Abstract.
Chikungunya fever (CHIKF) is a major public health concern and is caused by chikungunya virus (CHIKV). In 2005, the virus was reintroduced into India, resulting in massive outbreaks in several parts of the country. During 2010 and 2016 outbreaks, we recruited 588 patients from a tertiary care hospital in New Delhi, India, during the acute phase of CHIKF; collected their blood and clinical data; and determined their arthralgic status 12 weeks post-onset of fever. We evaluated IgM/IgG CHIKV-binding antibodies and their neutralizing capacity, sequenced complete genomes of 21 CHIKV strains, and correlated mutations with patient sequelae status. We also performed infections in murine models using representative strains from each outbreak to evaluate differences in pathogenesis. Our screening and analysis revealed that patients of the 2016 outbreak developed earlier IgM and neutralizing antibody responses that were negatively correlated with sequelae, compared with 2010 patients. Mutations that correlated with human disease progression were also correlated with enhanced murine virulence and pathogenesis. Overall, our study suggests that the development of early neutralizing antibodies and sequence variation in clinical isolates are predictors of human sequelae.
INTRODUCTION
Chikungunya fever (CHIKF) is a viral illness characterized by acute fever coupled with incapacitating, often chronic, arthralgia. Although the fever subsides in 7–10 days, the joint pain varies in intensity and can persist from 3 months to more than 2 years postinfection (reviewed in ref 1). Before 2005, CHIKF disease manifestations were poorly characterized and clinical features were characterized mainly based on reports documented in the 1970s.2 Since 2005, the etiologic agent, chikungunya virus (CHIKV), has spread and devastated millions throughout the Indian Ocean islands, Europe, India,3–5 and other parts of Asia, and more recently, the South Pacific region and Americas (reviewed in ref 6–8).
Chikungunya virus was endemic in India at least since 1958 and probably caused periodic outbreaks much earlier,9 causing epidemics every two to three decades10–12 with relatively few cases reported during interepidemic periods. The first Indian outbreak was documented in 1963 in Calcutta (now Kolkata),13 followed by epidemics in Tamil Nadu, Andhra Pradesh, and Maharashtra14,15; the last outbreak during the twentieth century was recorded in Maharashtra in 1973.16 Then, India again experienced major outbreaks from 2005 to 2010.17–20 After 2010, the country experienced a drastic decline in the number of reported cases,20,21 raising the question of whether this infection was at the end of its transmission wave in India. However, in 2016, India reeled under a massive outbreak, with 64,057 cases confirmed across the country.20
We conducted a prospective study to investigate the evolution of CHIKV in India since 2010. As part of this study, we previously performed a detailed analysis of the clinical, serological, and virological aspects of CHIKF in patients between 2010 and 2013.11 Clinical aspects of the CHIKF outbreak in 2016 have also been recently reported.12 During the analysis of the samples from the 2016 outbreak, we were intrigued by the distinctions in disease outcomes between 2010 and 2016 outbreaks.
To study these putative differences between the 2010 and 2016 outbreaks in greater detail, we conducted detailed, comparative analyses of the viremia, antibody development, neutralization patterns, and sequelae intensities. We also sequenced the complete genomes of several isolates collected during each outbreak and identified sequence variants that correlated with disease outcome. We then compared the pathogenesis of the 2010–2016 outbreak viruses in type I interferon receptor–deficient (A129) and immunocompetent C57BL/6J mice.
Our results demonstrate distinctive features in the neutralization patterns of human antibodies induced during the two outbreaks, which may have implications for pathogenesis. We also detected a correlation between strain-specific mutations and sequelae. Finally, pathogenicity studies using mouse models revealed a strain-dependent pattern in virulence.
MATERIALS AND METHODS
Study site.
A prospective study to evaluate the evolution of CHIKV in India was conducted in New Delhi (28.6139°N, 77.2090°E). Samples were collected at the Vardhman Mahavir Medical College and Safdarjung Hospital (VMMC and SH), a teaching institution and a multispecialty hospital with 1,600 beds that serves as a referral hospital during outbreak situations.
Study design, participants, and clinical assessment.
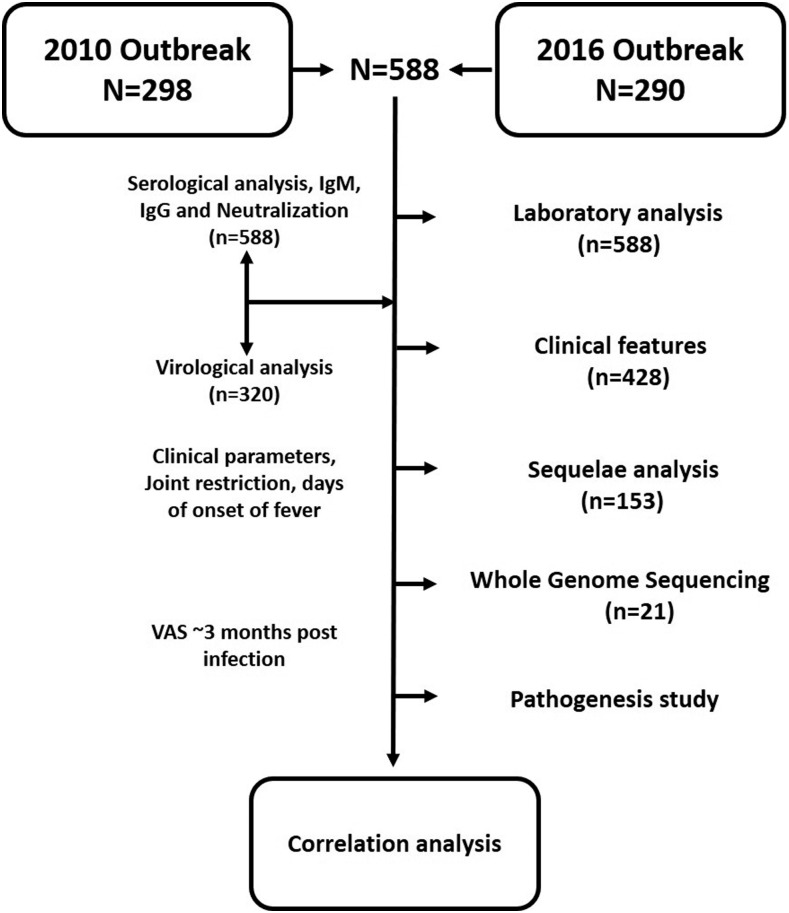
Samples were collected from a cohort of patients with laboratory-confirmed acute CHIKF, who sought care at the hospital during the 2010 and 2016 outbreaks during the prospective study.11,12 Confirmation of all recruited patients in various wards and outpatient departments was carried out on the basis of qualitative IgM ELISA and reverse transcription polymerase chain reaction (RT-PCR) for patient sera collected until day 5 post-onset of fever. Medical histories, including signs and symptoms, and laboratory findings during the acute phase of illness were documented in the clinical report form (CRF) used for downstream analyses. Wherever possible, a follow-up was performed 12 weeks after the acute episode by a qualified rheumatologist, and a visual analogue score (VAS) was assigned on a scale of 1–10 on the basis of questions related to the intensity of pain posed to the patients. Once confirmed and recruited, the patient sera were transferred to the laboratory for semiquantitative IgM and IgG analysis along with their neutralizing capacities, and Detection of CHIKV by reverse transcription–quantitative polymerase chain reaction (RT-qPCR) and further experiments is as detailed in Figure 1.
Figure 1.
Study design. Study design, inclusive of stepwise number of samples available and tests performed on these samples. VAS = visual analogue score.
Semiquantitative detection of CHIKV-specific IgM and IgG antibodies in patient sera samples using indirect ELISA.
