Abstract
The insect gustatory system senses taste information from environmental food substrates and processes it to control feeding behaviors. Drosophila melanogaster has been a powerful genetic model for investigating how various chemical cues are detected at the molecular and cellular levels. In addition to an understanding of how tastants belonging to five historically described taste modalities (sweet, bitter, acid, salt, and amino acid) are sensed, recent findings have identified taste neurons and receptors that recognize tastants of non-canonical modalities, including fatty acids, carbonated water, polyamines, H2O2, bacterial lipopolysaccharide (LPS), ammonia, and calcium. Analyses of response profiles of taste neurons expressing different suites of chemosensory receptors have allowed exploration of taste coding mechanisms in primary sensory neurons. In this review, we present the current knowledge of the molecular and cellular basis of taste detection of various categories of tastants. We also summarize evidence for organotopic and multimodal functions of the taste system. Functional characterization of peripheral taste neurons in different organs has greatly increased our understanding of how insect behavior is regulated by the gustatory system, which may inform development of novel insect pest control strategies.
Keywords: Drosophila gustation, Feeding behavior, Taste, Chemosensory receptors
Introduction
Animals continuously receive and process massive amounts of sensory information from the surrounding environment via different sensory systems, which direct appropriate behavioral responses. Specialized sensory organs in the body are specifically tuned to various types of sensory stimuli. Sensory information is then decoded in the central nervous system, mainly in the brain. In insects, contact chemosensory cues are sensed by the gustatory system, which is critical for mating, feeding, and oviposition behaviors. Drosophila melanogaster has been an excellent model organism for dissecting the genetic underpinnings of behaviors driven by gustatory systems in insects, including agricultural pests and disease vectors. A wealth of behavioral and functional assays, combined with the availability of genetic tools and reagents, offer the means to probe how chemical information is encoded at different levels of the gustatory pathway in Drosophila. Recent years have seen significant progress in understanding sensory coding in the periphery as well as in mapping of higher-order taste circuits in the fly brain. In this review, we focus on the adult Drosophila gustatory system and its role in detecting food-related cues that control feeding, oviposition, and hygiene behaviors. We provide a general overview of the adult Drosophila gustatory system and then present recent advances in our knowledge of chemosensory receptors and neurons underlying peripheral responses to various tastants. We also discuss evidence for multimodal taste sensing properties of Drosophila neurons, and for functional differences between neurons across taste organs towards operating different aspects of feeding behaviors.
Anatomical organization of the gustatory system in adult Drosophila
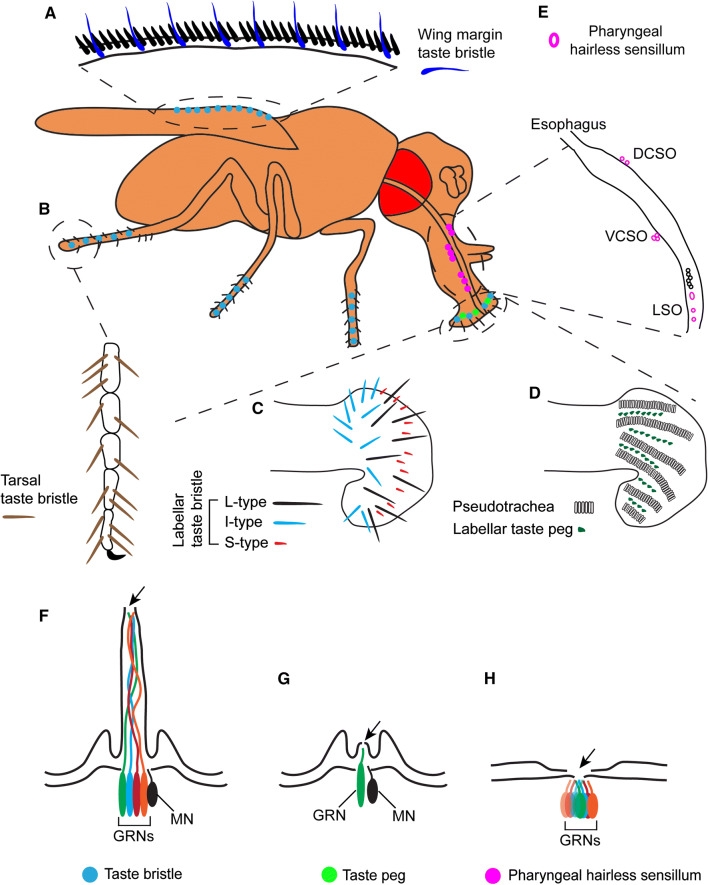
In adult Drosophila, taste organs are distributed in different parts of the body (Fig. 1). External taste organs include the anterior wing margins (Fig. 1a), distal segments of the legs (Fig. 1b), and the labellum (Fig. 1c, d). Internal taste organs include three pharyngeal taste organs located internally in the proboscis: labral sense organ (LSO), ventral cibarial sense organ (VCSO), and dorsal cibarial sense organ (DCSO) (Fig. 1e). Taste organs are covered by morphologically distinct taste sensilla, the basic functional units of taste detection (Fig. 1f–h). The Drosophila labellum, the most extensively characterized taste organ, consists of two types of taste sensilla: taste hairs (Fig. 1c) and taste pegs (Fig. 1d). Labellar taste hairs are located on the distal tip of the labellum. There are ~ 30 hairs on each half of the labellum that can be further divided into morphological subtypes based on the length of the hairs: L (long), I (intermediate), and S (short). Each taste hair has a single pore at the tip of the sensillum, which allows tastants to enter and make contact with the chemosensory neurons present within. All labellar taste hairs house a single mechanosensory neuron, but the number of chemosensory neurons that reside in them varies from two to four, depending on the subtype (i.e. four neurons in L- and S-hairs, two neurons in I-hairs) (Fig. 1f). Labellar taste pegs are hairless sensilla located between rows of pseudotrachea. The number of labellar taste pegs is sexually dimorphic, with females having more than males [1]. Each labellar taste peg is innervated by one mechanosensory neuron and one chemosensory neuron (Fig. 1g). During feeding, these taste pegs are thought to access food only when the flies open their labial palps. Besides the labellum, taste hairs are distributed on the five tarsal segments of all six legs as well as the anterior wing margins, all of which are innervated by one mechanosensory neuron and four chemosensory neurons [1] (Fig. 1f). Interestingly, the tarsal taste hairs on the forelegs are sexually dimorphic, with more hairs in males than in females. Perhaps not surprisingly, male-specific taste hairs on the forelegs are involved in pheromone detection during courtship behavior [2, 3]. Unlike external taste hairs, internal taste sensilla in the pharyngeal organs are hairless. They are innervated by one to eight chemosensory neurons, and may or may not be associated with mechanosensory neurons [4, 5] (Fig. 1h). In Drosophila, most sensory neurons are cholinergic [6], but recent studies showed that a small fraction of labellar and tarsal chemosensory neurons are glutamatergic [7, 8], suggesting neurochemical and functional heterogeneity within chemosensory neurons. However, further studies are required to systemically characterize neurotransmitters that are used in chemosensory neurons of all taste organs.
Fig. 1.
Organization of the adult Drosophila gustatory system. There are three types of taste sensilla in the different taste organs: taste bristles (blue dots), taste pegs (green dots), and pharyngeal hairless sensilla (magenta dots). The taste bristles are distributed in the anterior wing margins (blue in a), the distal segment of the legs (brown in b), and the labellum (black, blue and red in c). The taste pegs are located between pseudotrachea in the labellum (green in d). The hairless sensilla are located in the three internal pharyngeal taste organs: labral sense organs (LSO), ventral and dorsal cibarial sense organs (VCSO and DCSO) (magenta in e). f–h Schematic diagrams showing the structures of three types of taste sensilla. All of them have a terminal pore (arrows) that allows tastants to make contact with the taste neurons in each sensillum. The taste bristle has 2–4 gustatory receptor neurons (GRNs) (4 GRNs in this schematic example) whose dendrites extend up to the tip of the taste sensillum (f). The taste peg has one GRN (g). Both taste bristles and taste pegs have one mechanosensory neuron (MN) at the base of each sensillum (black in f, g). The pharyngeal hairless sensilla usually do not have mechanosensory neurons, except for the #8 and #9 LSO sensilla. The number of GRNs in the pharyngeal hairless sensilla can vary from 1 to 8 (8 GRNs in this schematic example) (h)
Physiological response profiles of chemosensory neurons
Single-sensillum extracellular tip recordings allow measurement of physiological responses of all chemosensory neurons in a single taste sensillum [9]. Recordings are obtained with tastant solutions in glass micropipette electrodes that are used to contact the tips of taste hairs. The stereotypical arrangement and accessibility of taste hairs in the labellum, tarsi, and wings have lent themselves to systematic surveys of tastant-evoked responses. In general, distinct responses have been recorded with stimuli representing distinct taste modalities, which include water, sugar, salt (high and low), acid, and bitter compounds [9–13]. Based on characteristic spike amplitudes and responses to tastants, neurons have been classified into water-, sweet-, salt-, and bitter-sensing populations. However, the extent to which each population is selectively tuned to tastants remains to be determined, and recent studies suggest that at least some taste neurons can respond to compounds of different taste categories (see below for details), hinting at multimodal taste detection properties in insect taste neurons. Moreover, gustatory coding information is incomplete because the same type of analysis has not been achieved for internal pharyngeal taste sensilla and hairless taste pegs of the oral surface, which are difficult to access as compared to external taste hairs.
