Hidden in plain sight: Methanotrophic feather duster worms redefine the methane seep sphere of influence.
Abstract
Deep-sea cold seeps are dynamic sources of methane release and unique habitats supporting ocean biodiversity and productivity. Here, we describe newly discovered animal-bacterial symbioses fueled by methane, between two species of annelid (a serpulid Laminatubus and sabellid Bispira) and distinct aerobic methane-oxidizing bacteria belonging to the Methylococcales, localized to the host respiratory crown. Worm tissue δ13C of −44 to −58‰ are consistent with methane-fueled nutrition for both species, and shipboard stable isotope labeling experiments revealed active assimilation of 13C-labeled methane into animal biomass, which occurs via the engulfment of methanotrophic bacteria across the crown epidermal surface. These worms represent a new addition to the few animals known to intimately associate with methane-oxidizing bacteria and may further explain their enigmatic mass occurrence at 150–million year–old fossil seeps. High-resolution seafloor surveys document significant coverage by these symbioses, beyond typical obligate seep fauna. These findings uncover novel consumers of methane in the deep sea and, by expanding the known spatial extent of methane seeps, may have important implications for deep-sea conservation.
INTRODUCTION
Methane seeps and hydrothermal vents globally host dense communities of unique organisms, mainly supported via symbioses with chemosynthetic microorganisms, which offer direct access to energy from reduced gases such as H2S, H2, and CH4. Most known chemosynthetic symbioses involve sulfide-oxidizing bacteria, while far less common are those that associate with methane-oxidizing bacteria (1, 2). Nevertheless, many methane-rich seeps exist along continental margins, thus providing the distinct possibility of as-yet unknown symbioses that may act as important sinks for this potent greenhouse gas. As an example, along the Pacific continental margin of northern Central Costa Rica, a vast series of seeps occurs (nearly one every 4 km) related to landslide scars, seamount subduction–related fractures, mounds, and faults (3, 4). Because of their unique community structure and significant cycling of carbon, sulfur, and nitrogen, it is increasingly important to understand the trophic interactions between these ubiquitous seep ecosystems and the chemosynthetic animals that they support.
One of the historically recognizable animals to inhabit the periphery of seeps and hydrothermal vents are tube-dwelling “fan worm” annelids placed in Sabellida (including Fabriciidae, Sabellidae, and Serpulidae) (5). Several species, in particular, achieve such high densities that they are sometimes referred to as “thickets” (200 to 400 individuals per m2) (6–8). Since the discovery of hydrothermal vents, serpulids have been specifically noted as stable and conspicuous indicators of reduced fluids, forming a virtually continuous zone around hydrothermal vent fields (9). The first serpulid species formally described from hydrothermal vents was Laminatubus alvini, discovered along the East Pacific Rise (10). Since then, three serpulid genera (Laminatubus, Protis, and Hyalopomatus) are known to be common at deep-sea chemosynthetic habitats (11). Closely related sabellids are usually found in shallow benthic marine habitats, but some have been reported from the deep sea and one species (Bispira wireni) has been described from hydrothermal vents (12, 13). Both serpulids and sabellids have generally been regarded as opportunistic inhabitants in these areas of high productivity, presumably suspension feeding on increased bacteria in the surrounding water, using their ciliated anterior appendages (7, 14).
Mass occurrences of serpulids in ancient methane seep deposits, known as “fossil serpulid seeps,” have perplexed scientists for decades, and the recognition of paleoseep indicator species continues to be of value for geologists and geobiologists alike. For serpulids, which permanently inhabit calcareous tubes, seven taxa (including Laminatubus) have been reported from early Cretaceous to Miocene seep communities ~150 to 200 million years ago (15–17). They appear attached to, or in layers immediately adjacent to, known chemosynthetic vesicomyid clams, lucinid clams, or bathymodiolin mussels, prompting the suggestion that serpulids found in fossil seep deposits were actually regular members of the chemosynthetic community, as opposed to arriving only after seepage had ceased (15). The suggestion that modern-day serpulids might be endemic to chemosynthetic habitats was proposed early on by other researchers (7, 18), calling into question the assumption of obligate suspension feeding (14). By contrast, annelids within Sabellidae inhabit soft mucous and sediment tubes and thus leave only a minor fossil record (19).
Here, by integrating microbial community profiling, ultrastructural analysis via microscopy, live animal stable isotope tracer experiments, and seafloor surveys, we describe two separate, but closely related, tube-dwelling annelids that demonstrate active methane-based nutrition via methanotrophic bacterial symbionts. These newly discovered methane-reliant animals are commonly found at seeps and vents worldwide and extend the boundaries of the “seep” habitat, a classification that is increasingly important for regulatory and stewardship efforts concerning fisheries and oil drilling in the deep sea (20).
RESULTS
At a seep site known as Jaco Scar at 1768 to 1887 m depth off the west coast of Costa Rica (9°7.1′N, 84°50.4′W) (3, 21), a single serpulid species (Laminatubus n. sp.) and a single species of sabellid (Bispira n. sp.) were abundant in zones of active seepage (Fig. 1). By visual estimation, both annelid species represented a large fraction of the animal community and were observed near and often attached to obligate seep fauna, including Lamellibrachia barhami and Bathymodiolus spp. mussels. Sampling and shipboard experiments of these two species were carried out with the human-occupied vehicle (HOV) Alvin during research vessel (R/V) Atlantis expeditions AT37-13 (May to June 2017) and AT42-03 (October to November 2018) (table S1), along with high-resolution seafloor surveys by the autonomous underwater vehicle (AUV) Sentry, owned and operated by Woods Hole Oceanographic Institution (WHOI).
Fig. 1. In situ images of the two annelid species featured in this study.

A new species of serpulid annelid (Laminatubus) and a new species of sabellid (genus Bispira), at a site known as Jaco Scar at 1768 to 1887 m depth (9°7.1′N, 84°50.4′W), along the convergent margin off the west coast of Costa Rica. Insets show the worm species in life position. Photo credit: HOV Alvin, Woods Hole Oceanographic Institute.
Isotopic evidence of methane-based nutrition
Tissue stable δ13C and δ15N isotopes of both Jaco Scar Laminatubus and Bispira species were distinct from nonseep members of the same families (Fig. 2). δ13C values for the crown appendages, called radioles, of both Jaco Scar worm species were highly negative [−56.9 ± 0.9‰ for Laminatubus and −45.3 ± 4.3‰ for Bispira (n = 4 to 6, ±1 SD)] compared to related known suspension feeders collected from the same general vicinity without active methane seepage (~−18 to 20‰, n = 5) (Fig. 2 and tables S1 and S2). Laminatubus and Bispira isotopic values were consistent among all major body parts, including radiolar crown, body wall, and digestive tissues [analysis of variance (ANOVA) P = 0.65; n = 12 combined for each category; table S2] and consistent with methane values reported for the water column above Jaco Scar seeps (−50 to −62‰) (21). Particulate organic matter had a δ13C of −25‰ in the Jaco Scar seep bottom water (n = 3; Fig. 2).
Fig. 2. Carbon and nitrogen isotope data for native and experimental animals.
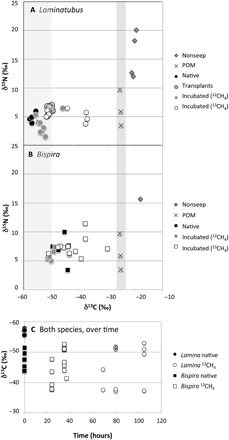
Biplots of δ13C and δ15N values for the crown radioles of (A) the serpulid Laminatubus n. sp. and (B) sabellid, Bispira n. sp. from the Jaco Scar seep, sampled directly upon collection from the active seep (“Native”), after an in situ transplant for 16 months to an area of no methane venting (“Transplants”), or after shipboard incubation with either 12CH4 (“controls”) or 13CH4 (example “13CH4-incubated”). “Nonseep” sabellids were also collected from an offshore seamount but were not identified beyond family. The right-hand vertical shaded bar highlights the measured values of particulate organic material (“POM”) [collected from CTD (conductivity/temperature/depth) casts over the area of active seepage]. The left-hand vertical shaded bar indicates the δ13C for methane from the water column above Jaco Scar seeps (−62 to −50‰) reported by Mau et al. (21). (C) Changes in δ13C over time are shown for both species. Two 13CH4-incubated Laminatubus that were observed to have a decreased relative abundance of putative methanotrophs based on 16S rRNA analysis (0.3 and 14%; see also Fig. 5) are shown with an asterisk in the symbol.
