Abstract
AKT inhibitors have promising activity in AKT1 E17K-mutant estrogen receptor (ER)-positive metastatic breast cancer, but the natural history of this rare genomic subtype remains unknown. Utilizing AACR Project GENIE, an international clinicogenomic data-sharing consortium, we conducted a comparative analysis of clinical outcomes of matched AKT1 E17K-mutant (n=153) and -wildtype (n=302) metastatic breast cancer patients. AKT1-mutant cases had similar adjusted overall survival (OS) compared with AKT1-wildtype controls (median OS, 24.1 vs 29.9, respectively; p=0.98). AKT1-mutant cases enjoyed longer durations on mTOR inhibitor therapy, an observation previously unrecognized in pivotal clinical trials due to the rarity of this alteration. Other baseline clinicopathologic features, as well as durations on other classes of therapy were broadly similar. In summary, we demonstrate the feasibility of using a novel and publicly accessible clincogenomic registry to define outcomes in a rare genomically defined cancer subtype, an approach with broad applicability to precision oncology.
Keywords: AACR Project GENIE, breast cancer, PI3K, AKT1 mutation, AKT1 E17K mutation
INTRODUCTION
Activation of the phosphatidylinositol 3-kinase (PI3K) pathway is common in estrogen receptor (ER)-positive (+) breast cancer, with approximately 40% of cases harboring mutations in PIK3CA, which encodes the PI3K alpha catalytic subunit p110 alpha (1). Providing further clinical validation of the oncogenic role of this pathway, the selective PI3K alpha inhibitor alpelisib when added to fulvestrant prolongs progression-free survival (PFS) compared to fulvestrant alone in PIK3CA mutant ER+ breast cancer and is now a standard of care in this indication (2). In an estimated 7% of ER+ breast cancers, the PI3K pathway is alternatively activated by mutation of AKT1 (1). In these cases, AKT1 E17K predominates as the most common (~80%) alteration and constitutively activates PI3K signalling by promoting pathological localization of AKT1 to the plasma membrane (3–5). Recent clinical studies have demonstrated promising activity of the ATP-competitive pan-AKT kinase inhibitor capivasertib in AKT1 E17K-mutant metastatic breast cancer and other cancers (6–8). Importantly, however, the natural history, namely the clinico-genomic characteristics and prognostic significance of the AKT1 E17K-mutation in ER+ breast cancer, remain unknown, leading to challenges in contextualizing available single-arm efficacy data.
The rarity of AKT1 E17K mutations in ER+ breast cancer necessitates the use of new approaches to define the prognostic and therapeutic significance of this biomarker. Despite expanding next-generation sequencing programs at many large centers, no single institution has a sufficient number of AKT1 E17K-mutant ER+ breast cancer patients for a robust analysis. Similarly, although large commercial sequencing laboratories have amassed abundant genomic data, most do not currently possess the requisite demographic, treatment, and clinical outcome data necessary for such an analysis. Consequently, the therapeutic implications of AKT1 mutations in ER+ breast cancer, including whether its presence leads to differential responses to currently approved standard treatments, remain unknown.
In 2015, the American Association for Cancer Research (AACR) launched the Project Genomics Evidence Neoplasia Information Exchange (GENIE), an international genomics registry and data-sharing consortium. The purpose of GENIE is to facilitate the sharing of clinical and genomic data through the academic and biopharma community. In its initial phase, GENIE established standards for the collation and harmonization of variant calls from clinical sequencing tests utilized across an international consortium of academic centers (9). Now in its 6th release, GENIE has made public the genomic records from over 72,000 samples. To more fully leverage the value of this dataset, the ability to clinically annotate these samples is essential. As a proof-of-concept pilot study, GENIE conducted an international, multi-center, retrospective and matched study to describe the clinical, pathologic, and genomic features, as well as survival and duration on standard therapies in AKT1 E17K-mutant versus -wildtype ER+ metastatic breast cancer patients. A consistent approach to clinical data curation was harmonized across treatment sites and was operationalized using a common Research Electronic Data Capture (REDCap) database.
RESULTS
Patient and Disease Characteristics
A total of 455 ER+ metastatic breast cancer patients were included in this study (Supplementary Figure 1A), specifically 302 AKT1-wildtype controls were matched to 153 AKT1-mutant cases by birth and sequencing year, center, and histologic subtype. The 2 patient cohorts ultimately had very similar baseline disease characteristics, including presenting age, stage, histologic subtype, grade, receptor subtype, and distribution of disease at metastatic presentation (Table 1 and Supplementary Table 1). Patients were predominantly white and presented with stage 2–3, intermediate/high-grade disease. Although clinico-pathological enrichments should be interpreted with relative caution in a case-control study, we observed similar rates of lobular (18% vs. 17%) and ductal (69% vs. 72%) histologies in AKT1-mutant and -wildtype cohorts, respectively, interestingly despite prior analyses suggesting AKT1 mutations predominate in lobular histologies (10). At the time of metastatic diagnosis, both cohorts had comparable rates of multiple disease sites involved. AKT1-mutant patients had a marginally higher rate of liver and lymph node metastases (33% vs. 23%, p<0.001; 31% vs. 25%, p=0.026, respectively).
Table 1.
Patient and disease characteristics
|
AKT1 mutant, n=153, N (%) |
AKT1 wildtype, n=302 N (%) |
All, n=455, N (%) |
|
|---|---|---|---|
| Age at diagnosis, median (range), years | 50(28–74) | 50(25–83) | 50(25–83) |
| Race | |||
| White | 124(81%) | 227(75%) | 351(77%) |
| Black | 9(6%) | 18(6%) | 27(6%) |
| Other/unknown | 20(13%) | 57(19%) | 77(17%) |
| Stage at primary diagnosis | |||
| 0 | 0(0%) | 5(2%) | 5(1%) |
| 1 | 23(15%) | 51(17%) | 74(16%) |
| 2 | 52(34%) | 82(27%) | 134(30%) |
| 3 | 37(24%) | 70(23%) | 107(24%) |
| 4 | 26(17%) | 77(26%) | 103(23%) |
| Unknown | 15(10%) | 17(6%) | 32(7%) |
| Histologic subtype | |||
| Ductal | 106(69%) | 218(72%) | 324(71%) |
| Lobular | 28(18%) | 52(17%) | 80(18%) |
| Other | 19(12%) | 32(11%) | 51(11%) |
| Overall tumor Grade | |||
| 1 | 7(5%) | 20(7%) | 27(6%) |
| 2 | 66(43%) | 118(39%) | 184(40%) |
| 3 | 43(28%) | 121(40%) | 164(36%) |
| Unknown | 37(24%) | 43(14%) | 80(18%) |
| HER2 status | |||
| Negative | 140(92%) | 273(90%) | 413(91%) |
| Positive | 3(2%) | 6(2%) | 9(2%) |
| Unknown | 10(7%) | 23(8%) | 33(7%) |
| Loco-regional recurrenc^ | 26(21%) | 48(21%) | 74(21%) |
HER2, human epidermal growth factor receptor 2
Among the 352 non-stage IV patients.
