Actin cytoskeletal asymmetry produces unbalanced hydrodynamic forces to drive spindle rotation during mouse meiotic division.
Abstract
Patterned cell divisions require a precisely oriented spindle that segregates chromosomes and determines the cytokinetic plane. In this study, we investigated how the meiotic spindle orients through an obligatory rotation during meiotic division in mouse oocytes. We show that spindle rotation occurs at the completion of chromosome segregation, whereby the separated chromosome clusters each define a cortical actomyosin domain that produces cytoplasmic streaming, resulting in hydrodynamic forces on the spindle. These forces are initially balanced but become unbalanced to drive spindle rotation. This force imbalance is associated with spontaneous symmetry breaking in the distribution of the Arp2/3 complex and myosin-II on the cortex, brought about by feedback loops comprising Ran guanosine triphosphatase signaling, Arp2/3 complex activity, and myosin-II contractility. The torque produced by the unbalanced hydrodynamic forces, coupled with a pivot point at the spindle midzone cortical contract, constitutes a unique mechanical system for meiotic spindle rotation.
INTRODUCTION
Asymmetric cell division is a widely occurring mechanism during organismal development for the production of daughter cells with different developmental fates. Studies in the past three decades have focused mainly on asymmetric divisions of mitotic cells and revealed mechanistic paradigms. Common to these processes, cell polarity, as often manifested as asymmetric cortical organization, serves to orient the mitotic spindle along the axis of distribution of cell-fate determinants, and the spindle orientation and position, in turn, determine the plane of cytokinesis. The ensuing daughter cells hereby inherit different fate determinants with a spatial relationship in accordance with the developmental body plan (1, 2). The mechanical system for mitotic spindle orientation commonly involves astral microtubules emanating from the poles of the spindle and interacting with cortical regulators of motor proteins such as cytoplasmic dynein (1, 3, 4). Asymmetric forces exerted by these motor proteins on astral microtubules both orient and position the spindle during the metaphase or anaphase of mitosis.
The development of a mammalian organism initiates with two highly asymmetric meiotic cell divisions: meiosis I and II (MI and MII) of oocytes. During each of these divisions, the cytoplasm divides unequally, giving rise to a large egg and two small polar bodies (5, 6). In contrast to mitotic cells, meiotic spindles in mammalian oocytes lack canonical centriole-containing centrosomes and are devoid of astral microtubules that could form long-distance cortical connections (7–9). Thus, how oocytes precisely orchestrate spindle positioning or orientation during the two rounds of meiotic divisions has not been fully understood. Studies in recent years have pointed to crucial roles for the actin cytoskeleton in the positioning of meiotic spindles in mouse oocytes (10–12). In MII-arrested oocytes, the metaphase II spindle is overlying by an F-actin cap, the formation of which is dependent on the Arp2/3 complex, surrounded by a ring of myosin-II (13, 14). The formation of this stereotypic actin and myosin-II organization is regulated by RanGTP emanating from the chromatin in a distance-dependent manner. In these oocytes, the cortical Arp2/3 complex generates a continuous actin flow that drives cytoplasmic streaming in a pattern leading to a net force on the spindle toward the cortical cap (13). The Arp2/3 complex also prevents contraction of the myosin-II–containing ring surrounding the cortical cap, which would generate a reverse cytoplasmic streaming that moves the spindle away from the cortex (13). Thus, the subcortical positioning of the metaphase II spindle is maintained by balanced activities of these two evolutionarily conserved actin cytoskeletal regulators.
Upon fertilization, the oocyte completes the second meiotic cell division to eliminate a set of the duplicate sister chromosomes. During this process, MII spindle must rotate from the parallel to a radial orientation relative to the cortex to complete cell division and extrude the second polar body (15–18). Abnormalities in MII spindle rotation lead to failure in this critical asymmetric cell division, resulting in embryo loss or malformation (19, 20). It was proposed that F-actin may be involved in the MII spindle rotation (21, 22), but the mechanism was unknown. In this study, we show that in mouse oocyte, rotation of the MII spindle relies on asymmetric hydrodynamic forces powered separately by the Arp2/3 complex and myosin-II. These forces are initially symmetric but become asymmetric after chromosome segregation due to the symmetry breaking in the distribution of Arp2/3 complex and myosin-II on the cortex above the spindle. The resulting asymmetric cytoplasmic flow produces a torque on the two poles of the spindle. This, coupled with a central pivot point from the centralspindlin-cortex linkage, rotates the spindle to complete the second meiotic division.
RESULTS
MII spindle rotates in random orientation relative to the sperm entry site at the end of anaphase
We first performed immunofluorescence analysis of in vitro fertilized (IVF) mouse oocytes at various time points to visualize the spindle and chromosomes (Fig. 1, A and B). In metaphase and early anaphase (before two chromatin clusters move to the opposite spindle poles), the spindle is roughly parallel to the cortex (Fig. 1A). Spindle rotation occurs after chromosomes reached the spindle poles, which is followed by second polar body (PB2) extrusion (Fig. 1A). The direction of spindle rotation is random relative to the site of sperm entry (Fig. 1B), suggesting that MII spindle rotation is likely to occur independent of the sperm chromatin. Because a mechanistic understanding of this dynamic process required high-resolution live imaging using fluorescently labeled components, which is challenging in IVF eggs due to the constant motion of the egg pushed by surrounding sperms, we used parthenogenetic egg activation to trigger anaphase onset (Fig. 1, C to H, and fig. S1). Live imaging of these eggs revealed a similar sequence of events to those in IVF eggs (comparing Fig. 1, A to C, and movie S1). After anaphase onset, a unilateral membrane furrow ingressed from the proximal cortex overlying the spindle (Fig. 1, A to D) and approached the anaphase spindle midzone, dividing the cortical domain into two equal parts that each protruded out slightly (Fig. 1D). Rotation of the anaphase II spindle started at the completion of chromosome segregation, with one spindle pole and the associated chromatin cluster remaining juxtaposed to the cortex (referred to hereafter as the outward pole), while the opposite pole moved toward the oocyte interior (referred to as the inward pole) (Fig. 1, C to H). Concurrently, the cortex overlying the outward pole protruded out as the spindle rotated, forming the polar body after cytokinesis, while the other cortical region overlying the inward pole receded and remained as a part of the egg’s plasma membrane (Fig. 1C).
Fig. 1. MII spindle rotates in random direction relative to the sperm entry site at the end of anaphase.
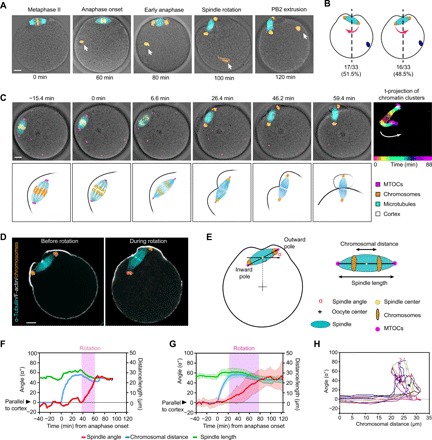
(A) The stages of MII completion after fertilization. Oocytes were fixed at different time points before (metaphase II) and after fertilization and immunostained for microtubules (α-tubulin) together with chromosomes (Hoechst). White arrows indicate the sperm chromatin. (B) Percentage of eggs that rotate in the opposite direction (left) or same direction (right) with respect to the position of sperm entrance. Position of the sperm chromatin is shown as the blue dot. Red arrows indicate the rotation direction. The quantification shows that the direction of spindle rotation is not biased to the site of sperm entry (P > 0.99, indicating no significant deviation from 50%, Fisher’s exact test). (C) Montage from time-lapse imaging of an oocyte expressing fluorescent markers: mCherry-MAP4 for microtubules (cyan), enhanced green fluorescent protein (EGFP)–CDK5RAP2 for microtubule-organizing centers (MTOCs) (magenta), and Hoechst for DNA (orange), merged with differential interference contrast (DIC) images of the oocyte. The panel on the far right shows time projection (t-projection) of sequential locations of the chromosomes that are colored as indicated in the color bar at the bottom to indicate the trajectories of two clusters of sister chromosomes during anaphase and spindle rotation. White arrow indicates the direction of spindle rotation. Time 0 corresponds to anaphase onset. The bottom row illustrates the sequence of events including chromosome segregation, spindle rotation, and polar body extrusion. (D) Immunofluorescence staining of F-actin (phalloidin), spindle (α-tubulin), and chromosomes (Hoechst) in oocytes before and during spindle rotation. (E) Schematics of parameters quantifying the spindle angle, length, and distance between chromatin clusters. (F and G) Spindle orientation, length, and the distance between chromatin clusters over time for (F) a single oocyte and (G) averaged for 21 oocytes (means ± SD) are shown. (H) Twelve example traces of spindle orientation (angle, y axis) as a function the distance of chromosome segregation (x axis). Scale bars, 10 μm (for all images).
Close tracking of spindle orientation relative to the distance of chromosome segregation by time-lapse confocal imaging shows that the angle between the MII spindle and the overlying cortex fluctuated around zero without directional bias before the decisive rotatory motion (Fig. 1H and fig. S1, A to C), which occurred at the completion of chromosome segregation and the spindle rotated on average 62° (fig. S1D).
MII spindle rotation requires Arp2/3 complex, myosin-II, and dynamic F-actin network
It was hypothesized previously that the spindle rotation in mouse oocyte is driven by an actin-dependent mechanism (21, 22). Disrupting actin polymerization by latrunculin A (Lat A) treatment completely inhibited spindle rotation (Fig. 2, A to C, and movie S2). Treatment of oocytes with jasplakinolide, an F-actin–stabilizing drug, also prevented spindle rotation (Fig. 2, A to C, and movie S2), suggesting that dynamic actin is required. Because the previous work revealed that the Arp2/3 complex–nucleated dynamic actin network and the myosin-II–based contractile network act on the metaphase II spindle in opposite ways (13), we tested how these actin regulators affect anaphase II spindle rotation. Inhibition of Arp2/3 complex activity with CK666, a small-molecule inhibitor of Arp2/3 complex (23), prevented spindle rotation, and inhibition of myosin-II motor activity with blebbistatin (24) had a similar effect (Fig. 2, A to C), suggesting that both the Arp2/3 complex and myosin-II–mediated actin assemblies are involved in MII spindle rotation. These findings were also confirmed in IVF eggs (fig. S2, A and B).
