Abstract
Background
Familial dilated cardiomyopathy (DCM) is genetically heterogeneous and is associated with mutations in at least 40 different genes. Apart from TTN encoding the giant protein Titin, none of these genes have an expected diagnostic yield of more than 5 % complicating genetic diagnosis. Whole exome sequencing (WES) is a powerful alternative for the identification of the causal gene, however variant interpretation remains challenging. We report on WES in a large family with autosomal dominant DCM complicated by end stage heart failure and non-sustained ventricular arrhythmias in whom no causative mutation was identified using a targeted gene panel including 28 genes.
Methods and results
WES was applied on 2 affected cousins. Stringent filtering of the identified genetic variants was performed including population variant frequencies, in silico analysis, orthologous and paralogous conservation. Subsequently Sanger sequencing was performed for 10 potential disease causing variants in order to confirm the presence of the variant and to evaluate co-segregation. Only one variant in exon 9 of the RBM20 gene (c.2714T>A, p.Met950Lys, NM_001334363) showed full co-segregation in the 7 affected family members resulting in a maximum 2-point LOD score of 2.1 and suggesting this as the pathogenic mutation responsible for the phenotype. Recently mutations in RBM20 have been linked to arrhythmogenic dilated cardiomyopathy caused by defective splicing of the giant sarcomere protein titin and abnormal calcium handling.
Conclusions
We report the identification of a novel mutation in RBM20 by WES in a large pedigree with DCM.
Keywords: RBM20, Whole Exome Sequencing, Dilated Cardiomyopathy
Introduction
Familial dilated cardiomyopathy (DCM) is a genetically heterogeneous disease associated with mutations in at least 40 different genes.(1) Apart from the giant protein Titin (TTN), none of these genes have an expected diagnostic yield of more than 5% complicating genetic diagnosis.(2) Genetic testing in clinical practice has evolved from gene by gene mutation scanning or Sanger sequencing to targeted gene panel testing. Whole exome sequencing (WES) is a powerful alternative for the identification of the causal gene in genetically heterogeneous diseases, however variant interpretation remains challenging. Identification of the causal gene in DCM helps in appropriately counseling the family, arrange proper follow-up for mutation positive family members and dismiss mutation negative family members from further follow-up. Some recent data even suggest gene or mutation specific risk stratification and treatment, for example in LMNA mutation carriers and in a Dutch cohort with a PLN founder mutation.(3, 4) We performed WES in a large Caucasian family with autosomal dominant DCM complicated by end stage heart failure and non-sustained ventricular arrhythmias. The index of this family was evaluated previously by means of targeted gene panel analysis including 28 genes, but no causal mutation was found.(5)
Methods
Sequencing
Whole exome sequencing was performed at the Human Genome sequencing center (HGSC) at Baylor College of Medicine through the Baylor-Hopkins Center for Mendelian Genomics initiative. Using 0.5 μg of DNA an Illumina paired-end pre-capture library was constructed according to the manufacturer’s protocol with modifications as described in the BCM-HGSC protocol.(6) Six pre-captured libraries were pooled and then hybridized in solution to the HGSC VCRome 2.1 design(7) (42Mb NimbleGen, Cat. No. 06266380001) according to the manufacturer’s protocol NimbleGen SeqCap EZ Exome Library SR User’s Guide with minor revisions. The sequencing run was performed in paired-end mode using the Illumina HiSeq 2000 platform, with sequencing-by-synthesis reactions extended for 101 cycles from each end and an additional 7 cycles for the index read. With a sequencing yield of 5.1 Gb, the samples achieved 89% of the targeted exome bases covered to a depth of 20X or greater. Illumina sequence analysis was performed using the HGSC Mercury analysis pipeline which moves data through various analysis tools from the initial sequence generation on the instrument to annotated variant calls.(8, 9) Detailed methodology is available upon request. Informed consent was obtained from all family members and the study was approved by the local ethics committee.
