Abstract
(Chymo)trypsin-like serine fold proteases belong to the serine/cysteine proteases found in eukaryotes, prokaryotes, and viruses. Their catalytic activity is carried out using a triad of amino acids, a nucleophile, a base, and an acid. For this superfamily of proteases, we propose the existence of a universal 3D structure comprising 11 amino acids near the catalytic nucleophile and base – Nucleophile-Base Catalytic Zone (NBCZone). The comparison of NBCZones among 169 eukaryotic, prokaryotic, and viral (chymo)trypsin-like proteases suggested the existence of 15 distinct groups determined by the combination of amino acids located at two “key” structure-functional positions 54T and 55T near the catalytic base His57T. Most eukaryotic and prokaryotic proteases fell into two major groups, [ST]A and TN. Usually, proteases of [ST]A group contain a disulfide bond between cysteines Cys42T and Cys58T of the NBCZone. In contrast, viral proteases were distributed among seven groups, and lack this disulfide bond. Furthermore, only the [ST]A group of eukaryotic proteases contains glycine at position 43T, which is instrumental for activation of these enzymes. In contrast, due to the side chains of residues at position 43T prokaryotic and viral proteases do not have the ability to carry out the structural transition of the eukaryotic zymogen-zyme type.
Keywords: (Chymo)trypsin-like proteases, Catalytic triad, Structural motif, Structural framework
1. Introduction
Previously, when analyzing the spatial structures of a superfamily of proteins with an α/β-hydrolases fold (SCOP ID: [53473]), characterized by the 3-layer α/β/α architecture with a mixed β-sheet of eight β-strands placed in 12435678 order, and with β-strand 2 being antiparallel to the rest of the β-strands [1], the existence of a small internal position with variable contents was discovered. This position is filled with a water molecule or with an oxygen atom from the side-chain group of the catalytic acid residue. The set of amino acids surrounding this position was termed the catalytic acid zone [2].
In addition to the acid residue, the active site of proteins with an α/β hydrolase fold includes a nucleophile, base, and two residues of the oxyanion hole that stabilize the tetrahedral intermediate during catalysis [3]. Together, these residues form the catalytic machinery necessary for performing the hydrolase function. Similarly to the catalytic acid zone, the nucleophile and oxyanion zones, which coordinate the catalytic nucleophile and the residues of the oxyanion hole, were also described [4]; and it was speculated that the catalytic triad zones together form a conserved structural motif [2].
There is another superfamily of hydrolases, (chymo)trypsin-like serine fold proteases (SCOP ID: [50493]) [5]. These are the all-β proteins comprised of two six-stranded Greek key β-barrels lying perpendicular to one another with the active site cleft located between them [1]. The hydrolases of this type also have the same catalytic triad [[5], [6], [7]], but the three amino acids are arranged in a different sequential order (Fig. 1 ). The nucleophile of α/β hydrolases is located at the turn, known as the “nucleophile elbow”, and is identified by the sequence motif Sm–X–Nuc–X–Sm (Sm, small residue; X, any residue; Nuc, nucleophile) [3]. The corresponding pentapeptide of (chymo)trypsin-like serine fold proteases has the consensus pattern G–[DE]–S–G–[GS] (https://prosite.expasy.org/; PROSITE documentation PDOC00124; TRYPSIN_SER, PS00135 [8]). In addition to this pattern, there is also a histidine active site pattern [LIVM]-[ST]-A-[STAG]-H-C (TRYPSIN_HIS, PS00134). In both families, the oxyanion hole is situated adjacent to the nucleophile and is mainly shaped by the main-chain nitrogen atoms of two residues, but again the sequential order is different.
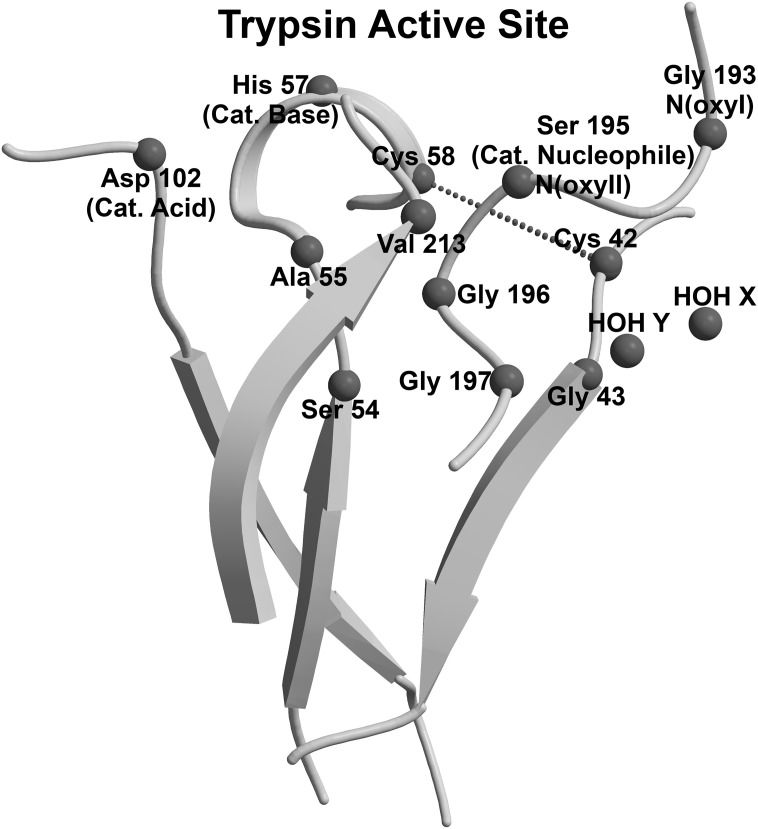
Fig. 1.
Structure of the active site in (chymo)trypsin-like serine fold proteases. Amino acid numbers are taken as in Trypsin (PDB ID: 4I8H_A). The catalytic triad includes Asp102 (the catalytic acid), His57 (the catalytic base) and Ser195 (the catalytic nucleophile). The PROSITE “TRYPSIN_SER” pattern (PS00135; G–[DE]–S–G–[GS]) includes Gly193-Asp194-Ser195(cat. nucleophile)-Gly196-Gly197. The PROSITE “TRYPSIN_HIS” pattern (PS00134; [LIVM]-[ST]-A-[STAG]-H-C) includes Val53-Ser54-Ala55-Ala56-His57(cat. base)-Cys58. Two main-chain nitrogens, N/Gly193 and N/Ser195, are the two canonical oxyanions “N(oxyI)” and “N(oxyII)”. Gly43 and Val213 simultaneously interact with both the TRYPSIN_SER and TRYPSIN_HIS pattern, and thus constitute the “43/213 Nucleophile-Base Catalytic Zone” (43/213-NBCZone) of (chymo)trypsin-like serine fold proteases. The disulfide bond Cys42-Cys58 joins the elements of the “42/43 Base Catalytic Zone”, which includes the TRYPSIN_HIS pattern and the Cys42-Gly43 dipeptide. Two conserved structural water molecules in positions X and Y, HOH X and HOH Y, interact with the TRYPSIN_SER pattern and form the “Nucleophile-Base Catalytic Zone Conserved Extension” in eukaryotic serine (chymo)trypsin-like fold proteases. Structural data were visualized and analyzed using Discovery Studio [76] and Bodil [77]. Figures were drawn with MolScript [78] and Raster 3D [79].
Knowing the importance of the existence of a catalytic acid, nucleophile, and oxyanion zones for the function of the α/β hydrolases fold enzymes and with the possible presence of similar structural formations in other types of hydrolases, we carried out a detailed analysis of the spatial structures of (chymo)trypsin-like serine fold proteases near the catalytic triad residues. As a result, fifteen variants of a unique structural Nucleophile-Base Catalytic Zone (NBCZone) were found that affect the conformations of the catalytic triad residue loops of all (chymo)trypsin-like serine fold proteases (eukaryotic, prokaryotic, and viral).
2. Results and discussion
2.1. Eukaryotic serine (chymo)trypsin-like fold proteases
2.1.1. Nucleophile-Base Catalytic Zone (NBCZone) of trypsin
The presentation of our results will begin with an analysis of the tertiary structure of bovine trypsin active site (Protein Data Bank (PDB: [9]), PDB ID: 4I8H, chain A, Fig. 1) [10]. Fig. 1 shows the catalytic triad that includes residues His57, Asp102 and Ser195. In addition to the base His57 and the nucleophile Ser195, the localization of Ser54, Ala55, and Cys58 of the PROSITE TRYPSIN_HIS pattern is shown, as well as amino acids of the PROSITE TRYPSIN_SER pattern: Gly193-Asp194-Ser195-Gly196-Gly197 and two main-chain nitrogens: N/Gly193 and N/Ser195, which are the canonical oxyanions OxyI and OxyII [11]. In trypsin, there are two amino acids that interact with both the PROSITE TRYPSIN_HIS pattern and the PROSITE TRYPSIN_SER pattern. The amino acids Gly43 and Val213 are in contact with the tripeptide Ser195-Gly196-Gly197 (Fig. 2A, Table 1 ). Main-chain atoms of Gly43 form a hydrogen bond with Ser195: N/Gly43-O/Ser195, and a weak hydrogen bond with Gly196: O/Gly43-CA/Gly196. Similar to Gly43, main-chain atoms of Val213 form two hydrogen bonds with the dipeptide Gly196-Gly197 (N/Val213-O/Gly197 and O/Val213-N/Gly196); the tetrapeptide Asp194-Gly197 forms a β-turn (data not shown). Main-chain atoms of Gly43 and Val213 are also in contact with the dipeptide Ser54-Ala55 from the PROSITE TRYPSIN_HIS pattern: contacts O/Gly43-OG/Ser54 and O/Val213-CB/Ala55 are shown (Fig. 2A). The group of connected amino acids presented in Fig. 2A we have named the 43/213 Nucleophile-Base Catalytic Zone (43/213-NBCZone) of trypsin.
Fig. 2.
