proteins, as well as organelle fission—reflecting the need to prepare for mitosis. Finally, the group of mRNA-protein pairs that are delayed over mitosis are enriched for GO terms related to RNA processing and ribonucleoprotein assembly (Fig. 2E). These three groups were also analyzed on the level of magnitude they were affected, and found to share a magnitude distribution with the total list of significantly changed proteins, except for the group which was up-regulated in G2 in both mRNA and Protein, which had a higher overall magnitude (supplemental Fig. S2F).
Fig. 2.
Comparing Transcriptomic and Proteomic Cell Cycle Dynamics. A, Venn diagram of significantly oscillating mRNA (Boström et al., 2017) and proteins (this study). B, Dynamic comparison of protein and mRNA oscillations across the cell cycle using the θ variable from TriComp. C, Detailed view of mRNA-protein pairs oscillating in both experiments. Arrow widths are normalized to proportion of each mRNA origin phase. D, Distribution of the delay from mRNA expression until enrichment of the corresponding protein, estimated as the difference between θ for protein and mRNA respectively. E, GSEA results of three major groups of oscillating mRNA-protein pairs and the twelve highest-magnitude protein changes in each group. F, Fraction of essential proteins among oscillating proteins. Essentiality is enriched among oscillating proteins. G, Proportion of essential and non-essential proteins in individual cell cycle phase patterns. Significance is calculated using Fisher's exact test or Log-Likelihood Ratio calculations. *: p < 0.05, ** p < 0.01, ***: p < 0.001, n.s: non-significant.
We compared our data set with a collection of ten different genome-wide CRISPR/Cas9 viability experiments (10) by using the number of cell lines out of ten affected in this collection as a broad “Essentiality Score” to denote importance for cellular proliferation. We found a clear positive correlation between cell cycle-dependent changes of proteins and their essentiality for cellular viability (Fig. 2F), the percentage of essential proteins was more than 70% higher in >2FC oscillators than non-oscillating proteins. Investigating the cell-cycle distribution of the group of essential-and-oscillating proteins we observed a significant decrease of protein enrichment in S phase compared with non-essential oscillating proteins (Fig. 2G).
Cell Cycle Dynamics of Phosphorylation Patterns
Analyzing the abundance of kinases and phosphatases in our data set using the human KinBase (18) and the Drug-Gene Interaction database v3 (DGIdb3) (19), we detected an enrichment of phosphatases up-regulated in S-phase, whereas kinases are significantly enriched in G2/M (Fig. 3A). This is also evident after comparison of protein and mRNA (Fig. 3B). To integrate the phosphorylation patterns for individual proteins across the cell cycle on a proteome-wide scale, we sorted cells in G1, S and G2/M phases and coupled a phospho-enrichment step to a TMT-based LC/MS/MS analysis (Fig. 3C). We identified 5829 phosphosites with a localization probability >75%. An ANOVA test was performed on 4833 unique phosphopeptides, out of which 3317 were significantly oscillating across the cell cycle after FDR correction (Fig. 3C; supplemental Fig. S2J–S2M). We applied TriComp to visualize the overall changes in phosphorylation over the cell cycle (Fig. 3D–3E). The most prominent patterns of phosphorylation enrichments were detected during S+G2/M phases of the cell cycle, which coincides with the increase in kinase abundance previously identified (Fig. 3A–3B) and overall increase in protein regulation during that phase (Fig. 1B). Among the top oscillating phosphopeptides we could identify many known cell cycle regulators including MKI67, MCM4, CDC20 and CDT1 (Fig. 3F). Highlighted phosphopeptides, which have corresponding genes not annotated with GO:7409 or child terms among the top fluctuating, are multiple phosphorylations on nuclear and nucleolar proteins; Nucleolin and multiple Histone 1 proteins (Fig. 3G).
Fig. 3.
The Cell Cycle of Phosphoproteomic Events. A, Cell cycle distribution frequency of annotated kinases and phosphatases. B, mRNA-protein dynamic comparison using TriComp θ, stratified on kinases and phosphatases. C, Workflow of cell cycle sorting and phosphoproteomic characterization over the cell cycle. D, Cell cycle distribution of all significantly oscillating and highly oscillating phosphosites. E Tricomp visualization of all phosphopeptides oscillating over the cell cycle. F–G, Top fluctuating phosphopeptide which associated Gene is annotated with GO:CellCycle or a child term or not. H, Comparison between protein oscillation patterns and corresponding phosphosite oscillation patterns using TriComp θ variable. I, Transcriptomic, proteomic and phosphoproteomic MKI67 abundance during the cell cycle. J, Cell cycle distribution of predicted Phosphopeptide Motifs from Perseus in the phosphoproteomic data set, analyzed for enrichment relative the background distribution of all significantly oscillating phosphopeptides. Significance measured with Log-likelihood-ratio testing. *: p < 0.05, **: p < 0.01, ***: p < 0.001.
