The molecular pathogenesis of Hidradenitis Suppurativa (HS) is purported to involve Notch dysregulation secondary to sequence variants in components of the gamma secretase complex (GSC)1. However, Notch dysregulation has also been identified in keratinocytes of other inflammatory dermatoses including psoriasis2 and atopic dermatitis2. Animal knockout models of components of the GSC with resultant Notch dysregulation result in the development of dermal cysts and histological features of follicular occlusion, suggesting that aberrant Notch signaling is linked to the unique clinical and histological manifestations of HS3. However, these models also rapidly develop multiple squamous cell carcinomas3 which is not consistent with the typical progression of HS. The precise role of Notch dysregulation as the primary driver in the molecular pathogenesis of HS is unclear. Dysregulated Notch signaling may be secondary to inflammation or other unknown molecular mechanisms, rather than an actual driver of HS2
We aimed to assess the quantitative changes between Notch signaling in publicly available genomic data from HS (GSE72702, n=30), Psoriasis (GSE13355, n=122), Atopic Dermatitis (GSE32924, n=22) and Alopecia Areata (GSE45512, n=10) patients. Comparison of lesional skin was made with healthy control skin (as opposed to non-lesional tissue) from affected patients in order to account of potential background inflammation. Statistical analysis was performed using R (R Core Team 2019). Expression values were modelled using a mixed-effects model with lesional categories as fixed factors and random effects for each patient. Fold Changes (FCH) were estimated under the general framework for linear models in the R LIMMA package. P-values from t-tests (LIMMA) were adjusted for multiple hypotheses using Benjamini‐Hochberg procedure, and p values ≤0.05 were considered significant. Differentially expressed genes (DEGs) were defined by FCH ≥1.5 or ≤−1.5 and false discovery rate (FDR ≤0.05).
No significant downregulation of Notch 1–4 was identified in HS lesional skin compared with control (Figure 1a). Amongst other inflammatory dermatoses, only Notch-1 downregulation reached statistical significance in Atopic Dermatitis. Downstream expression of the transcription factors HES1 and HEY1 was not significantly altered in HS or any of the other conditions examined (Figure 1b). Expression of NUMB, an endocytic inhibitor of the Notch intra-cellular-domain (which undergoes nuclear translocation and results in activation of the HES1 and HEY1 transcription factors), was also not significantly altered (Figure 1c), consistent with previous findings2. ADAM10 was downregulated in all inflammatory dermatoses, but ADAM 17 was upregulated in HS, in contrast with downregulation seen in psoriasis, atopic dermatitis and alopecia areata (Figure 1d). The significance of these findings were not altered when more stringent criteria (FCH≥2.0) were applied. ADAM10 and ADAM17 are involved in the cleavage and activation of GSC substrates (including Notch) extracellularly. ADAM17 (also known as tumor-necrosis-factor-α converting enzyme or TACE) is involved in the cleavage and activation of TNF- α on the cell surface4 and is upregulated in inflammatory bowel disease5 as well as involved in epidermal, ductal and hair follicle morphogenesis6. ADAM-17 also mediates the production of matrix metalloprotinases (MMP) implicated in HS as well as inflammatory arthropathies7. ADAM-17 activity is attenuated through the Nicastrin ectodomain, mutations in which are the most common sequence variant in inherited HS4.
Figure 1:
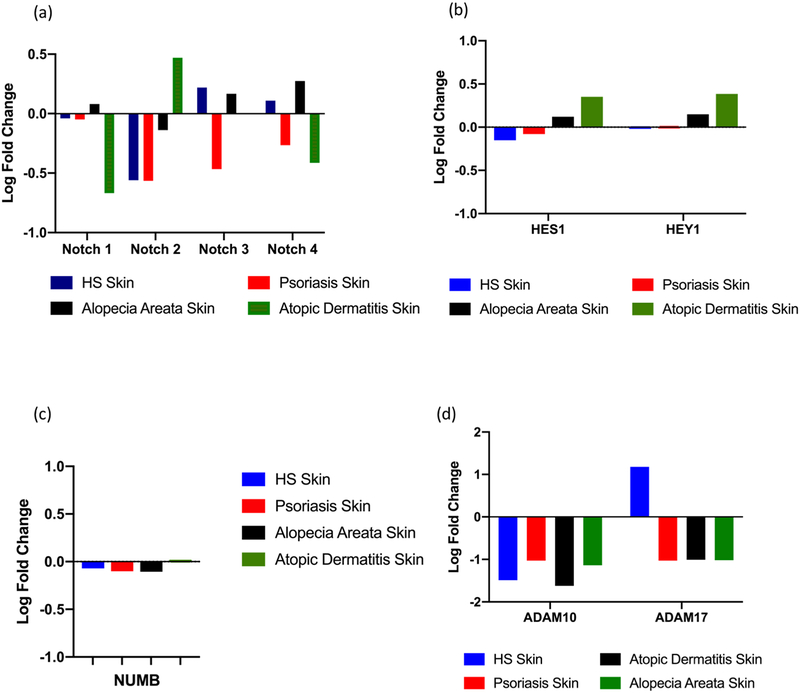
Log Fold change in the differential expression of specific genes in Hidradenitis Suppurativa (HS), Psoriasis, Alopecia Areata (AA) and Atopic Dermatitis (AD). (a) No significant differential expression of Notch 1–4 was identified in any condition. (b) HES1 and HEY1 and NUMB (c) expression was not significantly dysregulated across all four inflammatory dermatoses. (b) ADAM10 expression was non-significantly downregulated in HS, Psoriasis and AA but reached significance in AD (log fold change =1.51, p=0.05). ADAM17 expression was not significant but upregulated in HS and downregulated in all other conditions.