Chemosensory receptor gene expression in adult Drosophila taste neurons
Almost two decades ago, the Gustatory receptor (Gr) gene family was identified as a new family encoding transmembrane proteins as candidate taste receptors expressed in taste organs [14–16]. In D. melanogaster, there are 60 Gr genes encoding 68 proteins. Although Gr transcript expression was typically too low to be reliably detected by in situ hybridization, a series of transgenic reporter lines using the GAL4/UAS binary expression system were soon developed to analyze Gr expression [15, 16]. Receptor-to-neuron maps based on reporter analysis were constructed for the labellum [7, 12, 17], tarsi [10], and pharynx [18, 19]. Patterns of GAL4 reporter expression have been confirmed by independent means only in a few instances [13, 20]. Nevertheless, these reporter lines serve as excellent tools for functional analysis of molecularly defined taste neurons. In addition to members of the Gr gene family, recent studies have found that other chemosensory receptors, including those encoded by Ionotropic receptor (Ir), pickpocket (ppk), and Transient receptor potential (Trp) gene families, are involved in tastant detection [7, 21–47]. Transgenic reporter lines for many of these chemosensory genes, in particular the Ir genes, have also been constructed, and a significant fraction of them was found to be expressed in taste organs [21, 27]. In general, the expression of different chemosensory receptors showed some degree of overlap, especially in the pharynx where most pharyngeal taste neurons express more than one type of chemosensory receptor gene family [18]. In the following sections, we will discuss recent findings of chemosensory receptors involved in detecting tastants representing canonical taste categories as well as non-canonical taste modalities (Table 1). While we have attempted to provide information that is fairly extensive, readers are also encouraged to consult other recent reviews on the general function of these chemosensory receptors [48–50].
Table 1.
Receptors, neurons and taste responses in adult Drosophila
| Tastant | Receptor | Transgenic reporter | Example ligands | Taste organs | Physiological measurement | Behavioral measurement | References |
|---|---|---|---|---|---|---|---|
| Sweet | Gr5a, Gr43a, Gr61a, Gr64a-f | Trehalose, sucrose, glucose, fructose, glycerol and other sugars | Labellum | Tip recording | PER, food choice | [9, 13, 51, 52, 105–107] | |
| Gr5a, Gr61a, Gr64a-f | Labellum | None | PER | [17] | |||
| Gr5a-GAL4 | Labellum | Ca2+ imaging | Food choice | [108] | |||
| Gr5a-GAL4 | None | PER | [58] | ||||
| Gr43a | Brain | Ca2+ imaging | PER, CAFE | [17, 55] | |||
| Gr61a-GAL4 | Tarsi | Ca2+ imaging | PER | [57, 109] | |||
| Gr43a-GAL4 | Pharynx | Ca2+ imaging | Food choice, food consumption | [19] | |||
| Tub-GAL4 | Wing | Ca2+ imaging | Aggregation | [110] | |||
| Gr21a-GAL4, Gr63a-GAL4 | Ectopic expression system in olfactory sensilla | Tip recording | None | [111] | |||
| Tarsi | Tip recording | None | [10] | ||||
| Ir60b | Sucrose, glucose | Pharynx | Ca2+ imaging | Food consumption, FLIC | [63] | ||
| Bitter | Caffeine, quinine, denatonium, DEET, 7-tricosene, and other bitter compounds | Labellum, tarsi | Tip recording | Food choice | [9, 12, 112, 113] | ||
| Gr32a, Gr33a, Gr66a, Gr93a | Labellum | Tip recording | Food choice, courtship | [20, 64–66, 74] | |||
| Gr2a, Gr10a, Gr22b, Gr28a, Gr28b.a, Gr36a, Gr58c, Gr59c | Gr89a-GAL4 | Ectopic expression system in labellum | Tip recording | None | [75] | ||
| Gr66a-GAL4 | Labellum | Ca2+ imaging | Food choice | [108] | |||
| Gr66a-GAL4 | Strychnine, L-canavanine | Labellum, tarsi | Tip recording | Food choice, CAFE, PER | [114] | ||
| Gr66a-GAL4 | Caffeine, quinine, denatonium, berberine | None | PER | [58] | |||
| Gr8a, Gr66a, Gr98b | L-canavanine | Labellum | Tip recording | Food choice | [67] | ||
| Gr9a-GAL4 | L-canavanine | Pharynx | None | Food choice | [18] | ||
| Gr47a | Strychnine | Labellum | Tip recording | Food choice, PER | [71] | ||
| Gr22e | Strychnine, chloroquine | Labellum | Tip recording | Food choice, PER | [68] | ||
| Gr33a, Gr66a, Gr93a | Umbelliferone, coumarin | Labellum | Tip recording | Food choice, oviposition | [69, 70] | ||
| Gr28b | Saponin | Labellum | Tip recording | Food choice, PER | [72] | ||
| Gr10a | Nicotine | Labellum | Tip recording | PER | [73] | ||
| TrpA1 | N-methylmaleimide | Labellum | Tip recording | CAFE | [115] | ||
| TrpA1 | Aristolochic acid | Labellum | Tip recording | Food choice | [37] | ||
| TrpL | Camphor | Labellum | Tip recording | Food choice | [38] | ||
| Painless | Isothiocyanate | Labellum, tarsi, pharynx, wing | None | Food choice, PER | [116] | ||
| Salt | Ir76b | Sodium chloride | Labellum | Tip recording | Food choice | [23, 39] | |
| Ir76b-GAL4 | Tarsi | None | PER | [76] | |||
| Gr2a | Pharynx | None | Food choice | [77] | |||
| Labellum | Tip recording | Food choice | [9] | ||||
| Gr64f-GAL4, Gr66a-GAL4, ppk23-GAL4, Ir94e-GAL4 | Labellum | Ca2+ imaging | Food choice | [7] | |||
| Acid | Ir7a | Acetic acid | Labellum | Tip recording | Food choice, PER | [46] | |
| Gr64f-GAL4, Gr66a-GAL4 | Acetic acid | Labellum | Ca2+ imaging | PER | [78] | ||
| Ir25a, Ir76b | Carboxylic acids, HCl | Tarsi | Ca2+ imaging | Oviposition | [42] | ||
| Gr89a-GAL4 | Carboxylic acids, HCl | Labellum | Tip recording | Food choice, PER | [11] | ||
| Amino acids/Yeast | Ir76b | Serine, threonine, phenylalanine, alanine, glycine, yeast extract | Tarsi | Ca2+ imaging | Food choice | [25] | |
| Ir76b-GAL4 | Yeast | Labellum, taste pegs | Ca2+ imaging | FlyPAD | [24] | ||
| Gr66a-GAL4 | Tryptophan, phenylalanine | Labellum | Tip recording | Food choice | [86] | ||
| AstC-, Npf-, and Dh31-GAL4 (EE-GAL4) | Amino acids/yeast | Gut | CaLexA | None | [117] | ||
| Amino acids | Labellum, Tarsi | None | Food choice, PER, CAFE | [118] | |||
| Carbonated water | E409-GAL4 | Sodium bicarbonate, cesium bicarbonate (pH 5–6.5) | Taste pegs | Ca2+ imaging | Food choice | [91] | |
| Ir56d | Labellum, taste pegs | Ca2+ imaging | Food choice, Positional preference, PER, Expresso, FlyPAD | [21] | |||
| Fatty acids | Ir56d | Hexanoic acid, octanoic acid, and other fatty acids | Taste pegs, tarsi | Ca2+ imaging | PER | [43, 44] | |
| Gr64e | Labellum | Tip recording | PER | [90] | |||
| Gr33a[GAL4] | Hexanoic acid | Tarsi | Ca2+ imaging | PER | [43] | ||
| Polyamines | Ir76b | Gr66a-GAL4 | Putrescine, cadaverine | Labellum | Ca2+ imaging, tip recording | Oviposition | [45] |
| UV/H2O2 | TrpA1 | Gr66a-GAL4 | UV/H2O2 | Labellum | Ca2+ imaging | Oviposition | [34] |
| TrpA1 | Gr66a-GAL4 | UV/H2O2 | Labellum | Tip recording | Food choice | [36] | |
| LPS | TrpA1 | Gr66a-GAL4 | LPS | Pharynx | Ca2+ imaging | Food choice, PER, oviposition | [35] |
| Gr64f-GAL4, Gr5a-GAL4, Gr33a-GAL4, Gr66a-GAL4, Ir76b-GAL4 | LPS | Wing | None | Grooming | [95] | ||
| Gr33a-GAL4 | LPS | Wing, tarsi | Tip recording | Grooming | [96] | ||
| Ammonia | Gr66a-GAL4 | Ammonium chloride | Labellum | Tip recording | Food choice, food consumption | [98] | |
| Calcium | Ir62a | ppk23-GAL4 | Calcium chloride | Labellum | Tip recording | Food choice | [99] |
CAFE capillary feeder assay, CaLexA calcium-dependent nuclear import of LexA, DEET N,N-Diethyl-meta-toluamide, EE-GAL4 enteroendocrine-GAL4, Expresso an automated feeding assay for quantification of real time food ingestion, FLIC fly liquid food interaction counter, flyPAD fly proboscis and activity detector, LPS lipopolysaccharide, PER proboscis extension response
Recent research updates on taste detection by chemosensory neurons
Sweet
In Drosophila, eight Grs belong to a clade of conserved sweet taste receptors that include Gr5a, Gr61a, and Gr64a-f. Based on transgenic reporter techniques, subsets of sweet taste neurons were found to express distinct combinations of sweet Grs [13, 17, 19]. Mutant analyses showed that individual sweet Grs are required for sensing multiple sugars, and each sugar response appears to be dependent on multiple sweet Grs [13, 17, 51–53]. In addition, some sweet gustatory receptor neurons (GRNs) also express Gr43a, a highly conserved Gr that is outside of the sweet clade [17, 19, 54, 55]. Gr43a is also expressed in nutrient-sensing neurons in the brain, which monitor fructose levels in the hemolymph [55].