To detect midpoint and endpoint titers of anti-CHIKV IgG and IgM antibodies, indirect ELISA using purified, inactivated CHIKV as antigen was performed as previously reported.11 Briefly, purified CHIKV was coated onto microtiter plates and left undisturbed overnight at 4°C. Post-blocking, 2-fold serially diluted patient sera, starting at 1:100, were incubated at room temperature for 2 hours. After washing, the plates were developed using antihuman IgM–horseradish peroxidase (HRP) (Cat No: A4290; Sigma-Aldrich) and antihuman IgG–HRP antibodies (Cat No: AP113P; Sigma-Aldrich, St. Louis, MO) followed by TMB (Cat No: T0440; Sigma-Aldrich) as the substrate. The assay was terminated by a stop solution. Previously, well-characterized samples with neutralizing antibodies to CHIKV were pooled and used as a positive control to obtain a linear curve for absorbance and dilution.
Plaque reduction neutralization test (PRNT).
The neutralization capacity of sera from confirmed CHIKF patients was analyzed by PRNTs on Vero cells using standard protocols.11 The CHIKV strain used in the assay belongs to the ECSA (Indian Ocean Lineage) genotype. Briefly, sera were diluted 1:50, mixed with 50 plaque-forming units (PFU) of CHIKV, then 2-fold serially diluted in 5% complete Dulbecco’s Modified Eagle Medium (DMEM), and incubated at 37°C. Sixty minutes after addition of the virus/serum mix to Vero cell monolayers, the cells were overlaid with 2% carboxymethyl cellulose prepared in 10% complete DMEM. After incubation for 36 hours, the cells were washed and fixed with 4% paraformaldehyde, and visualized plaques were counted. Results were interpreted using GraphPad Prism 6, and PRNT endpoint titers were expressed as the reciprocal value of the maximum serum dilution inducing a 50% reduction (PRNT50) in plaque counts.
Reverse transcription–quantitative polymerase chain reaction detection of CHIKV viral RNA (vRNA).
Viral RNA was isolated directly from patient sera using the High Pure Viral Nucleic Acid kit (Product No: 11858874001; Roche, Mannheim, Germany) according to the manufacturer’s instructions. Viral RNA samples were screened by RT-qPCR (QuantiTect SYBR® Green RT-PCR Kit [Cat No: 204243; Qiagen, Hilden, Germany]) for CHIKV using previously described primers.19 Laboratory-generated CHIKV was used to extrapolate virus titers using a standard curve.
Illumina sequencing.
Viral RNA fragmentation, first and second strand synthesis and amplification, cluster formation, and paired-end 50-base sequencing on an Illumina HiSeq 1000 system were performed as previously described.22 Briefly, for samples with low threshold cycle (Ct) values (lesser than 20) in RT-qPCR assays, vRNA was fragmented by incubation at 94°C for eight minutes in 19.5 μL of fragmentation buffer (Illumina Inc., San Diego, CA). First and second strand synthesis, adapter ligation, and amplification of the library were performed using the Illumina TruSeq RNA Sample Preparation Kit v2 under conditions prescribed by the manufacturer (Illumina Inc.). Cluster formation of the library DNA templates was performed using the TruSeq PE Cluster Kit v3 (Illumina Inc., San Diego, CA) and the Illumina cBot workstation using conditions recommended by the manufacturer. Paired-end 50-base sequencing by synthesis was performed using TruSeq SBS Kit v3 (Illumina Inc.) on an Illumina HiSeq 1000 system using protocols defined by the manufacturer. Cluster density per lane was 820–940 k/mm2, and post-filter reads ranged from 148 to 218 million per lane.
Sequence analysis.
To assemble complete genomic sequences, we first assessed the quality of the paired-end reads using FastQC; low base-quality ends were trimmed using Trim Galore version 0.4.4. The remaining paired-end reads were aligned to an Indian strain isolated during an outbreak in 2009 (accession no. KJ796844), using BWA version 0.7.12 and default parameters. The single nucleotide polymorphism (SNP) variants were identified using bcftools version 0.1.19 23 and GATK HaplotypeCaller version 3.1.1. 24 The complete pipeline used to perform the sequence and variant analyses is presented in Supplemental Figure 1. Variants were visualized using Circos to facilitate the identification and analysis of similarities and differences in genome comparisons.25 Furthermore, hierarchical clustering was performed using the hclust package in R-Bioconductor statistical packages.26
In vivo pathogenesis.
Viruses were passaged once in C6/36 cells (ATCC® CRL-1660) for 72 hours to amplify the viruses and purified using the PEG purification method. Five-week-old C57/B6J (n = 8) and 8-week-old A129 (interferon type I receptor–deficient; n = 5 or 6) mice were inoculated intradermally in the footpad with a 10-µL inoculum containing 104 PFU of CHIKV strains representative of the 2010 and 2016 outbreaks, and well-characterized strains from all major CHIKV genotypes: West African (strain SH2380), East/Central/South African (strain La Reunion), and Asian/American (strain 99659). In a pilot study, doses in the range of 102–106 PFU were tested to determine which strain caused disease in C57BL/6J mice by monitoring for signs including body weight, morbidity, and footpad thickness. Every third day, changes in footpad thickness and morbidity according to the Morton and Griffiths scale27 were recoded as a surrogate for arthritis, the most differentiating feature of CHIKF. In a second study, Indian outbreak CHIKV strains were compared in 8-week-old C57/B6J mice at a dose of 106 PFU. Body weight and footpad thickness were measured daily for 14 days, and mice were bled on alternate days for the first 4 days post-challenge to measure viremia. C57/B6J mice were purchased from the Jackson Laboratory (Sacramento, CA), and A129 mice were bred locally at University of Texas Medical Branch (UTMB) for study. All animals were handled in accordance with the UTMB and ICGEB policies following IACUC- and CPCSEA-approved protocols.
Statistical analyses.
A Spearman rank test was used to determine the correlation between 1) absorbance of IgM (at optical density [OD]450) and days post-onset of fever, 2) absorbance of IgG (at OD450) and days post-onset of fever, 3) absorbance of IgM (at OD450) and neutralization capacity of the patient samples, and 4) absorbance of IgG (at OD450) and neutralization capacity of the patient samples. Unpaired t-test was performed to determine the correlation and level of significance between neutralization status and absorbance of IgM (at OD450). Two-way Analysis of Variance (ANOVA) was used to study the differences in pathogenicity in the animals. All tests were performed using GraphPad Prism 6.
Ethics statement.
Written informed consent was received from anonymized participants before inclusion in the study. All the ethical approvals were peer-reviewed and preapproved by the authors’ institutional ethical clearance committees, namely, ICGEB Institutional Ethical Committee and the Institutional Ethical Committee at VMMC and SH (ICGEB/IEC/2010/01, ICGEB/IEC/2014/01, and IEC/VMMC/SJH/Project/February-2016/575) and IRB at UTMB.
RESULTS
A total of 588 blood samples (298 samples from the 2010 outbreak and 290 samples from the 2016 outbreak) were collected from patients who were seen within 1–10 days since the onset of fever. A detailed interview of the patients at the time of recruitment revealed that all were recently infected with no prior clinical presentation due to either dengue or chikungunya. Comparative analyses between the outbreaks were performed in a case-matched manner based on days post-onset of fever. Completed CRFs (n = 428) were analyzed for differences in clinical presentations. Neutralization capacity of sera included in the study (n = 588) was determined. Visual analogue score was documented from those patients who participated in the follow-up after 12 weeks (n = 153). Whole genome sequencing and mutational analysis were performed on a subset of the CHIKV-positive sera (n = 21), and in vivo pathogenesis of representative isolates (n = 3) from each outbreak was assessed in C57BL/6J mice. Correlations were analyzed between neutralization titers, mutation patterns of virus strains, human disease outcome, and murine virus pathogenesis, as described in the following.
Clinical features of chikungunya during the two outbreaks.
