Abstract
Prokaryotic NaV channels are tetramers and eukaryotic NaV channels consist of a single subunit containing four domains. Each monomer/domain contains six transmembrane segments (S1-S6), S1-S4 being the voltage-sensor domain and S5-S6 the pore domain. A crystal structure of NaVMs, a prokaryotic NaV channel, suggests that the S4-S5 linker (S4-S5L) interacts with the C-terminus of S6 (S6T) to stabilize the gate in the open state. However, in several voltage-gated potassium channels, using specific S4-S5L-mimicking peptides, we previously demonstrated that S4-S5L/S6T interaction stabilizes the gate in the closed state. Here, we used the same strategy on another prokaryotic NaV channel, NaVSp1, to test whether equivalent peptides stabilize the channel in the open or closed state. A NaVSp1-specific S4-S5L peptide, containing the residues supposed to interact with S6T according to the NaVMs structure, induced both an increase in NaVSp1 current density and a negative shift in the activation curve, consistent with S4-S5L stabilizing the open state. Using this approach on a human NaV channel, hNaV1.4, and testing 12 hNaV1.4 S4-S5L peptides, we identified four activating S4-S5L peptides. These results suggest that, in eukaryotic NaV channels, the S4-S5L of DI, DII and DIII domains allosterically modulate the activation gate and stabilize its open state.
Subject terms: Ion transport, Skeletal muscle
Introduction
Voltage-gated sodium channels (NaV) are crucial in excitable as well as non-excitable cells and mutations in NaV1.x-subunits have been associated with muscular, neuronal and cardiac channelopathies in human1. Voltage-gated potassium (KV) channels and prokaryotic NaV channels are tetramers of subunits containing six transmembrane segments (S1 to S6). Each of the four subunits consists of one voltage-sensor domain (S1 to S4) and a pore domain (S5-S6). The four pore domains tetramerize to form a single pore module, which is regulated by the four voltage sensor domains. The arrangement of eukaryotic NaV channels is similar, with one major difference: the channel is made of a single subunit containing four homologous domains, rather than four identical subunits. Each domain in eukaryotic NaV channels is structurally equivalent to one subunit in KV or prokaryotic NaV channels, and consists of six transmembrane segments (S1 to S6).
Despite intensive work on the voltage-gating of KV and NaV channels, we still lack a clear picture describing the coupling between S4 voltage-sensor movement and S6 pore gating. Both structural and functional studies identified the linker between S4 and S5 (named S4-S5L) and the C-terminus of S6 (named S6T), as major actors in this coupling2–21. Different coupling mechanisms have been suggested. The crystal structure of KV1.2, and more recently the cryo-electron microscopy and crystal structures of both eukaryotic and prokaryotic NaV channels suggested that the four S4-S5L form a mechanical lever or a constriction ring intimately interacting with S6T when the activation gate is closed. Upon membrane depolarization, constriction is relieved, and channel activation gate can open10,12,15,19–21. On the other hand, other studies performed on the bacterial NaVMs (from Magnetococcus marinus) channel suggest that the S4-S5L may also be involved in an interaction motif stabilizing the channel open state8,16,17. So rather than only playing the role of a constriction ring (obligatory role, as described for Shaker22), S4-S5L may also allosterically modulate channel gating: the “up” or activated S4 conformation would favor but not impose the channel open state. Such allosteric regulation has been suggested for several channels, including hKV11.1 (hERG) and hKV7.1 (KCNQ1) channels23,24. In these channels, we elucidated the nature of this allosteric coupling: when S4 sensors are in the “down” or deactivated conformation, the four S4-S5L bind to S6T in the closed state, stabilizing this state25,26. Noteworthy, ATP has also been shown to stabilize the closed state of KATP channels27,28. In KV channels, S4-S5L can thus be seen as an inhibitor (like ATP) attached to the S4 voltage sensor. When the membrane is depolarized, S4 pulls S4-S5L out of its binding pocket, leading to channel opening. This is consistent with the observation that specific S4-S5L-mimicking peptides inhibit hKV7.1 and hKV11.1 channels, by replacing the endogenous segment in the binding pocket25,26. This mechanism was recently extended to hKV10.2 channels29.
In the case of NaV channels, such an allosteric model of the voltage-dependent gating mechanism has never been functionally tested. From the interaction motif observed in NaVMs channel8,16,17, we hypothesized that S4-S5L acts as a ligand binding to S6T and stabilizing the channel open-state and not the closed state. We used the same peptide approach previously used for hKV7.1, hKV11.1 and hKV10.2 channels25,26,29 to test whether S4-S5L peptides lead to a gain of function in both prokaryotic and eukaryotic NaV channels.
We designed three S4-S5L mimicking peptides specific for prokaryotic NaVSp1 (from Silicibacter pomeroyi), and three S4-S5L mimicking peptides specific for each of the four domains of hNaV1.4. None of these S4-S5L peptides had an inhibitory effect. One S4-S5L peptide from NaVSp1 and at least one S4-S5L peptide from DI, DII and DIII domains of hNaV1.4 promoted channel activity. Our results suggest that, as demonstrated in three KV channels, the ligand/receptor model of interaction between S4-S5L and S6T applies also to both NaVSp1 and hNaV1.4 channels, with one major difference: S4-S5L stabilizes the open state in NaV channels.
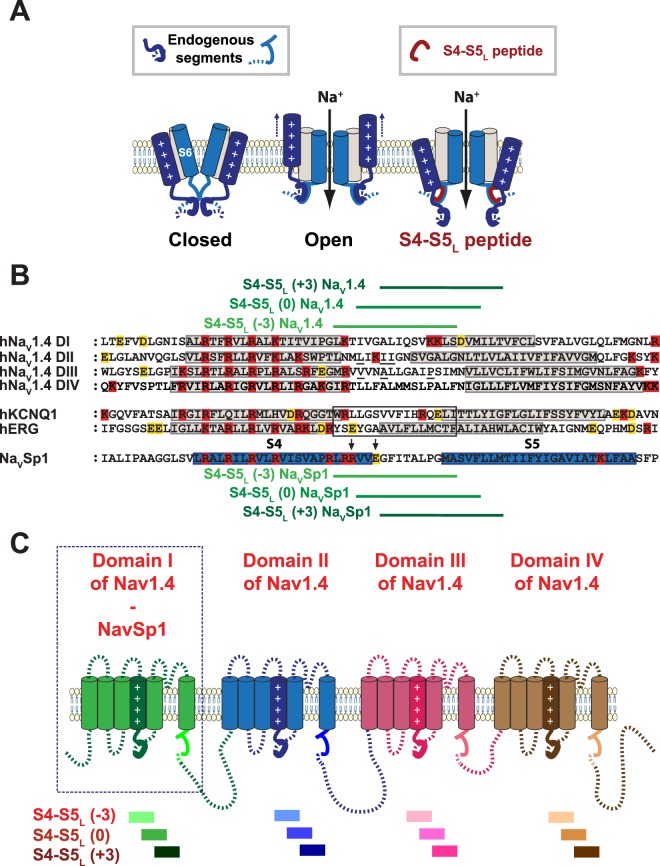
Results
A specific S4-S5L mimicking peptide activates the bacterial channel NaVSp1
First, we tested the ligand/receptor model on NaVSp1, a bacterial channel that is organized as a tetramer of identical subunits. If endogenous S4-S5L acts like a ligand that stabilizes the activation gate in the open state, then a peptide mimicking endogenous S4-S5L should increase NaVSp1 channel activity (Fig. 1A). Three peptides were designed. One peptide, S4-S5L(−3), is aligned with the active peptides for hKV7.125 and hKV11.126 channel (Fig. 1B). Noteworthy, this peptide includes the sequence that aligns with NaVMs RRVVQ motif. This RRVVQ motif engages a series of salt bridge and hydrogen-bonded interactions with S6T and S3, such interactions playing a major role in channel open state stabilization16. Two other NaVSp1 peptides, S4-S5L(0) and S4-S5L(+3) lack this sequence. Each NaVSp1 peptide, S4-S5L(−3), S4-S5L(0) or S4-S5L(+3), was functionally tested separately. One peptide-encoding plasmid was co-transfected with the NaVSp1-encoding plasmid. Results were compared to those from reference cells, co-transfected with NaVSp1 and an unrelated peptide (hKV11.1 S6 C-terminal part, I663-T675, Control 1). An additional negative control, also unrelated to NaV channels (hKV11.1 S4-S5L, A536-F551, Control 2) was used to confirm the absence of the Control 1 peptide effect. In all the following experiments, Control 2 did not show any significant difference, when compared to Control 1.
Figure 1.
Ligand/receptor model. Multiple alignment used to design NaVSp1 and NaV1.4 S4-S5L peptides. (A) scheme of the ligand/receptor model in which S4-S5L (endogenous segment, deep blue) binds to S6T (endogenous segment, light blue) to stabilize the channel in the open state, as suggested by works on NaVMs channel. The S4-S5L peptide (red) mimics endogenous S4-S5L, stabilizing the channel open conformation. (B) Multiple alignment used to design NaVSp1 and hNaV1.4 peptides from previously potent hKV7.1 and hKV11.1 S4-S5L peptides (framed). Starting from S4-S5L(−3) peptide, two others peptides were designed, by shifting toward the C-terminus by 3 (S4-S5L(0)) and 6 amino acids (S4-S5L(+3)). Red: basic residues, yellow: acidic residues. Colored boxes represent the S4 and S5 segments. Mutated residues in skeletal channelopathies are underlined (in NaV1.4 S4-S5L). Arrows point to NaVMs-corresponding residues interacting with S6T (text) C: Scheme of the hNaV1.4 and NaVSp1 channels showing the color used for each peptide/domain.