Study groups were also broadly similar for therapy received in the metastatic setting (Table 2 and Supplementary Table 2). The AKT1-mutant group had a slightly higher rate of chemotherapy treatment as first-line therapy for metastatic disease (35% vs. 28%, p=0.145). By comparison, the AKT1-wildtype group had received more lines of endocrine therapy for metastatic disease (median of 2 vs. 1 lines, p=0.006). This interesting observation may reflect a phenotypic difference in the study groups that prompted treating clinicians to preferentially use sequential endocrine-based therapy for the AKT1-wildtype group. Certainly the AKT1-wildtype group demonstrated a marginally higher rate of endocrine therapy sensitivity overall (78% vs. 69%, p=0.202). Approximately 30% overall had prior treatment with an mTOR-, CDK4/6-inhibitor, and an investigational agent on a clinical trial. Expectedly, more AKT1-mutant than -wildtype patients received an AKT inhibitor as part of an international basket clinical trial (NCT01226316) (15% vs. 1%, respectively; p<0.001).
Table 2.
Therapy exposure
|
AKT1 mutant, n=153, N (%) |
AKT1 wildtype, n=302 N (%) |
All, n=455, % | |
|---|---|---|---|
| Endocrine Therapy* | |||
| Tamoxifen | 96(63%) | 184(61%) | 280(62%) |
| Aromatase Inhibitor | 127(83%) | 266(88%) | 393(86%) |
| Fulvestrant | 79(52%) | 162(54%) | 241(53%) |
| Endocrine therapy sensitivity† | 106(69%) | 236(78%) | 342(75%) |
| Chemotherapy | 137(90%) | 269(89%) | 406(89%) |
| Endocrine therapies for metastatic disease, median (range)§ | 1 (0–6) | 2 (0–9) | 2 (0–9) |
| Chemotherapies for metastatic disease, median (range) | 2 (0–7) | 2 (0–9) | 2 (0–9) |
| Targeted Therapy | |||
| CDK4/6 inhibitor | 45(29%) | 87(29%) | 132(29%) |
| mTOR inhibitor | 49(32%) | 97(32%) | 146(32%) |
| ‡AKT inhibitor§ | 23(15%) | 4(1%) | 27(6%) |
| Therapeutic clinical trial | 52(34%) | 95(32%) | 147(32%) |
Any endocrine therapy in adjuvant or metastatic setting;
≥24 months of adjuvant endocrine therapy or ≥6 months treatment duration on any metastatic endocrine therapy;
p<0.05;
NCT01226316 was a first in human, phase I, multipart study of the AKT inhibitor, AZD5363, in advanced solid tumors. This study had many cohorts including individual molecularly selected cohorts for PIK3CA- AKT1-, and PTEN- mutant tumors. Four AKT1-wild type patients were treated on NCT01226316, 1 PIK3CA- and 2 PTEN -mutant breast cancers and 1 other patient for reasons unknown.
