Abstract
The bacterial communities that collectively inhabit our body are called the microbiome. Virtually all body surface harbors bacteria. Recent advances in next-generation sequencing that have provided insight into the diversity, composition of bacterial communities, and their interaction are discussed in this review, as well as the current knowledge of how the microbiome promotes ocular health. The ocular surface is a site of low bacterial load. Sjögren Syndrome is an autoimmune disease that affects the exocrine glands, causing dry mouth and dry eye. Systemic antibiotic treatment and germ-free mice have demonstrated that commensal bacteria have a protective role for the ocular surface and lacrimal gland. The existence of a gut-eye-lacrimal gland axis-microbiome is discussed.
Keywords: microbiome, Sjögren Syndrome, dysbiosis, dry eye, dry mouth
1. Introduction
The microbiome is the generic term to describe the bacterial communities that inhabit our body. Similarly, the virome and the fungal mycobiome have been proposed as terms to identify virus and fungi communities, respectively. Virtually all body surface harbors bacteria, and most of them are commensal bacteria, namely, those that live in harmony with the host without causing deleterious effects. The advent of the Human Microbiome Project has identified that the number of genes that belong to intestinal bacteria is 150 x more abundant than the human genes [1]. The highest concentration of bacteria can be found in the gastrointestinal tract [2, 3]. Microbial composition varies according to body sites, as bacterial communities in places such as skin, urinary tract, and gastrointestinal tract tend to be very different from one to another [4]. There is increasing recognition that intestinal commensal microbiota limits colonization by enteropathogens and maintains mucosal immune homeostasis throughout the body. Commensal symbionts produce factors that support intestinal barrier function, promote the generation of tolerogenic dendritic cells and regulatory T cells (Tregs), and modulate cytokine production by natural killer cells. Short-chain fatty acids (SCFA), primarily butyric acid (and to a lesser degree propionic and acetic acid) produced by fermentation of dietary starches by commensal taxa, have been found to have potent immunomodulatory activity. This review will focus on the current knowledge of the microbiome and its interaction regarding the ocular surface and lacrimal glands in health and in Sjögren Syndrome (SS).
2-. Current knowledge of the mechanism of action of microbiome promoting health
Awareness about the beneficial role of commensal bacteria in human health started long before the description of the human microbiome, with the knowledge about the nutritional and therapeutic role of fermented products such as bread, kumis, and cheese nearly 10.000 years ago [5]. Nowadays, evidence from the literature demonstrates that our microbial commensals affect behavior, modulate the endocrine and immune system, alter epigenetic markers, and produce bioactive components and energy metabolites. With the advent of the massive sequencing methodologies and the availability of germ-free animals, the knowledge about the impact of human-associated microbial communities in health and disease has expanded. Germ-free animals are born and raised in sterile incubators with no contact with microbes. The first generation is delivered through a cesarean section to prevent contamination with the birth canal. Germ-free mice are fertile, leaner, have immunological, behavior, and morphological changes (such as enlarged cecum and low goblet cells) than animals conventionally raised in the specific-pathogen-free vivarium [6-8].
Recent evidence points to the existence of a brain-intestine microbiome axis. Male germ-free animals exhibit social impairment, and conversely, reduced anxiety-like behavior [9, 10], and mice with chronic stimulation by LPS might exhibit hyperphagia [11]. Although many of these effects are due to the interplay between the microbes and both hormones and immune mediators in the intestine, several commensals also produce neurotransmitters such as gamma-aminobutyric acid, serotonin, and acetylcholine, among others [9]. Whether the amount of microbial neurotransmitters is sufficient to have direct systemic effects requires more investigation; however, circulating neuroendocrine peptides are altered in germ-free rats [12]. Microbes might also stimulate the secretion of neurotransmitters by host colonic cells, which in turn, have an effect through the autonomic nervous system and the hypothalamic-pituitary-adrenal axis [13-15]. Lack of microbiome selectively impaired adrenal catecholamine responses to insulin-induced hypoglycemia [16]. Germ-free mice exhibit lower fecal gonadal hormones and corticosterone, which correlates to the tendency of these female mice to prefer female instead of male odors [17]. When these mice are subjected to either restraint stress or during acclimatization, they exhibit an elevation in the plasma levels of hypothalamic-pituitary-adrenal axis hormones, which is restored after microbiota reconstitution at an early stage of development [10, 14]. These animals have also upregulation of several glucocorticoid receptor pathway genes, such as water channels and fatty acid transporters, in the hippocampus [18]. The presence of fecal Ruminococcus in young pigs independently predicts serum cortisol levels [19]. An investigation is ongoing regarding the role of the “estrobolome,” namely, the enteric bacterial genes whose products are capable of metabolizing estrogens in women’s health [20]. These products might act in endogenous as well as exogenous estrogens, since the metabolism of several phytoestrogens, natural non-steroid products that act through the estrogen receptors, depend on the gut microbiota [21]. In men and postmenopausal, as oppose to premenopausal women, levels of total urinary non-ovarian estrogens and most of their metabolites were associated with the fecal microbiome diversity [22].
Among the mechanisms that associate commensal colonization with immunity are the induction of antimicrobial peptides and the regulation of inflammation in different cell types at the mucosal surfaces. Conventionally raised mice show greater jejunal Paneth cell numbers and increased transcription expression of the antimicrobial peptide Reg3γ, as compared to their germ-free counterparts [23], as well as greater number of goblet cells [8]. Human sebocytes co-cultured with Propionibacterium acnes, a member of the normal microbiome in the skin, produce AMP and exhibit altered differentiation and viability [24]. Exposure of pigs to L. reuteri, a probiotic bacteria found in different body surfaces in humans, increases the expression of porcine beta-defensins in the gut [25]. Also, commensal skin microbiota regulates the expression of complement genes in the skin [26]. Accumulating evidence indicate that several species of Lactobacillus modulate NF-κB and STAT-3-signaling pathways [27, 28]. The outcome of the Leishmania major intradermal infection in germ-free mice is influenced by the mono-colonization of the skin with the commensal S. epidermidis, an effect dependent on the production of IL-1α and the downstream signaling molecules [29]. The adaptive immune system is also not a passive spectator of commensals but actively interacts with microbes and they are even imperative for immune cell maturation and function. It is widely known that lymphoid organs have unorganized B- and T-zones and reduced cellularity in the absence of microbes [30]. Segmented filamentous bacteria antigens, presented by intestinal dendritic cells, drive mucosal Th-17 differentiation [31]. Strikingly, Th-17 related cytokines such as IL-17 regulate the expression of tight junction proteins, which are key molecules for the maintenance of proper mucosal permeability in the epithelia [32], preventing antigen bloodstream access and sensitization. In addition, these cytokines also promote the production of AMP by epithelial cells and the secretion of granulopoietic factors that drives neutrophil recruitment, likely facilitating immunity to pathogens [33]. The gut of germ-free mice exhibits also reduced numbers and function of T regulatory cells [34]. In scurfy mice, which bears a mutation in the Foxp3 gene, early oral feeding with Lactobacillus reuteri ameliorates autoimmune manifestations, an effect mediated by the adenosine receptors [35]. Polysaccharide A from the commensal Bacteroides fragilis induces expression of Foxp3 in human naïve CD4+ T cells and potentiates its suppressive function [36].
Among the main bioactive components of commensals are SCFA, such as lactate, acetate, propionate, and butyrate [37]. They are the result of bacterial fermentation of high fiber foods and are one of the primary energy sources for enterocytes and colonocytes. Although most of the SCFA produced are metabolized in the gut, small amounts of SCFA can also either exit the colon to the portal vein or circulate systemically. These might explain the effects of SCFA in immune, adipose, and neuronal cells, among others located at distant sites. Effects of SCFA are exerted through the free fatty-acid receptors 2 and 3 (FFAR2/3), although butyrate also signals through GPR109a (and its transporter, Slc5a8) [38]. One of the mechanisms of action of SCFA relates to the inhibition of histone deacetylases (HDAC). They allow the loosening of chromatin, enabling transcription factor accessibility to the DNA backbone. Naive CD4+ T cells cultured in Treg differentiation conditions together with butyrate exhibit enhanced histone acetylation at the Foxp3 promoter, leading to increased Foxp3 induction and an enhanced regulatory capacity of Tregs [39]. In a seminal work by Yuille, human colorectal adenocarcinoma cells were exposed to cell-free supernatants of 79 human commensals to evaluate their HDAC inhibitory properties. The three most potent HDAC inhibitor strains were also butyrate-producers. Among them, Megasphaera massiliensis MRx0029 potentiates cell HDAC inhibition in a model microbial consortium [40]. Animal models suggest autoimmunity may be promoted by reduced diversity of the intestinal microbiome with loss of commensal flora that produces metabolites, such as butyrate that suppress inflammation by promoting the generation of tolerogenic dendritic and regulatory T (Treg) cells. Stool butyrate concentrations have been found to decrease in antibiotic-treated and germ-free mice [39]. Concentrations of SCFAs, including butyrate in the colon correlated with the number of Foxp3+Tregs in the caecum [41] and oral administration of butyrate increased Foxp3 expression by Tregs, the number of Tregs in mucosal tissues and enhanced ability of dendritic cells to induce Treg differentiation [39]. We have shown that butyrate, one of the SCFAs, can have anti-inflammatory properties on the ocular surface epithelium [42], suggesting an indirect effect from a gut metabolite.