Some differences in neuronal activation profiles of sweet GRNs in different taste organs have been reported. D- and L-arabinose, for example, have been found to activate tarsal and pharyngeal Gr43a GRNs differentially, but not Gr43a-expressing neurons in the brain in which both D- and L-arabinose evoke similar responses in terms of both magnitude and kinetics [56]. Instances of variation in physiological responses observed between different sweet GRNs have been attributed to distinct chemosensory receptor repertoires [13, 51, 53, 57].
Sweet GRNs originating from different organs exhibit distinct axonal projection patterns in the subesophageal zone (SEZ), the primary taste center in the central nervous system [18, 58, 59]. The organotopic map has been the basis for a model in which input from each taste organ is relayed to distinct higher-order neuronal circuits, which in turn regulate different aspects of feeding behavior. Notably, recent studies have found evidence for such differences in sweet GRN-controlled feeding behaviors. For example, two anatomically distinct classes of tarsal sweet GRNs, one that terminates in the ventral nerve cord (VNC) and a second that passes through the VNC and terminates in the SEZ, have been reported to regulate different behavioral responses to sugars. Those ending in the VNC are responsible for stopping the fly’s movements upon encountering sugar, while the ones that project to the SEZ are responsible for initiating feeding [60]. In addition, pharyngeal sweet GRNs, which project to a discrete region of the SEZ, are distinct from external sweet GRNs in terms of the behaviors they regulate [18, 19]. Another study reported that sugar detection can elicit local search behavior, and this appears to be mediated primarily by pharyngeal Gr43a GRNs and not external GRNs [61]. A finding that confirms the presence of discrete circuit elements for internal and external taste is the identification of IN1 interneurons that are connected with pharyngeal Gr43a GRNs but not with external sweet GRNs. [62]. IN1 neurons integrate information about pharyngeal sweet taste and hunger to control meal dynamics. Altogether, these findings suggest that sweet GRNs in different locations can sense ligands in different ways, convey input to different regions in the CNS, and thereby control different aspects of feeding behaviors in response to carbohydrate cues in food substrates.
Given the extended focus on studies of Gr involvement in sweet taste, it was a surprise when sugar-sensitivity was found in a pair of Ir60b-expressing neurons in the pharynx [63]. Ir60b GRNs are unique in that (1) they do not express sweet Grs, but rather a few Irs, including Ir60b, Ir94f, Ir94h, and Ir25a; (2) their activation restricts sugar consumption rather than promotes it; and (3) they appear to be selectively involved in cellular and behavioral responses to sucrose and glucose but not to other sugars such as trehalose and fructose. These results evoke several interesting questions for follow up studies. Are there other non-Gr expressing neurons that detect sugars, possibly those other than sucrose and glucose? How does Ir60b confer sugar responsiveness? Is it directly involved in detecting sucrose, either alone or in combination with other Irs? How does the activation of pharyngeal Ir60b GRNs limit sugar consumption—by directly inhibiting Gr-expressing sweet taste circuits or by conveying information for integration in higher-order circuits? Finally, the ethological relevance of such narrow tuning of sugar sensitivity in Ir60b pharyngeal GRNs also awaits future research.
Bitter
Bitter taste is mediated by members of the Gr family. Initial analyses of Gr mutants as well as Gr-GAL4 reporters revealed that bitter GRNs expressing several bitter Grs, including Gr32a, Gr33a, Gr66a, Gr89a, and Gr93a, are required for physiological and behavioral responses to bitter compounds [20, 64–66]. A number of observations also suggested that multiple Grs are likely to come together in heteromeric complexes to detect various bitter substances, however, a minimum Gr subunit composition remained unclear until 2015, when a combination of Gr8a, Gr66a, and Gr98b was reported as a full receptor repertoire for detection of L-canavanine [67]. All three receptors are required for L-canavanine response in bitter GRNs and co-expression of the three receptors is sufficient to confer L-canavanine response in sweet GRNs as well as in Drosophila S2 cells. Subsequently, several other members of Grs have been reported to be involved in detection of specific bitter compounds, such as strychnine, coumarin, umbelliferone, chloroquine, saponin, and nicotine [68–73]. Two recent studies have further elucidated the molecular basis of bitter detection by characterizing differences in responses of bitter GRNs that have distinct molecular profiles of bitter Gr expression [74, 75]. One study found that Gr32a, Gr59c, and Gr66a together are sufficient for sensing lobeline, berberine, and denatonium, whereas Gr22e, Gr32a, and Gr66a are sufficient for sensing the same three bitter compounds as well as strychnine. Given that the two combinations differ only in one Gr and show overlapping but distinct bitter response profiles, it was suggested that a selected bitter compound could activate molecularly distinct receptor complexes, and a selected heteromeric receptor complex could detect multiple bitter compounds. Thus, the observed heterogeneity of Gr expression in bitter GRNs would contribute to an even greater diversity in cellular responses to bitter tastants [74]. Consistent with these observations, the presence or absence of a single bitter Gr can alter endogenous responses of bitter GRNs by increasing or decreasing responses to selected bitter tastants or by conferring novel responses to bitter tastants [75]. These findings complicate evaluation of the functional roles of single Grs using mutant or ectopic expression analyses. Extensive studies have been focused on labellar bitter GRNs while leaving other taste organs unexplored, except one pharyngeal GRN labeled by Gr9a-GAL4 shown to be responsible for behavioral avoidance of L-canavanine [18]. An understanding of behavioral roles of various classes of bitter GRNs in different organs, and how inputs from various bitter GRNs are integrated to mediate selected behaviors, will be facilitated by further elucidation of the molecular profiles and cellular responses of bitter GRNs in different taste organs.
Salt
Salt is an essential nutrient for many physiological processes, including reproduction. However, salt elicits opposite behavioral responses depending on its concentration: low salt (< 100 mM) is attractive while high salt (> 200 mM) is aversive in binary choice assays [23]. The gustatory response to salt is also sexually dimorphic and mating status dependent—mated females show higher proboscis extension upon stimulation of either the labellum or the tarsi as compared to virgin females or males [76]. Ir76b was first identified as a salt receptor functioning in labellar taste neurons that mediate salt attraction [23] but was subsequently also reported to be involved in avoidance of high salt [39]. Besides Ir76b, Gr2a and Gr23a expressed in the pharyngeal L7-3 GRN of the LSO have been implicated in feeding avoidance of salt in a specific behavioral context, in which mildly starved flies were tested with a moderate level of salt (150–450 mM) [77]. The complex view of salt coding emerging from these studies was tackled by a recent comprehensive functional imaging analysis of salt responses in labellar GRNs [7]. To begin to decode taste responses to different concentrations of salts, the authors first gathered molecular tools for labeling subsets of taste neurons in all labellar hairs. First, the authors identified a driver, Ir94e-GAL4, which labels a single GRN that is distinct from previously characterized ppk28, Gr64f, or ppk23-expressing GRNs neurons in L-type hairs, thus completing a molecular genetic toolkit for accessing all four GRNs in these hairs. The authors then identified two subpopulations of ppk23-expressing neurons by labeling either glutamatergic or cholinergic neurons (ppk23glut and ppk23chat), which represented distinct taste neurons in the S-type labellar hairs. Imaging of salt responses in these GRN subpopulations revealed that most if not all types of GRNs respond to salt at some range of the tested concentrations. Specifically, weak calcium activity in response to low concentrations of salt was observed in Gr64f and Ir94e neurons, while response to high salt was observed in Gr64f, Gr66a, and ppk23 neurons. Notably, previous electrophysiological recordings had found high salt-induced activity in two neurons in labellar L-type hairs [9]. Since there are no Gr66a-labeled neurons in these hairs, one possibility is that the L-type responses are derived from Gr64f and ppk23 neurons. In I-type hairs, tip recordings have identified high salt sensitivity in both taste neurons that innervate them [9], which are labeled by Gr64f and Gr66a, respectively. Interestingly, only the salt response in Gr66a neurons is independent of Ir76b function, although it is partially dependent on Ir25a, suggesting potential heterogeneity among salt receptor complexes as well [7]. This appears to go hand-in-hand with functional diversity in salt-sensing circuits—although both ppk23glut and ppk23chat GRNs respond to high salt, only the ppk23glut subset is involved in mediating internal state-dependent modulation of high salt avoidance. Altogether, it is conceivable that different concentrations of salt activate distinct populations of GRNs, many of which express Ir76b, which explains the previously observed roles of this receptor in both low and high salt detection.