Fig. 2. MII spindle rotation requires Arp2/3 complex, myosin-II, and dynamic F-actin network.
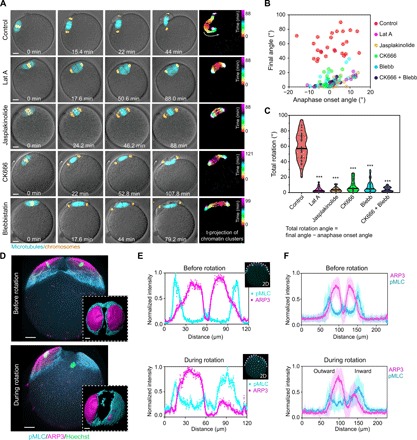
(A) Montages from live imaging showing that inhibition of actin polymerization (Lat A treatment), actin depolymerization (jasplakinolide treatment), Arp2/3 complex (CK666 treatment), or myosin-II (blebbistatin treatment) prevented spindle rotation. Time projections of chromatin clusters are shown on the right as in Fig. 1C. (B and C) Quantification of spindle angle at anaphase onset and final angle (B) of the spindle after various treatments as indicated. Total rotation angle (C) equals the final spindle angle minus that at anaphase onset. ***P < 0.001. (D) 3D projected images of immunofluorescence staining showing that ARP3 and active myosin-II [phosphorylated myosin light chain (pMLC)] changed from a symmetric distribution to an asymmetric distribution on the cortex overlying chromatin clusters during spindle rotation. Top views of 3D reconstructed ARP3 and myosin-II are shown in the bottom insets. (E) Fluorescence intensity profiles of ARP3 and pMLC in a middle optical section across the spindle proximal pole in the oocyte from (D), with colored curves displaying smoothened data. (F) Line profiles of ARP3 and pMLC fluorescence intensity from an optical section parallel to the spindle and cutting across the spindle proximal cortex in oocytes prerotation (averaged for 11 oocytes, means ± SD) and during rotation (averaged for 13 oocytes, means ± SD). Scale bars, 10 μm (for all images).
We next examined the organization of the Arp2/3 complex and active myosin-II, as marked by phosphorylated myosin light chain (pMLC) using three-dimensional (3D) immunofluorescence analysis (Fig. 2, D to F, and movie S3). In parthenogenically activated anaphase II eggs before spindle rotation, ARP3 was distributed symmetrically at two approximately equal-sized cortical caps above each chromosome cluster, and each Arp2/3 cortical cap was surrounded by a ring of myosin-II (Fig. 2, D to F). However, during spindle rotation, myosin-II became enriched in the cortical region overlying the inward pole and the associated chromatin cluster, whereas the outward pole remained in close proximity to the Arp2/3 cortical cap (Fig. 2, D to F, and movie S3). It is known that in fertilized eggs, subcortical sperm chromosomes could stimulate the formation of an actomyosin-rich cortical structure called the fertilization cone (FC). We found that FC formation occurs after MII spindle rotation (fig. S2C), consistent with a lack of effect on the direction of spindle rotation by the sperm chromosomes.
Asymmetric cortical organization of Arp2/3 complex and myosin-II correlates with asymmetric cytoplasmic streaming during spindle rotation
While observing spindle rotation, we noticed that the cytoplasm underwent stereotypic changes in the pattern of streaming. Before the start of spindle rotation, the cytoplasm flowed away from the proximal cortex and the spindle along the lateral cortex and back toward the oocyte pole proximal to the spindle at oocyte interior (Fig. 3A and movie S4). The prerotation flow pattern was roughly bilaterally symmetric, as shown with particle image velocimetry (PIV) from time-lapse recordings under the transmitted light {Fig. 3A and fig. S3, A and B; note the opposite vorticity [defined as the curl of the measured velocity field (∇ × v)/|(∇ × v∣)] associated with the right and left side of the oocyte in the plane roughly across the spindle long axis}. This flow pattern is equivalent of that generated by two clusters of chromatin, each activating a zone of Arp2/3 complex on its proximal cortex as shown in Fig. 2D. During spindle rotation, however, the cytoplasmic flow changed from the bilateral symmetry to an asymmetric pattern: Whereas the flow around the outward pole maintained a similar sign of vorticity, cytoplasmic flow around the inward pole was visibly reversed, whereby the cytoplasm flowed toward the proximal pole along the lateral cortex and toward oocyte interior on the cytoplasmic side (Fig. 3B, fig. S3, A and B, and movie S4). Blocking actin dynamics with Lat A and jasplakinolide completely attenuated the cytoplasmic streaming (fig. S3, C to F, and movie S5).
Fig. 3. Observed and model-predicted cytoplasmic streaming patterns under various conditions.
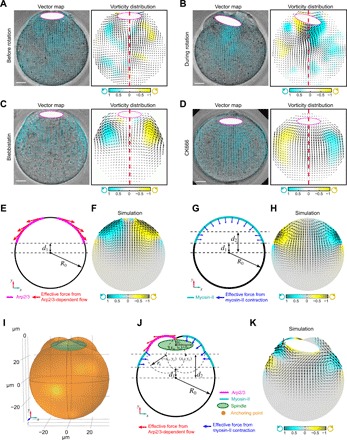
(A and B) Representative patterns of cytoplasm streaming analyzed using PIV, showing cytoplasmic flow before (A) and after spindle rotation (B). Notice that in (A), the flow pattern is roughly symmetric bilaterally as indicated by the opposite vorticity on opposite sides of the midline, but this symmetry is disrupted in (B) with regions of similar vorticity on the two sides. Left panel shows PIV map overlaid with DIC image; cyan arrows are velocity vectors. The color map in the right panel represents the normalized vorticity distribution, with cyan and yellow tones representing counterclockwise and clockwise vorticity, respectively. White ellipses with magenta outline show the positions of spindle. The flow patterns shown in (A) and (B) are time-averaged plots. The red dashed line shows the middle of the oocyte. Scale bars, 10 μm. (C and D) Representative pattern of cytoplasm flow in a myosin-II (C) (blebbistatin-treated) or Arp2/3 complex–inhibited (D) (CK666-treated) oocyte analyzed using PIV, shown similarly as described in (A) and (B). Notice that the flow patterns are bilaterally symmetric in both cases, although the vorticities were reversed between (C) and (D). (E and F) Diagram (E) of a 3D COMSOL model with cytoplasmic streaming driven solely by Arp2/3 complex–nucleated actin mimicking when myosin-II is inhibited. The xy plane is shown where z = 0. The forces generated by Arp2/3 complex are tangential to the cell boundary. Cytoplasmic flow pattern predicted from the model shown in (F). Black arrows in (F) indicate the velocity vectors of cytoplasmic flow; the color map represents the normalized vorticity distribution, with cyan and yellow tones representing counterclockwise and clockwise vorticity, respectively. (G and H) Diagram of a 3D COMSOL model with cytoplasmic streaming driven solely by myosin-II mimicking when Arp2/3 complex is inhibited (G). The xy plane is shown where z = 0. The forces generated by myosin-II are generally normal to the cell boundary. Cytoplasmic flow pattern predicted from the model shown in (H). Black arrows in (H) indicate the velocity vectors of cytoplasmic flow; the color map represents the normalized vorticity distribution, with cyan and yellow tones representing counterclockwise and clockwise vorticity, respectively. (I) A 3D view of the cytoplasmic flow-driven spindle rotation model. The geometry of the oocyte cortex during spindle rotation is modeled as the union of three sphere: one big circle centered at (0, 0, 0) with radius R0 representing the main body of the oocyte and two small spheres centered at (∓x0, y0, 0) with radius r0 representing two protrusions at the animal pole. The center of the spindle is positioned at (xs, ys), where xs = 0 at t = 0. The spindle has long semi-axis as and short semi-axis bs. The upper point of the spindle (0, ys|t = 0 + bs, 0) is anchored to the cortex as the pivot point for spindle rotation. (J) The xy plane of the 3D model showing the modeling details. The force generated by Arp2/3 complex is tangential to the cell boundary, and the force generated by myosin-II is generally normal to the cell boundary. (K) Predicted cytoplasmic flow and spindle rotation from the model shown in (I) and (J). The spindle rotates toward where Arp2/3 concentrates. Black arrows indicate the velocity vectors of cytoplasmic flow; the color map represents the normalized vorticity distribution, with cyan and yellow tones representing counterclockwise and clockwise vorticity, respectively.
Because, in MII-arrested oocytes, Arp2/3 and myosin-II generate cytoplasmic streaming in opposite patterns, we analyzed the individual contributions of the actin networks that they represent by inhibiting one or the other. Treatment with blebbistatin did not have any obvious effect on Arp2/3 complex distribution (fig. S3G) and resulted in a flow pattern reminiscent of the prerotational pattern (Fig. 3C and movie S6), suggesting that the latter is generated mostly by the Arp2/3 complex. Inactivation of the Arp2/3 complex by CK666 caused expanded myosin-II domains in anaphase II oocytes (fig. S3H) and reversed streaming on both sides of the oocyte [Fig. 3D, compared to Fig. 3, A (left) and B; and movie S6], as well as reminiscent of the observation in CK666-treated, metaphase II oocytes (13). The cytoplasmic flow is markedly reduced and lost directionality after inactivation of both Arp2/3 complex and myosin-II (fig. S3, E and F), suggesting that patterned cytoplasmic movements are driven mostly by these two actin cytoskeletal systems. Combining the observed opposite flow patterns associated with Arp2/3 complex and myosin-II and the asymmetric distribution of Arp2/3 complex and myosin-II during anaphase spindle rotation, we hypothesize that rotation of the anaphase II spindle results from asymmetric hydrodynamic forces produced by activities of the Arp2/3 complex and myosin-II.
