Abstract
Venetoclax-based therapy can induce responses in approximately 70% of older previously untreated acute myeloid leukemia (AML) patients. However, upfront resistance as well as relapse following initial response demonstrate the need for a deeper understanding of resistance mechanisms. In the present study, we report that responses to venetoclax + azacitidine in AML patients correlate closely with developmental stage, where phenotypically primitive AML is sensitive, but monocytic AML is more resistant. Mechanistically, resistant monocytic AML has a distinct transcriptome profile, loses expression of venetoclax target BCL2 and relies on MCL1 to mediate oxidative phosphorylation and survival. This differential sensitivity drives a selective process in patients which favors the outgrowth of monocytic subpopulations at relapse. Based on these findings, we conclude that resistance to venetoclax + azacitidine can arise due to biological properties intrinsic to monocytic differentiation. We propose that optimal AML therapies should be designed so as to independently target AML subclones that may arise at differing stages of pathogenesis.
Keywords: Acute myeloid leukemia, BCL2, MCL1, venetoclax, monocytic differentiation, OXPHOS
Introduction
With a median age at diagnosis of 68, AML is predominantly a disease of the elderly(1). For decades, intensive induction chemotherapy has been the standard of care for patients with AML. However, most elderly patients are poor candidates for this type of therapy, given their higher treatment-related mortality and lower response rates (2,3). The standard of care has therefore been to offer low-dose therapies such as hypomethylating agents (HMA). However, response to HMA therapy is limited and long-term overall survival is negligible (4,5). Recent clinical trials have reported that the addition of the highly specific BCL2 inhibitor venetoclax to the HMA backbone can greatly increase the response rates and potentially the overall survival (OS) for older, newly diagnosed AML patients who are unfit for conventional chemotherapy (6,7). These findings led to the recent United States Food and Drug Administration approval of this regimen for this population, and it is now considered to be the standard care.
The combination of venetoclax and the HMA azacitidine results in a remission rate of approximately 70% (6,7). However, a significant minority of patients do not achieve a remission and are refractory. In addition, the majority of patients who do achieve a remission ultimately relapse(6,7). It is therefore critical to understand mechanisms of venetoclax resistance. Historically, adverse disease features that predict response to conventional therapies have been defined in the setting of chemotherapy-based regimens, and particular chromosomal and genomic abnormalities strongly predict poor outcomes in this context(8). Notably though, with the exception of in vitro work which has demonstrated a link between TP53 and venetoclax resistance(9), analyses of clinical response data have for the most part not identified traditional adverse risk features as poor outcome predictors in AML patients treated with venetoclax-based therapies (6,7,10).
One explanation for this observation is that venetoclax with azacitidine has a novel mechanism of action in AML(10,11), necessitating a re-assessment of biological features associated with prognostication. To this end, we considered the relative impact of the developmental stages of AML on BCL2 mediated metabolism. Previously, we identified BCL2 as differentially expressed in subpopulations of AML cells that are enriched for malignant stem/progenitor cells compared to more differentiated tumor cells (12). In addition, recent in vitro studies show reduced venetoclax sensitivity in primary AML cells with a monocytic phenotype(13). Thus, we hypothesized that clinical features of AML that are indicative of myeloid differentiation status may correlate with reduced BCL2 dependence in AML patients. As reported herein, we discovered that more differentiated monocytic AML is much more likely to be refractory to venetoclax-based therapy. Further, we demonstrate that altered regulation of energy metabolism and survival is an inherent property of AML development and appears to underlie this resistance. Consequently, the selective pressure of venetoclax-based therapy mediates profound changes in the biology of leukemic cell populations that manifests in altered developmental and metabolic properties.
Results
AML patients with monocytic disease are more likely to be refractory to venetoclax + azacitidine
To test whether differentiation status may predict responsiveness to venetoclax + azacitidine (VEN+AZA) in the clinic, we retrospectively reviewed 100 consecutive, newly diagnosed, previously untreated AML patients who received VEN+AZA at the University of Colorado between January 2015 and October 2019 (all baseline characteristics are listed in Supplementary Table S1). We analyzed several baseline factors to determine the ability of each to predict disease that was refractory to treatment as defined by the European Leukemia Network (lack of complete remission [CR], CR with incomplete recovery of peripheral blood counts [CRi], partial remission [PR], or morphological leukemia free state [MLFS])(8). The median age of the cohort was 72; 20 (20%) had a documented antecedent hematological disorder; 64 (64%) had adverse risk disease by ELN criteria(8).
To specifically examine features associated with myeloid differentiation, we initially employed the FAB (French, American, British) classification system. Although this system is no longer employed for clinical purposes, it provides a well-described and clinically associated means to segregate AML patients by virtue of myeloid differentiation status. In our VEN+AZA treated patient cohort, 13 patients (13%) were identified as the FAB-M5 subtype, which is defined as a more differentiated phenotype of monocytic AML, and 77 (77%) were FAB-M0 or M1, indicative of a less differentiated phenotype (Supplementary Fig. S1A). Univariate analysis revealed sex (p=0.0495), presence of a RAS pathway mutation (p=0.0039) and FAB-M5 maturation state (p=<0.0001) to be associated with disease that was refractory to VEN+AZA (Table 1). A multivariate analysis revealed only the FAB-M5 maturation state (p=.0066) to be predictive of refractory response (Table 1). Specifically, 62% of FAB-M5 patients were refractory to VEN+AZA, whereas 0% of FAB-M4 and only 8% of non-FAB-M5 patients were refractory (Supplementary Fig. S1B). In addition, the median overall survival in FAB-M5 patients was 89 days, compared with 518 days for non-FAB-M5 patients (p=0.0039) (Supplementary Fig. S1C). These findings indicate a strong correlation between myeloid differentiation status and resistance to venetoclax-based therapy.
Table 1. Baseline Characteristics and univariate and multivariate logistic regression analysis of 100 consecutive patients with newly diagnosed, previously untreated AML who received venetoclax + azacitidine.
Patient characteristics are detailed in Supplementary Table S1.
NA = Not Available; NE = Not Estimable.
| Baseline Variables | Value | Univariate Analysis as a Predictor for Refractory Disease | Multivariate Analysis as a Predictor for Refractory Disease | ||||
|---|---|---|---|---|---|---|---|
| OR | 95% CI | p-value | OR | 95% CI | p-value | ||
| Age (median) | 71.5 (22-89) | 0.984 | 0.947, 1.022 | 0.4028 | |||
| Sex (female) | 51 (51%) | 3.401 | 1.002, 11.539 | 0.0495 | 2.096 | 0.417, 10.544 | 0.3694 |
| Antecedent hematologic disorder | 20 (20%) | 0.573 | 0.118, 2.772 | 0.4884 | |||
| Complex cytogenetics | 28 (28%) | 2.667 | 0.863, 8.237 | 0.0883 | |||
| ELN Prognostic Group | |||||||
| Favorable | 18 (18%) | ||||||
| Intermediate | 17 (17%) | 4.078 | 0.494, 33.642 | 0.0697 | |||
| Adverse | 64 (64%) | ||||||
| NA | 1 (1%) | ||||||
| RAS Pathway Mutations | 14 (14%) | 6.417 | 1.813, 22.708 | 0.0039 | 2.266 | 0.201, 25.522 | 0.5080 |
| TP53 | 10 (10%) | 1.481 | 0.282, 7.766 | 0.6424 | |||
| IDH1/IDH2 | 27 (27%) | NE | NE | 0.9521 | |||
| NPM1 | 27 (27%) | 0.162 | 0.020, 1.298 | 0.0865 | 0.488 | 0.034, 6.966 | 0.5967 |
| FLT3-ITD | 18 (18%) | 0.663 | 0.136, 3.273 | 0.6119 | |||
| ASXL1 | 24 (24%) | 1.182 | 0.339, 4.122 | 0.7932 | |||
| French-American-British (FAB) Classification | |||||||
| M0/M1 | 77 (77%) | 0.131 | 0.040 0.428 |
0.0008 | |||
| M2 | 1 (1%) | ||||||
| M4 | 8 (8%) | NE | NE | 0.9745 | |||
| M5 | 13 (13%) | 18.285 | 4.701, 71.129 | <0.0001 | 33.481 | 2.657, 421.90 | 0.0066 |
| M6a | 1 (1%) | ||||||
Monocytic AML is intrinsically resistant to venetoclax + azacitidine
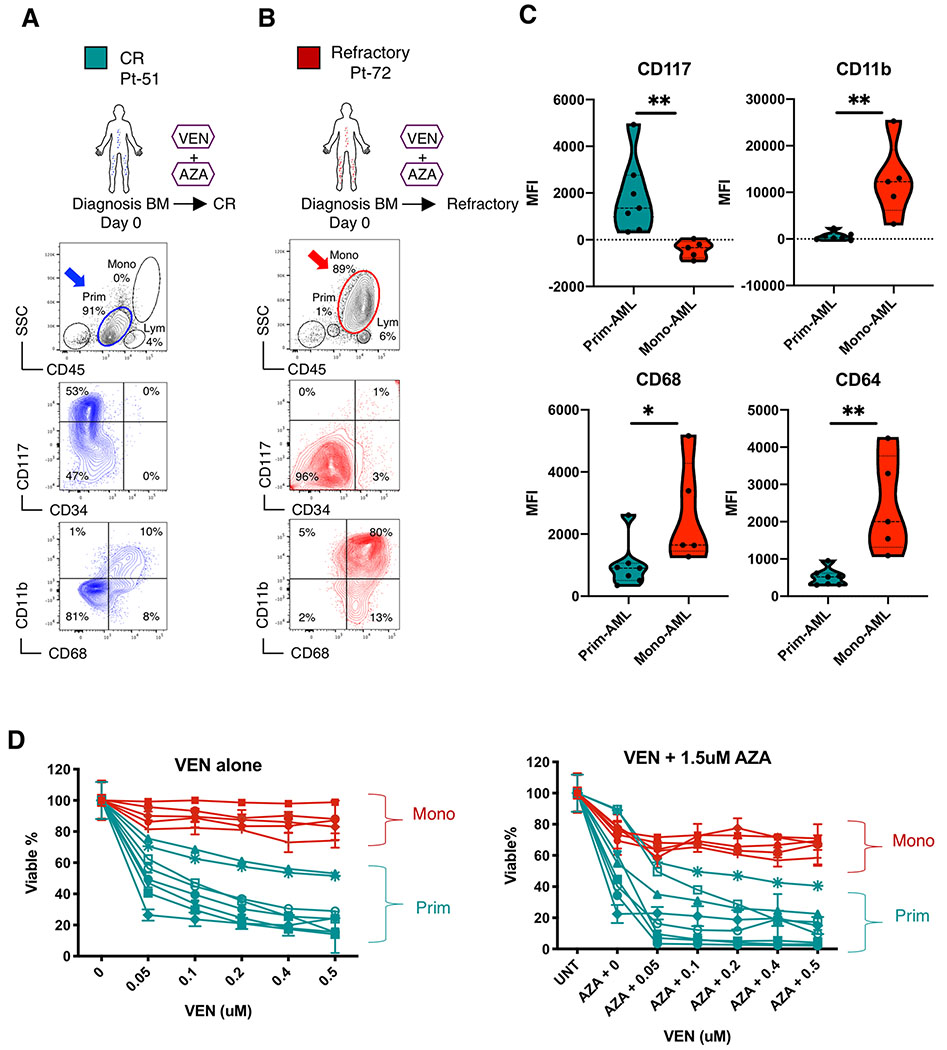
To understand if the lack of response by monocytic AML to VEN+AZA is driven by intrinsic mechanisms, we sought to directly evaluate VEN+AZA sensitivity in vitro, where protection from extrinsic factors such as the microenvironment is minimal. Because the FAB system is no longer used in the AML field, we employed phenotypic markers that would serve as a surrogate for the FAB-M5 subtype. Previous studies have shown that FAB-M5 patients lose expression of the primitive marker CD117 and up-regulate expression of the monocytic markers CD11b, CD68 and CD64(14–18). This expression pattern was verified in our own analysis of CD117, CD11b, CD68 and CD64 gene expression levels in all FAB subclasses in TCGA-AML dataset (19) (Supplementary Figure S1D). Therefore, we designed a multicolor flow cytometry panel including CD117, CD11b, CD68 and CD64 to distinguish patients with monocytic (FAB-M5) from primitive (FAB-M0/M1/M2) AML. As shown in Figure 1, this approach readily distinguishes two predominant cell populations within AML patients. For example, Pt-51 (a typical FAB-M0/M1/M2) presented with a single dominant disease population that was phenotypically primitive as evidenced by CD45-medium/SSC-low/CD117+/CD11b−/CD68− (Fig. 1A). This patient achieved CR with VEN+AZA treatment. In contrast, Pt-72 (a typical FAB-M5) was refractory to VEN+AZA and presented with dominant monocytic disease that was CD45-bright/SSC-high/CD117−/CD11b+/CD68+ (Fig. 1B). Analysis of an additional 12 primary AML specimens (Supplementary Table S2) confirmed the phenotypic profile for primitive vs. monocytic specimens (Fig. 1C and Supplementary Fig. S1E–F). Hereafter these AMLs are noted as “prim-AML” or “mono-AML” respectively.