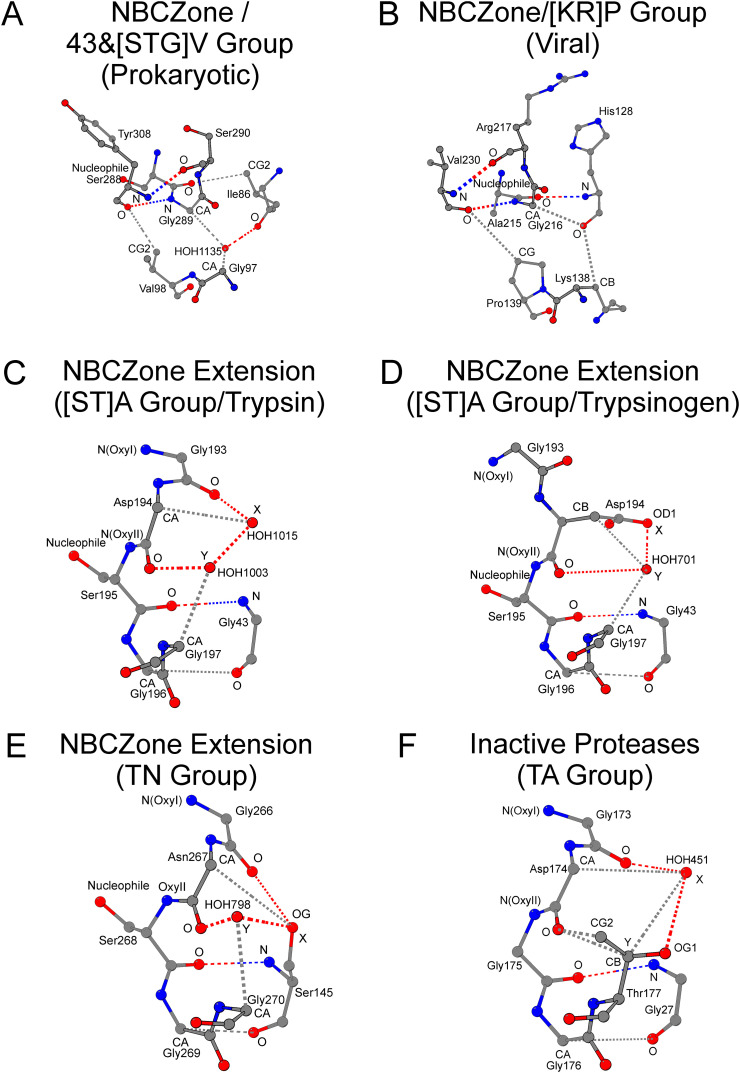
(A) shows the “43/213 Nucleophile-Base Catalytic Zone” (43/213-NBCZone) of the “[ST]A Group” of (chymo)trypsin-like serine fold proteases (see Table 1). The 43/213-NBCZone, shown in (A), together with the “42/43 Base Catalytic Zone” shown in (B) constitute the entire Nucleophile-Base Catalytic Zone (NBCZone) of trypsin, which is the representative structure of the “[ST]A Group” (see Table 1). Unlike the “[ST]A Group”, shown in (A), where the 55T position of the 43/213-NBCZone is occupied by an alanine (Ala55 in panel A), in the “TN Group” enzymes, shown in (C), (D), (E) and (F), the 55T position is occupied by an asparagine (Asn196 in panel C; Asn218 in panel D; Asn171 in panel E and Asn38 in panel F), whose conformation is different in four different groups, named Sets I to IV, of the TN Group enzymes. In Sets I and II, respectively shown in (C) and (D), the ND2 atom of Asn55T takes part in the formation of the 43/213-NBCZone, while the OD1 atom of Asn55T does either form an Asx-turn with the catalytic histidine (as in C) or interacts with the main-chain oxygen atom of the catalytic acid (as in D). In Sets III and IV, respectively shown in (E) and (F), the OD1 atom of Asn55T takes part in the formation of the 43/213-NBCZone, while the ND2 atom of Asn55T interacts with either the catalytic acid (as in E) or base (as in F).
Table 1.
Geometrical parameters of interactions within the amino acid sets forming NBCZones in representative structures of nine (chymo)trypsin-like serine fold proteases groups.
| Protein | Organism | PDB ID resolution | Hydrogen bonds of amino acid at position 43T | Hydrogen bonds of amino acid at position 213T | Interactions of amino acids at positions 42T&58T | Ref. |
|---|---|---|---|---|---|---|
| Eukaryotic proteases | ||||||
| [ST]A group | ||||||
| Trypsin | Bos taurus | 4I8H_A R = 0.75 Å |
N/G43-O/S195, 2.8 O/G43-CA/G196, 3.4 (2.7) O/G43-OG/S54, 2.8 |
N/V213-O/G197, 2.9 O/V213-N/G196, 2.9 O/V213-CB/A55, 4.0 (3.0) |
CB/C42-O/S195, 3.3 (2.7) SG/C58-O/S195, 3.6 C42-C58 |
10 |
| Trypsinogen | Bos taurus | 1TGT_A R = 1.70 Å |
N/G43-O/S195, 2.7 O/G43-CA/G196, 3.6 (2.8) O/G43-OG/S54, 2.8 |
N/V213-O/G197, 2.7 O/V213-N/G196, 2.9 O/V213-CB/A55, 4.1 (3.2) |
CA/C42-O/S195, 3.4 (2.8) SG/C58-O/S195, 3.6 C42-C58 |
12 |
| Mannan-binding lectin serine protease 2 | Homo sapiens | 3TVJ_B R = 1.28 Å |
N/A469-O/S633, 2.9 O/A469-CA/G634, 3.2 (2.3) O/A469-OG1/T480, 2.8 |
N/V653-O/G635, 2.9 O/V653-N/G634, 3.0 O/V653-CB/A481, 4.0 (3.0) |
CB/A468-O/S633, 3.7 (2.8) CB/A484-O/S633, 5.0 (4.3) CB/A468-CB/A484, 4.3 |
22 |
| TN group | ||||||
| Serine protease HTRA2, mitochondrial | Homo sapiens | 5M3N_A R = 1.65 Å Set I |
N/S183-O/S306, 3.0 O/S183-CA/G307, 3.2 (2.5) O/S183-OG1/T195, 2.7 |
N/N321-O/G308, 2.9 O/N321-N/G307, 2.8 O/N321-ND2/N196, 3.2 |
CA/G182-O/S306, 3.6 (2.9) CG2/V199-O/S306, 4.0 (3.2) CA/G182-CG2/V199, 4.4 |
27 |
| Serine protease HTRA1 | Homo sapiens | 3TJN_B R = 3.00 Å Set II |
N/S205-O/S328, 3.1 O/S205-CA/G329, 3.3 (2.4) O/S205-OG1/T217, 2.5 |
N/N343-O/G330, 3.0 O/N343-N/G329, 2.9 O/N343-ND2/N218, 3.0 |
CA/G204-O/S328, 4.0 (3.6) CG2/V221-O/S328, 3.5 (2.5) CA/G204-CG1/V221, 4.0 |
29 |
| Protease Do-like 1, chloroplastic | Arabi-dopsis thaliana | 3QO6_A R = 2.50 Å Set III |
N/S158-O/S282, 3.1 O/S158-CA/G283, 3.3 (2.3) O/S158-CG2/T170, 2.6 (2.0) |
N/N297-O/G284, 2.9 O/N297-N/G283, 2.9 O/N297-OD1/N171, 3.3 N/G283-OD1/N171, 2.9 |
CA/G157-O/S282, 4.1 (3.6) CG2/V174-O/S282, 4.0 (3.2) CA/G157-CG1/V174, 4.2 |
30 |
| Prokaryotic proteases | ||||||
| TN group | ||||||
| Serine protease Spl | Staphy-lococcus aureus | 2AS9_A R = 1.70 Å Set IV |
N/T26-O/S158, 2.8 O/T26-CA/G159, 3.2 (2.3) O/T26-OG1/T37, 2.9 |
N/V173-O/S160, 2.8 O/V173-N/G159, 3.0 O/V173-OD1/N38, 2.9 N/G159-OD1/N38, 3.1 |
CB/A25-O/S158, 3.6 (2.7) CG2/V41-O/S158, 4.3 (3.5) CB/A25-CG2/V41, 3.5 |
33 |
| 43&[STG]V group | ||||||
| Immunoglobulin A1 protease | Haemo-philus influ-enzae | 3H09_A R = 1.75 Å |
CG2/I86-O/S288, 3.4 (2.6) O/I86-HOH1135, 2.9 HOH1135-CA/G289, 3.4 (2.5) HOH1135-CA/G97, 3.4 (2.5) |
N/Y308-O/S290, 2.8 O/Y308-N/G289, 2.8 O/Y308-CG2/V98, 3.7 (2.9) |
CG2/V101-O/S288, 3.6 (2.6) CD1/I86-CG1/V101, 3.5 |
38 |
| Viral serine proteases | ||||||
| [KR]P group | ||||||
| Sindbis virus capsid protein | Sindbis virus | 1SVP_A R = 2.00 Å |
N/H128-O/S215A, 2.9 O/H128-CA/G216, 3.3 (2.4) O/H128-CB/K138, 3.8 (3.3) |
N/V230-O/R217, 3.0 O/V230-N/G216, 2.9 O/V230-CG/P139, 4.4 (3.8) |
CA/G127-O/S215A, 3.3 (2.7) CG2/V142-O/S215A, 4.4 (3.4) CA/G127-CG1/V142, 3.7 |
40 |
| Viral cysteine proteases | ||||||
| [TA]N group | ||||||
| Nuclear inclusion protein A | Tobacco etch virus | 1LVM_A R = 1.80 Å Set III |
N/Y33-O/C151, 2.8 O/Y33-CA/G152, 3.3 (2.3) O/Y33-OG1/T43, 2.7 |
N/H167-O/S153, 2.9 O/H167-N/G152, 2.9 O/H167-ND2/N44, 3.2 N/G152-OD1/N44, 2.9 |
CB/L32-O/C151, 3.5 (2.5) CD2/L32-CD1/L47, 3.7 |
45 |
| [ΨC][PQ] group | ||||||
| Hepatitis A protease 3C | Human hepatitis A virus | 2HAL_A R = 1.35 Å |
N/N30-O/C172, 3.0 O/N30-CA/G173, 3.4 (2.3) O/N30-CG2/V41, 4.1 (3.0) |
N/H191-O/G174, 2.9 O/H191-N/G173, 2.9 O/H191-CG/P42, 4.1 (3.5) |
CB/M29-O/C172, 3.5 (2.4) CE/M29-CB/A45, 3.7 |
46 |
| 3Cl protease | Alpha-mesoni-virus 1 | 5LAC_B R = 1.94 Å |
N/R35-O/C153, 3.0 O/R35-CA/G154, 3.4 (2.3) O/R35-HOH537, 2.7 HOH537-CA/I45, 3.3 (2.4) |
N/H168-O/G155, 2.7 O/H168-N/G154, 2.9 O/H168-NE2/Q46, 2.9 |
CB/L34-O/C153, 3.6 (2.5) CG/L34-CD1/L49, 4.0 |
47 |
| 43&[VR]N group | ||||||
| 2A proteinase | Coxsac-kievirus A16 | 4MG3_A R = 1.80 Å Set III |
N/A O/G8-CA/G111, 3.6 (3.1) O/G8-HOH303, 2.6 HOH303-CG2/V18, 2.8 (2.2) |
N/V124-O/G112, 2.8 O/V124-N/G111, 3.0 O/V124-ND2/N19, 2.8 HOH303-OD1/N19, 2.7 N/G111-OD1/N19, 3.0 |
CD1/L22-O/C110, 3.9 (3.6) CD1/L22-HOH303, 3.8 (2.9) |
48 |
| Inactive proteases | ||||||
| Eukaryotic proteases | ||||||
| T[TG] group | ||||||
| Propheno-loxidase activating factor-II | Holo-trichia diom-phalia | 2B9L_A R = 2.00 Å |
N/G186-O/G353, 2.6 O/G186-CA/G354, 3.7 (3.0) O/G186-OG1/T197, 2.9 |
N/V374-O/S355, 2.9 O/V374-N/G354, 2.8 O/V374-CA/G198, 5.4 (4.5) O/V374-CE1/H200, 3.5 (2.4) |
CB/C185-O/G353, 3.1 (2.5) SG/C201-O/G353, 3.8 C185-C201 |
53 |
Sets “I-IV” refer to four subgroups of TN groups proteases with different orientation of Asn55T. The values within the parentheses indicate distances to hydrogen atoms.