A total of 428 patients, 228 from 2010 and 200 from 2016, were included in this study for the determination of clinical features (Tables 1 and 2). The age of the patients ranged between 11 and 68 years; the male–female ratio was 1.3:1 from both years. At the time of assessment, 128 (56.14%) patients of 228 in 2010 and 194 (97%) patients of 200 in 2016 reported restricted joint movement related to CHIKV infection. Joint swelling was observed in 108 (47.36%) and 78 (39%) patients in 2010 and 2016, respectively, which was mainly observed in the small joints of the hand/wrist followed by the elbows. Rashes and itching were also important observations in 105 (52.5%) and 48 (24%) patients in 2016, as opposed to 46 (20.17%) and 14 (6.14%) patients in 2010. Although clinical signs/symptoms, such as restricted joint movements, rashes, and irritation, were majorly present in acute CHIKV infection in 2016 as opposed to 2010, ANOVA revealed that there were no significant differences between the clinical signs/symptoms exhibited in 2010 versus 2016 (P > 0.05), except for the VAS discussed below.
Table 1.
General observations and patient information for patients found positive by IgM and/or RT-PCR data
| General observations | 2010 (N = 298) | 2016 (N = 290) |
|---|---|---|
| Age (years), range | 17–68 | 11–68 |
| Age (years), median | 36 | 35 |
| Males | 145 | 131 |
| Females | 153 | 158 |
| Temperature range (°F) | 99–101 (98.6°F) | 99–101 (98.6°F) |
| Temperature median (°F) | 100 | 100 |
| Days of onset of fever (range) | 1–10 | 1–10 |
| Days of onset of fever (median) | 4 | 5 |
| Hemorrhagic symptoms,* n (%) | 33 (14.47) | 21 (10.5) |
| IgM+/RT-PCR−, n (%) | 101 (33.89) | 69 (23.67) |
| IgM−/RT-PCR+, n (%) | 141 (47.31) | 18 (6.33) |
| IgM+/RT-PCR+, n (%) | 56 (18.80) | 203 (70) |
| VAS (range) | 1–9 | 1–8 |
| VAS (median)† | 7 | 4 |
VAS = visual analogue score.
* Hemorrhagic symptoms include epistaxis, bleeding from the gums, positive Hess test, subconjunctival bleed, and petechial/purpuric rash.
† Statistically significant variable.
Table 2.
Clinical features and laboratory observations seen in patients suffering from chikungunya
| Clinical features | 2010 (N = 228) | % patients exhibiting the clinical feature | 2016 (N = 200) | % patients exhibiting the clinical feature |
|---|---|---|---|---|
| Lymphadenopathy | 38 | 16.7 | 4 | 2 |
| Vomiting/nausea | 4 | 1.8 | 23 | 11.5 |
| Diarrhea | 13 | 5.7 | 3 | 1.5 |
| Rashes | 46 | 20.17 | 105 | 52.5 |
| Itching | 14 | 6.14 | 48 | 24 |
| Muscoskeletal | ||||
| Joint movement restriction | 128 | 56.1 | 194 | 97 |
| Swelling | 108 | 47.4 | 78 | 39 |
| Morning stiffness | 69 | 30.7 | 43 | 21.5 |
| Rheumatoid arthritis-like illness | 85 | 37.3 | 4 | 2 |
| Undifferentiated inflammatory arthritis | 42 | 18.4 | 28 | 14 |
| Low backache | 76 | 33.3 | 5 | 2.5 |
| Body ache | 27 | 11.84 | 3 | 1.5 |
Antibody responses and correlation with disease outcome.
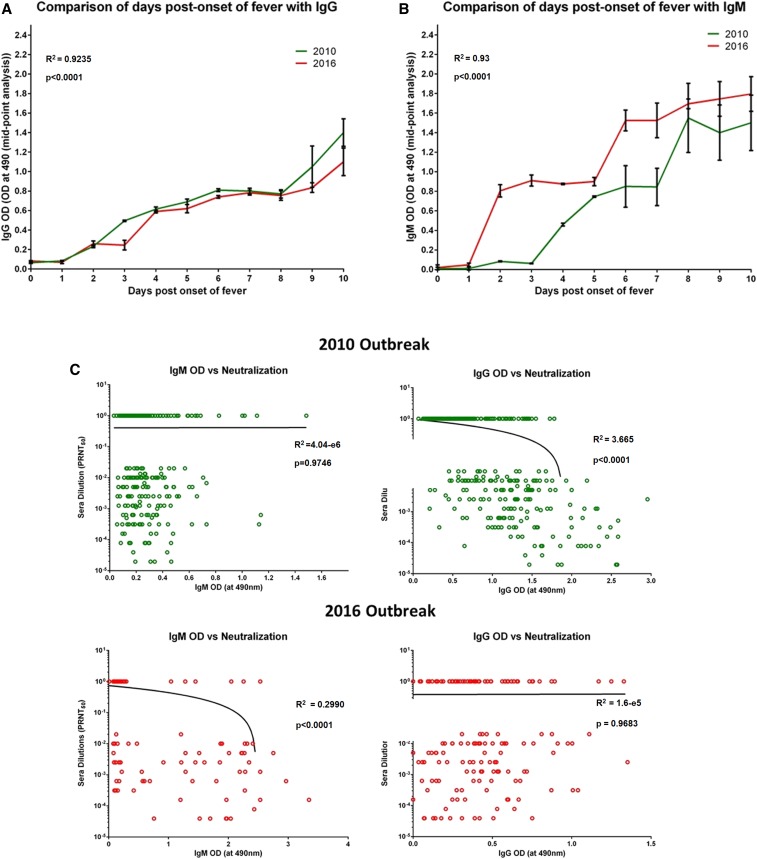
Analysis of IgM and IgG antibodies and neutralization titers in sera from the two outbreaks (n = 298 from 2010, n = 290 from 2016) revealed that 2016 patients developed higher mean titers of IgM than 2010 patients (Supplemental Figure 2), whereas this trend was inverted in the case of IgG titers (Supplemental Figure 2). Furthermore, we observed similar neutralizing capacities of sera from the two outbreaks, with 57.7% of 2010 sera and 52.3% of 2016 sera exhibiting neutralization activity (Supplemental Figure 2). In-depth analysis revealed no difference in the mean IgG titers of sera from the two outbreaks (Figure 2A). However, IgM antibodies developed 5–6 days after the onset of fever in 2010, whereas in 2016, they developed as early as 2 days post-onset of fever (Figure 2B). Furthermore, evaluation of the neutralizing capacity of these sera revealed that in the 2010 samples, neutralization titers correlated with the development of neutralizing IgG antibodies (62.5%) (r = −0.61, P < 0.05). However, among the 2016 samples, neutralization correlated better with IgM antibodies, with 89.1% of IgM-positive patients having neutralizing antibodies to Chikungunya (CHIKV) (r = −0.55, P < 0.05) (Figure 2C).
Figure 2.
Comparison of days post-onset of fever and development of binding antibodies along with their neutralization status. (A) Comparison of days post-onset of fever and IgG (OD at 450 nm) antibodies between the patient sera samples from 2010 and 2016 outbreaks (P < 0.0001). (B) Comparison of days post-onset of fever and IgM (OD at 450 nm) between the patient sera samples from 2010 and 2016 outbreaks: (C) Correlation of binding antibodies (IgM and IgG; OD at 450 nm) of the patient sera samples and their neutralizing capacity from the two CHIK outbreaks. Pooled negatives and positive sera samples were used as control. The error bars in A and B indicate SD. The straight and curved lines indicate the correlation status between binding antibodies and their neutralizing status. The dotted line in (A), (B), and (C) represents the cutoffs. P-values and R2 values are presented in the graphs. This figure appears in color at www.ajtmh.org.