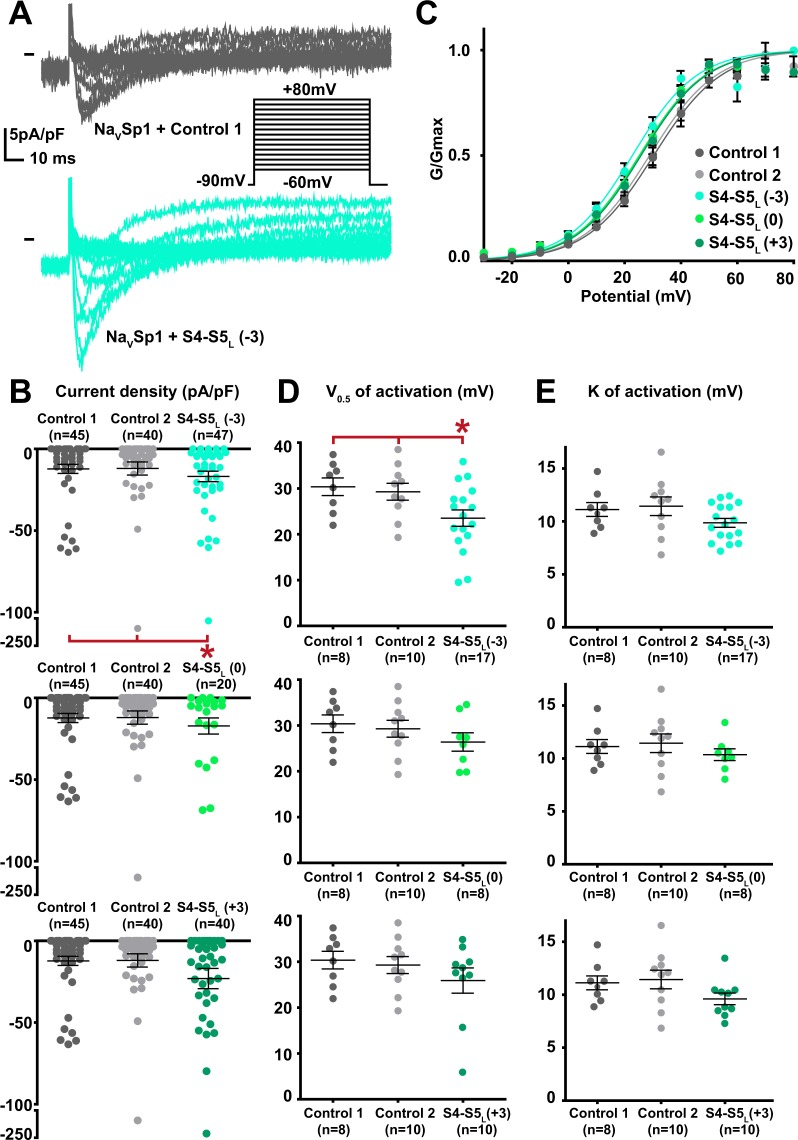
When co-expressed with NaVSp1, the S4-S5L(−3) peptide provoked a gain of function on the current density (Fig. 2, Supplemental Table 1). Moreover, the activation curve was shifted to more negative potential with no concurrent shift of the voltage-dependence of the activation/inactivation kinetics. This latter observation excludes a membrane charge screening, locally changing the potential detected by the voltage-sensor (Supplemental Fig. 1). It is possible that enhancement of the NaVSp1 current density of NaVSp1 at 30 mV stimuli is partially caused by the negative shift of voltage dependent activation. Since a gain of function may also lead to incomplete channel deactivation25, we also tested if the current measured at −90mV, in the presence of S4-S5L peptides, was greater than in ctrl1 and ctrl2 conditions. Our data shows that this is not the case (Supplemental Fig. 2), suggesting that channel deactivation is complete in the presence of peptides. The other two peptides, S4-S5L(0) and S4-S5L(+3) lacking the sequence aligning to the NaVMs RRVVQ motif had no effect. The gain of function, caused by NaVSp1 S4-S5L(−3) peptide suggests that NaVSp1 follows a ligand/receptor model of voltage-dependent gating, with S4-S5L stabilizing the channel in the open state.
Figure 2.
Effect of NaVSp1 S4-S5L mimicking peptides on NaVSp1 current density and activation curve. (A) representative, superimposed current recordings in COS-7 cells transfected with NaVSp1 and control 1 (top trace) or S4-S5L(−3) peptide (bottom trace). Inset: activation voltage protocol used (holding potential: −90 mV; 300-ms pulse at the indicated potentials; one sweep every 5 s). (B) Dot plot and mean ± sem of peak NaVSp1 current densities recorded in COS-7 cells co-transfected with NaVSp1 and the indicated peptide, at 30 mV. (C) Relative peak conductance versus membrane potential curves for NaVSp1 channels in COS-7 cells co-transfected with NaVSp1 and the indicated peptide. Lines are Boltzmann fits to the data. (D,E) Dot plot and mean ± sem of NaVSp1 half-activation potential (V0.5; D) and activation slope (K; E) in COS-7 cells co-transfected with NaVSp1 and the indicated peptide. *p value vs. both controls <0.05.
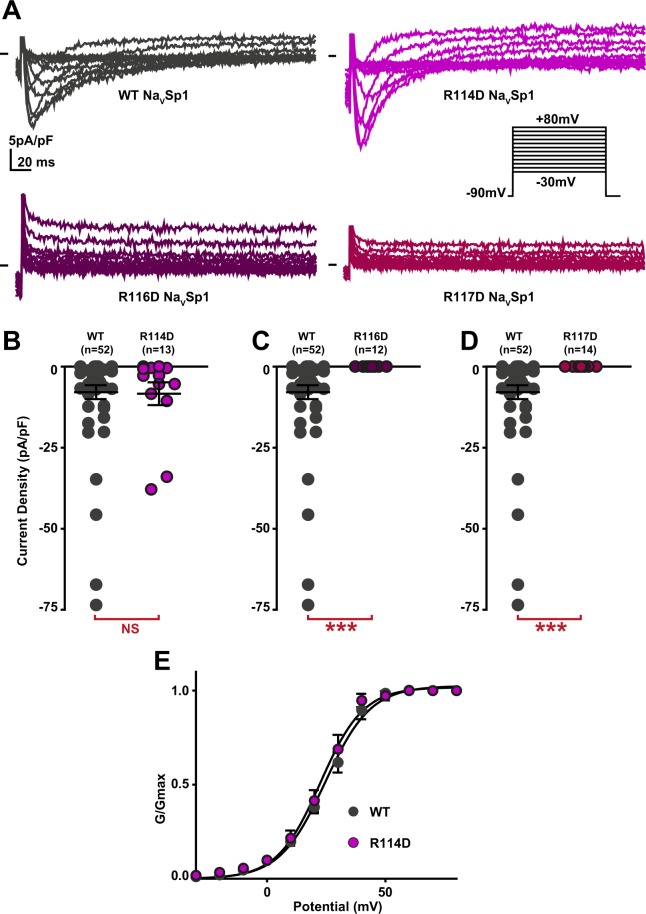
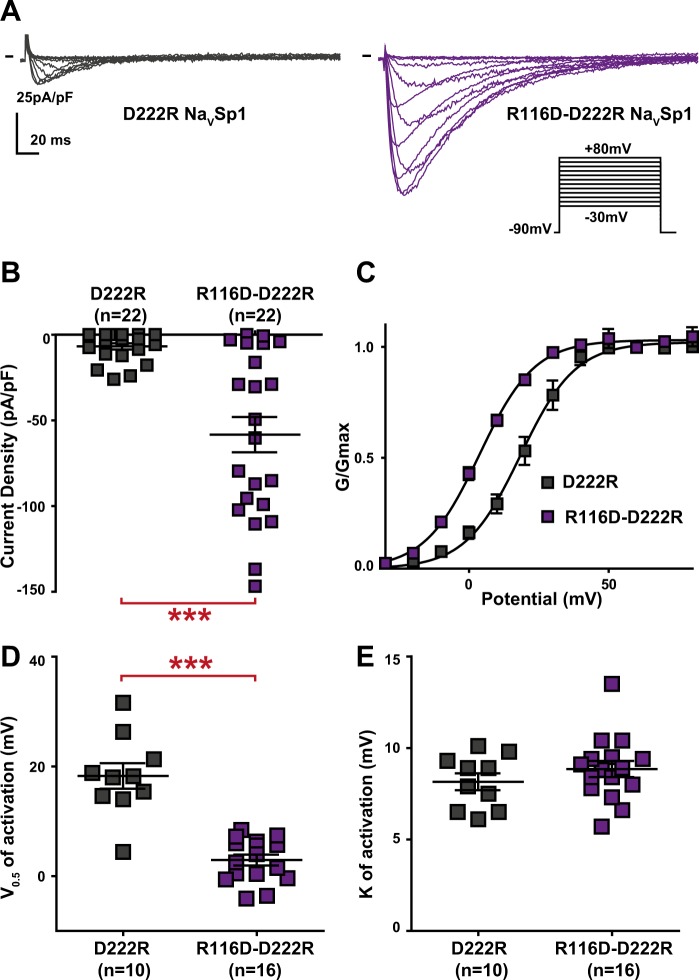
Because the active peptide S4-S5L(−3) contains two arginines (R116 and R117) that are absent in the inactive peptides (S4-S5L(0) and S4-S5L(+3)), we tested the role of these positively charged residues in NaVSp1 gating. Charged amino-acid distribution in S4-S5L and S6T is quite different between NaVSp1 and NaVMs, with an additional arginine at the start of NaVSp1 S4-S5L (R114, before R116 and R117, cf. black arrows pointing to R in Supplemental Fig. 3) and an additional aspartate at position 222 in S6T (black arrow pointing to D in Supplemental Fig. 3). In order to test the potential contribution of R114, R116 and R117 to the stabilization of the open state, we mutated one by one each arginine to an aspartate carrying the opposite charge. R114D did not have any effect on channel activity (Fig. 3A–B,E), but both R116D and R117D led to nonfunctional channels (Fig. 3A,C–D), consistent with a major role of both arginines in NaVSp1 open state stabilization by both the endogenous and exogenous peptides. But because of the absence of detectable current, we could not exclude that R116D and R117D were preventing channel trafficking to the membrane. In order to confirm electrostatic interaction between S4-S5L and S6T to stabilize NaVSp1 open state, we had to identify residues in S6T with which R116 and/or R117 interacts. D222 was, among other residues, a good candidate because it is in a region that aligns with NaVMs residues interacting with S4-S5L to stabilize the channel open state (Supplemental Fig. 3). Interestingly, the addition of the D222R mutation on top of the nonfunctional R116D mutant was not only able to restore channel activity but also led to a channel that is more prone to be open, as compared to when both amino acids at position 116 and 222 carry a positive charge (D222R): we observed an increase in current amplitude and a −20-mV shift in the activation curve (Fig. 4). Such activity restoration and gain of function when amino acids at position 116 and 222 carry opposite charges (R116D + D222R) suggest that both endogenous and exogenous S4-S5L peptides stabilize the channel open state through specific S4-S5L and S6T interaction.