Variant filtering
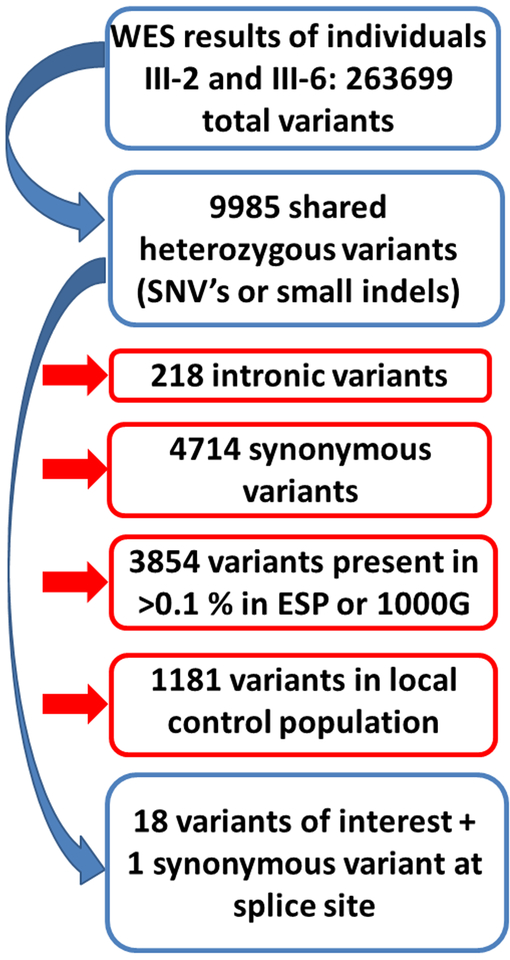
Two distant affected family members were chosen for whole exome sequencing. First, shared heterozygous variants (single nucleotide variants, small insertions and deletions) located inside the exon or at the exon/intron boundary (up to 3 nucleotides intronic) were selected. Synonymous variants were excluded, except if they were located at the exon/intron boundary. Variants with a minor allele frequency of >0.1% in publicly available exome databases (1000 Genomes(10), Exome Variant Server(11), ExAC(12)) were excluded. Furthermore, variants that were present in an in-house exome cohort (N = 80) performed at the same laboratory for other disease entities were also excluded since these probably represent local genetic variation or incorrect calling. The remaining variants were evaluated using a comprehensive scoring system specifically developed for variants in cardiomyopathy genes that includes different in silico analysis tools, orthologous and paralogous conservation and population frequencies.(13) Variants of unknown significance and (likely) pathogenic variants were further evaluated by Sanger sequencing for co-segregation analysis. Finally, the variants with matching co-segregation were also evaluated using the ACMG-AMP criteria.(14) Co-segregation in the ACMG-AMP criteria was assessed using the recommendations of Jarvik and Browning.(15)
Results
Clinical data
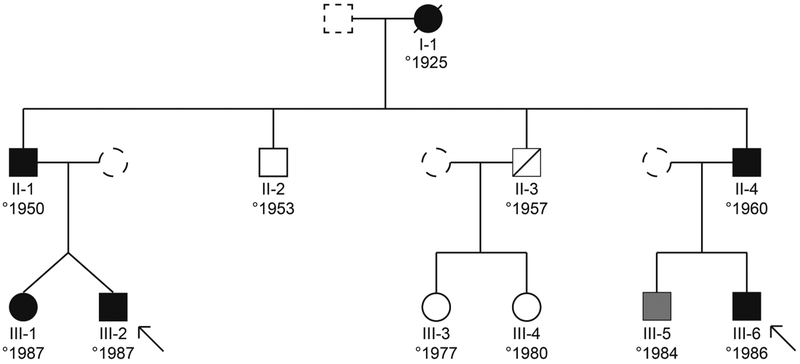
The family of interest concerns a 3 generation pedigree with dilated cardiomyopathy (figure 1 and table 1). They first came to our attention in 2001. The index of the family (III-2) is a male (°1987) who was initially admitted in another hospital with heart failure due to a dilated cardiomyopathy with a decreased ejection fraction. He was transferred to our center in cardiogenic shock, after an apical thrombus embolized to his right carotid artery and left coronary artery. The resulting left hemiplegia fortunately subsided within hours, but the occlusion of both his lateral branches caused a large area of persisting akinesia in the posterolateral part of his already diffusely weakened left ventricle. A temporary left ventricular assist device was inserted, but six days after the initial presentation an urgent heart transplantation had to be performed due to refractory cardiogenic shock. Histology of the explanted heart showed diffuse myocardial necrosis with a lymphocytic infiltrate without clear cause. An underlying myocarditis might have triggered the cardiomyopathy, however viral infection could not be proven.
Figure 1: Pedigree.