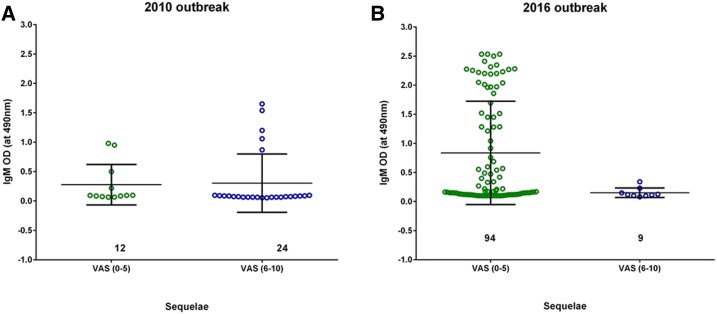
Sequelae analysis was carried out for 103 patients from the 2016 CHIK outbreak and 50 patients from 2010, 12 weeks post-onset of disease. From physical examination and interviews, a VAS was assigned on the basis of the clinical disease activity index and further rated on a scale of 1–10 on the basis of questions on the intensity of pain prepared by a rheumatologist10 and documented in the clinical records. Visual analogue score ranging between 0 and 5 were considered low, whereas a score between 6 and 10 was ranked as high. On examination, 91.3% of patients from the 2016 outbreak exhibited low VAS (mean = 2.8 and median = 2), whereas 70% of the patients from 2010 exhibited high VAS (mean = 7.2 and median = 4). Furthermore, correlation of VAS and IgM status was studied in 139 patients, for which complete data were available (n = 36 from 2010 and n = 103 from 2016) (Figure 3). Of the patients with low VAS, 88.9% developed high titers of IgM (P > 0.05). Furthermore, of the patients who developed high IgM titers, 86.9% of these sera also had neutralizing capacity. We segregated these high IgM titer samples on the basis of their neutralizing capacities into two groups: group 1: patients who developed IgM antibodies in the early days of infection (defined here as 2–6 days post-onset of fever) and group 2: patients who developed IgM antibodies later (> 6 days after the onset of fever). We observed that 61.5% of patients in group 1 developed early neutralizing antibodies, whereas only 38.5% in group 2 developed neutralizing antibodies, all > 6 days after onset. Furthermore, the first appearance of neutralizing antibodies during the acute phase (early or late) was correlated with the onset of fever; P-values for both groups were < 0.05. Therefore, we hypothesized that infections during the 2016 outbreak led to the generation of high IgM titers and that these had higher neutralizing capacity than the 2010 patients who developed low IgM titers (P < 0.05).
Figure 3.
Comparison of the status of neutralizing IgM antibodies with sequelae (visual analog score [VAS]). (A) Comparison of IgM and VAS in the 2010 CHIK outbreak: IgM (at 450 nm) was compared with VAS collected from patients 12 weeks pi. (B) Comparison of IgM and VAS in the 2016 CHIK outbreak: IgM (at 450 nm) was compared with VAS collected from patients 12 weeks pi. In both the analyses, VAS was divided into two groups (0–5) and (6–10) corresponding to low and high VAS, respectively. This figure appears in color at www.ajtmh.org.
Mutant analysis in CHIKV isolates from the two outbreaks.
Based on PRNT assays and associated clinical symptoms, we hypothesized that the CHIKV strains circulating during the two outbreaks differed in one or more mutations that affect pathogenesis, resulting in differential host responses and disease chronicity. Representative viral strains (n = 11 from 2010 and n = 10 from 2016) that showed maximum viral load, variation in neutralization capacity, and sequelae were subjected to genomic sequencing; 19 samples (n = 11 from 2010 and n = 8 from 2016) yielded sufficient depth (> 200 ×) and coverage (> 98%) (Accession numbers: MH124570–MH124589) for analyses (SupplementalTable 1).
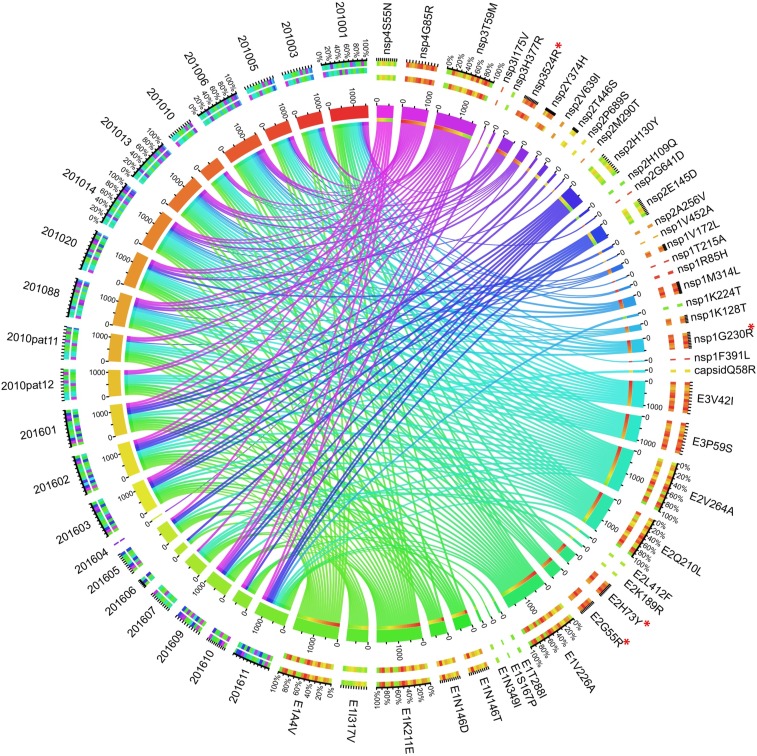
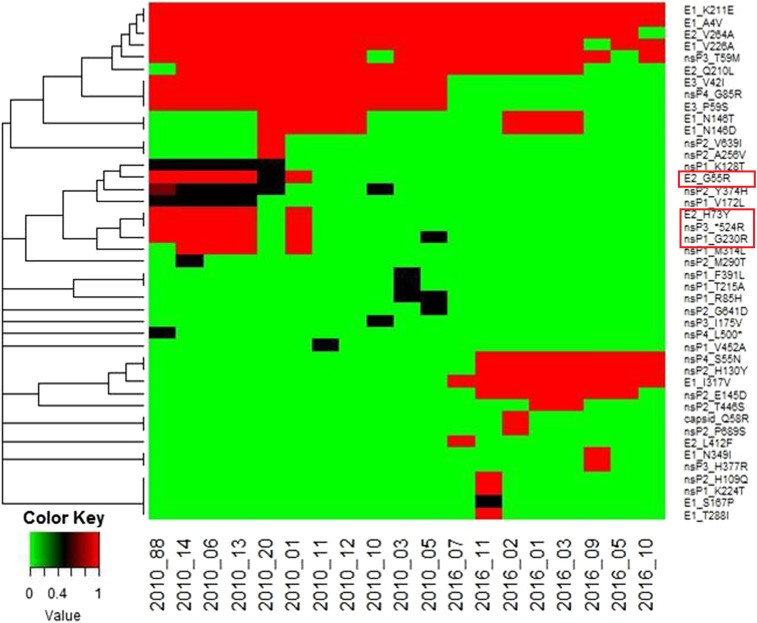
Mutation analysis of these sequences and a reference strain isolated in 2009 from India (GenBank Accession no. KJ796844) revealed a total of 43 nonsynonymous differences when the reference was compared with the 2010 and 2016 strains (Figure 4). Of these, 18 were present in structural protein genes and 25 in nonstructural genes. Furthermore, outbreak-specific hierarchical clustering revealed a total of 28 nonsynonymous differences (11 in structural proteins and 17 in nonstructural proteins) that distinguished all 2010 sequences from the 2009 reference. Twenty-four nonsynonymous differences (14 in structural proteins and 10 in nonstructural proteins) distinguished all 2016 CHIKV sequences from the 2009 reference (Figure 5). Of these nonsynonymous mutations, nine were common to both 2010 and 2016 strains. With respect to outbreak-specific mutations, 19 nonsynonymous mutations (four in structural and 15 in nonstructural protein genes) were unique to 2010, whereas 15 nonsynonymous mutations (seven in structural and eight in nonstructural protein genes) were unique to 2016 (Table 3).