Acid
Carboxylic acids are detected via both olfactory and gustatory systems in adult Drosophila to mediate appropriate selection of food and oviposition sites [11, 46, 78–83]. Although flies are attracted to vinegar, they avoid high concentrations of acetic acid detected via Ir64a neurons in olfactory sensilla in the antennae [81]. In the gustatory system, several carboxylic acids have been shown to activate labellar bitter GRNs and also to suppress sugar responses in sweet GRNs [11]. In contrast to the overlap between bitter and acid detection in labellar GRNs, acid sensing in tarsal hairs occurs via two separate groups of GRNs that do not respond to either sugars or bitter compounds: one is broadly tuned to various carboxylic acids, while the second is narrowly tuned to glycolic and malic acids and to high concentrations of salt [42]. Acid responses in both these classes of tarsal GRNs require two broadly expressed Irs, Ir25a and Ir76b. Given that Ir25a and Ir76b are widely expressed in both olfactory and gustatory neurons, the identity of additional Irs that may confer ligand specificity remains to be determined. Interestingly, one recent report identified another member of the Ir family, Ir7a, which is only expressed in a subset of labellar bitter GRNs as a receptor for acetic acid [46]. A high concentration of acetic acid (5%) was found to evoke feeding aversion in binary choice feeding assays. Although feeding avoidance of acetic acid was disrupted in Ir7a mutants, it was not dependent on Ir25a or Ir76b. The observed defects in feeding avoidance were selective for acetic acid and responses to other carboxylic acids were not affected in the absence of Ir7a, consistent with the idea that different receptors with distinct ligand-binding specificities may be involved in sensing various carboxylic acids. Ectopic expression of Ir7a in sweet GRNs conferred acetic acid response as measured with tip recordings [46], an observation that needs to be reconciled with acetic acid-evoked calcium activity in endogenous sweet GRNs [78]. Moreover, the restricted expression of Ir7a in bitter GRNs indicates that the molecular mechanism of acetic acid detection in sweet GRNs is yet to be determined.
Amino acids/yeast
Yeast is the primary source of dietary proteins and amino acids for Drosophila. Yeast feeding is modulated by mating status and prior yeast feeding experience [84, 85]. Recent reports suggest that amino acids are the principal gustatory cues in yeast extract [25], and cellular and behavioral responses to amino acids are mediated via Ir76b [25, 26], which is broadly expressed in peripheral GRNs. Although Ir76b may act alone for salt detection [23], it is likely to serve as a co-receptor for amino acid detection given that taste neurons in labellar hairs, many expressing Ir76b, have limited responses to amino acids [25, 86]. An RNAi screen identified one putative amino acid co-receptor, Ir20a. Ectopic expression of Ir76b and Ir20a together in labellar sweet GRNs conferred amino acid response but not salt response and expression of Ir20a in labellar Ir76b-expressing salt GRNs reduced salt responses but did not confer amino acid response, invoking the contribution of additional receptors/factors present in sweet GRNs but not in salt GRNs in mediating amino acid response. Since Ir20a shows a considerably limited domain of expression in comparison with Ir76b, and Ir20a mutants do not phenocopy the Ir76b mutant, it is expected that additional amino acid receptors in other GRNs will be involved in taste detection of amino acids.
Although amino acids might be salient components in yeast extract, another recent study indicates that flies might have distinct pathways for sensing amino acids and yeast [24]. Using yeast rather than yeast extract, the authors showed that yeast feeding requires Ir76b-expressing GRNs in labellar taste hairs and taste pegs but not in tarsal taste hairs. Further, Ir76b GRNs in labellar taste hairs are responsible for the initiation of yeast feeding (i.e. PER responses), while those in labellar taste pegs are involved in sustaining yeast feeding, providing additional insight into taste organ-specific roles in controlling feeding behavior. Interestingly, yeast-evoked activity in GRNs of both labellar hairs and pegs is modulated by internal amino acids, suggesting that consumption of amino acids and yeast is tightly integrated even though peripheral neuronal detection pathways may be distinct. Future experiments identifying receptors for yeast taste in the two types of labellar GRNs would provide the means to compare mechanisms of amino acid and yeast sensing in peripheral GRNs. In addition to taste-sensing mechanisms, there is evidence that three specific dietary amino acids are detected by brain DH44 neuroendocrine cells which innervate the gut [87, 88]. The proposed fast-acting, post-ingestive mechanism of amino acid detection is independent of Ir76b and requires putative amino acid transporters in the DH44 cells.
Fatty acids
Fatty acid taste elicits an appetitive or aversive response depending upon the concentration [43]. Recent studies have largely focused on the positive behavioral valence of low concentrations (< 1%) of short to medium chain fatty acids (hexanoic, octanoic), which is mediated by a subset of labellar and tarsal sweet GRNs [43, 44, 89]. Notably, a number of studies have found fatty acid taste to be dependent on several members of the Ir family, including Ir56d, Ir25a, and Ir76b. In the labellum, there are two subpopulations of Ir56d GRNs: one is a subset of sweet GRNs in taste hairs that responds to both sugars and fatty acids, and another is a subset of GRNs in taste pegs that responds to fatty acids but not to sugars. Fatty acid-stimulated proboscis extension requires Ir56d GRNs in the labellar taste hairs, but not in taste pegs [44], consistent with distinct behavioral roles for the two GRN populations. Tarsal stimulation-evoked proboscis extension response (PER) is also mediated by Ir56d-labeled sweet GRNs, whose function is dependent on Ir56d, Ir25a, and Ir76b [43]. Notably, tarsal PER to hexanoic and octanoic acids is significantly higher in octuple mutant flies lacking all 8 sweet Grs [43], indicating a possible role in for one or more of these receptors in regulating fatty acid response. Consistent with this idea, a recent study reported that one sweet Gr, Gr64e, is involved in mediating fatty acid taste in the labellum [90]. Whether Ir56d and Gr64e act independently or together for mediating fatty acid signaling is still unclear. However, all studies have found that NorpA, which encodes a phospholipase C, is essential for fatty acid signaling in sweet GRNs [43, 44, 89, 90].
Interestingly, hexanoic acid shows dose-dependent activation of tarsal GRNs that express Gr33a, a receptor that broadly marks bitter-sensing GRNs. At concentrations of hexanoic acid exceeding 1%, control flies exhibit a reduction in proboscis extension, which is not the case in flies in which Gr33a GRNs are functionally ablated [43], consistent with the idea that tarsal bitter GRNs mediate an aversive response to fatty acids. Whether or not labellar bitter GRNs also respond to fatty acids has not been reported. Notably, tarsal bitter GRN sensitivity to fatty acids does not require Ir25a and Ir76b, suggesting that other as yet unidentified receptors are involved in fatty acid taste aversion. As in the case of salt, which elicits opposing behaviors at low and high concentrations, it will be of interest to decipher fatty acid coding at the sensory level and dissect how appetitive and aversive fatty acid-sensing pathways are integrated to shape feeding behaviors.
Carbonated water
Gustatory responses to carbonated water in Drosophila were found to be mediated by E409-GAL4-labeled GRNs that innervate labellar taste pegs [91]. Surprisingly, a suite of chemosensory receptors involved in fatty acid taste (Ir56d, Ir25a, and Ir76b) is also required for sensing carbon dioxide dissolved in fluids [21]. Unlike fatty acids that can activate Ir56d GRNs in labellar hairs, labellar pegs, and tarsal hairs, carbonated water mainly activates GRNs in labellar taste pegs. GRNs in taste hairs of the labellum but not tarsi show a weaker response to carbonated water; however, Ir56d, Ir25a, and Ir76b are unlikely to be involved in these responses according to mutant and rescue analyses [21]. Although the three Irs are necessary for carbonated water detection in taste peg neurons, combined ectopic expression, which was tested in labellar bitter GRNs, did not confer carbonated water sensitivity, indicating that additional factors may be involved. How does carbonated water taste affect feeding behavior? It was first reported that carbonated solutions trigger mild behavioral attraction in a position-based preference assay [91]. However, no behavioral relevance for carbonated fluid has been observed in consumption-based feeding assays such as flyPAD (solid food) and Expresso (liquid food), in which several high-resolution micro-feeding parameters are monitored, including total number of sips, number of sips per feeding burst, feeding success, latency to the first bout, total consumption per fly, number of meal bouts and average bout volume [21]. Thus, carbonated water may be used as a gustatory cue for behaviors other than food ingestion.
Polyamines
Taste input is important not only for food consumption and choice but also for egg-laying site selection by female Drosophila. Polyamines, such as putrescine or cadaverine, are important nutrients for reproductive success and have been shown to activate both olfactory and gustatory pathways for long-range positional attraction and short-range oviposition site selection, respectively [45]. Interestingly, both short-range and long-range behaviors require a common chemosensory receptor, Ir76b. A more narrowly expressed antennal chemoreceptor, Ir41a, is also necessary for polyamine attraction. In fact, Ir76b expression in Ir41a olfactory neurons is sufficient to rescue polyamine attraction in Ir76b mutants. In the gustatory system, there are at least two classes of polyamine-sensing GRNs that mediate oviposition site selection: Ir76b GRNs in labellar taste hairs and taste pegs, and Gr66a GRNs in labellar taste hairs. Ir76b GRNs in taste pegs exhibit stronger responses to polyamines than those in taste hairs, but Ir76b is required for the responses in both. However, polyamine response in Gr66a GRNs is independent of Ir76b, invoking a distinct mechanism for polyamine detection in these GRNs. In dissecting the behavioral contributions of various polyamine-sensing GRNs in controlling egg laying, the authors found that polyamine avoidance during egg-laying behavior relied on labellar input. Silencing of Ir76b or Gr66a-expressing neurons reduced polyamine avoidance to different extents, implicating roles for both classes of neurons. In fact, silencing of Gr66a GRNs caused a slight attraction to polyamine substrate, which was lost upon silencing both Ir76b and Gr66a neurons, indicating some positive behavior component in the Ir76b pathway for egg-laying site selection. Functional heterogeneity in Ir76b and Gr66a neurons might provide multiple substrates for modulation, which could be important for the highly context-dependent egg-laying site selection behavior [92].