Figure 1. Monocytic AML is intrinsically resistant to venetoclax + azacitidine.
A, B, Treatment history of Pt-51, Pt-72 and flow analysis of their bone marrow specimens at diagnosis. In the CD45/SSC plots, Mono, Prim and Lym gates indicate monocytic, primitive and lymphocytic subpopulations, respectively. The CD34/CD117 and CD68/CD11b plots show immunophenotype of the gated primitive subpopulations in blue and monocytic subpopulations in red. Arrows highlight populations of interest. Clinical information for these patients is listed in Supplementary Table S1.
C, Violin plots showing median fluorescence intensity (MFI) of CD117, CD11b, CD68 and CD64 in mono-AML (N=5) and prim-AML (N=7) quantified by flow cytometry analysis. Each dot represents a unique AML. Mann-Whitney test was used to determine significance.
D, Viability of sorted ROSlow LSCs from mono-AML (N=5) and prim-AML (N=7) after 24 hours in vitro treatment with VEN alone or in combination with a fixed dose of 1.5 uM AZA. Mean +/− SD of technical triplicates. All viability data were normalized to untreated controls.
Multiple studies have suggested that leukemia stem cells (LSC) are an important target of AML therapies (20). Our previous studies have shown that a phenotype of low reactive oxygen species (ROS-low) enriches for functionally defined LSCs(12,21). Therefore, to more directly assess drug responsiveness in the LSC subpopulation, we isolated ROS-low cells from prim- and mono-AML specimens. Because mono-AML has never been directly characterized by ROS level, we confirmed using colony-forming unit (CFU) assays that the ROS-low phenotype enriches for stem/progenitor potential in mono-AML (Supplementary Fig. S1G). These data indicate the ROS-low phenotype strongly enriches for stem/progenitor potential in mono-AML, similar to what was reported for prim-AML (12,21). We then treated the ROS-low subpopulations from prim- or mono-AML with VEN+AZA in vitro. Our results show that ROS-low LSCs of the mono-AML specimens are significantly more resistant than those of the prim-AML specimens (Fig. 1D), suggesting the refractory responses seen in FAB-M5 patients can be at least partially attributed to intrinsic molecular mechanisms uniquely present in monocytic AML cells.
Monocytic AML is biologically distinct from primitive AML
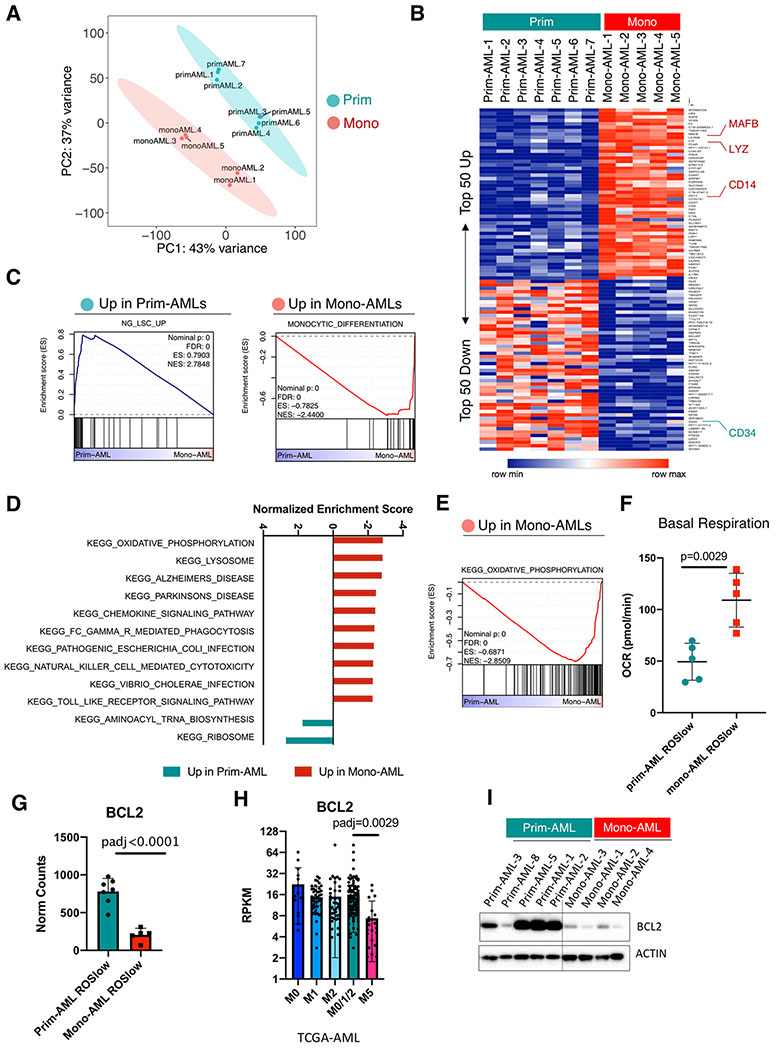
To identify intrinsic molecular mechanisms of mono-AML that may be responsible for resistance to VEN+AZA, we sorted ROS-low LSCs from mono- and prim-AML specimens and performed bulk RNA-seq analysis. After removal of low expressing genes and normalization (Supplementary Fig. S2A and S2B), Principle Component Analysis (PCA) showed that the mono-AMLs clustered separately from the prim-AMLs (Fig. 2A), highlighting distinct biologically features. Indeed, the top 50 up and down regulated genes in mono-AMLs include the monocytic markers MAFB, LYZ, CD14 and the primitive marker CD34 respectively (18,22–25) (Fig. 2B). More broadly, using Gene Set Enrichment Analysis (GSEA), we observed that prim-AMLs are enriched for multiple LSC gene sets, while the mono-AMLs are enriched for monocytic differentiation and AML lysosome gene sets (18,25–27), confirming their distinct transcriptome profiles (Fig. 2C and Supplementary Fig. S2C, S2D).
Figure 2. Monocytic AML is biologically distinct from primitive AML and loses expression of venetoclax target BCL2.
A, PCA analysis of the bulk RNA-seq data showing clear segregation of ROSlow mono-AML (N=5) from ROSlow prim-AML (N=7).
B, Heatmap showing expression of top 50 up- and down-regulated genes in ROSlow mono-AML (N=5) relative to ROSlow prim-AML (N=7), MAFB, LYZ and CD14 are highlighted as monocytic markers; CD34 is highlighted as a primitive marker.
C, GSEA enrichment plots showing up-regulated gene sets in prim- or mono-AML specimens.
D, Bar graphs showing normalized enrichment score (NES) of top-ranked gene sets produced by GSEA analysis of mono-AML versus prim-AML bulk RNA-seq data using the KEGG gene set collection.
E, A GSEA enrichment plot showing up-regulated OXPHOS gene sent in mono-AML.
F, Basal respiration rate in ROSlow prim-AML (N=5) versus ROSlow mono-AML (N=5). Each dot represents a unique AML. Mean +/− SD.
G, Bar graphs showing expression of BCL2 in ROSlow prim-AML (N=7) and ROSlow mono-AML (N=5). Each dot represents a unique AML. Mean +/− SD.
H. Bar graphs showing expression of BCL2 in FAB-M0 (N=16), M1 (N=44), M2 (N=40), M0/1/2 (N=100) and M5 (N=21) subclasses of AMLs from the TCGA dataset. Each dot represents a unique AML.
I, Western blot results showing protein level expression of BCL2 in prim-AML (N=5) and mono-AML (N=4). Actin is used as loading control.
Interestingly, our GSEA analysis also identified oxidative phosphorylation (OXPHOS) as the top up-regulated KEGG gene set in mono-AML (Fig. 2D, 2E) and Seahorse functional assays confirmed that the basal respiration rate of OXPHOS is indeed significantly higher in ROS-low of mono-AML (Fig. 2F). Together these data suggest mono-AML are transcriptionally distinct from prim-AML and exhibit elevated energy metabolism in the form of OXPHOS activity.