Figure 4.
Visualization of nonsynonymous mutations in the patient sera samples collected during the 2010 and 2016 outbreaks using the Circos Table Viewer. All samples are represented on the left side, and the mutations observed in these samples are indicated on the right side of the figure. The ribbons in between indicate the allelic frequency of a particular mutation as observed in each sample normalized on the scale of 100. The outer ring represents the individual mutations observed during two outbreaks along with the patient samples analyzed. The inner ring represents the frequency of mutations in relation to the particular sample IDs. Major mutations under discussion are marked with *. This figure appears in color at www.ajtmh.org.
Figure 5.
Hierarchical clustering of nonsynonymous mutations in 2010 and 2016 CHIK patient serum samples. A total of 41 nonsynonymous mutations were observed in the structural (E1, E2, E3, C, and 6K) and nonstructural (nsP1, nsP2, nsP3, and nsP4) regions of the chikungunya virus genome in 11 samples from the 2010 outbreak and eight patient sera samples from the 2016 outbreak. These mutations were clustered hierarchically using R-Bioconductor statistical packages. It was observed that 2010 and 2016 patients clustered separately of each other. Also, certain mutations were found to be linked. This figure appears in color at www.ajtmh.org.
Table 3.
List of nonsynonymous mutations unique to 2010 and 2016 samples: Whole genome sequencing was performed for 22 chikungunya virus–infected patient sera from the two outbreaks (11 each from 2010 and 2016)
| Protein | Nonsynonymous mutations | Percent occurrence |
|---|---|---|
| Mutations unique to 2010 (n = 11) | ||
| E2 | G55R | 54.54 |
| E2 | H73Y | 45.45 |
| E3 | V42I | 100 |
| E3 | P59S | 100 |
| nsP1 | G230R | 54.54 |
| nsP1 | M314L | 36.36 |
| nsP1 | R85H | 9.09 |
| nsP1 | T215A | 9.09 |
| nsP1 | F391L | 9.09 |
| nsP1 | K128T | 45.54 |
| nsP1 | V172L | 36.36 |
| nsP1 | V452A | 9.09 |
| nsP2 | G641D | 9.09 |
| nsP2 | M290T | 9.09 |
| nsP2 | A256V | 9.09 |
| nsP2 | V639I | 9.09 |
| nsP3 | I175V | 9.09 |
| nsP3 | STOP524R | 45.45 |
| nsP4 | G85R | 100 |
| Mutations unique to 2016 (n = 8) | ||
| C | Q58R | 12.5 |
| E1 | I317V | 100 |
| E1 | N349I | 12.5 |
| E1 | S167P | 12.5 |
| E1 | T288I | 12.5 |
| E2 | K189R | 12.5 |
| E2 | L412F | 12.5 |
| nsP1 | K224T | 12.5 |
| nsP2 | H130Y | 100 |
| nsP2 | E145D | 75 |
| nsP2 | T446S | 25 |
| nsP2 | P689S | 12.5 |
| nsP2 | H109Q | 12.5 |
| nsP3 | H377R | 12.5 |
| nsP4 | S55N | 87.5 |
Correlation between mutations and sequelae.
Clustering analysis between SNPs and arthralgia symptoms or VAS revealed that one or more mutations differing between the two outbreaks may have contributed to the host response. In 2010, two sequence clusters were identified, with viruses from six of 11 patients (54.5%) possessing mutations in the E2 (G55R, H73Y), nsP1 (G230R), and nsP3 (opal 524R) proteins that were unique to the 2010 outbreak. Although the mean VAS was 6.90 (± 2.92) for the 2010 outbreak, all six samples containing these mutations (hereafter called putative high pathogenicity genotype) showed exceptionally high VAS (> 7). By contrast, none of the 2016 outbreak samples with a low mean VAS of 2.25 (± 2.60) exhibited the aforementioned mutations.
Differences in in vivo pathogenesis between 2010 and 2016 CHIKV virus strains.
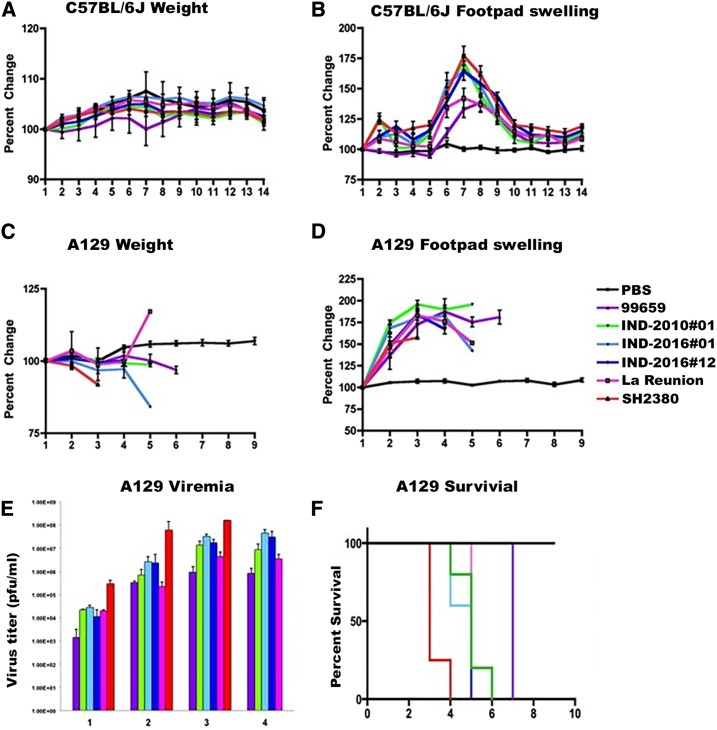
Given the observed differences in human immune response and disease progression between the 2010 and 2016 outbreaks, and the existence of outbreak-specific mutations, we evaluated murine pathogenicity among strains from the two outbreaks using footpad swelling as a proxy for arthritis. We included all major CHIKV lineages as comparators. As described in an earlier study,28 the most pathogenic strain belonged to the West African genotype, followed by the ECSA and Asian strains, with the most recently emergent Asian/American strain being the least virulent for footpad swelling (Figure 6). At lower doses (104 PFU) in C57BL/6J mice, representative strains of the 2010 versus 2016 Indian outbreaks showed no significant differences in footpad swelling. The study was repeated in A129 (a lethal IFNAR−/− model) mice with similar results (Figure 6).
Figure 6.
Comparison of pathogenicity among various chikungunya virus (CHIKV) strains in C57BL/6J and A129 mice models until day 10 postinfection. (A) Percent change in weight among the C57BL/6J mice post-challenge with 104 plaque-forming units (PFU) of CHIKV strains (n = 8). (B) Percent change in footpad thickness among the C57BL/6J mice post-challenge with 104 PFU of CHIKV strains (n = 8). (C) Percent change in weight in A129 mice post-challenge with 104 PFU of CHIKV strains (n = 5/6). (D) Percent change in footpad thickness in A129 mice post-challenge with 104 PFU of CHIKV strains (n = 5/6). (F) Viral load in A129 mice post-challenge with 104 PFU of CHIKV strains (n = 2/3). The limit of detection was 102 PFU/mL. All percent change data were estimated from the day 0 measurements. For all the results discussed earlier, error bars indicate SD. This figure appears in color at www.ajtmh.org.