H2O2/bacterial lipopolysaccharide
Recent research has uncovered functions for Gr66a bitter GRNs in detecting other types of aversive stimuli such as H2O2, which can be induced by UV [34] or by microbial infection [93, 94], for example, bacterial lipopolysaccharide (LPS) is a known substance from bacteria that induces H2O2 [35]. These chemicals are detected by TrpA1, one of the transient receptor potential (Trp) channels, which is expressed in a subset of Gr66a GRNs in labellar taste hairs [38] and in pharyngeal L8 and L9 GRNs of the LSO [35, 41]. UV-induced H2O2 -sensing bitter GRNs in the labellum were found to promote egg-laying avoidance of strong UV. In addition, the nucleophile-sensitive TrpA1 (A) isoform expressed in I-labellar hairs was found to play an important role in suppressing intake of food sources with reactive oxygen species produced by strong UV exposure [36]. Another study reported that pharyngeal L8 and L9 GRNs detect bacterial LPS via TrpA1 and mediate feeding aversion [35]. Flies also sense LPS via GRNs in the legs and wing margins that mediate grooming behaviors [95, 96], but a requirement of TrpA1 for LPS sensitivity in these organs has not been tested. Since TrpA1 is a highly conserved channel in many species, the recent observations raise the possibility that it may be an ancient chemoreceptor for various aversive stimuli.
Ammonia
Similar to acid, ammonia has been reported to activate both olfactory and gustatory neurons. While olfactory detection of ammonia as an attractive cue depends on Ir92a-expressing olfactory neurons [97], gustatory responses to ammonia depend on Gr66a GRNs in labellar hairs [98]. In addition, ammonia elicits weak responses in L-labellar hairs in which there are no Gr66a GRNs. Given that ppk23 GRNs that respond to high salt are the only known GRNs to detect aversive stimuli in L-labellar hairs, it is possible that they are the ones that sense ammonia. Experiments with ppk23-GAL4 would help to resolve this question. However, identification of the molecular basis of ammonia taste will need further investigation.
Calcium
High levels of calcium activate ppk23 GRNs in S- but not L-type labellar hairs and stimulate aversive behaviors [99]. At least three Irs, Ir25a, Ir62a, and Ir76b, were found to be required for the neuronal response to calcium but ectopic expression of the three in sweet GRNs did not confer calcium sensitivity, suggesting that additional factors may be involved. Similar to the activity of bitter compounds and acids, calcium also inhibits sweet GRNs, providing an additional mechanism for behavioral avoidance of calcium–laced mixtures. The report of calcium taste invites many interesting questions. For example, what is the ligand specificity of Ir62a, since the ppk23 GRNs in S-labellar hairs also respond to high salt (NaCl and KCl)? Do multiple neurons in S-type hairs respond to calcium? An Ir62a reporter is expressed in tarsal GRNs [27], which raises the question of whether GRNs in other organs respond to calcium, and if so, how they contribute to behavioral avoidance of calcium. Finally, is it possible that the mechanism underlying calcium detection is one common to various salts? The answers to these questions will provide insight into how flies distinguish different salts and mount appropriate feeding responses.
Concluding remarks and future perspectives
Taste neurons in adult Drosophila exhibit complex molecular signatures in terms of chemosensory receptor expression. Accumulating evidence suggests that members of Gr, Ir, ppk, and Trp gene families contribute to the detection of various tastants. Overlapping expression patterns of these different chemosensory receptors could be the underlying basis of multimodal taste sensing that has now been reported for many taste neurons (Table 1). In many cases, tastant-evoked responses rely on Ir25a and Ir76b, which might serve as co-receptors for various categories of tastants. Although transgenic chemosensory reporters have presented valuable tools for interrogating the functions and response profiles of taste neurons, it should be noted that there might be further functional sub-division within these molecularly defined groups of taste neurons.
For example, Ir76b-GAL4 and Ir56d-GAL4 label both labellar taste hairs and pegs that respond to polyamines and fatty acid, respectively. Projection patterns of GRNs originating in these two areas can be prominently distinguished by their positions in the SEZ (posterior vs anterior), but calcium activity observed in termini of GRNs from taste hairs cannot be assigned to one type (L-, I-, or S-), from among the types that are labeled. Development of genetic tools for further defining subgroups of GRNs, possibly at single-neuron resolution, will be helpful to understand the extent of molecular and functional heterogeneity in GRNs. Single sensillum recordings can be used to better target types of sensilla that are measured, but analysis can be complicated by the fact that this method simultaneously gathers activity from all neurons in a sensillum, and also that direct comparisons between tip recordings and calcium imaging are complicated by the presence of interneurons in the SEZ that modulate pre-synaptic activity from other taste input or internal state [100–104]. Finally, since GRNs appear to detect multiple compounds of distinct taste modalities, the idea that population coding mechanisms may be involved in discrimination between tastants has some appeal. In the future, not only will it be of interest to determine how taste neurons in different organs control different aspects of feeding behaviors and connect to different higher-order neuronal circuits, but to understand how input from GRNs is integrated and evaluated for more complex taste-associated behaviors.
Acknowledgements
We thank Lisa Baik and Vaibhav Menon for helpful comments on the manuscript. Work in A.D’s lab is funded by funds from the National Institutes of Health (R01DC013587, R21AI140065, and R01DC017390), DARPA (D18AC00026), and the University of California AESMF program. Y.-C.D.C. is a Howard Hughes Medical Institute International Student Research Fellow.
Author contributions
Conceptualization, Y-CDC and AD; Writing—Original Draft, Y-CDC, Writing—Review and Editing, Y-CDC and AD; Supervision, AD; Funding Acquisition, AD.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Shanbhag SR, Park SK, Pikielny CW, Steinbrecht RA. Gustatory organs of Drosophila melanogaster: fine structure and expression of the putative odorant-binding protein PBPRP2. Cell Tissue Res. 2001;304(3):423–437. doi: 10.1007/s004410100388. [DOI] [PubMed] [Google Scholar]
- 2.Thistle R, Cameron P, Ghorayshi A, Dennison L, Scott K. Contact chemoreceptors mediate male-male repulsion and male-female attraction during Drosophila courtship. Cell. 2012;149(5):1140–1151. doi: 10.1016/j.cell.2012.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bray S, Amrein H. A putative Drosophila pheromone receptor expressed in male-specific taste neurons is required for efficient courtship. Neuron. 2003;39(6):1019–1029. doi: 10.1016/S0896-6273(03)00542-7. [DOI] [PubMed] [Google Scholar]
- 4.Stocker RF. The organization of the chemosensory system in Drosophila melanogaster: a review. Cell Tissue Res. 1994;275(1):3–26. doi: 10.1007/BF00305372. [DOI] [PubMed] [Google Scholar]
- 5.Stocker RF, Schorderet M. Cobalt filling of sensory projections from internal and external mouthparts in Drosophila. Cell Tissue Res. 1981;216(3):513–523. doi: 10.1007/BF00238648. [DOI] [PubMed] [Google Scholar]
- 6.Salvaterra PM, Kitamoto T. Drosophila cholinergic neurons and processes visualized with Gal4/UAS-GFP. Brain Res Gene Expr Patterns. 2001;1(1):73–82. doi: 10.1016/S1567-133X(01)00011-4. [DOI] [PubMed] [Google Scholar]
- 7.Jaeger AH, Stanley M, Weiss ZF, Musso PY, Chan RC, Zhang H, Feldman-Kiss D, Gordon MD. A complex peripheral code for salt taste in Drosophila. eLife. 2018;7:e37167. doi: 10.7554/elife.37167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kallman BR, Kim H, Scott K. Excitation and inhibition onto central courtship neurons biases Drosophila mate choice. eLife. 2015;4:e11188. doi: 10.7554/elife.11188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hiroi M, Meunier N, Marion-Poll F, Tanimura T. Two antagonistic gustatory receptor neurons responding to sweet-salty and bitter taste in Drosophila. J Neurobiol. 2004;61(3):333–342. doi: 10.1002/neu.20063. [DOI] [PubMed] [Google Scholar]