Monocytic AML loses expression of the venetoclax target BCL2
We next focused on expression of apoptosis family genes given that VEN is a BCL2 specific inhibitor and several studies have shown that BCL2 expression strongly correlates with VEN sensitivity in vitro (28,29). Among genes related to apoptosis regulation (Supplementary Fig. S2E), our analysis revealed significant and consistent loss of BCL2 in mono-AMLs (N=5), compared to the prim-AMLs (N=7) (Fig. 2G). Analysis of the TCGA-AML dataset also showed progressive loss of BCL2 gene expression through stages of AML morphological maturation (FAB-M0 to FAB-M5, Supplementary Fig. S2F, S2G). As a result, significantly lower expression of BCL2 is observed in FAB-M5 relative to FAB-M0/M1/M2 in the TCGA-AML dataset (Fig. 2H). Further, reduced expression of BCL2 in mono-AML was confirmed at the protein level (Fig. 2I).
Interestingly, loss of BCL2 also occurs during normal monocytic development (30,31). We found consistent loss of BCL2 at the monocytic stage in both human and murine systems (Supplementary Fig. S2H–K). Together, these analyses indicate BCL2 loss is a conserved biological feature during both normal and malignant monocytic development. Further, the data suggest BCL2 loss in monocytic AML may drive resistance to venetoclax-based therapies.
Monocytic AML is preferentially reliant on MCL1 for energy metabolism and survival
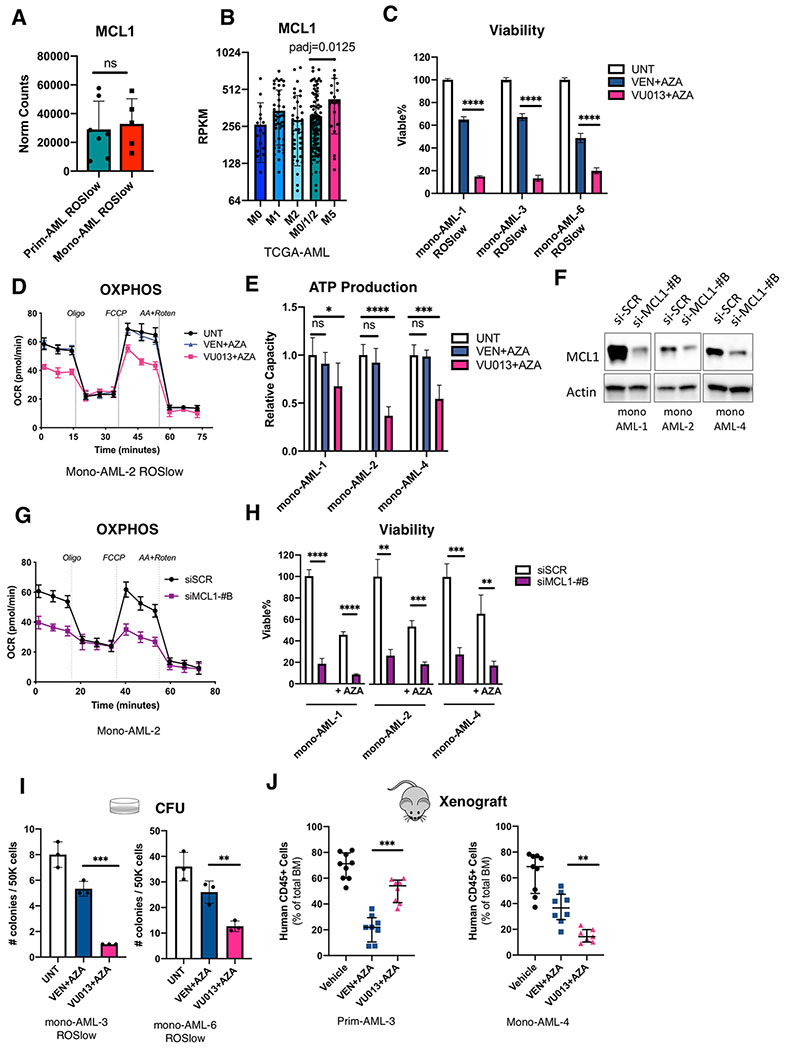
We have previously reported that BCL2 mediates OXPHOS in ROS-low AML cells (12). Given that monocytic AML has loss of BCL2 expression yet elevated OXPHOS activity (Fig. 2D–I), we hypothesized that other members of the BH3 family may become more active as a means to compensate for the role of BCL2. Notably, our bulk RNA-seq data show that expression of MCL1 is comparable in seven prim-AML versus five mono-AML (Fig. 3A). We then analyzed the TCGA-AML dataset and found that MCL1 expression is significantly higher in FAB-M5 patients than FAB-M0/M1/M2 patient (Fig. 3B and Supplementary Fig. S3A). These data suggest MCL1 may be relevant to the intrinsic biology of monocytic AML.
Figure 3. Monocytic AML is preferentially reliant on MCL1 for energy metabolism and survival.
A, Bar graphs showing expression of MCL1 in ROSlow prim-AML (N=7) and ROSlow mono-AML (N=5). Each dot represents a unique AML. Mean +/− SD.
B, Bar graphs showing expression of MCL1 in FAB-M0 (N=16), M1 (N=44), M2 (N=40), M0/1/2 (N=100) and M5 (N=21) subclasses of AMLs from the TCGA dataset. Each dot represents a unique AML. Mean +/− SD.
C, Relative viability of monocytic AML specimens treated 24 hours with 0.5uM VEN+ 1.5uM AZA or 0.5 uM VU013 (VU661013) + 1.5uM AZA. Technical triplicates per group. Mean +/− SD. Two-tailed, unpaired t-test.
D, Oxygen consumption rate (OCR) curves from Seahorse Mito Stress Assay comparing impact of 0.5uM VEN + 1.5uM AZA and 0.5uM VU013 (VU661013) + 1.5uM AZA on OXPHOS activity of monocytic AML. Technical replicates of five per data point. Mean +/− SD. Vertical dotted lines indicate injection times used in the Mito Stress Assay.
E, Relative ATP production capacity calculated from the Seahorse Mito Stress Assay in 0.5uM VEN + 1.5uM AZA or 0.5uM VU013 (VU661013) + 1.5uM AZA treated monocytic AML specimens. Technical replicates of five per group. Mean +/− SD. Two-tailed, unpaired t-test.
F, Western blot results showing siMCL1-#B-mediated knock down of MCL1 at protein level.
G, OCR curves comparing OXPHOS activity in siMCL1-#B vs siSCR (siScramble) control monocytic AML. Technical replicates of five per data point. Mean +/− SD. Vertical dotted lines show injection times used in the Mito Stress Assay.
H, Relative viability of monocytic AML cells with 48 hours exposure to siMCL1-#B or siSCR (siScramble) control, with or without presence of 1.5uM AZA. Technical triplicates per group. Mean +/− SD. Two-tailed, unpaired t-test.
I, Results of Colony Forming Unit (CFU) assay comparing the impact of 0.5uM VEN + 1.5uM AZA versus 0.5uM VU013 (VU661013) + 1.5uM AZA on the stem/progenitor function of mono-AML. Mean +/− SD. Two-tailed, unpaired t-test.
J, Percentage of engraftment in NSG-S mice after ex vivo treatment with 0.5uM VEN + 1.5uM AZA or 0.5uM VU013 (VU661013) + 1.5uM AZA. Each dot represents an individual mouse. Median +/− interquartile range. Mann-Whitney test was used to compare the treatment groups.
To evaluate our hypothesis, we first compared effects of the selective MCL1 inhibitor, VU661013 (32), to VEN. Three primary mono-AML specimens were treated side by side with VU661013+AZA or VEN+AZA. We observed that VU661013 alone or in combination with AZA induced strong cell death in all monocytic specimens evaluated and the effect was significantly greater than VEN alone or VEN+AZA (Fig. 3C and Supplementary Fig. S3B). Notably, VEN+AZA failed to inhibit OXPHOS in mono-AML, whereas VU661013+AZA did show significant inhibition of OXPHOS (Fig. 3D and Supplementary Fig. S3C, S3D). As expected, differential modulation of OXPHOS is reflected in production of ATP (Fig. 3E), demonstrating increased MCL1 dependency as a distinct feature of energy metabolism in monocytic AML.
To more directly interrogate the functional role of MCL1, we also employed siRNA-mediated knockdown of MCL1 in three primary mono-AML specimens. As shown, two independent siMCL1 sequences (siMCL-#B and siMCL-#C) successfully reduced MCL1 protein (Fig. 3F and Supplementary Fig. S3E), and impaired OXPHOS activity (Fig. 3G and supplementary Fig. S3F, S3G). In addition, production of TCA cycle intermediates citrate, alpha-ketoglutarate and malate were significantly lowered in a subset of monocytic AML specimens (Supplementary Fig. S3H). Notably, in using siRNAs to reduce MCL1 expression, significant cell death was observed with or without addition of AZA in all three mono-AML specimens tested (Fig. 3H and supplementary Fig. S3I). These data suggest that MCL1 is a key mediator of OXPHOS and viability for the mono-AML cell type.
Given that OXPHOS is critical for LSCs, as well as other types of cancer stem cells (12,33–35), we next examined whether preferential dependence on MCL1 is also reflected in the stem/progenitor function of monocytic AML. In two independent mono-AML specimens, we observed that treatment with VU661013+AZA was significantly better than VEN+AZA in reducing colony-forming units (CFU) of mono-AML (Fig. 3I). We repeated the same treatments on three normal specimens isolated from umbilical cord blood (CBMC-1, -2, and -3) and observed that VU661013+AZA had only a modest impact on CFU potential, similar to VEN+AZA (Supplementary Fig. S3J), implying preferential reliance on MCL1 in malignant cells.
To more directly assess the increased reliance of monocytic LSCs on MCL1, we performed xenograft studies using prim- and mono-AMLs treated with VEN+AZA or VU661013+AZA followed by transplantation into immune deficient NSG-S mice (21). Our results showed VEN+AZA effectively impaired the LSC engraftment potential in primitive AML as expected(10), but less so in monocytic AML (Fig. 3J, left). Conversely, the VU661013+AZA treatment was significantly better in reducing the LSC engraftment potential of monocytic AMLs (Fig. 3J, right). Together, these data demonstrate that monocytic AML displays greater reliance on MCL1 than BCL2 for energy production, stem/progenitor potential, and survival.