Molecular and microscopic evidence of methanotrophic symbioses
For Laminatubus, the radioles were arranged in typical serpulid semicircles, fusing to form a pair of lobes located on each side of the mouth (Fig. 3A). The radioles, which were coiled upon collection, were covered with rows of paired, densely ciliated pinnules. The dense ciliation, and the bacteria covering the radioles, gave the crown a fuzzy appearance (Fig. 3B). For Bispira, the radiolar crown was very long (in some cases as long as the body; crown-to-body ratio, 0.6:1.3; Fig. 4A), with elongate and delicate pinnules arranged along the radioles (Fig. 4B). Of the three tissues associated with Laminatubus and Bispira in this study, bacteria could only be reliably detected by polymerase chain reaction (PCR) from the distal regions of the crown radioles. Bacterial community analysis via 250–base pair (bp) 16S ribosomal RNA (rRNA) Illumina barcoding revealed limited diversity, with four putative methanotrophic bacterial operational taxonomic units (OTUs) within Methylococcales Marine Methylotrophic Group 2 (MMG-2; clustered at 99% similarity) dominating the community associated with each annelid species (Fig. 5). MMG-2 OTUs comprised 52 to 63% of the bacterial community in Laminatubus and 72 to 90% in native Bispira individuals from Jaco Scar, processed immediately following in situ collection (=“native”). The relative abundance of MMG-2 was 3.6 to 7.3% of the community recovered from adjacent deep sea in water samples but was distinct from the putative methanotrophic symbionts. Specific worm-associated MMG-2 OTUs were not detected in either the surrounding column or nearby sediment (at 0 to 3 cm depth) or in association with two nonseep sabellid specimens collected from nearby seamounts (Fig. 5 and table S1). This bacterial diversity was remarkably consistent among individuals of both Laminatubus and Bispira species collected 16 months apart and from both Jaco Scar (1800 m in depth) and another Costa Rican methane seep at 1000 m depth (Mound 12; Fig. 5 and fig. S1) (22). The co-occurring bathymodiolin mussels from Jaco Scar, which are capable of forming methane-based symbioses (23), notably did not have putative methanotrophic bacteria based on 16S rRNA amplicon sequencing (n = 3 different mussel species; table S3) and had a gill δ13C value of −36.6‰, suggesting a symbiosis with sulfide-oxidizing bacteria only. Similarly, nearby frenulate siboglinids, also known for methanotrophic symbioses (24), hosted a different MMG-2 OTU that comprised only 15% of their bacterial community (with a δ13C value of −39.1‰, n = 3).
Fig. 3. Microscopy of Laminatubus n. sp. crown radioles.
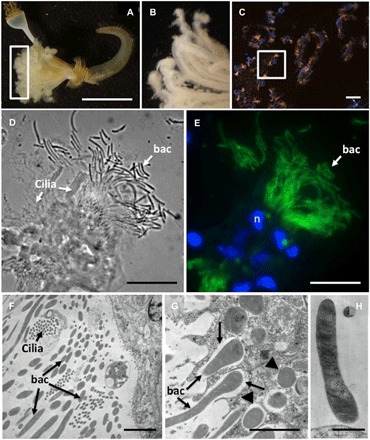
Light microscopy (A and B), fluorescence (C and E) plus corresponding bright field (D), and transmission electron (F to H) microscopy of Laminatubus n. sp. radioles. A FISH probe (MTC851) was designed to be an exact match to the putative methanotrophic symbionts [related to Methylococcales MMG-2, shown in orange (C) or green (E)]. DAPI-stained nuclei of host cells are shown in blue. TEM images show the bacteria, with dense internal membranes, on the epidermal surface of the host, and also possibly in the process of being engulfed (arrows) and completely engulfed [arrowheads in (F) and (G)] by host cells. (H) A close-up of a single filament, attached to the worm epidermis. n, nucleus; bac, bacteria. Scale bars, 1 cm (A), 10 μm (C), 15 μm (D and E), and 1 μm (F to H). Photo credit: Greg Rouse, Scripps Institution of Oceanography, and Shana Goffredi, Occidental College.
Fig. 4. Microscopy of the Bispira n. sp. crown radioles.
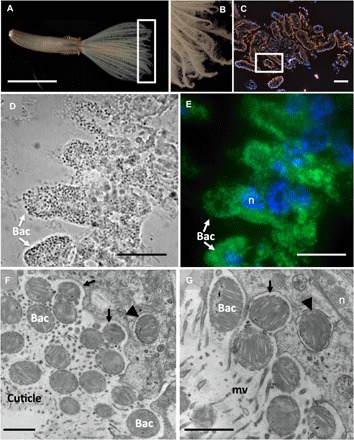
Light microscopy (A and B), fluorescence (C and E) plus corresponding bright field (D), and transmission electron (F and G) microscopy of Bispira n sp. radioles. A FISH probe (MTC851) was designed to be an exact match to the putative methanotrophic symbionts [related to Methylococcales MMG-2, shown in orange (C) or green (E)]. DAPI-stained nuclei of host cells are shown in blue. TEM images show the bacteria, with dense internal membranes, embedded in the cuticle of the host, and also possibly in the process of being engulfed (arrows) and completely engulfed [arrowheads in (F) and (G)] by host cells. n, nucleus; mv, microvilli; bac, bacteria. Scale bars, 0.5 cm (A), 10 μm (C), 15 μm (D and E), and 1 μm (F and G). Photo credit: Greg Rouse, Scripps Institution of Oceanography, and Shana Goffredi, Occidental College.
Fig. 5. Relative abundance of bacterial phylotypes, based on 16S rRNA.
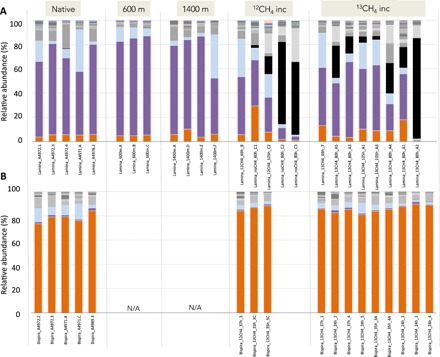
Bacterial community structure for crown radioles of (A) the serpulid Laminatubus n. sp. and (B) sabellid Bispira n. sp. from Jaco Scar seep, Costa Rica. Tissues were sampled directly upon worm collection from the active seep (native), after transplant for 16 months to areas of very little/no methane venting (“600 m” and “1400 m” away) or after shipboard incubation in either 12CH4 or 13CH4 (example “13CH4-incubated”). Bispira were not transplanted and thus do not have specimens for those categories (“N/A”). Each color on the graph represents a distinct genus-level phylotype or lowest level available. Phylotypes were grouped to 99% 16S rRNA sequence similarity. Dominant phylotypes within the Methylococcales Marine Group 2 are indicated by either purple or orange bars. Genera that were not putative aerobic methanotrophs are shown in light blue (Arenicella), black (Moritella) or gray (all others). The raw Illumina 16S rRNA barcode sequences and metadata collected in this study are available from the Dryad Digital Repository (https://doi.org/10.5061/dryad.wdbrv15jq) and the NCBI Small Read Archive (BioProject # PRJNA599018).
Targeted PCR amplification and sequencing of the particulate methane monooxygenase gene subunit A (pmoA, typically diagnostic of aerobic methanotrophic bacteria) and a clade-specific 16S rRNA gene was used to further characterize the bacterial methanotrophs in association with Laminatubus and Bispira. Analysis of a longer region of the 16S rRNA gene via direct sequencing was conducted using a MMG-2–specific reverse primer [MTC850R; based on (25)], revealing a single MMG-2 16S rRNA phylotype for each worm species (fig. S2A). These 16S rRNA phylotypes were 94% similar to each other and 97% similar to a closest relative (free-living bacteria from Gulf of Mexico seeps and East Pacific Rise hydrothermal vents, for the Laminatubus and Bispira phylotypes, respectively). Likewise, a single pmoA gene sequence was recovered from each host species via direct sequencing, with 83% nucleotide similarity between them (fig. S2B). These same pmoA sequences were not detected in the surrounding water column, suggesting that the worms hosted highly specific methanotrophic bacteria, relative to those commonly found in the environment. The worm-associated bacterial pmoA sequences, however, did share 99% amino acid identity to pmoA genes recovered from other seep environments, including Jaco Scar (26) and, notably, the frenulate Siboglinum from the Gulf of Cadiz (27).