Comparing the patterns of oscillations for the proteome and phosphoproteome, we observed many occurrences that are similar. However, there are also several phosphorylation events in other phases (Fig. 3H). Marker of Proliferation KI-67 (MKI67) provides an example of how protein regulation and activity can be decoupled from protein abundance. MKI67 is transcriptionally restrained to the G2/M phase, but the protein abundance is surprisingly stable throughout the cell cycle (Fig. 3I). Still, a characterization of phosphorylation patterns on MKI67 reveals a high degree of G2/M-phase phosphorylation of MKI67, with 11 unique phosphopeptides being enriched more than 2-fold in G2/M. This suggests a high degree of upstream kinase regulation on MKI67, which corresponds to its role in cellular proliferation (Fig. 3I).
We then used a motif-matching enrichment analysis to gain insights into which phosphopeptide patterns were cell-cycle dependent. We used a motif recognition algorithm in Perseus, with Perseus' integrated motif library which mostly contains known kinase motifs; both general motifs for kinase families, as well as specific kinase motifs (24). Predicted motifs were stratified by cell cycle phase oscillation, providing insights into the cell cycle dependence of specific kinases and enrichments of phosphopeptide motifs (Fig. 3J, supplemental Fig. S3A), supplemental Table S3). Out of 42 motifs in the Perseus library, 39 were significantly enriched either in the number of occurrences or in the specific cell cycle distribution relative to a background distribution of all significant hits.
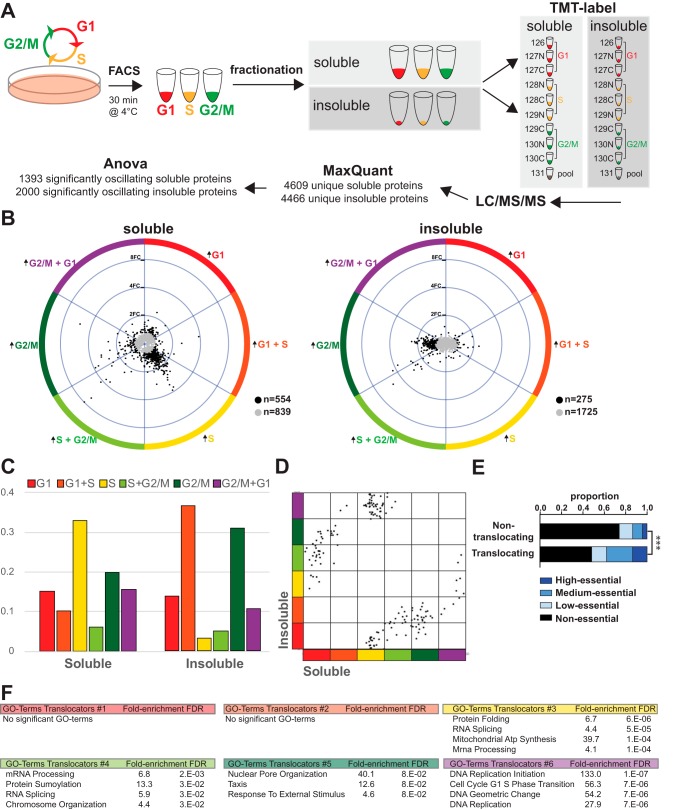
Subcellular Fractionation Reveals Cell Cycle-dependent Protein Relocalization
To analyze protein translocation events, we performed a simple subcellular fractionation on cell cycle-sorted HeLa-Fucci cells (Fig. 4A, supplemental Fig. S4A). Using digitonin as a mild detergent enabled us to separate the cytosol (soluble fraction) from cellular organelles, nuclei and membranes (insoluble fraction). Cellular components of the two fractions were validated using GSEA on GO cell cycle (GO_CC) Terms (supplemental Fig. S4B), supplemental Table S4). TMT-based proteomics analyses of both fractions detected ∼4500 proteins. ANOVA analysis retrieved 1393 significantly oscillating proteins in the soluble and 2000 significantly oscillating proteins in the insoluble fraction (Fig. 4A, supplemental Fig. S4C). Tricomp visualization of the two fractions revealed clearly distinct patterns. Soluble proteins were mostly enriched in S phase, whereas insoluble proteins were more prevalent in G1 and G2/M (Fig. 4B, 4C). We used these oscillation patterns to screen for possible translocation events by looking at proteins that oscillated diametrically opposite in the soluble and insoluble fraction but showed no extensive variation (<20% change) in the total proteome. This identified 169 potential cell cycle-dependent solubility-changing proteins (Fig. 4D). The essentiality score on this group of proteins revealed a strong enrichment for essential proteins (Fig. 4E), underscoring the functional significance of these events. Characterizing these 169 proteins disclosed multiple overarching trends with different kinds of soluble-insoluble relationships (Fig. 4F), some indicating a cell-cycle dependent translocation between subcellular compartments, and some indicating a change in binding configuration. We performed GSEA on a broad categorization of these distributions, using the phase they were enriched as soluble to denote their groups. The proteins enriched as soluble in G2/M phase are enriched in GO terms relating to the nuclear matrix, but also to external stimuli-responsive processes. The proteins soluble in S or S+G2/M phase are predominantly involved in mRNA processing, whereas those soluble in S phase are enriched in mitochondrial-related GO terms, including many mitochondrial ATP synthase proteins, suggesting that small mitochondrial fragments are being released during cell-cycle dependent mitochondrial fission events (25). Finally, proteins that have decreased abundance as soluble in S phase are 133-fold enriched for DNA replication initiation in GSEA, because of the sole presence of five different minichromosome maintenance (MCM) proteins, components of the replicative DNA helicase. In addition to the groups of proteins that relocate during the cell cycle, there are also many unique proteins with interesting phenotypes (supplemental Table S2).