Circles illustrate females, boxes illustrate males, filled symbols illustrate affected individuals, shaded symbols indicate borderline affected individuals, arrows indicate individuals that underwent whole exome sequencing
Table 1:
Clinical data
| Age | Phenotype | CHF | Treatment | ECG | LVEF (%) | LVEDD (mm) | nsVT | CAD | Alcohol | |
|---|---|---|---|---|---|---|---|---|---|---|
| I-1 | ± 30 | Peripartum CMP | + | NA | NA | NA | NA | NA | NA | NA |
| II-1 | 51 | DCM | + | ARB + BB | LAHB | 20 | 72 | + | − (CAG) | − |
| II-2 | 48 | neg | − | − | normal | 71 | 52 | − | NA | − |
| II-3 | † 45 | ischemic CMP | NA | NA | NA | NA | NA | NA | + | NA |
| II-4 | 39 | DCM | + | ICD + ACE-I + BB + digoxin | iLBBB | 10 | 59 | + | − (CAG) | NA |
| III-1 | 12 | DCM | + | ARB + BB | normal | 34 | 60 | + | NA | − |
| III-2 | 13 | DCM | + | urgent HTx | normal | 25 | 65 | − | + | − |
| III-3 | 24 | neg | − | − | normal | 60 | 45 | − | NA | − |
| III-4 | 21 | neg | − | − | normal | 60 | 44 | − | NA | − |
| III-5 | 17 | borderline DCM | − | BB | normal | 50 | 55 | + | NA | − |
| III-6 | 15 | DCM | − | BB | normal | 40 | 56 | − | − (CT) | + (20U/w) |
= age death; ACE-I = Angiotensin converting enzyme inhibitor; Age = age at diagnosis or first evaluation in the case of a negative phenotype; ARB = Angiotensin receptor blocker; BB = beta-blocker; CAD = coronary artery disease; CHF = congestive heart failure; HTx = heart transplant; ICD = implantable cardioverter defibrillator; iLBBB = incomplete left bundle branch block; LAHB = left anterior hemiblock; LVEDD = left ventricular end diastolic diameter; LVEF = left ventricular ejection fraction; NA = not available; nsVT = non sustained ventricular tachycardia
His twin sister presented 8 months later with an episode of congestive heart failure and a severely depressed left ventricular ejection fraction. Inotropic therapy and IV diuretics were temporarily started with good clinical response. Standard heart failure treatment was started including bisoprolol, enalapril and spironolactone. Since this first episode, she remained asymptomatic with recovery of the cardiac function (LVEF ± 50%). Their father (II-1) was diagnosed with DCM during screening after the cardiac transplant of his son. He had an asymptomatic mildly reduced LVEF (50%) with moderate dilatation (LVEDD 60mm) at presentation. Treatment with bisoprolol and losartan was started. During follow-up mildly symptomatic episodes of non-sustained ventricular tachycardia (polymorphic, up to 30 beats, up to 180 BPM) were documented on 24 holter recordings, however without syncope. The LVEF fluctuated between 50% and 25% during follow up. There was one episode of mild congestive heart failure necessitating the addition of hydrochlorothiazide to his drug regimen in 2006. One of the paternal uncles of the index (II-4) was diagnosed with DCM in 1999 in another center. At presentation he had a very severely reduced LVEF (10%) and was evaluated for cardiac transplant at that time. However his cardiac function partially recovered with standard heart failure therapy. His LVEF fluctuated between 30 and 45% and he was admitted several times for decompensated heart failure. Since 1999 episodes of non-sustained ventricular tachycardia have been documented, but without syncopal events. He received an ICD in 2007. Since implantation, antitachycardia pacing terminated 4 episodes of VT, no shock was delivered. His two sons have an asymptomatic borderline (III-5) and definite (III-6) DCM.
The grandmother (I.1) developed symptoms of heart failure shortly after she gave birth to her youngest son (II.4) and died many years later at age 72. Patient II-3 suffered a myocardial infarction at age 38 and died 7 years later. Unfortunately more information is lacking as he was treated at another hospital.
Filtering and interpretation of pathogenicity
WES was performed on 2 distant affected family members (III-2 and III-6 in the pedigree). Filtering of variants is illustrated in figure 2. After filtering, 19 variants were scored using the pre-specified protocol. Of these, 9 were classified as variants of unknown significance and 1 as likely pathogenic. These 10 variants were further evaluated with co-segregation analysis (table 2). Only the variant in RBM20 was present in all affected or borderline affected patients and not present in the unaffected individuals. The variant was classified as likely pathogenic according to the ACMG-AMP criteria due to strong co-segregation, absence from controls (1000G, ESP and ExAC) and the fact that missense variants in this gene cause disease. The LOD score for this variant was 2.1, the maximum score obtainable in this pedigree. A variant in CPN1 was present in all tested individuals apart from one who did not have DCM. CPN1 encodes kininase, which is not linked to cardiac phenotypes or expressed in cardiac tissue and therefore presumably has no effect on the DCM observed in this pedigree.