Unlike the 43/213-NBCZone, which incorporates only two amino acids of the PROSITE TRYPSIN_HIS pattern, Ser54 and Ala55, the entire PROSITE TRYPSIN_HIS pattern is included into the neighboring 42/43 Base Catalytic Zone, which contains the pentapeptide Ser54-Ala55-Ala56-His57-Cys58 and the dipeptide Cys42-Gly43 (Fig. 2B). Cys58 and Cys42 of this zone are linked by a disulfide bond, which maintains the conformation of the polypeptide chain near the active site. In addition, these two cysteines have contacts with the main-chain oxygen of the catalytic nucleophile (Table 1). In the remaining part of this article, the 43/213 Nucleophile-Base Catalytic Zone together with the 42/43 Base Catalytic Zone will be joined together and called the Nucleophile-Base Catalytic Zone (NBCZone) of trypsin. It is important to note that the side chain of the catalytic nucleophile Ser195, the side chain of the catalytic base His57 and the entire catalytic acid Asp102 are not part the NBCZone of trypsin.
The structure of trypsin may be either in active form [10] or inactive (zymogen) form [12] prior to proteolytic activation. Another name for the enzyme – trypsinogen – corresponds to the inactive form. However, in other proteases, the inactive form of the protein tertiary structure can be either similar or different from the zymogenic form of trypsin. Thus, in order to simplify the writing of the text of the article, we will use the structural term “zymogen” to describe only the zymogenic inactive form of trypsin and other proteases, while we will use the term “zyme” to collectively describe the remaining structural forms of proteases, which will include both non-zymogenic inactive forms (zymeinact) and active forms (zymeact) of the enzymes. In trypsin (PDB ID: 4I8H), the non-zymogenic form of the tertiary structure of the protein corresponds to zymeact.
Structural comparison shows that the NBCZones of bovine trypsin (zymeact; PDB ID: 4I8H) and trypsinogen (zymogen; PDB ID: 1TGT) are the same (Table 1). Indeed, the main structural differences between trypsin and trypsinogen lie in the activation domain [13], while the eleven amino acids of trypsin NBCZone lie outside of it.
2.1.2. [ST]A group
In the SCOP database, (chymo)trypsin-like fold serine proteins are divided into 4 families: eukaryotic, prokaryotic, viral serine, and viral cysteine proteases [1]. Looking at the tertiary structures among the eukaryotic (chymo)trypsin-like serine fold proteases in the PDB, SCOP, CATH [14], and MEROPS [15] databases we found, besides trypsin, another 14 proteins that have both zyme (either zymeinact or zymeact) and zymogen forms of the three-dimensional structures, 49 proteases without the zymogen form and three with only the zymogen form, totaling to 67 tertiary structures (Tables 1, S1 and S2). In this work, for each protease, we will use both the original numbering of the amino acid sequence and the canonical numbering of the amino acid sequence of trypsin which is shown by the index “T”. The 67 eukaryotic structures have either serine or threonine at position 54T, and alanine at position 55T. Therefore, the set of these 67 proteins was named the “[ST]A group” (Table S1).
The importance of alanine at position 55T for the catalytic activity of chymotrypsin-like serine proteases has been established [16,17]. Using the human plasmin model as an example, it was shown that replacing the alanine residue with a threonine leads to the formation of an unusual hydrogen bond between this threonine and the catalytic histidine [16]. The peculiarity of this interaction is that the catalytic histidine now adopts an inactive conformation. In contrast, with the bovine protein C as an example, replacing this alanine with a hydrophobic valine does not cause major changes in the conformation of the catalytic histidine [17].
The conservation of amino acids observed by us at the positions 54T and 55T fully complies with the definition of the PROSITE TRYPSIN_HIS pattern. Glycines at position 196T and 197T are also invariant. The same can be said about 42T-58T disulfide bond with the exception of four proteins: C1r, C1s, MASP-2 and MASP-3 (Table S1). C1r/C1s and MASP-1/-2/-3 form a family of mosaic serine proteases with identical domain organization [18], functioning as supramolecular complexes [19]. In each of the five mosaic serine proteases, two loops structurally corresponding to trypsin loop A (34T-41T) and loop B (56T-64T) [20] contact each other during the formation of such complexes [[21], [22], [23]]. In trypsin, cysteine at position 42T is located at the carboxy-terminal end of loop A, and cysteine at position 58T is placed at the amino-terminal end of loop B. Perhaps because of the need to form the supramolecular complexes, disulfide bonds are missing in all but MASP-1. As the analysis of the MASP-2 structure (PDB ID: 3TVJ, [22]) shows, despite the absence of a disulfide bond, contacts between amino acids at positions 42T, 58T and the catalytic nucleophile are conserved (Table 1). This indicates the existence of the NBCZones in these four proteins. In MASP-1 (PDB ID: 3GOV, [24]), unlike the other family members, there is a Cys475(42T)-Cys491(58T) disulfide bond. In addition, this protease has a very long loop B (Ala488-Asp513), compared with the corresponding loops in other proteases of this family [24]. It is possible that the structural and functional features of loop B, together with loop A and the amino acids adjacent to them, determine the presence of the Cys475(42T)-Cys491(58T) disulfide bond in MASP-1.
2.1.3. TN group
Although the probability is very high for alanine to be at position 55T of eukaryotic (chymo)trypsin-like serine fold proteases, the residue is not absolutely conserved. Asparagine is observed at position 55T in Homo sapiens HtrA and Arabidopsis thaliana Do-like serine proteases (8 proteases, Tables 1 and S1), joined together into HtrA family [25,26]. In these enzymes the 54T position is occupied by only threonine, and thus, this group of HtrA family proteases was named the “TN group”. Because in the TN group the 55T position is occupied by an asparagine and not by an alanine like in the [ST]A group, the 55T-213T interaction (Fig. 2A) is modified such that instead of the CB atom of alanine, the main chain oxygen of amino acid at position 213T interacts with the ND2 atom of asparagine (e.g. ND2/Asn196–O/Asn321 in human mitochondrial serine protease HtrA2) (PDB ID: 5M3N, [27]; Table 1, Fig. 2C).
The catalytic triad of HtrA2 is found in a catalytically incompetent conformation [27]. The distance between the ND1/His198 (base) and OG/Ser306 (nucleophile) atoms is 6.2 Å. There are also no hydrogen bonds between His198 (catalytic base) and Asp228 (catalytic acid). Asn196 is, however, directly involved in the mutual separation of the base and the nucleophile from each other, forming two hydrogen bonds: ND2/Asn196-OG/Ser306 = 2.8 Å and OD1/Asn196-N/His198 = 3.1 Å (Fig. 2C). In particular, the tripeptide Asn196-Ala197-His198 forms an Asx-turn [28].
The more complicated networks of interactions within the NBCZones are observed in the case of human HtrA1 (PDB ID: 3TJN, [29]). The PDB file 3TJN, 3 Å resolution, contains coordinates for the A, B, and D chains; chain D is relatively poorly ordered overall. Chain A has an incompetent conformation of active site: ND1/His220-OG/Ser328 = 7.6 Å, that is essentially the same as seen for the HtrA2 structure. The active site residues of chain B: Ser328 (nucleophile), His220 (base) and Asp250 (acid) are properly positioned for catalytic activity (Fig. 2D, Table 1). As in HtrA2, the contacts O/Asn343-ND2/Asn218 are present in chains A and B of HtrA1; however, atom OD1/Asn218, instead of forming the Asx-turn OD1/Asn218-N/His220 as in HtrA2, is now involved in the contacts with a catalytic acid Asp250.
There is no essential difference between the NBCZones of HtrA3 and HtrA2 that belong to set I (Table S1, column 4). Further analysis of prokaryotic and viral proteases will show that all these proteases of set I have an incompetent conformation of catalytic histidine, and as the result, the tripeptide Asn55T-Xaa56T-His57T forms an Asx-turn. Thus, the analysis of the structures of HtrA1, HtrA2, and HtrA3 proteases demonstrates that Asn55T is characterized by large conformational differences between the incompetent and competent conformations for substrate binding active site regions.
Although the HtrA and Do-like proteases are within the same TN group, there are some structural differences in their NBCZones. For example, in the proteases HtrA1 and Do-like 5 (set II, Table S1) atom ND2/Asn55T forms the hydrogen bond in the NBCZone and atom OD1/Asn55T interacts with the catalytic acid Asp102T (Fig. 2D). However, in protease Do-like 1, atom OD1/Asn171 forms the hydrogen bond in the NBCZone and atom ND2/Asn171 plays a key role in the interactions with the catalytic acid Asp102T (PDB ID: 3QO6 [30], Fig. 2E, Table 1). Similar NBCZones occur in the proteases Do-like 2, Do-like 8 and Do-like 9: set III (Table S1). Further analysis of prokaryotic and viral proteases also show that all proteases of set II have the catalytically competent or incompetent conformation of catalytic histidine, but all proteases of set III have only the catalytically competent conformation of the catalytic histidine (Table S1, column 4). The structural diversity of the side chain of Asn55T agrees well with the conformational changes in the active sites of the HtrA family proteases [[25], [26], [27],29].
The HtrA family proteases are multidomain proteins, which besides a proteolytic domain also contain at least one C-terminal PDZ domain [25,26]. The functional unit of the HtrA family proteases ranges from a trimer to a dodecamer. Loops A and B play important structural and regulatory roles in the HtrA multimer complexes [25,31]. It is possible that for the implementation of these functions, loops A and B require a certain mobility. The observed presence of asparagine at position 55T, the lack of the disulfide bond Cys42T-Cys58T and the substitution of the cysteine at position 42T for the small amino acid glycine (Table S1) contribute to this requirement.
2.2. Prokaryotic serine (chymo)trypsin-like fold proteases
2.2.1. TA and TN groups
As in the case of eukaryotic (chymo)trypsin-like serine fold proteases, most prokaryotic proteases fall into the TA and TN groups (Tables 1 and S1). Only now the TA and TN groups are almost equal in terms of the number of the available tertiary structures that have fallen into them. The presence (TA group) or absence (TN group) of a disulfide bond Cys42T-Cys58T is also similar in prokaryotic and eukaryotic proteases. Furthermore, when the disulfide bond is missing in the TN group, then there is a small amino acid – either glycine or alanine – at position 42T of the prokaryotic proteases, as is the case with the eukaryotic proteases.
Most prokaryotic proteases of the TN group fall into set III. In Table S1, there are also examples of tertiary structures included in set I and set II, and even one structure: Staphylococcus aureus serine protease SplE, PDB ID: 5MM8 [32], in which Asn37 has two conformations corresponding to sets I and III.
Prokaryotic proteases of the TN group, without the Cys42T-Cys58T disulfide bond, belong mostly to three subgroups: the HtrA family, Spl proteases, and exfoliative toxins (Table S1). Fig. 2F shows structural details of the NBCZone (TN group) for serine protease Spl from Staphylococcus aureus, PDB ID: 2AS9 [33]. The Spl protease is not in an active form due to a rotation of the side chain of catalytic His40, and its NBCZone is not similar to other NBCZones formed with the participation of asparagine at position 55T. The tripeptide Asn38-Lys39-His40 forms a modified Asx-turn. Therefore, the structure of this protease belongs to a separate set IV.
