Abstract
The increasing incidence of carbapenemase-producing K. pneumoniae strains (CP-Kps) in the last decade has become a serious global healthcare problem. Therapeutic options for the treatment of emerging hospital clones have drastically narrowed and therefore novel approaches must be considered. Here we have isolated and characterized a lytic bacteriophage, named vB_KpnS_Kp13, that was effective against all Verona integron-encoded metallo-β-lactamase (VIM) producing K. pneumoniae isolates originating from hospital samples (urine, blood, sputum and faeces), belonging to the ST15 clonal lineage and expressing the K24 capsule. Morphological characterization of vB_KpnS_Kp13 showed that the newly identified phage belonged to the Siphoviridae family, and phylogenetic analysis showed that it is part of a distinct clade of the Tunavirinae subfamily. Functional analysis revealed that vB_KpnS_Kp13 had relatively short latent period times (18 minutes) compared to other K. pneumoniae bacteriophages and could degrade biofilm by more than 50% and 70% in 24 and 48 hours respectively. Complete in vivo rescue potential of the new phage was revealed in an intraperitoneal mouse model where phages were administered intraperitoneally 10 minutes after bacterial challenge. Our findings could potentially be used to develop specific anti-CP-Kps bacteriophage-based therapeutic strategies against major clonal lineages and serotypes.
Subject terms: Bacteriophages, Bacterial infection
Introduction
Klebsiella pneumoniae is one of the most important opportunistic nosocomial pathogens, able to colonize the skin and mucosae of humans and cause urinary tract infections, septicaemia and pneumonia, predominantly in immunocompromised individuals1,2. In hospitals, colonization leads to patients becoming reservoirs and a source of cross infection for other patients3.
Dissemination of antibiotic resistance, particularly to carbapenem, is associated with highly diverse isolates of K. pneumoniae. In this process the importance of certain clonal groups (CGs) such as CG258 and CG15 has been demonstrated. These CGs contain several high-risk international clones including ST11, ST258 and ST154,5. Although recent studies have not described ST15 as a widespread clonal lineage6, other data indicates that ST15 is a pan-drug resistant widespread clone, frequently associated with carbapenemase production7 and nosocomial infections. ST15 was recently identified in the USA8, Brazil9, China10,11, Taiwan12, and several European countries13 including Portugal14, Belgium15, Bulgaria16 and Hungary4. Some epidemiological surveys suggested that the K24 is a frequent capsular type linked to ST159,17,18.
One potential option to combat these pathogens is to use bacteriophages as they have well defined target spectrums, characterized by host range specificity19. This feature makes them suitable therapeutic agents without influencing the normal microbiota of the patient20. Certain bacteriophages also have the capacity to target and destroy defined capsular types and weaken biofilm typically produced by Klebsiella pneumoniae21.
The most important virulence factors of K. pneumoniae are the capsular polysaccharides (CPS) and lipopolysaccharides (LPS) typically used for K-, and O-serotyping respectively. Over 80 capsular types have been defined, including 77 types from reference strains identified between 1926 and 1977 using serological tests, and 5 new types (KN1 to KN5) characterized by molecular genotyping and structural analyses in recent years22–24.
Here we isolated and characterized a lytic bacteriophage (vB_KpnS_Kp13), which was effective against all the carbapenemase-producing Klebsiella pneumoniae strains (CP-Kps) originating from the same teaching hospital and belonging to ST15. Phage vB_KpnS_Kp13 was specific for K24 capsular type. The K24 serotype was recently reported to be frequently associated with ST159,17,18 and frequently reported from other CP-Kps25. Our results may aid the development of anti CP-Kps bacteriophage-based therapeutic strategies targeting major clonal lineages.
Results
Morphology, host range and efficiency of plating (EOP)
Altogether 13 bacteriophages were isolated from wastewater against K. pneumoniae hospital isolates. Four of them were effective against all Klebsiella pneumoniae isolates with the K24 capsule. Based on their restriction profiles (EcoRI and HindIII; data not shown) one proved to be distinct from the other three and this single one always showed a clear lytic zone (around 10 mm diameter) without the emergence of phage-resistant colonies and showed a characteristic wide halo (~ 4 mm) (Supplementary Fig. 1). This phage was vB_KpnS_Kp13 and characterized in this study.
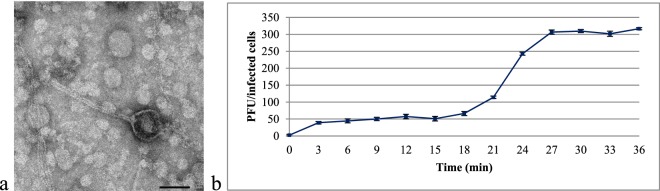
Transmission electron microscopy (TEM) analysis indicated that phage vB_KpnS_Kp13 comprised a ~60 nm diameter head and a flexible, non-contractile, ~200 nm tail (Fig. 1a). Based on these traits, phage vB_KpnS_Kp13 was classified as Caudovirales in the Siphoviridae family26.
Figure 1.
Characteristics of vB_KpnS_Kp13. (a) Electron micrograph of phage vB_KpnS_Kp13 shows the typical features of the Siphoviridae family. Phage was stained with 1.5% w/v phospho-tungstic acid. Scale bar represents 50 nm. (b) One-step growth curve of phage vB_KpnS_Kp13. The plaque forming unit (PFU) per infected cell at different times are shown. Data are the mean of 3 independent experiments. Error bars represent standard deviation.
Host range tests with 89 Klebsiella spp. isolates showed that phage vB_KpnS_Kp13 was only effective on K. pneumoniae isolates with the K24 capsule, causing lysis in 100% (40/40) of these strains (Table 1). Other Klebsiella spp. isolates with either known (K1, K2, K3, K6, K9, K11, K12, K13, K17, K19, K20, K21, K24, K27, K30, K33, K47, K52 and K64) or unknown, but non-K24 capsular types were not sensitive to vB_KpnS_Kp13.