Figure 2: Filtering of whole exome sequencing data.
Filtering steps applied to the variants that were discovered by whole exome sequencing. Red boxes indicate variants that were removed.
Table 2:
Identified variants of unknown significance or likely pathogenic variants after filtering and applying the variant interpretation score system
| Phenotype | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Positive | Borderline | Negative | |||||||||||||||
| Gene | Peptide | Exon | C. | P. | Transcript | Variant score | ExAC | I-1 | II-1 | II-4 | III-1 | III-2 | III-6 | III-5 | II-2 | III-3 | III-4 |
| ALPPL2 | Alkaline phosphatase | 11 | 1385del | Gly462Alafs*16 | NM_031313.2 | Class 3 (59%) | 35/105460 | Not confirmed by Sanger sequencing | |||||||||
| CPN1 | Kininase | 6 | 939_940insA | Phe314Ilefs*14 | NM_001308.2 | Class 3 (64%) | 0/121392 | + | + | + | + | + | + | + | − | + | + |
| GNAT2 | Cone specific transducin | 6 | 698T>C | Val233Ala | NM_005272.3 | Class 3 (46%) | 1/121408 | − | + | + | + | + | + | − | + | + | + |
| IRX2 | Iroquois homeobox protein 2 | 3 | 1226G>A | Arg409Gln | NM_001134222.1 | Class 3 (59%) | 0/112390 | + | + | + | − | + | + | + | + | − | − |
| KIAA0895 | Unknown | 5 | 1015C>T | Arg339* | NM_001100425.1 | Class 3 (64%) | 0/119518 | − | + | + | + | + | + | − | + | + | − |
| NBN | Nibrin | 2 | 93_94del | Ala32Hisfs*4 | NM_002485.4 | Class 3 (64%) | 0/121378 | + | + | + | − | + | + | − | + | − | + |
| RBM20 | RNA binding motif protein 20 | 11 | 2714T>A | Met905Lys | NM_001134363.1 | Class 4 (73%) | 0/21696 | + | + | + | + | + | + | + | − | − | − |
| ST3GAL4 | Sialyltransferase 4 | 7 | 404G>A | Gly135Glu | NM_001254757.1 | Class 3 (68%) | 0/121184 | − | + | + | − | + | − | − | − | − | − |
| TNS1 | Tensin 1 | 13 | 671T>A | Ile224Asn | NM_022648.5 | Class 3 (68%) | 2/121390 | − | + | + | + | + | + | − | + | − | − |
| ZNF135 | Zin Finger protein 135 | 4 | 714C>A | Ser238Arg | NM_001164530.1 | Class 3 (64%) | 31/121358 | − | + | + | − | + | − | + | − | − | − |
Discussion
Comprehensive gene panel testing is the current cornerstone of clinical DNA testing for cardiogenetic diseases. Especially in DCM, which is genetically very heterogeneous, this approach makes clinical DNA testing feasible.(2) However, a definite mutation can only be identified in about 40% of the cases.(2) Whole exome sequencing is a powerful alternative, since it covers the whole protein coding sequence of the genome. Therefore, it might unravel a mutation, which is located in a region, that is not yet known to be associated with a specific disease and thus not included in a dedicated disease panel. Since WES results in an enormous amount of genetic variants, identifying the disease-causing mutation remains similar to looking for a needle in a haystack. Therefore, we applied WES on 2 distant affected family members, thereby reducing the number of variants of interest to only 3.8% of the initial number. After some additional filtering steps a limited number of variants remained, and co-segregation analysis by Sanger sequencing of these variants became feasible. The RBM20 variant was the only that fully co segregated with the affected family members. At the time the proband of the current pedigree underwent genetic testing, RBM20 was not yet established as a definite DCM causing gene and was not included on the gene panel that was performed earlier.