- 10.Ling F, Dahanukar A, Weiss LA, Kwon JY, Carlson JR. The molecular and cellular basis of taste coding in the legs of Drosophila. J Neurosci. 2014;34(21):7148–7164. doi: 10.1523/JNEUROSCI.0649-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Charlu S, Wisotsky Z, Medina A, Dahanukar A. Acid sensing by sweet and bitter taste neurons in Drosophila melanogaster. Nat Commun. 2013;4:2042. doi: 10.1038/ncomms3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weiss LA, Dahanukar A, Kwon JY, Banerjee D, Carlson JR. The molecular and cellular basis of bitter taste in Drosophila. Neuron. 2011;69(2):258–272. doi: 10.1016/j.neuron.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dahanukar A, Lei YT, Kwon JY, Carlson JR. Two Gr genes underlie sugar reception in Drosophila. Neuron. 2007;56(3):503–516. doi: 10.1016/j.neuron.2007.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clyne PJ, Warr CG, Carlson JR. Candidate taste receptors in Drosophila. Science. 2000;287(5459):1830–1834. doi: 10.1126/science.287.5459.1830. [DOI] [PubMed] [Google Scholar]
- 15.Scott K, Brady R, Jr, Cravchik A, Morozov P, Rzhetsky A, Zuker C, Axel R. A chemosensory gene family encoding candidate gustatory and olfactory receptors in Drosophila. Cell. 2001;104(5):661–673. doi: 10.1016/S0092-8674(01)00263-X. [DOI] [PubMed] [Google Scholar]
- 16.Dunipace L, Meister S, McNealy C, Amrein H. Spatially restricted expression of candidate taste receptors in the Drosophila gustatory system. Curr Biol. 2001;11(11):822–835. doi: 10.1016/S0960-9822(01)00258-5. [DOI] [PubMed] [Google Scholar]
- 17.Fujii S, Yavuz A, Slone J, Jagge C, Song X, Amrein H. Drosophila sugar receptors in sweet taste perception, olfaction, and internal nutrient sensing. Curr Biol CB. 2015;25(5):621–627. doi: 10.1016/j.cub.2014.12.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen YD, Dahanukar A. Molecular and cellular organization of taste neurons in adult Drosophila pharynx. Cell Rep. 2017;21(10):2978–2991. doi: 10.1016/j.celrep.2017.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.LeDue EE, Chen YC, Jung AY, Dahanukar A, Gordon MD. Pharyngeal sense organs drive robust sugar consumption in Drosophila. Nat Commun. 2015;6:6667. doi: 10.1038/ncomms7667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moon SJ, Lee Y, Jiao Y, Montell C. A Drosophila gustatory receptor essential for aversive taste and inhibiting male-to-male courtship. Curr Biol. 2009;19(19):1623–1627. doi: 10.1016/j.cub.2009.07.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanchez-Alcaniz JA, Silbering AF, Croset V, Zappia G, Sivasubramaniam AK, Abuin L, Sahai SY, Munch D, Steck K, Auer TO, Cruchet S, Neagu-Maier GL, Sprecher SG, Ribeiro C, Yapici N, Benton R. An expression atlas of variant ionotropic glutamate receptors identifies a molecular basis of carbonation sensing. Nat Commun. 2018;9(1):4252. doi: 10.1038/s41467-018-06453-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Croset V, Rytz R, Cummins SF, Budd A, Brawand D, Kaessmann H, Gibson TJ, Benton R. Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genet. 2010;6(8):e1001064. doi: 10.1371/journal.pgen.1001064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang YV, Ni J, Montell C. The molecular basis for attractive salt-taste coding in Drosophila. Science. 2013;340(6138):1334–1338. doi: 10.1126/science.1234133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steck K, Walker SJ, Itskov PM, Baltazar C, Moreira JM, Ribeiro C. Internal amino acid state modulates yeast taste neurons to support protein homeostasis in Drosophila. eLife. 2018;7:e31625. doi: 10.7554/elife.31625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ganguly A, Pang L, Duong VK, Lee A, Schoniger H, Varady E, Dahanukar A. A molecular and cellular context-dependent role for Ir76b in detection of amino acid taste. Cell Rep. 2017;18(3):737–750. doi: 10.1016/j.celrep.2016.12.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Croset V, Schleyer M, Arguello JR, Gerber B, Benton R. A molecular and neuronal basis for amino acid sensing in the Drosophila larva. Sci Rep. 2016;6:34871. doi: 10.1038/srep34871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koh TW, He Z, Gorur-Shandilya S, Menuz K, Larter NK, Stewart S, Carlson JR. The Drosophila IR20a clade of ionotropic receptors are candidate taste and pheromone receptors. Neuron. 2014;83(4):850–865. doi: 10.1016/j.neuron.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu T, Wang Y, Tian Y, Zhang J, Zhao J, Guo A (2018) The receptor channel formed by ppk25, ppk29 and ppk23 can sense the Drosophila female pheromone 7,11-heptacosadiene. Gene Brain Behav. 10.1111/gbb.12529 [DOI] [PubMed]
- 29.Vijayan V, Thistle R, Liu T, Starostina E, Pikielny CW. Drosophila pheromone-sensing neurons expressing the ppk25 ion channel subunit stimulate male courtship and female receptivity. PLoS Genet. 2014;10(3):e1004238. doi: 10.1371/journal.pgen.1004238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Toda H, Zhao X, Dickson BJ. The Drosophila female aphrodisiac pheromone activates ppk23(+) sensory neurons to elicit male courtship behavior. Cell Rep. 2012;1(6):599–607. doi: 10.1016/j.celrep.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 31.Lu B, LaMora A, Sun Y, Welsh MJ, Ben-Shahar Y. ppk23-Dependent chemosensory functions contribute to courtship behavior in Drosophila melanogaster. PLoS Genet. 2012;8(3):e1002587. doi: 10.1371/journal.pgen.1002587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Z, Wang Q, Wang Z. The amiloride-sensitive epithelial Na+ channel PPK28 is essential for Drosophila gustatory water reception. J Neurosci. 2010;30(18):6247–6252. doi: 10.1523/JNEUROSCI.0627-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cameron P, Hiroi M, Ngai J, Scott K. The molecular basis for water taste in Drosophila. Nature. 2010;465(7294):91–95. doi: 10.1038/nature09011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guntur AR, Gou B, Gu P, He R, Stern U, Xiang Y, Yang CH. H2O2-sensitive isoforms of Drosophila melanogaster TRPA1 act in bitter-sensing gustatory neurons to promote avoidance of UV during egg-laying. Genetics. 2017;205(2):749–759. doi: 10.1534/genetics.116.195172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soldano A, Alpizar YA, Boonen B, Franco L, Lopez-Requena A, Liu G, Mora N, Yaksi E, Voets T, Vennekens R, Hassan BA, Talavera K (2016) Gustatory-mediated avoidance of bacterial lipopolysaccharides via TRPA1 activation in Drosophila. eLife 5:e13133. 10.7554/elife.13133 [DOI] [PMC free article] [PubMed]
- 36.Du EJ, Ahn TJ, Wen X, Seo DW, Na DL, Kwon JY, Choi M, Kim HW, Cho H, Kang K. Nucleophile sensitivity of Drosophila TRPA1 underlies light-induced feeding deterrence. Elife. 2016;5:e18425. doi: 10.7554/eLife.18425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang YV, Raghuwanshi RP, Shen WL, Montell C. Food experience-induced taste desensitization modulated by the Drosophila TRPL channel. Nat Neurosci. 2013;16(10):1468–1476. doi: 10.1038/nn.3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim SH, Lee Y, Akitake B, Woodward OM, Guggino WB, Montell C. Drosophila TRPA1 channel mediates chemical avoidance in gustatory receptor neurons. Proc Natl Acad Sci USA. 2010;107(18):8440–8445. doi: 10.1073/pnas.1001425107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee MJ, Sung HY, Jo H, Kim HW, Choi MS, Kwon JY, Kang K. Ionotropic receptor 76b is required for gustatory aversion to excessive Na + in Drosophila. Mol Cell. 2017;40(10):787–795. doi: 10.14348/molcells.2017.0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kang K, Panzano VC, Chang EC, Ni L, Dainis AM, Jenkins AM, Regna K, Muskavitch MA, Garrity PA. Modulation of TRPA1 thermal sensitivity enables sensory discrimination in Drosophila. Nature. 2011;481(7379):76–80. doi: 10.1038/nature10715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kang K, Pulver SR, Panzano VC, Chang EC, Griffith LC, Theobald DL, Garrity PA. Analysis of Drosophila TRPA1 reveals an ancient origin for human chemical nociception. Nature. 2010;464(7288):597–600. doi: 10.1038/nature08848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen Y, Amrein H. Ionotropic receptors mediate Drosophila oviposition preference through sour gustatory receptor neurons. Curr Biol CB. 2017;27(18):2741–2750.e2744. doi: 10.1016/j.cub.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ahn JE, Chen Y, Amrein H (2017) Molecular basis of fatty acid taste in Drosophila. eLife 6:e30115. 10.7554/elife.30115 [DOI] [PMC free article] [PubMed]
- 44.Tauber JM, Brown EB, Li Y, Yurgel ME, Masek P, Keene AC. A subset of sweet-sensing neurons identified by IR56d are necessary and sufficient for fatty acid taste. PLoS Genet. 2017;13(11):e1007059. doi: 10.1371/journal.pgen.1007059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hussain A, Zhang M, Ucpunar HK, Svensson T, Quillery E, Gompel N, Ignell R, Grunwald Kadow IC. Ionotropic chemosensory receptors mediate the taste and smell of polyamines. PLoS Biol. 2016;14(5):e1002454. doi: 10.1371/journal.pbio.1002454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rimal S, Sang J, Poudel S, Thakur D, Montell C, Lee Y (2019) Mechanism of acetic acid gustatory repulsion in Drosophila. Cell Rep 26(6):1432–1442 e1434. 10.1016/j.celrep.2019.01.042 [DOI] [PMC free article] [PubMed]