In support of the 16S rRNA–based analysis, abundant bacteria were observed via fluorescence in situ hybridization (FISH) attached to and surrounding the radioles of both Laminatubus and Bispira from Jaco Scar (Figs. 3 and 4). Via transmission electron microscopy (TEM), nearly the entire volume of each putative symbiont cell was composed of intracytoplasmic membranes (Figs. 3, G and H, and 4, G to I), which are characteristic of methanotrophs, and the typical site of methane oxidation (28). A Methylococcales MMG-2–specific FISH probe (MTC851) was designed and used to determine the localization and morphology of the worm-associated MMG-2. For Laminatubus, numerous long (~10+ μm long × 0.5 μm wide), segmented, filamentous bacteria were observed in rosettes or tufts emerging from the tips of each radiole (Fig. 3, C to E). Despite the presence of a second Arenicella gammaproteobacteria phylotype (8 to 35% of the recovered bacterial community, based on 16S rRNA barcoding) (Fig. 5), no other bacteria were observed via FISH microscopy on the Laminatubus radioles (fig. S3).
Via TEM, numerous filaments with densely packed intracellular membranes were observed directly attached to the epidermis of Laminatubus (Fig. 3, F to H). For Bispira, the symbiont morphology was distinct with numerous small (~0.6 μm), membranous, cocci-shaped bacterial cells (Fig. 4, C to E) embedded in an extracellular collagenous cuticle, penetrated by microvilli (Fig. 4F). TEM analysis of both species showed that putative methanotrophs were not only attached to the epidermal surface of the worms but appeared to be in the process of engulfment by host tissue (Figs. 3G and 4G). Bacterial cells with compacted and disorganized membranes appeared deep in the worm tissues, surrounded by intracellular structures interpreted as digestive vacuoles (fig. S4, D to G).
Further, hybridization with the lipophilic dye FM4-64 revealed lipid-rich, presumably host-sourced, structures near the sites of bacterial attachment (fig. S4, B and C). Examination of Bispira revealed a cuticle matrix composed primarily of mannose, as stained by Hippeastrum hybrid Amaryllis lectin (HHA; fig. S4A). Mannose is a constituent of the cuticle in some annelids (29) and is the lock-and-key sugar used in other symbioses (30). In agreement with molecular analysis, fluorescence microscopy did not reveal obvious bacteria in the digestive system for either species (fig. S5).
In situ transplantation of methanotroph-bearing serpulids
Laminatubus specimens were transplanted by submersible in situ from an area of active seepage to two inactive muddy areas 600 to 1400 m away from the active site, in the general direction of methane plume flow (south-southwest, based on deployed current meters). These worms survived for 16 months, and those examined had retained their respective putative methanotroph phylotypes (which comprised 73 to 83% of the bacterial community, based on 16S rRNA amplicon sequencing; Fig. 5) and light tissue δ13C values (−50.6 ± 0.7‰; Fig. 2A). Tissue δ13C values of the transplanted worms, however, were slightly heavier than the δ13C values of individuals that remained in the active seep area (ANOVA P < 0.00001; Fig. 2A and table S2), potentially related to a shift in methane availability.
13CH4 tracer evidence of active methane incorporation by host worms
To examine the possibility of methane oxidation and assimilation of methane-derived carbon into host tissues, whole worms with intact symbionts were incubated onboard ship for 24 to 105 hours in the presence of 13C-labeled CH4 (table S4). The generation of 13C-enriched dissolved inorganic carbon in the surrounding seawater was detectable within 15 hours of incubation, confirming active CH4 conversion to CO2 (fig. S6). This included a set of incubations where live worms were removed from their tubes before incubation to minimize the potential for methanotrophic activity associated with bacteria on the tube surface. Within 24 hours of incubation, clear assimilation of 13C-labeled CH4 was observed within animal tissues (crown radioles; Fig. 2). For Laminatubus, δ13C values increased from −52.9 ± 3.0‰ (n = 8; for control worms exposed to 12CH4) to −44.4 ± 6.9‰ for 13CH4-incubated animals (n = 7; ANOVA P = 0.012, F = 7.847), with the tissues of three individuals revealing a shift to ~−37‰ δ13C (Fig. 2). Similarly, 13CH4-incubated Bispira had heavier δ13C values (−42.2 ± 5.9‰, n = 11) compared to control animals (−48.9 ± 2.9‰, n = 6; ANOVA P = 0.021, F = 6.6471), again with four individuals showing δ13C values as heavy as −29 to −39‰ within 37 hours of 13CH4 incubation (Fig. 2).
The variable degree of 13CH4 incorporation by some Laminatubus individuals (Fig. 2) is likely explained by the variable proportion of methane-oxidizing bacteria observed for 13CH4-incubated Laminatubus (0.3 to 44%; Fig. 5). For example, several Laminatubus individuals (3 of 13) that exhibited low 13CH4 uptake (δ13C values ~ −52‰; Fig. 2A) were found to be overgrown during the incubation period by the opportunistic heterotrophic bacteria Moritella (example up to 61 to 84%, based on 16S rRNA sequencing; Fig. 5), a bacterium not observed in native worms. FISH microscopy confirmed the lack of methane-oxidizing bacteria on the crowns of these individuals. Individuals that showed the greatest 13CH4 assimilation (a positive δ13C shift of ~16‰) maintained a dominance of the MMG-2 bacterial phylotype throughout the incubations (~44%; Fig. 5).
Mapping serpulid community scale at the Jaco Scar seep
Using the AUV Sentry, the presence of serpulids, with their easily observed white calcareous tubes, was mapped at the Jaco Scar seep region (Fig. 6). The AUV Sentry traveled a total of ~44 km in a grid pattern (during four dives in 2017 and 2018), at an altitude of ~7 m, taking high-resolution digital georeferenced photographs (~1 image every 3 s). In photos with clear visibility, the presence or absence of carbonate hard substrate and obligate seep organisms—bacterial mats, vestimentiferan tubeworms, vesicomyid clams, and bathymodiolin mussels—was noted. These species and substrate occurrences were resampled into 5-m grid cells in the survey area and mapped over the corresponding multibeam bathymetric data, using ArcGIS. Serpulids were categorized as “seep-associated” if obligate seep organisms were observed in the same grid cell. This autonomous approach revealed the seascape-scale distribution of these organisms, as compared to the limited range of observations that can be made visually from a remotely operated vehicle (ROV) or HOV. Overall, serpulids were observed to cover at least 65,000 m2 at the Jaco Scar seep site (~15 acres) and importantly extended the boundary, in some areas, ~300 m beyond the observed obligate seep organisms (Fig. 6). In particular, the distribution of serpulids extended the furthest to the southwest, which was the prevailing direction of water current flow from the active seep during the study period from 2017 to 2018. Only a single species of serpulid was recovered from the Jaco Scar seep site; thus, it was assumed that the serpulids visualized by Sentry were mostly, if not all, Laminatubus. Similar analyses were not possible for Bispira, because their sediment-coated tubes are not obvious in Sentry images, but they were most often found co-occurring with Laminatubus.
Fig. 6. Bathymetric map of the Jaco Scar seep region, as surveyed by the AUV Sentry.
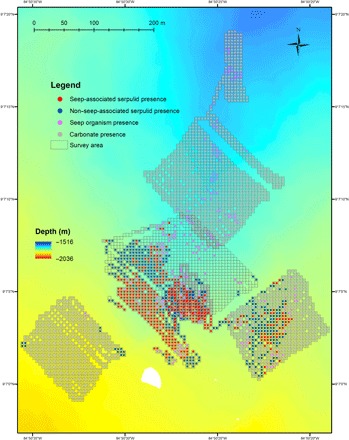
Location of the Jaco Scar seep study site, off the west coast of Costa Rica, and the 44-km track surveyed by the AUV Sentry, equipped with high-resolution mapping tools (sidescan sonar, multibeam echosounder, and digital still camera), traversed during four dives in 2017 and 2018. Moving at an average speed of 0.54 m/s, Sentry traveled at an average altitude of 6.9 m and took ~20 photos/min. Out of a total of 42,272 downward-facing photos, 28,962 photos were annotated with serpulids. Serpulid presence was categorized by the occurrence of other obligate seep fauna within the same 5-m grid cell (seep-associated) and the absence of other obligate seep fauna within the same grid cell (non–seep-associated). The distribution of seep fauna and carbonate rock without serpulids is also shown.