Figure 3.
Effect of charge reversal in amino acids present in NaVSp1 S4-S5L(−3) activating peptide on NaVSp1 current density and activation curve. (A) representative, superimposed recordings of WT and mutant NaVSp1 current. Activation voltage protocol used is the same as in Fig. 2. (B–D) Dot plot and mean ± sem of peak current densities recorded in COS-7 cells transfected with WT or mutant NaVSp1, at 30 mV. (E) Relative peak conductance versus membrane potential curves for WT or mutant NaVSp1 channels transfected in COS-7. Lines are Boltzmann fits to the data. ***p value vs. WT < 0.001.
Figure 4.
Opposite charges at position 116 (NaVSp1 S4-S5L) and 222 (NaVSp1 S6T) stabilizes the NaVSp1 channel open state. (A) representative, superimposed current recordings of single NaVSp1 mutant D222R and double mutant D222R/R116D. Activation voltage protocol used is the same as in Fig. 2. (B) Dot plot and mean ± sem of peak current densities recorded in COS-7 cells transfected with D222R or D222R/R116D NaVSp1, at 30 mV. (C) Relative peak conductance versus membrane potential curves for D222R or D222R/R116D NaVSp1 channels transfected in COS-7 cells. Lines are Boltzmann fits to the data. (D,E) Dot plot and mean ± sem of NaVSp1 half-activation potential (V0.5; D) and activation slope (K; E) in COS-7 cells transfected with D222R or D222R/R116D NaVSp1. ***p value vs. D222R < 0.001.
hNaV1.4 S4-S5L peptides activate hNaV1.4
We also tested the ligand/receptor model on the hNaV1.4 voltage-gated channel that is organized as a single subunit of four homologous domains11,15,20,21. Again, three S4-S5L-encoding plasmids were designed for each domain, based on sequence alignment with hKV7.1 and hKV11.1 (Fig. 1B). Each of the 12 designed S4-S5L peptides was tested separately: each hNaV1.4 S4-S5L peptide-encoding plasmid was co-transfected with hNaV1.4 and hNaVβ1-encoding plasmids.
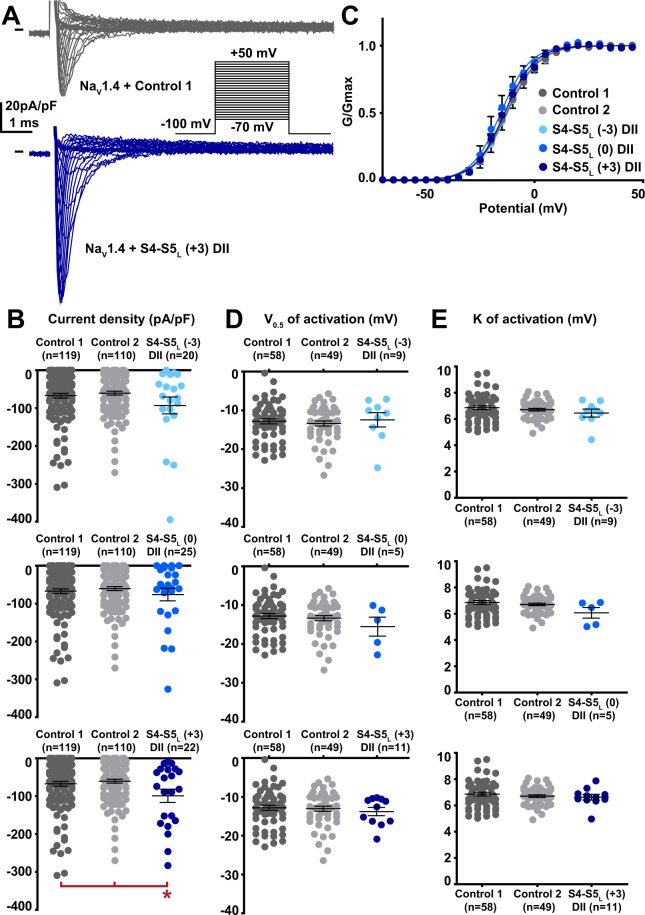
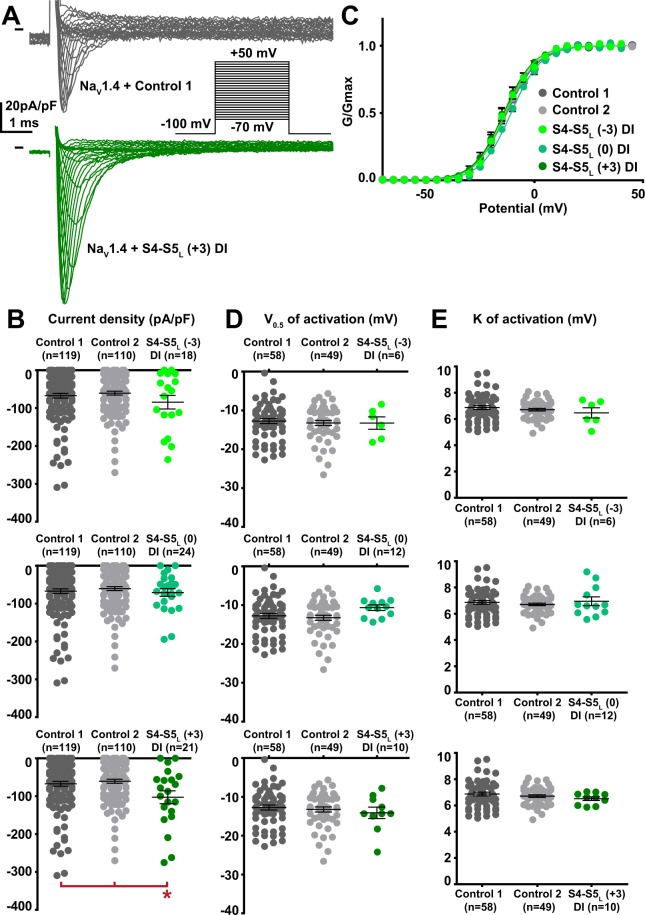
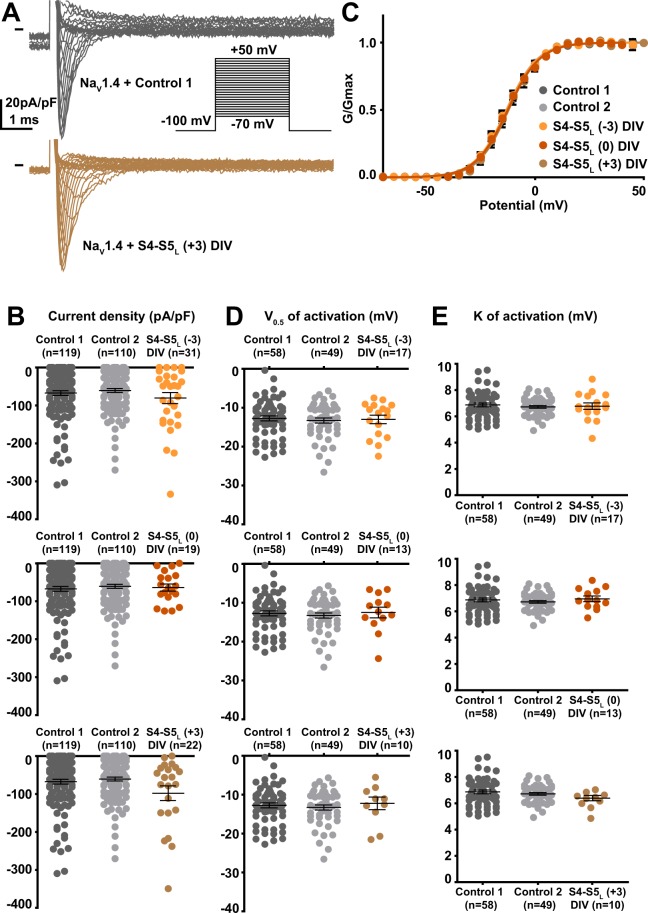
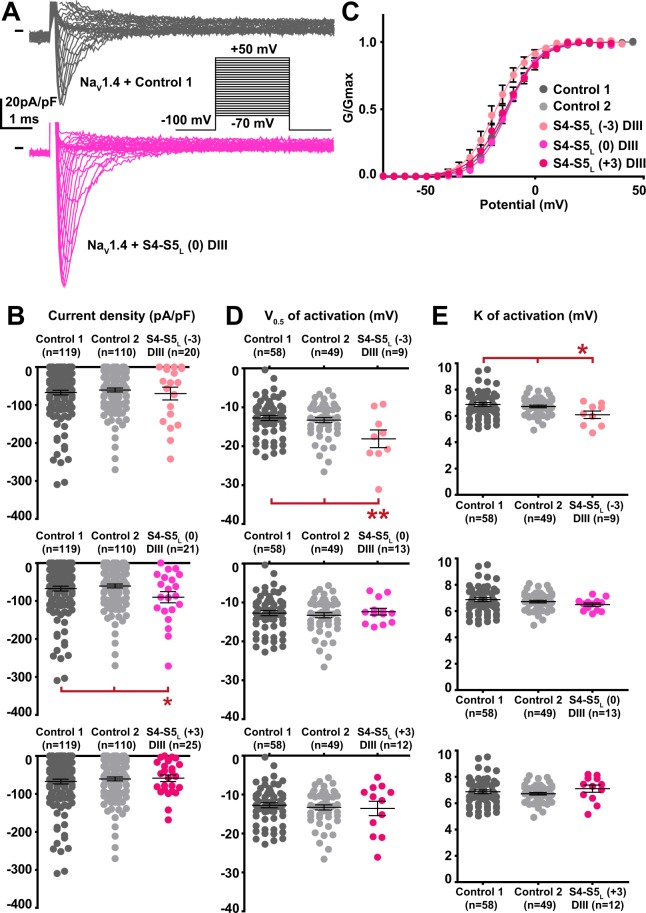
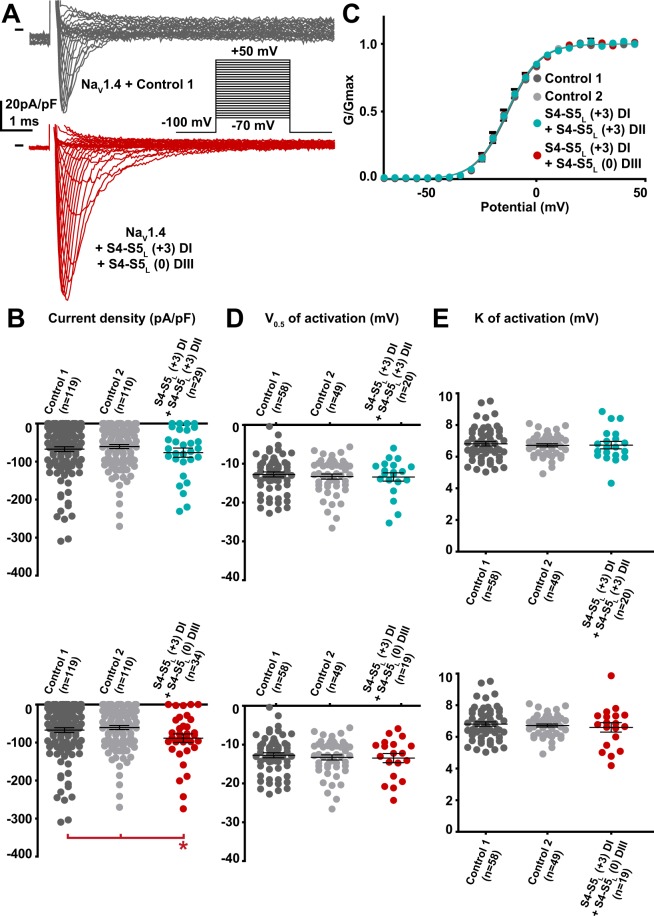
Among the 12 tested hNaV1.4 S4-S5L peptides, three peptides increased the hNaV1.4 current density. These activating peptides mimic three of the four hNaV1.4 S4-S5 linkers, in domain I (S4-S5L(+3)), domain II (S4-S5L(+3)) and domain III (S4-S5L(0)) of hNaV1.4 (Figs. 5B–8B, Supplemental Table 2).
Figure 6.
Effect of NaV1.4 S4-S5L mimicking peptides of domain II on NaV1.4 current density and activation curve. (A) representative, superimposed current recordings in COS-7 cells co-transfected with NaV1.4, NaVß1, and control 1 (top trace) or domain II S4-S5L(+3) peptide (bottom trace). Inset: activation voltage protocol used (holding potential: −100 mV; 30-ms pulse; one sweep every 2 s). (B) Dot plot and mean ± sem of peak NaV1.4 current densities recorded in COS-7 cells co-transfected with NaV1.4, NaVß1, and the indicated peptide, at 0 mV. (C) Relative peak conductance versus membrane potential curves for NaV1.4 channels in the same cell groups as in (B). Lines are Boltzmann fits to the data. (D,E) Dot plot and mean ± sem of NaV1.4 half-activation potential (V0.5; D) and activation slope (K; E) in the same cells group as in (B). *p value vs. both controls <0.05.