Table 1.
Host range of vB_KpnS_Kp13 tested on Klebsiella spp. isolates.
| Bacterial strain | Number of isolates | Sequence type | K-serotype | Lysis | Reference/Source |
|---|---|---|---|---|---|
| Klebsiella pneumoniae subsp. pneumoniae | 1 | ST23 | K1 | − | NTUH-K2044 |
| Klebsiella pneumoniae subsp. pneumoniae | 1 | ST66 | K2 | − | CIP 52.145* |
| Klebsiella pneumoniae subsp. rhinoscleromatis | 1 | ND | K3 | − | CIP 80.51* |
| Klebsiella quasipneumoniae | 1 | ST489 | K6 | − | ATCC 700603 |
| Klebsiella pneumoniae subsp. pneumoniae | 1 | ND | K9 | − | CIP 52.207* |
| Klebsiella pneumoniae subsp. pneumoniae | 1 | ND | K11 | − | CIP 52.215* |
| Klebsiella pneumoniae subsp. pneumoniae | 1 | ND | K12 | − | CIP 52.216* |
| Klebsiella pneumoniae subsp. pneumoniae | 1 | ND | K13 | − | CIP 52.217* |
| Klebsiella pneumoniae subsp. pneumoniae | 1 | ND | K17 | − | CIP 52.221* |
| Klebsiella pneumoniae subsp. pneumoniae | 1 | ND | K19 | − | CIP 52.223* |
| Klebsiella pneumoniae subsp. pneumoniae | 1 | ND | K20 | − | CIP 52.224* |
| Klebsiella pneumoniae subsp. pneumoniae | 1 | ND | K21 | − | CIP 52.225* |
| Klebsiella pneumoniae subsp. pneumoniae | 1 | ST59 | K24 | + | CIP 52.229* |
| Klebsiella pneumoniae subsp. pneumoniae | 39 | ST15 | K24 | + | Clinical isolates, CCH** |
| Klebsiella pneumoniae | 30 | ND | not K24 | − | Clinical isolates, own collection |
| Klebsiella pneumoniae subsp. pneumoniae | 1 | ND | K27 | − | CIP 52.232* |
| Klebsiella pneumoniae subsp. pneumoniae | 1 | ND | K30 | − | CIP 52.235* |
| Klebsiella pneumoniae subsp. pneumoniae | 1 | ND | K33 | − | CIP 53.8* |
| Klebsiella pneumoniae subsp. pneumoniae | 1 | ND | K47 | − | CIP 53.23* |
| Klebsiella pneumoniae subsp. pneumoniae | 1 | ST38 | K52 | − | MGH 78578 |
| Klebsiella pneumoniae subsp. pneumoniae | 1 | ND | K64 | − | CIP 80.47* |
Details of each bacterial strains are reported (+: clear plaque formation; −: no plaque formation; ND: not defined).
*Strains with CIP prefixes were purchased from the Institut Pasteur (France).
**CCH: Countess of Chester Hospital, Department of Microbiology, Chester, Cheshire, UK.
The EOP analysis revealed a high productive (EOP ≥ 0.5) infection of phage vB_KpnS_Kp13 for 37 of the 40 (92.5%) sensitive strains. Medium production occurred in 3 of the 40 (7.5%) strains sensitive to vB_KpnS_Kp13 in the double-layer agar (DLA) assay (Supplementary Table 1).
One-step phage growth experiments – burst-size determination
The one-step growth experiment was performed to determine the latent time period and burst size of the phage. A triphasic curve was obtained showing the latent period, log or rise period, and plateau period. The phage vB_KpnS_Kp13 showed relatively short latency period (18 min) followed by a rise period of 10 min and a growth plateau starting at 27 min (Fig. 1b). The burst size of vB_KpnS_Kp13 was ~220 phage particles per infected bacteria.
Killing dynamic of vB_KpnS_Kp13 against K. pneumoniae 533 strain and phage-resistant clone screening
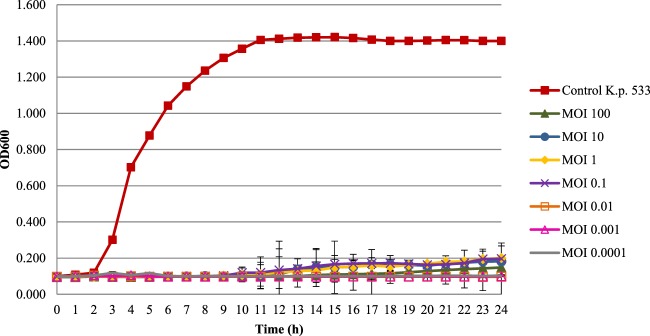
Phage vB_KpnS_Kp13 inhibited the proliferation of K. pneumoniae 533 in a concentration dependent manner. At multiplicity of infection (MOI) between 0.0001 and 100, phage vB_KpnS_Kp13 effectively inhibited the growth of K. pneumoniae 533 in LB broth for 24 h (Fig. 2).
Figure 2.
Time-kill assay of phage vB_KpnS_Kp13 against K. pneumoniae 533. Overnight bacterial culture was diluted (105 CFU/ml) and infected with phage vB_KpnS_Kp13 at MOI of 100 (green, ▲), at MOI of 10 (blue, ●), at MOI of 1 (yellow, ◆), at MOI of 0.1 (purple, ×), at MOI of 0.01 (orange, □), at MOI of 0.001 (pink, Δ) and at MOI of 0.0001 (grey). Non-phage treated bacterial cultures (control K.p. 533, red, ■) were used as a positive control. Optical density at 600 nm was measured every hour up to 24 h. Each data point is the mean from 3 experiments. Standard deviations (±SD) are shown as vertical lines.
Several spot testing of the phage vB_KpnS_Kp13 on the lawn of different Klebsiella pneumoniae isolates, indicated that no resistant bacterium clones emerged against the newly isolated phage. In the liquid culture screening at different MOIs, we could not isolate phage-resistant clones.