It has been shown that germ-free rats have lower IgA and IgM in their lacrimal glands than rats raised in conventional conditions [43]. Recently, a study by Kugadas and colleagues showed that gut colonization of germ-free mice with B. acidifaciens, a strict gut anaerobe, alters IgA transcript levels in the lacrimal glands [44]. This study provides functional evidence for the connection between gut microbiota and lacrimal gland-IgA transcript levels, postulating circulation of gut-derived B cells. The latter has been recently verified by Rojas and colleagues, which demonstrated that commensal-reactive gut-derived IL-10-producing IgA+ plasmablasts recirculate and suppress neuroinflammation [45]. It has also been shown that antibiotic dysbiosis-induced impaired corneal neurogenesis and corneal CCR2 negative macrophage distribution and activities in post-natal mice, and decreased corneal wound healing responses, demonstrating a strong connection between the eye and the microbiome [46, 47].
3-. Methods to evaluate the microbiome
The technical and computational tools for data collection and analysis of microbiomes have rapidly advanced over the last decade. The Human Microbiome Project, launched in 2007 by the NIH, has been instrumental in driving the development of these tools, including large-scale sequencing technologies, software and statistical tools, and the assembly of large reference sequence databases [48-50]. Initially, microbiome analysis was performed using bacterial 16S rRNA sequences, which contain alternating highly conserved and variable regions. The field has since expanded to include metagenomes and analysis of function by looking at microbiome transcriptional activity and translational products such as metabolites.
Initially, studies of microbial community members were limited to those which could be cultured in the laboratory; 16S sequencing allows identification of microbial community members that would otherwise be inaccessible. Bacterial 16S rRNA sequencing has been used for over four decades for the characterization of microbial communities, initially for environmental samples such as soil and wastewater and later for many other ecological niches, including the human body [51, 52]. The bacterial 16S rRNA molecule has unique features useful for characterizing microbial community members. Regions that are highly conserved across all bacteria alternate with variable regions, allowing for the design of universal primers that flank these variable regions. In addition, evolutionary relationships between bacterial genera can be characterized through sequence comparison of these more quickly-evolving regions, and the development of large annotated sequence reference databases such as the Ribosomal Database Project has provided essential tools [53-55]. The resulting phylogenetic framework generated using 16S sequences constitutes the foundation for most microbial ecology analyses today.
Next-generation sequencing technologies that arose 15 years ago allowed for high-throughput analysis of microbial communities at a much lower cost and time investment. This enabled microbial ecologists to identify rare members of communities and to gain a complete understanding of the composition of commensal communities. This advance was especially crucial for studying communities in niches like the ocular surface, which harbor far fewer bacteria than the gastrointestinal tract or oral cavity [56-58].
While 16S data allows us to identify bacterial genera associated with various health and disease states, it has also shown that the number of types of microbes associated with the human body is astoundingly large and diverse. The taxonomic composition can differ within body sites in the same person both spatially (such as in different points along the small intestine [59, 60]) and temporally (such as in the vagina during pregnancy [61]). Researchers have often focused on identifying specific genera associated with a particular condition, but even defining an overall microbial community signature can be very difficult because of the diversity from person to person, which can be influenced by many factors including diet, host genetics, and early microbial exposure and antibiotic use [62, 63]. Increasingly, studies are focusing on the metagenomes of these communities. A metagenome is the collective genome of a microbial community, or rather all the genes that are present in a community as opposed to all the species that are present. One major disadvantage of 16S sequencing is that since the highly conserved regions are conserved but not identical from species to species, universal primers favor amplification of specific sequences over others, and some organisms may be missed entirely. Shotgun sequencing of the entire metagenome avoids this problem. Shotgun-metagenomics uses next-generation sequencing to sequence the total DNA contained in a sample and allows insight into the functional aspects of a community [64, 65]. While the individual microbial species may differ from person to person, the functions carried out by those microbes could be very similar. Microbial products and metabolites have been shown in several instances to impact the functioning of the host [22-24]. Understanding the microbiome will require a multipronged approach that includes characterizing species content, gene content, and microbial protein content. This will help to inform much-needed functional studies to elucidate the interactions and functional impacts between commensal microbial communities and their human hosts.
4-. The Gut-Eye-Lacrimal Gland-Microbiome Axis in Sjögren Syndrome.
SS is an autoimmune disease that affects the exocrine glands, such as the salivary and lacrimal gland, and it is associated with significant morbidity. Patients often complain clinically of dry mouth and dry eye, fatigue, and other non-specific symptoms. It affects 9 times more women than men, and it can be either idiopathic (primary SS) or associated with systemic diseases such as rheumatoid arthritis and systemic lupus erythematosus. SS patients exhibit cellular infiltration of lymphocytes in the lacrimal and salivary glands, resulting in loss of the acinar cell function. However, the factors contributing to the misbalanced recruitment of the immune cells to the exocrine glands are not fully understood.
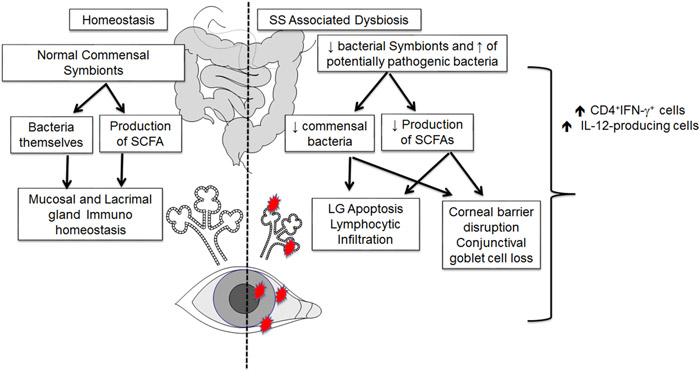
In mice, antibiotic-induced intestinal dysbiosis worsens the dry eye response to desiccating stress and increases the recruitment of effector T cells to the ocular surface [56]. Interestingly, we observed that germ-free C57BL/6 mice spontaneously develop SS-like disease, showing goblet cell loss, and cornea barrier dysfunction [66], hallmarks of dry eye in humans. Similarly, germ-free CD25 knock out mice (which exhibit severe SS disease), have an early onset and worse dacryoadenitis than conventional CD25 knock-out with complex murine microbiota [67]. Furthermore, we showed that the dry eye phenotype could be recapitulated after the adoptive transfer of CD4+ T cells into immunodeficient hosts and that fecal transplantation reversed both the spontaneous disease in the donor as well it decreased the pathogenicity of CD4+ T cells [66, 67]. These results are exciting because CD25 knock-out mice are devoid of Tregs, and therefore, unable to benefit from the intestinal Treg promotion after fecal transplant, as previously shown [68, 69]. These findings also highlight the existence of a gut-eye-lacrimal gland-microbiome axis (Figure 1).
Figure 1:
Proposed gut-ocular surface-lacrimal gland-microbiome axis.
Another evidence that supports the contribution of microbes-mediated signaling in ocular surface health is the MyD88-dependency of corneal epithelial barrier function and dry-eye induced damage [70, 71]. After oral antibiotics-induced dysbiosis, mice with LPS stimulation on the ocular surface have increased inflammatory response compared to vehicle-treated animals [72]. Interestingly, in the model of experimental autoimmune uveitis, antibiotic therapy, which induces dysbiosis, results in eye-protection [73]. These findings strongly suggest that commensals can be either beneficial or deleterious for the maintenance of the ocular surface tolerance.