- 47.He Z, Luo Y, Shang X, Sun JS, Carlson JR. Chemosensory sensilla of the Drosophila wing express a candidate ionotropic pheromone receptor. PLoS Biol. 2019;17(5):e2006619. doi: 10.1371/journal.pbio.2006619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rimal S, Lee Y. The multidimensional ionotropic receptors of Drosophila melanogaster. Insect Mol Biol. 2018;27(1):1–7. doi: 10.1111/imb.12347. [DOI] [PubMed] [Google Scholar]
- 49.Joseph RM, Carlson JR. Drosophila chemoreceptors: a molecular interface between the chemical world and the brain. Trends Genet TIG. 2015;31(12):683–695. doi: 10.1016/j.tig.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Freeman EG, Dahanukar A. Molecular neurobiology of Drosophila taste. Curr Opin Neurobiol. 2015;34:140–148. doi: 10.1016/j.conb.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiao Y, Moon SJ, Wang X, Ren Q, Montell C. Gr64f is required in combination with other gustatory receptors for sugar detection in Drosophila. Curr Biol. 2008;18(22):1797–1801. doi: 10.1016/j.cub.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiao Y, Moon SJ, Montell C. A Drosophila gustatory receptor required for the responses to sucrose, glucose, and maltose identified by mRNA tagging. Proc Natl Acad Sci USA. 2007;104(35):14110–14115. doi: 10.1073/pnas.0702421104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Slone J, Daniels J, Amrein H. Sugar receptors in Drosophila. Curr Biol. 2007;17(20):1809–1816. doi: 10.1016/j.cub.2007.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miyamoto T, Amrein H. Diverse roles for the Drosophila fructose sensor Gr43a. Fly (Austin) 2014;8(1):19–25. doi: 10.4161/fly.27241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miyamoto T, Slone J, Song X, Amrein H. A fructose receptor functions as a nutrient sensor in the Drosophila brain. Cell. 2012;151(5):1113–1125. doi: 10.1016/j.cell.2012.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McGinnis JP, Jiang H, Agha MA, Sanchez CP, Lange J, Yu Z, Marion-Poll F, Si K. Immediate perception of a reward is distinct from the reward’s long-term salience. eLife. 2016;5:e22283. doi: 10.7554/elife.22283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miyamoto T, Chen Y, Slone J, Amrein H. Identification of a Drosophila glucose receptor using Ca2+ imaging of single chemosensory neurons. PLoS One. 2013;8(2):e56304. doi: 10.1371/journal.pone.0056304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Z, Singhvi A, Kong P, Scott K. Taste representations in the Drosophila brain. Cell. 2004;117(7):981–991. doi: 10.1016/j.cell.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 59.Thorne N, Chromey C, Bray S, Amrein H. Taste perception and coding in Drosophila. Curr Biol. 2004;14(12):1065–1079. doi: 10.1016/j.cub.2004.05.019. [DOI] [PubMed] [Google Scholar]
- 60.Thoma V, Knapek S, Arai S, Hartl M, Kohsaka H, Sirigrivatanawong P, Abe A, Hashimoto K, Tanimoto H. Functional dissociation in sweet taste receptor neurons between and within taste organs of Drosophila. Nat Commun. 2016;7:10678. doi: 10.1038/ncomms10678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Murata S, Brockmann A, Tanimura T. Pharyngeal stimulation with sugar triggers local searching behavior in Drosophila. J Exp Biol. 2017;220(Pt 18):3231–3237. doi: 10.1242/jeb.161646. [DOI] [PubMed] [Google Scholar]
- 62.Yapici N, Cohn R, Schusterreiter C, Ruta V, Vosshall LB. A taste circuit that regulates ingestion by integrating food and hunger signals. Cell. 2016;165(3):715–729. doi: 10.1016/j.cell.2016.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Joseph RM, Sun JS, Tam E, Carlson JR. A receptor and neuron that activate a circuit limiting sucrose consumption. eLife. 2017;6:e24992. doi: 10.7554/elife.24992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee Y, Moon SJ, Montell C. Multiple gustatory receptors required for the caffeine response in Drosophila. Proc Natl Acad Sci USA. 2009;106(11):4495–4500. doi: 10.1073/pnas.0811744106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moon SJ, Kottgen M, Jiao Y, Xu H, Montell C. A taste receptor required for the caffeine response in vivo. Curr Biol. 2006;16(18):1812–1817. doi: 10.1016/j.cub.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 66.Lee Y, Kim SH, Montell C. Avoiding DEET through insect gustatory receptors. Neuron. 2010;67(4):555–561. doi: 10.1016/j.neuron.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shim J, Lee Y, Jeong YT, Kim Y, Lee MG, Montell C, Moon SJ. The full repertoire of Drosophila gustatory receptors for detecting an aversive compound. Nat Commun. 2015;6:8867. doi: 10.1038/ncomms9867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Poudel S, Kim Y, Gwak JS, Jeong S, Lee Y. Gustatory receptor 22e is essential for sensing chloroquine and strychnine in Drosophila melanogaster. Insect Biochem Mol Biol. 2017;88:30–36. doi: 10.1016/j.ibmb.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 69.Poudel S, Lee Y. Gustatory receptors required for avoiding the toxic compound coumarin in Drosophila melanogaster. Mol Cells. 2016;39(4):310–315. doi: 10.14348/molcells.2016.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Poudel S, Kim Y, Kim YT, Lee Y. Gustatory receptors required for sensing umbelliferone in Drosophila melanogaster. Insect Biochem Mol Biol. 2015;66:110–118. doi: 10.1016/j.ibmb.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 71.Lee Y, Moon SJ, Wang Y, Montell C. A Drosophila gustatory receptor required for strychnine sensation. Chem Sens. 2015;40(7):525–533. doi: 10.1093/chemse/bjv038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sang J, Rimal S, Lee Y (2019) Gustatory receptor 28b is necessary for avoiding saponin in Drosophila melanogaster. EMBO Rep. 10.15252/embr.201847328 [DOI] [PMC free article] [PubMed]
- 73.Rimal S, Lee Y. Molecular sensor of nicotine in taste of Drosophila melanogaster. Insect Biochem Mol Biol. 2019;111:103178. doi: 10.1016/j.ibmb.2019.103178. [DOI] [PubMed] [Google Scholar]
- 74.Sung HY, Jeong YT, Lim JY, Kim H, Oh SM, Hwang SW, Kwon JY, Moon SJ. Heterogeneity in the Drosophila gustatory receptor complexes that detect aversive compounds. Nat Commun. 2017;8(1):1484. doi: 10.1038/s41467-017-01639-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Delventhal R, Carlson JR. Bitter taste receptors confer diverse functions to neurons. eLife. 2016;5:e11181. doi: 10.7554/elife.11181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Walker SJ, Corrales-Carvajal VM, Ribeiro C. Postmating circuitry modulates salt taste processing to increase reproductive output in Drosophila. Curr Biol CB. 2015;25(20):2621–2630. doi: 10.1016/j.cub.2015.08.043. [DOI] [PubMed] [Google Scholar]
- 77.Kim H, Jeong YT, Choi MS, Choi J, Moon SJ, Kwon JY. Involvement of a Gr2a-expressing Drosophila pharyngeal gustatory receptor neuron in regulation of aversion to high-salt foods. Mol Cells. 2017;40(5):331–338. doi: 10.14348/molcells.2017.0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Devineni AV, Sun B, Zhukovskaya A, Axel R. Acetic acid activates distinct taste pathways in Drosophila to elicit opposing, state-dependent feeding responses. eLife. 2019 doi: 10.7554/eLife.47677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Deshpande SA, Yamada R, Mak CM, Hunter B, Soto Obando A, Hoxha S, Ja WW. Acidic food pH increases palatability and consumption and extends Drosophila lifespan. J Nutr. 2015;145(12):2789–2796. doi: 10.3945/jn.115.222380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen Y, Amrein H. Enhancing perception of contaminated food through acid-mediated modulation of taste neuron responses. Curr Biol. 2014;24(17):1969–1977. doi: 10.1016/j.cub.2014.07.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ai M, Min S, Grosjean Y, Leblanc C, Bell R, Benton R, Suh GS. Acid sensing by the Drosophila olfactory system. Nature. 2010;468(7324):691–695. doi: 10.1038/nature09537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Joseph RM, Devineni AV, King IF, Heberlein U. Oviposition preference for and positional avoidance of acetic acid provide a model for competing behavioral drives in Drosophila. Proc Natl Acad Sci USA. 2009;106(27):11352–11357. doi: 10.1073/pnas.0901419106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Abuin L, Bargeton B, Ulbrich MH, Isacoff EY, Kellenberger S, Benton R. Functional architecture of olfactory ionotropic glutamate receptors. Neuron. 2011;69(1):44–60. doi: 10.1016/j.neuron.2010.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vargas MA, Luo N, Yamaguchi A, Kapahi P. A role for S6 kinase and serotonin in postmating dietary switch and balance of nutrients in D. melanogaster. Curr Biol. 2010;20(11):1006–1011. doi: 10.1016/j.cub.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ribeiro C, Dickson BJ. Sex peptide receptor and neuronal TOR/S6 K signaling modulate nutrient balancing in Drosophila. Curr Biol. 2010;20(11):1000–1005. doi: 10.1016/j.cub.2010.03.061. [DOI] [PubMed] [Google Scholar]