Modeling hydrodynamic forces on the spindle before and during spindle rotation
We developed a 3D fluid-structure interaction model that investigates the relationship between the observed cortical distribution of Arp2/3 complex and myosin-II, cytoplasmic streaming pattern, and forces on the spindle (Fig. 3, E to K). Cytoplasmic streaming was modeled as linear Stoke’s flow, in which the resulting flow pattern and the prescribed boundary force have a one-to-one correspondence. We first analyzed the flow pattern due to only Arp2/3 complex or myosin-II. Assuming that continuous actin nucleation by Arp2/3 complex leads to actin treadmilling along the cortex of the oocyte (13), we anticipated that when the oocyte is treated with blebbistatin, the boundary force due to the presence of the Arp2/3 complex is mainly along the tangential direction, providing an effective shear stress on the cell boundary (Fig. 3E). With this implementation, the model predicted a symmetric rotational flow pattern in the same directions as observed (compare Fig. 3F to Fig. 3, A and C). On the other hand, myosin-II provides contraction of the cortex that acts on the cytoplasmic fluid as an elevated pressure term, as suggested by the Young-Laplace law (25). For this reason, we anticipated that when the oocyte is treated with CK666, the boundary force due to the presence of myosin-II is mainly along the normal direction, providing an effective compressive stress on the cell boundary (Fig. 3G). With this implementation, the model predicted a symmetric rotational flow pattern in the same direction as those observed in the experiments (compare Fig. 3H with Fig. 3D) but in the opposite direction from that produced by the Arp2/3 complex (compare Fig. 3H with Fig. 3C). In neither case, the spindle was predicted not to rotate due to the symmetric flow pattern and, thus, a balanced hydrodynamic force on the spindle.
We next examined the flow pattern generated by an asymmetric distribution of Arp2/3 complex and myosin-II and the resulting dynamics of the spindle in untreated oocytes (Fig. 3, I to K). On the basis of the observed distribution of Arp2/3 complex and myosin-II during spindle rotation (Fig. 2, D to F), we applied higher normal stress coming from myosin-II on the side of the oocyte with enriched myosin-II localization (right side in Fig. 3J) and maintained tangential stress coming from Arp2/3 on the other side (left side of Fig. 3J). With this asymmetric boundary force implementation, we observed a downward flow on the right side of the spindle and an upward flow on the left side (Fig. 3K and movie S7). The combined asymmetric flow pattern (Fig. 3K) resembles that in untreated oocytes during spindle rotation (Fig. 3B). Cytoplasmic flows in opposite directions at the two ends of the spindle provide a net torque on the spindle causing it to rotate toward the cortical side enriched for the Arp2/3 complex (Fig. 3, J and K), as observed experimentally (Fig. 2, D to F).
Nested feedback loops bring about symmetry breaking in the distribution of cortical Arp2/3 complex and myosin-II
A key question was how the hydrodynamic system transitions from a symmetric (prerotation) to an asymmetric organization (during rotation). This transition is correlated with symmetry breaking in the distribution of the Arp2/3 complex relative to myosin-II on the cortex (Fig. 2D). Feedback loops are known to be instrumental to systems that can spontaneously break symmetry (26, 27). On the basis of the known players and their interactions in the system, we envisioned that two feedback loops could exist and contribute to the emergence of spatial asymmetry between cortical Arp2/3 complex and myosin-II during spindle rotation (Fig. 4A). The first is a positive feedback loop between RanGTP-induced activation of cortical Arp2/3 complex and increasing interaction of the RanGTP gradient with the cortex as the associated chromatin cluster moves toward the cortex. The second is a double-negative feedback loop based on the proposed mutual inhibition of Arp2/3 and myosin-II–mediated actin assemblies that helps determine the front-back polarity of migratory cells (28, 29).
Fig. 4. Evidence for two feedback loops that could drive symmetry breaking during spindle rotation.
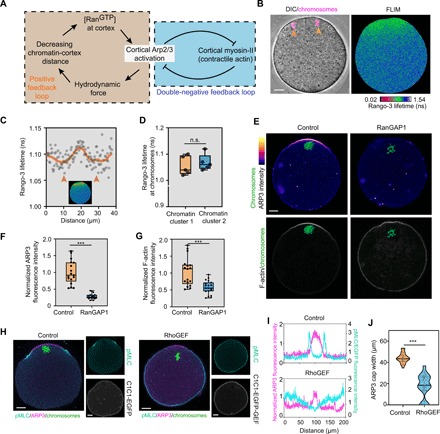
(A) Schematic diagram showing feedback loops between RanGTP gradient and Arp2/3 complex and between Arp2/3 complex and myosin-II that may drive symmetry breaking during spindle rotation. (B and C) Representative images from fluorescence lifetime imaging microscopy (FLIM) imaging of live oocytes (B) and line profiles across chromatin clusters (arrowheads) (C) showing the presence of a RanGTP gradient around each of the chromatin clusters. RanGTP gradient was detected by FLIM/fluorescence resonance energy transfer (FRET) imaging using the Rango-3 biosensor for RanGTP-released importin β cargos. Note that lower Rango-3 lifetime indicates higher RanGTP. (D) Violin plots showing quantification of the Rango-3 lifetime around each chromatin clusters. n.s., not significant. For detailed data processing and quantification, see the Supplementary Materials. (E) Representative images showing that RanGAP1 (Ran guanosine triphosphatase activating protein 1) overexpression disrupted the cortical localization of ARP3 and the actin cap. (F and G) Box plots showing quantification of averaged cortical ARP3 (F) and actin cap (G) intensities in RanGAP1 overexpressing and control oocytes. ***P < 0.001. (H) Immunofluorescence staining showing that artificial cortical myosin-II activation by cortical RhoGEF (Rho guanine nucleotide exchange factor) (C1C1-EGFP-GEF) reduced the size of the cortical Arp2/3 cap compared with oocytes expressing the control (C1C1-EGFP). (I) Fluorescence intensity profiles of ARP3 (magenta lines) and pMLC/EGFP (cyan lines) in the oocytes from (H). (J) Box plots showing quantification of cortical ARP3 cap width in C1C1-EGFP-GEF overexpression and control oocytes. ***P < 0.001. Scale bars, 10 μm (for all images).
To gain further evidence for these feedback loops, we first examined the existence of RanGTP gradient in anaphase II oocytes using a biosensor, Rango-3, which has a high fluorescence resonance energy transfer (FRET) signal when it is liberated from importin β by RanGTP (Fig. 4, B to D, and fig. S4, A and B) (30, 31). As expected, a gradient corresponding to RanGTP activity could be observed symmetrically around each chromatin cluster (Fig. 4, B to D); the half-decay distance was on an average of 9.3 μm (fig. S4B). Given that the difference of the pole-to-cortex distance changes roughly this amount during rotation, spindle rotation is expected to lead to a considerable difference of RanGTP concentration at the cortex above the two spindle poles. We also confirmed that inhibition of RanGTP activity by RanGAP1 (Ran guanosine triphosphatase activating protein 1) overexpression disrupted the ARP3 cortical localization and the actin cap in MII oocytes (Fig. 4, E to G).
To test the mutual inhibitory relationship between the Arp2/3 complex and myosin-II in oocytes, we first performed experiments to enhance cortical myosin-II activation in MII oocytes. To this end, we fused the RhoGEF (Rho guanine nucleotide exchange factor) domain of LARG (Leukemia-associated RhoGEF) (32), a RhoA guanine nucleotide exchange factor, to two tandem C1 domain that binds plasma membrane (33, 34), which would allow activation of myosin-II through RhoA-ROCK (Rho-associated protein kinase)–myosin-II pathway specifically at the oocyte cortex (Fig. 4, H and I, and fig. S4C). Targeting the RhoGEF to the plasma membrane led to active myosin-II localizing broadly on the cortex and a reduced size of the Arp2/3 cortical cap (Fig. 4, I and J). To examine whether the Arp2/3 complex could inhibit myosin-II cortical activation, we targeted the WCA (WH2-Connector-Acidic) domain of Wave2, a nucleating promoting factor that activates Arp2/3 (35, 36), to plasma membrane via the CAAX prenylation motif. This construct was introduced into germinal vesicle oocytes where active myosin-II localizes to the entire oocyte cortex. This experiment showed that targeting WCA domain to the plasma membrane reduced active myosin-II on the cortex (fig. S4, D and E).
To investigate the impact of the two feedback loops on symmetry breaking of cortical Arp2/3 complex and myosin-II during spindle rotation, we developed a dynamic model for cortical concentrations of Arp2/3 complex (a) and myosin-II (m)
where D values are the diffusion coefficients on the membrane; kaf and kmf are the off rates of Arp2/3 complex and myosin-II, respectively; kar is the actin nucleation rate of Arp2/3 complex activated by the Ran signal Ran(x,t), which changes in space and time as the RanGTP gradient interacts with the cortex (Fig. 5A); kmo is the binding rate of myosin-II from the cytoplasmic pool, mc, assumed to be a constant; and kam is the inhibitory coefficient between Arp2/3 complex and myosin-II. Simulations show that this model exhibits symmetry breaking in the cortical distribution of Arp2/3 complex and myosin-II that was consistent with experimental observations (Fig. 5B, compared to Fig. 2E, and movie S8). The resulting peaks of the Arp2/3 complex and myosin-II above the two opposite spindle poles lead to the production of a torque that rotates the spindle. As long as the positive feedback mechanism is implemented, symmetry breaking and, thus, spindle rotation are robust predictions of the model (fig. S5; for detailed model description, simulations, and parameter analysis, see the Supplementary Materials).
Fig. 5. Modeling symmetry breaking of the cortical organization of Arp2/3 complex and myosin-II.
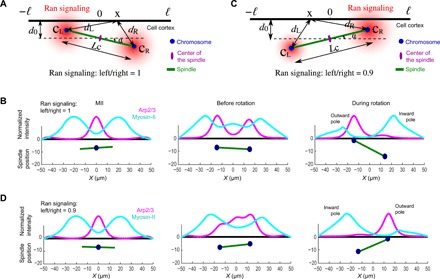
(A) Diagram of the symmetry breaking model. The cell cortex around the spindle proximal pole is approximated as a 1D line space across −ℓ ≤ x ≤ ℓ, where x = 0 align with the center of the spindle in the vertical direction. During spindle rotation, the distance from the center of the spindle to the cortex, d0, is a constant. The distance between the separating chromosomal clusters, Lc(t), is given on the basis of the observed rate of chromosome separation rate. The RanGTP gradient is assumed to decay exponentially from the center of each set of chromosome. (B) Model simulations showing three stages of Arp2/3 and myosin-II distributions: (left) Before chromosome separation, Arp2/3 has a peak in the middle, and myosin-II distributes outside of the Arp2/3 complex peak; (middle) after chromosome separation and before spindle rotation, Arp2/3 complex has two symmetric peaks above the two sets of chromosomes, and myosin-II distributes outside of the Arp2/3 peaks; (right) during spindle rotation, Arp2/3 complex peaks above the chromosomal cluster close to the cortex (outward pole), and myosin-II peaks where the chromosome is away from the cortex (inward pole). (C) Diagram of the symmetry-breaking model when Ran signaling at the left side of the chromosome is partially inhibited to 90%. The rest of the setup is identical to (A). Notice that the side of lower RanGTP was predicted to rotate away from the cortex. (D) Arp2/3 and myosin-II distributions when Ran signal at the left side of the chromosome is partially inhibited. After chromosomes segregation, the distribution of Arp2/3 is concentrated on the right side of the cortex, leading to counterclockwise spindle rotation.