Specific commensal molecules might cross-react with ocular antigens, activating ocular-specific T cells in the lamina propria that, in turn, migrate to the eye inducing the pathological damage. In accordance with this hypothesis, several peptides derived from commensal bacteria activate Ro60-reactive T cell hybridomas [74]. Ro60/SSA is one of the major autoantigens in SS and systemic lupus erythematosus. Immunization of NZM2758 mice with recombinant Ro52 protein caused loss of tear and salivary secretion that were independent of glandular involvement [75, 76]. It has been reported shared fecal microbiota composition in SS and systemic lupus erythematosus that differs from that of healthy controls [77]. Repeated injections of the outer membrane protein A of E. coli induce extra-intestinal gland inflammation (in the Harderian gland) and the production of SS-related autoantibodies [78]. On the other hand, it has been shown that commensals license regulatory cells in a specific or unspecific manner to perform active surveillance and prevent inflammation in distant sites of the body [35, 45, 69]. Since cornea, conjunctiva and lacrimal gland are highly innervated tissues, and loss of corneal axon density occurs after desiccation stress in mice [79], it is also possible that commensals not only regulate immune tolerance but also contributes to the proper development of nerve fibers in the ocular surface [80]. This considering the role of gut microbial factors in the development and homeostasis of the enteric nervous system [81].
4.1. Ocular Microbiome
The existence of a resident ocular surface microbiome remains in question. Conjunctival cultures performed on healthy eyes have recovered a limited number of organisms in up to 70% of eyes [82]. Staphylococcus epidermidis and Propionibacterium acnes are the most commonly recovered organisms in aerobic cultures. Gene sequencing methods have detected bacterial sequences on the ocular surface; however, these could represent contaminants, live or dead commensals, or skin bacteria transiently residing, but not colonizing the conjunctiva. Because of the paucity of bacterial genomic sequences on the ocular surface compared to other mucosal surfaces such as the large intestine, low abundant sequences from contaminating organisms could be misinterpreted as commensals. Because DNA is long-lived, the use of probes to identify live bacteria on the ocular surface has been proposed [83].
Similar to culture-based studies, metagenomic studies evaluating the ocular surface have found a limited number of bacterial taxa in low abundance. Doan et al. compared the relative abundance of bacterial/human DNA on the ocular surface with other body surfaces [57]. They calculated a 0.1 ratio of 16S bacterial rRNA/human actin on the ocular surface compared to a ratio >10 in samples taken from the facial skin or buccal mucosa. De Paiva and associates found the number of 16S genomic sequences in conjunctival swab samples was slightly higher (average of 216 mapped sequences per sample) than the number obtained from the collection swabs alone [56]. Ozkan et al. evaluated the stability of the ocular surface microbiome by culture-dependent and independent methods by sequentially sampling the ocular surface at baseline, 1 month, and 3 months [84]. A total of nine bacterial species or genera were cultured from 76.7% of cultures, typically with a low number of colony-forming units (<20 CFU in over 70% of samples). Cultured bacteria were from three phyla: Firmicutes, Actinobacteria, and Proteobacteria. No species were present in all subjects at all times or in all subjects at any given time point. By 16S rRNA sequencing, there was an average of 16 operational taxonomic units (OTUs) at each time point. There were significant differences in the number of OTUs identified between time points, but there were no significant differences between individuals or between different age or sex groups. Over 90% of the OTUs were in three phyla: Proteobacteria (64.4%), Firmicutes (15.5%), and Actinobacteria (15%). Cavuoto and colleagues found a significantly higher number of OTUs with greater diversity on the conjunctiva of children under the age of 18, compared to adults [85]. It has been shown that the composition of the ocular microbiome may change depending on the area that it was collected (conjunctiva vs. lid margin, for example), and the biogeography may be influenced by age and sex [86]. The findings of these studies at the genus or phyla level are listed in Table 1.
Table 1:
Ocular Microbiome in Adult Healthy Conjunctiva.
| Authors/Year | Detection Method | Taxa |
|---|---|---|
| Perkins/1975[82] | Culture (aerobic and anaerobic) | Aerobic: S. epidermidis, Streptococcus sp., Micrococcus sp., S. aureus, Gram neg rods, Corynebacterium sp., Bacillus sp. Anaerobic: Propionibacterium acnes, Peptostreptococcus sp., Lactobacillus sp., Clostridium sp., Eubacterium sp. |
| Graham/2007[92] | Culture and 16S rRNA sequencing | Coagulase negative Staphylococcus sp. Staphylococcus epidermidis Rhodococcus erythropolis, Uncultured bacterium, Corynebacterium sp., Propionibacterium acnes, Klebsiella sp., Klebsiella sp, Erwinia sp |
| Dong Q/2011[58] | 16S rRNA sequencing | Pseudomonas, Propionibacterium, Bradyrhizobium, Corynebacterium, Acinetobacter, Brevundimonas, Staphylococci, Aquabacterium, Streptococcus |
| Lee/2012[94] | 16S rRNA sequencing | Propionibacterium, Staphylococcus, Streptophyta, Corynebacterium, and Enhydrobacter |
| Doan/2016[57] | 16S rRNA sequencing | Corynebacteria, Proprionibacteria and coagulase negative Staphylococci |
| De Paiva/2016[56] | 16S rRNA sequencing | Firmicutes, Acinobacteria, Proteobacteria, Bacteroides phyla |
| Ozkan/2017[84] | Culture (aerobic and anaerobic) and 16S rRNA sequencing | Firmicutes, Acinobacteria, and Proteobacteria phyla by both methods |
| Wen/2017[95] | Shotgun sequencing | Propionibacterium acnes, Staphylococcus epidermidis, Escherichia coli, micrococcus luteus, Ochrobactrum anthropic, Acidovorarx sp, Acidovorarx ebreus, Actinobacter baumannii, Pseudomononas aeruginosa, S. haemolyticus, Neisseria meningitides and other sp. |
| Shimizu/2019[93] | Culture (aerobic and anaerobic) | S. epidermidis, Propionibacterium acnes and other sp. |
| Ozkan/2019[86] | 16S rRNA sequencing | Pseudomonas, Corynebacterium, Neisseriaceae, Staphylococcus, Acinetobacter, Aeribacillus, Streptococcus, Acetobacter, Bacillus, Veillonella, Thermoanaerobacterium, Deinococcus, Geobacillus, Sphingomonas, |
Overall, the metagenomic studies are consistent with culture studies in finding a low abundance, minimally diverse ocular surface microbiome. Neither the culture nor the genomic studies have addressed the physiological role of the ocular surface microbiome in maintaining homeostasis or suppressing inflammation. Studies in mice have pointed out for a protective role of conjunctival commensals in the resistance to Pseudomonas and C. albicans keratitis [44, 87, 88], but the lack of widespread bacterial colonization of conjunctiva from vendor C57BL/6J mice have made these studies very challenging [88]. However, commercially available Swiss Webster mice have detectable commensal species, such as Coagulase Negative Staphylococcus sp, indicating that the isolation and functional assays of murine conjunctival bacteria are feasible [87].
Changes in the conjunctival microbiome have been reported with contact lens wear [89] and with ocular and systemic diseases, such as diabetes [90, 91]. The ocular surface microbiome has been compared between healthy and dry eyes, including those with dry eye associated with the autoimmune disease Sjögren’s syndrome. Graham et al. cultured bacteria from the surfaces of most normal and dry eyes (75% of normal and 97% of dry eye)[92]. A higher number of bacterial colony-forming units grew from dry eyes (26 dry eye vs. 18 normal). There was no difference in the number of samples with PCR positive 16S rRNA sequences between normal and dry eyes. Genera specific gene sequences were also similar between groups. Conjunctival goblet cell density was inversely correlated with a number of cultured bacteria. Shimizu and colleagues found a much more abundant and diverse microbiome in the conjunctiva of patients with GVHD, and the number of species isolated correlated with the severity of ocular surface disease [93]. We compared the conjunctival microbiome in normal eyes and those of patients with SS keratoconjunctivitis sicca using 16S rRNA gene sequencing [56]. As opposed to the intestinal microbiome, which showed significant differences between groups, taxonomic units recovered from the ocular surface were similar between groups.
4.2. Oral microbiome
The oral cavity has a high bacterial load compared to the ocular surface. The actual role of oral microbiota in the pathogenesis of SS is not completed understood, but metagenomic changes have been identified. Bacterial mimicry has been proposed as one of the mechanisms that the microbiome may participate in disease induction. For example, mice immunized with Ro52 protein have decreased tear secretion and salivary flow rate than animals immunized with control proteins, and this effect was independent of tissue infiltration. [75, 76]. Another example is the high degree of homology of aquaporin Z of human-associated oral bacteria with human aquaporin 5, a major water channel protein involved in saliva secretion. Autoantibodies against aquaporin 5 are present in the sera of SS patients [96]. It is plausible that aquaporin 5 autoantibodies develop during the immune response to oral bacteria, and they actively participate in sicca pathogenesis. Recently, we detected bacteria within the ductal cells and in the area of infiltration in minor salivary gland biopsies from SS patients by in situ hybridization. Bacterial infection of ductal cells and its spread into neighboring acinar cells may explain the pattern of periductal lymphocytic infiltration in SS (Dysbiotic Oral Microbiota and Infected Salivary Glands in Sjӧgren Syndrome; Alam, J. manuscript under review).