The structural significance of the amino acid at position 55T for the stabilization of the catalytic triad of prokaryotic (chymo)trypsin-like serine fold proteases is analyzed in detail in several publications [34,35]. Using V8 protease and glutamyl-endopeptidase as examples, it was assumed that accommodation of an asparagine instead of alanine in position 55T is impossible without some rearrangement of interactions between the catalytic histidine and the catalytic acid. In particular, weakening of the interactions of the catalytic acid D102T with the amides of residues 56T and 57T were observed. In addition, it was predicted that in glutamate-specific endopeptidase from Bacillus subtilis, the replacement of the conserved Gly193T (Fig. 1) with a cysteine could lead to the formation of a new disulfide bond that stabilizes the conformation of the oxyanion hole.
2.2.2. 43&[STG][AV] group
Another noticeable difference between the NBCZones of eukaryotic and prokaryotic proteases is the appearance of the 43&[STG]V group in prokaryotic proteins (Tables 1 and S1). These proteases belong to the SPATE family [36,37]. The presence of number 43 in the group name is due to the fact that the six proteins of this group have the amino acid main chain conformation at position 43T different compared to the amino acid main chain conformation at position 43T of all proteins analyzed so far. As an example, in the Haemophilus influenzae immunoglobulin A1 protease (Fig. 3A, PDB ID: 3H09, [38]), the contact with O/Ser288 is formed not by the N/Ile86 atom, but by CG2/Ile86. A change in the course of the polypeptide chain at position 43T leads to a complete impossibility of forming the Cys42T-Cys58T disulfide bond. Nevertheless, the contact of atom CD1/Ile86(43T) with atom CG1/Val101(58T) indicates the presence of the NBCZone (Table 1).
Fig. 3.
(A) shows the 43/213-NBCZone of the “43&[STG]V Group” of (chymo)trypsin-like serine fold proteases, which is not found in eukaryotes (example of immunoglobulin A1 protease; see Tables 1 and S1). In these prokaryotic and viral proteins, the change in the course of the polypeptide chain at the position 43T leads to the replacement of the “key” canonical NH...O hydrogen bond (N/Gly43-O/cat.nucleophile in Fig. 2A) with a weak CH...O hydrogen bond (CG2/Ile86-O/cat.nucleophile in panel A), and the impossibility of forming a Cys42T-Cys58T disulfide bridge within the 42/43 Base Catalytic Zone. In (B), the viral “[KR]P group” is shown, where at the position 54T, a lysine or an arginine is found instead of a threonine or a serine. In (C) and (D), extension of the NBCZone in trypsin and trypsinogen, respectively, is shown due to either inclusion of two conserved structural water molecules at the positions X and Y of trypsin (as in C), or a side-chain oxygen atom (OD1 atom of Asp194 in trypsinogen) and one water molecule at the same spatial positions X and Y (as in D). (E) shows the extension of the NBCZone in the “TN Group” of (chymo)trypsin-like serine fold proteases (example of the chloroplastic protease Do-like 2; PDB ID: 5ILB), where the OG atom of Ser43T is located at position X instead of the structural water molecule found in the NBCZone extension of trypsin. (F) shows the NBCZone extension in the five “inactive” proteases (example of Heparin binding protein; PDB ID: 1A7S; see Tables S1 and S2), where instead of a glycine at position 197T there is a serine, threonine, or aspartate (Thr177 in F), whose side-chain OG1 atom substitutes the HOH Y water molecule at the NBCZone extension.
It is possible that the formation of a truncated loop A (34T-41T) is directly related to the specificity of the catalytic activity of SPATE family proteases. Instead, a conservative tyrosine (Tyr239 in immunoglobulin A1 protease) of the unique functional loop D (143T-149T) is located in place of the loop A bend of the polypeptide chain of these proteases [38].
Another structural feature observed in the 43&[STG]V group is the presence of valine at position 55T (Val98 in immunoglobulin A1 protease). Interesting amino acid variability is also observed at position 54T, with serine or threonine found in five proteins and glycine in one protein. In the immunoglobulin A1 protease that has glycine at position 54T, instead of the OG atom of the side chain of Ser54T, there is a water molecule HOH1135, which completely replaces the OG atom in the construction of the NBCZone (Fig. 3A). The possible structural and functional role of the existence of a water molecule near the amino acid at position 54T is discussed below using the viral cysteine proteases as examples.
2.3. Viral serine (chymo)trypsin-like fold proteases
2.3.1. TA, [ST]Ψ and [KR]P groups
SCOP divides viral (chymo)trypsin-like fold proteases into two superfamilies: serine and cysteine proteases [1]. None of the viral proteases in Table S1 have a Cys42T-Cys58T disulfide bond. In turn, viral serine (chymo)trypsin-like fold proteases are divided into three groups: TA, [ST]Ψ and [KR]P (Ψ – amino acids with large aliphatic side chains: V, I, L; [39]) (Tables 1 and S1). In all proteases that fall into these three groups, the amino acids at position 55T do not show new structural features in the formation of the NBCZone compared to Val98 of the immunoglobulin A1 protease (Table 1 and Fig. 3B). Instead of cysteine, glycine is located at position 42T, with the exception of phenylalanine in the HCV NS3 protease (Table S1). Since the [KR]P group has lysine or arginine instead of threonine or serine at position 54T, as pointed out above, only the corresponding representative structure of this group (Sindbis virus capsid protein, PDB ID: 1SVP, [40]) is presented in Table 1 and Fig. 3B. The inclusion of lysine and proline at positions 54T and 55T, respectively, does not cause any major steric problems in the construction of the NBCZone.
Substitutions of Thr54 and Val55 in the HCV NS3 protease/helicase ([ST]Ψ group) affect the level of drug resistance of this virus [[41], [42], [43], [44]]. The Thr54Ala mutation changes the type of the hydrogen bond with Leu44(43T), as a result of which the conformations of amino acids Leu44(43T) and Phe43(42T) may change, and thus the protease/helicase binding to the inhibitor is weakened. The Thr54/Ser mutation was associated with medium level drug resistance. The Val55Ala, Arg155(214T)Lys/Thr and Ala156(215T)Thr resistant variants have been also identified.
2.4. Viral cysteine (chymo)trypsin-like fold proteases
Viral cysteine (chymo)trypsin-like fold proteases are divided into four groups: [TA]N, T[TSA], [ΨC][PQ], and 43&[VR]N (Tables 1 and S1, [[45], [46], [47], [48]]). Perhaps, to accommodate the cysteine nucleophile, the active site of the viral cysteine (chymo)trypsin-like fold proteases is larger than that of the serine proteases [49]. This structural result is consistent with the observation that the contact between the amino acid at position 58T and the catalytic cysteine has disappeared (Table 1).
The NBCZone of the nuclear inclusion protein A from Tobacco vein mottling virus ([TA]N group) demonstrates a previously found feature in immunoglobulin A1 protease (Table S1, 43&[STG][AV] group): the amino acid Ala43 at position 54T for contact with amino acid Phe33 at position 43T uses a water molecule HOH246 as an intermediary. Perhaps this presence of a water molecule is caused by the existence of large hydrophobic Leu32 and Leu47 instead of cysteines at the positions 42T and 58T (Table S1). The largest number of these viral protease structures belong to the [ΨC][PQ] group.
There are two more noticeable differences between the NBCZones of serine and cysteine proteases. The first difference is the presence of glutamate instead of aspartate (catalytic acid) at position 102T of the 3C and 3C-like proteases (T[TSA] and [ΨC][PQ] groups). The second difference is that half of the proteases from the [ΨC][PQ] group do not have a catalytic acid at position 102T at all; i.e., they have a catalytic dyad in the active site instead of the catalytic triad. However, all these dyad proteases, instead of the missing catalytic acid, have a water molecule (Table S1, column 10), which forms several hydrogen bonds with the residues surrounding it, including the catalytic histidine [50].
With one exception, the group of proteases with a catalytic dyad has cysteine and proline at positions 54T and 55T, respectively (Table S1). The alignment of the primary structures of such proteases, given in the work of Kanitz et al. [47], shows that the number of dyad proteases with the dipeptide Ile-Gln at positions 54T and 55T is approximately equal to the number of dyad proteases with the dipeptide Cys-Pro [47]. Table 1 lists the structural parameters of the 3Cl protease from Alphamesonivirus 1 with the participation of the dipeptide Ile-Gln in the active site (PDB ID: 5LAC, [47]). Gln46, located in position 55T, is the largest amino acid of all structurally similar amino acids listed in Table S1. Therefore, despite the large size of Ile45 located at position 54T, Ile45 required a water molecule HOH537 to contact with Arg35 (position 43T) (Table 1). The existence of water HOH537 correlates with the presence of two leucine residues at positions 42T and 58T, as it was shown earlier for the nuclear inclusion protein A. It is possible that the presence of a water molecule (Table S1, column 7) in the 2A proteinase from the 43&[VR]N group is also associated with the existence of leucine at position 58T and structural specificity of 43&[VR]N group. The connection between the structural specificity of a group and the presence of a water molecule is also supported by the existence of a similar water molecule in immunoglobulin A1 protease (43&[STG][AV] group), which was described earlier. However, the structural reasons for the presence of the water near the residues at positions 54T and 43T in these six proteins are not entirely clear. It can only be assumed that the replacement of a direct contact of amino acids at positions 54T and 43T with water-mediated contacts demonstrates a weakening of the interaction of the nucleophilic loop with a β-sheet containing the catalytic base and the catalytic acid. This weakening is somehow related to the functional characteristics of these six proteins. In conclusion, we note that the presence of water molecules in a similar place in (chymo)trypsin-like serine fold proteases has not been previously established.
In the [TA]N and 43&[VR]N groups of cysteine proteases, rotation of the side chain of asparagine at position 55T is observed, as has already been noted for the representative of the TN group of eukaryotic and prokaryotic serine proteases.
Unlike the HCV NS3 protease/helicase, the Thr27(54T)Ala/Ser/Val mutations of 3C-like norovirus protease (T[TSA] group) do not affect the catalytic activity of this protease. Rather, it was suggested that Thr27 is involved in stabilizing the conformation [51]. Mutation Leu19(58T)Ser in the HRV2 2A proteinase (43&[VR]N group) leads to a similar result [52]. However, the Asn16(55T)Ala mutation inhibits proteolytic activity completely. Mutations of residues around the nucleophilic cysteine: Pro103(192T)Gly and Asp105(194T)Thr/Asn, also impair the proteinase activity.
2.5. Inactive (chymo)trypsin-like fold proteases
In addition to 161 protease structures, 8 proteins with the (chymo)trypsin-like fold were found that are not proteases (Table S1). These 8 proteins do not have a catalytic nucleophile, and five of them do not have a catalytic base either. Seven structures belong to the eukaryotic proteins (TA and T[TG] groups) and one structure (TT group) is a prokaryotic protein. Six out of the seven eukaryotic proteins have a Cys42T-Cys58T disulfide bond.
The active site of Holotrichia diomphalia prophenoloxidase activating factor-II demonstrates the zymogenic conformation (PDB ID: 2B9L, [53]). In addition, due to the lack of a side chain on glycine 55T (Gly198), the contact O/Val374-CA/Gly198 = 5.4 (4.5) Å is weak (Table 1). However, in this protein, the catalytic serine 195T is also replaced by glycine (Gly353). Perhaps for this reason, the CE1 atom of the catalytic histidine 57T (His200) forms the contact O/Val374-CE1/His200 = 3.5 (2.4) Å. According to the canonical rule of the Derewenda et al. [54], the CE1 atom of a catalytic histidine should form a weak hydrogen bond with the main-chain oxygen of Ala375 (O/Ala375-CE1/His200, 3.1 (3.0) Å), the residue following Val374 in the amino acid sequence. Therefore, for the main-chain oxygen of the amino acid at position 213T (Val374), a new structure-catalytic role is discovered as a fixator of a catalytic histidine. Despite the loss of protease activity, all 8 proteins have a characteristic NBCZone.