Fig. 4.
Subcellular Fractionation Reveals Protein Translocation During the Cell Cycle. A, Workflow of proteomic characterization of the cell cycle after separation in soluble and insoluble fractions. B, TriComp visualizations of the soluble and insoluble fractions over the cell cycle. C, Distribution of soluble and insoluble patterns. D, Identification of possible cell cycle translocators using a TriComp comparison of the θ value for proteins that have a diametrically opposite pattern (θDiff > 120°) in insoluble and soluble fractions. E, Essentiality distribution of possible translocating proteins. F, GSEA results of six different groups of possible translocators.
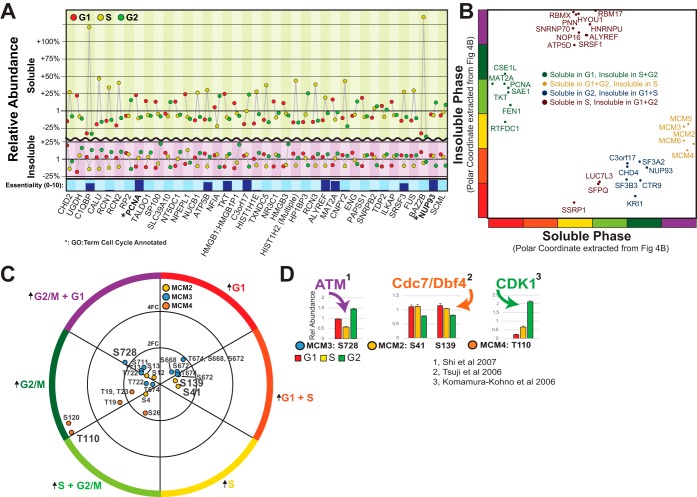
The top possible translocating proteins, sorted on maximum change in relative abundance from the smaller change out of the soluble or the insoluble phase, are shown in Fig. 5A. Compared with Fig. 1B, the prevalence of GO terms denoting Cell Cycle is much smaller. Some of the top possible translocators are involved in chromatin remodeling (CHD2 and CHD4, although not in the top-list), and calcium-dependent signaling in ER (CALU, RCN1, RCN2). Other interesting translocators that become insoluble during S-phase are TALDO1 (Transaldolase involved in PPP), NT5DC1 (5′nucleotidase) and TKT (Transketolase in the hexosamine pathway). All three are involved in nucleoside biosynthesis which would concur with local nuclear regulation of nucleotides during replication. We can also see NUP93, a nuclear-membrane-associated protein becoming more soluble in G2, possibly because of a fragmentation of the nuclear membrane in mitosis.
Fig. 5.
The MCM Family Translocates to the Nucleus in S-phase and has Extensive Phospho-Proteomic Regulation. A, Top soluble and insoluble oscillators with diametrically opposite patterns, suggesting a cell-cycle translocating phenotype. B, High-essential possible translocators, divided into four major groups. C, All phosphopeptides identified as oscillating during the cell cycle belonging to the MCM family. D, References on previously identified kinase pathways affecting five specific phosphosites.
The shortlist of potential translocating proteins that are also classified as essential can be subdivided into four broad groups, depending on specific phases (Fig. 5B). One of these groups, which becomes more insoluble during S-phase, is composed of just five proteins, MCM components MCM2,3,4,5 and 6, the majority of the members of the MCM family, which are components of the MCM double hexamer responsible for unwinding DNA during DNA replication (supplemental Fig. S5A). The DNA polymerase clamp Proliferating Cell Nuclear Antigen (PCNA) also shifts to the insoluble fraction in S-phase, but in contrast to the MCM proteins, PCNA does not revert to the soluble fraction during G2/M phase. A detailed look at the significant phosphorylation events on the MCM family members emphasizes the complexity of PTM regulation of the cell cycle. Purely based on protein dynamics and translocation data, the MCM proteins show very similar phenotypes but they exhibit dissimilar phosphorylation patterns (Fig. 5C). For example, MCM3 is dephosphorylated on many different serine sites during S-phase. This dephosphorylation is in accordance with a previous report describing negative regulation of MCM3 activity by the cell cycle regulator Chk1 (26). The highest regulation of MCM3 was on Ser728, a site which has been described by (27) to be phosphorylated by ATM in response to DNA damage (Fig. 5D). Similarly, we can find the highest regulations of MCM2 and MCM4 in the literature (28, 29) (Fig. 5D).