The distribution of sequencing time relative to first metastatic diagnosis was 13.4 months (−5.23–197.04) in the overall study population, without a significant difference observed between the study groups [9.51 months (−2.04–297.04) in AKT1-mutant cases versus 14.62 months (−5.23–207.73 months) in AKT1-wildtype controls, p=0.171]. Patients were identified by a variety of different sequencing platforms, most commonly using hybridization capture-based (50–60%) followed by amplicon-based next-generation sequencing (NGS) (Supplementary Tables 3A & B).
Overall Survival
Median duration of follow-up among survivors was 35.4 months (range, 0.2–232.8 months). Approximately 60% of the sequencing tests utilized to identify patients for this study were obtained within 2 years following metastatic diagnosis (Figure 1A, Table 3). To account for this selection bias, overall survival (OS) estimates were analyzed using left truncation methodology, in which patients enter the risk set based on time of sequencing. After this adjustment, the median OS between the AKT1-mutant and -wildtype cohorts was not statistically significant (24.1 vs. 29.9 months, respectively; HR 1.0; 95% CI, 0.74–1.36; p=0.98) (Figure 1B). The uncorrected median OS in the AKT1-mutant and -wildtype cohorts, estimated using the Kaplan-Meier method and ignoring bias, was statistically similar at 64.6 vs. 52.3 months, respectively (HR, 1.16; 95% CI, 0.86–1.57; p=0.331) (Figure 1C, Table 3).
Figure 1.
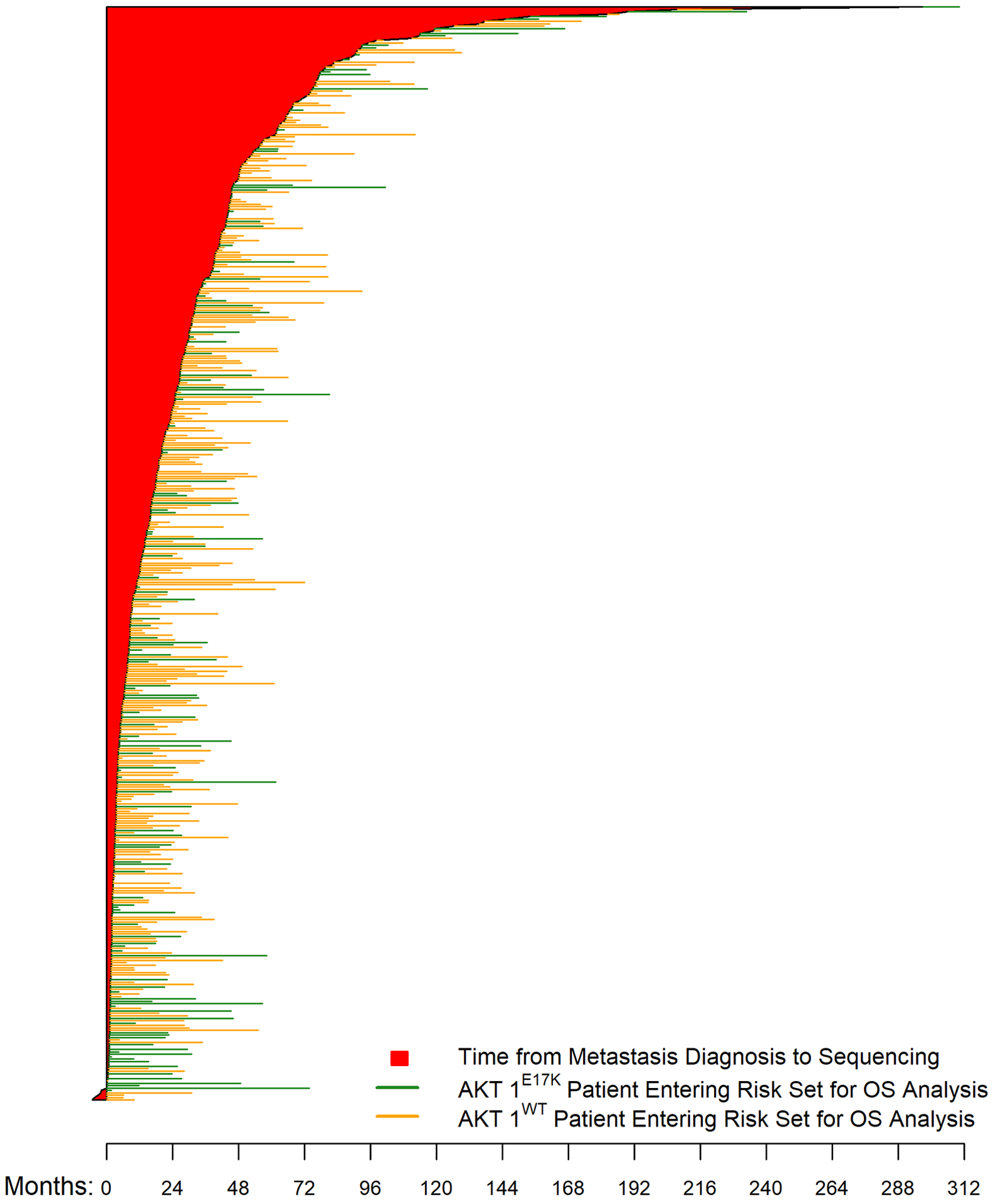
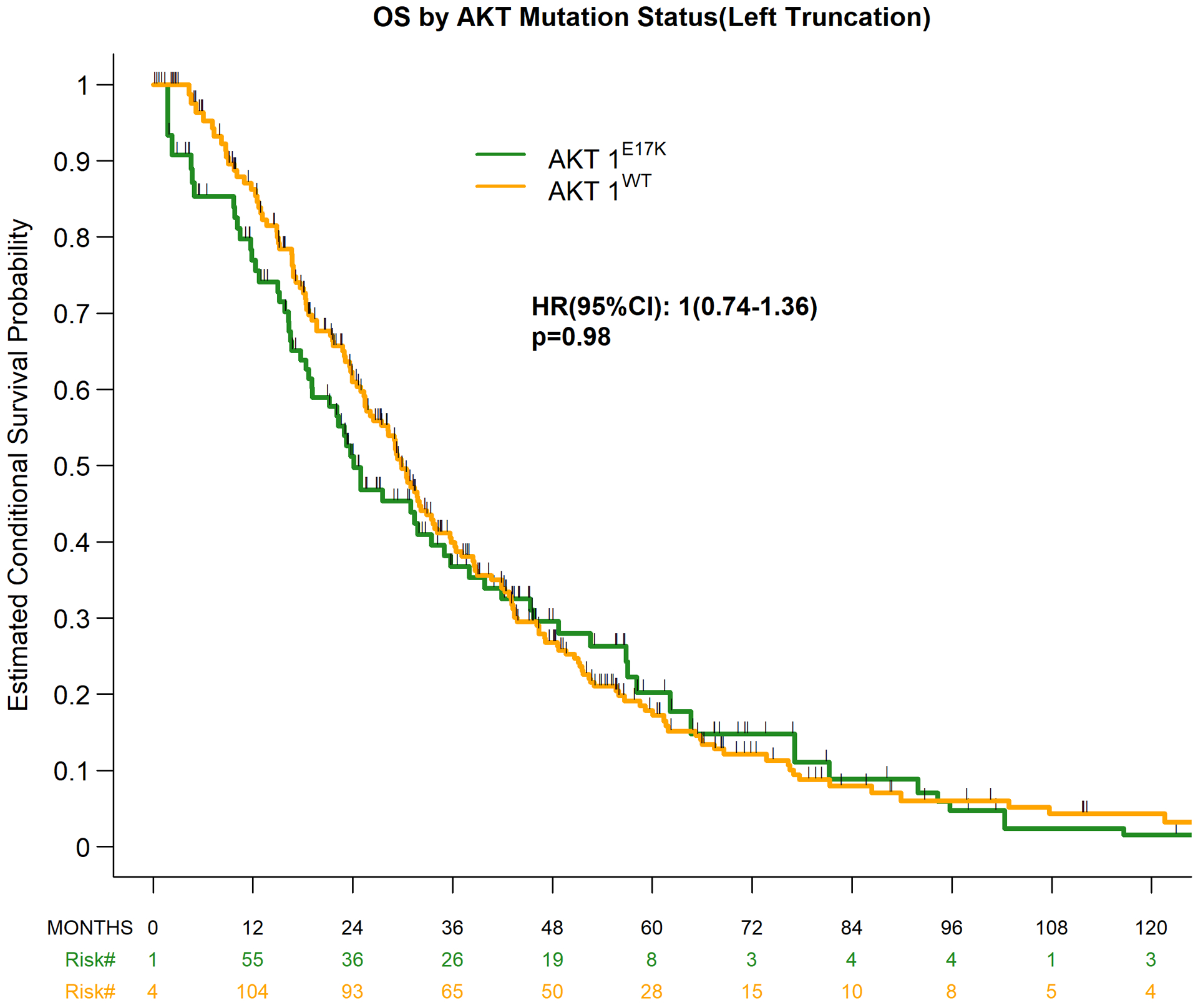
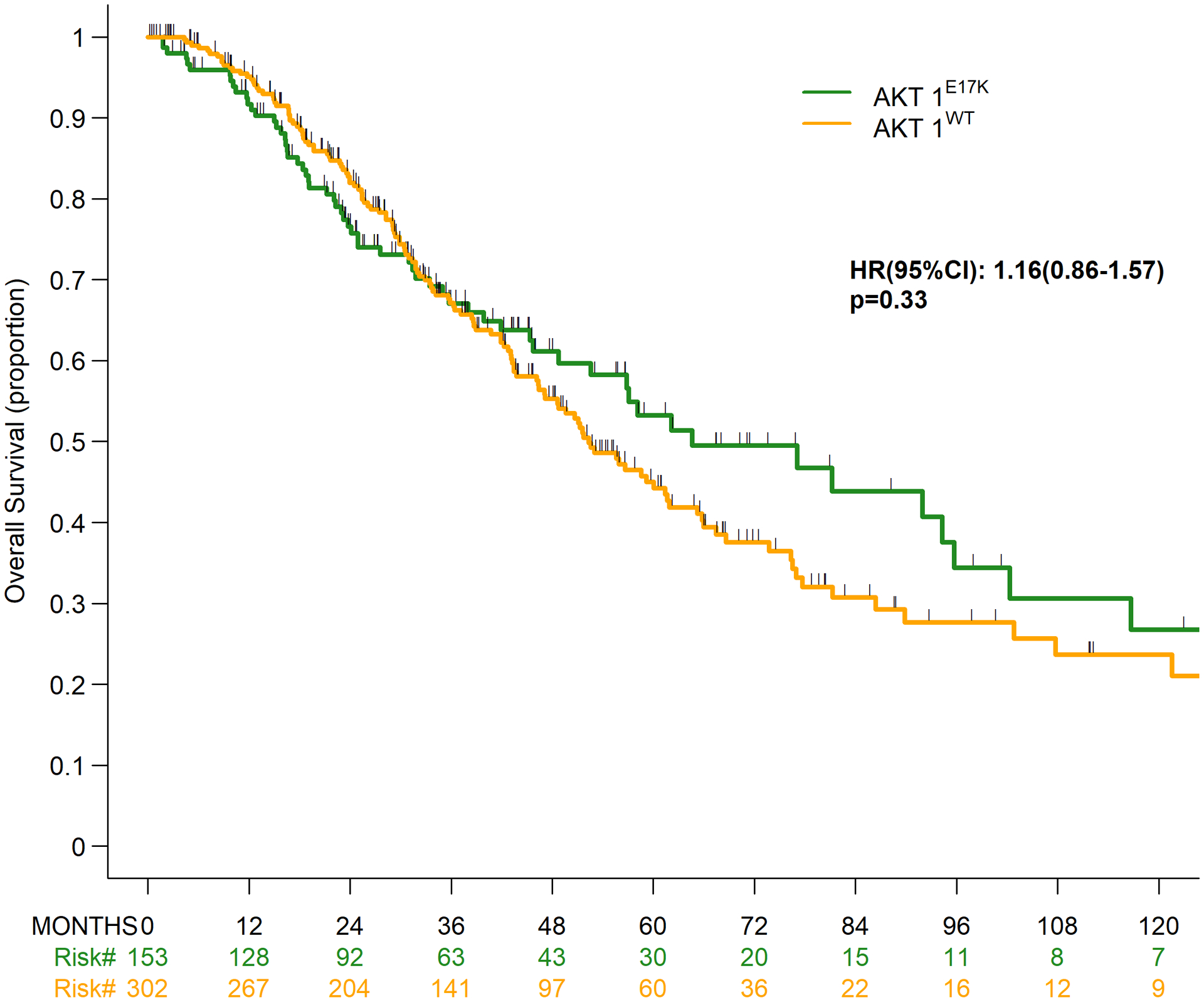
Overall survival by AKT1 mutation status. (A) Time from metastatic diagnosis to death or last follow-up, as well as tumor sequencing*; (B) Overall survival using left truncation; (C) Overall survival without left truncation.
*5 patients underwent tumor sequencing before their metastatic diagnosis
Table 3.
Overall survival and treatment outcomes
|
AKT1 mutant, n=153, N (%) |
AKT1 wildtype, n=302 N (%) |
p | |
|---|---|---|---|
| Unadjusted Overall Survival | |||
| Median, months (95% CI) | 64.6(48.8–95.7) | 52.3(46.4–61.6) | .33 |