Biofilm degradation assay and Confocal Laser Scanning Microscopy (CLSM) analysis
The K. pneumoniae 533 isolate is a strong biofilm producer (mean OD = 4.4 ± 0.1) based on the formerly established OD cut-off (ODc) values27. Here, pre-established biofilms were degraded by phage vB_KpnS_Kp13 in a MOI and time dependent manner. A 51.8% loss in biomass was detected after 2 h incubation at MOI 10. Biomass reduction was further increased to 54.2%, 57.5%, and 72.9% after 12, 24 and 48 h respectively if compared to the OD values of the untreated biofilm control. These values were also reduced if the applied phage:bacterium ratios were 1:1 (MOI 1) and 1:10 (MOI 0.1), but still led to marked biofilm degradation (Fig. 3).
Figure 3.
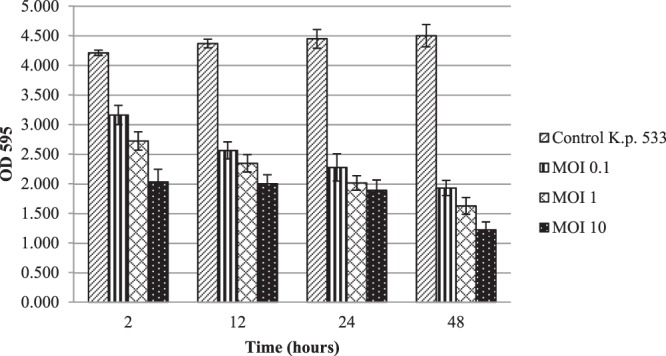
Effect of vB_KpnS_Kp13 on K. pneumoniae 533-produced biofilm degradation. The established biofilm was infected with vB_KpnS_Kp13 at MOI of 0.1 (striped), at MOI of 1 (gridded) and MOI of 10 (spotted). Non-phage treated biofilm (side striped) was used as a positive control. OD at 595 nm was measured at 2, 12, 24 and 48 h. Each data point is the mean from 3 experiments. Error bars represent standard deviation.
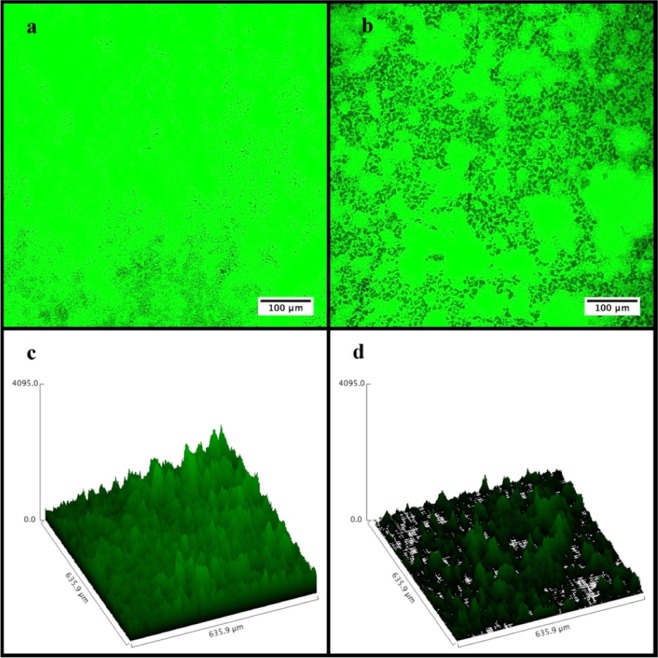
Biofilm degradation efficacy of phage vB_KpnS_Kp13 was also analysed with Confocal Laser Scanning Microscopy (CLSM). CLSM images showed that phage vB_KpnS_Kp13 firmly reduced the biomass and thickness of the biofilm architecture (Fig. 4a,b). Confocal micrographs show the non-infected control (only bacterium, no phage) displayed a highly structured matrix formation (Fig. 4c). The phage vB_KpnS_Kp13-treated biofilm showed disintegrated clumps with scattered microflora that resulted in the collapse of the pre-formed biofilm (Fig. 4d). The structural difference in the control biofilm and phage-treated biofilm, showed the efficacy of phage vB_KpnS_Kp13 on biofilm degradation. The biomass of the control K. pneumoniae 533 biofilm was 13.12 μm3/μm2, in contrast to the vB_KpnS_Kp13 treated biofilm where this value was 4.76 μm3/μm2 (Supplementary Table 2).
Figure 4.
Visualization of Klebsiella pneumoniae 533 biofilm by CLSM. (a) Control (only bacterium, no phage). (b) Treated with phage vB_KpnS_Kp13. Scale bar represents 100 µm. Surface plot was generated using CLSM microscopy software (Olympus FV1000). (c) Control K. pneumoniae 533 biofilm surface plot. (d) Phage vB_KpnS_Kp13-treated 533 biofilm surface plot.
Genome characteristics
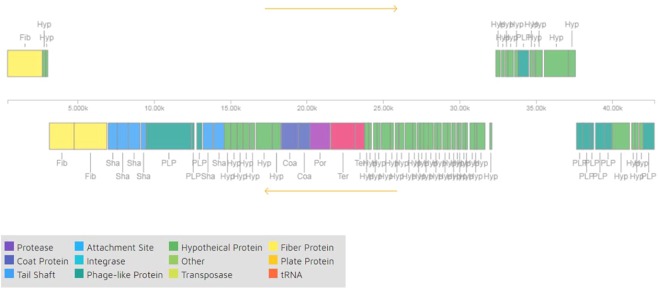
The phage vB_KpnS_Kp13 genome measured 43,094 bp (accession number MK170446) with a 50.6% G + C content. It contains 75 predicted open reading frames (ORFs), while no tRNA gene was predicted by GeneMark program. The orientation of genome annotation showed that 61 genes are on the minus strand and 14 are on the plus strand. A linear genome map of phage vB_KpnS_Kp13 was obtained from a PHASTER analysis (Fig. 5).