MAT2A Nuclear Localization Is Enriched in S- and G2/M-phase
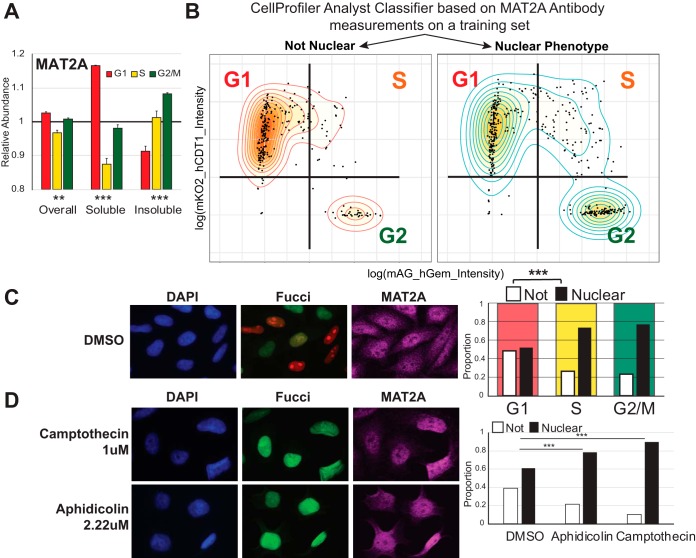
MAT2A is one of the proteins that do not oscillate over the cell cycle in total abundance but undergoes significant changes in oscillation patterns of the soluble and insoluble fractions. This protein is involved in methyl donor production and was previously found to have a dynamic nuclear localization (30). We identified increasingly insoluble MAT2A in S-phase and G2/M-phase cells, with a corresponding decline of soluble MAT2A in these phases (Fig. 6A). We confirmed these findings by independent immunofluorescence experiments, using a machine-learning-based classifier in CellProfiler Analyst (v2.2.1) to categorize MAT2A localization. We identified a strong correlation between MAT2A nuclear localization and S+G2 cell cycle phase using Fucci categorization (Fig. 6B–6D). Arresting cells prior to replication (but post G1/S-checkpoint) using two inhibitors of DNA replication (Aphidicolin and Camptothecin), increased the proportion of cells with nuclear MAT2A (Fig. 6D, supplemental Fig. S5B–S5C). We can thus conclude that passing the G1/S checkpoint is sufficient to facilitate the nuclear translocation of MAT2A, without active replication or entry into G2 phase.
Fig. 6.
MAT2A Use Nuclear Translocation for Localized Cell-Cycle Dependent Functions. A, Bar graph of MAT2A protein abundance pattern. Significance denotes identifying the pattern to be oscillating. B–C, Immunofluorescence-based classification of nuclear and non-nuclear MAT2A localizations corresponds to cell cycle phase according to Fucci categorization. Nuclear localization was determined using a machine learning classifier using a separate training set. D, Arrest in replication leads to an accumulation of nuclear MAT2A phenotype. **: p < 0.01, ***: p < 0.001.
DISCUSSION
The importance of understanding the cell cycle on a proteomic level has only recently become the focus of intense research, as methods to measure protein stability have been employed on cell cycle separated cell lines (31, 32). Additionally, the complexity of the relationship between transcriptomics and proteomics has been highlighted, and in many cases, transcriptomic data cannot confidently be used to predict protein dynamics (33). With the current study, we provide a system-level understanding of the cell cycle in unperturbed cells using combined proteomics, transcriptomics, phosphoproteomics and functional genomics approaches (9, 10). Of note, the HeLa cell line contains inactivated Rb because of its HPV-positive status, and E2F regulation of the cell cycle can be dysregulated (34, 35).
The algorithm TriComp enabled us to compare and visualize large three-dimensional data sets by breaking down the dimensions into a relationship-explanatory variable and a relationship-intensity-variable. This enabled the comparison of patterns from multiple experiments and yielded novel insights into the relationship between transcriptional activity and proteomic regulation on a global level.
Besides classic cell cycle regulatory systems, we also find many proteins whose abundance changes are significantly delayed from their respective mRNA, suggesting that cells prepare for the next G1 phase during G2/M. During the late phases of mitosis, the cell is in a transcriptionally paused state because of the condensation of chromatin (36). Significant delays between late G2/M transcription and corresponding protein accumulation in the following G1 phase suggest that cells buffer mRNA production for subsequent translation during this period of transcriptional pause. Enrichment analysis of gene-protein pairs exhibiting such a delay in our data shows heavy involvement in RNA processing and splicing, possibly suggesting additional regulatory functions. This extent of influence of the parental cells' transcriptional regulation on the protein content of the divided cells raises the interesting possibility of a high parental control of cell fate determination in developmental biology and stem-cell regulation. Additionally, this delay implies that a mRNA identified as strongly up-regulated in G2 might not actually be relevant for the G2-phase but might be preparatory for G1-requisite protein translation instead. Overall, with a mixed cell cycle population such as with tissues or cell samples, the mRNA abundance is a reliable predictor of steady-state protein content (1). However, for the fine-tuned temporal dynamics of the cell cycle, the delay between transcription and translation is oscillatory. As such, the cell will start synthesizing an mRNA in one cell cycle phase in preparation for protein translation in the next phase, especially during the transcriptional hiatus of mitosis. This highlights the need to re-assess the cell cycle classifications of proteins using protein-based techniques, such as Western blotting or mass spectrometry, rather than relying solely on transcriptomics data.