Venetoclax + azacitidine selects monocytic disease at relapse
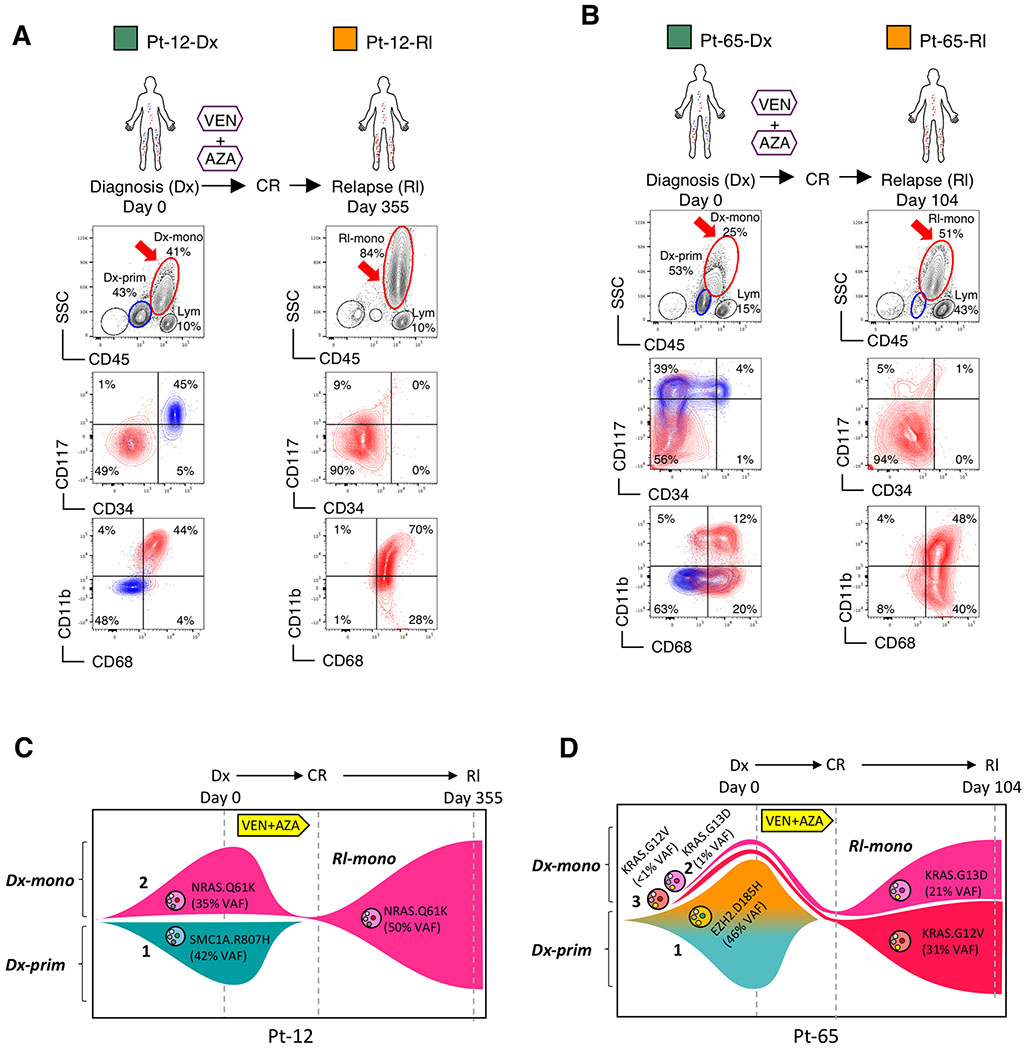
Based on the above findings, we next investigated the extent to which monocytic disease is evident in patients who initially responded but then relapsed on VEN+AZA therapy. In analyzing AML patients prior to VEN+AZA treatment, we noted that the majority of patients actually present with tumors showing a mixture of the monocytic and primitive phenotype, which we term “MMP-AML” (for Mixed Monocytic/Primitive-AML). We analyzed the characteristics of two MMP-AML patients (Pt-12, Pt-65) during the course of treatment (Fig. 4A and 4B). Upon relapse after an initial CR, both patients showed almost complete loss of the primitive subpopulation, and emergence of a dominant monocytic phenotype (CD45-bright/SSC-high/CD117−/CD11b+/CD68+). Thus, VEN+AZA treatment appeared to induce striking in vivo selection for the monocytic subpopulation in each patient (Fig. 4A and 4B).
Figure 4. Monocytic disease arising from venetoclax + azacitidine treatment is derived from pre-existing monocytic subclones.
A, B, Treatment history of Pt-12, Pt-65 and flow analysis of their diagnosis (Dx) and relapse (Rl) specimens. In the CD45/SSC plots, Mono, Prim and Lym gates identify monocytic, primitive and lymphocytic populations, respectively. The CD34/CD117 and CD68/CD11b plots show immunophenotype of the gated primitive subpopulations in blue and monocytic subpopulations in red. Arrows highlight populations of interest. Clinical information of these patients is listed in Supplementary Table S1.
C, D, Fish plots showing clonal dynamics in paired diagnosis (Dx) and relapse (Rl) specimens of Pt-12 and Pt-65. Genetic subclones are illustrated by distinct shapes accompanied by their clonal number (1, 2, or 3). Phenotypic subpopulations are illustrated by color as follows: teal indicates primitive phenotype; brown, pink or red indicate monocytic phenotype. For Pt-12, clone 1 presents a primitive phenotype; clone 2 presents a monocytic phenotype. For Pt-65, clone 1 presents a mixed monocytic and primitive phenotype; clone 2 presents a monocytic phenotype; clone 3 is inferred to have a monocytic phenotype as well.
Of note, this monocytic selection phenotype seems to be a unique clinical characteristic of VEN+AZA therapy. Indeed, previous analyses of patients treated with conventional chemotherapy has shown consistent enrichment of more primitive LSC phenotypes(36). To further corroborate this finding, we analyzed RNA-seq data of 11 pairs of diagnostic and relapsed specimens after conventional chemotherapy from a separate study by Shlush et al(37). In this setting, we observed a gain of the LSC gene expression signature, and loss of monocytic markers (CD11b and CD68) and a monocytic gene expression signature at relapse, suggesting suppression of the myeloid phenotype following chemotherapy (Supplementary Fig. S4A and S4B). Lastly, we compared paired diagnosis vs. relapse specimens from six AML patients treated with conventional chemotherapy at our institution. In no case was a monocytic phenotype evident at relapse. In fact, for two patients with monocytic characteristics at diagnosis, conversion to a more primitive phenotype at relapse was observed (Supplementary Fig. S4C). Together, these data suggest that relapse following conventional chemotherapy strongly favors a primitive phenotype, and that selection of a monocytic phenotype at relapse appears to be a distinct characteristic of VEN+AZA therapy.
Monocytic disease arising from venetoclax + azacitidine treatment is derived from pre-existing monocytic subclones
To investigate the origin of relapsed monocytic subpopulations in Pt-12 and Pt-65, we performed whole exome sequencing (WES) analysis on sorted primitive and monocytic subpopulations from paired diagnosis and relapse specimens (sorting strategy outlined in Supplementary Fig. S4D). For each patient, three subpopulations of cells were isolated: diagnosis primitive (“Dx-prim”), diagnosis monocytic (“Dx-mono”) and relapse monocytic (“Rl-mono”). In Pt-12, 400X WES detected five unique non-synonymous cancer-related mutations in 49 commonly mutated genes in AML (Supplementary Table S3). While the variant allele frequency (VAF) for three of five mutations are similar among all three sorted subpopulations, two mutations SMC1A.R807H and NRAS.Q61K presented unique patterns (Supplementary Fig. S4E). The SMC1A.R807H mutation was only detected in Dx-prim, suggesting that the Dx-prim subpopulation represents a genetically distinct subclone that was effectively eradicated by VEN+AZA treatment (Fig. 4C, clone 1 in teal). In contrast, the NRAS.Q61K mutation was only detected in Dx-mono and Rl-mono, suggesting the monocytic subpopulation that emerges at relapse represents a genetically distinct subclone that pre-existed at diagnosis (Fig. 4C, clone 2 in pink).
In Pt-65, our analysis detected three unique non-synonymous cancer-related mutations (Supplementary Fig. S4F). Interestingly, the mutation profile between the Dx-prim and Dx-mono subpopulations was identical, except for a low level (~1%) RAS G13D mutation in the Dx-mono subpopulation. These data suggest the majority of cells in the Dx-mono subpopulation arose from the same genetic subclone from which Dx-prim was derived, despite their phenotypic differences. In particular, an EZH2.D185H mutation presented at an identical VAF of 46% in Dx-prim and Dx-mono but 0% in Rl-mono, suggesting the Dx-prim and Dx-mono cells containing the EZH2.D185H mutation arose from the same genetic subclone and were eradicated by VEN+AZA treatment (Fig. 4D, clone 1 in teal and brown, the gradual transition of color from teal to brown illustrates phenotypic progression from primitive to monocytic). Intriguingly, two KRAS mutations G13D and G12V emerged in Rl-mono at a VAF of 21% and 31%, respectively. The KRAS.G13D mutation was confirmed at 1% VAF in Dx-mono, demonstrating that this subclone pre-existed at a low level in the monocytic subpopulation at diagnosis (Fig. 4D, clone 2 in pink). Although not confirmed due to limitation of sequencing depth, we infer that the KRAS.G12V mutation likely also pre-existed as a phenotypically monocytic subclone at diagnosis (Fig. 4D, clone 3 in red). Therefore, overall, our WES analyses of Pt-12 and Pt-65 suggest dominant monocytic disease at relapse pre-existed within the monocytic subpopulation of cells at diagnosis.