Metagenomic changes in oral microbiota have been reported in SS patients [56, 97-101]. The results vary based on the type of samples that were collected (saliva, tongue and buccal swab), geographical area, ethnicity, diet, and the different variable regions of the 16S rRNA gene sequenced for analysis (Table 2). Nonetheless, there are few common findings: dysbiosis of oral microbiota is associated with hypo-salivation, and SS patients share their oral microbiome with an individual with sicca symptoms. Bacterial species richness estimated by Chao 1 index did not differ among healthy individuals and patients with dry mouth [56, 99, 102] while Shannon diversity either decreased [56, 97, 98, 102] or did not change [99]. Overall, the bacterial community composition of SS and non-SS sicca patients are comparable but revealed clear separation from healthy control communities by the UniFrac-based principal coordinate plot [56, 102]. As previously defined, >99% of core healthy oral microbiome belongs to the predominant taxa Firmicutes, Proteobacteria, Actinobacteria, Bacteroidetes, Fusobacteria, TM7, and Spirochaetes [103, 104]. Phylum Firmicutes increases while phylum Proteobacteria, TM7, and Spirochaetes decrease in SS [97-99]. Similarly, on the genus level, Haemophilus, Neisseria, and Porphyromonas were decreased in most of the studies [97-99, 102].
Table 2:
Oral microbiome and Sjögren Syndrome.
| Authors/Year | Detection Method |
Sample | Phyla or genus level comparison in SS | |
|---|---|---|---|---|
| Increased in SS | Decreased in SS | |||
| Almstah, 2003 [110] | Culture | Saliva | Candida species and Streptococcus | Fusobacterium nucleatum |
| De Paiva, 2016 [56] | V4 region of 16S rRNA | Tongue swab | Streptococcus | Leptotrichia, Fusobacterium, Bergeyella, Peptococcus, Butyrivibrio |
| Li M, 2016 [97] | V1-V3 region of 16S rRNA | Buccal mucosa swabs | Leucobacter, Delftia, Pseudochrobactrum, Ralstonia, Mitsuaria | Haemophilus, Neisseria, Comamona, Granulicatella, Limnohabitans, |
| Siddiqui H, 2016 [98] | V1-V2 region of 16S rRNA | Whole unstimulated saliva | Streptococcus Veillonella | Treponema,Peptostreptococcaceae Bacteroidaceae, Moryella, Catonella Fretibacterium, Porphyromonas Tannerella, |
| Van der Meulen T, 2018 [99] | V4 region of 16S rRNA | Buccal swab | Gemella, Dialister, Anaeroglobus, Lactobacillus, Parvimonas Peptostreptococcaceae Atopobium, Scardovia Bifidobacterium Alloscardovia | Streptococcus, Haemophilus, Neisseria,Lautropia, Ruminococcaceae, Parvimonas, Proteobacteria, Enterococcus, Granulicatella, Abiotrophia Bergeyella, Alloprevotella |
| Zhou Z, 2018 [102] | V3–V4 regions of 16S rRNA | Mouth rinse | Veillonella | Actinomyces, Haemophilus, Neisseria, Rothia, Porphyromonas, Peptostreptococcus |
| Zhou S, 2018[101] | V4-V5 region of 16S rRNA | Saliva | Bacteroidetes and actinobacteria | Proteobacteria |
| Rusthen S, 2019 [100] | V3–V5 regions of 16S rRNA | Saliva | Prevotella Veillonella | Streptococcus, Haemophilus Neisseria |
4.3. Intestinal microbiome
The intestinal microbiome is one of the more diverse niches in the human body. Members of the phyla Bacteroidetes and Firmicutes dominate the intestinal microbiome, and the microbiome of an individual is more similar to self over time than to others [105]. Bacteroides is generally the most abundant genus, but healthy humans exhibit a wide range of Bacteroides spp. abundance. Faecalibacterium prausnitzii is another abundant microbe that is found in the Clostridium cluster IV (phylum Firmicutes) and is a high butyrate producer [106].
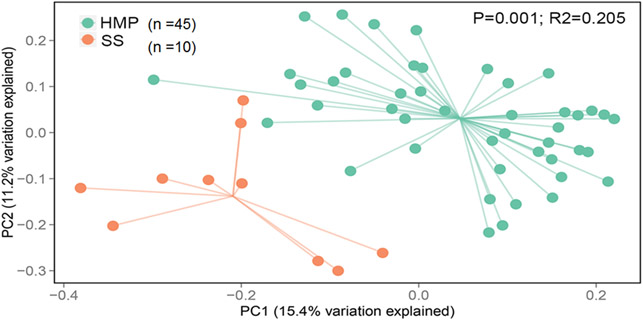
There are few studies that investigated the intestinal microbiome of SS patients. We were the first group to show that SS patients have dysbiosis [56], which was later confirmed by other groups. (Table 3) [77, 107, 108]. The principal component analysis showed that SS patients had segregation of microbial communities, and this was different from the healthy controls (Fig. 2, R2=0.025, P=0.001). Our own group has described differences in the intestinal microbiome taxa in SS patients in comparison to healthy controls with a significant reduction in Parabacteriodes and Faecalibacterium and an increase in Streptococcus, Blautia, Escherichia, and Pseudobutyrivibrio. In addition, the intestinal microbiome diversity inversely correlates with combined ocular and systemic disease severity index [56]. A similar finding was reported by Mandl, who also found decreased Bifidobacterium, Alistipes, and again, Faecalibacterium in SS, and reported that greater dysbiosis was associated with worse clinical scores [107]. In a study that compared primary SS and systemic lupus erythematosus patients, van der Meulen and colleagues observed that the intestinal microbiome did not differ significantly from both diseased groups but was different from healthy controls [77]. At least two studies identified that greater dysbiosis score or reduced number of OTUs correlated with greater clinical severity, suggesting a role of the microbiome in disease pathogenesis [56, 107]. Because SS is also a heterogeneous disease, it is possible that SS patients with dry eye may have a different microbiota than SS patients who have predominantly dry mouth. In a cohort of 1028 subjects, Whitcher and colleagues showed that patients with ocular surface disease might have extensive ocular staining without other features of SS [109]. Further functional studies are needed to dissect the individual contributions of the intestinal microbiome to SS [107].
Table 3:
Intestinal Microbiome and Sjögren Syndrome.
| Authors/Year | Detection Method |
Sample | Genus level comparison in SS | ||
|---|---|---|---|---|---|
| Increased in SS | Decreased in SS | Observations | |||
| de Paiva, 2016 [56] | V4 hypervariable region of 16S rRNA | Stools | Bilophila Bifidobacterium Moryella Lachnospira Anaerostipes Streptococcus Blautia Escherichia/Shigella Pseudobutyrivibrio | Bacteroides Parabacteroides Faecalibacterium Prevotella Odoribacter Haemophilus | Inverse correlation of OTUs and systemic severity score |
| Mandl/2017[107] | The GA-map™ Dysbiosis Test (16S) | Stools | N/A | Bifidobacterium Alistipes | Clinical disease activity scores were correlated with greater dysbiosis |
| Van der Meulen, 2019[77] | V2 and V4 hypervariable region of 16S rRNA | Stools | Bacteroides Ruminococcus Faecalibacterium Alistipes Proteobacteria Lachnoclostridium Barnesiella | Turicibacter Romboutsia Enterorhabdus FamilyXIIIAD3011 FamilyXIIIUCG Senegalimassilia Slackia Unknown genus | Intestinal microbiome of primary SS was similar to SLE but different from healthy controls |
Fig. 2:
Principal component analysis showing B diversity in intestinal microbiome from SS patients and subjects from the Human Microbiome Project [56].