2.6. NBCZones based conclusions
Summarizing the results on a structural comparison of the NBCZones for 169 (chymo)trypsin-like fold proteases, we can conclude:
-
1)
For the majority of eukaryotic and prokaryotic proteins, the presence of a Cys42T-Cys58T disulfide bond and the location of alanine at position 55T are interrelated. In those cases where the analyzed proteins lack the Cys42T-Cys58T disulfide bond, position 42T is predominantly occupied by glycine, position 55T by asparagine, and position 58T by valine.
-
2)
Viral proteases do not have the Cys42T-Cys58T disulfide bond. In these proteases, position 42T is predominantly glycine, phenylalanine, or leucine (cysteine proteases), position 55T can be occupied by 9 different amino acids (predominantly hydrophobic residues or proline in serine proteases and hydrophilic uncharged residues or proline in cysteine proteases), and the amino acid at position 58T is either valine or leucine (cysteine proteases).
It was shown that the presence or absence of the Cys42T-Cys58T disulfide bond affects the overall thermal stability of trypsin [55]. Moreover, mutations Cys42Ala, Cys58Ala/Val and Ser195Thr convert the serine protease trypsin to a functional threonine protease.
2.7. Eukaryotic serine (chymo)trypsin-like fold proteases
2.7.1. Extension of trypsin NBCZone
As aforementioned, NBCZones of trypsin and trypsinogen do not differ from each other. An additional visual inspection of the trypsin tertiary structure showed that near the nucleophile-oxyanion loop Gly193-Gly197 there are two water molecules HOH1015 (position X) and HOH1003 (position Y) that form two hydrogen bonds with the main-chain oxygen atoms of Gly193 and Asp194, and two weak hydrogen bonds with the Cα-atoms of Asp194 and Gly197 (Fig. 3C, Table 2 ). Water molecules HOH1015 and HOH1003 are located at a distance comparable to the distance of a hydrogen bond. This conformation of the nucleophile-oxyanion loop corresponds to the zyme type trypsin conformation. The contacts HOH1015-O/Gly193 and HOH1015-CA/Asp194 dispose the N/Asp194 atom to the position required for the formation of an important β-turn contact (O/Cys191-N/Asp194; not shown). The contacts HOH1003-O/Asp194 and HOH1003-CA/Gly197 are important for the correct orientation of the nucleophile Ser195 and its N(OxyII) oxyanion atom. Consequently, HOH1015 and HOH1003 do not affect the position of the N(OxyI) oxyanion atom. In trypsin, the nitrogen atom N/Gly193 is located at the position (type II β-turn) that ensures the activity of this protease [10]. However, in other proteases with the same zyme pattern contacts, as shown in Fig. 3C, nitrogen N(OxyI) may not be appropriate for the activity of the protein (type I β-turn; see below). Therefore, the zyme structural organization is necessary, but it is not sufficient to conclude whether the particular tertiary structure corresponds to the active or inactive state of the protease.
Table 2.
Geometrical parameters of interactions within the positions X and Y of active sites in representative structures of (chymo)trypsin-like serine fold proteases.
| Protein | Organism | PDB ID resolution | Nuc195 Xaa43 Xaa197 |
Hydrogen bonds of water molecule or amino acid at position X | Hydrogen bonds of water molecule or amino acid at position Y | Ref. |
|---|---|---|---|---|---|---|
| Eukaryotic proteases | ||||||
| [ST]A group | ||||||
| Trypsin | Bos taurus | 4I8H_A R = 0.75 Å |
Ser195 Gly43 Gly197 |
HOH1015-O/G193, 2.8 HOH1015-CA/D194, 3.7 (2.7) HOH1015-HOH1003, 2.8 |
HOH1003-O/D194, 2.9 HOH1003-CA/G197, 3.3 (2.2) |
10 |
| Trypsinogen | Bos taurus | 1TGT_A R = 1.70 Å |
Ser195 Gly43 Gly197 |
OD1/D194-HOH701, 2.9 | HOH701-CB/D194, 3.4 (2.5) HOH701-O/D194, 3.7 HOH701-CA/G197, 3.1 (2.4) |
12 |
| Mannan-binding lectin serine protease 2 | Homo sapiens | 3TVJ_B R = 1.28 Å |
Ser633 Ala469 G635 |
HOH6-O/G631, 2.8 HOH6-CA/D632, 3.8 (2.8) HOH6-CB/A469, 3.5 (2.6) HOH6-HOH10, 2.9 |
HOH10-O/D632, 2.9 HOH10-CA/G635, 3.6 (2.7) HOH10-CB/A469, 3.2 (2.7) |
22 |
| Kallikrein-4 | Homo sapiens | 4K8Y_A R = 1.00 Å |
S195 S43 G197 |
HOH549-O/G193, 2.8 HOH549-CA/D194, 3.6 (2.6) HOH549-OG/S43, 2.7 HOH549-HOH301, 2.8 |
HOH301-O/D194, 2.8 HOH301-CA/197, 3.4 (2.4) HOH301-CB/S43, 3.4 (2.6) |
56 |
| Complement factor C2 | Homo sapiens | 2ODP_A R = 1.90 Å |
Ser659 Arg473 Gly661 |
NH1/R473-O/G657, 3.1 CG/R473-O/G657, 3.6 (2.5) CG/R473-CA/E658, 4.0 CB/R473-HOH960, 3.7 (3.1) CG/R473-HOH960, 3.7 (3.0) |
HOH960-O/E658, 2.8 HOH960-CA/G661, 3.4 (2.4) |
57 |
| TN group | ||||||
| Protease Do-like 2, chloroplastic | Arabidopsis thaliana | 5ILB_A R = 1.85 Å |
Ser268 Ser145 Gly270 |
OG/S145-O/G266, 2.7 OG/S145-CA/N267, 4.0 (3.3) OG/S145-HOH798, 4.6 |
HOH798-O/N267, 2.8 HOH798-CA/G270, 3.7 (2.8) |
60 |
| Prokaryotic proteases | ||||||
| TA group | ||||||
| Trypsin | Saccharopolyspora erythraea | 5KWM_A R = 0.78 Å |
Ser179 Gly28 Gly181 |
HOH490-O/G177, 2.9 HOH490-CA/D178, 3.7 (2.8) HOH490-HOH480, 2.8 |
HOH480-O/D194, 2.9 HOH480-CA/G197, 3.3 (2.4) |
62 |
| VESB protease | Vibrio cholerae | 4LK4_A R = 2.40 Å |
S221A Gly64 Gly223 |
OD1/D194-HOH455, 3.0 | HOH455-O/D220, 3.3 HOH455-CA/G223, 4.6 (3.6) |
64 |
| 43&[STG][AV] group | ||||||
| Immunoglobulin A1 protease | Haemophilus influenzae | 3H09_A R = 1.75 Å |
S288 I86 S290 |
HOH1038-O/G286, 2.9 HOH1038-CA/D287, 3.6 (2.6) HOH1038-CG2/I86, 3.6 (3.1) HOH1038-CB/S290, 3.7 (2.7) |
OG/S290-O/D287, 2.6 | 38 |
| Viral serine proteases | ||||||
| TA group | ||||||
| Putative serine protease | Human astrovirus-1 | 2W5E_A R = 2.00 Å |
S551 T448 A553 |
HOH2022-O/G549, 3.2 HOH2022-CA/M550, 3.6 (2.9) HOH2022-OG1/T448, 2.7 HOH2022-CB/A553, 5.5 (4.5) |
CB/A553-O/M550, 3.1 (2.7) CB/A553-CB/T448, 4.5 |
69 |
| Viral cysteine proteases | ||||||
| [ΨC][PQ] group | ||||||
| EV71 3C protease | Enterovirus A71 | 3R0F_A R = 1.31 Å |
C147 T26 G149 |
HOH186-O/G145, 2.8 HOH186-CA/Q146, 3.3 (2.7) |
No HOH | 66 |
| Inactive proteases | ||||||
| Eukaryotic proteases | ||||||
| TA group | ||||||
| Heparin binding protein | Homo sapiens | 1A7S_A R = 1.12 Å |
G175 Gly27 Thr177 |
HOH451-O/G173, 2.9 HOH451-CA/D174, 4.0 (3.0) HOH451-OG1/T177, 4.5 HOH451-CB/T177, 4.6 (3.8) |
CB/T177-O/D174, 3.3 (2.7) CG2/T177-O/D174, 3.2 (2.5) |
75 |
For Asp194, the amino acid of the neighboring nucleophile, significant zyme-zymogen conformational changes are observed [13]. The result of these changes is that the side-chain oxygen OD1 of Asp194 of trypsinogen occupies the position of water molecule HOH1015 (position X) that is found in the structure of trypsin (Fig. 3D, Table 2). Atom OD1/Asp194 has no contact with the main chain oxygen of Gly193. Tetrapeptide Cys191-Asp194 no longer forms a β-turn conformation in the trypsinogen structure (not shown). Position Y of the trypsinogen structure is still occupied by a water molecule HOH701. Contacts of HOH701 with O/Asp194 and CA/Gly197 atoms are conserved, but there is also an additional contact with CB/Asp194 atom.
Due to the catalytic importance of the described structural differences between trypsin and trypsinogen, it is necessary to extend the NBCZone of proteases by including the atomic contents of positions X and Y.
2.7.2. [ST]A group
In 58 eukaryotic (chymo)trypsin-like serine fold proteases of the [ST]A group there are two water molecules that are structurally similar to the waters HOH1015 (position X) and HOH1003 (position Y) in trypsin (Table S2). Once again, we emphasize here that the presence in these 58 proteins of the identical zyme pattern contacts does not automatically mean that their full catalytic center is in the active configuration, as in trypsin. The structural zymogen subgroup consists of 18 proteases (Tables 2 and S2).
Glycine at position 193T and aspartic acid at position 194T are absolutely conserved in these eukaryotic (chymo)trypsin-like serine fold proteases, and glycine is the most commonly found amino acid at positions 43T and 197T. However, of eleven proteases (Tables 2 and S2), five have alanine, four have serine, one has arginine, and one methionine instead of glycine at position 43T [56,57]. In addition, we found one example of glycine replaced at position 197T by another amino acid, namely serine, in granzyme A (PDB ID: 1ORF, [58]), and alanine is found at position 43T instead of the conserved glycine. However, this does not affect the presence of a water molecule at position X. Water is also present at position X in the remaining four proteases, all of which have alanine at position 43T. In the case of serine at position 43T, a water molecule may be present or absent at position X. In the structure of granzyme A, there is no water molecule at position Y. Therefore, in 6 out of 11 proteases, the amino acid substitutions at position 43T do not change the zyme or zymogen (2 proteases) contact diagrams, which are shown in Fig. 3C and D, respectively.
At position 43T of eukaryotic proteases, one can find not only small amino acids, such as glycine, alanine, or serine, but also large residues, e.g., arginine or methionine. These large amino acids at position 43T are seen in complement factors C2 and B (PDB IDs: 2ODP and 1RRK, respectively [57,59]); in these cases a pair of carbon atoms of the side chains at position 43T are located at position X (Tables 2 and S2).