- 86.Park J, Carlson JR. Physiological responses of the Drosophila labellum to amino acids. J Neurogen. 2018;32(1):27–36. doi: 10.1080/01677063.2017.1406934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yang Z, Huang R, Fu X, Wang G, Qi W, Mao D, Shi Z, Shen WL, Wang L. A post-ingestive amino acid sensor promotes food consumption in Drosophila. Cell Res. 2018;28(10):1013–1025. doi: 10.1038/s41422-018-0084-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dus M, Lai JS, Gunapala KM, Min S, Tayler TD, Hergarden AC, Geraud E, Joseph CM, Suh GS. Nutrient sensor in the brain directs the action of the brain-gut axis in Drosophila. Neuron. 2015;87(1):139–151. doi: 10.1016/j.neuron.2015.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Masek P, Keene AC (2013) Drosophila fatty acid taste signals through the PLC pathway in sugar-sensing neurons. Plos Genet. 10.1371/journal.pgen.1003710 [DOI] [PMC free article] [PubMed]
- 90.Kim H, Kim H, Kwon JY, Seo JT, Shin DM, Moon SJ. Drosophila Gr64e mediates fatty acid sensing via the phospholipase C pathway. PLoS Genet. 2018;14(2):e1007229. doi: 10.1371/journal.pgen.1007229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fischler W, Kong P, Marella S, Scott K. The detection of carbonation by the Drosophila gustatory system. Nature. 2007;448(7157):1054–1057. doi: 10.1038/nature06101. [DOI] [PubMed] [Google Scholar]
- 92.Yang CH, Belawat P, Hafen E, Jan LY, Jan YN. Drosophila egg-laying site selection as a system to study simple decision-making processes. Science. 2008;319(5870):1679–1683. doi: 10.1126/science.1151842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zug R, Hammerstein P. Wolbachia and the insect immune system: what reactive oxygen species can tell us about the mechanisms of Wolbachia-host interactions. Front Microbiol. 2015;6:1201. doi: 10.3389/fmicb.2015.01201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Apel K, Hirt H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Ann Rev Plant Biol. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- 95.Yanagawa A, Couto A, Sandoz JC, Hata T, Mitra A, Ali Agha M, Marion-Poll F. LPS perception through taste-induced reflex in Drosophila melanogaster. J Insect Physiol. 2019;112:39–47. doi: 10.1016/j.jinsphys.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 96.Yanagawa A, Guigue AM, Marion-Poll F. Hygienic grooming is induced by contact chemicals in Drosophila melanogaster. Front Behav Neurosci. 2014;8:254. doi: 10.3389/fnbeh.2014.00254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Min S, Ai M, Shin SA, Suh GS. Dedicated olfactory neurons mediating attraction behavior to ammonia and amines in Drosophila. Proc Natl Acad Sci USA. 2013;110(14):E1321–E1329. doi: 10.1073/pnas.1215680110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Delventhal R, Menuz K, Joseph R, Park J, Sun JS, Carlson JR. The taste response to ammonia in Drosophila. Sci Rep. 2017;7:43754. doi: 10.1038/srep43754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lee Y, Poudel S, Kim Y, Thakur D, Montell C. Calcium taste avoidance in Drosophila. Neuron. 2018;97(1):67–74.e64. doi: 10.1016/j.neuron.2017.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.LeDue EE, Mann K, Koch E, Chu B, Dakin R, Gordon MD. Starvation-induced depotentiation of bitter taste in Drosophila. Curr Biol CB. 2016;26(21):2854–2861. doi: 10.1016/j.cub.2016.08.028. [DOI] [PubMed] [Google Scholar]
- 101.Youn H, Kirkhart C, Chia J, Scott K. A subset of octopaminergic neurons that promotes feeding initiation in Drosophila melanogaster. PLoS One. 2018;13(6):e0198362. doi: 10.1371/journal.pone.0198362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Inagaki HK, Panse KM, Anderson DJ. Independent, reciprocal neuromodulatory control of sweet and bitter taste sensitivity during starvation in Drosophila. Neuron. 2014;84(4):806–820. doi: 10.1016/j.neuron.2014.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Inagaki HK, Ben-Taboude-Leon S, Wong AM, Jagadish S, Ishimoto H, Barnea G, Kitamoto T, Axel R, Anderson DJ. Visualizing neuromodulation in vivo: TANGO-mapping of dopamine signaling reveals appetite control of sugar sensing. Cell. 2012;148(3):583–595. doi: 10.1016/j.cell.2011.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang QP, Lin YQ, Zhang L, Wilson YA, Oyston LJ, Cotterell J, Qi Y, Khuong TM, Bakhshi N, Planchenault Y, Browman DT, Lau MT, Cole TA, Wong AC, Simpson SJ, Cole AR, Penninger JM, Herzog H, Neely GG. Sucralose promotes food intake through NPY and a neuronal fasting response. Cell Metab. 2016;24(1):75–90. doi: 10.1016/j.cmet.2016.06.010. [DOI] [PubMed] [Google Scholar]
- 105.Dahanukar A, Foster K, van der Goes van Naters WM, Carlson JR. A Gr receptor is required for response to the sugar trehalose in taste neurons of Drosophila. Nat Neurosci. 2001;4(12):1182–1186. doi: 10.1038/nn765. [DOI] [PubMed] [Google Scholar]
- 106.Hiroi M, Marion-Poll F, Tanimura T. Differentiated response to sugars among labellar chemosensilla in Drosophila. Zool Sci. 2002;19(9):1009–1018. doi: 10.2108/zsj.19.1009. [DOI] [PubMed] [Google Scholar]
- 107.Wisotsky Z, Medina A, Freeman E, Dahanukar A. Evolutionary differences in food preference rely on Gr64e, a receptor for glycerol. Nat Neurosci. 2011;14(12):1534–1541. doi: 10.1038/nn.2944. [DOI] [PubMed] [Google Scholar]
- 108.Marella S, Fischler W, Kong P, Asgarian S, Rueckert E, Scott K. Imaging taste responses in the fly brain reveals a functional map of taste category and behavior. Neuron. 2006;49(2):285–295. doi: 10.1016/j.neuron.2005.11.037. [DOI] [PubMed] [Google Scholar]
- 109.Yavuz A, Jagge C, Slone J, Amrein H. A genetic tool kit for cellular and behavioral analyses of insect sugar receptors. Fly (Austin) 2014;8(4):189–196. doi: 10.1080/19336934.2015.1050569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Raad H, Ferveur JF, Ledger N, Capovilla M, Robichon A. Functional gustatory role of chemoreceptors in Drosophila wings. Cell Rep. 2016;15(7):1442–1454. doi: 10.1016/j.celrep.2016.04.040. [DOI] [PubMed] [Google Scholar]
- 111.Freeman EG, Wisotsky Z, Dahanukar A. Detection of sweet tastants by a conserved group of insect gustatory receptors. Proc Natl Acad Sci USA. 2014;111(4):1598–1603. doi: 10.1073/pnas.1311724111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Meunier N, Marion-Poll F, Rospars JP, Tanimura T. Peripheral coding of bitter taste in Drosophila. J Neurobiol. 2003;56(2):139–152. doi: 10.1002/neu.10235. [DOI] [PubMed] [Google Scholar]
- 113.Lacaille F, Hiroi M, Twele R, Inoshita T, Umemoto D, Maniere G, Marion-Poll F, Ozaki M, Francke W, Cobb M, Everaerts C, Tanimura T, Ferveur JF. An inhibitory sex pheromone tastes bitter for Drosophila males. PLoS One. 2007;2(7):e661. doi: 10.1371/journal.pone.0000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.French AS, Sellier MJ, Ali Agha M, Guigue A, Chabaud MA, Reeb PD, Mitra A, Grau Y, Soustelle L, Marion-Poll F. Dual mechanism for bitter avoidance in Drosophila. J Neurosci. 2015;35(9):3990–4004. doi: 10.1523/jneurosci.1312-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Du EJ, Ahn TJ, Choi MS, Kwon I, Kim HW, Kwon JY, Kang K. The mosquito repellent citronellal directly potentiates Drosophila TRPA1, facilitating feeding suppression. Mol Cells. 2015;38(10):911–917. doi: 10.14348/molcells.2015.0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Al-Anzi B, Tracey WD, Jr, Benzer S. Response of Drosophila to wasabi is mediated by painless, the fly homolog of mammalian TRPA1/ANKTM1. Curr Biol. 2006;16(10):1034–1040. doi: 10.1016/j.cub.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 117.Park JH, Chen J, Jang S, Ahn TJ, Kang K, Choi MS, Kwon JY. A subset of enteroendocrine cells is activated by amino acids in the Drosophila midgut. FEBS Lett. 2016;590(4):493–500. doi: 10.1002/1873-3468.12073. [DOI] [PubMed] [Google Scholar]
- 118.Toshima N, Tanimura T. Taste preference for amino acids is dependent on internal nutritional state in Drosophila melanogaster. J Exp Biol. 2012;215(Pt 16):2827–2832. doi: 10.1242/jeb.069146. [DOI] [PubMed] [Google Scholar]