4.4. Lessons learned from SS Mouse Animal Models
Because metagenomic analysis offers little insight into the physiologic or pathogenic role of bacteria on the ocular surface, oral cavity, and lacrimal gland, animal studies have been invaluable in trying to understand the in vivo relationship between bacteria and host. Experimental approaches have varied among topical application of bacterial ligands, administration of topical and systemic antibiotics, generation of germ-free mice, and colonization of germ-free with single or complex bacterial communities [46, 47, 56, 66, 67, 71, 87, 88, 111]. The education of the immune system by bacteria is well documented [112-114]. Antibiotic-induced dysbiosis is generally obtained after a short course of oral antibiotics and allow the investigation of acute ablation of microbes and its effects on the host, while germ-free studies allow the investigation of the chronical effects of a sterile environment.
Compared to mice that received regular water, antibiotic administration prior to desiccating stress worsens the dry eye phenotype in C57BL/6 mice [56]. This effect can be reversed by fecal material transplant [66]. Similarly, germ-free C57BL/6 mice spontaneously develop an SS-like syndrome, with female sex predilection, increased corneal staining, goblet cell loss, and production of inflammatory cytokines, such as IFN-γ and IL-12 [66]. Germ-free CD25 knock-out mice have an early onset and worse dacryoadenitis than conventional CD25 knock-out counterparts [67].
Conventionalization of the gut microbiome by fecal material transplant decreased the dry eye phenotype and decreased the pathogenicity of CD4+ T cells in both C57BL/6 and germ-free CD25 knock-out mice [66, 67]. These studies suggest a protective role for the microbiome in the lacrimal gland and ocular surface.
The effects of the microbiome in SS on the salivary glands are different than on lacrimal glands. For example, there is no difference in the lymphocytic infiltration of salivary glands between germ-free and conventional CD25 knock-out mice [67]. In the non-obese diabetic mice, a widely used strain to study SS [117, 125, 126], antibiotic usage alleviates sialadenitis while sialadenitis scores improve in a germ-free environment [118, 119]. Bacterial mimicry has been shown as a potential mechanism for microbiota-induced salivary gland inflammation after immunization [76, 127].
Table 4 summarizes the above-mentioned studies as well as other animal models that have been published regarding the known effects of the microbiome in the pathogenesis of SS.
Table 4:
Animals studies related to microbiome in Sjögren Syndrome.
| Strain/ condit ion |
Conventional microbiome |
Antibiotic-induced dysbiosis |
Germ-Free | Fecal material transplant or monocolonization studies in germ- free mice |
|---|---|---|---|---|
| Periodontal tissue in C57BL/6 mice | Normal |
|
|
|
| NOD | Severe sialadenitis (females) and dacryoadenitis (males) [117] |
|
|
|
| C57BL/6J | Minimal inflammation in the salivary and lacrimal glands |
|
|
|
| IL-2KO | Spontaneous colitis, dacryoadenitis and sialadenitis. [120] |
|
|
|
| CD25 KO (IL-2rα KO) | Spontaneous colitis, dacryoadenitis and sialadenitis.[120, 124] |
|
|
|
NR=not reported
5. Conclusions and implications for future research
The beneficial effects of the microbiome are just now beginning to be understood. We are making significant advances regarding bacterial identification. Data on pre-clinical models of SS suggest that fecal material transplantation might be an alternative therapeutic approach for severe SS. In fact, a clinical trial using fecal microbial transplant for SS is ongoing (PI: Anat Galor, University of Miami; www.clinicaltrials.gov, Identifier: NCT03926286) but most recently, the FDA suspended all fecal material transplant clinical trials due to serious adverse effects. A few pilot studies have investigated the use of probiotics in dry eye and SS patients with promising results [128-130], but more extensive, placebo-controlled, randomized clinical trials have not yet been performed.
We envisage that the identification of distinctive intestinal microbial consortia or single microorganisms with effects on the ocular surface will open new possibilities to cure autoimmune diseases such as SS with the use of probiotics. For this to happen, the field has to move from metagenomics to functional studies, so physiological and pathological interactions between microbes and human cells can be better understood.
Acknowledgments:
We would like to express our cordial gratitude to Leiqi Zhang and Zhiyuan Yu for the expert help with animal care and husbandry.
Support: This work was supported by NIH/NEI EY026893 (CSDP), Alkek Center for Metagenomics and Microbiome Research (CSDP), NIH/NEI EY002520 (Core Grant for Vision Research Department of Ophthalmology), Biology of Inflammation Center (CSDP), Research to Prevent Blindness Stein Innovation Award (RAB), Research to Prevent Blindness (Dept. Of Opthalmology), The Oshman Foundation, William Stamps Farish Fund, The Hamill Foundation, The Sid Richardson Foundation, Pathology Core (P30CA125123) and by Baylor Cytometry and Cell Sorting Core [CPRIT Core Facility Support Award (CPRIT-RP180672), P30 Cancer Center Support Grant (NCI-CA125123), NIH-RR024574, and NIH S10 OD025251 (Union BioMetrica BioSorter)]. No foreign funds were used.
Footnotes
Declarations of interest: Baylor College of Medicine has filed a patent related to some of the points in this review.
References
- [1].Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Belkaid Y, Harrison OJ. Homeostatic Immunity and the Microbiota. Immunity. 2017;46:562–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].He J, Li Y, Cao Y, Xue J, Zhou X. The oral microbiome diversity and its relation to human diseases. Folia microbiologica. 2015;60:69–80. [DOI] [PubMed] [Google Scholar]
- [4].Whiteside SA, Razvi H, Dave S, Reid G, Burton JP. The microbiome of the urinary tract--a role beyond infection. Nat Rev Urol. 2015;12:81–90. [DOI] [PubMed] [Google Scholar]
- [5].Ozen M, Dinleyici EC. The history of probiotics: the untold story. Beneficial microbes. 2015;6:159–65. [DOI] [PubMed] [Google Scholar]
- [6].Tazume S, Umehara K, Matsuzawa H, Yoshida T, Hashimoto K, Sasaki S. Immunological function of food-restricted germfree and specific pathogen-free mice. Jikken Dobutsu. 1991;40:523–8. [DOI] [PubMed] [Google Scholar]
- [7].Al-Asmakh M, Zadjali F. Use of Germ-Free Animal Models in Microbiota-Related Research. J Microbiol Biotechnol. 2015;25:1583–8. [DOI] [PubMed] [Google Scholar]
- [8].Kandori H, Hirayama K, Takeda M, Doi K. Histochemical, lectin-histochemical and morphometrical characteristics of intestinal goblet cells of germfree and conventional mice. Exp Anim. 1996;45:155–60. [DOI] [PubMed] [Google Scholar]
- [9].Johnson KV, Foster KR. Why does the microbiome affect behaviour? Nature reviews Microbiology. 2018;16:647–55. [DOI] [PubMed] [Google Scholar]
- [10].Neufeld KM, Kang N, Bienenstock J, Foster JA. Reduced anxiety-like behavior and central neurochemical change in germ-free mice. Neurogastroenterol Motil. 2011;23:255–64, e119. [DOI] [PubMed] [Google Scholar]
- [11].de La Serre CB, de Lartigue G, Raybould HE. Chronic exposure to low dose bacterial lipopolysaccharide inhibits leptin signaling in vagal afferent neurons. Physiology & behavior. 2015;139:188–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Husebye E, Hellstrom PM, Sundler F, Chen J, Midtvedt T. Influence of microbial species on small intestinal myoelectric activity and transit in germ-free rats. American journal of physiology Gastrointestinal and liver physiology. 2001;280:G368–80. [DOI] [PubMed] [Google Scholar]
- [13].Ge X, Ding C, Zhao W, Xu L, Tian H, Gong J, et al. Antibiotics-induced depletion of mice microbiota induces changes in host serotonin biosynthesis and intestinal motility. Journal of translational medicine. 2017;15:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Sudo N, Chida Y, Aiba Y, Sonoda J, Oyama N, Yu XN, et al. Postnatal microbial colonization programs the hypothalamic-pituitary-adrenal system for stress response in mice. The Journal of physiology. 2004;558:263–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Cawthon CR, de La Serre CB. Gut bacteria interaction with vagal afferents. Brain research. 2018;1693:134–9. [DOI] [PubMed] [Google Scholar]