DISCUSSION
Deep-sea cold seeps are dynamic point sources for methane in the environment. Most studies in these habitats have emphasized environmental methane-oxidizing bacteria and archaea as important methane sinks; however, certain animals can also enrich for and host dense communities of methanotrophs. Here, we demonstrate active methane consumption by two new species of annelid (a serpulid Laminatubus sp. and sabellid Bispira sp.) and distinct methane-oxidizing gammaproteobacteria within the MMG-2 group (Methylococcales).
In both modern and paleo-ecosystems, isotope analysis is frequently used as an indicator of reduced chemical utilization, whether via methane or sulfide oxidation coupled to carbon fixation. The highly negative δ13C values in Laminatubus and Bispira tissues (−44 to −58‰) suggest a significant contribution of methane-derived carbon to their biomass. Two possibilities for the relatively light animal δ13C values involve aerobic methane-oxidizing bacteria, which occur ubiquitously in areas with exposure to both oxygen and methane, and have been described from the water column near Jaco Scar and other seep areas (26, 28). Either the worms selectively filter feed on methanotrophic bacteria from the overlying water or they form specific methane-based symbioses with methanotrophs. Previous evidence exists for selective feeding on aerobic methanotrophs by annelids in seeps, particularly sediment dwelling ampharetids that form crater-like depressions in the sediments as they errantly deposit feed (31, 32). For Laminatubus and Bispira, however, selective feeding does not seem likely given that water column methanotrophs are only a small fraction of the total bacteria (4, 33), and therefore presumably unable to support the very high biomass of these worms, which cannot leave their tubes. Furthermore, the δ13C of Jaco Scar particulate organic material (POM) from the surrounding water (−25‰) was not reflected in the tissues of either species, and we found no evidence for bacteria in their digestive systems. Alternatively, aerobic methanotrophs are known to live in intimate symbioses with marine invertebrates, including siboglinid tubeworms, bathymodiolin mussels, provannid snails, and cladorhizid sponges [reviewed in (2)]. Certain Bathymodiolus mussels have methanotrophic bacteria intracellularly in gill tissues, with corresponding negative δ13C values (−40 to −80‰) (2, 34)—a similar resemblance of Laminatubus and Bispira tissues to water column CH4 values from Jaco Scar (−50 to −62‰) (21).
Molecular evidence supported a specific and persistent symbiotic association between both Laminatubus and Bispira and distinct putative methanotrophs within the Methylococcales MMG-2 group. These particular bacteria were present in individuals collected 16 months apart and conversely not detected in the surrounding water column, underlying sediments, or in association with closely related worms from nearby inactive areas. Microscopy was marked in that abundant long filamentous MMG-2 bacteria covered the radioles of Laminatubus, while numerous smaller cocci-shaped bacteria were embedded in the collagenous cuticle of Bispira radioles. The MMG-2 has recently been shown to associate with asphalt seep sponges (35). The bathymodiolin mussels, which are known to form symbioses with methanotrophs, have not done so at the Jaco Scar methane seep sites. Thus, it appears that serpulids and sabellids have assumed an important symbiotic niche enabling the exploitation of methane for animal nutrition.
Transplanted Laminatubus survived for 16 months, and those examined retained their respective MMG-2 bacterial phylotypes and had light tissue δ13C values suggestive of persistent CH4 metabolism and assimilation. Bathymodiolus mussels with methanotrophic symbionts have likewise been shown to assimilate methane concentrations far below detection limits (36), but in the case of Laminatubus, the affinity for methane by the methanotrophic symbionts is not yet known. Tissue δ13C values of the transplanted worms, however, were significantly heavier than individuals that remained in the active seep area, perhaps indicating breakdown of tissue biomass (the transplanted worms appeared more translucent in nature), a lack of carbon fractionation during starvation, or increased suspension feeding activity, which has also been observed in mixotrophic Bathymodiolus mussels (37).
Whole worms with intact symbionts, incubated in the presence of 13C-labeled CH4, revealed active oxidation of methane and assimilation into host tissues. This appears to be the first example of marine invertebrates hosting a nutritional methane-oxidizing bacterial symbiont externally. This raises the important question of how the worms acquire carbon, or other nutrients, from their bacterial symbionts. Typically, organic carbon is passed from symbiont to host by either translocation of small organic molecules or by host intracellular digestion of the symbionts (38). The distinction between these two mechanisms in methanotrophic symbioses is often determined by ultrastructural observations and incubations with isotopically labeled methane. In bathymodiolin mussels, a time frame of carbon transfer to symbiont-free tissues on the order of hours has been attributed to translocation, whereas transfer in 1 to 5 days, consistent with the time frame of our experiments, is indicative of digestion (38). TEM analysis of both annelid species showed that putative methanotrophs were not only attached to the epidermal surface of the worms but were in the process of engulfment by host tissue. Bacterial cells with compacted and disorganized membranes appeared deep in the worm tissues, surrounded by intracellular structures interpreted as digestive vacuoles. Further, hybridization with a lipophilic dye revealed lipid-rich structures near the sites of bacterial attachment, supporting internalization and possible host endocytosis of bacteria (39). To our knowledge, the only other example of this mechanism for nutrient acquisition is in thyasirid clams, whose extracellular symbionts are periodically engulfed and digested by host gill epithelial cells (40). It remains a possibility that Laminatubus and Bispira also recover specific small carbon molecules from the bacteria (e.g., acetate, succinate, lactate, and pyruvate). Follow-up investigations with live specimens are required to determine the specific mechanism for carbon exchange and exactly how much of the nutrition of both annelids is supported by methane.
CONCLUSION
Two species of tube-dwelling annelids from within Sabellida have independently established highly specific, persistent, nutritional symbioses with two closely related aerobic methanotrophic bacteria. The assumption, until now, was that most sabellids and serpulids acquire nutrients via suspension feeding (14). We are reminded that heterotrophy involving suspension feeding was also initially proposed for the giant hydrothermal vent tubeworm Riftia pachyptila, which we now know relies entirely on sulfide-oxidizing chemoautotrophic symbionts for nutrition (41–43). Very little is known about the ecology of extant serpulids and sabellids in chemosynthetic ecosystems, and many species reported from hydrothermal vents and cold seeps remain identified only to the family level [example (18)]. Laminatubus, in particular, is known from other seeps (e.g., Pescadero Transform fault) and vents [e.g., Alarcon Rise; (8)], and additional serpulids have been observed at hydrothermal vents with significant methane values in end member fluids, including Logatchev, Rainbow, Menez Gwen, and Lucky Strike (44). Thus, the occurrence of methanotrophic symbionts associated with these ubiquitous animals is likely to extend beyond the Costa Rica margin seeps and may even extend beyond species of Laminatubus and Bispira. Further, these results also explain the unusually high abundance of serpulids at ancient hydrocarbon seeps worldwide [up to ~50% of estimated total animal biomass; (17)].
Using the AUV Sentry, the regional distribution of serpulids was mapped at Jaco Scar. Despite the power of this approach, to our knowledge, few publications have similarly used an autonomous vehicle for high-resolution spatial characterization of cold seep communities (45, 46). The general expanse of these worms beyond obvious observable vent or seep features is in agreement with past studies (6–7) and provides ecological support for the concept that they significantly broaden the influence of methane in deep-sea ecosystems. Given their presence at seeps along the Costa Rica margin and high densities at other known vent and seep sites, Laminatubus and Bispira may provide a potentially valuable and previously unrecognized service in this particular ecosystem—that is, the oxidation and sequestration of methane, a potent greenhouse gas. Although not yet quantified, these observations appear to expand the “sphere of influence” [sensu (47)] of methane in the deep sea, including the transfer of methane-based carbon across areas of high seepage to the periphery of seeps and into the background deep sea.
Continental margins at bathyal depths, where methane seeps typically occur, are targets for increased exploitation (48). Seeps, as essential fish habitats, contribute to the productivity of bottom fisheries (49) and generally often co-occur with areas of interest to the oil and gas industry (50). Current national and international spatial management prohibits particular activities from certain areas, for example, exclusion zones where sensitive species or habitats, including cold seeps, are present (50). This, of course, depends on an informed understanding of how to locate and define the seep ecosystem footprint, not just via the presence of methane bubbles and traditionally large visible, symbiont-bearing animals. The boundaries between seep ecosystems and the surrounding deep sea are not as sharp in space or time as often depicted (47). A new recognition of abundant methanotrophic serpulid and sabellid assemblages as an expansion of the traditional notion of the methane seep ecosystem brings us closer to accurate and knowledgeable use of the spatial influence of seepage to guide regulatory actions along the world’s active continental margins.