Monocytic disease at relapse has activated MLL-specific LSC programs and sustained reliance on MCL1
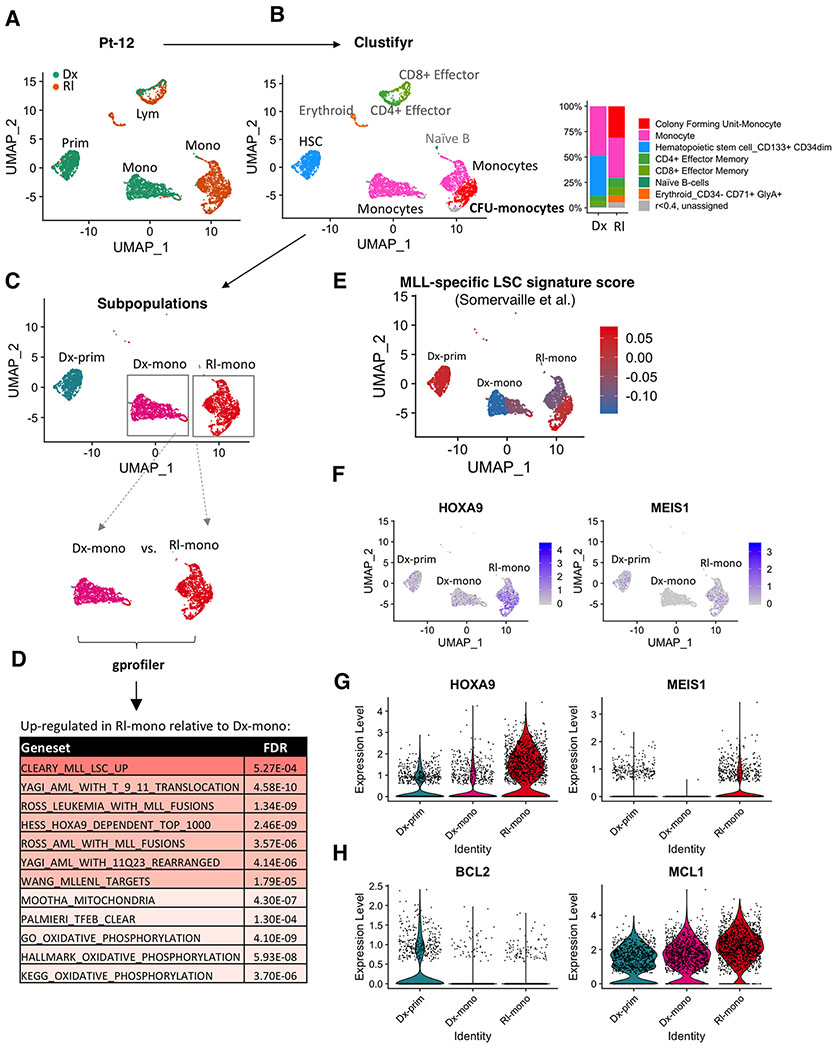
For patient 12 (Pt-12), the WES analysis shows the same monocytic clone at diagnosis and relapse (Fig. 4C, clone 2). However, based on the clinical course of the patient (i.e. initial CR following by relapse) we inferred that biological properties of the monocytic subclone had at least partially evolved between diagnosis to relapse. To investigate further we performed CITE-seq (38) to simultaneously profile surface antigen expression and transcriptomics at single cell resolution. Analysis of the CITE-seq data revealed three major clusters at diagnosis and two major clusters at relapse for Pt-12 (Figure 5A and Supplementary Fig. S5A). Shown by surface antigen expression, the three clusters at diagnosis consisted of a lymphocytic cluster (CD3+/CD33−), a primitive myeloid cluster (CD34+/CD33+), and a monocytic myeloid cluster (CD11b+/CD33+) (Supplementary Fig. S5A left and S5B). In contrast, the relapse specimen was comprised of a lymphocytic cluster (CD3+/CD33−) and a monocytic myeloid cluster (CD11b+/CD33+), but no primitive cluster, consistent with our flow cytometric analysis (Supplementary Fig. S5A right and S5B). Importantly, the transcriptome of lymphoid clusters observed at diagnosis and relapse were quite similar, as depicted by their close proximity in the UMAP projection (Fig. 5A). In contrast, the mono-AML populations at diagnosis versus relapse were completely distinct (Fig. 5A), suggesting a significant change in the transcriptional profile of relapsed disease.
Figure 5. Monocytic disease at relapse has activated MLL-specific LSC programs and sustained reliance on MCL1.
A, UMAP plots of single cell RNA-seq data generated from CITE-seq analysis of paired diagnosis (Dx) and relapse (Rl) specimens from Pt-12. Each cluster represents a subpopulation of biologically similar cells clustered by their transcriptome similarity. Each dot within each cluster represents a single cell. Teal indicates cells from diagnosis, Brown indicates cells from relapse. Also see supplementary Fig. S5A,B.
B, Clustifyr analysis assigning each cluster to its closest normal hematopoietic lineage counterpart according to their transcriptome similarity. Bar graph comparing relative percentage of each subcluster in diagnosis and relapse specimens.
C, Major myeloid subpopulations analyzed in subsequent analyses.
D, Gprofiler analysis results showing significantly upregulated gene sets in the “Rl-mono” cluster relative to the “Dx-mono” cluster.
E, A heatmap showing relative expression of MLL-specific LSC gene expression signature at single cell resolution. Red indicates strong positive expression of MLL-specific LSC signature; Blue indicates a negative expression pattern suggesting non-LSC nature.
F, A heatmap showing relative expression of HOXA9 and MEIS1 at single cell resolution.
G,H, Violin plots showing relative expression of HOXA9, MEIS1, BCL2 and MCL1 in different clusters. Each dot represents a single cell.
To characterize the transcriptional changes at relapse, we first analyzed the CITE-seq data using “clustifyr” (bioRxiv doi: 10.1101/855064), a program that assigns each cluster to its closest normal hematopoietic lineage counterpart according to transcriptomic similarity. The clustifyr analysis showed that the primitive cluster at diagnosis is indeed transcriptionally similar to HSC and the monocytic cluster at diagnosis is most similar to mature monocytes (Fig. 5B, blue and pink). Interestingly, within the relapse monocytic cluster, clustifyr identified two subpopulations including one that is most similar to mature monocytes and one that is closer to monocytic progenitors (CFU-monocytes) (Fig. 5B, pink and red). These data suggest that a significant subpopulation of monocytic cells at relapse have acquired unique biology that clearly differs from the monocytic cells at diagnosis.
Next, using “gprofiler” pathway analysis, we observed significant up-regulation of an LSC signature in the RI-mono population (Fig. 5C, 5D). Intriguingly, the signature is derived specifically from MLL-rearranged leukemia (MLL+). This is notable because unlike most other previously reported LSC signatures, MLL-specific LSCs have a myeloid phenotype (39,40). Thus, in comparison to Dx-mono, the RI-mono population appears to have dedifferentiated to a more stem-like phenotype, while retaining myeloid characteristics. Furthermore, the RI-mono subpopulation shows increased HOXA9 and OXPHOS signatures, characteristics of MLL+ leukemia and VEN+AZA resistant leukemias respectively (Fig. 5D). Similar results were observed when the gprofiler analysis was used to compare the “CFU-monocytes” subpopulation to the “Monocytes” subpopulation within the parent Rl-mono cluster (Supplementary Fig. S5C). Of note, Dx-prim uniquely enriches for HSC (normal stem cells) and non-MLL LSC gene expression signatures including the GOODELL_HSC, EPPERT_HSC, and NG_LSC gene sets relative to Dx-mono, consistent with their expected non-MLL LSC nature (Supplementary Fig. S5D). Further, we plotted the enrichment score of a curated MLL-specific LSC signature obtained by Somervaille et al. and two other non-MLL LSC signatures obtained by Eppert et al. and Ng et al. (Supplementary Table S4)(25,26,39). This analysis showed that although the Rl-mono cluster loses non-MLL LSC gene expression signatures (Supplementary Fig. S5E), the “CFU-monocytes” subpopulation highly expresses the MLL-specific LSC signature (Fig. 5E). Consistently, expression of the HOXA9 and MEIS1 genes were up in the Rl-mono cluster relative to Dx-mono and Dx-prim (Fig. 5F and G), further suggesting activation of MLL-specific LSC program in VEN+AZA relapsed monocytic AML.
Finally, we asked if relapsed monocytic AML retains MCL1 dependence. Despite loss of BCL2 in both Dx-mono and Rl-mono clusters, MCL1 expression is sustained in both and presents a trend of increase at relapse (Fig. 5H). This result was corroborated by analysis of the single cell RNA-seq data from van Galen et al. showing simultaneous loss of BCL2 and gain of MCL1 expression in monocytic AML subpopulations (Supplementary Fig. S5F)(41). Importantly, viability assays showed that VU661013+AZA performed significantly better than VEN+AZA in eradicating relapsed monocytic cells from Pt-12 and Pt-65 (Supplementary Fig. S5G). Further, CFU assays showed that the VU661013+AZA regimen is significantly better than VEN+AZA in reducing the CFU capacity of relapsed monocytic AML from Pt-69 (Supplementary Fig S5H, S5I). Together, these findings indicate that relapsed monocytic AML retains dependence on MCL1.
Discussion
Our analysis of patients treated with venetoclax-based therapy has revealed several previously unrecognized characteristics of AML. First, genetic subclones can manifest in distinct developmental stages, where the intrinsic properties of monocytic differentiation mediate venetoclax resistance. Second, leukemic cells at different developmental stages rely on distinct mechanisms to mediate energy metabolism. In particular, regulation of OXPHOS through BCL2 is more prevalent in primitive AML(12,42–44), while reliance on MCL1 appears to be more important in monocytic AML. Third, intra-patient heterogeneity amongst LSCs underlies therapeutic resistance. Specifically, primitive or monocytic LSC phenotypes can co-exist in the same AML patient (i.e. MMP-AML) and demonstrate differential response trajectories to venetoclax-based therapy. Together, these data clearly indicate that to improve outcomes, AML therapies must be designed so as to target unique biological properties found in distinct AML subclones, developmental stages, and LSC subpopulations.
A notable aspect of these findings is the observation that AML cells can apparently switch from BCL2 to MCL1 dependence to drive energy metabolism as cells acquire a more differentiated developmental state. This finding suggests developmental heterogeneity is a previously underappreciated factor in determining therapeutic response. Based on our findings in AML, we note that similar considerations may be relevant in other forms of cancer. As we have shown, loss of BCL2 and sustained MCL1 dependency is a conserved property of monocytic development in both normal and malignant hematopoiesis. Others have shown similar switches can be found in the context of neuronal development and hematopoietic T/B cell development(45,46). Therefore, it is possible that in cancer types where differing developmental stages are evident, the relative reliance on BCL2 or MCL1 may vary in a manner analogous to what we have observed in AML.
While the correlation between myeloid differentiation and venetoclax resistance seems clear, the underlying genetic events that may drive this process are as yet not well characterized. Intriguingly, the two patients profiled in this manuscript had relapse disease arising from RAS mutations and we report here that RAS pathway mutations are a univariate, but not multivariate, predictor of refractory response to VEN+AZA (unlike FAB-M5 being highly significant in both univariate and multivariate analyses). These data suggest a possible link between the RAS pathway and myeloid development. To this point, analysis of data from the BEAT-AML study(47), shows co-enrichment between FAB-M5 and RAS mutations (data not shown). In addition, mutations in RAS pathway members such as PTPN11 have been strongly associated with FAB-M5 in previous studies (48). Further assessment of the RAS pathway and other genetic factors is ongoing, but evidence thus far suggests that genetic factors such as RAS mutations may contribute to venetoclax resistance through either epistatic mechanisms and/or by driving monocytic differentiation.