Figure 5.
Effect of NaV1.4 S4-S5L mimicking peptides of domain I on NaV1.4 current density and activation curve. (A) representative, superimposed current recordings in COS-7 cells co-transfected with NaV1.4, NaVß1, and control 1 (top trace) or domain I S4-S5L(+3) peptide (bottom trace). Inset: activation voltage protocol used (holding potential: −100 mV; 30-ms pulse; one sweep every 2 s). (B) Dot plot and mean ± sem of peak NaV1.4 current densities recorded in COS-7 cells co-transfected with NaV1.4, NaVß1, and the indicated peptide, at 0 mV. C: Relative peak conductance versus membrane potential curves for NaV1.4 channels in the same cell groups as in (B). Lines are Boltzmann fits to the data. (D,E) Dot plot and mean ± sem of NaV1.4 half-activation potential (V0.5; D) and activation slope (K; E) in the same cells group as in (B). *p value vs. both controls <0.05.
Figure 8.
Effect of NaV1.4 S4-S5L mimicking peptides of domain IV on NaV1.4 current density and activation curve. (A) representative, superimposed current recordings in COS-7 cells co-transfected with NaV1.4, NaVß1, and control 1 (top trace) or domain IV S4-S5L(+3) peptide (bottom trace). Inset: activation voltage protocol used (holding potential: −100 mV; 30-ms pulse; one sweep every 2 s). (B) Dot plot and mean ± sem of peak NaV1.4 current densities recorded in COS-7 cells co-transfected with NaV1.4, NaVß1, and the indicated peptide, at 0 mV. (C) Relative peak conductance versus membrane potential curves for NaV1.4 channels in the same cell groups as in (B). Lines are Boltzmann fits to the data. (D,E) Dot plot and mean ± sem of NaV1.4 half-activation potential (V0.5; D) and activation slope (K; E) in the same cells group as in (B).
One additional peptide in domain III shifted the activation curve to more negative potentials (S4-S5L(−3), Supplemental Table 2; Fig. 7C–E), also leading to a gain of function. This S4-S5L(−3) peptide is different from the S4-S5L(0) peptide that increased the current density in the same domain: it is shifted by three amino acids toward the N-terminus. We did not observe any alteration of the activation/inactivation kinetics by any of the peptides (Supplemental Fig. 4).
Figure 7.
Effect of NaV1.4 S4-S5L mimicking peptides of domain III on NaV1.4 current density and activation curve. (A) representative, superimposed current recordings in COS-7 cells co-transfected with NaV1.4, NaVß1, and control 1 (top trace) or domain III S4-S5L(0) peptide (bottom trace). Inset: activation voltage protocol used (holding potential: −100 mV; 30-ms pulse; one sweep every 2 s). (B) Dot plot and mean ± sem of peak NaV1.4 current densities recorded in COS-7 cells co-transfected with NaV1.4, NaVß1, and the indicated peptide, at 0 mV. C: Relative peak conductance versus membrane potential curves for NaV1.4 channels in the same cell groups as in (B). Lines are Boltzmann fits to the data. (D,E) Dot plot and mean ± sem of NaV1.4 half-activation potential (V0.5; D) and activation slope (K; E) in the same cells group as in (B). *p value vs. both controls <0.05. **p value vs. both controls <0.01.