Mutations in RBM20 (RNA Binding Motif 20) have been first identified in patients from 2 families with familial dilated cardiomyopathy in 2009.(16) In the following years, several mutations have been described.(17, 18) Most of these were located in an arginine serine rich mutation hotspot region in exon 9. In 2012, a comprehensive evaluation of the RBM20 gene in a cohort of 283 individuals with DCM revealed a mutation in RBM20 in 2.8%.(19) In contrast to the first reports, these mutations were equally distributed across the gene. Functional studies of mutations in the RBM20 gene showed the importance of the gene in TTN splicing.(20) Missense mutations in RBM20 result in increased expression of a longer titin isoform by acting as a splicing repressor.(21) However, RBM20 not only regulates TTN splicing, it is also involved in splicing of several other genes that have been implicated in DCM.(20) Furthermore, it was recently shown that mutations in RBM20 also lead to aberrant splicing of the calcium homeostasis related genes CAMK2D and RYR2 and subsequent activation of L type Calcium channels, intracellular calcium overload and increased sarcoplasmic reticulum calcium content.(22) This leads to more pro-arrhythmic spontaneous calcium releases from the sarcoplasmic reticulum. The combination of disturbed titin splicing and function with disturbed calcium handling explains the more severe phenotype of RBM20 mutation carriers compared to TTN mutation carriers.(22) Indeed, a recent meta-analysis showed that RBM20 mutation carriers were transplanted at a significant younger age compared to other gene related DCM.(23) Also in our pedigree, the index patient presented with a refractory cardiogenic shock necessitating urgent heart transplantation, however he suffered a second ischemic insult on top of his genetic cardiomyopathy.
DCM is a heterogeneous disease with variable expression of disease severity in patients with the same disease causing mutation.(24) This is also evident from this pedigree, where one patient suffered severe heart failure necessitating urgent heart transplantation. Reasons for this variable expression have been explored and include environmental triggers (eg arterial hypertension, toxic exposure, cardiac inflammation, viral infection) but also co-inheritance of disease modulating genetic variants.(25) In contrast to disease causing mutations, these modulating variants might be more frequently observed in healthy population. We cannot exclude that some of the identified rare variants in this pedigree play a modulating role in disease expression and severity. However to unravel these more complex inheritance patterns, large multicenter genotype-phenotype studies are deemed necessary.
Conclusion
Whole exome sequencing of two distant affected family members with DCM is a powerful tool to identify a disease-causing mutation. This approach simplifies variant filtering and interpretation. RBM20 should be routinely evaluated in patients with familial DCM.
Funding Sources:
This work was supported in part by the National Institutes of Health, and a jointly funded National Human Genome Research Institute (NHGRI), and National Heart, Lung, and Blood Institute (NHLBI) grant to the Baylor-Hopkins Center for Mendelian Genomics [UM1 HG006542].
Footnotes
Disclosure of interests:
James R Lupski has stock ownership in 23andMe and Lasergen, is a paid consultant for Regeneron, and is a co-inventor on multiple United States and European patents related to molecular diagnostics for inherited neuropathies, eye diseases, and bacterial genomic fingerprinting. Other authors have no potential conflicts to report.
References
- 1.Wilde AA, Behr ER. Genetic testing for inherited cardiac disease. Nat Rev Cardiol. 2013;10(10):571–83. [DOI] [PubMed] [Google Scholar]
- 2.Haas J, Frese KS, Peil B, Kloos W, Keller A, Nietsch R, et al. Atlas of the clinical genetics of human dilated cardiomyopathy. Eur Heart J. 2015;36(18):1123–35a. [DOI] [PubMed] [Google Scholar]
- 3.van Rijsingen IA, Arbustini E, Elliott PM, Mogensen J, Hermans-van Ast JF, van der Kooi AJ, et al. Risk factors for malignant ventricular arrhythmias in lamin a/c mutation carriers a European cohort study. J Am Coll Cardiol. 2012;59(5):493–500. [DOI] [PubMed] [Google Scholar]
- 4.van Rijsingen IA, van der Zwaag PA, Groeneweg JA, Nannenberg EA, Jongbloed JD, Zwinderman AH, et al. Outcome in phospholamban R14del carriers: results of a large multicentre cohort study. Circ Cardiovasc Genet. 2014;7(4):455–65. [DOI] [PubMed] [Google Scholar]