| 5-year survival rate, % (95% CI) | 53 (43–63) | 45 (38–52) | |
| Adjusted Overall Survival*, months (95% CI) | |||
| Median, months (95% CI) | 24.1 (11.4–35) | 29.9(25.4–33.8) | .97 |
| 5-year survival rate, % (95% CI) | 20 (11–31) | 18 (12–24) | |
| 1st Line^, median DOT, months (95% CI) | |||
| Endocrine Therapy (n=289) | 13(8.2–15.3) | 9.4(7.9–11) | |
| Fulvestrant (n=65) | 13.2(3.6–16.2) | 9.3 (6–12.9) | |
| Chemotherapy (n=135) | 3.9 (3–4.6) | 4.1(3.1–4.7) | |
| 2nd Line^, median DOT, months (95% CI) | |||
| Endocrine Therapy (n=219) | 6 (5–9) | 5.3(4.1–7.3) | |
| Fulvestrant (n=84) | 4 (3–8.4) | 4.6(3.7–5.5) | |
| Chemotherapy (n=137) | 4.2(2.5–5) | 4.1(3.2–4.8) | |
| Received in any treatment line after metastatic diagnosis, median DOT, months (95% CI) | |||
| CDK4/6 inhibitor (n=131) | 5.3 (4–6.2) | 5.3(3.4–8.1) | |
| mTOR inhibitor (n=145)§ | 7.8(4.3–9) | 4.5(3.5–5.3) | |
| Fulvestrant (n=237)§ | 5.8 (4–8.4) | 4.8 (4–5.8) |
Applying left-truncation technique, see Methods for details.
Refers only to treatment given after metastatic diagnosis;
p<0.05; DOT, Duration on treatment.
Treatment outcomes
Treatment outcomes across lines and classes of therapy were also analyzed (Table 3). Given the challenges of retrospectively determining date of progression, duration of treatment (DOT) was selected as a surrogate of benefit received. The DOT for first-line endocrine-containing therapy, and fulvestrant specifically, was longer in the AKT1-mutant compared to -wildtype group, although this difference did not reach statistical significance. DOT for endocrine therapy and chemotherapy was similar for second-line treatment of metastatic disease. In total, 131 patients (44 AKT1-mutant, 87 AKT1-wildtype) received CDK4/6 inhibitor-containing therapy in any line of therapy, with a median DOT of 5.3 months in both groups. Interestingly, the median DOT for mTOR inhibitor (everolimus)-containing therapy in any line of therapy (49 AKT1-mutant, 97 AKT1-wildtype) was longer in the AKT1-mutant cohort (7.8 vs. 4.5 months, p=0.032). A similar trend was observed for fulvestrant-containing therapy when evaluated in any line of therapy, although the absolute numerical differences here was smaller (5.8 vs. 4.8 months, p=0.045).