The thymine DNA glycosylase TDG is not cell cycle associated using gene ontology (GO) enrichment analysis, however it was described to be degraded by the UPS in cells entering S-phase. The authors argue that this maintains a separation of function between TDG and the uracil DNA glycosylase UNG2 which peaks during S-phase (37). Although UNG2 was not detected in our data set, we can confirm the G1 prevalence of TDG, and UNG is up-regulated in S and G2/M phases. However, TDG was also linked to DNA demethylation raising the possibility that it is also implicated in regulating gene expression (38). Recently, TDG was suggested as a novel target for melanoma with promising anti-cancer effects through the transcriptional regulation of genes important for cell cycle progression and senescence (39).
Investigating the essentiality of protein oscillations, we could identify a significant lack of essential proteins in the groups S and S+G2/M. Although this observation could suggest an aversion to regulate protein levels in S phase, it might alternatively indicate that essential proteins which are controlled in S-phase, are more likely to be redundant, and therefore not be identified as essential in single-gene knockout CRISPR/Cas9 survivability studies. One of these S-G2/M-induced cell cycle markers, MKI67, which only exhibited a phenotype in one out of ten cell lines in Bertoume et al. (10), exhibited both extensive cell cycle-dependent phosphorylation and high transcriptional regulation. Although its protein levels oscillated significantly, the difference among the phases was relatively small. We hypothesize that this is because of a relatively slow degradation of MKI67, and the fast proliferation rate of HeLa cells reaching the next transcriptomic burst before degradation of MKI67 is complete. Although Sobecki et al. shows that MKI67 undergoes APC/C-mediated degradation (40), quantifications in continuously proliferating cells by Miller et al. highlights that KI67 is continuously increasing in S/G2/M and continuously degraded in G1 (41), and its degradation is a slower process than for example APC/C-mediated degradation of Geminin, which takes place within minutes at the end of mitosis (8).
We furthermore detected many proteins that oscillate over the cell cycle while their transcripts remain stable, indicating widespread post-translational regulation, possibly through the ubiquitin-proteasome system. The major functional difference between transcriptional regulation and regulation via protein degradation is the speed. An additional consideration for the cell to actively degrade a protein, is the future cost in energy and ribosome occupancy of rebuilding it the next time it is needed. PTM-based regulation offers an even faster and more energy-efficient way to control protein activity. We provide information regarding the phosphorylation events over the cell cycle, and a method of comparing these to protein and transcript level dynamics.
Besides the post-translational regulation of protein activity, the intracellular localization plays a major role and the compartmentalization of the cell serves as a well-recognized way to separate protein functions without altering abundance. Our method to compare the soluble and insoluble proteomic cell cycle while controlling for the overall cell cycle revealed several interesting patterns. We identify three major reasons for why proteins show up in this hit list; a change in structural integrity of a larger structure, a change in binding confirmation to a larger structure, or a translocation event between subcellular compartments with different solubility.
The first reason can account for the enrichment of nuclear matrix and mitochondrial GO-terms, as both complexes are known to undergo cell-cycle-induced conformational changes; the breakdown of the nuclear membrane in mitosis, and the mitochondrial fission respectively. Indeed, the largest unique group of proteins that appeared as oscillating in the soluble cell cycle was a group of 22 mitochondrial proteins, confirming the extensive cell-cycle-mediated fission of large mitochondrial complexes that occurs during S-phase (42, 43). Release of mitochondrial material into the cytosol during the fission process would lead to a markedly higher amount of mitochondrial proteins in the cytoplasm during S-phase until they are naturally broken down.
The second reason is related to binding configurations to structures inducing a solubility shift of the proteins themselves. A prominent group that becomes insoluble during S-phase but is soluble throughout the remaining cell cycle phases are five MCM family members, highlighting the specificity of the prediction algorithm. The MCM proteins are part of the helicase responsible for unwinding duplex DNA during replication, and is stabilized on DNA in S-phase, explaining the solubility shift.
The last reason is related to translocation events between subcellular compartments. We identified and confirmed a distinct translocating phenotype of the S-adenosylmethionine synthase MAT2A. This protein was previously reported to be localized to the nucleus as well as to the cytosol, but the functional significance was unknown (30). We show here that MAT2A is enriched in the nucleus of cells undergoing replication and during the subsequent G2-phase. We suggest that the role for this translocation is the result of the high methylation requirement inside the nucleus during S-phase for both DNA and histone methylation processes. Although the highest requirement of methyl donor is during S-phase, recent evidence (44) suggests that the copying of histone methylation patterns is performed throughout G2 phase. In addition, methylation is required during G2 phase to facilitate heterochromatin formation (45). MAT2A and methionine metabolism was just recently found to be a critical feature of tumor initiating cells (46). Together, this provides a plausible explanation as to why MAT2A nuclear localization persists until mitosis. We speculate that localized nuclear S-adenosylmethionine is essential to provide materials for the copying of epigenetic methylation patterns in proliferating cells.