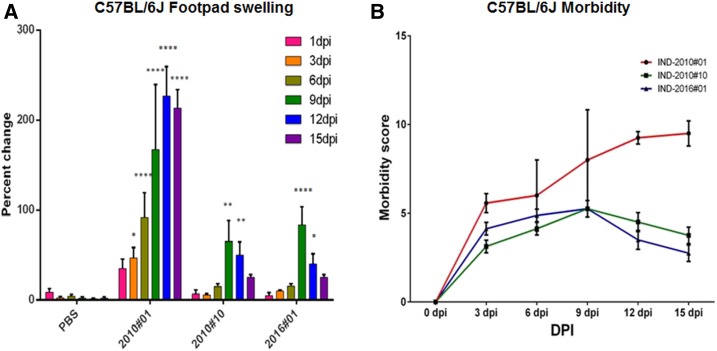
Our genetic analysis revealed that certain unique mutations in 2010 samples, specifically in proteins E2, nsP1, and nsP3, separated these 2010 CHIKV strains into two major categories: 1) CHIKV strains carrying possible pathogenic mutations (E2-G55R, E2-H73Y, nsP1-G230R, and nsP3-STOP524R), associated with high VAS (6–10) during the post-acute phase (2010#01), and 2) CHIKV isolates without these mutations, associated with low VAS (2010#10). By contrast, no such genetic distinction was seen among the 2016 CHIKV strains, as all had similar clinical associations and exhibited lower VAS at convalescence; because of this, only one 2016 isolate was selected at random for murine studies (2016#01). To understand the implication of this observation for pathogenesis, we used one strain of each category from the 2010 outbreak and one strain from the 2016 outbreak for a comparative analysis. Previous reports have emphasized that initial viremia plays a decisive role in disease progression and intensity of CHIKF in the post-acute phase.11,29 Furthermore, a recent murine pathogenesis study from our group showed that better distinction of clinical presentation with respect to limb thickness and morbidity was observed at a viral dose of 106 for footpad injections30; hence, the same dose was used to study mutation-specific pathogenesis in the wild-type C57BL/6J mice. After intradermal challenge with 106 PFU, significant differences in footpad swelling and morbidity were observed. The strain with mutations E2-G55R, E2-H73Y, nsP1-G230R, and nsP3-STOP524R (2010#01) showed a continuous increase in thickness until day 12 post-infection (Figure 7A), after which all animals died. Footpad swelling was positively correlated with the VAS for the patient from whom this strain was isolated (Figure 7B). On the other hand, sample 2010#10, the CHIKV strain from the 2010 outbreak not containing the mutations mentioned earlier, exhibited a continuous but gradual increase in the footpad swelling until day 9 pi, with a slight reduction in the footpad thickness by day 12 pi; the trend was similar to that produced by strain 2016#01 from the 2016 outbreak (Figure 7A). It was observed that the VAS for both the cases associated with these strains was < 5, and was not considered significant according to the Morton and Griffiths scale31 (Figure 7B). These results suggested that although all animals developed footpad swelling, the magnitude of disease caused by 2016#01 was not comparable to that produced by CHIKV strain 2010#1 bearing the four mutations, and animals did not suffer restricted movement at any point during the study.
Figure 7.
Chikungunya virus (CHIKV) strains were used for this study, viz., CHIK#01, CHIK#10, and CHIK#16. CHIK#01 was a 2010 strain that contained the mutations E2 G55R, E2 H73Y, nsP1 G230R, and nsP3 STOP524R. CHIK#10 was a 2010 strain that did not contain these mutations. 2016#01 was 2016 strains that contained the mutation E1 I317V. (A) Change in the footpad thickness in C57BL/6J mice when infected with various CHIKV strains. Maximum footpad thickness was observed in the mice infected with strain 2010#01 starting day 6 pi and peaked at day 12 pi. Strains 2010#10 and 2016#01 showed maximum footpad thickness on day 9 pi, which reduced gradually until the termination of study (day 15 pi). (B) Evaluation of morbidity score in C57BL/6J mice when infected with CHIKV strains collected from India during CHIK outbreaks. Maximum morbidity score was recorded for strain 2010#01 (morbidity score = 9.2 ± 1.2) at day 15 pi. Two-way ANOVA was used to perform statistical analysis. P-value is indicated as asterisks (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001). This figure appears in color at www.ajtmh.org.
DISCUSSION
Our study compared the 2016 Indian CHIKF outbreak dynamics with those of the 2010 outbreak that affected the population in similar numbers, but with different clinical manifestations. Our detailed comparison of immunological responses revealed that the neutralization capacity observed in the 2010 patients was due to the development of IgG antibodies, whereas in 2016 outbreak it was due to the development of early IgM antibodies. Furthermore, sequence analysis of these strains identified distinct sets of mutations unique to each outbreak. Preliminary in vivo murine studies with representative CHIKV strains from both outbreaks correlated with our clinical observations. These findings suggest that disease dynamics of the 2010 and 2016 outbreaks were different owing to the neutralizing capacities induced by circulating viral strains that may be determined by mutations in these viruses affecting their overall pathogenicity.
Host response leads to differences in the disease progression.
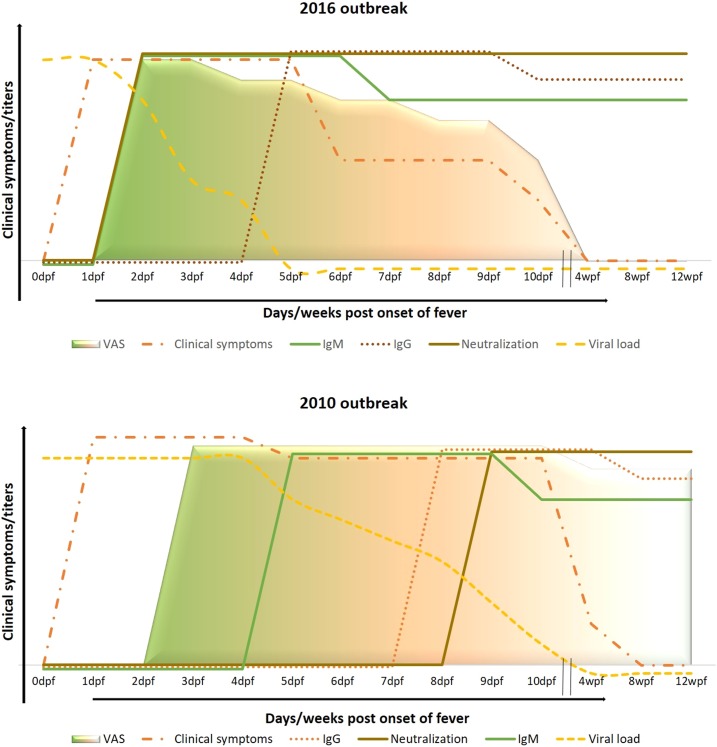
Comparison of the clinical findings between the two CHIKF outbreaks revealed differences with respect to clinical presentation, viremia, and neutralization antibody induction that correlated with disease outcome, as represented in Figure 8. One of the main differences was the manner in which the disease progressed. Patients in 2016 recovered faster than those infected during 2010, possibly because of the virus being neutralized earlier in the former. Correlations between the timing of secretion of antibody isotypes suggested that the 2010 neutralization capacity was primarily attributed to IgG, whereas in 2016, early neutralization was attributed to IgM. Although patients in 2010 and 2016 initially exhibited similar viral loads (data not shown) and suffered similar initial arthralgia, the chronic phase lasted only for a few weeks in most 2016 patients. By contrast, a longer chronic phase, lasting for more than 2 months on average, was observed in 2010 patients. Earlier reports proposed that IgG mainly contributes to the neutralization of CHIKV and determines the risk for chronic CHIKF.11,32 However, another study recently reported the presence of neutralizing IgM in CHIKF patients and demonstrated independent contributions of IgM and IgG toward neutralizing activity during the acute phase. This suggested that IgM may play an integral and complementary role in IgG-mediated neutralization in preventing chronicity.33,34 We observed that our 2016 patients developed neutralizing IgM within the first 4 days of illness. When IgM-positive and -negative samples were assayed for neutralization activity, samples with higher IgM OD values showed a significant, positive correlation with neutralization titer and a negative correlation with earlier viral load (data not shown). Furthermore, we observed that IgM not only acted in a complementary manner with the early neutralizing IgG antibodies but was also capable of neutralizing the virus as early as 2 days post-onset of fever, presumably reducing viremia and shortening the arthralgic phase of the disease.