Figure 5.
Linear genomic map of vB_KpnS_Kp13 obtained with PHASTER. Different functional groups of phage vB_KpnS_Kp13 are denoted by different colors, according to their function.
The ORF analysis (GeneMark) of the complete vB_KpnS_Kp13 genome revealed 5 major functional clusters21, hypothetically responsible for coding determinants of (i) DNA replication/modification/transcriptional regulations: DNA N-6-adenine methyltransferase (ORF72), ssDNA-binding protein (ORF1), DNA terminase (large subunit = ORF24 and small subunit = ORF 25), (ii) structure and packaging: portal protein (ORF23), major capsid protein (ORF21), membrane protein (ORF32 and ORF73) and head morphogenesis protein (ORF22), (iii) host lysis: holin (ORF65), endolysin (ORF64), (iv) tail structure: tail fiber proteins (ORF4, ORF5 and ORF6), tail assembly (ORF10) and tail length tap measure proteins (ORF9) and (v) hypothetical or unknown functions.
The hypothetical capsule depolymerase gene, responsible for the degradation of the K24 capsule, was encoded by ORF2 (2289 bp), that shared different rates of amino-acid sequence similarities to other known phage depolymerases: Klebsiella phage KOX1 (77.67%), Klebsiella phage JY917 (75.23%), Klebsiella phage KP36 (68.08%), Klebsiella phage GH-K3 (66.54%), Klebsiella phage KLPN1 (58.13%), Klebsiella phage Sushi (46.32%) and Klebsiella phage K5-2 (31.82%).
Phylogenetic relations
A phylogenetic analysis of the complete vB_KpnS_Kp13 genome was performed using VICTOR web service. It was found that the Klebsiella JY917 phage is the closest relative of vB_KpnS_Kp13 with 90% similarity (Fig. 6). The Genome BLAST Distance Phylogeny (GBDP) tree inferred using the formula D4 and yielding an average support of 3%. This data is consistent with the obtained and current International Committee on Taxonomy of Viruses (ICTV) taxonomy, confirming that vB_KpnS_Kp13 forms a distinct clade of the Tunavirinae subfamily and Kp36virus genus.
Figure 6.
Whole genome based phylogenetic relations of phage vB_KpnS_Kp13 generated with VICTOR. The bacteriophages and their accession numbers are: MezzoGao (MF612072.1), KpCol1 (MG552615.1), N141 (MF415412.1), Sushi (KT001920.1), TAH8 (MH633484.1), NJS3 (MH633486.1), NJS2 (MH633485.1), NJS1 (MH445453.1), 1513 (KP658157.1), KP36 (JF501022.1), KPK126 (KR269719.1), KLPN1 (KR262148.1), JY917 (MG894052.1), TSK1 (MH688453) and vB_KpnS_Kp13 (MK170446).
Mouse intraperitoneal (IP) phage rescue model
The therapeutic potential of vB_KpnS_Kp13 was revealed in an intraperitoneal (IP) mouse model where the effect of vB_KpnS_Kp13 against K. pneumoniae 533 was strongly dependent on the time that passed between the bacterial infection and phage administration.
In the positive control group, 8 mice were challenged with 2 × 108 CFU of K. pneumoniae 533 in suspension per mouse, and all of them died within 24 h (LD50 = 12 h). No effect was observed when the phage suspension (1.75 × 108 PFU/mice) was administered to uninfected mice (negative control group, n = 8). Phage administration within 10 min of bacterial challenge prevented death in all mice (n = 8), and by this assuring 100% survival without showing any signs of infection. This therapeutic effect was not observed if the phage suspension was administered 1 h after bacterial challenge. In this case, the survival rate was still 100% at 24 h, but the general condition of the mice drastically deteriorated, and their survival decreased to 12.5% (1/8 mice) 48 h after infection. No rescuing effect of vB_KpnS_Kp13 was detected if it was administered 3 h after bacterial challenge (Fig. 7). Ten days after bacterial challenge and subsequent phage administrations, surviving mice were sacrificed and active phage particles were isolated from the lungs, spleen and blood. No active phage particles were detected.
Figure 7.
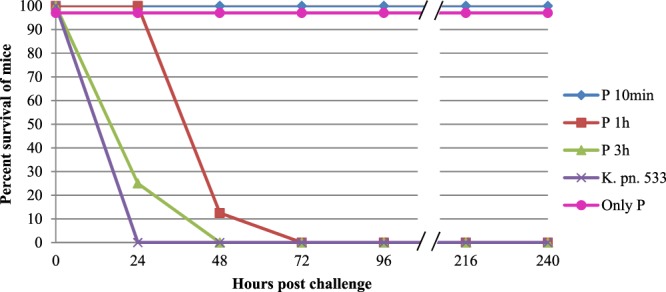
Therapeutic effect of vB_KpnS_Kp13 in an intraperitoneal (IP) mouse infection model. Phage administration occurred 10 min (P 10 min, blue, ♦), 1 hour (P 1 h, red, ■) and 3 hours (P 3 h, green, ▲) post-infection, while the K. pneumoniae 533 (K. pn. 533, purple, ×) and the phage vB_KpnS_Kp13 (only P, pink, ●) suspensions were used as positive and negative controls, respectively. The graphs show percent survival of mice from the start of IP infection over 240 h. Each data point is the mean from two experiments (altogether 8 mice in one group). Error bars represent standard deviation. For better visibility we artificially lowered the values of the phage control (only P, pink, ●) from 100% to 97%.
Discussion
The appearance and dissemination of carbapenem-resistant Klebsiella pneumoniae isolates in hospital environments is one of the biggest challenges of today’s healthcare system. Therapeutic options are limited, with the aim being to treat colonized patients and remove endemic strains from the hospital environment. Due to their high specificity and antibacterial features, bacteriophages are promising candidates for these tasks25.