Genomic Comparison
Broader genomic data on 428 (126 AKT1-mutant, 302 AKT1-wildtype) of the total 455 patients were available for analysis (reasons for exclusion are detailed in Supplementary Figure 1B). To evaluate whether the presence of AKT1 mutations was associated with other oncogenic alterations, the total number of variants was compared across cohorts, with no significant difference observed (Figure 2A). There were a total of 198 patients (64 AKT1-mutant, 134 AKT1-wildtype) with available copy number alteration (CNA) data. We observed that the fraction of genome altered was significantly lower in AKT1-mutant tumors compared to AKT1-wildtype tumors (median, 19.4% vs. 24.3%, respectively; p=0.01; Figure 2B). To better understand the basis of this apparent difference, we compared the genome-wide CNA frequencies of AKT1-mutant vs. AKT1-wildtype tumors. We found that chromosome 8q, containing the known MYC oncogene, was more frequently affected by copy number gain in AKT1-wildtype compared to AKT1-mutant tumors. Of interest, a locus including AKT1, at the very end of chromosome 14 (14q32.33), was more frequently gained in AKT1-mutant vs. AKT1-wildtype tumors. We found that other focal regions containing known oncogenes, including PIK3CA, FGFR1 and GATA3, were also more frequently gained in AKT1-wildtype tumors. Finally, two regions containing known tumor suppressors (ARID1B and PTEN) were more frequently lost in AKT1-wildtype compared to AKT1-mutant tumors (Figure 2C). To investigate differences at the gene level, we identified 72 recurrent oncogenic alterations, including 39 mutated genes, 23 amplified genes, and 10 genes with homozygous deletion (Supplementary Table S4). We found that PIK3CA and PTEN mutations were more frequent in AKT1-wildtype patients (39.4% vs. 5.6%, false discovery rate [FDR] = 2.5×10−12 and 6.3% vs. 0%, FDR = 0.054, respectively) (Figure 2D and Supplementary Table S4). Notably, only 7.1% of AKT1-mutant tumors had co-occurring alterations in the PI3K pathway (Supplementary Figure 2). Next, we interrogated the alteration frequency of 9 canonical oncogenic signaling pathways: cell cycle, Hippo, Myc, Notch, Nrf2, RTK-RAS, TGFβ signaling, p53, and β-catenin/Wnt. The cell cycle and RTK-RAS pathways were significantly more frequently altered in AKT1-wildtype patients compared to AKT1-mutant patient (FDR < 0.05, Figure 2E).
Figure 2.
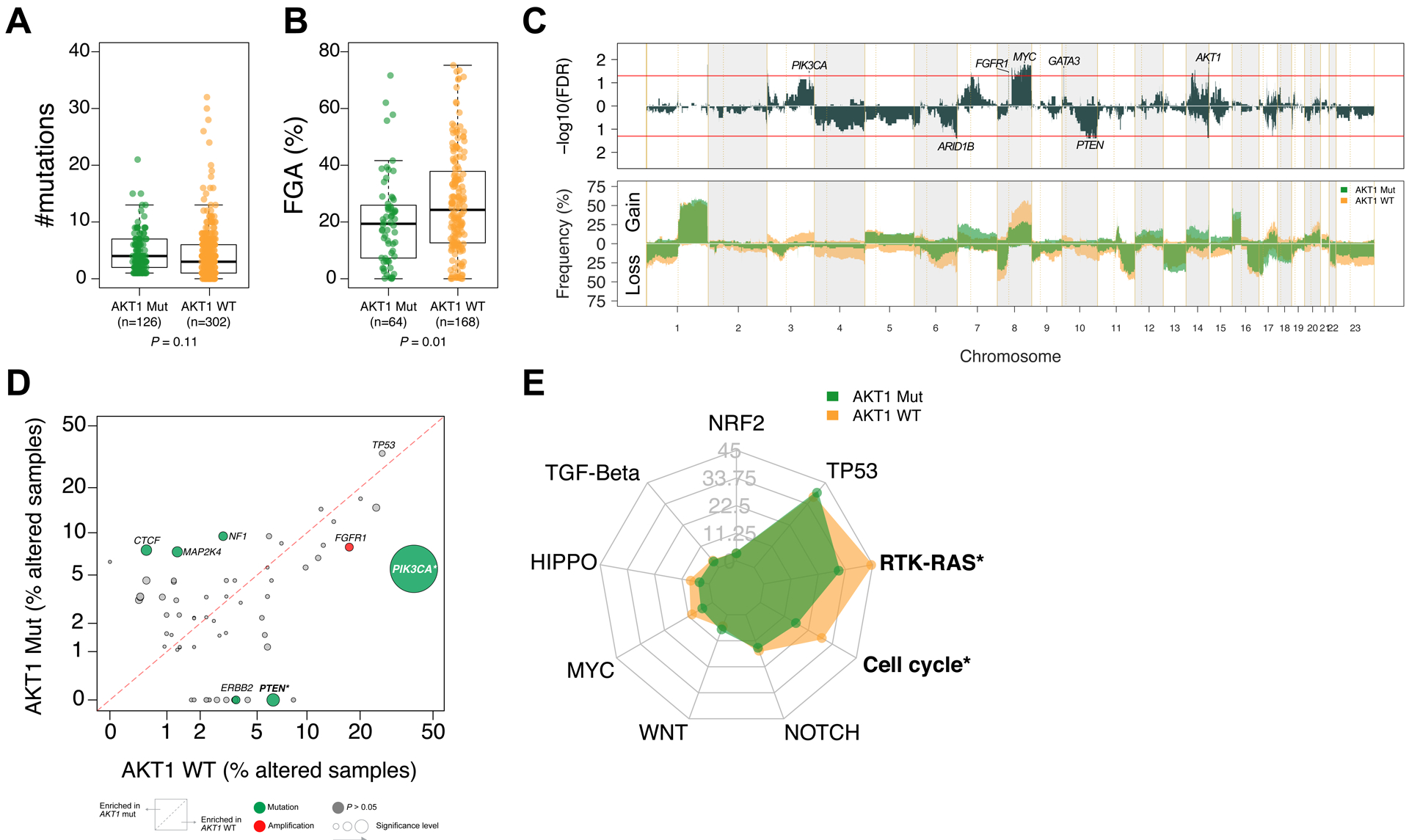
Genomic landscape of breast cancer according to AKT1 mutations. Comparison of (A) mutation number; (B) fraction of genome altered (FGA); (C) copy number alteration (CNA) frequencies in AKT1-mutant (MT, green) vs AKT1-wildtype (WT, orange). False discovery rate (FDR) derived from the Fisher exact test. The red lines indicate significance FDR < 0.05; (D) Comparison of the alteration frequency for 72 genomic alterations in AKT1-MT vs. AKT1-WT tumors. *FDR < 0.1; (E) Radar plots showing the percentage of patients with alterations in the corresponding canonical oncogenic signaling pathways according to AKT1 mutation. Statistically significant pathways associated with AKT1 mutation are indicated with an asterisk (*; FDR < .05).