S4-S5L peptides do not modify hNaV1.4 channel trafficking
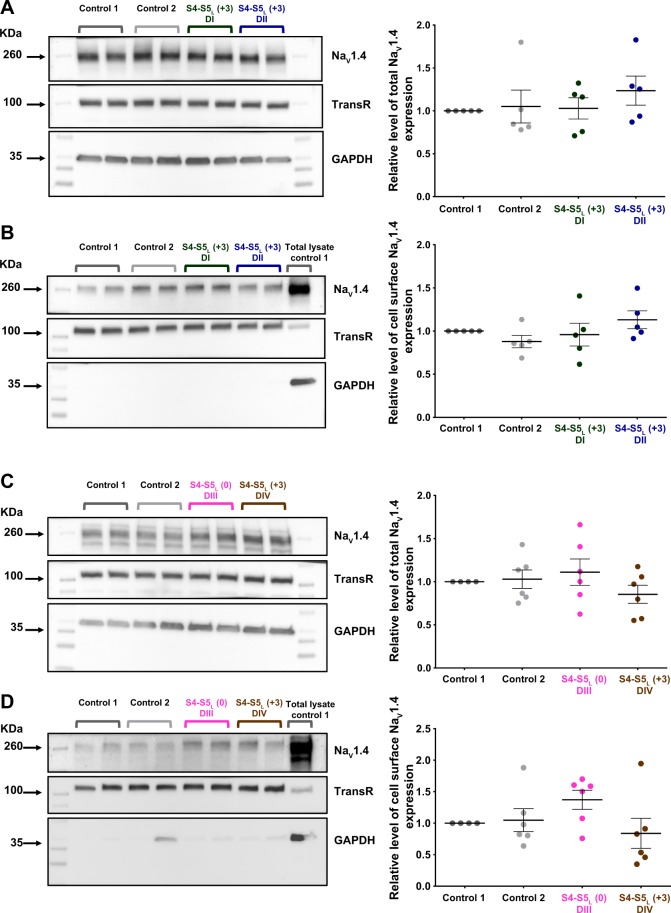
Cell surface biotinylation experiments were performed in order to verify whether increased current densities were due to gating alteration or to an increased channel trafficking. These experiments were done using the three peptides in domain I, II, III of hNaV1.4 that were causing an increase in current density, and using the S4-S5L(+3) peptide in domain IV of hNaV1.4 that was showing a trend of increased current density, although not significant. Neither the total nor the biotinylated fraction (plasma membrane) of hNaV1.4 protein was increased by any of the peptides, suggesting that domain I S4-S5L(+3), domain II S4-S5L(+3) and domain III S4-S5L(0) peptides increase hNaV1.4 current density through an alteration of channel gating and not its trafficking (Fig. 9; Supplemental Fig. 5).
Figure 9.
Effect on NaV1.4 channel expression of S4-S5L mimicking peptides associated with an increased current density. Left: (A,C) representative western blots of total NaV1.4, transferrin receptor (TransR) and GAPDH from transfected COS-7 cells, in the presence of various control and S4-S5L peptides as indicated. (B,D) representative western blots of the cell surface fraction of NaV1.4, transferrin receptor (TransR) and GAPDH from transfected COS-7 cells, in the presence of various control and S4-S5L peptides. Right: corresponding quantifications of normalized mean ± sem intensities. Band intensities are first normalized to the intensity of the corresponding TransR bands, and ratios are then normalized to control 1 condition. In all condition, p > 0.05. In A–D, the three blots, realized on the same membrane, are cropped. Full-length blots of each tested protein are reported in Supplemental Fig. 5.
Thus, out of the 12 tested peptides, four led to a gain of function of hNaV1.4 channel through an effect on channel gating.
Effects of combination of peptides
Since in hNaV1.4 the S4-S5 linker sequences of the four domains differ, we explored if co-transfecting two activating peptides exerts a stronger effect on hNaV1.4 current density than the individual peptides, as two domains will be stabilized open instead of only one. In order to keep the same expression level of the channel, we needed to keep the same total DNA quantity in all conditions. Thus, to combine two peptides we added half quantity of each peptide-encoding plasmid, as compared to conditions with only one peptide. We did not observe any increase in current density when DI-S4-S5L(+3) and DII-S4-S5L(+3) peptides were co-expressed. This observation suggests that combination of the two peptides in lesser quantity was not as potent as when only one peptide was expressed (Fig. 10). It is possible that the presence of (i) smaller quantity of peptides in addition to (ii) some steric hindrance prevent the activating effect. Noteworthy, domains I and II are adjacent, consistent with the hypothetical steric hindrance. To limit the effect of steric hindrance, we selected activating peptides from two non-adjacent domains, namely DI-S4-S5L(+3) and DIII-S4-S5L(0). Indeed, co-expression of these DI-S4-S5L(+3) and DIII-S4-S5L(0) peptides caused an increase in the hNaV1.4 current density. Such an increase was similar but not greater than when only one peptide was expressed, probably because each of the peptides was present in lesser quantity. This observation highlights a limit of the model in which S4-S5 effects are not strong enough to potentially quantify the synergistic effect of the combination of peptides.
Figure 10.
Effect of combination of two NaV1.4 S4-S5L mimicking peptides that both had an effect on NaV1.4 current density when expressed alone. (A) representative, superimposed current recordings in COS-7 cells co-transfected with NaV1.4, NaVß1, and control 1 (top trace) or the combination of domain I S4-S5L(+3) peptide and domain III S4-S5L(0) peptide (bottom trace). Inset: activation voltage protocol used (holding potential: −100 mV; 30-ms pulse; one sweep every 2 s). (B) Dot plot and mean ± sem of peak NaV1.4 current densities recorded in COS-7 cells co-transfected with NaV1.4, NaVß1, and the indicated peptides, at 0 mV. C: Relative peak conductance versus membrane potential curves for NaV1.4 channels in the same cell groups as in (B). Lines are Boltzmann fits to the data. (D,E) Dot plot and mean ± sem of NaV1.4 half-activation potential (V0.5; D) and activation slope (K; E) in the same cells group as in (B). *p value vs. both controls <0.05.
S4-S5L peptides modify hNaV1.4 channel inactivation
Since mutations in domains I, III and IV S4-S5L have been associated with a large modification of the NaV1.4 channel fast inactivation30–32, we also tested the effect of the peptides on channel inactivation. We observed an increase in the slope factor of the inactivation curve when DI-S4-S5L(+3) or DIII-S4-S5L(+3) peptide was expressed (Supplemental Figs. 6–9; Supplemental Table 2). Also, and consistent with the effect of the combination of peptides on the activation curve, effect of DI-S4-S5L(+3) was still observed when it was co-expressed with the peptide corresponding to the non-adjacent domain (III), but not with the peptide corresponding to the adjacent domain (II) (Supplemental Fig. 10).
Discussion
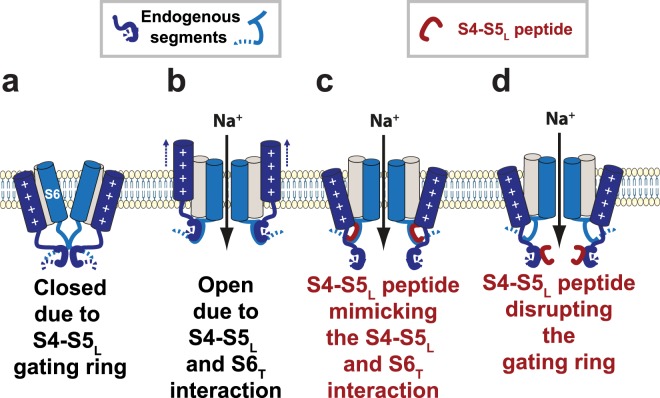
In this work, we used a S4-S5L mimicking peptide approach to test whether voltage-gated sodium channels follow the ligand/receptor model previously proposed for hKV7.125, hKV11.126 and hKV10.229 channels. We identified one activating S4-S5L peptide in NaVSp1 and four in hNaV1.4, suggesting that NaV channels follow a ligand/receptor model of voltage-dependent gating (Fig. 11c): when the membrane is depolarized, endogenous S4-S5L stabilizes the open state of NaV channels, as indicated by the NaVMs structure captured in the open state8,16,17. This contrasts with what is happening with KV channels: when the membrane is polarized, endogenous S4-S5L stabilizes the closed state of KV channels, as suggested by several studies25,26,29.
Figure 11.
Summarizing schemes. (a,b) Two mechanisms of voltage-dependent gating. (a) The gating ring model in which the four S4-S5L (endogenous segments, deep blue, only two are shown) constitute a constriction ring preventing S6 (endogenous segment, light blue) iris-like dilation. (b) Scheme of the ligand/receptor model in which S4-S5L binds to S6T to stabilize the channel in the open state. (c) The S4-S5L peptides (red) mimic the effect of endogenous S4-S5L described in (b), stabilizing the open conformation. (d) Alternatively, the S4-S5L peptides (red), interact with endogenous S4-S5L, destabilizing the gating ring described in (a) and hence lead to channel opening.
The various peptide effects, either on current density or on the activation voltage dependence, suggest that peptides are acting on different conformational transitions leading to channel opening. Due to the multi-state process of channel voltage-dependent gating, implying several conformational changes, peptides affinity may be high enough for the alteration of one parameter (e.g., current density), but not for the alteration of the other one (e.g., V0.5, slope factor). The peptides effects on current density but not on the activation voltage dependence have already been observed in hKV11.1 channel26 and hKV7.1 channels25 and were also described in a kinetic model of the peptide effect on KV10.2 channels29. In hKV11.1 channel, we show that a S6T mimicking peptide has only an effect on the current density when affinity is low but can also drastically change the activation curve when its affinity is increased by a specific disulfide bridge26.