Optogenetic control of RanGTP signaling or Arp2/3 complex activation biases the direction of spindle rotation
A key prediction of our model is that biasing the level of RanGTP signal or Arp2/3 complex activation would bias the direction of spindle rotation (Fig. 5, C and D, and movie S9). To test this, we used an optogenetic system involving the light-induced interaction between improved light-induced dimer (iLID) and stringent starvation protein B (SspB) (37) to spatially manipulate the RanGTP gradient. RanGAP1 protein was fused to SspB, whereas iLID was fused with histone 2B (H2B) and, thus, targeted to the chromatin (Fig. 6A and fig. S6A). We first used fluorescence staining to confirm that optogenetic chromosome accumulation of RanGAP1 led to a decrease in ARP3 and F-actin in the cortical cap in MII oocytes (fig. S6, B to E). We then performed light-induced recruitment of RanGAP1 to one of the chromatin clusters in anaphase II oocytes, which resulted in this pole of the spindle to rotate inwardly in 17 of 19 oocytes, compared with 8 of 19 in control experiments where only mCherry-SspB was recruited to the chromatin (Fig. 6, B and C, fig. S6F, and movie S10).
Fig. 6. Biasing spindle rotation through optogenetic manipulation of RanGTP gradient and Arp2/3 activation.
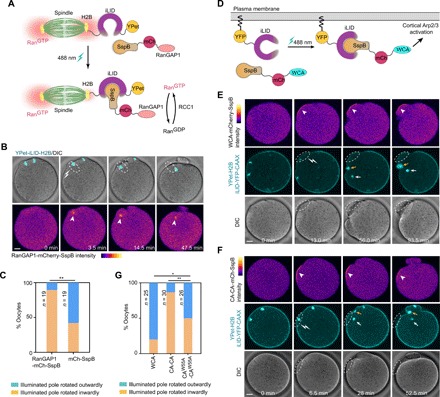
(A) Schematic depiction of optogenetic targeting of RanGAP1 to chromosomes via iLID-SspB dimerization. (B) Live imaging show optogenetic recruitment of RanGAP1 (arrowheads) to one of the chromatin clusters (dashed spheres) induced this spindle pole to rotate inward. (C) Quantification of the direction of spindle rotation after recruiting RanGAP1-mCherry-SspB or mCherry-SspB (control) to one chromosomal cluster. **P < 0.01 (Fisher’s exact test). (D) Schematic depiction of optogenetic targeting of WCA-mCherry-SspB to the plasma membrane. (E) Live imaging show optogenetic recruitment of WCA domain (white arrowheads) to the cortical region (white dashed spheres) led this side of the spindle to rotate outwardly. Oocytes were injected with WCA-mCherry-SspB, iLID-YFP-CAAX, and YPet-H2B (to label chromosomes). Note that in this sequence, the outward pole, circled in white dotted lines, became invisible due to moving out of the focal plane, while the inward pole is pointed by the white arrow. The cyan cluster pointed by the orange arrow resulted from the iLID-YFP-CAAX signal at furrow membrane. In addition, see movie S11. (F) Recruitment of CA-CA domain (arrowheads) by illumination with a 488-nm laser to a cortical region (white dashed spheres) led this side of the spindle and chromatin cluster (white arrows) to move inwardly. Oocytes were injected with CA-CA–mCherry-SspB, iLiD-YFP-CAAX, and YPet-H2B (to label chromosomes). Note that the cyan cluster pointed with yellow arrows resulted from the furrow membrane labeled with iLID-YFP-CAAX. In addition, see movie S12. (G) Quantification of spindle rotation direction after optogenetic membrane targeting of WCA-mCherry-SspB, CA-CA–mCherry-SspB, or CAW55A-CAW55A–mCherry-SspB. *P < 0.05, **P < 0.01 (Fisher’s exact test). Scale bars, 10 μm (for all images).
We also fused WCA with SspB and tethered iLID to the plasma membrane via the CAAX prenylation motif (Fig. 6D). Light-induced recruitment of WCA to the cortical region adjacent to a chromatin cluster led this side of the spindle to move outwardly in 20 of 25 oocytes (Fig. 6, E and G, and movie S11). By contrast, recruiting the dominant-negative Arp2/3 complex–inhibiting construct CA-CA (13, 35, 38, 39) to the cortical region near a chromatin cluster led this side to move inwardly in 26 of 30 oocytes, whereas recruitment of the W55A mutant (CAW55A-CAW55A, negative control), which does not bind Arp2/3 complex (13, 38, 39), did not bias the direction of spindle rotation (Fig. 6, F and G, fig. S6F, and movie S12).
Spindle midzone cortex interaction provides the pivot crucial for spindle rotation
The second prediction of the model was that, in the absence of an anchor/pivot point, asymmetric hydrodynamic forces would only cause the spindle to move along the cortex rather than to rotate (Fig. 7, A and B, fig. S7A, and movie S13). We reasoned that this pivot point is likely to be provided by the ingressed furrow from the proximal cortex that anchors the spindle midzone (Fig. 1, A and D). In mitotic cells, the centralspindlin subunit CYK4 (also known as RACGAP1 or MgcRacGAP) links the central spindle to the plasma membrane during cytokinesis (33). Immunostaining showed that CYK4 accumulated at the spindle midzone and the contacting furrow membrane after anaphase onset (Fig. 7, C and D, and fig. S7B). The same localization was also observed for enhanced green fluorescent protein (EGFP)–tagged CYK4 expressed through mRNA injection (fig. S7C).
Fig. 7. Spindle midzone cortex interaction determines spindle rotation.
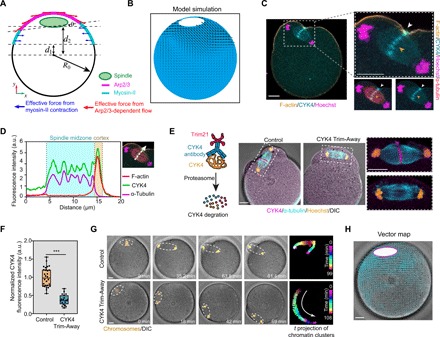
(A) Diagram of a 3D COMSOL model of an oocyte without pivot point. The xy plane is shown where z = 0. The spindle is not anchored at the top and is free to move with the cytoplasmic flow. The forces generated by Arp2/3 complex are tangential to the cell boundary, and the forces generated by myosin-II are generally normal to the cell boundary. (B) Simulated flow pattern and the motion of the spindle contributed by Arp2/3 complex and myosin-II–generated flow without the spindle pivot point. The spindle drifts away from the center toward the side where Arp2/3 is dominant. The color map shows the strength of vorticity. Arrows indicate the velocity vectors of cytoplasmic flow. (C) Representative image of immunofluorescence staining showing that CYK4 accumulated at the spindle midzone (orange arrowhead) and the contacting furrow membrane (white arrowheads). (D) Line scans of CYK4 immunofluorescence intensity across the spindle midzone and the contacting furrow membrane. (E) Immunofluorescence staining showing reduced endogenous CYK4 at the spindle midzone and the contacting furrow membrane after CYK4 Trim-Away. Schematics of CYK4 Trim-Away experiments are shown in the left panel. (F) Box plots showing quantification of CYK4 fluorescence signal in the spindle midzone. ***P < 0.001. a.u., arbitrary units. (G) Montage of time-lapse imaging showing anaphase spindle (outlined with white dash) moved laterally along the cortex (arrow) after CYK4 Trim-Away. Oocytes were injected with mCherry-Trim21 mRNA and anti-CYK4 antibody (CYK4 Trim-Away) or control immunoglobulin G (IgG) antibody (control). Chromosomes were stained with Hoechst (orange). Time projections of chromatin clusters are shown on the right as in Fig. 1C. Scale bars, 10 μm. (H) Characterization of cytoplasm flow in the CYK4 Trim-Away oocyte shown in (G) analyzed using PIV. Arrows are velocity vectors. White ellipsis with magenta outline shows the positions of spindle. Note that the image is rotated 90° to the right for the better comparison with the model simulation. Scale bars, 10 μm (for all images).
To test the importance of this connection acting as the pivot point required for spindle rotation, we used the recently developed “Trim-Away” method (40) to acutely deplete endogenous CYK4 by microinjecting Trim21 (Tripartite motif containing-21) mRNA and CYK4 antibody into MII oocytes (Fig. 7, E and F, and fig. S7D). After activation of these oocytes, CYK4 was no longer observed at the spindle midzone, which was detached from the cortex, and the membrane furrow did not form (Fig. 7E). Consistent with the model prediction, the anaphase spindle moved laterally along the cortex without rotation in the absence of the pivot point formed by the spindle midzone cortex contact (Fig. 7G and movie S14), and the flow pattern was qualitatively consistent with that predicted by the model (Fig. 7H, compared to Fig. 7B). In some oocytes, the anaphase spindle collapsed, and the segregated chromatin clusters recongressed. In these oocytes, the spindle either did not move or ceased the lateral movement, and the chromatin clusters induced a single cortical protrusion (fig. S7E). Confirming the specificity of CYK4 Trim-Away, CYK4-EGFP expression via mRNA injection rescued furrow formation, spindle rotation, and PB2 extrusion (fig. S7, F and G, and movie S14).
DISCUSSION
The results revealed above have revealed a mechanism of spindle rotation during oriented cell division that is distinct from all other known examples that operate in mitotic cells or meiotic cells of nonmammalian organisms. Perhaps, because of the lack of astral microtubules that directly connect spindle poles with cortical anchors, spindle rotation in mouse eggs relies on hydrodynamic forces due to asymmetric cytoplasmic streaming powered by two actin-based cytoskeletal systems involving the Arp2/3 complex and myosin-II. To be able to produce the torque through these forces, the force distribution along the spindle must be asymmetric. Chromosome segregation first distributes the Ran signaling centers to the two poles of the spindle. Symmetry breaking then occurs through a signaling and cytoskeletal network that comprises at least two feedback loops involving RanGTP, Arp2/3 complex, and myosin-II. Our model shows that these feedback loops are sufficient to cause spontaneous polarization of the two actin cytoskeletal domains. The resulting asymmetric cytoplasmic streaming pattern produces opposite forces on the two poles of the spindle. This, together with a central pivot point from the centralspindlin-cortex linkage, brings about spindle rotation as an exquisite example of self-organizing biological mechanics.