The antibacterial potential of the newly isolated phage vB_KpnS_Kp13 against the clonally disseminated, VIM carbapenemase-producing Klebsiella pneumoniae strains (CP-Kps), possessing the K24 capsule in an English Tertiary Hospital demonstrated the viability of phage vB_KpnS_Kp13 as a possible alternative method to manage this local problem.
Phage vB_KpnS_Kp13 was characterized by several favourable and unique features. Notably, no phage-resistant bacterium clones emerged in this study, a problem that can lead to therapeutic failure during phage therapy28. DNA N-6-adenine methyl transferase, encoded by ORF72 (729 bp), might be a useful feature of phage vB_KpnS_Kp13 since it could protect the injected phage DNA against bacterial nucleases29. It is unknown if this gene is functional in vB_KpnS_Kp13, thereby supporting phage infection. Similar methyltransferases have been described from the Sushi (98%), Kp1513 (98%) and JY917 (97%) K. pneumoniae phages, but no data were available concerning the resistance ratios against these phages. The other crucial aspect of the phage action is the receptor itself. Although we currently have no data about the receptor of vB_KpnS_Kp13, lack of resistance against this bacteriophage strongly suggests that target of this phage contributes to survival of strain 533. Due to its seemingly specific action on K24 capsular type K. pneumoniae isolates, it was reasonable to question if K24 was the only receptor of vB_KpnS_Kp13 or if another surface associated structure was also necessary. Recent studies showed that capsule itself could be the receptor itself30. In contrast spontaneous capsule mutants of K. pneumoniae 533 remained sensitive to vB_KpnS_Kp13. Receptor structure of vB_KpnS_Kp13 is still under investigation, but similarly to other previously reported K. pneumoniae bacteriophages KP3431 and KP 151332, our phage also showed a narrow host range. Therefore, it could be applied in phage therapy in combination with other bacteriophages or to treat local spread of K24 K. pneumoniae.
Inability of vB_KpnS_Kp13 to lyse and/or weaken the lawn of other non-K24 serotype K. pneumoniae strains (Table 1) referred to the specific feature of the capsule depolymerase encoded by ORF2 of vB_KpnS_Kp13. Only one study focuses on the functional analysis of a K24 specific phage33. Beyond that another bacteriophage (K5-2) was recently reported as lytic against a K5 serotype K. pneumoniae strain by forming a characteristic halo and showing different rates of lytic activity against 7 other serotypes including K24 and K3034. Lack of marked sequence homology on both genomic and protein level and – in contrast to K5-2 – inability of vB_KpnS_Kp13 to weaken K30 (Table 1) suggest the different origins of the two capsule depolymerases of K5-2 and vB_KpnS_Kp13. The K24 depolymerase of vB_KpnS_Kp13 was highly specific with limited homologies to other known capsule depolymerases.
The latent period time (18 min) of phage vB_KpnS_Kp13 was longer that of Klebsiella phage φBO1E (Podoviridae; 10 min)35. The burst size of vB_KpnS_Kp13 (~220 phage particles/infected cell) was not extremely high compared with two phages in the Siphoviridae36 and Podoviridae35 families, but they assured an effective bactericidal potential as vB_KpnS_Kp13 could prevent proliferation of its target bacterium from an MOI of 0.01. Compared to phage TSK121, this value was lower by 2 orders of magnitude and therefore was more effective. There is no relevant efficacy studies published in vivo or in vitro using K. pneumoniae phages such as JY917 (accession MG894052.1), MezzoGao (accession MF612072.1), Sushi (accession KT001920.1), NJS1 (accession MH445453.1), 1513 (accession KP658157.1), KP36 (accession JF501022.1), and NJR15 (accession MH633487.1). vB_KpnS_Kp13 has shown its characteristic antibacterial potential without the emergence of resistant mutants during the one-week test periods.
To best of our knowledge this is the first described phage active against K24 producing K. pneumoniae strains. A characteristic biofilm degradation potential was also visualized by the classical assay and Confocal Laser Scanning Microscopy (CLSM). Few works detailing biofilm degradation are available, but similarly to phage Z36 and TSK121, the phage vB_KpnS_Kp13 showed drastically degraded biofilm, reducing the biomass by ~73% 48 h post-treatment. Analysis of biofilm formation is essential as it is an important survival strategy of bacteria on biotic and abiotic surfaces, potentially inhibiting antibiotic efficacy and protecting bacteria from host responses21. The biofilm disruptor capacity of vB_KpnS_Kp13 could solve this problem in the future.
Rescue experiments with vB_KpnS_Kp13 showed the potency of this phage in vivo. Survival of 100% of the mice, if treatment occurred 10 min after bacterial challenge, indicated the efficacy of vB_KpnS_Kp13 in vivo. Failure of the 1 h treatment suggested that 533 gained advantage and by this vB_KpnS_Kp13 treatment could only delay the death of the experimental animals. These results were comparable with the relevant (MOI 1) results of a recent study37 where however a slowly escalating wound infection model was investigated using the K2 serotype K. pneumoniae strain B5055 and the lytic phage Kpn5. Effective therapeutic potential was also demonstrated with phage SS38 but reduction of colonization of the K. pneumoniae strain B5055 in the lungs could only be reached if MOI 100 was applied. From that point the efficacy of vB_KpnS_Kp13 was revealed in such a mouse model where bacterial challenge caused the death of infected mice in 24 hours without phage treatment.
Conclusion
Phage vB_KpnS_Kp13 was effective against the locally disseminated Klebsiella pneumoniae isolates, possessing the K24 capsule type and belonging to the ST15 clonal linage. As this phage targeted a high-risk international clone our results may aid the development of anti-CP-Kps bacteriophage-based therapeutic strategies targeting major clonal lineages.
Materials and methods
Bacterial strains and growth conditions
The VIM-producing carbapenem-resistant clinical Klebsiella pneumoniae 533 isolate belonging to the ST15 clonal lineage and expressing the K24 capsule was used for bacteriophage isolation and for phage propagation. All ST15 K24 strains were isolated from the Microbiology Laboratories of the University Hospital in Chester, England. Samples from urine, blood, sputum and faeces were collected from different wards (Supplementary Table 3).