An intriguing finding revealed by the WES analysis is the possibility that genetically distinct subclones with independent LSC populations may co-occur in some patients. This phenomenon was observed for both patients in our study (Figure 4), albeit with differing presentations. For one patient (Pt-12), the diagnostic specimen had two predominant genetically independent subclones, each with its own LSC population (clone 1 and 2, Fig. 4C). Upon treatment with VEN+AZA, the monocytic clone was strongly selected. However, it is important to note that the patient did initially respond to therapy, implying that the relapse population must have evolved to some degree. In fact, CITE-seq analysis in this patient confirmed that the transcriptional profile of monocytic cells at relapse had markedly changed, with the selection and/or acquisition of a more stem-like profile. In contrast, at presentation the second patient (Pt-65) had primitive and monocytic populations with identical genetic characteristics (clone 1, Fig. 4D), implying a parent-progeny relationship. Notably, VEN+AZA therapy appears to have been highly effective in eradicating the clone 1 but resulted in selection for rare genetically distinct monocytic subclones 2 and 3 that were resistant. Thus, in both patients the data clearly indicate at least two genetically distinct LSC populations, one of which was ultimately resistant to venetoclax-based therapy. The prevalence of this type of dual LSC pathogenesis remains to be determined, and of course additional resistance mechanisms may emerge as relapse to VEN+AZA continues to be characterized in the future.
Our data suggest that prospective identification of VEN+AZA resistant subpopulations will be of clinical value. Analogous to the previously employed FAB classification system, our approach thus far has been to use flow cytometry to define stages of differentiation, and we utilized SSC, CD45, CD117, CD11b, CD64 and CD68 to distinguish monocytic AML versus primitive AML. This preliminary panel provides a starting point towards developing a rapid tool to make treatment decisions related to the use of VEN+AZA. A more refined method will likely include additional features such as genetic mutations and/or metabolic properties.
Lastly, our studies have additional implications for the design of improved AML therapies. For elderly or otherwise unfit patients, VEN+AZA is well tolerated and provides deep and durable remissions for the majority of patients. For patients who respond well, we believe that identifying and targeting the VEN+AZA resistant subpopulations is key to improved remission duration. To this end, well-tolerated consolidation regimens that selectively target VEN+AZA are an important future objective. Based on the findings presented here, MCL1 inhibition may be one such strategy. Similarly, we have recently reported that inhibition of fatty acid oxidation may also be a strategy to enhance the efficacy of VEN+AZA (11). Regardless of the specific strategy, treating patients prior to the emergence of resistant disease is likely to be highly beneficial. For the problem of disease that is refractory to VEN+AZA, our findings suggest it should be possible to prospectively identify such individuals and treat them with alternative therapies that enhance, or perhaps replace, venetoclax.
Methods
Patients, Treatment and Responses
One hundred consecutive patients with newly diagnosed AML and no prior therapy who were prescribed venetoclax + azacitidine at our institution and had taken at least one dose of either therapy were identified. Patients were treated over a period from January 2015 to October 2019. Six patients were not included in the analysis because they had no documented follow up. Of the 100 evaluable patients, all received the same regimen of venetoclax + azacitidine, as previously reported(6); some were treated in the context of a clinical trial, while others were treated outside of a clinical trial, with off-label use (prior to approval of venetoclax in November 2018) or on-label use after approval. Baseline details of all 100 patients are summarized in Supplementary Table S1. The University of Colorado Institutional Review Board approved a request to retrospectively analyze these patients (#19-0115). A single experienced hematopathologist (J.S.) categorized all patients into appropriate FAB groups by retrospectively reviewing the microscopic descriptions of all baseline pathology reports, including all of the flow cytometry histograms and the immunohistochemical studies, if performed. Responses were assessed in accordance with the European Leukemia Network(8). Non-responding patients were defined as those who failed to achieve a CR, CRi, PR or MLFS as described previously(6).
Clinical statistics
Logistic regression was used to determine the effects of age, sex, prior hematologic disorder, complex cytogenetics, ELN risk group, RAS pathway mutation, TP53, IDH1/IDH2, NPM1, FLT3 ITD, ASXL1, and FAB-M5 classification on refractory status. Univariate logistic regression was used to assess the effect of each predictor on its own while multivariate logistic regression was used to assess their effects in aggregate. All univariate predictors with a p≤0.10 were included in a multivariate model.
Primary AML specimens
Primary human AML specimens were obtained from apheresis product, peripheral blood, or bone marrow of AML patients who gave written informed consent for sample procurement on the University of Colorado tissue procurement protocols (Colorado Multiple Institutional Review Board Protocol #12-0173 & #06-0720). See Supplementary Table S2 for clinical information of primary AML specimens.
Cell culture and reagents
Primary human AML cells were stored in freezing media composed of 50% FBS (Corning), 10% DMSO (Sigma) and 40% IMDM media (Gibco) and then cryo-preserved in liquid nitrogen. Freshly thawed or sorted cells were cultured in cytokine added Serum Free Media (cytokine+ SFM) at 37°C, in 5% CO2 incubator. SFM is composed of IMDM (Gibco), 20% BIT-9500 (Stemcell Technologies), 10ug/ml LDL (Low Density Lipoprotein, Millipore), 55uM 2-Mercaptoethanol (GIBCO) and 1% Pen/Strep (Gibco). Cytokine+ SFM were made by supplementing the SFM with FLT-3, IL-3 and SCF cytokines (PeproTech), each at 10 ng/ml. All reagents are listed in Supplementary Table S5.
Phenotyping primary AML
0.5-1e6 freshly thawed primary AML cells were stained with our phenotyping panel containing antibodies against human CD45, CD34, CD117, CD11b, CD64 at 4°C for 15 mins, washed with ice cold FACS buffer, fix/perm for 20 mins using the BD Cytofix/Cytoperm kit (BD), washed with perm/wash, stained with intracellular antibody against human CD68 in perm/wash buffer for 30 mins, washed with FACS buffer, and resuspended in FACS buffer and analyzed on BD FACSCelesta flowcytometry (BD). FCS files were analyzed on Flowjo 10.5.3 (Flowjo).
Cell sorting
Primary AML cells were stained with anti-CD45, CD3 and CD19 in FACS buffer (PBS + 0.5% FBS) at 4°C for 15 minutes. The cells were then washed in FACS buffer and stained with 5uM CellRox-DeepRed dye (Life Technologies) in FACS buffer at 37°C in CO2 incubator for 30 minutes. After CellRox staining, the cells were washed twice and resuspended in FACS buffer containing 10uM DAPI. The BD FACSAria II Cell Sorter was used to analyze ROS profile and sort. For sorting ROS-low LSCs from primitive AML cells, we first gated on live (DAPI-), non-lymphocytic (CD3-, CD19-) and primitive subpopulation (CD45-medium, SSC-low), and then gated on ROS-low cells (lower 20% of CellRox histogram). For sorting ROS-low from monocytic AML cells, we first gated on live (DAPI-), non-lymphocytic (CD3-, CD19-) and monocytic subpopulation (CD45-bright, SSC-high), and then gated on ROS-low cells (lower 20% of CellRox histogram). ROS-high was sorted similarly except for higher 20% of CellRox was gated in the second step. The primitive and monocytic phenotype were validated using our phenotyping panel on BD FACSCelesta (BD).
Drug treatment
Freshly thawed and/or sorted AML cells were preincubated in cytokine+ SFM for 1 hour before treatment with various regimens prior to functional or viability assays. All drugs in the combo regimens were added simultaneously. Stock concentration of Venetoclax and VU661013 are 10mM in DMSO; Azacitidine is 40mM in Saline.
Viability assays
All viability assays were performed after 24 hours of drug treatment unless otherwise specified. Cells were pelleted and stained in 1 x AnnexinV staining buffer containing Anti-AnnexinV antibody for 15 minutes in 4°C. Stained cells were then resuspended in 1 x AnnexinV buffer containing DAPI and 0.5% FBS and analyzed on BD FACSCelesta (BD). Viability was determined by percentage of AnnexinV-, DAPI- cells within the parent population.
siRNA
The two independent siMCL1 sequences were purchased directly from Dharmacon™’s ON-TARGETplus siRNA Reagents collection: siMCL1-#B (J-004501-16) and siMCL1-#C (J-004501-15). The lyophilized siRNA products were resuspended in RNAse/DNAse free water at 5uM and used as stock solution. Electroporation of siRNA was performed using the Neon Electroporation Transfection System (Thermo) according to the manufacturer’s protocol. Specifically, for every 2e6 primary AML cells, the cells were spun and resuspended in 80ul Buffer T with addition of 20ul siRNA stock solution in an Eppendorf tube. Electroporation was then performed using microtips (10ul capacity) in a cuvette containing 3ml of Buffer E with the following electroporation settings: 1600 V, 10ms, 3 pulses. To complete the 100ul volume containing 2e6 cells, 10 continuous repeats of 10ul each were performed, the cells from all 10 runs were sequentially combined into one well containing media, and the electroporated cells were then cultured for subsequent analysis.
Seahorse Mito Stress assays
Extracellular flux assay XF96 kit was used to measure OCR. Drug-treated or siRNA exposed primary AML cells were plated in Cell-Tak (Corning) coated XF96 cell culture microplates at 200K cells /well in five replicates and measured according to the manufacture’s protocol. OCR was measured at basal and after injection of 5ug/ml oligomycin (Sigma), 2uM FCCP (Sigma), 5uM Antimycin A (Sigma) and 5uM Rotenone (Sigma).
Immunoblotting
Cells were harvested and lysed in 1x Laemmli Sample Buffer (Bio-Rad) at 5 e6/ml. About 50-200K cells per lane were loaded and resolved on 6–12% SDS-PAGE gels, transferred to PVDF membranes, blocked in 5% milk in TBS with 0.1% Triton X-100 (Sigma). After incubation, the PVDF membranes were incubated with primary antibodies at 4°C overnight, washed, incubated with secondary antibodies at room temperature for 2 hours, subjected for imaging on the ChemiDoc Imaging System (Bio-Rad).
Colony-forming Unit Assay
Primary AML or normal CBMC cells were seeded in human methylcellulose complete media (R&D Systems) at 50-250K cells/ml or 10K/ml, respectively, and grown in 37°C, 5% CO2 incubator for 2-4 weeks before counting on stereomicroscope. For drug treatment, primary AML cells were treated in culture dish overnight with various inhibitors in cytokine+ SFM.
Ex vivo treatment and xenograft studies
One day prior to transplant, freshly thawed primary AML cells were treated in culture dish overnight with various inhibitors in cytokine+ SFM. NSG-S mice were conditioned with 25 mg/kg busulfan via IP injection. Second day at injection, overnight treated primary AML cells were washed with FACS buffer and resuspended in Saline at 50 e6/ml concentration. Anti-human CD3 antibody (BioXCell) was added at a final concentration of 1ug/e6 cells to avoid potential graft versus host disease. Per mouse, 5 e6 cells in 0.1ml saline were tail vein injected. 8-10 mice per experiment group. Mice engrafted with primary AML cells were sacrificed after 5-8 weeks. All animal experiments were approved by the University of Colorado Anschutz (IACUC) under protocol number 0308.