MATERIALS AND METHODS
Sampling
All Laminatubus and Bispira specimens were collected from active seep sites with the HOV Alvin during R/V Atlantis expeditions AT37-13 (May to June 2017) and AT42-03 (October to November 2018) from Jaco Scar (~1800 m depth) and Mound 12 (~1000 m depth), off the coast of Costa Rica (table S1). Targeted water samples (2 to 5 liters) were collected via Niskin bottle, also via HOV Alvin. Laminatubus specimens were transplanted, via HOV Alvin (during R/V Atlantis expedition AT37-13), to two separate areas 600 and 1400 m from the sites of active venting at Jaco Scar. They were recovered, via HOV Alvin (during R/V Atlantis expedition AT42-03), 16 months later. Additional nonseep species were collected during ROV SubBastian dives SO224 and SO227 (in January 2019) near sites known as Coco South and Seamount 6 (320 to 530 m depth; table S1). Upon recovery immediately following each dive, worms were subsampled for various analyses or shipboard experiments.
Isotope analysis
Tissue samples were dissected at sea, rinsed in Milli-Q water, and frozen at −20°C until shipment to the stable isotope laboratory (at Washington State University). Carbon and nitrogen isotope values of worm tissues and particulate organic carbon, 4 liters of water pulled onto glass fiber filters, were determined by isotope ratio mass spectrometry. Freeze-dried samples (0.3 to 0.7 mg dry weight) were packaged in tin capsules for mass spectrometry and analyzed using a Costech (Valencia, CA, USA) elemental analyzer interfaced with a continuous-flow GV Instruments (Manchester, UK) IsoPrime isotope ratio mass spectrometer (EA-IRMS) for 15N/14N and 13C/12C ratios. Measurements are reported in δ notation [per mil (‰) units], and a protein hydrolysate calibrated to National Institute of Standards and Technology (NIST) reference materials was used as a standard. Precision for δ13C and δ15N was generally ±0.2 and ±0.4‰, respectively.
Molecular analysis
Specimens for molecular analysis were preserved either immediately upon collection or immediately following shipboard experiments in ~90% ethanol and stored at 4°C. Total genomic DNA was extracted from tissues (50 to 130 mg) using the Qiagen DNeasy Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. Water samples taken by HOV Alvin were filtered (2 liters) onto a 0.22-μm Sterivex-GP polyethersulfone filter (MilliporeSigma, St. Louis, MO, USA) and frozen at −80°C until DNA analysis. DNA extraction was also performed using the Qiagen DNeasy Kit, according to the manufacturer’s instructions, with the exception of the first step, where 2 ml of ATL lysis buffer was added to the Sterivex filter, via luer lock and syringe, and rotated at 56°C for 12 hours. This solution was recovered from the filter, also via luer lock and syringe, and processed as usual. A 478-bp region of the gene coding for pmoA (subunit of particulate methane monooxygenase enzyme) was amplified using the primers pmoA189f (5′-GGNGACTGGGACTTCTGG-3′) and pmoA661r (5′-CCGGMGCA-ACGTCYTTACC-3) (51). An 823-bp fragment of the 16S rRNA gene was amplified using the primers 27F and MTC850R, originally designed as a FISH probe targeting the MMG-2 group (25). Annealing conditions of 56°C and 54°C were used for pmoA and 16SrRNA, respectively, for 25 cycles. Amplification products were sequenced directly using Sanger sequencing, via Laragen Inc., and submitted to GenBank (accession numbers MN416048 and MN416065). Close environmental and cultured relatives were chosen using top hits based on BLAST (www.ncbi.nlm.nih.gov).
The V4-V5 region of the 16S rRNA gene was amplified using bacterial primers with Illumina (San Diego, CA, USA) adapters on 5′ end 515F (5′-TCGTCGGC-AGCGTCAGATGTGTATAAGAGACAGGTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGGACTACHV-GGGTWTCTAAT-3′) (52). PCR mix was set up in duplicate for each sample with Q5 Hot Start High-Fidelity 2× Master Mix (New England Biolabs, Ipswich, MA, USA) with annealing conditions of 54°C for 25 cycles. Duplicate PCR samples were then pooled, and 2.5 μl of each product was barcoded with Illumina NexteraXT Index 2 Primers that include unique 8-bp barcodes (P5, 5′-AATGATACGGCGACCACCGAG-ATCTACAC-XXXXXXXX-TCGTCGGCAGCGTC-3′; P7, 5′-CAAGCAGAA-GACGGCATACGAGAT-XXXXXXXX-GTCTCGTGGGCTCGG-3′). Amplification with barcoded primers used conditions of 66°C annealing temperature and 10 cycles. Products were purified using MilliporeSigma (St. Louis, MO, USA) MultiScreen Plate MSNU03010 with vacuum manifold and quantified using Thermo Fisher Scientific (Waltham, MA, USA) Quant-iT PicoGreen dsDNA Assay Kit P11496 on the Bio-Rad CFX96 Touch Real-Time PCR Detection System. Barcoded samples were combined in equimolar amounts into single tube and purified with the Qiagen PCR Purification Kit (28104) before submission to Laragen (Culver City, CA, USA) for 2 × 250–bp paired-end analysis on the Illumina MiSeq platform with PhiX addition of 15 to 20%.
MiSeq 16S rRNA sequence data were processed in Quantitative Insights Into Microbial Ecology (QIIME) (v1.8.0). Raw sequence pairs were joined and quality-trimmed using the default parameters in QIIME. Sequences were clustered into de novo OTUs with 99% similarity using UCLUST open reference clustering protocol, and then, the most abundant sequence was chosen as representative for each de novo OTU. Taxonomic identification for each representative sequence was assigned using the Silva 119 database, clustered at 99% similarity. A threshold filter was used to remove any OTU that occurred below 0.01% in the combined samples dataset. The sequence data were further rarified by random subsampling to equal the sample with the least amount of sequence data (4006 sequences). The raw and processed sequence data, as well as representative sequences, are available in data file S1. The raw Illumina 16S rRNA barcode sequences and metadata collected in this study are available from the Dryad Digital Repository (https://doi.org/10.5061/dryad.wdbrv15jq) and the National Center for Biotechnology Information (NCBI) Small Read Archive (SRA) (BioProject no. PRJNA599018).
Fluorescence microscopy
Specimens for FISH microscopy were initially preserved in 4% sucrose-buffered paraformaldehyde (PFA) and kept at 4°C. These PFA-preserved specimens were rinsed with 2× phosphate-buffered saline (PBS), transferred to 70% ethanol, and stored at −20°C. Tissues were dissected and embedded in Steedman’s wax [1 part cetyl alcohol:9 parts polyethylene glycol (400) distearate, mixed at 60°C]. An ethanol:wax gradient of 3:1, 2:1, and 1:1, and eventually full strength wax, was used to embed the samples (1 hour each treatment). Embedded samples were sectioned at 2- to 5-μm thickness using a Leica RM2125 microtome and placed on SuperFrost Plus slides. Sections were dewaxed in 100% ethanol rinses. Hybridization buffers and wash buffers were prepared according to (53), using 35% formamide in the hybridization buffer and 450 mM NaCl in the wash solution, and fluorescent probes were at a final concentration of 5 μg/ml. Initially, we used the MTC850 probe (5′-ACGTTAGCTCCGCCACTA-3′; labeled with Cy3) (Figs. 3C and 4C), designed to target MMG-2 methanotrophs, with 1-bp mismatch to the Laminatubus and Bispira symbionts (25). Ultimately, a probe that was an exact match to both serpulid and sabellid symbionts [MTC851; 5′-ATACGTTAGCTCCACCACT-3′ labeled with fluorescein isothiocyanate (FITC); Figs. 3E and 4E] was designed and had 5-bp mismatches to Arenicella; the other bacterial phylotype recovered via amplicon sequencing from Laminatubus. A universal bacterial probe mix (Eub338˙ I-III) (54) was also used. Probes were hybridized at 46°C for 4 to 8 hours, followed by a 15-min wash at 48°C. To visualize the extracellular polymer structure, the sections were stained for 15 min with the mannose-specific lectin stain HHA labeled with tetramethyl rhodamine isothiocyanate (TRITC) at a final concentration of 100 μg/ml in PBS and then rinsed for 10 min with water. To investigate endocytosis, the sections were stained for 15 min with a lipophilic FM4-64 dye at a final concentration of 10 μg/ml in PBS and then rinsed for 10 min with water. Sections were counterstained with 4′,6-diamidino-2-phenylindole (DAPI; 5 mg/ml) for 1 min, rinsed, and mounted in either Citifluor or Vectorshield. Tissues were examined by epifluorescence microscopy using either a Nikon E80i epifluorescence microscope with a Nikon DS-Qi1Mc high-sensitivity monochrome digital camera or a Zeiss Elyra microscope with an ANDOR-iXon EMCCD camera.