- [16].Giri P, Hu F, La Gamma EF, Nankova BB. Absence of gut microbial colonization attenuates the sympathoadrenal response to hypoglycemic stress in mice: implications for human neonates. Pediatric research. 2019;85:574–81. [DOI] [PubMed] [Google Scholar]
- [17].Kamimura I, Watarai A, Takamura T, Takeo A, Miura K, Morita H, et al. Gonadal steroid hormone secretion during the juvenile period depends on host-specific microbiota and contributes to the development of odor preference. Developmental psychobiology. 2019. [DOI] [PubMed] [Google Scholar]
- [18].Luo Y, Zeng B, Zeng L, Du X, Li B, Huo R, et al. Gut microbiota regulates mouse behaviors through glucocorticoid receptor pathway genes in the hippocampus. Translational psychiatry. 2018;8:187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Mudd AT, Berding K, Wang M, Donovan SM, Dilger RN. Serum cortisol mediates the relationship between fecal Ruminococcus and brain N-acetylaspartate in the young pig. Gut microbes. 2017;8:589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Baker JM, Al-Nakkash L, Herbst-Kralovetz MM. Estrogen-gut microbiome axis: Physiological and clinical implications. Maturitas. 2017;103:45–53. [DOI] [PubMed] [Google Scholar]
- [21].Kolatorova L, Lapcik O, Starka L. Phytoestrogens and the intestinal microbiome. Physiological research. 2018;67:S401–s8. [DOI] [PubMed] [Google Scholar]
- [22].Flores R, Shi J, Fuhrman B, Xu X, Veenstra TD, Gail MH, et al. Fecal microbial determinants of fecal and systemic estrogens and estrogen metabolites: a cross-sectional study. Journal of translational medicine. 2012;10:253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Schoenborn AA, von Furstenberg RJ, Valsaraj S, Hussain FS, Stein M, Shanahan MT, et al. The enteric microbiota regulates jejunal Paneth cell number and function without impacting intestinal stem cells. Gut microbes. 2019;10:45–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Nagy I, Pivarcsi A, Kis K, Koreck A, Bodai L, McDowell A, et al. Propionibacterium acnes and lipopolysaccharide induce the expression of antimicrobial peptides and proinflammatory cytokines/chemokines in human sebocytes. Microbes and infection. 2006;8:2195–205. [DOI] [PubMed] [Google Scholar]
- [25].Liu H, Hou C, Wang G, Jia H, Yu H, Zeng X, et al. Lactobacillus reuteri I5007 Modulates Intestinal Host Defense Peptide Expression in the Model of IPEC-J2 Cells and Neonatal Piglets. Nutrients. 2017;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Chehoud C, Rafail S, Tyldsley AS, Seykora JT, Lambris JD, Grice EA. Complement modulates the cutaneous microbiome and inflammatory milieu. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:15061–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kumar A, Wu H, Collier-Hyams LS, Hansen JM, Li T, Yamoah K, et al. Commensal bacteria modulate cullin-dependent signaling via generation of reactive oxygen species. The EMBO journal. 2007;26:4457–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Lim SM, Jang HM, Jang SE, Han MJ, Kim DH. Lactobacillus fermentum IM12 attenuates inflammation in mice by inhibiting NF-kappaB-STAT3 signalling pathway. Beneficial microbes. 2017;8:407–19. [DOI] [PubMed] [Google Scholar]
- [29].Naik S, Bouladoux N, Wilhelm C, Molloy MJ, Salcedo R, Kastenmuller W, et al. Compartmentalized control of skin immunity by resident commensals. Science (New York, NY). 2012;337:1115–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Bauer H, Horowitz RE, Levenson SM, Popper H. The response of the lymphatic tissue to the microbial flora. Studies on germfree mice. The American journal of pathology. 1963;42:471–83. [PMC free article] [PubMed] [Google Scholar]
- [31].Goto Y, Panea C, Nakato G, Cebula A, Lee C, Diez MG, et al. Segmented filamentous bacteria antigens presented by intestinal dendritic cells drive mucosal Th17 cell differentiation. Immunity. 2014;40:594–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].de Paiva CS, Chotikavanich S, Pangelinan SB, Pitcher JD III, Fang B, Zheng X, et al. IL-17 disrupts corneal barrier following desiccating stress. Mucosal Immunol. 2009;2:243–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Abusleme L, Moutsopoulos NM. IL-17: overview and role in oral immunity and microbiome. Oral diseases. 2017;23:854–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Ostman S, Rask C, Wold AE, Hultkrantz S, Telemo E. Impaired regulatory T cell function in germ-free mice. European journal of immunology. 2006;36:2336–46. [DOI] [PubMed] [Google Scholar]
- [35].He B, Hoang TK, Wang T, Ferris M, Taylor CM, Tian X, et al. Resetting microbiota by Lactobacillus reuteri inhibits T reg deficiency-induced autoimmunity via adenosine A2A receptors. J Exp Med. 2017;214:107–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Telesford KM, Yan W, Ochoa-Reparaz J, Pant A, Kircher C, Christy MA, et al. A commensal symbiotic factor derived from Bacteroides fragilis promotes human CD39(+)Foxp3(+) T cells and Treg function. Gut microbes. 2015;6:234–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Nagpal R, Wang S, Ahmadi S, Hayes J, Gagliano J, Subashchandrabose S, et al. Human-origin probiotic cocktail increases short-chain fatty acid production via modulation of mice and human gut microbiome. Scientific reports. 2018;8:12649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Bourassa MW, Alim I, Bultman SJ, Ratan RR. Butyrate, neuroepigenetics and the gut microbiome: Can a high fiber diet improve brain health? Neurosci Lett. 2016;625:56–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Yuille S, Reichardt N, Panda S, Dunbar H, Mulder IE. Human gut bacteria as potent class I histone deacetylase inhibitors in vitro through production of butyric acid and valeric acid. PloS one. 2018;13:e0201073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–50. [DOI] [PubMed] [Google Scholar]
- [42].Hernandez H, de Souza RG, Yu Z, Britton RA, de Paiva CS. Anti-inflammatory properities of butyrate on the ocular surface epithelium. Investigative Ophthalmology & Visual Science 2019. 60 (9):2818. [Google Scholar]
- [43].Allansmith MR, Gudmundsson OG, Hann LE, Keys C, Bloch KJ, Taubman MA, et al. The immune response of the lacrimal gland to antigenic exposure. Curr Eye Res. 1987;6:921–7. [DOI] [PubMed] [Google Scholar]
- [44].Kugadas A, Wright Q, Geddes-McAlister J, Gadjeva M. Role of Microbiota in Strengthening Ocular Mucosal Barrier Function Through Secretory IgA. Invest Ophthalmol Vis Sci. 2017;58:4593–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Rojas OL, Probstel AK, Porfilio EA, Wang AA, Charabati M, Sun T, et al. Recirculating Intestinal IgA-Producing Cells Regulate Neuroinflammation via IL-10. Cell. 2019;176:610–24.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Liu J, Wu M, He J, Xiao C, Xue Y, Fu T, et al. Antibiotic-Induced Dysbiosis of Gut Microbiota Impairs Corneal Nerve Regeneration by Affecting CCR2-Negative Macrophage Distribution. Am J Pathol. 2018;188:2786–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Wu M, Liu J, Li F, Huang S, He J, Xue Y, et al. Antibiotic-induced dysbiosis of gut microbiota impairs corneal development in postnatal mice by affecting CCR2 negative macrophage distribution. Mucosal Immunol. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Knight R, Callewaert C, Marotz C, Hyde ER, Debelius JW, McDonald D, et al. The Microbiome and Human Biology. Annu Rev Genomics Hum Genet. 2017;18:65–86. [DOI] [PubMed] [Google Scholar]