For phage host range testing an additional 88 K. pneumoniae and 1 K. quasipneumoniae isolates were used (Supplementary Table 3). All bacterial strains were grown under aerobic conditions in Luria-Bertani (LB) broth or agar (1.5%) at 37 °C.
Phage isolation and purification
Klebsiella pneumoniae-specific bacteriophages were isolated from sewage samples from the local wastewater treatment station (Pellérd, Hungary). Briefly, 10 ml sewage was added to 200 ml log phase K. pneumoniae 533 culture and incubated at 37 °C for 24 h shaking at 120 RPM. Residual bacterial cells were removed with centrifugation (5000 RPM, 8 min) and supernatant was sterile filtered (0.2 µm pore size filter; Sarstedt, Germany). The phage suspensions were serially diluted in phosphate-buffered saline (PBS) and purified in two consecutive steps by using the double-layer agar (DLA) technique39 with 0.4% agar. The resulting high titre (109 PFU/ml) phage stocks were stored at +4 °C and used for further characterization.
Transmission electron microscopy (TEM)
One drop from the purified high titre (109 PFU/ml) phage stock was deposited onto formvar-coated copper grids (Pelco Grids, Canada) and negatively stained with 1.5% w/v phospho-tungstic acid (Merck, Germany). Phages were examined using a JEM-1400 Flash TEM (JEOL USA Inc.) transmission electron microscope operated at 120 kV acceleration voltage. Based on their morphology, phages were classified according to the guidelines of the International Committee on Taxonomy of Viruses (ICTV)40.
Phage host range testing and K24 specific PCR
Spot testing41 of the purified phage on the lawn (108 CFU/plate) of different Klebsiella spp. isolates belonging to different clonal lineages and possessing known or unknown capsular types (Supplementary Table 3) were used to estimate host range of the newly isolated bacteriophage. Lysis characteristics were evaluated after 18 h incubation at 37 °C. Genetic determinant of the K24 capsular type was revealed with the following primer pair: K24 Fw: 5′ AGATAATAGGCAACAGCGTTCT 3′ and K24 Rev: 5′ GATACGTTAAACGCCTCAAGTA 3′. Primers were self-designed based on the available sequence data of the wzi gene of the K. pneumoniae strain 533. These primers were specific to the gene of K24 capsular type and targeted a tyrosine-protein kinase. Validation for positivity was performed on the K24 reference strain CIP 52.229 (Institut Pasteur), and on the strain 533, the sequence of which was confirmed to possess the genetic determinant for the K24 capsule (http://kaptive.holtlab.net/). On the other hand 18 non-K24 capsular type possessing K. pneumoniae strains were used as negative controls (Table 1). Conditions of the PCR were as follows: initial denaturation, 95 °C for 2 min; denaturation: 95 °C for 30 s; annealing at 62 °C for 40 s; elongation 72 °C for 1 min; 34 cycles. At the end a one-step termination was applied at 72 °C for 10 min. Primers were evaluated on K. pneumoniae 533 and another three − K24 possessing − K. pneumoniae strains.
Efficiency of plating (EOP)
All K. pneumoniae isolates sensitive to phage vB_KpnS_Kp13 in the spot test (n = 40) were selected for the determination of EOP as described previously41, with some modifications. All bacterial isolates (n = 40) to be tested were grown overnight (18 h) at 37 °C and 100 µl of each culture was used in DLA assay39 with 100 µl of diluted phage vB_KpnS_Kp13 lysate. The phage lysate was 106 times diluted from the phage stock. The plates were incubated overnight (18 h) at 37 °C and the number of plaque forming units (PFU) was enumerated. Finally, the EOP was calculated (average PFU on target bacteria / average PFU on host bacteria) along with the standard deviation (± SD) for the three measurements.
The average EOP value for a phage-bacterium ratio was classified according to Mirzaei and Nilsson41: highly productive (EOP ≥ 0.5), medium productive (0.1 ≤ EOP < 0.5), low productive (0.001 <EOP < 0.1) or inefficient (EOP ≤ 0.001). The assay was performed in triplicate and results are reported as the mean of three observations (Supplementary Table 1).
One-step phage growth curve
One-step growth experiments for determination of latent period, log or rise period, plateau phase and burst size was carried out according to a previously published protocol42, with some modifications. Five ml of exponential-growth-phase culture of K. pneumoniae 533 (108 CFU/ml) and 50 µl of phage suspension (107 PFU/ml) were mixed (MOI 0.1). Phages were allowed to adsorb for 10 min at 37 °C in a shaking incubator (120 RPM), and then the mixture was centrifuged twice at 12,000 RPM for 2 min in order to eliminate the non-adsorbed phages. The pellet was then suspended in 5 ml of fresh LB medium and incubated at 37 °C in a shaking incubator (120 RPM). 200 µl samples were taken at 3 min intervals between 0- and 36-min. Phage titres were estimated by spotting their dilutions on top agar (0.4%) using the DLA method39 and incubated overnight (18 h) at 37 °C. The latency period was defined as the time between infection and the shortest incubation time allowing the production of phages43. The burst size was calculated as the ratio between the number of phage particles released at the plateau level and the initial number of infected bacterial cells. Experiments were performed three times.