Bulk RNA-seq
mRNA isolation, quality check, library construction and sequencing were performed according to same protocol used previously(21). Single end reads of 100nt were generated for each sample on the Illumina HiSeq2500 platform. For analysis, raw reads were demultiplexed using bcl2fastq version 2.19.0. Quality filtering and adapter removal were performed using Trimmomatic-0.36 with the following parameters: “TRAILING:13 LEADING:13 ILLUMINACLIP:adapters.fasta:2:30:10 SLIDINGWINDOW:4:20 MINLEN:35”. Processed/cleaned reads were then mapped to the Homo sapiens reference sequence (GRCh38, hg38) with STAR-2.6.0c given the following parameters: “--twopassMode Basic --runMode alignReads --genomeDir $ --readFilesIn ${SAMPLE} --outSAMtype BAM SortedByCoordinate --outSAMstrandField intronMotif --outFilterIntronMotifs RemoveNoncanonical”. The subread-1.6.1 package (featureCounts) was used to derive gene counts given the following parameters: “-s 2 -t exon -g gene_name”. Differential expression analysis and data normalization was performed using DESeq2-1.16.1 with an adjusted p-value threshold of 0.05 within an R-3.4.1 environment. A batch factor was given to the differential expression model in order to control for batch differences.
Single-cell CITE-seq
Design of oligo-antibody panel is detailed in Supplementary Table S6. For constructing CITE-seq libraries, following modifications were added to the standard 10x Genomics V3 protocol. During cDNA amplification an additive primer was added at 0.2 μM to selectively amplify the antibody tags. In the following purification the supernatant is removed in the first clean-up step to be further purified while the beads are cleaned to generate the gene expression libraries. The supernatant is cleaned with 1.4x Ampure Select beads (Agencourt) and eluted in 40 μl of EB. CITE-seq libraries are amplified with Illumina compatible primers for 14 cycles and purified with 1.2x Ampure Select beads. All libraries were normalized by Qubit (Invitrogen) and Tapestation (Agilent) analysis. Libraries were sequenced on a NovaSeq 6000 (Illumina) for a depth of 5000 reads per cell for CITE-seq libraries. For analysis, CITE-seq data were first processed through the 10x Genomics Cell Ranger pipeline (v3.1), and then analyzed in R (v3.6 with the Seurat (v3.1.1) package 1. Cell barcodes with 25% or more mitochondrial RNA content were excluded from analysis. RNA counts were normalized using scran to estimate normalization size factors and ADT counts were normalized using centered log-ratio normalization in Seurat. Cell type similarity and gene signature analyses were conducted using R package “clustifyr” (https://github.com/rnabioco/clustifyR). Cell clusters from CITE-seq data were assigned to cell-type identities based on Spearman correlation against a reference dataset generated from microarray data of purified populations of human hematopoietic cells (GSE24759), comparing the expression of variable genes determined by the VST method in Seurat. Gene signature scoring for both positive and negative markers was calculated by clustifyr, first on a per-cell basis by Spearman correlation, and then averaged over each cluster. Comparisons between curated gene sets were performed using hypergeometric tests or the Jaccard index. Gene set and pathway enrichment analysis was performed in R via the “gprofiler2” package, an interface to the gProfiler web suite, using previously curated AML gene sets.
WES
WES libraries were generated using the Agilent Sure Select XT Exome prep kit with 200 ng of input as per protocol (Agilent). The probe used was SureSelect XT Human All Exon V7 (Agilent). Libraries were normalized by Qubit (Invitrogen) and Tapestation (Agilent) and sequenced on a Novaseq 6000(Illumina) to obtain 400x coverage. For analysis, we adopted IMPACT (Integrating Molecular Profiles with Actionable Therapeutics) pipeline that we previously developed(49). IMPACT links variants detected from WES to actionable therapeutics. Briefly, IMPACT takes sequence data as input and outputs a VCF file containing predicted deleterious mutations. The sequencing reads were mapped to the human hg19 reference exome using the Burrows-Wheeler Aligner. SAMTools and BCFtools (v1.1) were utilized to detect variants from the BAM file and output into a VCF file. In the IMPACT pipeline, we used ANNOVAR (v2014-11-12) to annotate the variants. Synonymous and intronic variants were removed. Variants were further analyzed by deleterious prediction tools such as SIFT and PolyPhen2. We also focused on 49 genes commonly mutated in AML (http://raindancetech.com/thunderbolts-myeloid-panel/) and cancer-related mutations reported in COSMIC (https://cancer.sanger.ac.uk/cosmic) to infer clonal dynamics in paired diagnosis and relapse specimens for VEN+AZA trial patients.
GSEA
GSEA analysis was performed using GSEA version 3.0 (Broad). Normalized count matrix produced from DESeq2 analysis were formatted into GCT files containing expression values. CLS files were built to label biological stats. When performing GSEA analysis, several gene set databases were used including: c2.cp.kegg.v6.0.symbols.gmt, c2.cp.reactome.v6.0.symbols.gmt and an in-house database containing a collection of 791 gene sets of our interest. Following parameters were used: Number of permutations = 1000, permutation type = gene_set. Other parameters were left at default values.
Metabolomics
After 16 hours of knock down with siSCR control or siMCL1, primary AML cells were washed in ice cold PBS and snap frozen for metabolomic analysis. Technical replicates of four were used per condition, 100K cells per replicate. Metabolomic methods are detailed previously(11,50). Metabolite levels were normalized to protein quantification.
Statistical analysis
Methods used for statistical analyses were detailed in figure legends. In all figures, ns indicates a not significant p value of >0.05; *, **, ***, **** indicate p<0.05, p<0.01, p<0.001, p<0.0001 respectively.
Data Availability
The data that support the findings of this study are available from the corresponding author upon reasonable request. Patient-related clinical data are included as Supplementary tables. Bulk RNA-seq (GSE132511), CITE-seq (GSE143363) and WES (PRJNA600769) data have been uploaded to public databases.
Supplementary Material
Statement of Significance.
Identifying characteristics of patients who respond poorly to venetoclax-based therapy and devising alternative therapeutic strategies for such patients are important topics in AML. We show that venetoclax resistance can arise due to intrinsic molecular/metabolic properties of monocytic AML cells, and that such properties can potentially be targeted with alternative strategies.
Acknowledgements
We thank all of the patients and their families, as well as the nurses, pharmacists and advanced practice practitioners who were involved in their care. C.T.J. is generously supported by the Nancy Carroll Allen Chair in Hematology Research and NIH (R01CA200707). D.A.P is supported by the University of Colorado Department of Medicine Outstanding Early Career Scholar Program, the Robert H. Allen MD Chair in Hematology Research and the Leukemia and Lymphoma Society Scholar in Clinical Research Award. M.R.S. is a Leukemia and Lymphoma Society Clinical Scholar and is supported by an E.P. Evans Foundation Discovery Research Grant and the Biff Ruttenberg Foundation. A.I. is supported by NIH MSTP T32 (GM008497). A.D. is supported by funds from the Boettcher Foundation (Webb-Waring Early Career 2017). C.L.J. is supported by a Leukemia and Lymphoma Society Special Fellow award and the American Cancer Society (25A5072). We appreciate bioinformatic support provided by the Informatics Fellows of the RNA Bioscience Initiative at the University of Colorado School of Medicine. We also appreciate funding supports for the Biostatistics and Bioinformatics Shared Resource and the Genomics Shared Resource of the University of Colorado Cancer Center (P30CA046934).
Footnotes
Conflict of interest: D.A.P. receives research funding from and serves as an advisor to AbbVie
References
- 1.Almeida AM, Ramos F. Acute myeloid leukemia in the older adults. Leuk Res Rep 2016;6:1–7 doi 10.1016/j.lrr.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buchner T, Berdel WE, Haferlach C, Haferlach T, Schnittger S, Muller-Tidow C, et al. Age-related risk profile and chemotherapy dose response in acute myeloid leukemia: a study by the German Acute Myeloid Leukemia Cooperative Group. J Clin Oncol 2009;27(1):61–9 doi 10.1200/JCO.2007.15.4245. [DOI] [PubMed] [Google Scholar]
- 3.Pollyea DA, Kohrt HE, Medeiros BC. Acute myeloid leukaemia in the elderly: a review. Br J Haematol 2011;152(5):524–42 doi 10.1111/j.1365-2141.2010.08470.x. [DOI] [PubMed] [Google Scholar]
- 4.Fenaux P, Mufti GJ, Hellstrom-Lindberg E, Santini V, Finelli C, Giagounidis A, et al. Efficacy of azacitidine compared with that of conventional care regimens in the treatment of higher-risk myelodysplastic syndromes: a randomised, open-label, phase III study. Lancet Oncol 2009;10(3):223–32 doi 10.1016/S1470-2045(09)70003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kantarjian H, Issa JP, Rosenfeld CS, Bennett JM, Albitar M, DiPersio J, et al. Decitabine improves patient outcomes in myelodysplastic syndromes: results of a phase III randomized study. Cancer 2006;106(8):1794–803 doi 10.1002/cncr.21792. [DOI] [PubMed] [Google Scholar]
- 6.DiNardo CD, Pratz K, Pullarkat V, Jonas BA, Arellano M, Becker PS, et al. Venetoclax combined with decitabine or azacitidine in treatment-naive, elderly patients with acute myeloid leukemia. Blood 2019;133(1):7–17 doi 10.1182/blood-2018-08-868752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DiNardo CD, Pratz KW, Letai A, Jonas BA, Wei AH, Thirman M, et al. Safety and preliminary efficacy of venetoclax with decitabine or azacitidine in elderly patients with previously untreated acute myeloid leukaemia: a non-randomised, open-label, phase 1b study. Lancet Oncol 2018;19(2):216–28 doi 10.1016/S1470-2045(18)30010-X. [DOI] [PubMed] [Google Scholar]