- 5.Robyns T, Kuiperi C, Breckpot J, Devriendt K, Souche E, Van Cleemput J, et al. Repeat genetic testing with targeted capture sequencing in primary arrhythmia syndrome and cardiomyopathy. Eur J Hum Genet. 2017;25(12):1313–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baylor College Of Medicine. BCM-HGSC protocols for sequencing library construction. https://www.hgsc.bcm.edu/content/protocols-sequencing-library-construction Accessed 16/03/2018
- 7.Bainbridge MN, Wang M, Wu Y, Newsham I, Muzny DM, Jefferies JL, et al. Targeted enrichment beyond the consensus coding DNA sequence exome reveals exons with higher variant densities. Genome Biol. 2011;12(7):R68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Challis D, Yu J, Evani US, Jackson AR, Paithankar S, Coarfa C, et al. An integrative variant analysis suite for whole exome next-generation sequencing data. BMC Bioinformatics. 2012;13:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reid JG, Carroll A, Veeraraghavan N, Dahdouli M, Sundquist A, English A, et al. Launching genomics into the cloud: deployment of Mercury, a next generation sequence analysis pipeline. BMC Bioinformatics. 2014;15:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, et al. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Exome Variant Server, NHLBI GO Exome Sequencing Project (ESP), Seattle, WA: (URL: http://evs.gs.washington.edu/EVS/) [February, 2018 accessed]. [Google Scholar]
- 12.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Spaendonck-Zwarts KY, van Rijsingen IA, van den Berg MP, Lekanne Deprez RH, Post JG, van Mil AM, et al. Genetic analysis in 418 index patients with idiopathic dilated cardiomyopathy: overview of 10 years’ experience. Eur J Heart Fail. 2013;15(6):628–36. [DOI] [PubMed] [Google Scholar]
- 14.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jarvik GP, Browning BL. Consideration of Cosegregation in the Pathogenicity Classification of Genomic Variants. Am J Hum Genet. 2016;98(6):1077–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brauch KM, Karst ML, Herron KJ, de Andrade M, Pellikka PA, Rodeheffer RJ, et al. Mutations in ribonucleic acid binding protein gene cause familial dilated cardiomyopathy. J Am Coll Cardiol. 2009;54(10):930–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li D, Morales A, Gonzalez-Quintana J, Norton N, Siegfried JD, Hofmeyer M, et al. Identification of novel mutations in RBM20 in patients with dilated cardiomyopathy. Clin Transl Sci. 2010;3(3):90–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Millat G, Bouvagnet P, Chevalier P, Sebbag L, Dulac A, Dauphin C, et al. Clinical and mutational spectrum in a cohort of 105 unrelated patients with dilated cardiomyopathy. Eur J Med Genet. 2011;54(6):e570–5. [DOI] [PubMed] [Google Scholar]
- 19.Refaat MM, Lubitz SA, Makino S, Islam Z, Frangiskakis JM, Mehdi H, et al. Genetic variation in the alternative splicing regulator RBM20 is associated with dilated cardiomyopathy. Heart Rhythm. 2012;9(3):390–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo W, Schafer S, Greaser ML, Radke MH, Liss M, Govindarajan T, et al. RBM20, a gene for hereditary cardiomyopathy, regulates titin splicing. Nat Med. 2012;18(5):766–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maatz H, Jens M, Liss M, Schafer S, Heinig M, Kirchner M, et al. RNA-binding protein RBM20 represses splicing to orchestrate cardiac pre-mRNA processing. J Clin Invest. 2014;124(8):3419–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van den Hoogenhof MMG, Beqqali A, Amin AS, van der Made I, Aufiero S, Khan MAF, et al. RBM20 Mutations Induce an Arrhythmogenic Dilated Cardiomyopathy Related to Disturbed Calcium Handling. Circulation. 2018;138(13):1330–42. [DOI] [PubMed] [Google Scholar]
- 23.Kayvanpour E, Sedaghat-Hamedani F, Amr A, Lai A, Haas J, Holzer DB, et al. Genotype-phenotype associations in dilated cardiomyopathy: meta-analysis on more than 8000 individuals. Clin Res Cardiol. 2017;106(2):127–39. [DOI] [PubMed] [Google Scholar]
- 24.Hershberger RE, Hedges DJ, Morales A. Dilated cardiomyopathy: the complexity of a diverse genetic architecture. Nat Rev Cardiol. 2013;10(9):531–47. [DOI] [PubMed] [Google Scholar]
- 25.Verdonschot JAJ, Hazebroek MR, Derks KWJ, Barandiarán Aizpurua A, Merken JJ, Wang P, et al. Titin cardiomyopathy leads to altered mitochondrial energetics, increased fibrosis and long-term life-threatening arrhythmias. Eur Heart J. 2018;39(10):864–73. [DOI] [PubMed] [Google Scholar]