A key finding of our study is that an asymmetric organization of Arp2/3 complex and myosin-II on the cortex above the anaphase II spindle orchestrates the hydrodynamic forces that drive spindle rotation. Most of the evidence was obtained by live imaging of parthenogenically activated eggs, raising a potential question of whether, in fertilized eggs, the sperm chromatin could interfere with the cytoplasmic streaming due to its ability to induce an actin cap (FC) (22, 41). However, we found that the sperm-induced actin cap became apparent only after the PB2 extrusion. A possible explanation is that the mature mammalian sperm chromatin is highly compacted with the sperm-specific histone protamine (42–44). After entry into the egg cytoplasm, the paternal chromosomes are remodeled into the decondensed chromatin by replacing protamines with maternal histones (42, 45, 46), which are required for the recruitment of the RanGEF regulator of chromosome condensation 1 (RCC1) to establish the RanGTP gradient (31, 47). As the chromatin-associated RanGTP gradient is required to induce cortical Arp2/3 activation and the actin cap, the time it takes for the repackaging of sperm chromatin is likely to explain the delay in FC formation relative to spindle rotation. Thus, the sperm chromatin is unlikely to interfere with our proposed mechanism for anaphase II spindle rotation, which is also consistent with the observation that the sperm entry site does not bias the direction of spindle rotation.
We have also considered an alternative model in which a spindle pole may be physically tethered or pulled by a connection with the extruding cortex, while the midzone of the spindle is pushed toward oocyte interior from cortical contraction preceding cytokinesis. However, this model is not supported by our experiment disrupting the spindle midzone anchor point by Trim-Away of CYK4, as the remaining presence of a physical tether at one of the spindle poles would not predict the spindle to “swim” along the cortex. By contrast, this behavior of the spindle is generated by our model-predicted hydrodynamic force due to the asymmetric flow pattern in the absence of the anchor point. Another question is whether cytoplasmic streaming could be a result rather than the cause of spindle rotation. Observations arguing against this scenario are that in CK666- or blebbistatin-treated oocytes, while spindle rotation failed, cytoplasmic streaming continued, although it reversed in direction in the case of CK666 treatment. In addition, we have further tested in our 3D fluid-structure interaction model the flow pattern if the spindle rotation is driven by an external force. The predicted flow pattern caused by spindle rotation was different from that observed experimentally (fig. S8). We therefore conclude that the observed flow pattern was not a result of spindle rotation.
Our experiment using optogenetics to bias the direction of spindle rotation by modulating the RanGTP level at one of the two chromosome clusters also predicts that the dose of DNA could affect the direction of spindle rotation. Thus, if the two DNA masses are not equivalent at the two anaphase spindle poles, then the larger mass is expected to accumulate more RanGTP and activate more Arp2/3 complex on the proximal cortex, leading to its rotation outwardly. To this end, we previously reported experiments in which DNA beads or sperm chromatin was used to induce spindle formation in MII oocytes (48). This sperm chromatin spindle or DNA bead spindle underwent an anaphase whereby the whole sperm chromatin mass or all DNA beads moved together to one of the spindle poles. Consistent with our model prediction, the spindle pole with DNA beads or sperm chromatin was found to be expelled into the ectopic polar body in most cases (48).
Last, our model assumed two possible feedback loops, one between the RanGTP gradient and Arp2/3 complex activation and the other a mutual inhibitory interaction between the Arp2/3 protrusive actin network and the myosin-II contractile network. While our experimental evidence, as well as findings reported in previous studies in oocytes or other systems (12, 13, 28, 29, 36), supports the existence of these feedback loops and the model shows that these feedback loops are sufficient for symmetry breaking under reasonable ranges of parameter, we cannot rule out the presence of other feedback loops, such as a possible effect of asymmetric cytoplasmic streaming to concentrate other actin cytoskeletal regulators (49, 50). Much remains to be learned in future study about the molecular pathways through which RanGTP activates the Arp2/3 complex and the potentially multifaceted relationships between the actin network regulated by Arp2/3 complex and that controlled by myosin-II.
MATERIALS AND METHODS
Mouse oocyte collection and culture
All animals used in this research were handled in accordance with guidelines defined by the Institutional Animal Care and Use Committee of the Johns Hopkins University School of Medicine. CF-1 female mice were superovulated by injection of pregnant mare serum gonadotropin, followed 48 hours later by injection of human chorionic gonadotropin. Fourteen hours later, MII-arrested eggs were collected from oviducts. Resumption of MII was induced by IVF performed in human tubal fluid medium (Sigma-Aldrich) or activating with 10 mM SrCl2 in M16 medium containing 2 mM EGTA as described previously (51).
Microinjection and confocal microscopy
Capped mRNA was synthesized from linearized plasmid templates using the T7 or T3 mMESSAGE mMACHINE Transcription Kit (Ambion), polyadenylated by using a Poly(A) tailing kit (Ambion) and purified with the RNeasy MinElute Cleanup Kit (QIAGEN). Approximately 5 to 10 pl of mRNAs was microinjected into the cytoplasm of oocytes using a micromanipulator (IM-300 microinjector, Narishige). After injection, the oocytes were cultured at 37°C with 5% CO2 in M16 medium for at least 3 hours to allow protein expression. Oocytes were microinjected with the following RNAs: mCherry-Trim21, YPet-H2B, H2B-mCherry, and yellow fluorescent protein (YFP)–CAAX at 500 ng/μl; mEGFP-MAP4, mCherry-MAP4, and mEGFP-CDK5RAP2 at 800 ng/μl; CYK4-EGFP, Lifeact-mCherry, WCA-mCherry-SspB, CA-CA–mCherry-SspB, and CAW55A-CAW55A–mCherry-SspB at 200 ng/μl; mCherry-SspB and RanGAP1-mCherry-SspB at 150 ng/μl; iLID-YFP-CAAX at 600 ng/μl; RanGAP1-mCherry and C1C1-EGFP-GEF at 2000 ng/μl; YPet-iLID-H2B at 1000 ng/μl; and Rango-3 at 800 ng/μl.
For live oocyte imaging, images were acquired with a Zeiss LSM780 microscope using a 40× C-Apochromat 1.2–numerical aperture water-immersion objective and a Pecon XL multi S1 incubator box, which maintained oocytes at 37°C with 5% CO2. Typically, a region of 27 μm (nine confocal z sections every 3 μm) surrounding the chromosomes signal in the z dimension was imaged for a total of ~3 hours following oocyte activation. For fixed oocytes, images were acquired using the Zeiss LSM800 or LSM880 microscopes and processed after acquisition using ZEN. In some images, shot noise was reduced with a Gaussian filter.
Drug treatments
For drug treatment experiments, the IVF or activation medium was supplemented with 30 μM Lat A (Cayman Chemical), 5 μM jasplakinolide (Sigma-Aldrich), 100 μM CK666 (Sigma-Aldrich), or 100 μM blebbistatin (Sigma-Aldrich). A corresponding amount of dimethyl sulfoxide (the solvent of the chemicals) was used in control groups.
Immunofluorescence
For immunofluorescence staining, the oocytes were fixed for 30 min in 4% paraformaldehyde in phosphate-buffered saline (PBS) and then transferred to a membrane permeabilization solution (0.5% Triton X-100) for 20 min. After blocking with 2% bovine serum albumin in PBS, the oocytes were stained with the first antibody overnight and then subjected to a secondary antibody for 1 hour. Primary antibodies used were rat anti–α-tubulin (MCA77G, Bio-Rad; 1:1000), mouse anti–α-tubulin–fluorescein isothiocyanate (F2168, Sigma-Aldrich; 1:500), mouse anti-ARP3 (ab49671, Abcam; 1:100), rabbit anti–phospho-MLC2 (Ser19, Cell Signaling Technology; 1:100), and mouse anti-CYK4 (sc-271110, Santa Cruz Biotechnology; 1:50). For secondary antibodies, Alexa Fluor 488–labeled anti-rat (Invitrogen), Alexa Fluor 488–labeled anti-mouse (Invitrogen), Alexa Fluor 488–labeled anti-rabbit (Sigma-Aldrich), Alexa Fluor Plus 647–labeled anti-mouse (Invitrogen), and DyLight 550–labeled anti-rabbit (Thermo Fisher Scientific) antibodies were used. F-actin was stained with rhodamine phalloidin (Invitrogen). DNA was stained with Hoechst 33342 (10 μg/ml).
Optogenetics experiments
For the optogenetic recruitment of RanGAP1 to the chromatin clusters, MII oocytes were injected with YPet-iLID-H2B (1000 ng/μl) and RanGAP1-mCherry-SspB (150 ng/μl) or mCherry-SspB (150 ng/μl; control). For the optogenetic recruitment of WCA domain to the cortical region, MII oocytes were injected with iLID-YFP-CAAX (600 ng/μl) and WCA-mCherry-SspB (200 ng/μl). For the optogenetic inhibition Arp2/3 complex, oocytes were injected with iLID-YFP-CAAX (600 ng/μl) and CA-CA–mCherry-SspB (200 ng/μl). For negative controls, oocytes were injected with iLID-YFP-CAAX (600 ng/μl) and CAW55A-CAW55A–mCherry-SspB (200 ng/μl). Using the bleaching function of the LSM780 confocal microscope (Zeiss), oocytes were stimulated by 488-nm light at 10% of laser output to induce interaction between iLID and SspB. The 514-nm and 564-nm lasers were used to image YFP/YPet and mCherry, respectively.
For the immunostaining of ARP3, we used a custom-made blue light light-emitting diode illuminator (provided from T. Inoue’s group) for stimulation. Oocytes were injected with RanGAP1-mCherry-SspB and YPet-iLID-H2B, incubated for 3 to 4 hours, and stimulated by the illuminator in the incubator for 30 min. Oocytes were then fixed and immunostained as described above.