- 8.Dohner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Buchner T, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 2017;129(4):424–47 doi 10.1182/blood-2016-08-733196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nechiporuk T, Kurtz SE, Nikolova O, Liu T, Jones CL, D’Alessandro A, et al. The TP53 Apoptotic Network Is a Primary Mediator of Resistance to BCL2 Inhibition in AML Cells. Cancer Discov 2019;9(7):910–25 doi 10.1158/2159-8290.CD-19-0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pollyea DA, Stevens BM, Jones CL, Winters A, Pei S, Minhajuddin M, et al. Venetoclax with azacitidine disrupts energy metabolism and targets leukemia stem cells in patients with acute myeloid leukemia. Nature medicine 2018;24(12):1859–66 doi 10.1038/s41591-018-0233-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones CL, Stevens BM, D’Alessandro A, Reisz JA, Culp-Hill R, Nemkov T, et al. Inhibition of Amino Acid Metabolism Selectively Targets Human Leukemia Stem Cells. Cancer Cell 2018;34(5):724–40 e4 doi 10.1016/j.ccell.2018.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lagadinou ED, Sach A, Callahan K, Rossi RM, Neering SJ, Minhajuddin M, et al. BCL-2 inhibition targets oxidative phosphorylation and selectively eradicates quiescent human leukemia stem cells. Cell stem cell 2013;12(3):329–41 doi 10.1016/j.stem.2012.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuusanmaki H, Leppa AM, Polonen P, Kontro M, Dufva O, Deb D, et al. Phenotype-based drug screening reveals association between venetoclax response and differentiation stage in acute myeloid leukemia. Haematologica 2019. doi 10.3324/haematol.2018.214882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu Y, McKenna RW, Wilson KS, Karandikar NJ, Schultz RA, Kroft SH. Immunophenotypic identification of acute myeloid leukemia with monocytic differentiation. Leukemia 2006;20(7):1321–4 doi 10.1038/sj.leu.2404242. [DOI] [PubMed] [Google Scholar]
- 15.Garcia C, Gardner D, Reichard KK. CD163: a specific immunohistochemical marker for acute myeloid leukemia with monocytic differentiation. Appl Immunohistochem Mol Morphol 2008;16(5):417–21 doi 10.1097/PAI.0b013e31815db477. [DOI] [PubMed] [Google Scholar]
- 16.Cascavilla N, Musto P, D’Arena G, Melillo L, Carella AM, Petrilli MP, et al. CD117 (c-kit) is a restricted antigen of acute myeloid leukemia and characterizes early differentiative levels of M5 FAB subtype. Haematologica 1998;83(5):392–7. [PubMed] [Google Scholar]
- 17.Di Noto R, Lo Pardo C, Schiavone EM, Manzo C, Vacca C, Ferrara F, et al. Stem cell factor receptor (c-kit, CD117) is expressed on blast cells from most immature types of acute myeloid mallignancies but is also a characteristic of a subset of acute promyelocytic leukaemia. British journal of haematology 1996;92(3):562–4. [DOI] [PubMed] [Google Scholar]
- 18.Naeim F Atlas of hematopathology : morphology, immunophenotype, cytogenetics, and molecular approaches. 1st ed. London: Academic Press,; 2013. p xi, 743 p. [Google Scholar]
- 19.Cancer Genome Atlas Research N, Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. The New England journal of medicine 2013;368(22):2059–74 doi 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pollyea DA, Jordan CT. Therapeutic targeting of acute myeloid leukemia stem cells. Blood 2017;129(12):1627–35 doi 10.1182/blood-2016-10-696039. [DOI] [PubMed] [Google Scholar]
- 21.Pei S, Minhajuddin M, Adane B, Khan N, Stevens BM, Mack SC, et al. AMPK/FIS1-Mediated Mitophagy Is Required for Self-Renewal of Human AML Stem Cells. Cell stem cell 2018;23(1):86–100 e6 doi 10.1016/j.stem.2018.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelly LM, Englmeier U, Lafon I, Sieweke MH, Graf T. MafB is an inducer of monocytic differentiation. EMBO J 2000;19(9):1987–97 doi 10.1093/emboj/19.9.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ziegler-Heitbrock HW, Ulevitch RJ. CD14: cell surface receptor and differentiation marker. Immunol Today 1993;14(3):121–5 doi 10.1016/0167-5699(93)90212-4. [DOI] [PubMed] [Google Scholar]
- 24.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nature medicine 1997;3(7):730–7. [DOI] [PubMed] [Google Scholar]
- 25.Ng SW, Mitchell A, Kennedy JA, Chen WC, McLeod J, Ibrahimova N, et al. A 17-gene stemness score for rapid determination of risk in acute leukaemia. Nature 2016;540(7633):433–7 doi 10.1038/nature20598. [DOI] [PubMed] [Google Scholar]
- 26.Eppert K, Takenaka K, Lechman ER, Waldron L, Nilsson B, van Galen P, et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nature medicine 2011;17(9):1086–93 doi 10.1038/nm.2415. [DOI] [PubMed] [Google Scholar]
- 27.Sukhai MA, Prabha S, Hurren R, Rutledge AC, Lee AY, Sriskanthadevan S, et al. Lysosomal disruption preferentially targets acute myeloid leukemia cells and progenitors. J Clin Invest 2013;123(1):315–28 doi 10.1172/JCI64180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Souers AJ, Leverson JD, Boghaert ER, Ackler SL, Catron ND, Chen J, et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nature medicine 2013;19(2):202–8 doi 10.1038/nm.3048. [DOI] [PubMed] [Google Scholar]
- 29.Pan R, Hogdal LJ, Benito JM, Bucci D, Han L, Borthakur G, et al. Selective BCL-2 inhibition by ABT-199 causes on-target cell death in acute myeloid leukemia. Cancer Discov 2014;4(3):362–75 doi 10.1158/2159-8290.CD-13-0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Novershtern N, Subramanian A, Lawton LN, Mak RH, Haining WN, McConkey ME, et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell 2011;144(2):296–309 doi 10.1016/j.cell.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lara-Astiaso D, Weiner A, Lorenzo-Vivas E, Zaretsky I, Jaitin DA, David E, et al. Immunogenetics. Chromatin state dynamics during blood formation. Science 2014;345(6199):943–9 doi 10.1126/science.1256271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramsey HE, Fischer MA, Lee T, Gorska AE, Arrate MP, Fuller L, et al. A Novel MCL1 Inhibitor Combined with Venetoclax Rescues Venetoclax-Resistant Acute Myelogenous Leukemia. Cancer Discov 2018;8(12):1566–81 doi 10.1158/2159-8290.CD-18-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roesch A, Vultur A, Bogeski I, Wang H, Zimmermann KM, Speicher D, et al. Overcoming intrinsic multidrug resistance in melanoma by blocking the mitochondrial respiratory chain of slow-cycling JARID1B(high) cells. Cancer Cell 2013;23(6):811–25 doi 10.1016/j.ccr.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Viale A, Pettazzoni P, Lyssiotis CA, Ying H, Sanchez N, Marchesini M, et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature 2014;514(7524):628–32 doi 10.1038/nature13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vlashi E, Lagadec C, Vergnes L, Matsutani T, Masui K, Poulou M, et al. Metabolic state of glioma stem cells and nontumorigenic cells. Proceedings of the National Academy of Sciences of the United States of America 2011;108(38):16062–7 doi 10.1073/pnas.1106704108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ho TC, LaMere M, Stevens BM, Ashton JM, Myers JR, O’Dwyer KM, et al. Evolution of acute myelogenous leukemia stem cell properties after treatment and progression. Blood 2016;128(13):1671–8 doi 10.1182/blood-2016-02-695312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shlush LI, Mitchell A, Heisler L, Abelson S, Ng SWK, Trotman-Grant A, et al. Tracing the origins of relapse in acute myeloid leukaemia to stem cells. Nature 2017;547(7661):104–8 doi 10.1038/nature22993. [DOI] [PubMed] [Google Scholar]
- 38.Stoeckius M, Hafemeister C, Stephenson W, Houck-Loomis B, Chattopadhyay PK, Swerdlow H, et al. Simultaneous epitope and transcriptome measurement in single cells. Nat Methods 2017;14(9):865–8 doi 10.1038/nmeth.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Somervaille TC, Matheny CJ, Spencer GJ, Iwasaki M, Rinn JL, Witten DM, et al. Hierarchical maintenance of MLL myeloid leukemia stem cells employs a transcriptional program shared with embryonic rather than adult stem cells. Cell stem cell 2009;4(2):129–40 doi 10.1016/j.stem.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krivtsov AV, Twomey D, Feng Z, Stubbs MC, Wang Y, Faber J, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature 2006;442(7104):818–22 doi 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- 41.van Galen P, Hovestadt V, Wadsworth Ii MH, Hughes TK, Griffin GK, Battaglia S, et al. Single-Cell RNA-Seq Reveals AML Hierarchies Relevant to Disease Progression and Immunity. Cell 2019;176(6):1265–81 e24 doi 10.1016/j.cell.2019.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Skrtic M, Sriskanthadevan S, Jhas B, Gebbia M, Wang X, Wang Z, et al. Inhibition of mitochondrial translation as a therapeutic strategy for human acute myeloid leukemia. Cancer Cell 2011;20(5):674–88 doi 10.1016/j.ccr.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sriskanthadevan S, Jeyaraju DV, Chung TE, Prabha S, Xu W, Skrtic M, et al. AML cells have low spare reserve capacity in their respiratory chain that renders them susceptible to oxidative metabolic stress. Blood 2015;125(13):2120–30 doi 10.1182/blood-2014-08-594408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cole A, Wang Z, Coyaud E, Voisin V, Gronda M, Jitkova Y, et al. Inhibition of the Mitochondrial Protease ClpP as a Therapeutic Strategy for Human Acute Myeloid Leukemia. Cancer Cell 2015;27(6):864–76 doi 10.1016/j.ccell.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carrington EM, Zhan Y, Brady JL, Zhang JG, Sutherland RM, Anstee NS, et al. Anti-apoptotic proteins BCL-2, MCL-1 and A1 summate collectively to maintain survival of immune cell populations both in vitro and in vivo. Cell Death Differ 2017;24(5):878–88 doi 10.1038/cdd.2017.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Opferman JT, Kothari A. Anti-apoptotic BCL-2 family members in development. Cell Death Differ 2018;25(1):37–45 doi 10.1038/cdd.2017.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tyner JW, Tognon CE, Bottomly D, Wilmot B, Kurtz SE, Savage SL, et al. Functional genomic landscape of acute myeloid leukaemia. Nature 2018;562(7728):526–31 doi 10.1038/s41586-018-0623-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goemans BF, Zwaan CM, Martinelli S, Harrell P, de Lange D, Carta C, et al. Differences in the prevalence of PTPN11 mutations in FAB M5 paediatric acute myeloid leukaemia. British journal of haematology 2005;130(5):801–3 doi 10.1111/j.1365-2141.2005.05685.x. [DOI] [PubMed] [Google Scholar]
- 49.Hintzsche J, Kim J, Yadav V, Amato C, Robinson SE, Seelenfreund E, et al. IMPACT: a whole-exome sequencing analysis pipeline for integrating molecular profiles with actionable therapeutics in clinical samples. J Am Med Inform Assoc 2016;23(4):721–30 doi 10.1093/jamia/ocw022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nemkov T, D’Alessandro A, Hansen KC. Three-minute method for amino acid analysis by UHPLC and high-resolution quadrupole orbitrap mass spectrometry. Amino acids 2015;47(11):2345–57 doi 10.1007/s00726-015-2019-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request. Patient-related clinical data are included as Supplementary tables. Bulk RNA-seq (GSE132511), CITE-seq (GSE143363) and WES (PRJNA600769) data have been uploaded to public databases.