Here, all the data obtained on hNaV1.4 suggest that S4-S5L in domain I, II and III play a significant role in the channel voltage dependence of activation. In the neuronal channel NaV1.2, mutations of S4 gating charges in all four domains were found to affect the activation33. Nevertheless, of the mutations that shifted the V0.5 of activation, the most pronounced effects were observed when the fourth charge in each of domains I, II, and III was neutralized. This suggests that domains I to III play a critical role in coupling the voltage sensor with the activation gate of NaV1.2, consistent with our results on hNaV1.4. Moreover, voltage-clamp fluorimetry experiments performed on NaV1.4 S4 segments showed that domain I, II and III play a significant role in the channel voltage-dependence of activation, also consistent with our results on hNaV1.434.
Although consistent with a ligand/receptor model, S4-S5L peptides effects on NaVSp1 and hNaV1.4 are moderate. It is worth mentioning that these effects are nevertheless in the range of those observed in previous studies on three different voltage-gated potassium channels25,26,29. In the previous studies, we interpreted that the S4-S5L/S6T interaction has to be loose, probably due to the low affinity between native S4-S5L and S6T, which is necessary for S4-S5L ligand unbinding and channel opening during membrane depolarization26. In the channel, this low affinity is compensated by the imposed proximity of the two segments. Experimentally, this can be compensated only partly by high peptide concentration. In two KV channels, we found a way to reinforce S4-S5L peptide binding to S6T via a specific disulfide bridge between two cysteines, and hence, increase their effects on the channel26,29. It would be interesting to identify such a pair of cysteines in NaV channels.
Because of the moderate peptides effects, we cannot be sure that the ligand/receptor is the major actor of signal transduction between S4 movement and pore opening in NaV channels. Other mechanisms such as the constriction ring mechanism, suggested by several NaV channels structures, certainly play a major role10,12,15,20,21. The S4-S5L peptides may also disrupt the constriction ring and by this way lead to channel opening (Fig. 11d). But the fact that NaVSp1 most potent peptides contain amino-acids that play a major role in channel open state stabilization (Figs. 3 and 4), strongly supports the hypothesis that these peptides (endogenous or exogenous peptides) bind to the C-terminal end of S6 and stabilize the channel open state (Fig. 11c).
Both mechanisms (Constriction ring in Fig. 11a and ligand-receptor in Fig. 11b) may coexist: the interaction between S4-S5L and S6T suggested in the closed state (recent work of Wisedchaisri et al. on the bacterial channel NaVAb19) may completely reconfigure in the open state (studies on NavMs8,16,17, and also the present study on NaVSp1 and NaV1.4). Interestingly, S4-S5L residues implicated in the interaction with S6T in the NaVAb closed state (highlighted in Supplemental Fig. 3) do not align with the S4-S5L residues implicated in the interaction with S6T in the NaVMs open state, suggesting the aforementioned reconfiguration of the interaction between S4-S5 linker and S6T. In addition, effects may be moderate because the fast kinetics of NaV channel do not give the S4-S5L peptides enough time to outcompete the endogenous S4-S5L from interacting with S6T.
Regarding peptides effect on hNaV1.4, we took the option to co-express the pore-forming subunit with the auxiliary subunit hNaVß1 to be closer to physiological conditions. Because of the presence of hNaVß1, we cannot exclude that peptides effects on hNaV1.4 are mediated by this subunit. However, (i) the similitude of the peptide effects independently of the domain (I, II or III), (ii) the absence of domain IV peptide effect and (iii) the similitude of hNaV1.4 results to those observed on NaVSp1 channel, lacking auxiliary subunit, suggest that the effect is rather specific to the pore-forming subunit.
From the present and previous studies, it seems that the coupling between voltage sensor movement and pore gating falls into two categories:
the mechanical lever model: an obligatory coupling in which S4 resting state directly translates into S6 gate closed state. This obligatory coupling may be described as a simple mechanical work. At rest, S4-S5 linker helices compress the S6 helices and maintain the pore closed. Upon membrane depolarization, an outward displacement of S4 relieves the compression, allowing pore opening10. This model of electromechanical coupling is likely for Shaker-like channels in which open probability is very close to zero at hyperpolarizing potentials22;
the ligand/receptor model: the obligatory coupling cannot hold if the S6 gate is able to open, even if S4 segments are in the resting state, as shown for hKV11.1 and hKV7.1 channels23,24,35. In this case, coupling is allosteric rather than obligatory: in these KV channels, S4 resting state favors rather than forces channel closing. This allosteric coupling is realized through a ligand/receptor mechanism between S4-S5L and S6T in hKV7.1, hKV11.1 and hKV10.2 channels25,26,29. At rest, S4-S5L binds to S6T and stabilizes the channel in a closed state. If S4-S5L affinity to S6T is low enough, S4-S5L and S6T interaction is not permanent in S4 resting state, allowing transient pore opening. This is consistent with mutations in S4-S5L and S6T increasing the fraction of constitutively active current9,23,35,36.
Together, structural studies and the present study suggest that NaV channels combine both mechanical lever (obligatory) and ligand/receptor (allosteric) models: (i) Structural data of various NaV channel point to an electromechanical (obligatory) coupling model in which the four S4-S5 linkers are organized in a constriction ring that forces the channel gate to close when membrane is polarized. Upon membrane depolarization, movement of the S4 induces a lateral dilation of the S4–S5 linker, leading to a rotation and bending of the pore-lining S6 segments, which ultimately open the activation gate10,12,15,20,21,37. (ii) The present study, associated with studies on NaVMs8,16,17, suggests an allosteric coupling: when S4 segments are in the activated conformation, S4-S5 linkers bind to S6T and stabilize the channel in the open state. Various NaV1.4 structures, all showing S4 in the activated state but showing the S6 gate either closed15 or open11,21, are consistent with this allosteric mechanism in which S4, when activated, is not strongly coupled to S6T, but rather develop interactions with S6T favoring the channel open state.
Many mutations of the hNaV1.4-encoding gene, SCN4A, are linked to muscular channelopathies38. The role of the S4-S5 linker as a modulator of the channel open state is consistent with the identification of several mutations in the area corresponding to the activating peptides: L689F, I692M, I693T in domain II and V1149L, A1152D and P1158S in domain III (underlined in Fig. 1B). Noteworthy, mutations in and around these sites (L689F, I692M, N1151C, A1152C, A1160C, P1158S) impair activation kinetics39–45.
Finally, mimicking peptides engineered for hKV7.1, hKV11.1, hKV10.2 and now hNaV1.4 may lead to a new therapeutic strategy for cardiac, neuronal and muscular channelopathies46.
Methods
Similar methods have been used in previous studies25,26,29.
Plasmid constructs
For NaVSp1 and hNaV1.4, S4-S5L plasmids were designed from the alignment with hKV11.1 and hKV7.1 S4-S5L peptides. First, multiple-sequence alignment was realized with Cobalt47. This program aligned the predicted/observed S4 and S5 transmembrane domains of hKV11.1 (Uniprot Q12809), hKV7.1 (P51787), NaVSp114,48, and the four domains of hNaV1.4 (Uniprot P35499). We designed a NaVSp1 S4-S5L peptide and also a NaV1.4 S4-S5L peptide in each domain, based on the aligned most potent hKV7.1 and hKV11.1 S4-S5L peptides, as shown in Fig. 1B25,26. Since a peptide shifted by three amino acids toward the C-terminus also inhibited hKV7.1 (L251-L266), we also selected the corresponding peptide and also the next one in case of slight differences in binding sites. Thus, for NaVSp1 and each hNaV1.4 domain, three different S4-S5L plasmids were designed. All the peptides had the same length (16 amino-acids). Names of the peptides were given according to their position along the sequence: S4-S5L (−3), S4-S5L (0), and S4-S5L (+3) (Fig. 1B). Two peptides, corresponding to hKv11.1 S4-S5 linker (A536-F551) and hKv11.1 C-terminus of S6 (I663-T675) were used as two negative controls. Oligonucleotides encoding hNaV1.4 and NaVSp1 peptides were synthesized by TOP Gene Technologies and contained a XhoI restriction enzyme, a methionine (ATG) for translation initiation, and a glycine (GGA) to protect the ribosome binding site during translation and the nascent peptide against proteolytic degradation49. A BamHI restriction enzyme site was synthesized at the 3′ end immediately following the translational stop codon (TGA). These oligonucleotides were then cloned into pIRES2-EGFP (Clontech) and sequenced. Mutant NaVSp1 were generated by using the QuikChange site-directed mutagenesis kit (Stratagene).