Killing efficacy of vB_KpnS_Kp13 against K. pneumoniae 533 strain and phage-resistant clone screening
Lytic ability of vB_KpnS_Kp13 in different MOIs was demonstrated on exponential-growth-phase cultures of K. pneumoniae 533 (108 CFU/ml). A 96-well tissue culture plate (Sarstedt, Germany) was partitioned into 4 parallel sections, each containing 3 columns. Sections 1 and 2 contained 105 CFU/ml (180 μl) of K. pneumoniae 533. LB broth without bacteria was used as negative control. Inoculated bacteria without phages served as positive control. All other rows contained 20 μl of bacteriophage suspensions from the phage dilutions of 107 (MOI 100), 106 (MOI 10), 105 (MOI 1), 104 (MOI 0.1), 103 (MOI 0.01), 102 (MOI 0.001) and 10 (MOI 0.0001). Cultures were incubated at 37 °C for 24 h shaking at 180 RPM. Bacterial growth was monitored in a Synergy HT multimode reader (BioTek, Winooski, VT). OD600 was measured every 5 min44. Experiments were performed in three replicates and reproduced in three independent trials.
The ratio of phage-resistant mutant colonies was assessed by spot testing and liquid culture screening. Phage-resistant clones was assessed on 39 clinical K. pneumoniae isolates from the Microbiology Laboratories of the University Hospital in Chester (Supplementary Table 3), through standard spot test41. Briefly, 10 µl of the phage dilutions (102, 104, 106) were added to the lawn of K. pneumoniae isolates to estimate the phage resistant clones. Following overnight incubation (18 h) at 37 °C, plates were observed for a clear spot in the bacterial lawn. In liquid culture, 3 ml of exponential-growth-phase culture of K. pneumoniae 533 (108 CFU/ml) and 3 ml of phage suspensions were mixed at different MOIs (MOI 100, MOI 10, MOI 1, MOI 0.1, MOI 0.01, MOI 0.001 and MOI 0.0001). Cultures were incubated at 37 °C for 7 days, shaking at 120 RPM. 200 µl samples were taken at 24 h intervals. Phage titres and phage resistant isolates were monitored by spot testing.
Biofilm degradation assay and visualization with CLSM
Biofilm degradation activity of vB_KpnS_Kp13 on the K. pneumoniae 533 biofilm was determined using the traditional biofilm assay with slight modifications45. Aliquots (200 µl) of K. pneumoniae 533 (2 × 106 CFU/ml) were incubated (24 h, 37 °C) in 96-well polystyrene microtitre plates (Sarstedt, Germany). Unattached planktonic cells were carefully removed. Using different MOIs (0.1, 1 and 10) 1 day after biofilm establishment, 200 μl aliquots of vB_KpnS_Kp13 diluted in LB broth were added to each well for 2, 12, 24 and 48 h. LB broth without bacteria was used as a negative control, while inoculated bacteria without phage served as a positive control. The biomass of preformed biofilm was quantified with 1% w/v crystal violet (Sigma, USA) staining for 20 min at RT46. After removing the excess dye with PBS, the crystal violet was solubilized in 1% w/v SDS. The absorbance was measured using an ELISA reader (BioRad, USA) at OD595. Experiments were performed in triplicate and reproduced in three independent trials.
Structure of K. pneumoniae 533 biofilms with or without vB_KpnS_Kp13 biofilm matrix was observed by Confocal Laser Scanning Microscope (CLSM) FV1000 (Olympus Corporation, Japan). The protocol was adapted from a previous publication47. Biofilm grew on glass slide pieces (1 × 1 cm) placed in 24-well polystyrene plates (Sarstedt, Germany) at 37 °C for 48 h. The glass pieces were washed with PBS, stained with 0.1% acridine orange (Sigma-Aldrich, Germany) and inspected under CLSM. The biofilm was then infected with vB_KpnS_Kp13 and further incubated for 24 h. The biofilm stack images of slides with vB_KpnS_Kp13 and control biofilm slides without phage infection were analyzed separately using COMSTAT software (MATLAB 5.1, The MathWorks Inc., Natick, MA). The thickness (µm) of the biofilm and biovolume (µm3) of bacteria per µm2 were recorded.
Phage genome sequence determination and analysis
Phage DNA was isolated from phage stocks with a concentration ≥109 PFU/ml. Phage DNA was extracted and purified from phage lysate using a QIAGEN Lambda Midi Kit (QIAGEN Inc., CA, USA) and following the manufacturer’s protocol. At the end of the extraction process, DNA samples were dissolved in 100 μl of sterile nuclease free H2O. Genomic DNA sequencing libraries were prepared using the Nextera XT Library Preparation kit (Illumina, California, USA). Sequencing was performed using MiSeq Reagent Kit v2 (2 × 150 bp) on an Illumina MiSeq (Illumina, California, USA). Assembly of the pure sequence was performed with the MyPro pipeline48. The genome was annotated using the RAST server49. After that, the qualified sequence reads were represented using CLC Sequence Viewer v.8 (CLC Bio). Homology searches were conducted with the BLAST tools available at NCBI website (https://www.ncbi.nlm.nih.gov/blast). Identification of open reading frames (ORFs) and gene predictions were used the GeneMark program50.
Genome map was illustrated with PHASTER (PHAge Search Tool Enhanced Release, www.phaster.ca).
Function of protein coding sequences (CDSs) in whole phage genome, was predicted by using the UniProtKB database (https://genome.ucsc.edu).
The nucleotide sequence of vB_KpnS_Kp13 was deposited in the GenBank database under the accession number MK170446.
Phylogenetic analysis
Whole genome based phylogenetic analysis was conducted with VICTOR51. All pairwise comparisons of the nucleotide sequence were conducted using the Genome-BLAST Distance Phylogeny (GBDP) method, under settings recommended for prokaryotic viruses. Branch support was inferred from 100 pseudo-bootstrap replicates each. Tree was rooted at the midpoint and visualized with FigTree52. Taxon boundaries at the species, genus and family level were estimated with the OPTSIL program with the recommended clustering thresholds and F value (fraction of links required for cluster fusion) of 0.551.
Rescue experiments in an intraperitoneal (IP) mouse model of infection
For in vivo rescue tests, 6–7-week-old (18–21 g) female BALB/c mice (Charles and Rivers, Hungary) were used.