Transmission electron microscopy
For TEM, radioles of several individuals of each species were fixed in 2.5% glutaraldehyde buffered in 0.05 M phosphate buffer with 0.3 M NaCl (2 to 24 hours at 4°C). Ruthenium red (~0.5%) was added to the fixative. The specimens were rinsed in the same buffer and postfixed with 1% OsO4 for 30 min. The radioles were dehydrated in an ascending alcohol series and embedded in Spurr’s resin. Silver interference–colored sections (65 to 70 nm) were prepared using a diamond knife (Diatome Ultra 45°) on a Leica Ultracut S ultramicrotome. The sections were placed on Formvar-covered, single-slot copper grids and stained with 2% uranyl acetate and lead citrate in an automated TEM stainer (QG-3100, Boeckeler Instruments). The sections were examined using a Zeiss EM10 transmission electron microscope with digital imaging plates (DITABIS Digital Biomedical Imaging Systems, Germany).
Shipboard isotope labeling experiments and analysis
To assess interactions between the methanotrophic bacteria and host annelids, short-term stable isotope incubation experiments with 13C-labeled methane (13CH4) were set up at sea using either Laminatubus or Bispira individuals removed from tubes (to avoid activity caused by bacteria transiently associated with the tubes) or worm-colonized rocks from the Jaco Scar seep. Unlabeled control incubations were included in each incubation series. All treatments were incubated at 4°C in sealed Mylar bags with 0.2-μm filtered bottom seawater from the collection site. Methane was added to each incubation to represent approximately one-quarter of the total headspace. For one series of incubations, 0.2-μm filtered air was added at 15- and 37-hour incubation time. Initial conditions for all incubations, assuming equilibrium between gases and seawater and no starting methane in the filtered seawater, are displayed in table S4. At the end of each incubation, worms were picked from the rocks, tissues were dissected, rinsed in Milli-Q water, and frozen −20°C until analysis, according to the isotopic analyses section above. In addition, water samples for dissolved inorganic carbon measurements were filtered through a 0.2-μm polyethersulfone filter into helium (He)–flushed, 12-ml Exetainer vials (Labco Ltd., Lampeter, UK), following the addition of 100 μl of ~40% phosphoric acid. Samples were stored upright at room temperature. Vials were sampled using a GC-PAL autosampler (CTC Analytics, Zwingen, Switzerland) equipped with a double-holed needle that transferred headspace using a 0.5 ml min−1 continuous flow of He to a 50-μm sample loop before separation by a PoraPLOT Q fused silica column (25 m; inside diameter, 0.32 mm) at 72°C. CO2 was then introduced to a Delta V Plus IRMS using a ConFlo IV interface (Thermo Fisher Scientific, Bremen, Germany) in the Caltech Stable Isotope Facility. A sample run consisted of three reference CO2 gas peaks, 10 replicate sample injections, and two final reference CO2 peaks. δ13C values were corrected for sample size dependency and then normalized to the VPDB (Vienna Pee Dee Belemnite) scale with a two-point calibration. Using NBS-19 and a previously calibrated laboratory carbonate as internal standards. The overall precision of this technique is estimated to be better than ±0.2‰, based on multiple analysis of independent standards as samples.
Seafloor surveys with the AUV Sentry
In the current study, we used WHOI’s AUV Sentry, equipped with sidescan sonar, a Reson 7125 multibeam echosounder, and a down-looking digital color camera, to collect bathymetric and photographic data. In 2017 and 2018, the AUV Sentry was deployed off of R/V Atlantis, during four dives at Jaco Scar, analyzed in this study. In the photo survey portion of each dive, the AUV Sentry moved back and forth on tightly spaced tracks (i.e., “mowing the lawn”) following a preset navigation plan. Moving at an average speed of 0.54 m/s, Sentry traveled a total of 44.10 km across four dives (#433, 436, 501, and 503), at an average altitude of 6.92 m, and took ~20 photos/min. Out of a total of 42,272 downward-facing photos, 28,962 photos were annotated with serpulids. For photos with clear visibility, the presence or absence of seep foundation species—bacterial mats, vestimentiferan tubeworms, vesicomyid clams, bathymodiolin mussels, and serpulids—were noted. In addition, the presence or absence of hard substrate in the form of carbonate rock was noted for each photo. In ArcGIS 10.6.1, these species and substrate presences were resampled into 5-m grid cells in the survey area to include only spatially explicit presence points, and they were mapped over multibeam bathymetric data collected by the AUV Sentry. The presence of serpulids was categorized as seep-associated or “non–seep-associated,” where seepage was indicated by the presence of seep-related foundation species found in the same grid cell.
Statistical analysis
Quantification and statistical analyses are described in Results and figure legends. Comparisons were performed using ANOVA, and statistical significance was declared at P < 0.05. Statistical analyses of beta diversity were performed with Primer E.
Supplementary Material
Acknowledgments
We thank the captains and crew of the R/V Atlantis, HOV Alvin pilots and technicians, AUV Sentry team, as well as scientific participants of AT37-13 and AT42-03, especially O. Pereira, L. McCormick, C. Seid, and A. Durkin, for their assistance at sea. We also thank the captain and crew of the R/V Falkor, ROV SuBastian pilots and technicians, scientific participants of FK190106, as well as J. Gonzalez, A. Crémière, J. Magyar, and S. Connon for their assistance and contributions to the isotope, ion chromatography, nanoSIMS, and microbial community analysis, respectively. C. Roman (University of Rhode Island, as co–principal investigator) and J. Cortes Nunez (University of Costa Rica, international collaborator) played significant roles as co–principal investigators during both expeditions and contributed current meter data. In addition, Occidental College undergraduates M. Cazin, K. Ruis, and C. Brzechffa assisted with microbial community and microscopy analysis, sponsored by the Oxy Undergraduate Research Center. Funding: Support for S.K.G. was provided by a Faculty Enrichment Grant through Occidental College. Support for E.T. was provided, in part, by a postdoctoral fellowship from the German Research Foundation (DFG TI 973/1-1). Support for V.J.O. was provided, in part, by the Gordon and Betty Moore Foundation (grant no. 3780). The research was primarily supported by U.S. NSF grants OCE 1635219 (to E.E.C), OCE 1634172 (to L.A.L. and G.W.R.), and OCE 1634002 (to V.J.O.). Sample collection and export permits were acquired through the Costa Rican Ministry of the Environment and Energy (SINAC-CUSBSE-PI-R-032-2018 and Academic License SINAC-SE-064-2018). Author contributions: S.K.G. conducted DNA analysis, including 16S rRNA barcoding and fluorescent microscope analyses, analyzed experimental data, wrote the manuscript with input from coauthors, and participated in both expeditions. E.T. performed electron microscopy analyses and participated in AT42-03. A.K. performed Sentry data analysis and wrote the manuscript. K.S.D. designed and performed the incubation experiments, participated in both expeditions, and managed the SRA submission. S.W.M. designed and performed the incubation experiments and participated in both expeditions. F.W. performed fluorescent microscopy analyses. R.W.L. performed the isotope analyses. G.W.R. was a principal investigator on the NSF-funded project, fixed the specimens for study (including seamount worms), coordinated and interpreted the electron microscopy analyses, identified and is naming the worm species, and participated in all expeditions. L.A.L. was a principal investigator on the NSF-funded project, coordinated isotope analyses, wrote the manuscript, and participated in both expeditions. E.E.C. was a principal investigator on the NSF-funded project and chief scientist on both expeditions. V.J.O. was a principal investigator on the NSF-funded project, designed the incubation experiments, and participated in both expeditions. All authors contributed to data interpretation and editing of the paper. Competing interests: All authors declare that they have no competing interests. Data and materials availability: 16S rRNA sequences for bacterial isolates are available from GenBank under accession numbers MN416048 through MN416065. The raw Illumina 16S rRNA barcode sequences and metadata collected in this study are available from the Dryad Digital Repository (https://doi.org/10.5061/dryad.wdbrv15jq) and the NCBI Small Read Archive (BioProject no. PRJNA599018). Animal images and specimens were vouchered (Laminatubus catalog no. A9589 and Bispira catalog no.A9598) for long-term archiving into the Benthic Invertebrate Collection at Scripps Institution of Oceanography (https://sioapps.ucsd.edu/collections/bi/).