2.7.3. TN group
As we showed above, although the probability is very high for the existence of a glycine residue at position 43T of eukaryotic (chymo)trypsin-like serine fold proteases, this position is not absolutely conserved. The HtrA and Do-like families of Homo sapiens and Arabidopsis thaliana serine proteases also have a serine at position 43T (Tables 2 and S2) that leads to structural changes whereby the OG atom of Ser43T is locates to position X instead of a water molecule. The example of the chloroplastic protease Do-like 2 (PDB ID: 5ILB, [60]) shows the zyme contacts near the nucleophile-oxyanion loop (Fig. 3E).
While analyzing the tertiary structures of the eukaryotic (chymo)trypsin-like serine fold proteases, we never encountered an enzyme in which, as a result of activation, the side chain atom of the residue at position 194T would be located in position X instead of the side chain atom of the residue at position 43T. In particular, at position 194T of TN group proteases there is asparagine instead of aspartate found at this position in the [ST]A group. Given the conservative changes in amino acids at positions 43T, 55T, and 194T and the absence of the disulfide bond 42T-58T, it seems that proteases of the TN group do not have the ability to undergo a structural transition of the zymogen-zyme type. It is possible that this rule applies also to the complement factors C2 and B. This assumption is fully supported by the literature [29,57,59,61].
2.8. Prokaryotic serine (chymo)trypsin-like fold proteases
2.8.1. TA and TN groups
Only two examples of the NBCZone extension in prokaryotic TA group proteases with the zyme (chymo)trypsin-like form and two examples of the NBCZone extension with the zymogen form were found in the PDB: trypsins from Saccharopolyspora erythraea (PDB ID: 5KWM, [62]) and Streptomyces griseus (PDB ID: 1SGT, [63]), VESB and VesC proteases from Vibrio cholerae (PDB ID: 4LK4, [64] and PDB ID: 6BQM, [65]) (Tables 2 and S2). In terms of the organization of (chymo)trypsin-like zyme and zymogen forms, eukaryotic and prokaryotic NBCZone extensions of these four proteases are not different. Therefore, in the eukaryotic TA group proteases, the (chymo)trypsin-like form of NBCZone extension is dominant but in the prokaryotic proteases of this group it is auxiliary.
Most prokaryotic TA group proteases have the amino acid serine or threonine at position 43T, and possess glycine at position 197T (Table S1). In the corresponding TN group, all proteases have serine or threonine at position 43T, and glycine or serine at position 197T. As a result, in the cases where glycine is not located at position 43T and 197T, there are no water molecules at positions X and Y, but atoms of the side chains of serine or threonine are located there instead.
Among the prokaryotic TN group proteases, there is one exception from the viewpoint of building a zyme NBCZone extension pattern. With the serine protease SplE (Table S1, PDB ID: 5MM8, [32]) the dipeptide 193T-194T has a different conformation compared to the conformation of the corresponding dipeptide in the remaining proteases. As a result, the contact of the side-chain atom of the amino acid at position 43T with the main-chain oxygen of amino acid at position 193T is absent (Thr25 and Gly153, respectively, in the serine protease SplE). Earlier, we stated that, apparently, only members of eukaryotic and prokaryotic [ST]A groups can demonstrate two alternative conformations of the NBCZone extension pattern: zyme and zymogen. The NBCZone extension pattern of SplE shows that, sometimes, neither zyme nor zymogen form is possible. Instead a third form is formed, which is an atypical variant. Therefore, the letter A (Atypical) is added to the number of the structures of this protease and to the numbers of all similar structures in Table S1. This structural feature of the prokaryotic SplE protein is discussed in detail in the work [32].
2.8.2. 43&[STG][AV] group
Six (chymo)trypsin-like fold structures belonging to the 43&[STG][AV] group have five different amino acids, other than glycine, at position 43T (Tables 2 and S2). However, all of these proteases have a water molecule at position X. This is due to the removal of the side chain of amino acid 43T from position X as a result of a break in the course of the polypeptide chain in this place. Currently there are no data on the existence of the activation structure changes near the catalytic site in SPATE family proteases. Their N-terminal amino acid often forms a hydrogen bond with aspartate, an amino acid preceding the catalytic nucleophile [38].
2.9. Viral serine/cysteine (chymo)trypsin-like fold proteases
As aforementioned, a significant number of prokaryotic serine (chymo)trypsin-like fold proteases have amino acids, other than glycine, at positions 43T and 197T (Table S1, 22 structures), and only one such protease was observed among eukaryotic proteases (granzyme A). Analysis of the tertiary structures of viral proteases possessing such a fold showed that there are 37 such structures. With the exception of two proteases from the 43&[VR]N group, all viral proteases lack a glycine at position 43T (Tables 1, S1 and S2). As a result, there is no water molecule in position X, except among the four viral cysteine 3C proteases from the [ΨC][PQ] group (Tables 2 and S2) [66].
Besides a greater number of such viral structures, there is another difference between prokaryotic and viral proteases. The prokaryotic proteases show conservation of the amino acids occupying positions 43T (Thr) and 197T (Ser), while the viral proteases demonstrate a sequence variability at these positions, since 14 and 8 different amino acids are located at the positions 43T and 197T, respectively (Table S1). Mutations Asn28(43T) and Ser147(197T) to alanine modulate the dimerization (active form of enzyme) and completely inactivate the main viral 3C-like proteinase from human SARS coronavirus [67,68]. Interestingly, mutation Ser139(189T)/Ala had a slight effect on the activity and dimer stability of the proteinase. Mutation Ser144(194T)/Ala showed a two-fold decrease in catalytic efficiency compared to that of the wild type, but maintained a similar dimeric state. Ser139, Ser144 and Ser147 form a cluster of conserved serine residues near the catalytic nucleophile Cys145 of 3C-like proteinase.
2.10. Viral proteases with an atypical NBCZone
As with the members of the prokaryotic TN group, viral serine/cysteine (chymo)trypsin-like fold proteases also have several members with an atypical NBCZone (Tables 2 and S2). They are: a putative serine protease (PDB ID: 2W5E, [69]), whose active site residues are not in the typical (chymo)trypsin-like conformation, and 4 additional proteins, non-structural protein NSP4 (PDB IDs: 5Y4L and 3FAN, [70,71]), 3C-like protease (PDB IDs: 5E0G and 5E0J, [72]), 3C-like proteinase (PDB IDs: 5C5O and 2QCY, [50,73]), and infectious bronchitis virus (IBV) main protease (PDB IDs: 2Q6F and 2Q6D, [74]), all of which demonstrate both a typical and atypical conformation of the dipeptide, residues 193T-194T.
2.11. Inactive proteases
As aforementioned, in eukaryotic serine (chymo)trypsin-like fold proteases, only glycine (with one exception) is found at the 197T position of the nucleophile-oxyanion loop 193T-197T. We found five inactive proteins, in which this requirement is not the case (Tables S1 and S2). In all these five proteins, instead of glycine in position 197T there is serine, threonine, or aspartate amino acid. The presence of any residue other than glycine at position 197T leads to a structural change in which a water molecule is replaced by the atom(s) of the side chain of the amino acid of residue 197T. Fig. 3F shows an example of a zyme contact pattern for such a protein: Heparin binding protein (PDB ID: 1A7S, [75]) has Thr177 at a position equivalent to position 197T in trypsin (Table 2). The CB and CG2 atoms of the side chain of Thr177 have the same structural role in this protein as the water molecule HOH1003 in the structure of trypsin (see Fig. 3C). In position 43T there is Gly27. Therefore, a water molecule HOH451 is located at the X position. The other four eukaryotic proteins also have water molecule at position X because they have glycine at position 43T (Table S2).
2.12. Extension of NBCZone based conclusions
Summarizing the results of a structural comparison of the extension of the NBCZone for 169 (chymo)trypsin-like fold proteases, we can conclude:
-
1)
The vast majority of eukaryotic zyme type proteases have glycine residues at positions 43T and 197T. As a result, one water molecule is located at position X and another at Y.
-
2)
Eukaryotic zymogen type proteases place the side-chain oxygen of Asp194T at position X and water molecule at position Y.
-
3)
In almost all prokaryotic and viral proteases, the amino acid at position 43T is not glycine. This leads to the displacement of a water molecule at position X by the atom(s) of the side chain of the amino acid at position 43T.
-
4)
Due to the presence of a side chain in the amino acid at position 43T, prokaryotic and viral proteases do not have the ability to undergo a structural transition from the zymogen to zyme type.
CRediT authorship contribution statement
Alexander I. Denesyuk:Formal analysis, Methodology, Visualization, Writing - original draft, Writing - review & editing, Conceptualization.Mark S. Johnson: Formal analysis, Methodology, Writing - original draft. Outi M.H. Salo-Ahen: Conceptualization, Writing - review & editing. Vladimir N. Uversky: Formal analysis, Methodology, Writing - original draft, Writing - review & editing. Konstantin Denessiouk: Formal analysis, Methodology, Visualization, Investigation, Writing - original draft, Writing - review & editing, Conceptualization.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgements
This work is supported by a grant from the Sigrid Jusélius Foundation and Joe, Pentti, and Tor Borg Memorial Fund. We thank the Biocenter Finland Bioinformatics Network (Dr. Jukka Lehtonen) and CSC IT Center for Science for computational support for the project. The Structural Bioinformatics Laboratory is part of the Drug Development and Diagnostics Platform of Åbo Akademi University.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijbiomac.2020.03.025.