Throughout our characterizations of protein abundance, phosphorylation and translocation patterns, we investigated the essentiality of the hit lists using CRISPR/Cas9 survival screening data, and we could identify a clear correlation between the level of regulation over the cell cycle and essentiality for cellular proliferation. This supports the argument that essential genes are more likely to have specific cell cycle-dependent functions. Thus, the essentiality score itself is a useful tool to narrow large hit lists and identify proteins that are also proven to be functionally necessary for cell division, and to filter out candidates that are regulated but not functionally essential.
DATA AVAILABILITY
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (47) with the data set identifier PXD011836. Spectras for the phospho-proteomic experiment are viewable on MS-Viewer using the search key “bhiwr0fv8r”.
Supplementary Material
Acknowledgments
We thank LifeSensors for material and technical support and Jaromir Mikes at the Mass Cytometry facility at Science for Life Laboratory, Stockholm for cell sorting. We thank Dr Nathan Wall for valuable comments on the manuscript. We acknowledge Prof. Sonia Lain and Prof. David Lane and their groups for technical support and granting us permission to access their Orbitrap Fusion.
Footnotes
* M.A. has been awarded grants from the Hållsten Foundation and the SciLifeLab Technology Development Project Grant. G.M. has been awarded grants from: O. E. and Edla Johanssons foundation (5310-7132) and G.M., J.L. Swedish Cancer Society (Radiumhemmets) (154202, 141243), J.L. from Swedish Research Council (2015-04622), the Swedish Foundation for Strategic Research (RIF14-0046), the Swedish Childhood Cancer Foundation and the Swedish Cancer Society (CAN 2017/685). S.G.R. is supported by a grant from the Swedish Cancer Society (19-0056-JIA). The authors declare that they have no conflicts of interest with the contents of this article.
 This article contains supplemental Figures, Tables, and Material.
This article contains supplemental Figures, Tables, and Material.
1 The abbreviations used are:
- UPS
- Ubiquitin-Proteasome-System
- PTM
- post-translational modifications
- FACS
- fluorescence-activated cell sorting.
REFERENCES
- 1. Edfors F., Danielsson F., Hallström B. M., Käll L., Lundberg E., Pontén F., Forsström B., and Uhlén M. (2016) Gene-specific correlation of RNA and protein levels in human cells and tissues. Mol. Syst. Biol. 12, 883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vogel C., and Marcotte E. M. (2012) Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 13, 227–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Orre L. M., Vesterlund M., Pan Y., Arslan T., Zhu Y., Fernandes Woodbridge A., Frings O., Fredlund E., and Lehtiö J.. SubCellBarCode: Proteome-wide mapping of protein localization and relocalization. Mol. Cell. [DOI] [PubMed] [Google Scholar]
- 4. Ly T., Ahmad Y., Shlien A., Soroka D., Mills A., Emanuele M. J., Stratton M. R., and Lamond A. I. (2014) A proteomic chronology of gene expression through the cell cycle in human myeloid leukemia cells. Elife. 3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ly T., Whigham A., Clarke R., Brenes-Murillo A. J., Estes B., Madhessian D., Lundberg E., Wadsworth P., and Lamond A. I. (2017) Proteomic analysis of cell cycle progression in asynchronous cultures, including mitotic subphases, using PRIMMUS. Elife. 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dephoure N., Zhou C., Villén J., Beausoleil S. A., Bakalarski C. E., Elledge S. J., and Gygi S. P. (2008) A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 105, 10762–10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Olsen J. V., Vermeulen M., Santamaria A., Kumar C., Miller M. L., Jensen L. J., Gnad F., Cox J., Jensen T. S., Nigg E. A., Brunak S., and Mann M. (2010) Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 3, ra3. [DOI] [PubMed] [Google Scholar]
- 8. Sakaue-Sawano A., Kurokawa H., Morimura T., Hanyu A., Hama H., Osawa H., Kashiwagi S., Fukami K., Miyata T., Miyoshi H., Imamura T., Ogawa M., Masai H., and Miyawaki A. (2008) Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 132, 487–498 [DOI] [PubMed] [Google Scholar]