Notch is one of more than 70 GSC substrates and the non-significant differential expression of Notch in the HS transcriptome as well in other inflammatory dermatoses suggest that Notch dysregulation is not a unique feature in the pathogenesis of HS. This is supported by the lack of significant alteration in downstream HES1 and HEY1 transcription factors, and validation of the previously described lack of alteration in NUMB2. A number of other GSC substrates were significantly differentially expressed in HS include CX3CL1, EphB2, IFNaR2, KCNE3, NRG1 and PTK7. CX3CL1, ephB2 and IFNaR2 were only significant in HS (unpublished data Frew et al). IFNaR2 is also cleaved by ADAM17 and has been validated as upregulated in a Nicastrin knockdown keratinocyte cell line8.
The implication of these findings is that inherited and acquired defects in keratinocyte Notch signaling may be an over-simplistic explanation for the molecular pathogenesis of HS. ADAM17 has potential as a central mediator in the molecular pathogenesis of HS. The downstream effects of ADAM17 upregulation may contribute to effects upon multiple gamma secretase substrates other than notch involved in inflammation, fibrosis and scarring. ADAM17 is also targetable with pre-clinical trials of at-least two monoclonal antibodies in development5,7. Further experimental investigation into our in silico results and the role of ADAM17 and its effect upon gamma secretase substrates other than Notch are needed to further explore the molecular mechanisms in HS.
Funding Sources:
Supported in part by grant # UL1 TR001866 from the National Center for Advancing Translational Sciences (NCATS), National Institutes of Health (NIH) Clinical and Translational Science Award (CTSA) program, and T32GM007739 grant from the NIH.
References:
- 1).Melnik BC, Plewig G Impaired Notch signaling: the unifying mechanism explaining the pathogenesis of hidradenitis suppurativa Br J Dermatol 2013;168(4):876–878 [DOI] [PubMed] [Google Scholar]
- 2).Scala E, Balato A, Marasca C et al. New insights into mechanism of Notch signalling in hidradenitis suppurativa. G Ital Dermatol Venereol 2018. 10.23736/S0392-0488.18.06083-2 [DOI] [PubMed] [Google Scholar]
- 3).Vossen ARJV van der Zee HH Prens EP Hidradenitis Suppurativa: A Systematic Review Integrating Inflammatory Pathways into a Cohesive Pathogenic Model Front Immunol 2018;2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4).Gooz M ADAM-17: The Enzyme That Does It All Crit Rev Biochem Mol Biol 2010;45(2):146–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5).Moss ML, Minond D Recent Advances in ADAM17 Research: A Promising Target for Cancer and Inflammation Mediators INflamm 2017; 9673537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6).Peschon JJ, Slack JL, Reddy P, Stocking KL, Sunnaborg SW Lee DC et al. An essential role for ectodomain shedding in Mammalian development Science 1998;282(5392):1281–1284 [DOI] [PubMed] [Google Scholar]
- 7).Malemud CJ Inhibition of MMPs and ADAM/ADAMTS Biochem Pharmacol 2019;165:33–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8).Cao L, Morales-Heil DJ, Roberson EDO Nicastrin haploinsufficiency alters expression of type 1 interferon-stimulated genes: the relationship to familial hidradenitis suppurativa. Clin Exp Dermatol 2019; doi: 10.1111/ced.13906 [DOI] [PMC free article] [PubMed] [Google Scholar]