- [49].Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Team NIHHMPA. A review of 10 years of human microbiome research activities at the US National Institutes of Health, Fiscal Years 2007–2016. Microbiome. 2019;7:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Zhou J, Xia B, Huang H, Palumbo AV, Tiedje JM. Microbial diversity and heterogeneity in sandy subsurface soils. Appl Environ Microbiol. 2004;70:1723–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, et al. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014;42:D633–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Maidak BL, Cole JR, Lilburn TG, Parker CT Jr., Saxman PR, Farris RJ, et al. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 2001;29:173–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Olsen GJ, Overbeek R, Larsen N, Marsh TL, McCaughey MJ, Maciukenas MA, et al. The Ribosomal Database Project. Nucleic Acids Res. 1992;20 Suppl:2199–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].de Paiva CS, Jones DB, Stern ME, Bian F, Moore QL, Corbiere S, et al. Altered Mucosal Microbiome Diversity and Disease Severity in Sjogren Syndrome. Sci Rep. 2016;6:23561–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Doan T, Akileswaran L, Andersen D, Johnson B, Ko N, Shrestha A, et al. Paucibacterial Microbiome and Resident DNA Virome of the Healthy Conjunctiva. Invest Ophthalmol Vis Sci. 2016;57:5116–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Dong Q, Brulc JM, Iovieno A, Bates B, Garoutte A, Miller D, et al. Diversity of bacteria at healthy human conjunctiva. Invest Ophthalmol Vis Sci. 2011;52:5408–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Donaldson GP, Lee SM, Mazmanian SK. Gut biogeography of the bacterial microbiota. Nat Rev Microbiol. 2016;14:20–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Mu C, Yang Y, Su Y, Zoetendal EG, Zhu W. Differences in Microbiota Membership along the Gastrointestinal Tract of Piglets and Their Differential Alterations Following an Early-Life Antibiotic Intervention. Front Microbiol. 2017;8:797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Gajer P, Brotman RM, Bai G, Sakamoto J, Schutte UM, Zhong X, et al. Temporal dynamics of the human vaginal microbiota. Sci Transl Med. 2012;4:132ra52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Luca F, Kupfer SS, Knights D, Khoruts A, Blekhman R. Functional Genomics of Host-Microbiome Interactions in Humans. Trends Genet. 2018;34:30–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Wang C, Zaheer M, Bian F, Quach D, Swennes AG, Britton RA, et al. Sjogren-Like Lacrimal Keratoconjunctivitis in Germ-Free Mice. Int J Mol Sci. 2018;19:pii: E565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Zaheer M, Wang C, Bian F, Yu Z, Hernandez H, de Souza RG, et al. Protective role of commensal bacteria in Sjogren Syndrome. J Autoimmun. 2018;pii: S0896–8411:45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–6. [DOI] [PubMed] [Google Scholar]
- [69].Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Metruccio MME, Tam C, Evans DJ, Xie AL, Stern ME, Fleiszig SMJ. Contributions of MyD88-dependent receptors and CD11c-positive cells to corneal epithelial barrier function against Pseudomonas aeruginosa. Sci Rep. 2017;7:13829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Reins RY, Lema C, Courson J, Kunnen CME, Redfern RL. MyD88 Deficiency Protects Against Dry Eye-Induced Damage. Invest Ophthalmol Vis Sci. 2018;59:2967–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Wang CB F; Simmons KT; Zaheer M; Pflugfelder SC; de Paiva CS. . Commensal Bacteria Modulate Ocular Surface Inflammatory Response to Liposaccharide. Invest Ophthalmol Vis Sci. 2017;58(8):3916. [Google Scholar]
- [73].Horai R, Zarate-Blades CR, Dillenburg-Pilla P, Chen J, Kielczewski JL, Silver PB, et al. Microbiota-Dependent Activation of an Autoreactive T Cell Receptor Provokes Autoimmunity in an Immunologically Privileged Site. Immunity. 2015;43:343–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Szymula A, Rosenthal J, Szczerba BM, Bagavant H, Fu SM, Deshmukh US. T cell epitope mimicry between Sjogren’s syndrome Antigen A (SSA)/Ro60 and oral, gut, skin and vaginal bacteria. Clin Immunol. 2014;152:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Trzeciak M, Bagavant H, Papinska J, Deshmukh US. Immune Response Targeting Sjogren’s Syndrome Antigen Ro52 Suppresses Tear Production in Female Mice. Int J Mol Sci. 2018;19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Sroka M, Bagavant H, Biswas I, Ballard A, Deshmukh US. Immune response against the coiled coil domain of Sjogren’s syndrome associated autoantigen Ro52 induces salivary gland dysfunction. Clin Exp Rheumatol. 2018;36 Suppl 112:41–6. [PMC free article] [PubMed] [Google Scholar]
- [77].van der Meulen TA, Harmsen HJM, Vila AV, Kurilshikov A, Liefers SC, Zhernakova A, et al. Shared gut, but distinct oral microbiota composition in primary Sjogren’s syndrome and systemic lupus erythematosus. J Autoimmun. 2019;97:77–87. [DOI] [PubMed] [Google Scholar]
- [78].Yanagisawa N, Ueshiba H, Abe Y, Kato H, Higuchi T, Yagi J. Outer Membrane Protein of Gut Commensal Microorganism Induces Autoantibody Production and Extra-Intestinal Gland Inflammation in Mice. International journal of molecular sciences. 2018;19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Stepp MA, Pal-Ghosh S, Tadvalkar G, Williams A, Pflugfelder SC, de Paiva CS. Reduced intraepithelial corneal nerve density and sensitivity accompany desiccating stress and aging in C57BL/6 mice. Exp Eye Res. 2018;169:91–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].McMonnies CW. The potential role of neuropathic mechanisms in dry eye syndromes. Journal of optometry. 2017;10:5–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Obata Y, Pachnis V. The Effect of Microbiota and the Immune System on the Development and Organization of the Enteric Nervous System. Gastroenterology. 2016;151:836–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Perkins RE, Kundsin RB, Pratt MV, Abrahamsen I, Leibowitz HM. Bacteriology of normal and infected conjunctiva. J Clin Microbiol. 1975;1:147–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Wan SJ, Sullivan AB, Shieh P, Metruccio MME, Evans DJ, Bertozzi CR, et al. IL-1R and MyD88 Contribute to the Absence of a Bacterial Microbiome on the Healthy Murine Cornea. Front Microbiol. 2018;9:1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Ozkan J, Nielsen S, Diez-Vives C, Coroneo M, Thomas T, Willcox M. Temporal Stability and Composition of the Ocular Surface Microbiome. Scientific reports. 2017;7:9880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Cavuoto KM, Mendez R, Miller D, Galor A, Banerjee S. Effect of clinical parameters on the ocular surface microbiome in children and adults. Clin Ophthalmol. 2018;12:1189–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Ozkan J, Willcox M, Wemheuer B, Wilcsek G, Coroneo M, Thomas T. Biogeography of the human ocular microbiota. The ocular surface. 2019;17:111–8. [DOI] [PubMed] [Google Scholar]
- [87].Kugadas A, Christiansen SH, Sankaranarayanan S, Surana NK, Gauguet S, Kunz R, et al. Impact of Microbiota on Resistance to Ocular Pseudomonas aeruginosa-Induced Keratitis. PLoS Pathog. 2016;12:e1005855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].St Leger AJ, Desai JV, Drummond RA, Kugadas A, Almaghrabi F, Silver P, et al. An Ocular Commensal Protects against Corneal Infection by Driving an Interleukin-17 Response from Mucosal gammadelta T Cells. Immunity. 2017;47:148–58.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Shin H, Price K, Albert L, Dodick J, Park L, Dominguez-Bello MG. Changes in the Eye Microbiota Associated with Contact Lens Wearing. mBio. 2016;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Li S, Yi G, Peng H, Li Z, Chen S, Zhong H, et al. How Ocular Surface Microbiota Debuts in Type 2 Diabetes Mellitus. Front Cell Infect Microbiol. 2019;9:202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Ham B, Hwang HB, Jung SH, Chang S, Kang KD, Kwon MJ. Distribution and Diversity of Ocular Microbial Communities in Diabetic Patients Compared with Healthy Subjects. Curr Eye Res. 2018;43:314–24. [DOI] [PubMed] [Google Scholar]
- [92].Graham JE, Moore JE, Jiru X, Moore JE, Goodall EA, Dooley JS, et al. Ocular pathogen or commensal: a PCR-based study of surface bacterial flora in normal and dry eyes. Invest Ophthalmol Vis Sci. 2007;48:5616–23. [DOI] [PubMed] [Google Scholar]
- [93].Shimizu E, Ogawa Y, Saijo Y, Yamane M, Uchino M, Kamoi M, et al. Commensal microflora in human conjunctiva; characteristics of microflora in the patients with chronic ocular graft-versus-host disease. The ocular surface. 2019;17:265–71. [DOI] [PubMed] [Google Scholar]
- [94].Lee SH OD, Jung JY, Kim JC, Jeon CO. Comparative Ocular Microbial Communities in Humans with and without Blepharitis. Invest Ophthalmol Vis Sci 2012;53:5585–93. [DOI] [PubMed] [Google Scholar]
- [95].Wen X, Miao L, Deng Y, Bible PW, Hu X, Zou Y, et al. The Influence of Age and Sex on Ocular Surface Microbiota in Healthy Adults. Invest Ophthalmol Vis Sci. 2017;58:6030–7. [DOI] [PubMed] [Google Scholar]