CYK4 Trim-Away
To induce the degradation of CYK4 by the Trim-Away, mouse MII oocytes were coinjected with mCherry-Trim21 mRNA (final concentration of 500 ng/μl) and CYK4 antibody (final concentration of 100 ng/μl) 3 to 4 hours before imaging. For the control experiment, normal mouse immunoglobulin G (IgG) antibody (final concentration of 100 ng/μl; sc-2025, Santa Cruz Biotechnology) was used instead of CYK4 antibody. For the rescue experiment, CYK4-EGFP mRNA (final concentration of 200 ng/μl) was coinjected with mCherry-Trim21 mRNA (final concentration of 500 ng/μl) and CYK4 antibody (final concentration of 100 ng/μl) into mouse oocytes.
Image analysis
3D visualizations and reconstructions of oocytes were performed using Imaris software (Bitplane AG). The fluorescence intensity profiles for ARP3 and pMLC were measured in ImageJ using a line width of 15 pixels along the oocyte cortex in a 2D plane. For CYK4 Trim-Away oocytes, we used a line width of 25 pixels along the spindle midzone and measured the mean intensity. To generate kymographs of Lifeact-mCherry dynamics, a region of interest (ROI) was defined along the cortical cap domain in a 2D plane and straightened using ImageJ. Time projection of chromatin clusters was performed with the Temporal-Color Code plugin on ImageJ.
For tracking spindle orientation, spindle length, and chromosomal distance, the movements of microtubule-organizing centers (MTOCs) and chromosomes were quantified through image processing in Mathematica. For the position of chromosomes, the fluorescence images of chromosomes were applied with a Gaussian filter to remove noise, the resultant images were then binarized using a threshold, and some small debris were removed through DeleteSmallComponents algorithm. The centroids of the components left in the binarized images (mostly only chromosomes) were obtained by ComponentsMeasurements algorithm. For the positions of spindle pole, fluorescence images of MTOCs were identified automatically by applying Gaussian filter first and then binarizing the image. In some frames, the positions of spindle pole MTOCs were identified manually if the disturbing noise was too high and could not be identified automatically. To extract the oocyte outline, the fluorescence images of the membrane (labeled with YFP-CAAX) or background from the spindle staining channel were used. After applying Gaussian filter to remove noise, as well as binarizing, the resultant images were subjected to FillingTransform algorithm to avoid holes, and the Erosion and DeleteSmallComponents algorithms were applied to remove small branches and debris. The outline of oocyte was then obtained by EdgeDetect algorithm. The lines connecting oocyte center with MTOCs were extended to cross with oocyte outline to locate the closest points on the oocyte outline to spindle pole MTOCs, and then, the lengths from these points to spindle pole MTOCs are considered as the distance from spindle pole to oocyte shell.
Fluorescence lifetime imaging microscopy data processing and quantification
Confocal fluorescence lifetime imaging microscopy (FLIM) of oocytes expressing the Rango-3 was performed with the use of Zeiss LSM780 microscope equipped with the PicoQuant PicoHarp 300 time-correlated single-photon counting module, two hybrid PMA-04 detectors, and Sepia II laser control module. During imaging, cells were maintained in Pecon environmental chamber at 37°C with 5% CO2. Before the FLIM, a single 1024-pixel square confocal scan capturing the SiR-DNA (Cytoskeleton) and the 633-nm laser backscatter differential interference contrast (DIC) images were acquired with Apochromat 40×/1.1 W Corr 27 lens (Zeiss). The same lens, frame size, position, and z thickness were used to collect the FLIM data while using the 485-nm diode laser (PicoQuant) that was pulsed at 32.5 MHz and set to 70% of the maximum output power. The emission light was reflected by the 80/20 mirror (Zeiss) and passed through a transparent plate, and emission below 560 nm was reflected by a dichroic filter into the PicoQuant PMA-04 hybrid detector. The pinhole size was individually set to acquire data from 1.5- to 5-μm z sections to limit the emission photon count rate below 10% of the laser excitation rate. The single 1024-pixel square FLIM images were acquired with 58.2-μs pixel dwell time (61 s of total scan time).
FLIM data were processed with SymPhoTime 64 v.2.3 (PicoQuant) software, using the customized script developed by PicoQuant for the calculation of the internal response function from 100 data points with no smoothing. The acquired data were binned to obtain at least 400 photons per binned pixel (bin 4-6), and the out-of-cell fluorescence was removed by applying a cell-specific threshold. The three-exponential reconvolution was used to fit the fluorescence decays into every binned pixel. The SymPhoTime 64 v.2.3 was used to calculate the average amplitude-weighted fluorescence lifetime averages in different areas of the oocytes. The Free ROI tool was used to manually select image areas corresponding to the chromosomes. The line profiles of FLIM images were measured in ImageJ using a line width of 8 μm across two chromatin clusters or from the chromatin clusters to the oocyte center. The one-phase decay nonlinear fit of the curve was calculated using GraphPad Prism.
PIV analysis
Transmitted light images were used to visualize the dynamics of the cytoplasmic flow. Images were preprocessed first by alignment using the Image Stabilizer plugin written for ImageJ. The cytoplasmic flow was then analyzed with the PIVlab package (52). The specific parameters were as follows: CLAHE window size, 20 pixels; high-pass filter size, 16 pixels; algorithm, fast Fourier transform window deformation; pass 1 interrogation area, 50 pixels (step: 25); pass 2 interrogation area, 25 pixels (step: 13); window deformation interpolator, linear; subpixel estimator, Gauss 2 × 3 point. After flow analysis, outliners were adjusted using the vector validation function. After obtaining a series of instant velocity fields, we calculated mean flow field by averaging the consecutive frames to obtain a time-averaged vector plot and normalized vorticity by the maximum to denote the orientation of the flow. A positive value indicates counterclockwise flow, whereas a negative value indicates clockwise flow. Mirror symmetry of the flow vorticity on the two sides of the axis perpendicular to and bisecting the spindle axis was represented as the absolute value of the sum of the mean vorticity on each side. The closer this value is to zero, the more mirror symmetric are the flow patterns. To extract mean vorticity in specified area as shown in Fig. 3A, the PIVlab draw tool was used to create ROIs. Using the Extracting Parameters from Area in the Extractions menu of PIVlab, the Area Mean Value for the Vorticity (1 per frame) parameter was obtained.
Statistics
Statistical analyses were performed with GraphPad Prism. Average (mean), SD, and statistical significance based on two-tailed Student’s t test or Fisher’s exact test were calculated in GraphPad Prism. All box plots show median (horizontal lines), 25th and 75th percentiles (small boxes), and minimum and maximum values (whiskers). Violin plot shows median (horizontal lines) and quartiles (dot lines). Nonlinear regression and Gaussian fit curves were calculated using Prism software. n represents oocyte numbers.
Modeling force production by actin-driven cytoplasmic flow
A 3D model of spindle rotation with an anchoring point is implemented in the finite element software COMSOL. The geometry of the oocyte cortex during spindle rotation is described as the union of three spheres (Fig. 3, I and G): one large sphere centered at (0, 0) with radius R0 representing the main body of the oocyte and two smaller spheres centered at (∓x0, y0) with radius r0 representing two protrusions at the animal pole. The center of the spindle is positioned at (xs, ys), where xs = 0 at t = 0. The spindle has long semi-axis as and short semi-axis bs. The upper point of the spindle (0, ys|t = 0 + bs) is anchored to the cortex so that the spindle can only rotate around the anchoring point. Here, for simplicity, we have omitted the coordinate of the third dimension, which is perpendicular to the page. All the objects are centered at z = 0.
The actin-driven cytoplasmic flow is modeled as a Newtonian fluid that satisfies Navier-Stokes equation. The spindle is modeled as a rigid body that interacts with the cytoplasmic flow, modeled using the fluid-structure interaction module in COMSOL, whereby the cytoplasmic flow on spindle satisfies nonslip and nonpenetration conditions. The latter is justified assuming the presence of a spindle matrix (53); however, our previous analysis found that cytoplasmic streaming pattern and the force on the spindle are not qualitatively affected when assuming the spindle being partially penetrant by the flow (13). The parameters in the model are listed in table S1.
The chromosome-proximal cortex of the oocyte is rich in Arp2/3 complex and myosin-II. Continuous nucleation of Arp2/3 leads to an actin flow in the cytoplasm (13). This flow is close to the cortex and generates an effective stress along the cortex. Myosin-II provides contractile stress that effectively increases the local cytoplasm pressure to set off the elevated cortical tension. Thus, the actions of Arp2/3 complex and myosin-II can be incorporated as boundary forces in the actin-driven flow. The normal component of the boundary stress (which is also a boundary force density) is F = [−pI + μ(∇v + (∇v)T)]·n, where p is the hydrostatic pressure of the cytoplasm, v is the cytoplasmic velocity, μ is the dynamic viscosity of the cytoplasm, I is the identity tensor, and n is the unit norm of the cortex boundary. In COMSOL, the boundary forces can be explicitly incorporated as boundary conditions.
We first study the cytoplasmic flow pattern generated by Arp2/3 complex or myosin-II–based actin force alone. In the case of Arp2/3 complex alone, we let the effective boundary force generated by Arp2/3 complex be in the tangential direction along the cortex (Fig. 3E). The resulting flow and vortex pattern (Fig. 3F) match well with the experimental observation when myosin-II was inhibited by blebbistatin (Fig. 3C). In the case of myosin-II alone, we let the effective boundary force generated by myosin-II to be in the normal direction along the cortex (Fig. 3G). The resulting flow and vortex pattern (Fig. 3H) match well with the experimental observation (Fig. 3D). Since the cytoplasmic flow pattern is determined by the boundary condition, these results suggest that the implemented boundary forces from Arp2/3 complex (tangential) and myosin-II (mainly normal) are appropriate and reflect the forces associated with these proteins.
We next used a 3D model to examine the flow pattern generated by an asymmetric distribution of Arp2/3 complex and myosin-II and the resulting dynamics of the spindle in untreated oocytes. On the basis of the observed distribution of Arp2/3 complex and myosin-II during spindle rotation (Fig. 2, D to F), we applied effective normal stress coming from myosin-II on the side of the oocyte with enriched myosin-II localization (right side in Fig. 3J) and increased tangential stress coming from Arp2/3 complex on the other side (left side of Fig. 3J). With this asymmetric boundary force implementation, we observed a mainly downward flow near the right side of the spindle and an upward flow on the left side (Fig. 3K). The direction of the vorticities associated with the Arp2/3 versus myosin-II side of the oocyte in the spindle vicinity is consistent with those observed experimentally (Fig. 3B). Cytoplasmic flows in opposite directions at the two ends of the spindle provide a net torque on the spindle, causing it to rotate toward the cortical side enriched for the Arp2/3 complex, as observed experimentally (Fig. 2, D to F).