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/14/eaay8562/DC1
REFERENCES AND NOTES
- 1.Dubilier N., Bergin C., Lott C., Symbiotic diversity in marine animals: The art of harnessing chemosynthesis. Nat. Rev. Microbiol. 6, 725–740 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Petersen J. M., Dubilier N., Methanotrophic symbioses in marine invertebrates. Environ. Microbiol. Rep. 1, 319–335 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Sahling H., Masson D. G., Ranero C. R., Hühnerbach V., Weinrebe W., Klaucke I., Bürk D., Brückmann W., Suess E., Fluid seepage at the continental margin offshore Costa Rica and southern Nicaragua. Geochem. Geophys. Geosyst. 9, Q05S05 (2008). [Google Scholar]
- 4.Levin L. A., Orphan V. J., Rouse G. W., Rathburn A. E., Ussler W. III, Cook G. S., Goffredi S. K., Perez E. M., Waren A., Grupe B. M., Chadwick G., Strickrott B., A hydrothermal seep on the Costa Rica margin, middle ground in a continuum of reducing ecosystems. Proc. R. Soc. B 279, 2580–2588 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kupriyanova E. K., Rouse G. W., Yet another example of paraphyly in Annelida: Molecular evidence that Sabellidae contains Serpulidae. Mol. Phylogenet. Evol. 46, 1174–1181 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Hessler R. R., Smithey W. M., Boudrias M. A., Keller C. H., Lutz R. A., Childress J. J., Temporal change in megafauna at the Rose Garden hydrothermal vent (Galapagos Rift; eastern tropical Pacific). Deep Sea Res. Part A Oceanogr. Res. Pap. 35, 1681–1709 (1988). [Google Scholar]
- 7.Olu K., Duperret A., Sibuet M., Foucher J.-P., Fiala-Médoni A., Structure and distribution of cold seep communities along the Peruvian active margin: Relationship to geological and fluid patterns. Mar. Ecol. Prog. Ser. 132, 109–125 (1996). [Google Scholar]
- 8.Goffredi S. K., Johnson S., Tunnicliffe V., Caress D., Clague D., Escobar E., Lundsten L., Paduan J. B., Rouse G., Salcedo D. L., Soto L. A., Spelz-Madero R., Zierenberg R., Vrijenhoek R., Hydrothermal vent fields discovered in the southern Gulf of California clarify role of habitat in augmenting regional diversity. Proc. R. Soc. B Biol. Sci. 284, 20170817 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.R. R. Hessler, W. M. Smithey Jr., The distribution and community structure of megafauna at the Galapagos rift hydrothermal vents, in Hydrothermal Processes at Seafloor Spreading Centers, P. A. Rona, K. Boström, L. Laubier, K. L. Smith Jr., Eds. (Springer, 1983), pp. 735–770. [Google Scholar]
- 10.ten Hove H. A., Ztbrowius H., Laminatubus alvini gen. et sp.n. and Protis hydrothermica sp.n. (Polychaeta, Serpulidae) from the bathyal hydrothermal vent communities in the eastern Pacific. Zool. Scr. 15, 21–31 (1986). [Google Scholar]
- 11.Kupriyanova E. K., Nishi E., Kawato M., Fujiwara Y., New records of Serpulidae (Annelida, Polychaeta) from hydrothermal vents of North Fiji, Pacific Ocean. Zootaxa 2389, 57–68 (2010). [Google Scholar]
- 12.Knight-Jones P., Contributions to the taxonomy of Sabellidae (Polychaeta). Zool. J. Linn. Soc. 79, 245–295 (1983). [Google Scholar]
- 13.Capa M., Nishi E., Tanaka K., Fujikura K., First record of a Bispira species (Sabellidae, Polychaeta) from a hydrothermal vent. Mar. Biodivers. Rec. 6, e68 (2013). [Google Scholar]
- 14.Jumars P. A., Dorgan K. M., Lindsay S. M., Diet of worms emended: An update of polychaete feeding guilds. Annu. Rev. Mar. Sci. 7, 497–520 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Vinn O., Kupriyanova E. K., Kiel S., Serpulids (Annelida, Polychaeta) at Cretaceous to modern hydrocarbon seeps: Ecological and evolutionary patterns. Palaeogeogr. Palaeoclimatol. Palaeoecol. 390, 35–41 (2013). [Google Scholar]
- 16.Kiel S., Sami M., Taviani M., A serpulid-Anodontia-dominated methane-seep deposit from the Miocene of northern Italy. Acta Palaeontol. Pol. 63, 569–577 (2018). [Google Scholar]
- 17.Georgieva M. N., Paull C. K., Little C. T. S., McGann M., Sahy D., Condon D., Lundsten L., Pewsey J., Caress D. W., Vrijenhoek R. C., Discovery of an extensive deep-sea fossil serpulid reef associated with a cold seep, Santa Monica Basin, California. Front. Mar. Sci. 6, 115 (2019). [Google Scholar]
- 18.Olu K., Sibuet M., Harmegnies F., Foucher J.-P., Fiala-Médoni A., Spatial distribution of diverse cold seep communities living on various diapiric structures of the southern Barbados prism. Prog. Oceanogr. 38, 347–376 (1996). [Google Scholar]
- 19.Georgieva M. N., Little C. T. S., Watson J. S., Sephton M. A., Ball A. D., Glover A. G., Identification of fossil worm tubes from Phanerozoic hydrothermal vents and cold seeps. J. Syst. Palaeontol. 17, 287–329 (2019). [Google Scholar]
- 20.M. C. Baker, E. Z. Ramirez-Llodra, P. A. Tyler, C. R. German, A. Boetius, E. E. Cordes, N. Dubilier, C. R. Fisher, L. A. Levin, A. Metaxas, A. A. Rowden, R. S. Santos, T. M. Shank, C. L. Van Dover, C. M. Young, A. Warén, Biogeography, ecology, and vulnerability of chemosynthetic ecosystems in the deep sea, in Life in the World’s Oceans: Diversity, Distribution, and Abundance, A. McIntyre, Ed. (John Wiley & Sons, 2010), pp. 161–183. [Google Scholar]
- 21.Mau S., Rehder G., Sahling H., Schleicher T., Linke P., Seepage of methane at Jaco Scar, a slide caused by seamount subduction offshore Costa Rica. Int. J. Earth Sci. 103, 1801–1815 (2014). [Google Scholar]
- 22.Mau S., Rehder G., Arroyo I. G., Gossler J., Suess E., Indications of a link between seismotectonics and CH4 release from seeps off Costa Rica. Geochem. Geophys. Geosyst. 8, Q04003 (2007). [Google Scholar]
- 23.Childress J. J., Fisher C. R., Brooks J. M., Kennicutt M. C. II, Bidigare R., Anderson A. E., A methanotrophic marine molluscan (Bivalvia, Mytilidae) symbiosis: Mussels fueled by gas. Science 233, 1306–1308 (1986). [DOI] [PubMed] [Google Scholar]
- 24.Schmaljohann R., Flügel H. J., Methane-oxidizing bacteria in Pogonophora. Sarsia 72, 91–98 (2011). [Google Scholar]
- 25.Ruff S. E., Arnds J., Knittel K., Amann R., Wegener G., Ramette A., Boetius A., Microbial communities of deep-sea methane seeps at Hikurangi continental margin (New Zealand). PLOS ONE 8, e72627 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tavormina P. L., Ussler W. III, Steele J. A., Connon S. A., Klotz M. G., Orphan V. J., Abundance and distribution of diverse membrane-bound monooxygenase (Cu-MMO) genes within the Costa Rica oxygen minimum zone. Environ. Microbiol. Rep. 5, 414–423 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Rodrigues C. F., Hilário A., Cunha M. R., Weightman A. J., Webster G., Microbial diversity in Frenulata (Siboglinidae, Polychaeta) species from mud volcanoes in the Gulf of Cadiz (NE Atlantic). Antonie Van Leeuwenhoek 100, 83–98 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Hanson R. S., Hanson T. E., Methanotrophic bacteria. Microbiol. Mol. Biol. Rev. 60, 439–471 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spiro R. G., Bhoyroo V. D., Studies on the carbohydrate of collagens. Characterization of a glucuronic acid-mannose disaccharide unit from Nereis cuticle collagen. J. Biol. Chem. 255, 5347–5354 (1980). [PubMed] [Google Scholar]