- 9. Boström J., Sramkova Z., Salašová A., Johard H., Mahdessian D., Fedr R., Marks C., Medalová J., Souček K., Lundberg E., Linnarsson S., Bryja V., Sekyrova P., Altun M., Andäng M. (2017). Comparative cell cycle transcriptomics reveals synchronization of developmental transcription factor networks in cancer cells. PLoS ONE 12, e0188772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bertomeu T., Coulombe-Huntington J., Chatr-Aryamontri A., Bourdages K. G., Coyaud E., Raught B., Xia Y., and Tyers M. (2018) A high-resolution genome-wide CRISPR/Cas9 viability screen reveals structural features and contextual diversity of the human cell-essential proteome. Mol. Cell. Biol. 38, pii: e00302–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Maldonado L. Y., Arsene D., Mato J. M., and Lu S. C. (2018) Methionine adenosyltransferases in cancers: Mechanisms of dysregulation and implications for therapy. Exp. Biol. Med. 243, 107–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu X., and Fagotto F. (2011) A method to separate nuclear, cytosolic, and membrane-associated signaling molecules in cultured cells. Sci. Signal. 4, pl2. [DOI] [PubMed] [Google Scholar]
- 13. Elia A. E. H., Wang D. C., Willis N. A., Boardman A. P., Hajdu I., Adeyemi R. O., Lowry E., Gygi S. P., Scully R., and Elledge S. J. (2015) RFWD3-dependent ubiquitination of RPA regulates repair at stalled replication forks. MOLCEL. 60, 280–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Matheron L., van den Toorn H., Heck A. J. R., and Mohammed S. (2014) Characterization of biases in phosphopeptide enrichment by Ti(4+)-immobilized metal affinity chromatography and TiO2 using a massive synthetic library and human cell digests. Anal. Chem. 86, 8312–8320 [DOI] [PubMed] [Google Scholar]
- 15. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 16. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., and Mann M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 17. Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., Harris M. A., Hill D. P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J. C., Richardson J. E., Ringwald M., Rubin G. M., and Sherlock G. (2000) Gene Ontology: tool for the unification of biology. Nat. Genet. 25, 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Manning G., Whyte D. B., Martinez R., Hunter T., and Sudarsanam S. (2002) The protein kinase complement of the human genome. Science 298, 1912–1934 [DOI] [PubMed] [Google Scholar]
- 19. Cotto K. C., Wagner A. H., Feng Y.-Y., Kiwala S., Coffman A. C., Spies G., Wollam A., Spies N. C., Griffith O. L., and Griffith M. (2018) DGIdb 3.0: a redesign and expansion of the drug–gene interaction database. Nucleic Acids Res. 46, D1068–D1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rauniyar N., Yates JRIII. (2014) Isobaric labeling-based relative quantification in shotgun proteomics. J. Proteome Res. 13, 5293–5309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zanin E., Desai A., Poser I., Toyoda Y., Andree C., Moebius C., Bickle M., Conradt B., Piekny A., and Oegema K. (2013) A conserved RhoGAP limits M phase contractility and coordinates with microtubule asters to confine RhoA during Cytokinesis. Dev. Cell. 26, 496–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Emanuele M. J., Ciccia A., Elia A. E. H., and Elledge S. J. (2011) Proliferating cell nuclear antigen (PCNA)-associated KIAA0101/PAF15 protein is a cell cycle-regulated anaphase-promoting complex/cyclosome substrate. Proc. Natl. Acad. Sci. U.S.A. 108, 9845–9850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kauffman M. G., and Kelly T. J. (1991) Cell cycle regulation of thymidine kinase: residues near the carboxyl terminus are essential for the specific degradation of the enzyme at mitosis. Mol. Cell. Biol. 11, 2538–2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M. Y., Geiger T., Mann M., and Cox J. (2016) The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 13, 731–740 [DOI] [PubMed] [Google Scholar]
- 25. Mishra P., and Chan D. C. (2014) Mitochondrial dynamics and inheritance during cell division, development and disease. Nat. Rev. Mol. Cell Biol. 15, 634–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Han X., Mayca Pozo F., Wisotsky J. N., Wang B., Jacobberger J. W., and Zhang Y. (2015) Phosphorylation of minichromosome maintenance 3 (MCM3) by checkpoint kinase 1 (Chk1) negatively regulates DNA replication and checkpoint activation. J. Biol. Chem. 290, 12370–12378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shi Y., Dodson G. E., Mukhopadhyay P. S., Shanware N. P., Trinh A. T., and Tibbetts R. S. (2007) Identification of carboxyl-terminal MCM3 phosphorylation sites using polyreactive phosphospecific antibodies. J. Biol. Chem. 282, 9236–9243 [DOI] [PubMed] [Google Scholar]
- 28. Tsuji T., Ficarro S. B., and Jiang W. (2006) Essential role of phosphorylation of MCM2 by Cdc7/Dbf4 in the initiation of DNA replication in mammalian cells. Mol. Biol. Cell. 17, 4459–4472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Komamura-Kohno Y., Karasawa-Shimizu K., Saitoh T., Sato M., Hanaoka F., Tanaka S., and Ishimi Y. (2006) Site-specific phosphorylation of MCM4 during the cell cycle in mammalian cells. FEBS J. 273, 1224–1239 [DOI] [PubMed] [Google Scholar]