Model for symmetry breaking and spindle rotation
We approximate the cell cortex around the animal pole as a 1D line space across −ℓ ≤ x ≤ ℓ, where x = 0 aligns with the center of the spindle in the vertical direction (Fig. 5A). We assume that during spindle rotation, the distance from the center of the spindle to the cortex, d0, is a constant.
Let Lc(t) be the distance between the separating chromosomes. This is given on the basis of the observed rate of chromosome separation, i.e.
| (1) |
where Ts0 is the starting time of chromosome separation, Ts is the time course of chromosome separation, and ℓs is the maximum separation distance of the two sets of chromosomes. With the prescribed Lc(t), the coordinates of the left and right chromosomes, cL and cR, respectively, can be expressed as
| (2) |
where α(t) is the rotational angle of the spindle in radius to be determined by the dynamics of the system.
We assume that the RanGTP concentration in space decays exponentially from each chromosome cluster (31, 54) with a characteristic length λ, which we measured from experiments. Therefore, the Ran signal, Ran(x,t), received on any point x = (x, 0) on the cortex is given by
| (3) |
where dL = |cL − x| and dR = |cR − x|, β is a constant, and Ran0 is the Ran signal intensity when dL,R = 0. In the case that the Ran signal is practically inhibited at the left side of the chromosome, the first term in the bracket in Eq. 3 is reduced to 90% of the original value (a 10% reduction, as suggested from experiments).
The levels of Arp2/3 complex, a(x,t), and myosin-II, m(x,t), on the cortex are governed by reaction-diffusion equations. For computational purposes, a and m are set to be dimensionless and scaled to 1. The governing equations of a and m are
| (4) |
| (5) |
where D values are the effective diffusion coefficients of a and m on the membrane. Note that these diffusion terms arise from the diffusion of actin and myosin in the cytoplasm and the exchange of their cytoplasmic pools with the cortical pool. Integrating out the cytoplasmic pool generates the effective diffusion term in the equation above. The symbols kaf and kmf are the off rates of Arp2/3 complex and myosin-II, respectively; kmo is the binding rate of myosin-II from the myosin-II pool in the cytoplasm, mc, which is assumed to be a constant; kar is the activation rate for Arp2/3 due to the Ran signal, and we have assumed that there is no cytoplasmic pool of Arp2/3 complex; and kam is the inhibitory coefficient between Arp2/3 and myosin. The binding rate of myosin-II kmo is defined on −ℓm ≤ x ≤ ℓm and is 0 elsewhere, where ℓm is the myosin-binding domain given by ℓm = (36 –t/18) μm, where t is time in minutes. When t ≤ 1.5(Ts0 + Ts), kmo = k0mo is a constant; when t > 1.5(Ts0 + Ts), kmo = k0mo(|x|/ℓm)2. This temporal and spatial choice of kmo is made such that the resulting Arp2/3 and myosin-II distribution on the cortex from the model resembles that from experimental observations. The boundary conditions for Arp2/3 and myosin-II at x = ∓ℓ are a = 0 and m = 0.
From the previous section, we showed from hydrodynamic simulations that the cytoplasmic flow due to the asymmetry distribution of Arp2/3 complex and myosin-II provides an effective torque that rotates the spindle toward the cortex where Arp2/3 is concentrated. We therefore include the torque contributions from both Arp2/3 complex and myosin-II, where myosin-II provides an effective torque that rotates the spindle away from it and Arp2/3 complex provides an effective torque that rotates the spindle toward it. Phenomenologically, the net torque generated by the cortical Arp2/3 complex and myosin-II on the spindle can be expressed as
| (6) |
where kaq and kmq are the coefficients of effective force generated by the Arp2/3-induced and myosin-II–induced cytoplasmic flow, respectively. Since the distribution of a(x,t) is affected by Ran(x,t) (Eq. 4), the torque q(t) is also a function of Ran(x,t), which determines the dynamics of the angle, α, via
| (7) |
where η is the rotational viscosity of the spindle. R(t) = r(t)/krandom is a random fluctuating torque on the spindle, where r(t) is uniformly distributed between −0.5 and 0.5 μN·μm and krandom is a scaling factor. The rest of the parameters are listed in table S2. The parameters, especially the diffusion coefficients, the on and off rates, and the inhibitory coefficients, were adjusted such that the predicted temporal and spatial distributions of cortical Arp2/3 and myosin-II are consistent with those from experimental observations.
Simulations of the model in Eqs. 4, 5, and 7 showed that random fluctuations of the spindle angle and the feedback loops as modeled in the governing equations are able to break symmetry and generate three stages of Arp2/3 and myosin-II distributions (Fig. 5B) that are consistent with the experimental observation (Fig. 2, D to F): (i) Before chromosome separation, Arp2/3 complex has a single peak in the cortex center, and myosin-II distributes outside of the Arp2/3 peak (13). (ii) After chromosome separation and before spindle rotation, Arp2/3 complex has two symmetric peaks above the two sets of chromosomes, and myosin-II distributes outside of the Arp2/3 peaks. (iii) During spindle rotation, Arp2/3 complex has a peak where the chromosome is close to the cortex, and myosin-II has a peak where the chromosome is away from the cortex (Fig. 5B). The model suggests that the inhibitory relation between Arp2/3 and myosin-II induces the respective peaks at different locations, leading to a coordinated flow pattern that rotates the spindle.
When Ran is partially inhibited at one side of the chromosome, the Ran signal at the other side generates a strong cortical Arp2/3 localization close to the other chromosome cluster. This leads to a biased rotation of the spindle away from the side where Ran is inhibited (Fig. 5, C and D).
Symmetry breaking and spindle rotation are robust predictions from the model due to the underlying biophysical mechanism. Below, we provide a sensitivity analysis of the major parameters used in the model to demonstrate the robustness of the model. Because parameters related to Arp2/3 complex and myosin-II are relative, we varied those associated with the former while keeping the latter constant.
Multiple dynamic outputs can potentially be investigated through parameter study. Here, we focus on the maximum rotational angle of the spindle within the simulation time because the degree of spindle rotation is a key outcome of the model. Figure S5 shows the dependence of the maximum spindle rotation angle within the simulation time on each parameter. As expected, increasing the coefficient of effective Arp2/3 complex–mediated force (kaq) and the Ran-dependent activation of Arp2/3 complex (kar) increases the spindle rotation angle. Consistently, reducing the diffusion coefficient of Arp2/3 on the cortex (Da) and the off rate of Arp2/3 (kaf) also increases the spindle rotation angle. Since we examined the maximum angle within the simulation time, the sensitivity analysis also indicates the rotation speed. If we had increased the simulation time, then the maximum angle would be larger than the current maximum output. Thus, as long as the positive feedback mechanism is implemented, symmetry breaking and, thus, spindle rotation are robust predictions from the model.
Spindle dynamics in the absence of the anchor (pivot) point
The main difference from the normal case is that, here, the spindle does not have a pivot point for rotation. The center position of the spindle, cs, is given by
where Rs is a random vector in both x and y directions. Simulation shows that under this case, the resulting asymmetry is subtle with the myosin-II peak slightly larger on one side than that on the other (fig. S7A). On the basis of this distribution, we simulated cytoplasmic flow and spindle dynamics using the 3D COMSOL model without an anchoring point. A sphere centered at (0, 0, 0) with radius R0 represents the oocyte (Fig. 7A); the spindle is centered at (xs, ys), where xs = 0 μm and ys = 29.5 μm at t = 0. The result shows that without the anchoring between the spindle and the cortex, the spindle drifts laterally along the cortex due to the asymmetric flow (Fig. 7B). This prediction was validated experimentally (Fig. 7, E to H).