- 30.Bulgheresi S., Schabussova I., Chen T., Mullin N. P., Maizels R. M., Ott J. A., A new C-type lectin similar to the human immunoreceptor DC-SIGN mediates symbiont acquisition by a marine nematode. Appl. Environ. Microbiol. 72, 2950–2956 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levin L. A., Michener R. H., Isotopic evidence for chemosynthesis-based nutrition of macrobenthos: The lightness of being at Pacific methane seeps. Limnol. Oceanogr. 47, 1336–1345 (2002). [Google Scholar]
- 32.Sommer S., Linke P., Pfannkuche O., Niemann H., Treude T., Benthic respiration in a seep habitat dominated by dense beds of ampharetid polychaetes at the Hikurangi Margin (New Zealand). Mar. Geol. 272, 223–232 (2010). [Google Scholar]
- 33.Hansman R. L., Thurber A. R., Levin L. A., Aluwihare L. I., Methane fates in the benthos and water column at cold seep sites along the continental margin of Central and North America. Deep-Sea Res. I Oceanogr. Res. Pap. 120, 122–131 (2017). [Google Scholar]
- 34.MacAvoy S. E., Carney R. S., Morgan E., Macko S. A., Stable isotope variation among the mussel Bathymodiolus childressi and associated heterotrophic fauna at four cold-seep communities in the Gulf of Mexico. J. Shellfish. Res. 27, 147–151 (2008). [Google Scholar]
- 35.Rubin-Blum M., Antony C. P., Sayavedra L., Martínez-Pérez C., Birgel D., Peckmann J., Wu Y.-C., Cardenas P., MacDonald I., Marcon Y., Sahling H., Hentschel U., Dubilier N., Fueled by methane: Deep-sea sponges from asphalt seeps gain their nutrition from methane-oxidizing symbionts. ISME J. 13, 1209–1225 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kochevar R. E., Childress J. J., Fisher C. R., Minnich E., The methane mussel : Roles of symbiont and host in the metabolic utilization of methane. Mar. Biol. 112, 389–401 (1992). [Google Scholar]
- 37.Page H. M., Fisher C. R., Childress J. J., Role of filter-feeding in the nutritional biology of a deep-sea mussel with methanotrophic symbionts. Mar. Biol. 104, 251–257 (1990). [Google Scholar]
- 38.Fisher C. R., Childress J. J., Organic carbon transfer from methanotrophic symbionts to the host hydrocarbon-seep mussel. Symbiosis 12, 221–235 (1992). [Google Scholar]
- 39.Fischer-Parton S., Parton R. M., Hickey P. C., Dijksterhuis J., Atkinson H. A., Read N. D., Confocal microscopy of FM4-64 as a tool for analysing endocytosis and vesicle trafficking in living fungal hyphae. J. Microsc. 198, 246–259 (2000). [DOI] [PubMed] [Google Scholar]
- 40.Laurich J. R., Batstone R. T., Dufour S. C., Temporal variation in chemoautotrophic symbiont abundance in the thyasirid bivalve Thyasira cf. gouldi. Mar. Biol. 162, 2017–2028 (2015). [Google Scholar]
- 41.Corliss J. B., Dymond J., Gordon L. I., Edmond J. M., von Herzen R. P., Ballard R. D., Green K., Williams D., Bainbridge A., Crane K., van Andel T. H., Submarine thermal springs on the galápagos rift. Science 203, 1073–1083 (1979). [DOI] [PubMed] [Google Scholar]
- 42.Felbeck H., Chemoautotrophic potential of the hydrothermal vent tube worm, Riftia pachyptila Jones (Vestimentifera). Science 213, 336–338 (1981). [DOI] [PubMed] [Google Scholar]
- 43.Cavanaugh C. M., Gardiner S. L., Jones M. L., Jannasch H. W., Waterbury J. B., Prokaryotic cells in the hydrothermal vent tube worm Riftia pachyptila Jones: Possible chemoautotrophic symbionts. Science 213, 340–342 (1981). [DOI] [PubMed] [Google Scholar]
- 44.Charlou J. L., Donval J. P., Fouquet Y., Jean-Baptiste P., Holm N., Geochemistry of high H2 and CH4 vent fluids issuing from ultramafic rocks at the rainbow hydrothermal field (36°14′N, MAR). Chem. Geol. 191, 345–359 (2002). [Google Scholar]
- 45.Wagner J. K. S., McEntee M. H., Brothers L. L., German C. R., Kaiser C. L., Yoerger D. R., Van Dover C. L., Cold-seep habitat mapping: High-resolution spatial characterization of the Blake Ridge Diapir seep field. Deep-Sea Res. II Top. Stud. Oceanogr. 92, 183–188 (2013). [Google Scholar]
- 46.Valentine D. L., Fisher G. B., Pizarro O., Kaiser C. L., Yoerger D., Breier J. A., Tarn J., Autonomous marine robotic technology reveals an expansive benthic bacterial community relevant to regional nitrogen biogeochemistry. Environ. Sci. Technol. 50, 11057–11065 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Levin L. A., Baco A. R., Bowden D. A., Colaco A., Cordes E. E., Cunha M. R., Demopoulos A. W. J., Gobin J., Grupe B. M., Le J., Metaxas A., Netburn A. N., Rouse G. W., Thurber A. R., Tunnicliffe V., Van Dover C. L., Vanreusel A., Watling L., Hydrothermal vents and methane seeps: Rethinking the sphere of influence. Front. Mar. Sci. 3, 72 (2016). [Google Scholar]
- 48.Mengerink K. J., Van Dover C. L., Ardron J., Baker M., Escobar-Briones E., Gjerde K., Koslow J. A., Ramirez-Llodra E., Lara-Lopez A., Squires D., Sutton T., Sweetman A. K., Levin L. A., A call for deep-ocean stewardship. Science 344, 696–698 (2014). [DOI] [PubMed] [Google Scholar]
- 49.Levy E. M., Lee K., Potential contribution of natural hydrocarbon seepage to benthic productivity and the fisheries of Atlantic Canada. Can. J. Fish. Aquatic Sci. 45, 349–352 (1988). [Google Scholar]
- 50.Cordes E. E., Jones D. O. B., Schlacher T. A., Amon D. J., Bernardino A. F., Brooke S., Carney R., DeLeo D. M., Dunlop K. M., Escobar-Briones E. G., Gates A. R., Génio L., Gobin J., Henry L.-A., Herrera S., Hoyt S., Joye M., Kark S., Mestre N. C., Metaxas A., Pfeifer S., Sink K., Sweetman A. K., Witte U., Environmental impacts of the deep-water oil and gas industry: A review to guide management strategies. Front. Environ. Sci. 4, 58 (2016). [Google Scholar]
- 51.Costello A. M., Lidstrom M. E., Molecular characterization of functional and phylogenetic genes from natural populations of methanotrophs in lake sediments. Appl. Environ. Microbiol. 65, 5066–5074 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Caporaso J. G., Lauber C. L., Walters W. A., Berg-Lyons D., Lozupone C. A., Turnbaugh P. J., Fierer N., Knight R., Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U.S.A. 108, 4516–4522 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pernthaler A., Pernthaler J., Simultaneous fluorescence in situ hybridization of mRNA and rRNA for the detection of gene expression in environmental microbes. Methods Enzymol. 397, 352–371 (2005). [DOI] [PubMed] [Google Scholar]
- 54.Daims H., Brühl A., Amann R., Schleifer K.-H., Wagner M., The domain-specific probe EUB338 is insufficient for the detection of all bacteria: Development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22, 434–444 (1999). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/14/eaay8562/DC1