Appendix A. Supplementary data
Supplementary tables
References
- 1.Murzin A.G., Brenner S.E., Hubbard T., Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247(4):536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 2.Dimitriou P.S., Denesyuk A., Takahashi S., Yamashita S., Johnson M.S., Nakayama T., Denessiouk K. Alpha/beta-hydrolases: a unique structural motif coordinates catalytic acid residue in 40 protein fold families. Proteins. 2017;85(10):1845–1855. doi: 10.1002/prot.25338. [DOI] [PubMed] [Google Scholar]
- 3.Nardini M., Dijkstra B.W. Alpha/beta hydrolase fold enzymes: the family keeps growing. Curr. Opin. Struct. Biol. 1999;9(6):732–737. doi: 10.1016/s0959-440x(99)00037-8. [DOI] [PubMed] [Google Scholar]
- 4.Dimitriou P.S., Denesyuk A.I., Nakayama T., Johnson M.S., Denessiouk K. Distinctive structural motifs co-ordinate the catalytic nucleophile and the residues of the oxyanion hole in the alpha/beta-hydrolase fold enzymes. Protein Sci. 2019;28(2):344–364. doi: 10.1002/pro.3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Di Cera E. Serine proteases. IUBMB Life. 2009;61(5):510–515. doi: 10.1002/iub.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dodson G., Wlodawer A. Catalytic triads and their relatives. Trends Biochem. Sci. 1998;23(9):347–352. doi: 10.1016/s0968-0004(98)01254-7. [DOI] [PubMed] [Google Scholar]
- 7.Hedstrom L. Serine protease mechanism and specificity. Chem. Rev. 2002;102(12):4501–4524. doi: 10.1021/cr000033x. [DOI] [PubMed] [Google Scholar]
- 8.Sigrist C.J., de Castro E., Cerutti L., Cuche B.A., Hulo N., Bridge A., Bougueleret L., Xenarios I. New and continuing developments at PROSITE. Nucleic Acids Res. 2013;41(Database issue):D344–D347. doi: 10.1093/nar/gks1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The protein data bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liebschner D., Dauter M., Brzuszkiewicz A., Dauter Z. On the reproducibility of protein crystal structures: five atomic resolution structures of trypsin. Acta Crystallogr. D Biol. Crystallogr. 2013;69(Pt 8):1447–1462. doi: 10.1107/S0907444913009050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Menard R., Storer A.C. Oxyanion hole interactions in serine and cysteine proteases. Biol. Chem. Hoppe Seyler. 1992;373(7):393–400. doi: 10.1515/bchm3.1992.373.2.393. [DOI] [PubMed] [Google Scholar]
- 12.Walter J., Steigemann W., Singh T.P., Bartunik H., Bode W., Huber R. On the disordered activation domain in trypsinogen. Chemical labelling and low-temperature crystallography. Acta Crystallogr. 1982;B38:1462–1472. [Google Scholar]
- 13.Huber R., Bode W. Structural basis of the activation, action and inhibition of trypsin, H-S. Z. Physiol. Chem. 1979;360(4):489. [Google Scholar]
- 14.Dawson N.L., Lewis T.E., Das S., Lees J.G., Lee D., Ashford P., Orengo C.A., Sillitoe I. CATH: an expanded resource to predict protein function through structure and sequence. Nucleic Acids Res. 2017;45(D1):D289–D295. doi: 10.1093/nar/gkw1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rawlings N.D., Alan J., Thomas P.D., Huang X.D., Bateman A., Finn R.D. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2018;46(D1):D624–D632. doi: 10.1093/nar/gkx1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takeda-Shitaka M., Umeyama H. Elucidation of the cause for reduced activity of abnormal human plasmin containing an Ala55-Thr mutation: importance of highly conserved Ala55 in serine proteases. FEBS Lett. 1998;425(3):448–452. doi: 10.1016/s0014-5793(98)00280-4. [DOI] [PubMed] [Google Scholar]
- 17.Takeda-Shitaka M., Umeyama H. Effect of exceptional valine replacement for highly conserved alanine-55 on the catalytic site structure of chymotrypsin-like serine protease. Chem. Pharm. Bull.(Tokyo) 1998;46(9):1343–1348. doi: 10.1248/cpb.46.1343. [DOI] [PubMed] [Google Scholar]
- 18.Gal P., Barna L., Kocsis A., Zavodszky P. Serine proteases of the classical and lectin pathways: similarities and differences. Immunobiology. 2007;212(4–5):267–277. doi: 10.1016/j.imbio.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 19.Gal P., Dobo J., Zavodszky P., Sim R.B. Early complement proteases: C1r, C1s and MASPs. A structural insight into activation and functions. Mol. Immunol. 2009;46(14):2745–2752. doi: 10.1016/j.molimm.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 20.Perona J.J., Craik C.S. Evolutionary divergence of substrate specificity within the chymotrypsin-like serine protease fold. J. Biol. Chem. 1997;272(48):29987–29990. doi: 10.1074/jbc.272.48.29987. [DOI] [PubMed] [Google Scholar]
- 21.Kardos J., Harmat V., Pallo A., Barabas O., Szilagyi K., Graf L., Szabo G.N., Goto Y., Zavodszky P., Gal P. Revisiting the mechanism of the autoactivation of the complement protease C1r in the C1 complex: structure of the active catalytic region of C1r. Mol. Immunol. 2008;45(6):1752–1760. doi: 10.1016/j.molimm.2007.09.031. [DOI] [PubMed] [Google Scholar]
- 22.Heja D., Harmat V., Fodor K., Wilmanns M., Dobo J., Kekesi K.A., Zavodszky P., Gal P., Pal G. Monospecific inhibitors show that both mannan-binding lectin-associated serine protease-1 (MASP-1) and-2 are essential for lectin pathway activation and reveal structural plasticity of MASP-2. J. Biol. Chem. 2012;287(24):20290–20300. doi: 10.1074/jbc.M112.354332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaboriaud C., Gupta R.K., Martin L., Lacroix M., Serre L., Teillet F., Arlaud G.J., Rossi V., Thielens N.M. The serine protease domain of MASP-3: enzymatic properties and crystal structure in complex with ecotin. PLoS One. 2013;8(7) doi: 10.1371/journal.pone.0067962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dobo J., Harmat V., Beinrohr L., Sebestyen E., Zavodszky P., Gal P. MASP-1, a promiscuous complement protease: structure of its catalytic region reveals the basis of its broad specificity. J. Immunol. 2009;183(2):1207–1214. doi: 10.4049/jimmunol.0901141. [DOI] [PubMed] [Google Scholar]
- 25.Clausen T., Southan C., Ehrmann M. The HtrA family of proteases: implications for protein composition and cell fate. Mol. Cell. 2002;10(3):443–455. doi: 10.1016/s1097-2765(02)00658-5. [DOI] [PubMed] [Google Scholar]
- 26.Zurawa-Janicka D., Wenta T., Jarzab M., Skorko-Glonek J., Glaza P., Gieldon A., Ciarkowski J., Lipinska B. Structural insights into the activation mechanisms of human HtrA serine proteases. Arch. Biochem. Biophys. 2017;621:6–23. doi: 10.1016/j.abb.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 27.Merski M., Moreira C., Abreu R.M.V., Ramos M.J., Fernandes P.A., Martins L.M., Pereira P.J.B., Macedo-Ribeiro S. Molecular motion regulates the activity of the mitochondrial serine protease HtrA2. Cell Death Dis. 2017;8(10):e3119. doi: 10.1038/cddis.2017.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wan W.Y., Milner-White E.J. A natural grouping of motifs with an aspartate or asparagine residue forming two hydrogen bonds to residues ahead in sequence: their occurrence at alpha-helical N termini and in other situations. J. Mol. Biol. 1999;286(5):1633–1649. doi: 10.1006/jmbi.1999.2552. [DOI] [PubMed] [Google Scholar]
- 29.Eigenbrot C., Ultsch M., Lipari M.T., Moran P., Lin S.J., Ganesan R., Quan C., Tom J., Sandoval W., Campagne M.V., Kirchhofer D. Structural and functional analysis of HtrA1 and its subdomains. Structure. 2012;20(6):1040–1050. doi: 10.1016/j.str.2012.03.021. [DOI] [PubMed] [Google Scholar]
- 30.Kley J., Schmidt B., Boyanov B., Stolt-Bergner P.C., Kirk R., Ehrmann M., Knopf R.R., Naveh L., Adam Z., Clausen T. Structural adaptation of the plant protease Deg1 to repair photosystem II during light exposure. Nat. Struct. Mol. Biol. 2011;18(6):728–731. doi: 10.1038/nsmb.2055. [DOI] [PubMed] [Google Scholar]
- 31.Wenta T., Glaza P., Jarzab M., Zarzecka U., Zurawa-Janicka D., Lesner A., Skorko-Glonek J., Lipinska B. The role of the LB structural loop and its interactions with the PDZ domain of the human HtrA3 protease. BBA-Proteins Proteom. 2017;1865(9):1141–1151. doi: 10.1016/j.bbapap.2017.06.013. [DOI] [PubMed] [Google Scholar]
- 32.Stach N., Kalinska M., Zdzalik M., Kitel R., Karim A., Serwin K., Rut W., Larsen K., Jabaiah A., Firlej M., Wladyka B., Daugherty P., Stennicke H., Drag M., Potempa J., Dubin G. Unique substrate specificity of SplE serine protease from Staphylococcus aureus. Structure. 2018;26(4):572–579. doi: 10.1016/j.str.2018.02.008. [DOI] [PubMed] [Google Scholar]
- 33.Popowicz G.M., Dubin G., Stec-Niemczyk J., Czarny A., Dubin A., Potempa J., Holak T.A. Functional and structural characterization of Spl proteases from Staphylococcus aureus. J. Mol. Biol. 2006;358(1):270–279. doi: 10.1016/j.jmb.2006.01.098. [DOI] [PubMed] [Google Scholar]
- 34.Barbosa J.A.R.G., Saldanha J.W., Garratt R.C. Novel features of serine protease active sites and specificity pockets: sequence analysis and modelling studies of glutamate-specific endopeptidases and epidermolytic toxins. Protein Eng. 1996;9(7):591–601. doi: 10.1093/protein/9.7.591. [DOI] [PubMed] [Google Scholar]
- 35.Meijers R., Blagova E.V., Levdikov V.M., Rudenskaya G.N., Chestukhina G.G., Kostrov S.V., Lamzin V.S., Kuranova I.P. The crystal structure of glutamyl endopeptidase from Bacillus intermedius reveals a structural link between zymogen activation and charge compensation. Biochemistry-Us. 2004;43(10):2784–2791. doi: 10.1021/bi035354s. [DOI] [PubMed] [Google Scholar]
- 36.Nishimura K., Tajima N., Yoon Y.H., Park S.Y., Tame J.R.H. Autotransporter passenger proteins: virulence factors with common structural themes. J. Mol. Med. 2010;88(5):451–458. doi: 10.1007/s00109-010-0600-y. [DOI] [PubMed] [Google Scholar]
- 37.Ruiz-Perez F., Nataro J.P. Bacterial serine proteases secreted by the autotransporter pathway: classification, specificity, and role in virulence. Cell. Mol. Life Sci. 2014;71(5):745–770. doi: 10.1007/s00018-013-1355-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson T.A., Qiu J.Z., Plaut A.G., Holyoak T. Active-site gating regulates substrate selectivity in a chymotrypsin-like serine protease: the structure of Haemophilus influenzae immunoglobulin A1 protease. J. Mol. Biol. 2009;389(3):559–574. doi: 10.1016/j.jmb.2009.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aasland R., Abrams C., Ampe C., Ball L.J., Bedford M.T., Cesareni G., Gimona M., Hurley J.H., Jarchau T., Lehto V.P., Lemmon M.A., Linding R., Mayer B.J., Nagai M., Sudol M., Walter U., Winder S.J. Normalization of nomenclature for peptide motifs as ligands of modular protein domains. FEBS Lett. 2002;513(1):141–144. doi: 10.1016/s0014-5793(01)03295-1. [DOI] [PubMed] [Google Scholar]
- 40.Lee S., Owen K.E., Choi H.K., Lee H., Lu G.G., Wengler G., Brown D.T., Rossmann M.G., Kuhn R.J. Identification of a protein binding site on the surface of the alphavirus nucleocapsid and its implication in virus assembly. Structure. 1996;4(5):531–541. doi: 10.1016/s0969-2126(96)00059-7. [DOI] [PubMed] [Google Scholar]