Intrigued by the AKT1-mutant cohort’s comparitive benefit to mTOR inhibitor therapy, we went on to evaluate further genomic determinants of MTOR inhibitor benefit by looking at the impact of co-incident PI-3-Kinase pathway mutation/activation (PI3KPW_MT) in 137 patients (41 AKT1_MT; 96 AKT1_WT) with comprehensive genomic data available and who received an mTOR inhibitor. Interestingly, we found that in a small number of patients (7/41,17%) with a “double hit” within the pathway, namely an AKT1 mutation along with another PI3K pathway alteration, median MTOR inhibitor DOT was longest at 13 months (3.2-NA/infinity) (Supplementary Table S5 and Supplementary Figure 3). This thought-provoking observation, while only hypothesis generating at this point given the small sample size, does echo one recently observed with PI3K inhibitors, where patients with double mutations in the same allele of PIK3CA resulting in hyperactivation of PI3K signaling and increased oncogenicity had enhanced sensitivity to therapy targeting PI3K (11). In our analysis, the presence of an AKT1 mutation remained important however, as patients with PI3K pathway activation in the absence of a co-incident AKT1 mutation, seemed to fair the same on an MTOR inhibitor as patients without any hit in the pathway.
DISCUSSION
Leveraging the AACR Project GENIE framework, we assembled and deeply clinically annotated a large, multi-institutional, international cohort of AKT1-mutant ER+ metastatic breast cancer patients and matched controls. In doing so, we found that the initial presentation, outcome to standard therapy, and OS was broadly similar between the groups, with a few notable exceptions. For example, we found that AKT1-mutant patients had longer DOT when treated with mTOR inhibitors. Interestingly, limited enrollment of AKT1-mutant patients into the BOLERO-2 trial (phase 3, randomized trial that showed improved PFS with the addition of everolimus to exemestane in 724 ER+ metastatic breast cancer patients) precluded analysis of this biomarker within the context of this practice-changing study, demonstrating how even previous pivotal datasets can be underpowered to evaluate the impact of rare genomic alterations (12). Collectively, these data provide important context to the efficacy data generated with AKT inhibitors in AKT1-mutant breast cancer while also demonstrating the unique power of an academic-led data sharing consortium to address emerging needs in drug development.
This analysis has several important limitations. Although we found similar outcomes and survival regardless of AKT1 mutational status, we recognize that this analysis does not definitely exclude the possibility that such differences may exist. While the study cohorts were ultimately well balanced for baseline disease characteristics, we lacked data on ECOG performance- and co-morbid -states, variables that could potentially impact outcome, and therefore we cannot outrule imbalances in these factors among the study groups. Additionally our chosen study endpoints are limited by the nature of retrospective EHR interrogation, in that firstly, survival data can often lag in recency at institutions and secondly, while we used duration on therapy as a surrogate for PFS, therapy may have been discontinued for reasons other than cancer progression, such as toxicity. Finally, there is an inherent potential bias associated with patient selection solely from GENIE consortium institutions with access to broad tumor sequencing, testing that at the time was not yet broadly available or standard of care for breast cancer management across the world.
Our experience also demonstrates some of the potential challenges associated with the use of real-world datasets assembled on the basis of prospectively obtained clinical sequencing. Specifically, a meaningful proportion of patients were sequenced long after their metastatic diagnosis. Nonetheless, this delay did balance between cohorts, suggesting that it may not have impacted our ability to detect differences in outcome between groups. However, this source of ascertainment bias has an impact on survival estimates, as clearly demonstrated by different median OS estimates obtained by different statistical techniques. Conversely, evolution in standards of care, such as the recent approval of alpelisib for PIK3CA-mutant breast cancer (2), are likely to diminish this source of bias over time, as more patients with metastatic breast cancer undergo tumor sequencing earlier in their metastatic disease course. Appreciation of these and other related issues will be important as we continue to expand our use of real-world evidence to aid in clinical and regulatory decision making. Indeed, analyses of real-world data sets have already managed to clearly replicate therapeutic associations previously described in prospective randomized clinical trials (13).
Precision medicine in oncology is now largely focused on targeting increasingly rare genomically defined subpopulations (14). By necessity, interventions in these orphan populations are often single-arm, non-comparative studies (15). When this approach yields dramatic efficacy, the need to rigorously define the natural history of these orphan patient populations to existing standards may be viewed as less urgent. However, when the effect size is somewhat more modest but still promising, as is arguably the case with AKT inhibition in AKT1-mutant cancers, the successful advancement of novel therapies in these populations will increasingly rely on our ability to define how this population is likely to have faired with existing standards. Thus, providing a (synthetic) control group that can be of value in comparative interpretation of these non-randomized studies. In these situations, the ability to efficiently and seamlessly generate high-quality linked genomic and phenomic data will be increasingly important if opportunities to benefit patients are not to be lost. This approach has been specifically endorsed by global health authorities. One recent demonstration is the approval of palbociclib for male breast cancer using real-world data (16). Finally, like many consortium approaches, data-sharing initiatives such as these require the understanding and collaboration of industry partners and academic investigators alike, recognizing that the eligibility restrictions of traditional clinical trial platforms are unlikely to rigorously answer questions about the clinico-genomic implications of rare genomic events such as AKT1.
In conclusion, we demonstrate that AACR GENIE can be used to successfully define the natural history of rare genomic subsets such as AKT1-mutant breast cancer. Although as a genomic biomarker, AKT1-E17K did not appear to have obvious prognostic implications, it did nonetheless appear predictive of benefit to mTOR inhibitor therapy. Linkage of genomic, therapeutic, and phenomic data enables the discovery of predictive biomarkers with clinical utility in therapy selection. Harmonization of standard ontologies and use of a common curation platform enable inter-institutional collaborations, thereby accelerating the potential for discovery of actionable findings from molecular tumor profiles and more broadly permitting an understanding of the clinical phenotypes associated with alterations in the cancer genome.
METHODS
Patients
Eligible patients had radiologically confirmed distant metastatic (de novo or relapsed), hormone-positive, human epidermal growth factor receptor 2 (HER2)-negative breast cancer and had undergone tumor sequencing at one of the AACR GENIE consortium institutions. This consortium included 6 international academic medical centers with large-scale tumor sequencing initiatives.
Study Design
This was a multi-center, retrospective and observational, matched study of AKT1 E17K (AKT1)-mutant and AKT1 E17K-wildtype ER+ metastatic breast cancer patients, conducted between November 2016 and November 2017.
After Institutional Review Board approval was obtained at each member institution, genomic data linked to core patient-level and specimen-level clinical data were submitted to Sage Bionetworks on all patients undergoing tumor sequencing at these institutions, through a secure web-based platform (Synapse), as previously described (9). Core clinical data attributes submitted included sex, race, ethnicity, birth year, age at sequencing, primary cancer diagnosis (using the OncoTree cancer type ontology), and sample type (primary/metastatic).