Supplementary Material
Acknowledgments
We thank T. Inoue for the SspB and iLID constructs, M. Schuh for the Trim21 and MAP4 constructs, P. Solc for the CDK5RAP2 constructs, and M. H. Verlhac for the WCA constructs. We thank M. Wang for assistance with schematics drawing and K. Yi (Stowers Institute for Medical Research) for helpful discussion. Funding: This work was supported by NIH RO1 HD086577. Author contributions: H.W. performed experiments with assistance from X.D. and P.K. Y.L. performed PIV analysis and model simulations. H.W. and J.Y. analyzed imaging data. H.W., Y.L., S.X.S., and R.L. prepared the manuscript. S.X.S. and R.L. supervised the study. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/14/eaaz5004/DC1
REFERENCES AND NOTES
- 1.Morin X., Bellaïche Y., Mitotic spindle orientation in asymmetric and symmetric cell divisions during animal development. Dev. Cell 21, 102–119 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Li R., The art of choreographing asymmetric cell division. Dev. Cell 25, 439–450 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Pearson C. G., Bloom K., Dynamic microtubules lead the way for spindle positioning. Nat. Rev. Mol. Cell Biol. 5, 481–492 (2004). [DOI] [PubMed] [Google Scholar]
- 4.di Pietro F., Echard A., Morin X., Regulation of mitotic spindle orientation: An integrated view. EMBO Rep. 17, 1106–1130 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li R., Albertini D. F., The road to maturation: Somatic cell interaction and self-organization of the mammalian oocyte. Nat. Rev. Mol. Cell Biol. 14, 141–152 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Clift D., Schuh M., Restarting life: Fertilization and the transition from meiosis to mitosis. Nat. Rev. Mol. Cell Biol. 14, 549–562 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Szollosi D., Calarco P., Donahue R. P., Absence of centrioles in the first and second meiotic spindles of mouse oocytes. J. Cell Sci. 11, 521–541 (1972). [DOI] [PubMed] [Google Scholar]
- 8.Clift D., Schuh M., A three-step MTOC fragmentation mechanism facilitates bipolar spindle assembly in mouse oocytes. Nat. Commun. 6, 7217 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holubcová Z., Blayney M., Elder K., Schuh M., Human oocytes. Error-prone chromosome-mediated spindle assembly favors chromosome segregation defects in human oocytes. Science 348, 1143–1147 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaigne A., Campillo C., Gov N. S., Voituriez R., Azoury J., Umaña-Diaz C., Almonacid M., Queguiner I., Nassoy P., Sykes C., Verlhac M.-H., Terret M.-E., A soft cortex is essential for asymmetric spindle positioning in mouse oocytes. Nat. Cell Biol. 15, 958–966 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Schuh M., An actin-dependent mechanism for long-range vesicle transport. Nat. Cell Biol. 13, 1431–1436 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yi K., Rubinstein B., Unruh J. R., Guo F., Slaughter B. D., Li R., Sequential actin-based pushing forces drive meiosis I chromosome migration and symmetry breaking in oocytes. J. Cell Biol. 200, 567–576 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yi K., Unruh J. R., Deng M., Slaughter B. D., Rubinstein B., Li R., Dynamic maintenance of asymmetric meiotic spindle position through Arp2/3-complex-driven cytoplasmic streaming in mouse oocytes. Nat. Cell Biol. 13, 1252–1258 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng M., Suraneni P., Schultz R. M., Li R., The Ran GTPase mediates chromatin signaling to control cortical polarity during polar body extrusion in mouse oocytes. Dev. Cell 12, 301–308 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Wang Q., Racowsky C., Deng M., Mechanism of the chromosome-induced polar body extrusion in mouse eggs. Cell Div 6, 17 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matson S., Markoulaki S., Ducibella T., Antagonists of myosin light chain kinase and of myosin II inhibit specific events of egg activation in fertilized mouse eggs. Biol. Reprod. 74, 169–176 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Halet G., Carroll J., Rac activity is polarized and regulates meiotic spindle stability and anchoring in mammalian oocytes. Dev. Cell 12, 309–317 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Dehapiot B., Carrière V., Carroll J., Halet G., Polarized Cdc42 activation promotes polar body protrusion and asymmetric division in mouse oocytes. Dev. Biol. 377, 202–212 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaufman M. H., Sachs L., The early development of haploid and aneuploid parthenogenetic embryos. J. Embryol. Exp. Morphol. 34, 645–655 (1975). [PubMed] [Google Scholar]
- 20.Rosenbusch B. E., Mechanisms giving rise to triploid zygotes during assisted reproduction. Fertil. Steril. 90, 49–55 (2008). [DOI] [PubMed] [Google Scholar]
- 21.Zhu Z.-Y., Chen D.-Y., Li J.-S., Lian L., Lei L., Han Z.-M., Sun Q.-Y., Rotation of meiotic spindle is controlled by microfilaments in mouse oocytes. Biol. Reprod. 68, 943–946 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Maro B., Johnson M. H., Pickering S. J., Flach G., Changes in actin distribution during fertilization of the mouse egg. J. Embryol. Exp. Morphol. 81, 211–237 (1984). [PubMed] [Google Scholar]
- 23.Nolen B. J., Tomasevic N., Russell A., Pierce D. W., Jia Z., McCormick C. D., Hartman J., Sakowicz R., Pollard T. D., Characterization of two classes of small molecule inhibitors of Arp2/3 complex. Nature 460, 1031–1034 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Straight A. F., Cheung A., Limouze J., Chen I., Westwood N. J., Sellers J. R., Mitchison T. J., Dissecting temporal and spatial control of cytokinesis with a myosin II Inhibitor. Science 299, 1743–1747 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Tao J., Sun S. X., Active biochemical regulation of cell volume and a simple model of cell tension response. Biophys. J. 109, 1541–1550 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li R., Gundersen G. G., Beyond polymer polarity: How the cytoskeleton builds a polarized cell. Nat. Rev. Mol. Cell Biol. 9, 860–873 (2008). [DOI] [PubMed] [Google Scholar]
- 27.Goryachev A. B., Leda M., Many roads to symmetry breaking: Molecular mechanisms and theoretical models of yeast cell polarity. Mol. Biol. Cell 28, 370–380 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu J., Wang F., Van Keymeulen A., Herzmark P., Straight A., Kelly K., Takuwa Y., Sugimoto N., Mitchison T., Bourne H. R., Divergent signals and cytoskeletal assemblies regulate self-organizing polarity in neutrophils. Cell 114, 201–214 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Lomakin A. J., Lee K.-C., Han S. J., Bui D. A., Davidson M., Mogilner A., Danuser G., Competition for actin between two distinct F-actin networks defines a bistable switch for cell polarization. Nat. Cell Biol. 17, 1435–1445 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soderholm J. F., Bird S. L., Kalab P., Sampathkumar Y., Hasegawa K., Uehara-Bingen M., Weis K., Heald R., Importazole, a small molecule inhibitor of the transport receptor importin-β. ACS Chem. Biol. 6, 700–708 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kalab P., Heald R., The RanGTP gradient–A GPS for the mitotic spindle. J. Cell Sci. 121, 1577–1586 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wagner E., Glotzer M., Local RhoA activation induces cytokinetic furrows independent of spindle position and cell cycle stage. J. Cell Biol. 213, 641–649 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lekomtsev S., Su K.-C., Pye V. E., Blight K., Sundaramoorthy S., Takaki T., Collinson L. M., Cherepanov P., Divecha N., Petronczki M., Centralspindlin links the mitotic spindle to the plasma membrane during cytokinesis. Nature 492, 276–279 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Cho W., Membrane targeting by C1 and C2 domains. J. Biol. Chem. 276, 32407–32410 (2001). [DOI] [PubMed] [Google Scholar]
- 35.Campellone K. G., Welch M. D., A nucleator arms race: Cellular control of actin assembly. Nat. Rev. Mol. Cell Biol. 11, 237–251 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chaigne A., Campillo C., Gov N. S., Voituriez R., Sykes C., Verlhac M. H., Terret M. E., A narrow window of cortical tension guides asymmetric spindle positioning in the mouse oocyte. Nat. Commun. 6, 6027 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Guntas G., Hallett R. A., Zimmerman S. P., Williams T., Yumerefendi H., Bear J. E., Kuhlman B., Engineering an improved light-induced dimer (iLID) for controlling the localization and activity of signaling proteins. Proc. Natl. Acad. Sci. U.S.A. 112, 112–117 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Higgs H. N., Blanchoin L., Pollard T. D., Influence of the C terminus of Wiskott-Aldrich syndrome protein (WASp) and the Arp2/3 complex on actin polymerization. Biochemistry 38, 15212–15222 (1999). [DOI] [PubMed] [Google Scholar]
- 39.Padrick S. B., Cheng H.-C., Ismail A. M., Panchal S. C., Doolittle L. K., Kim S., Skehan B. M., Umetani J., Brautigam C. A., Leong J. M., Rosen M. K., Hierarchical regulation of WASP/WAVE proteins. Mol. Cell 32, 426–438 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clift D., McEwan W. A., Labzin L. I., Konieczny V., Mogessie B., James L. C., Schuh M., A method for the acute and rapid degradation of endogenous proteins. Cell 171, 1692–1706.e18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ajduk A., Ilozue T., Windsor S., Yu Y., Seres K. B., Bomphrey R. J., Tom B. D., Swann K., Thomas A., Graham C., Zernicka-Goetz M., Rhythmic actomyosin-driven contractions induced by sperm entry predict mammalian embryo viability. Nat. Commun. 2, 417 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McLay D. W., Clarke H. J., Remodelling the paternal chromatin at fertilization in mammals. Reproduction 125, 625–633 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Braun R. E., Packaging paternal chromosomes with protamine. Nat. Genet. 28, 10–12 (2001). [DOI] [PubMed] [Google Scholar]
- 44.Sassone-Corsi P., Unique chromatin remodeling and transcriptional regulation in spermatogenesis. Science 296, 2176–2178 (2002). [DOI] [PubMed] [Google Scholar]
- 45.Perreault S. D., Chromatin remodeling in mammalian zygotes. Mutat. Res. 296, 43–55 (1992). [DOI] [PubMed] [Google Scholar]
- 46.Nonchev S., Tsanev R., Protamine-histone replacement and DNA replication in the male mouse pronucleus. Mol. Reprod. Dev. 25, 72–76 (1990). [DOI] [PubMed] [Google Scholar]
- 47.Nemergut M. E., Mizzen C. A., Stukenberg T., Allis C. D., Macara I. G., Chromatin docking and exchange activity enhancement of RCC1 by histones H2A and H2B. Science 292, 1540–1543 (2001). [DOI] [PubMed] [Google Scholar]
- 48.Deng M., Li R., Sperm chromatin-induced ectopic polar body extrusion in mouse eggs after ICSI and delayed egg activation. PLOS One 4, e7171 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goldstein B., Macara I. G., The PAR proteins: Fundamental players in animal cell polarization. Dev. Cell 13, 609–622 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mittasch M., Gross P., Nestler M., Fritsch A. W., Iserman C., Kar M., Munder M., Voigt A., Alberti S., Grill S. W., Kreysing M., Non-invasive perturbations of intracellular flow reveal physical principles of cell organization. Nat. Cell Biol. 20, 344–351 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Kishigami S., Wakayama T., Efficient strontium-induced activation of mouse oocytes in standard culture media by chelating calcium. J. Reprod. Dev. 53, 1207–1215 (2007). [DOI] [PubMed] [Google Scholar]
- 52.Thielicke W., Stamhuis E. J., PIVlab-towards user-friendly, affordable and accurate digital particle image velocimetry in MATLAB. J. Open Res. Softw. 2, e30 (2014). [Google Scholar]
- 53.Zheng Y., A membranous spindle matrix orchestrates cell division. Nat. Rev. Mol. Cell Biol. 11, 529–535 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Caudron M., Bunt G., Bastiaens P., Karsenti E., Spatial coordination of spindle assembly by chromosome-mediated signaling gradients. Science 309, 1373–1376 (2005). [DOI] [PubMed] [Google Scholar]
- 55.Kim T., Gardel M. L., Munro E., Determinants of fluidlike behavior and effective viscosity in cross-linked actin networks. Biophys. J. 106, 526–534 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zaner K. S., Stossel T. P., Some perspectives on the viscosity of actin filaments. J. Cell Biol. 93, 987–991 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Satcher R. L. Jr., Dewey C. F. Jr., Theoretical estimates of mechanical properties of the endothelial cell cytoskeleton. Biophys. J. 71, 109–118 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.R. Milo, R. Phillips, N. Orme, Cell Biology by the Numbers (Garland Science, Taylor & Francis Group, 2016). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/14/eaaz5004/DC1