Figure 8.
Representation of progression of CHIK disease in 2016 when compared with the 2010 CHIK outbreak. During the 2016 outbreak, most patients suffered clinical symptoms of the disease; had viral load till day 5 post-onset of fever; and exhibited the presence of IgM- and IgG-binding antibodies starting days 2 and 7 post-onset of fever, respectively; and virus neutralization was due to development of early neutralizing IgM that worked in collaboration with neutralizing IgG antibodies. On the contrary, during the 2010 CHIK outbreak, patients suffered clinical symptoms for as long as 10–12 days post-onset of fever and exhibited the presence of IgM and IgG starting days 4 and 7 post-onset of fever, respectively; and neutralization was due to the development of early neutralizing IgG antibodies. This figure appears in color at www.ajtmh.org.
Outbreak-specific mutations correlated with disease progression.
Earlier reports have stressed the role of mutations in mosquito vector transmission potential35,36 and altered pathogenesis of CHIKV.36–39 Our genomic analysis of 21 representative viruses revealed several mutations unique to each outbreak, which may have a direct impact on disease outcome. Our genomic analysis also revealed two distinct CHIKV genotypes that circulated during the 2010 outbreak. With respect to the 2010 genotype associated with high pathogenicity, substitutions E2-G55R and E2-H73Y form a part of an epitope critical for viral pathogenesis40,41 and have been implicated in hindering the binding of neutralizing IgM. Furthermore, the relevance of mutations in the E2 gene for establishing infection of the joints and possibly increasing the severity of the arthritic phase has been recently proposed.37,42 Specific substitutions in the nonstructural proteins, namely, nsP1-G230R and nsP3-STOP524R, have been reported to directly impact viral replication, collectively increasing viral pathogenicity under experimental conditions.38 These reports are consistent with our observation that the four mutations we identified from the 2010 outbreak also had a positive correlation with disease progression and establishment of sequelae. These findings could have a direct implication on disease prognosis and predictive value in patient management. Analysis of the clinical data from patients with these mutations showed high VAS (> 7, P < 0.05), associated with a chronic phase exceeding 12 months. In 2016, the average VAS of patients was 2.25 ± 2.6, as opposed to a higher overall VAS of 6.9 ± 2.92 during 2010. One mutation, E1-I317V, was present in all 2016 samples, and its relevance in disease progression warrants in-depth studies, owing to its predicted role in decreasing protein stability based on informatic analyses (data not shown).
Viruses of the two outbreaks differed in pathogenesis based on genetic makeup.
Our genomic analysis of the outbreak samples revealed that two CHIKV genotypes circulated during the 2010 outbreak (one with putative pathogenic mutations and one without) and that there were also mutations unique to the 2016 viruses. Reports have suggested that sequence variations directly impact CHIKV pathogenicity, which could be lineage-specific, genotype-specific, and strain-specific.28,43,44 Furthermore, our preliminary evaluation of the pathogenicity of these viruses in a murine model suggests that the 2010 and 2016 viruses vary in their virulence; although some strains of viruses from both outbreaks revealed similar pathogenicity, another type of virus from the 2010 outbreak exhibited higher pathogenicity, which could be attributed to these mutations, and warrants further in-depth analyses. Furthermore, evaluation of neutralization responses to infection showed development of early neutralizing antibodies in mice infected with a 2016 CHIKV strain without the putative virulence mutations, compared with the 2010 strain with the mutations, in which neutralization onset was observed as late as day 9 pi (data not shown). However, an earlier study involving immune serum from humans infected with CHIKV of either ECSA or Asian genotypes also reported differences in neutralization responses.34 In-depth studies involving more virus strains and animals and reverse genetic approaches are required to validate our findings.
LIMITATIONS AND CONCLUSION
Taken together, our study compared two major CHIKF outbreaks (2010 and 2016) in the Indian subcontinent to evaluate the changes that can occur during outbreaks, both with respect to host response and pathogenesis. Although our study has provided potential determinants for the severity of a CHIKF outbreak, its limited scope with respect to the number of cases studied and animals infected prevented more direct testing of these important hypotheses, such as the role of developing early neutralizing antibodies in disease progression and the specific impacts of each mutation in this process. Our study design also did not permit nationwide sampling, limiting our understanding of the geographic origins and patterns of spread for the mutations identified. Although some of the mutations we identified have been studied previously, reverse genetic studies are required to understand the individual or combined effects on pathogenicity and potentially on spread of the mutations found in the putative high-pathogenicity 2010 genotype. Despite these caveats, we provide further evidence that the timing of the development of neutralizing antibodies may affect pathogenesis and chronic arthralgia. Considering the major, direct impact CHIKF has on disability, this information can play an important role in understanding disease burden at a population level.
Supplemental table and figures
Acknowledgments:
We would like to thank Archita Biswas for her assistance with amino acid variation visualization via Circos. Acknowledgments are also due to Gajinder Pal Singh for his inputs in whole genome sequence analysis. We would like to thank all the patients who agreed to be a part of this study.
Note: Supplemental table and figures appear at www.ajtmh.org.
REFERENCES
- 1.Staples JE, Breiman RF, Powers AM, 2009. Chikungunya fever: an epidemiological review of a re-emerging infectious disease. Clin Infect Dis 49: 942–948. [DOI] [PubMed] [Google Scholar]
- 2.Fourie E, Morrison J, 1979. Rheumatoid arthritic syndrome after chikungunya fever. S Afr Med J 56: 130–132. [PubMed] [Google Scholar]
- 3.Gérardin P, Guernier V, Perrau J, Fianu A, Le Roux K, Grivard P, Michault A, De Lamballerie X, Flahault A, Favier F, 2008. Estimating chikungunya prevalence in La Reunion Island outbreak by serosurveys: two methods for two critical times of the epidemic. BMC Infect Dis 8: 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mavalankar D, Shastri P, Raman P, 2007. Chikungunya epidemic in India: a major public-health disaster. Lancet Infect Dis 7: 306–307. [DOI] [PubMed] [Google Scholar]
- 5.Rezza G, et al. CHIKV Study Group , 2007. Infection with chikungunya virus in Italy: an outbreak in a temperate region. Lancet 370: 1840–1846. [DOI] [PubMed] [Google Scholar]
- 6.Weaver SC, 2014. Arrival of chikungunya virus in the new world: prospects for spread and impact on public health. PLoS Negl Trop Dis 8: e2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weaver SC, Charlier C, Vasilakis N, Lecuit M, 2018. Zika, chikungunya, and other emerging vector-borne viral diseases. Annu Rev Med 69: 395–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gibney KB, Fischer M, Prince HE, Kramer LD, St George K, Kosoy OL, Laven JJ, Staples JE, 2011. Chikungunya fever in the United States: a fifteen year review of cases. Clin Infect Dis 52: e121–e126. [DOI] [PubMed] [Google Scholar]