For the purpose of this study, all AKT1-mutant cases were first identified at each center. AKT1-wildtype controls were then matched in a 2:1 ratio to AKT1-mutant cases from within each center. The control population was selected from SAGE data provided by each center, using the Tier 1A variables linked to each patient’s genomic record, specifically: year of birth, year of tumor sequencing and oncotree code (histology). The control selection process applied frequency matching of birth year (10-year range) and sequencing year (4-year range) and breast cancer oncotree code (histologic breast cancer subtype), where available. In order to be eligible for complete data curation in this study, both cases and controls first required confirmation of ER+, HER2- and metastatic state. If patients were found to be ineligible at this point, no further data was entered. Controls found to be ineligible at this step were replaced by the same selection process until a final complete eligible dataset of 2:1, cases: controls, were available for complete data curation at each site.
Detailed clinical annotation was then performed on the identified study population at each center using available electronic health records (EHRs), with study data collected and managed using REDCap electronic data capture tools (17,18). REDCap is a secure, web-based software platform designed to support data capture for research studies, providing 1) an intuitive interface for validated data capture; 2) audit trails for tracking data manipulation and export procedures; 3) automated export procedures for seamless data downloads to common statistical packages; and 4) procedures for data integration and interoperability with external sources.
In efforts to harmonize data collection across study sites, a data collection guide (Appendix file 1) with directives on EHR interpretation and data entry and data dictionary (Appendix file 2) was created, data abstractor training was provided, and a common REDCAP data curation tool (Appendix file 3) was designed and shared across each of the 6 study centers. Data fields in the REDCAP database captured information about baseline patient attributes, baseline tumor attributes including stage, histology at primary diagnosis and metastatic disease characteristics, and included all therapeutic exposures to antineoplastic therapy with start and stop dates.
Information on standard of care biomarkers (ER, PR, HER2) were collected at 3 points in the database; the initial eligibilty section and the primary- and metastatic- diagnosis sections, where “positive” for each biomarker was defined as: ER: >1% (IHC); PR: >1% (IHC); HER2: 3+ (IHC)/ >2 (FISH Ratio). For the 336 patients who had a biopsy at metastatic diagnosis, data was collected on receptor discordance compared to the primary tumor. (Supplementary Table 1).
The EHR (medications lists and provider notes) were interrogated for details on anti-cancer therapies received and were recorded in 3 different sections according to disease state (primary, loco-regionally recurrent and distant metastatic diagnoses) with each single drug entered separately, even if it was administered as part of a regimen. When drugs formed part of a combination therapy regimen, this was specifically captured/tagged as such within the database and considered, at the time of analysis, as one line of therapy. If the exact/complete start/stop date of a therapy was not known, guidance was provided to estimate the date, using known data components.
Survival data were derived from institutional EHR data, which in some centers was linked to the National Death Index (NDI). Patient records were curated from date of diagnosis to death and were censored at their last known follow-up at the study site, if alive.
An independent data quality review/monitoring plan (Appendix file 4) was employed at each site, whereby source data verification (SDV) was required on all study patients for core eligibility data (ER, HER2, AKT1, and metastatic status and tumor histology). Complete SDV was then additionally required on all first patients entered per data abstractor at each site and on a minimum of 10% of patients at each site chosen at random. All data queries were resolved internally with local PI’s and where required, escalated to the lead PI for review. Documentation relating to data quality reviews were tracked and kept on file at the local site.
Statistical Considerations
The data cutoff for this report was August 2018, when the database was locked to further data updates. Patients were excluded from the final analyses dataset if insufficient core clinical data were available. Analyses were specified in a statistical analysis plan. Number of lines of endocrine-therapy/ chemo-therapy was determined by only considering each specific class of therapy and counting each new agent received by the patient as a line of therapy in that class. Endocrine therapy received as part of a combination regimen with for e.g a CDK4/6- or mTOR- inhibitor or ovarian function suppression was considered as 1 line of therapy. Descriptive statistics, including medians and ranges, were calculated. All statistical analyses and all plots were performed and generated using R 3.5.1. Associations between variables and AKT1 status were analyzed using Wilcoxon-rank sum test for continuous variables and chi-square test/Fisher exact test (for count <5) for categorical variables. The primary endpoint, OS, was calculated from the date of (radiologically determined) distant metastatic diagnosis to the date of death. Patients still alive were censored at last follow-up date. To account for the method of sample selection (selection bias), i.e., the fact that patients must have survived long enough to undergo tumor sequencing, Kaplan-Meier method with left truncation was used to estimate the median survival and 5-year survival rate. Cox Proportional Hazards model with left truncation at the date of the sequencing report, was applied to obtain the p value (19). Additionally, all AKT1- mutant patients enrolled on the international clinical trial (NCT01226316) with the AKT inhibitor AZD5363 were censored at the time of starting this therapy. The secondary endpoint, DOT, was analyzed as a time-to-event variable and was calculated from the start to stop date of therapy or death, with patients remaining on therapy censored at the last follow-up date. In the DOT analyses, we did not censor patients at AZD5363 start date (i.e., they were considered events). DOT was analyzed using Kaplan-Meier method, and comparisons between the various groups were obtained using the log-rank test. For the DOT analyses, the definition of an event was met by the therapy stop date or if this date was missing in a patient who died within 3 months of starting this therapy. Patients with a missing therapy stop date who died beyond 3 months of starting therapy were censored at 3 months from therapy start. Similarly, patients with a missing therapy stop date who were lost to follow-up, were censored at 3 months from therapy start or the patient’s last known follow-up date, whichever came first. All statistical endpoints were compared across the AKT1-mutant and AKT1-wildtype patient cohorts. Reported p values were two-tailed, and differences were considered significant when the p was less than 0.05.
Notably the power in this study is limited by the rarity of this alteration and the small event rate (deaths= 205) occurring in the study population during the observed study period. Nonetheless, all AKT1-mutant breast cancers identified at each consortium institution between 1990–2017 were included and the AKT1- wildtype control group was then doubled to decrease the variance in that group.
Genomic analyses
OncoKB was used to define oncogenic driver variants (20). For the alteration level analysis, recurrent oncogenic alterations were defined as oncogenic as per OncoKB annotation and present in at least 1% in the whole cohort. Segmented copy number data were processed using CNtools package v1.4. The canonical oncogenic pathway level alterations were computed using the curated pathway templates described by Sanchez-Vega et al. (21). All genomic analyses were performed using Rv3.5.2 (https://www.R-project.org) and Bioconductor version 3.4.
Data Availability
All patient-level clinical outcome and genomic data are available on the cBioPortal.org (http://genie.cbioportal.org/study?id=brca_akt1_genie_2019)
Data from this manuscript were partially reported at the American Association for Cancer Research (AACR) Annual Meeting, April 2019, Atlanta, Georgia, USA.
Supplementary Material
SIGNIFICANCE.
We delineate the natural history of a rare genomically distinct cancer, AKT1 E17K-mutant estrogen receptor-positive breast cancer, using a publicly accessible registry of real world patient data, thereby illustrating the potential to inform drug registration through synthetic control data.