- 41.Zhou Y., Bartels D.J., Hanzelka B.L., Muh U., Wei Y., Chu H.M., Tigges A.M., Brennan D.L., Rao B.G., Swenson L., Kwong A.D., Lin C. Phenotypic characterization of resistant Val36 variants of hepatitis C virus NS3-4A serine protease. Antimicrob. Agents Chemother. 2008;52(1):110–120. doi: 10.1128/AAC.00863-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Welsch C., Domingues F.S., Susser S., Antes I., Hartmann C., Mayr G., Schlicker A., Sarrazin C., Albrecht M., Zeuzem S., Lengauer T. Molecular basis of telaprevir resistance due to V36 and T54 mutations in the NS3-4A protease of the hepatitis C virus. Genome Biol. 2008;9(1):R16. doi: 10.1186/gb-2008-9-1-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thompson A.J., McHutchison J.G. Antiviral resistance and specifically targeted therapy for HCV (STAT-C) J. Viral Hepat. 2009;16(6):377–387. doi: 10.1111/j.1365-2893.2009.01124.x. [DOI] [PubMed] [Google Scholar]
- 44.Zeminian L.B., Padovani J.L., Corvino S.M., Silva G.F., Pardini M.I., Grotto R.M. Variability and resistance mutations in the hepatitis C virus NS3 protease in patients not treated with protease inhibitors. Mem. Inst. Oswaldo Cruz. 2013;108(1):13–17. doi: 10.1590/S0074-02762013000100002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Phan J., Zdanov A., Evdokimov A.G., Tropea J.E., Peters H.K., 3rd, Kapust R.B., Li M., Wlodawer A., Waugh D.S. Structural basis for the substrate specificity of tobacco etch virus protease. J. Biol. Chem. 2002;277(52):50564–50572. doi: 10.1074/jbc.M207224200. [DOI] [PubMed] [Google Scholar]
- 46.Yin J., Cherney M.M., Bergmann E.M., Zhang J., Huitema C., Pettersson H., Eltis L.D., Vederas J.C., James M.N. An episulfide cation (thiiranium ring) trapped in the active site of HAV 3C proteinase inactivated by peptide-based ketone inhibitors. J. Mol. Biol. 2006;361(4):673–686. doi: 10.1016/j.jmb.2006.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kanitz M., Blanck S., Heine A., Gulyaeva A.A., Gorbalenya A.E., Ziebuhr J., Diederich W.E. Structural basis for catalysis and substrate specificity of a 3C-like cysteine protease from a mosquito mesonivirus. Virology. 2019;533:21–33. doi: 10.1016/j.virol.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun Y., Wang X., Yuan S., Dang M., Li X., Zhang X.C., Rao Z. An open conformation determined by a structural switch for 2A protease from coxsackievirus A16. Protein Cell. 2013;4(10):782–792. doi: 10.1007/s13238-013-3914-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bergmann E.M., James M.N.G. In: Proteases as Target for Therapy. Von der Helm K., Korant B., Cheronis J.C., editors. Springer-Verlag; Berlin Heidelberg: 2000. The 3C proteinases of picornaviruses and other positive-sense, single-stranded RNA viruses; pp. 117–143. [Google Scholar]
- 50.Shi J., Sivaraman J., Song J. Mechanism for controlling the dimer-monomer switch and coupling dimerization to catalysis of the severe acute respiratory syndrome coronavirus 3C-like protease. J. Virol. 2008;82(9):4620–4629. doi: 10.1128/JVI.02680-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Someya Y., Takeda N. Functional consequences of mutational analysis of norovirus protease. FEBS Lett. 2011;585(2):369–374. doi: 10.1016/j.febslet.2010.12.018. [DOI] [PubMed] [Google Scholar]
- 52.Sommergruber W., Seipelt J., Fessl F., Skern T., Liebig H.D., Casari G. Mutational analyses support a model for the HRV2 2A proteinase. Virology. 1997;234(2):203–214. doi: 10.1006/viro.1997.8595. [DOI] [PubMed] [Google Scholar]
- 53.Piao S., Song Y.L., Kim J.H., Park S.Y., Park J.W., Lee B.L., Oh B.H., Ha N.C. Crystal structure of a clip-domain serine protease and functional roles of the clip domains. EMBO J. 2005;24(24):4404–4414. doi: 10.1038/sj.emboj.7600891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Derewenda Z.S., Derewenda U., Kobos P.M. (His)C epsilon-H...O=C < hydrogen bond in the active sites of serine hydrolases. J. Mol. Biol. 1994;241(1):83–93. doi: 10.1006/jmbi.1994.1475. [DOI] [PubMed] [Google Scholar]
- 55.Baird T.T., Jr., Wright W.D., Craik C.S. Conversion of trypsin to a functional threonine protease. Protein Sci. 2006;15(6):1229–1238. doi: 10.1110/ps.062179006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Riley B.T., Ilyichova O., Costa M.G., Porebski B.T., de Veer S.J., Swedberg J.E., Kass I., Harris J.M., Hoke D.E., Buckle A.M. Direct and indirect mechanisms of KLK4 inhibition revealed by structure and dynamics. Sci. Rep. 2016;6 doi: 10.1038/srep35385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krishnan V., Xu Y., Macon K., Volanakis J.E., Narayana S.V. The crystal structure of C2a, the catalytic fragment of classical pathway C3 and C5 convertase of human complement. J. Mol. Biol. 2007;367(1):224–233. doi: 10.1016/j.jmb.2006.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bell J.K., Goetz D.H., Mahrus S., Harris J.L., Fletterick R.J., Craik C.S. The oligomeric structure of human granzyme A is a determinant of its extended substrate specificity. Nat. Struct. Biol. 2003;10(7):527–534. doi: 10.1038/nsb944. [DOI] [PubMed] [Google Scholar]
- 59.Ponnuraj K., Xu Y., Macon K., Moore D., Volanakis J.E., Narayana S.V. Structural analysis of engineered Bb fragment of complement factor B: insights into the activation mechanism of the alternative pathway C3-convertase. Mol. Cell. 2004;14(1):17–28. doi: 10.1016/s1097-2765(04)00160-1. [DOI] [PubMed] [Google Scholar]
- 60.Ouyang M., Li X., Zhao S., Pu H., Shen J., Adam Z., Clausen T., Zhang L. The crystal structure of Deg9 reveals a novel octameric-type HtrA protease. Nat. Plants. 2017;3(12):973–982. doi: 10.1038/s41477-017-0060-2. [DOI] [PubMed] [Google Scholar]
- 61.Clausen T., Kaiser M., Huber R., Ehrmann M. HTRA proteases: regulated proteolysis in protein quality control. Nat. Rev. Mol. Cell Biol. 2011;12(3):152–162. doi: 10.1038/nrm3065. [DOI] [PubMed] [Google Scholar]
- 62.Blankenship E., Lodowski D.T.S. 2017. S. erythraea Trypsin Long Construct. [Google Scholar]
- 63.Read R.J., James M.N. Refined crystal structure of Streptomyces griseus trypsin at 1.7 A resolution. J. Mol. Biol. 1988;200(3):523–551. doi: 10.1016/0022-2836(88)90541-4. [DOI] [PubMed] [Google Scholar]
- 64.Gadwal S., Korotkov K.V., Delarosa J.R., Hol W.G., Sandkvist M. Functional and structural characterization of Vibrio cholerae extracellular serine protease B, VesB. J. Biol. Chem. 2014;289(12):8288–8298. doi: 10.1074/jbc.M113.525261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rule C.S., Park Y.J., Korotkov K.V., Delarosa J.R., Turley S., DiMaio F., Hol W.G.J., Sandkvist M. 2018. Secondary Mutations in Type II Secretion Mutants of Vibrio Cholerae: Inactivation of VesC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang J., Fan T., Yao X., Wu Z., Guo L., Lei X., Wang J., Wang M., Jin Q., Cui S. Crystal structures of enterovirus 71 3C protease complexed with rupintrivir reveal the roles of catalytically important residues. J. Virol. 2011;85(19):10021–10030. doi: 10.1128/JVI.05107-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Barrila J., Gabelli S.B., Bacha U., Amzel L.M., Freire E. Mutation of Asn28 disrupts the dimerization and enzymatic activity of SARS 3CL(pro) Biochemistry-Us. 2010;49(20):4308–4317. doi: 10.1021/bi1002585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Barrila J., Bacha U., Freire E. Long-range cooperative interactions modulate dimerization in SARS 3CLpro. Biochemistry-Us. 2006;45(50):14908–14916. doi: 10.1021/bi0616302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Speroni S., Rohayem J., Nenci S., Bonivento D., Robel I., Barthel J., Luzhkov V.B., Coutard B., Canard B., Mattevi A. Structural and biochemical analysis of human pathogenic astrovirus serine protease at 2.0 A resolution. J. Mol. Biol. 2009;387(5):1137–1152. doi: 10.1016/j.jmb.2009.02.044. [DOI] [PubMed] [Google Scholar]
- 70.Shi Y., Lei Y., Ye G., Sun L., Fang L., Xiao S., Fu Z.F., Yin P., Song Y., Peng G. Identification of two antiviral inhibitors targeting 3C-like serine/3C-like protease of porcine reproductive and respiratory syndrome virus and porcine epidemic diarrhea virus. Vet. Microbiol. 2018;213:114–122. doi: 10.1016/j.vetmic.2017.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tian X., Lu G., Gao F., Peng H., Feng Y., Ma G., Bartlam M., Tian K., Yan J., Hilgenfeld R., Gao G.F. Structure and cleavage specificity of the chymotrypsin-like serine protease (3CLSP/nsp4) of Porcine Reproductive and Respiratory Syndrome Virus (PRRSV) J. Mol. Biol. 2009;392(4):977–993. doi: 10.1016/j.jmb.2009.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Weerawarna P.M., Kim Y., Galasiti Kankanamalage A.C., Damalanka V.C., Lushington G.H., Alliston K.R., Mehzabeen N., Battaile K.P., Lovell S., Chang K.O., Groutas W.C. Structure-based design and synthesis of triazole-based macrocyclic inhibitors of norovirus protease: structural, biochemical, spectroscopic, and antiviral studies. Eur. J. Med. Chem. 2016;119:300–318. doi: 10.1016/j.ejmech.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shimamoto Y., Hattori Y., Kobayashi K., Teruya K., Sanjoh A., Nakagawa A., Yamashita E., Akaji K. Fused-ring structure of decahydroisoquinolin as a novel scaffold for SARS 3CL protease inhibitors. Bioorg. Med. Chem. 2015;23(4):876–890. doi: 10.1016/j.bmc.2014.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xue X., Yu H., Yang H., Xue F., Wu Z., Shen W., Li J., Zhou Z., Ding Y., Zhao Q., Zhang X.C., Liao M., Bartlam M., Rao Z. Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design. J. Virol. 2008;82(5):2515–2527. doi: 10.1128/JVI.02114-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Karlsen S., Iversen L.F., Larsen I.K., Flodgaard H.J., Kastrup J.S. Atomic resolution structure of human HBP/CAP37/azurocidin. Acta Crystallogr. D Biol. Crystallogr. 1998;54(Pt 4):598–609. doi: 10.1107/s0907444997016193. [DOI] [PubMed] [Google Scholar]
- 76.Biovia D.S. Dassault Systèmes; San Diego: 2016. Discovery Studio Modeling Environment. [Google Scholar]
- 77.Lehtonen J.V., Still D.J., Rantanen V.V., Ekholm J., Bjorklund D., Iftikhar Z., Huhtala M., Repo S., Jussila A., Jaakkola J., Pentikainen O., Nyronen T., Salminen T., Gyllenberg M., Johnson M.S. BODIL: a molecular modeling environment for structure-function analysis and drug design. J. Comput. Aided Mol. Des. 2004;18(6):401–419. doi: 10.1007/s10822-004-3752-4. [DOI] [PubMed] [Google Scholar]
- 78.Kraulis P.J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 1991;24:946–950. [Google Scholar]
- 79.Merritt E.A., Bacon D.J. Raster3D: photorealistic molecular graphics. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary tables