Cell culture and transfection
The African green monkey kidney-derived cell line, COS-7, was obtained from the American Type Culture Collection (CRL-1651) and cultured in Dulbecco’s modified Eagle’s medium (GIBCO) supplemented with 10% fetal calf serum and antibiotics (100 IU/ml penicillin and 100 µg/ml streptomycin) at 5% CO2 and 95% air, maintained at 37 °C in a humidified incubator. Cells were transfected in 35-mm Petri dishes when the culture reached 50–60% confluence, with DNA (2 µg total DNA) complexed with FuGENE-6 (Roche Molecular Biochemical) according to the standard protocol recommended by the manufacturer. For hNaV1.4 experiments, COS-7 cells were co-transfected with 0.4 µg pRC-hNaV1.4, 0.4 µg pRC-hNaVß1 (kind gifts of AL George, Northwestern University, Feinberg School of Medicine) and 1.2 µg pIRES2-EGFP plasmid (Clontech) encoding control or test peptides. For the experiments with the combination of peptides, COS-7 cells were co-transfected with 0.4 µg pRC-hNaV1.4, 0.4 µg pRC-hNaVß1 and 0.6 µg of each of the two peptides encoding plasmid. For NaVSp1 experiments, COS-7 cells were co-transfected with 0.8 µg pIRES2-EGFP-NaVSp1 in which EGFP was removed and 1.2 µg pIRES2-EGFP plasmid encoding a control or a test peptide. Plasmid quantities were optimized to maximize the quantity of peptides, as assessed by the amount of fluorescence, and to keep current amplitudes in such a range that (i) undetectable currents were rare, and (ii) large currents inducing incorrect voltage-clamp were also rare. In pIRES2-EGFP plasmids, the second cassette (EGFP) is less expressed than the first cassette, guaranteeing high level of peptide expression in fluorescent cells25. For experiments with mutant NaVSp1, COS-7 cells were transfected with 2 µg pIRES2-EGFP-NaVSp1. Cells were re-plated onto 35-mm Petri dishes the day after transfection for patch-clamp experiments.
Electrophysiology
The day after splitting, COS-7 cells were mounted on the stage of an inverted microscope and constantly perfused by a Tyrode solution (cf. below) at a rate of 1–3 ml/min. The bath temperature was maintained at 22.0 ± 2.0 °C. Stimulation and data recording were performed with pClamp 10, an A/D converter (Digidata 1440A) and an Axopatch 200B amplifier (all Molecular Devices). Patch pipettes (tip resistance: 1.5–2.2 MOhms) were pulled from soda lime glass capillaries (Kimble-Chase) and coated with dental wax to decrease capacitive currents. Currents were acquired in the whole-cell configuration, filtered at 10 kHz and recorded at a sampling rate of 20 kHz. Series resistance were compensated to 70–80%. To measure the NaVSp1 currents, from a holding potential of −90 mV, the membrane was depolarized to 30 mV for 300 ms every 5 s. NaVSp1 current was calculated after leak subtraction. To generate the activation curve, from a holding potential of −90 mV, the membrane was depolarized to values between −60 mV and +80 mV (+10 mV increment) for 300 ms, every 5 s. To measure the NaV1.4 current density after complete recovery from inactivation at −100 mV, a single step protocol was used to monitor current increase during recovery. From a holding potential of −100 mV, membrane was depolarized to 0 mV for 30 ms every 2 s. To generate the activation curve, from a holding potential of −100 mV, the membrane was depolarized to values between −70 mV and +50 mV (+5 mV increment) for 30 ms, every 2 s. As for NaVSp1, NaV1.4 current was calculated after leak subtraction. To generate the inactivation curve, from a holding potential of −100 mV, membrane was depolarized to values between −110 mV and +25 mv (+5 mV increment) for 500 ms, followed by a 20-ms test pulse to 0 mV, every 4 s. Activation and inactivation curves were fitted by Boltzmann equations. G/V curves are obtained as follows: GNa was calculated from GNa = INa/(V − Vrev), where INa is the peak sodium current, V is the membrane potential and Vrev is the reversal potential estimated for each cell by linear regression of the linear rectification of I/V curve, when channels are fully activated. GV curves were subsequently obtained by dividing at each potential the peak current by the corresponding value of the linear regression curve.
Solutions
The cells were continuously superfused with a HEPES-buffered Tyrode solution containing (in mmol/L): NaCl 145, KCl 4, MgCl2 1, CaCl2 1, HEPES 5, glucose 5, pH adjusted to 7.4 with NaOH. Patch pipettes were filled with the following solution (in mmol/L): KCl 90, Kgluconate 45, NaCl 10, HEPES 10, pH adjusted to 7.2 with KOH.
Cell surface biotinylation assays
Surface biotinylation of transfected COS-7 cells (same condition as for patch-clamp experiments) was completed as described previously50. Briefly, cells were incubated with 0.5 mg/ml EZ-Link Sulfo-NHS-SS-Biotin (Pierce) in PBS, pH 7.4, for 30 min on ice. The biotinylation reaction was quenched with Tris-saline solution (10 mmol/L Tris, pH 7.4, 120 mmol/L NaCl), and detergent-soluble cell lysates were prepared. Biotinylated cell surface proteins were affinity-purified using Streptavidin-conjugated agarose beads (Pierce), and analyzed by western blot as described previously50. Bands corresponding to hNaV1.4 were normalized to bands corresponding to TransR from the same sample. hNaV1.4 protein expression (total or biotinylated fraction) in cells co-transfected with test peptides is expressed relative to hNaV1.4 protein expression (total or biotinylated fraction) in cells co-transfected with Control 1 peptide-encoding plasmid. Antibodies used were anti-NaVPAN mouse monoclonal antibody (Sigma, S8809), mouse monoclonal antibody against the transferrin receptor (Invitrogen, 13-6890), and a mouse monoclonal antibody against GAPDH (Santa Cruz Biotechnology, sc-32233). Anti‐mouse horseradish peroxidase–conjugated secondary antibody was purchased from Santa Cruz Biotechnology.
Statistics
All data are expressed as mean ± sem. Statistical differences between samples were determined using Student’s t-tests when data were normally distributed (biophysical parameters) and rank-sum tests (Mann Whitney test) when data were not normally distributed (current densities). A value of p < 0.05 versus both controls was considered significant.
Supplementary information
Acknowledgements
We thank Daniel L. Minor, Jr. and Isabelle Baró for careful reading of the manuscript. The project was funded by the Association Française contre les Myopathies - Téléthon (16495), the 7th European Community Framework Programme and the Marie Curie European Actions (PIOF-GA-2011-298280) to Gildas Loussouarn, and the Agence Nationale de la Recherche (ANR-15-CE14-0006-01) to Céline Marionneau. Olfat Malak was laureate of the Line Pomaret-Delalande prize of the Fondation pour la Recherche Médicale (PLP20141031304; FRM). Olfat Malak wishes to personally thank Mrs. Line Pomaret for her generous support. Olfat Malak and Fayal Abderemane Ali were supported by the Fondation Génavie. Fabien Coyan and Fayal Abderemane Ali were recipients of a grant from the French Ministère de la Recherche. We thank Aurore Girardeau and Béatrice Ollivier for their technical support.
Author contributions
O.A.M. carried out the patch-clamp experiments on NaV1.4 activation. O.A.M., F.A.A., Y.W. carried out patch-clamp experiments on NaV1.4 inactivation. F.C.C., F.A.A., Y.W. and G.P carried out preliminary test in patch-clamp experiments. F.A.A. and D.S. carried out molecular biology of NaVSp1 channel. O.A.M. carried out the patch-clamp experiments on NaVSp1. O.A.M. carried out the western blot and biotinylation experiments, under C.M. supervision. O.A.M. analyzed the patch-clamp and biotinylation experiments. O.A.M. prepared the figures. G.L. wrote the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Olfat A. Malak and Fayal Abderemane-Ali.
Supplementary information
is available for this paper at 10.1038/s41598-020-62615-6.