- 9.Halstead SB, 2015. Reappearance of chikungunya, formerly called dengue, in the Americas. Emerg Infect Dis 21: 557–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yergolkar PN, Tandale BV, Arankalle VA, Sathe PS, Sudeep AB, Gandhe SS, Gokhle MD, Jacob GP, Hundekar SL, Mishra AC, 2006. Chikungunya outbreaks caused by African genotype, India. Emerg Infect Dis 12: 1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jain J, Nayak K, Tanwar N, Gaind R, Gupta B, Shastri JS, Bhatnagar RK, Kaja MK, Chandele A, Sunil S, 2017. Clinical, serological, and virological analysis of 572 chikungunya patients from 2010 to 2013 in India. Clin Infect Dis 65: 133–140. [DOI] [PubMed] [Google Scholar]
- 12.Kaur N, Jain J, Kumar A, Narang M, Zakaria MK, Marcello A, Kumar D, Gaind R, Sunil S, 2017. Chikungunya outbreak in Delhi, India, 2016: report on coinfection status and comorbid conditions in patients. New Microbes New Infect 20: 39–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shah KV, Gibbs CJ, Jr., Banerjee G, 1964. Virological investigation of the epidemic of haemorrhagic fever in Calcutta: isolation of three strains of chikungunya virus. Indian J Med Res 52: 676–683. [PubMed] [Google Scholar]
- 14.Rao TR, Carey DE, Pavri KM, 1965. Preliminary isolation and identification of chikungunya virus from cases of dengue-like illness in Madras city. Indian J Med Res 53: 689–693. [PubMed] [Google Scholar]
- 15.Rodrigues FM, Patankar MR, Banerjee K, Bhatt PN, Goverdhan MK, Pavri KM, Vittal M, 1972. Etiology of the 1965 epidemic of febrile illness in Nagpur city, Maharashtra State, India. Bull World Health Organ 46: 173–179. [PMC free article] [PubMed] [Google Scholar]
- 16.Padbidri VS, Gnaneswar TT, 1979. Epidemiological investigations of chikungunya epidemic at Barsi, Maharashtra state, India. J Hyg Epidemiol Microbiol Immunol 23: 445–451. [PubMed] [Google Scholar]
- 17.Arankalle VA, Shrivastava S, Cherian S, Gunjikar RS, Walimbe AM, Jadhav SM, Sudeep AB, Mishra AC, 2007. Genetic divergence of chikungunya viruses in India (1963–2006) with special reference to the 2005–2006 explosive epidemic. J Gen Virol 88: 1967–1976. [DOI] [PubMed] [Google Scholar]
- 18.Kumar NP, Joseph R, Kamaraj T, Jambulingam P, 2008. A226V mutation in virus during the 2007 chikungunya outbreak in Kerala, India. J Gen Virol 89: 1945–1948. [DOI] [PubMed] [Google Scholar]
- 19.Shrinet J, Jain S, Sharma A, Singh SS, Mathur K, Rana V, Bhatnagar RK, Gupta B, Gaind R, Deb M, 2012. Genetic characterization of chikungunya virus from New Delhi reveal emergence of a new molecular signature in Indian isolates. Virol J 9: 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.National-Guidelines-Clinical-Management-Chikungunya , 2016. Program DoNVBDC, New Delhi, India: Directorate General of Health Services. [Google Scholar]
- 21.Muniaraj M, 2014. Fading chikungunya fever from India: beginning of the end of another episode? Indian J Med Res 139: 468–470. [PMC free article] [PubMed] [Google Scholar]
- 22.Sahadeo N, Mohammed H, Allicock OM, Auguste AJ, Widen SG, Badal K, Pulchan K, Foster JE, Weaver SC, Carrington CV, 2015. Molecular characterisation of chikungunya virus infections in trinidad and comparison of clinical and laboratory features with dengue and other acute febrile cases. PLoS Negl Trop Dis 9: e0004199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Narasimhan V, Danecek P, Scally A, Xue Y, Tyler-Smith C, Durbin R, 2016. BCFtools/RoH: a hidden Markov model approach for detecting autozygosity from next-generation sequencing data. Bioinformatics 32: 1749–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA, 2009. Circos: an information aesthetic for comparative genomics. Genome Res 19: 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.R Core Team , 2016. R: A language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 27.Morton D, Griffiths P, 1985. Guidelines on the recognition of pain, distress and discomfort in experimental animals and an hypothesis for assessment. Vet Rec 116: 431–436. [DOI] [PubMed] [Google Scholar]
- 28.Langsjoen RM, Haller SL, Roy CJ, Vinet-Oliphant H, Bergren NA, Erasmus JH, Livengood JA, Powell TD, Weaver SC, Rossi SL, 2018. Chikungunya virus strains show lineage-specific variations in virulence and cross-protective ability in murine and nonhuman primate models. MBio 9: e02449-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dutta SK, Pal T, Saha B, Mandal S, Tripathi A, 2014. Copy number variation of chikungunya ECSA virus with disease symptoms among Indian patients. J Med Virol 86: 1386–1392. [DOI] [PubMed] [Google Scholar]
- 30.Jain J, Narayanan V, Kumar A, Shrinet J, Srivastava P, Chaturvedi S, Sunil S, 2019. Establishment and comparison of pathogenicity and related neurotropism in two age groups of immune competent mice, C57BL/6J using an Indian isolate of chikungunya virus (CHIKV). Viruses 11: 578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morton Da, Griffiths P, 1985. Guidelines on the recognition of pain, distress and discomfort in experimental animals and an hypothesis for assessment. Vet Rec 116: 431–436. [DOI] [PubMed] [Google Scholar]
- 32.Kam YW, et al. 2012. Early neutralizing IgG response to chikungunya virus in infected patients targets a dominant linear epitope on the E2 glycoprotein. EMBO Mol Med 4: 330–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chua CL, Sam IC, Chiam CW, Chan YF, 2017. The neutralizing role of IgM during early chikungunya virus infection. PLoS One 12: e0171989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chua CL, Sam IC, Merits A, Chan YF, 2016. Antigenic variation of East/Central/South African and Asian chikungunya virus genotypes in neutralization by immune sera. PLoS Negl Trop Dis 10: e0004960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Agarwal A, Sharma AK, Sukumaran D, Parida M, Dash PK, 2016. Two novel epistatic mutations (E1:K211E and E2:V264A) in structural proteins of chikungunya virus enhance fitness in Aedes aegypti. Virology 497: 59–68. [DOI] [PubMed] [Google Scholar]
- 36.Tsetsarkin KA, Vanlandingham DL, McGee CE, Higgs S, 2007. A single mutation in chikungunya virus affects vector specificity and epidemic potential. PLoS Pathog 3: e201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hawman DW, Carpentier KS, Fox JM, May NA, Sanders W, Montgomery SA, Moorman NJ, Diamond MS, Morrison TE, 2017. Mutations in the E2 glycoprotein and the 3' untranslated region enhance chikungunya virus virulence in mice. J Virol 91: e00816-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mounce BC, Cesaro T, Vlajnic L, Vidina A, Vallet T, Weger-Lucarelli J, Passoni G, Stapleford KA, Levraud JP, Vignuzzi M, 2017. Chikungunya virus overcomes polyamine depletion by mutation of nsP1 and the opal stop codon to confer enhanced replication and fitness. J Virol 91: e00344-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones JE, et al. 2017. Disruption of the opal stop codon attenuates chikungunya virus-induced arthritis and pathology. MBio 8: e01456-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin J, et al. 2015. Neutralizing monoclonal antibodies block chikungunya virus entry and release by targeting an epitope critical to viral pathogenesis. Cell Rep 13: 2553–2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fong RH, et al. 2014. Exposure of epitope residues on the outer face of the chikungunya virus envelope trimer determines antibody neutralizing efficacy. J Virol 88: 14364–14379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Selvarajah S, Sexton NR, Kahle KM, Fong RH, Mattia K-A, Gardner J, Lu K, Liss NM, Salvador B, Tucker DF, 2013. A neutralizing monoclonal antibody targeting the acid-sensitive region in chikungunya virus E2 protects from disease. PLoS Negl Trop Dis 7: e2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gorchakov R, et al. 2012. Attenuation of chikungunya virus vaccine strain 181/clone 25 is determined by two amino acid substitutions in the E2 envelope glycoprotein. J Virol 86: 6084–6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang SW, Cheng HL, Hsieh HY, Chang CL, Tsai HP, Kuo PH, Wang SM, Liu CC, Su IJ, Wang JR, 2014. Mutations in the non-structural protein region contribute to intra-genotypic evolution of enterovirus 71. J Biomed Sci 21: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.