Funding:
This research was supported by the American Association for Cancer Research (AACR) Project GENIE Consortium and by AstraZeneca. MSK authors are supported in part by the National Institutes of Health/National Cancer Institute Memorial Sloan Kettering Cancer Center Support Grant P30 CA008748. This work was supported in part by The Sheikh Khalifa Bin Zayed Al Nahyan Institute for Personalized Cancer Therapy (F. Meric-Bernstam) and the Cancer Prevention and Research Institute of Texas (CPRIT) Precision Oncology Decision Support Core RP150535 (F. Meric-Bernstam).
Footnotes
Conflict of Interest Statement: Dr. Hyman reports research funding and consulting/advisory fees from AstraZeneca. Outside the submitted work, Dr. Hyman reports stock/other ownership interest in Fount; consulting/advisory role for Chugai Pharma, Boehringer Ingelheim, Pfizer, Bayer, Genentech, and Fount; travel/accommodations from Genentech and Chugai Pharma; and research funding from Puma Biotechnology, Loxo, and Bayer. Dr. F. Meric-Bernstam reports receiving commercial research grants from Novartis, AstraZeneca, Calithera, Aileron, Bayer, Jounce, CytoMx, eFFECTOR, Zymeworks, PUMA Biotechnology, Curis, Millennium, Daiichi Sankyo, Abbvie, Guardant Health, Takeda, and GlaxoSmithKline, as well as grants and travel-related fees from Taiho and Seattle Genetics. She also served as a consultant to Pieris, Dialectica, Sumitomo Dainippon, Samsung Bioepis, Aduro, OrigiMed, Xencor, Jackson Laboratory, Zymeworks, Kolon Life Science, and Parexel International, and advisor to Inflection Biosciences, GRAIL, Darwin Health, Spectrum, Mersana, Seattle Genetics, and Immunomedics. L.M.S. has received research grants/funding (to her institution) from AstraZeneca, Puma Biotechnology Inc., and Roche Genentech. L.M.S has received payment for consultancy or advisory roles from AstraZeneca, Roche Genentech, Pfizer, and Novartis, honoraria from AstraZeneca, Roche Genentech and Pfizer, and support covering travel, accommodations, and expenses from Pfizer, Puma Biotechnology Inc., and Roche Genentech.
The other authors have no relevant conflict of interests to disclose.
REFERENCES
- 1.Razavi P, Chang MT, Xu G, Bandlamudi C, Ross DS, Vasan N, et al. The Genomic Landscape of Endocrine-Resistant Advanced Breast Cancers. Cancer Cell 2018;34:427–38.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.André F, Ciruelos E, Rubovszky G, Campone M, Loibl S, Rugo HS, et al. Alpelisib for PIK3CA-Mutated, Hormone Receptor-Positive Advanced Breast Cancer. N Engl J Med 2019;380:1929–40. [DOI] [PubMed] [Google Scholar]
- 3.Carpten JD, Faber AL, Horn C, Donoho GP, Briggs SL, Robbins CM, et al. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature 2007;448:439–44. [DOI] [PubMed] [Google Scholar]
- 4.Kim MS, Jeong EG, Yoo NJ, Lee SH. Mutational analysis of oncogenic AKT E17K mutation in common solid cancers and acute leukaemias. Br J Cancer 2008;98:1533–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012;490:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hyman DM, Smyth LM, Donoghue MTA, Westin SN, Bedard PL, Dean EJ, et al. AKT inhibition in solid tumors with AKT1 mutations. J Clin Oncol 2017;35:2251–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smyth LM, Oliveira M, Ciruelos E, Tamura K, El-Khoueiry A, Mita A, et al. AZD5363 in combination with fulvestrant in AKT1-mutant ER-positive metastatic breast cancer. American Association for Cancer Research Annual Meeting; April 14–18, 2018; Chicago, IL [abstract P5-21-32]. [Google Scholar]
- 8.Capivasertib Active against AKT1-Mutated Cancers. Cancer Discov 2019;9:OF7. [DOI] [PubMed] [Google Scholar]
- 9.AACR Project GENIE Consortium. AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov 2017;7:818–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desmedt C, Zoppoli G, Gundem G, Pruneri G, Larsimont D, Fornili M, et al. Genomic characterization of primary invasive lobular breast cancer. J Clin Oncol 2016;34:1872–81. [DOI] [PubMed] [Google Scholar]
- 11.Vasan N, Razavi P, Johnson JL, Shao H et al. Double PIK3CA mutations in cis increase oncogenicity and sensitivity to PI3Kα inhibitors. Science 2019. November 8;366(6466):714–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hortobagyi GN, Chen D, Piccart M, Rugo HS, Burris HA, Pritchard KI, et al. Correlative Analysis of Genetic Alterations and Everolimus Benefit in Hormone Receptor-Positive, Human Epidermal Growth Factor Receptor 2-Negative Advanced Breast Cancer: Results From BOLERO-2. J Clin Oncol 2016;34:419–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singal G, Miller PG, Agarwala V et al. Association of Patient Characteristics and Tumor Genomics With Clinical Outcomes Among Patients With Non-Small Cell Lung Cancer Using a Clinicogenomic Database. JAMA. 2019. April 9;321(14):1391–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hyman DM, Taylor BS, Baselga J. Implementing Genome-Driven Oncology. Cell 2017;168:584–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tao JJ, Schram AM, Hyman DM. Basket Studies: Redefining Clinical Trials in the Era of Genome-Driven Oncology. Annu Rev Med 2018;69:319–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartlett CH, Mardekian J, Yu-Kite M, Cotter MJ, Kim S, Decembrino J, et al. Real-world evidence of male breast cancer (BC) patients treated with palbociclib (PAL) in combination with endocrine therapy (ET). J Clin Oncol 2019;37:1055–1055. [Google Scholar]
- 17.Harris PA, Taylor R, Minor BL, Elliott V, Fernandez M, O’Neal L, et al. The REDCap consortium: Building an international community of software platform partners. J Biomed Inform 2019;95:103208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 2009;42:377–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klein JP, & Moeschberger ML Survival analysis: Techniques for censored and truncated data. Dietz K, Gail M, Krickeberg K, Samet J, Tsiatis A, editors. New York: Springer; 2003. [Google Scholar]
- 20.Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, et al. Oncokb: A precision oncology knowledge base. JCO Precis Oncol 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanchez-Vega F, Mina M, Armenia J, Chatila WK, Luna A, La KC, et al. Oncogenic signaling pathways in the cancer genome atlas. Cell 2018;173:321–37.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All patient-level clinical outcome and genomic data are available on the cBioPortal.org (http://genie.cbioportal.org/study?id=brca_akt1_genie_2019)
Data from this manuscript were partially reported at the American Association for Cancer Research (AACR) Annual Meeting, April 2019, Atlanta, Georgia, USA.