References
- 1.Huang W, Liu M, Yan SF, Yan N. Structure-based assessment of disease-related mutations in human voltage-gated sodium channels. Protein Cell. 2017;8:401–438. doi: 10.1007/s13238-017-0372-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choveau FS, et al. Opposite Effects of the S4-S5 Linker and PIP(2) on Voltage-Gated Channel Function: KCNQ1/KCNE1 and Other Channels. Front. Pharmacol. 2012;3:125. doi: 10.3389/fphar.2012.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arrigoni C, et al. Unfolding of a Temperature-Sensitive Domain Controls Voltage-Gated Channel Activation. Cell. 2016;164:922–936. doi: 10.1016/j.cell.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bagneris C, et al. Role of the C-terminal domain in the structure and function of tetrameric sodium channels. Nat. Commun. 2013;4:2465. doi: 10.1038/ncomms3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bagneris C, Naylor CE, McCusker EC, Wallace BA. Structural model of the open-closed-inactivated cycle of prokaryotic voltage-gated sodium channels. J. Gen. Physiol. 2015;145:5–16. doi: 10.1085/jgp.201411242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferrer T, Rupp J, Piper DR, Tristani-Firouzi M. The S4-S5 linker directly couples voltage sensor movement to the activation gate in the human ether-a’-go-go-related gene (hERG) K+ channel. J. Biol. Chem. 2006;281:12858–12864. doi: 10.1074/jbc.M513518200. [DOI] [PubMed] [Google Scholar]
- 7.Irie K, Shimomura T, Fujiyoshi Y. The C-terminal helical bundle of the tetrameric prokaryotic sodium channel accelerates the inactivation rate. Nat. Commun. 2012;3:793. doi: 10.1038/ncomms1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ke S, Ulmschneider MB, Wallace BA, Ulmschneider JP. Role of the Interaction Motif in Maintaining the Open Gate of an Open Sodium Channel. Biophys. J. 2018;115:1920–1930. doi: 10.1016/j.bpj.2018.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Labro AJ, et al. The S4-S5 linker of KCNQ1 channels forms a structural scaffold with the S6 segment controlling gate closure. J. Biol. Chem. 2011;286:717–725. doi: 10.1074/jbc.M110.146977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Long SB, Campbell EB, Mackinnon R. Voltage sensor of Kv1.2: structural basis of electromechanical coupling. Science. 2005;309:903–908. doi: 10.1126/science.1116270. [DOI] [PubMed] [Google Scholar]
- 11.Pan X, et al. Structure of the human voltage-gated sodium channel Nav1.4 in complex with. Science. 2018;362:beta1. doi: 10.1126/science.aau2486. [DOI] [PubMed] [Google Scholar]
- 12.Payandeh J, Scheuer T, Zheng N, Catterall WA. The crystal structure of a voltage-gated sodium channel. Nature. 2011;475:353–358. doi: 10.1038/nature10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanguinetti MC, Xu QP. Mutations of the S4-S5 linker alter activation properties of HERG potassium channels expressed in Xenopus oocytes. J. Physiol. 1999;514(Pt 3):667–675. doi: 10.1111/j.1469-7793.1999.667ad.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shaya D, et al. Structure of a prokaryotic sodium channel pore reveals essential gating elements and an outer ion binding site common to eukaryotic channels. J. Mol. Biol. 2014;426:467–483. doi: 10.1016/j.jmb.2013.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen,H. et al. Structure of a eukaryotic voltage-gated sodium channel at near-atomic resolution. Science 355 (2017). [DOI] [PubMed]
- 16.Sula A, et al. The complete structure of an activated open sodium channel. Nat. Commun. 2017;8:14205. doi: 10.1038/ncomms14205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sula A, Wallace BA. Interpreting the functional role of a novel interaction motif in prokaryotic sodium channels. J. Gen. Physiol. 2017;149:613–622. doi: 10.1085/jgp.201611740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tristani-Firouzi M, Chen J, Sanguinetti MC. Interactions between S4-S5 linker and S6 transmembrane domain modulate gating of HERG K+ channels. J. Biol. Chem. 2002;277:18994–19000. doi: 10.1074/jbc.M200410200. [DOI] [PubMed] [Google Scholar]
- 19.Wisedchaisri G, et al. Resting-State Structure and Gating Mechanism of a Voltage-Gated Sodium Channel. Cell. 2019;178:993–1003. doi: 10.1016/j.cell.2019.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu H, et al. Structural Basis of Nav1.7 Inhibition by a Gating-Modifier Spider Toxin. Cell. 2019;176:702–715. doi: 10.1016/j.cell.2018.12.018. [DOI] [PubMed] [Google Scholar]
- 21.Yan Z, et al. Structure of the Nav1.4-beta1 Complex from Electric Eel. Cell. 2017;170:470–482. doi: 10.1016/j.cell.2017.06.039. [DOI] [PubMed] [Google Scholar]
- 22.Lu Z, Klem AM, Ramu Y. Coupling between voltage sensors and activation gate in voltage-gated K+ channels. J. Gen. Physiol. 2002;120:663–676. doi: 10.1085/jgp.20028696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma LJ, Ohmert I, Vardanyan V. Allosteric features of KCNQ1 gating revealed by alanine scanning mutagenesis. Biophys. J. 2011;100:885–894. doi: 10.1016/j.bpj.2010.12.3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vardanyan V, Pongs O. Coupling of voltage-sensors to the channel pore: a comparative view. Front. Pharmacol. 2012;3:145. doi: 10.3389/fphar.2012.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choveau FS, et al. KCNQ1 channels voltage dependence through a voltage-dependent binding of the S4-S5 linker to the pore domain. J. Biol. Chem. 2011;286:707–716. doi: 10.1074/jbc.M110.146324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malak OA, Es-Salah-Lamoureux Z, Loussouarn G. hERG S4-S5 linker acts as a voltage-dependent ligand that binds to the activation gate and locks it in a closed state. Sci. Rep. 2017;7:113. doi: 10.1038/s41598-017-00155-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Enkvetchakul D, Loussouarn G, Makhina E, Nichols CG. ATP interaction with the open state of the K(ATP) channel. Biophys. J. 2001;80:719–728. doi: 10.1016/S0006-3495(01)76051-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loussouarn G, et al. Phosphatidylinositol-4,5-bisphosphate, PIP2, controls KCNQ1/KCNE1 voltage-gated potassium channels: a functional homology between voltage-gated and inward rectifier K+ channels. EMBO J. 2003;22:5412–5421. doi: 10.1093/emboj/cdg526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malak OA, et al. Voltage-dependent activation in EAG channels follows a ligand-receptor rather than a mechanical-lever mechanism. J. Biol. Chem. 2019;294:6506–6521. doi: 10.1074/jbc.RA119.007626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Popa MO, Alekov AK, Bail S, Lehmann-Horn F, Lerche H. Cooperative effect of S4-S5 loops in domains D3 and D4 on fast inactivation of the Na+ channel. J. Physiol. 2004;561:39–51. doi: 10.1113/jphysiol.2004.065912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Desaphy JF, et al. Translational approach to address therapy in myotonia permanens due to a new SCN4A mutation. Neurology. 2016;86:2100–2108. doi: 10.1212/WNL.0000000000002721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsujino A, et al. Myasthenic syndrome caused by mutation of the SCN4A sodium channel. Proc. Natl. Acad. Sci. USA. 2003;100:7377–7382. doi: 10.1073/pnas.1230273100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kontis KJ, Rounaghi A, Goldin AL. Sodium channel activation gating is affected by substitutions of voltage sensor positive charges in all four domains. J. Gen. Physiol. 1997;110:391–401. doi: 10.1085/jgp.110.4.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chanda B, Bezanilla F. Tracking voltage-dependent conformational changes in skeletal muscle sodium channel during activation. J. Gen. Physiol. 2002;120:629–645. doi: 10.1085/jgp.20028679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Osteen JD, et al. Allosteric gating mechanism underlies the flexible gating of KCNQ1 potassium channels. Proc. Natl. Acad. Sci. USA. 2012;109:7103–7108. doi: 10.1073/pnas.1201582109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boulet IR, Labro AJ, Raes AL, Snyders DJ. Role of the S6 C-terminus in KCNQ1 channel gating. J. Physiol. 2007;585:325–337. doi: 10.1113/jphysiol.2007.145813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lenaeus MJ, et al. Structures of closed and open states of a voltage-gated sodium channel. Proc. Natl. Acad. Sci. USA. 2017;114:E3051–E3060. doi: 10.1073/pnas.1700761114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loussouarn G, et al. Physiological and Pathophysiological Insights of Nav1.4 and Nav1.5 Comparison. Front. Pharmacol. 2015;6:314. doi: 10.3389/fphar.2015.00314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fan C, Mao N, Lehmann-Horn F, Burmann J, Jurkat-Rott K. Effects of S906T polymorphism on the severity of a novel borderline mutation I692M in Nav 1.4 cause periodic paralysis. Clin. Genet. 2017;91:859–867. doi: 10.1111/cge.12880. [DOI] [PubMed] [Google Scholar]
- 40.Trip J, et al. In tandem analysis of CLCN1 and SCN4A greatly enhances mutation detection in families with non-dystrophic myotonia. Eur. J. Hum. Genet. 2008;16:921–929. doi: 10.1038/ejhg.2008.39. [DOI] [PubMed] [Google Scholar]
- 41.Yoshinaga H, et al. Phenotypic variability in childhood of skeletal muscle sodium channelopathies. Pediatr. Neurol. 2015;52:504–508. doi: 10.1016/j.pediatrneurol.2015.01.014. [DOI] [PubMed] [Google Scholar]
- 42.Bouhours M, et al. A1152D mutation of the Na+ channel causes paramyotonia congenita and emphasizes the role of DIII/S4-S5 linker in fast inactivation. J. Physiol. 2005;565:415–427. doi: 10.1113/jphysiol.2004.081018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Popa MO, Alekov AK, Bail S, Lehmann-Horn F, Lerche H. Cooperative effect of S4-S5 loops in domains D3 and D4 on fast inactivation of the Na+ channel. J. Physiol. 2004;561:39–51. doi: 10.1113/jphysiol.2004.065912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ghovanloo MR, Abdelsayed M, Peters CH, Ruben PC. A Mixed Periodic Paralysis & Myotonia Mutant, P1158S, Imparts pH-Sensitivity in Skeletal Muscle Voltage-gated Sodium Channels. Sci. Rep. 2018;8:6304. doi: 10.1038/s41598-018-24719-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Plassart-Schiess E, Lhuillier L, George AL, Jr., Fontaine B, Tabti N. Functional expression of the Ile693Thr Na+ channel mutation associated with paramyotonia congenita in a human cell line. J. Physiol. 1998;507(Pt 3):721–727. doi: 10.1111/j.1469-7793.1998.721bs.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kadam RU, et al. Potent peptidic fusion inhibitors of influenza virus. Science. 2017;358:496–502. doi: 10.1126/science.aan0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Papadopoulos JS, Agarwala R. COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics. 2007;23:1073–1079. doi: 10.1093/bioinformatics/btm076. [DOI] [PubMed] [Google Scholar]
- 48.D’Avanzo N, et al. Differential lipid dependence of the function of bacterial sodium channels. PLoS. One. 2013;8:e61216. doi: 10.1371/journal.pone.0061216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gilchrist A, Li A, Hamm HE. G alpha COOH-terminal minigene vectors dissect heterotrimeric G protein signaling. Sci. STKE. 2002;2002:l1. doi: 10.1126/stke.2002.118.pl1. [DOI] [PubMed] [Google Scholar]
- 50.Marionneau C, et al. The sodium channel accessory subunit Navbeta1 regulates neuronal excitability through modulation of repolarizing voltage-gated K(+) channels. J. Neurosci. 2012;32:5716–5727. doi: 10.1523/JNEUROSCI.6450-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.