- [96].Alam J, Koh JH, Kim N, Kwok SK, Park SH, Song YW, et al. Detection of autoantibodies against aquaporin-5 in the sera of patients with primary Sjögren’s syndrome. Immunol Res. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Li M, Zou Y, Jiang Q, Jiang L, Yu Q, Ding X, et al. A preliminary study of the oral microbiota in Chinese patients with Sjogren’s syndrome. Arch Oral Biol. 2016;70:143–8. [DOI] [PubMed] [Google Scholar]
- [98].Siddiqui H, Chen T, Aliko A, Mydel PM, Jonsson R, Olsen I. Microbiological and bioinformatics analysis of primary Sjogren’s syndrome patients with normal salivation. J Oral Microbiol. 2016;8:31119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].van der Meulen TA, Harmsen HJM, Bootsma H, Liefers SC, Vich Vila A, Zhernakova A, et al. Dysbiosis of the buccal mucosa microbiome in primary Sjogren’s syndrome patients. Rheumatology (Oxford). 2018;57:2225–34. [DOI] [PubMed] [Google Scholar]
- [100].Rusthen S, Kristoffersen AK, Young A, Galtung HK, Petrovski BE, Palm O, et al. Dysbiotic salivary microbiota in dry mouth and primary Sjogren’s syndrome patients. PLoS One. 2019;14:e0218319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Zhou S, Cai Y, Wang M, Yang WD, Duan N. Oral microbial flora of patients with Sicca syndrome. Mol Med Rep. 2018;18:4895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Zhou Z, Ling G, Ding N, Xun Z, Zhu C, Hua H, et al. Molecular analysis of oral microflora in patients with primary Sjogren’s syndrome by using high-throughput sequencing. PeerJ. 2018;6:e5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Zaura E, Keijser BJ, Huse SM, Crielaard W. Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol. 2009;9:259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Crielaard W, Zaura E, Schuller AA, Huse SM, Montijn RC, Keijser BJ. Exploring the oral microbiota of children at various developmental stages of their dentition in the relation to their oral health. BMC Med Genomics. 2011;4:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Human Microbiome Project C. A framework for human microbiome research. Nature. 2012;486:215–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Miquel S, Martin R, Bridonneau C, Robert V, Sokol H, Bermudez-Humaran LG, et al. Ecology and metabolism of the beneficial intestinal commensal bacterium Faecalibacterium prausnitzii. Gut microbes. 2014;5:146–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Mandl T, Marsal J, Olsson P, Ohlsson B, Andreasson K. Severe intestinal dysbiosis is prevalent in primary Sjogren’s syndrome and is associated with systemic disease activity. Arthritis Res Ther. 2017;19:237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Mendez R, Watane A, Farhangi M, Cavuoto KM, Leith T, Budree S, et al. Gut microbial dysbiosis in individuals with Sjögren’s disease. bioRxiv. 2019:645739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Whitcher JP, Shiboski CH, Shiboski SC, Heidenreich AM, Kitagawa K, Zhang S, et al. A simplified quantitative method for assessing keratoconjunctivitis sicca from the Sjogren’s Syndrome International Registry. Am JOphthalmol. 2010;149:405–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Almstah IA, Wikstrom M, Stenberg I, Jakobsson A, Fagerberg-Mohlin B. Oral microbiota associated with hyposalivation of different origins. Oral Microbiol Immunol. 2003;18:1–8. [DOI] [PubMed] [Google Scholar]
- [111].Redfern RL, Reins RY, McDermott AM. Toll-like receptor activation modulates antimicrobial peptide expression by ocular surface cells. ExpEye Res. 2011;92:209–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Haghikia A, Jorg S, Duscha A, Berg J, Manzel A, Waschbisch A, et al. Dietary Fatty Acids Directly Impact Central Nervous System Autoimmunity via the Small Intestine. Immunity. 2015;43:817–29. [DOI] [PubMed] [Google Scholar]
- [113].Cho SY, Kim J, Lee JH, Sim JH, Cho DH, Bae IH, et al. Modulation of gut microbiota and delayed immunosenescence as a result of syringaresinol consumption in middle-aged mice. Sci Rep. 2016;6:39026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Luczynski P, McVey Neufeld KA, Oriach CS, Clarke G, Dinan TG, Cryan JF. Growing up in a Bubble: Using Germ-Free Animals to Assess the Influence of the Gut Microbiota on Brain and Behavior. Int J Neuropsychopharmacol. 2016;19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Irie K, Tomofuji T, Ekuni D, Morita M, Shimazaki Y, Darveau RP. Impact of Oral Commensal Bacteria on Degradation of Periodontal Connective Tissue in Mice. J Periodontol. 2015;86:899–905. [DOI] [PubMed] [Google Scholar]
- [116].Wu YY, Westwater C, Xiao E, Dias Correa J, Xiao WM, Graves DT. Establishment of oral bacterial communities in germ-free mice and the influence of recipient age. Mol Oral Microbiol. 2018;33:38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Makino S, Kunimoto K, Muraoka Y, Katagiri K. Effect of castration on the appearance of diabetes in NOD mouse. Jikken Dobutsu. 1981;30:137–40. [DOI] [PubMed] [Google Scholar]
- [118].Hansen CHF, Larsen CS, Petersson HO, Zachariassen LF, Vegge A, Lauridsen C, et al. Targeting gut microbiota and barrier function with prebiotics to alleviate autoimmune manifestations in NOD mice. Diabetologia. 2019;62:1689–700. [DOI] [PubMed] [Google Scholar]
- [119].Hansen CH, Yurkovetskiy LA, Chervonsky AV. Cutting Edge: Commensal Microbiota Has Disparate Effects on Manifestations of Polyglandular Autoimmune Inflammation. J Immunol. 2016;197:701–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Sharma R, Zheng L, Guo X, Fu SM, Ju ST, Jarjour WN. Novel animal models for Sjogren’s syndrome: expression and transfer of salivary gland dysfunction from regulatory T cell-deficient mice. J Autoimmun. 2006;27:289–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Contractor NV, Bassiri H, Reya T, Park AY, Baumgart DC, Wasik MA, et al. Lymphoid hyperplasia, autoimmunity, and compromised intestinal intraepithelial lymphocyte development in colitis-free gnotobiotic IL-2-deficient mice. J Immunol. 1998;160:385–94. [PubMed] [Google Scholar]
- [122].Waidmann M, Allemand Y, Lehmann J, di GS, Bucheler N, Hamann A, et al. Microflora reactive IL-10 producing regulatory T cells are present in the colon of IL-2 deficient mice but lack efficacious inhibition of IFN-gamma and TNF-alpha production. Gut. 2002;50:170–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Waidmann M, Bechtold O, Frick J-s, Lehr H-a, Schubert S, Dobrindt U, et al. Bacteroides vulgatus protects against escherichia coli-induced colitis in gnotobiotic interleukin-2-deficient mice. Gastroenterology. 2003;125:162–77. [DOI] [PubMed] [Google Scholar]
- [124].Willerford DM CJ, Ferry JA, Davidson L, Ma A, Alt FW. Interleukin-2 receptor alpha chain regulates the size and content of the peripheral lymphoid compartment. Immunity. 1995;3:521–30. [DOI] [PubMed] [Google Scholar]
- [125].Barr JY, Wang X, Kreiger PA, Lieberman SM. Salivary gland-protective regulatory T cell dysfunction underlies female-specific sialadenitis in the nonobese diabetic mouse model of Sjogren syndrome. Immunology. 2018;55:225–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Tellefsen S, Morthen MK, Richards SM, Lieberman SM, Rahimi Darabad R, Kam WR, et al. Sex Effects on Gene Expression in Lacrimal Glands of Mouse Models of Sjogren Syndrome. Invest Ophthalmol Vis Sci. 2018;59:5599–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Szczerba BM, Kaplonek P, Wolska N, Podsiadlowska A, Rybakowska PD, Dey P, et al. Interaction between innate immunity and Ro52-induced antibody causes Sjögren’s syndrome-like disorder in mice. Ann Rheum Dis. 2016;75:617–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Chisari G, Chisari EM, Borzi AM, Ozyalcin E, Chisari CG. Aging eye microbiota in Dry Eye Syndrome in patients treated with Enterococcus faecium and Saccharomyces boulardii. Curr Clin Pharmacol. 2017;12:99–105. [DOI] [PubMed] [Google Scholar]
- [129].Chisari G, Chisari EM, Francaviglia A, Chisari CG. The mixture of bifidobacterium associated with fructo-oligosaccharides reduces the damage of the ocular surface. Clin Ter. 2017;168:e181–e5. [DOI] [PubMed] [Google Scholar]
- [130].Kawashima M, Nakamura S, Izuta Y, Inoue S, Tsubota K. Dietary Supplementation with a Combination of Lactoferrin, Fish Oil, and Enterococcus faecium WB2000 for Treating Dry Eye: A Rat Model and Human Clinical Study. The ocular surface. 2016;14:255–63. [DOI] [PubMed] [Google Scholar]