- 30. Katoh Y., Ikura T., Hoshikawa Y., Tashiro S., Ito T., Ohta M., Kera Y., Noda T., and Igarashi K. (2011) Methionine adenosyltransferase II serves as a transcriptional corepressor of Maf oncoprotein. Mol. Cell. 41, 554–566 [DOI] [PubMed] [Google Scholar]
- 31. Becher I., Andrés-Pons A., Romanov N., Stein F., Schramm M., Baudin F., Helm D., Kurzawa N., Mateus A., Mackmull M.-T., Typas A., Müller C. W., Bork P., Beck M., and Savitski M. M. (2018) Pervasive protein thermal stability variation during the cell cycle. Cell. 173, 1–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dai L., Zhao T., Bisteau X., Sun W., Prabhu N., Lim Y. T., Sobota R. M., Kaldis P., and Nordlund P. (2018) Modulation of protein-interaction states through the cell cycle. Cell. 173, 1–28 [DOI] [PubMed] [Google Scholar]
- 33. Liu Y., Beyer A., and Aebersold R. (2016) On the dependency of cellular protein levels on mRNA abundance. Cell 165, 535–550 [DOI] [PubMed] [Google Scholar]
- 34. Popescu N. C., DiPaolo J. A., and Amsbaugh S. C. (1987) Integration sites of human papillomavirus 18 DNA sequences on HeLa cell chromosomes. Cytogenet. Genome Res. 44, 58–62 [DOI] [PubMed] [Google Scholar]
- 35. Yim E.-K., and Park J.-S. (2005) The role of HPV E6 and E7 oncoproteins in HPV-associated cervical carcinogenesis. Cancer Res. Treat. 37, 319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fan H., and Penman S. (1970) Regulation of protein synthesis in mammalian cells: II. Inhibition of protein synthesis at the level of initiation during mitosis. J. Mol. Biol. 50, 655–670 [DOI] [PubMed] [Google Scholar]
- 37. Hardeland U., Kunz C., Focke F., Szadkowski M., and Schär P. (2007) Cell cycle regulation as a mechanism for functional separation of the apparently redundant uracil DNA glycosylases TDG and UNG2. Nucleic Acids Res. 35, 3859–3867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kohli R. M., and Zhang Y. (2013) TET enzymes, TDG and the dynamics of DNA demethylation. Nature 502, 472–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mancuso P., Tricarico R., Bhattacharjee V., Cosentino L., Kadariya Y., Jelinek J., Nicolas E., Einarson M., Beeharry N., Devarajan K., Katz R. A., Dorjsuren D. G., Sun H., Simeonov A., Giordano A., Testa J. R., Davidson G., Davidson I., Larue L., Sobol R. W., Yen T. J., and Bellacosa A. (2019) Thymine DNA glycosylase as a novel target for melanoma. Oncogene 38, 3710–3728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sobecki M., Mrouj K., Camasses A., Parisis N., Nicolas E., Llères D., Gerbe F., Prieto S., Krasinska L., David A., Eguren M., Birling M. C., Urbach S., Hem S., Déjardin J., Malumbres M., Jay P., Dulic V., Lafontaine D. L. J., Feil R., and Fisher D. (2016) The cell proliferation antigen Ki-67 organises heterochromatin. Elife 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Miller I., Min M., Yang C., Tian C., Gookin S., Carter D., and Spencer S. L. (2018) Ki67 is a graded rather than a binary marker of proliferation versus quiescence. Cell Rep. 24, 1105–1112.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Salazar-Roa M., and Malumbres M. (2017) Fueling the cell division cycle. Trends Cell Biol. 27, 69–81 [DOI] [PubMed] [Google Scholar]
- 43. Horbay R., and Bilyy R. (2016) Mitochondrial dynamics during cell cycling. Apoptosis 21, 1327–1335 [DOI] [PubMed] [Google Scholar]
- 44. Pesavento J. J., Yang H., Kelleher N. L., and Mizzen C. A. (2008) Certain and progressive methylation of histone H4 at lysine 20 during the cell cycle. Mol. Cell. Biol. 28, 468–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Heit R., Rattner J. B., Chan G. K. T., and Hendzel M. J. (2009) G2 histone methylation is required for the proper segregation of chromosomes. J. Cell Sci. 122, 2957–2968 [DOI] [PubMed] [Google Scholar]
- 46. Wang Z., Yip L. Y., Lee J. H. J., Wu Z., Chew H. Y., Chong P. K. W., Teo C. C., Ang H. Y.-K., Peh K. L. E., Yuan J., Ma S., Choo L. S. K., Basri N., Jiang X., Yu Q., Hillmer A. M., Lim W. T., Lim T. K. H., Takano A., Tan E. H., Tan D. S. W., Ho Y. S., Lim B., and Tam W. L. (2019) Methionine is a metabolic dependency of tumor-initiating cells. Nat. Med. 25, 825–837 [DOI] [PubMed] [Google Scholar]
- 47. Vizcaíno J. A., Côté R. G., Csordas A., Dianes J. A., Fabregat A., Foster J. M., Griss J., Alpi E., Birim M., Contell J., O'Kelly G., Schoenegger A., Ovelleiro D., Pérez-Riverol Y., Reisinger F., Ríos D., Wang R., and Hermjakob H. (2012) The Proteomics Identifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41, D1063–D1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (47) with the data set identifier PXD011836. Spectras for the phospho-proteomic experiment are viewable on MS-Viewer using the search key “bhiwr0fv8r”.