After phage propagation bacterial cells were pelleted with centrifugation. Supernatants were sterile filtered. Phage suspensions were centrifuged (15,000 g, 30 min) and washed 2-times in PBS. As a result, phages became 30x concentrated. PFUs were determined and diluted in order to reach the required 7 × 108 PFU/ml. K. pneumoniae suspensions were established from logarithmic cultures. After centrifugation, cells were washed 3-times in PBS and the optical densities were set to OD600 = 10 (1 × 109 CFU/ml).
In vivo experiments were performed as previously described53 with some modifications. Animals were cared for in accordance with the guidelines of the European Federation for Laboratory Animal Science Associations (FELASA) and all procedures, care and handling of the animals were approved by the Animal Welfare Committee of University of Pécs (Permit Number: BA02/2000-37/2015). Five groups (4 mice per group) were used. Four groups were injected intraperitoneally (IP) with 200 µl of K. pneumoniae 533 at OD600 = 10 (2 × 108 CFU/mice). One group served as a negative or bacteriophage control, receiving 250 µl phage suspensions (1.75 × 108 PFU/mice). The positive control group (only bacterium, no phage) received only 200 µl OD600 = 10 bacteria (2 × 108 CFU/mice), while 250 µl bacteriophage suspensions (1.75 × 108 PFU/ mice) were administered to all other groups 10 min, 1 h and 3 h post-infection. General conditions and survival rates of the mice were monitored for 10 days. After 10 days the mice were euthanised by cervical dislocation, lungs and spleen were aseptically removed, and tissue homogenates were subjected to phage count using DLA technique. K. pneumoniae 533 was also measured from blood by spot testing. Experiments were performed twice.
Accession number
The nucleotide sequence of vB_KpnS_Kp13 was deposited in GenBank database under accession number MK170446.
Supplementary information
Acknowledgements
The authors wish to thank Dr. Zoltán Tigyi for the hospital-derived K. pneumoniae isolates. We also thank the Medical School of University of Pécs for supporting our work (KA-2019-01).
Author contributions
M. Horváth and Gy. Schneider were involved in the experimental design. M. Horváth performed basic phage characterization (host range, growth experiments, killing dynamic, classical biofilm degradation assay, phylogenetic analysis and mouse IP model), summarized the data and wrote the manuscript. T. Kovács performed the sequencing and the bioinformatic analysis. S. Koderivalappil and G. Rákhely performed the biofilm degradation assay with the CLSM. H. Ábrahám performed the TEM analysis. Gy. Schneider conceived and coordinated the whole work.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-62691-8.
References
- 1.Vuotto C, Longo F, Balice MP, Donelli G, Varaldo PE. Antibiotic resistance related to biofilm formation in Klebsiella pneumoniae. Pathogens. 2014;3:743–758. doi: 10.3390/pathogens3030743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Podschun R, Ullmann U. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clinical Microbiology Reviews. 1998;11:589–603. doi: 10.1128/CMR.11.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford PA. Extended-spectrum β-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clinical Microbiology Reviews. 2001;14:933–951. doi: 10.1128/CMR.14.4.933-951.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Damjanova I, et al. Expansion and countrywide dissemination of ST11, ST15 and ST147 ciprofloxacin-resistant CTX-M-15-type β-lactamase-producing Klebsiella pneumoniae epidemic clones in Hungary in 2005 – the new ‘MRSAs’? Journal of Antimicrobial Chemotherapy. 2008;62:978–985. doi: 10.1093/jac/dkn287. [DOI] [PubMed] [Google Scholar]
- 5.Ewers C, et al. Clonal spread of highly successful ST15-CTX-M-15 Klebsiella pneumoniae in companion animals and horses. Journal of Antimicrobial Chemotherapy. 2014;69:2676–2680. doi: 10.1093/jac/dku217. [DOI] [PubMed] [Google Scholar]
- 6.Lee C-R, et al. Global dissemination of carbapenamase-producing Klebsiella pneumoniae: Epidemiology, genetic context, treatment options, and detection methods. Frontiers in Microbiology. 2016;7:895. doi: 10.3389/fmicb.2016.00895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esteban-Cantos A, et al. The carbapenemase-producing Klebsiella pneumoniae population is distinct and more clonal than the carbapenem-susceptible population. Antimicrobial Agents and Chemotherapy. 2017;61:e02520–16. doi: 10.1128/AAC.02520-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Man TJB, et al. Genomic analysis of a pan-resistant isolate of Klebsiella pneumoniae, United States 2016. American Society for. Microbiology. 2018;9:e00440–18. doi: 10.1128/mBio.00440-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andrade LN, et al. Virulence genes, capsular and plasmid types of multidrug-resistant CTX-M (-2, -8, -15) and KPC-2-producing Klebsiella pneumoniae isolates from four major hospitals in Brasil. Diagnostic Microbiology and Infectious Disease. 2018;91:164–168. doi: 10.1016/j.diagmicrobio.2018.01.007. [DOI] [PubMed] [Google Scholar]
- 10.Ma Y, et al. Microbiological characterization of Klebsiella pneumoniae isolates causing bloodstream infections from five tertiary hospitals in Bejing, China. Journal of Global Antimicrobial Resistance. 2018;12:162–166. doi: 10.1016/j.jgar.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 11.Hu L, et al. Emergence of blaNDM-1 among Klebsiella pneumoniae ST15 and novel ST1031 clinical isolates in China. Diagnostic Microbiology and Infectious Disease. 2013;75:373–376. doi: 10.1016/j.diagmicrobio.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 12.Yan JJ, Wang MC, Zheng PX, Tsai LH, Wu JJ. Associations of the major international high-risk resistant clones and virulent clones with specific ompK36 allele groups in Klebsiella pneumoniae in Taiwan. New Microbe and New Infections. 2015;5:1–4. doi: 10.1016/j.nmni.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodrigues C, Machado E, Ramos H, Peixe L, Novais Â. Expansion of ESBL-producing Klebsiella pneumoniae in hospitalized patients: A successful story of international colnes (ST15, ST147, ST336) and epidemic plasmids (IncR, IncFIIκ). International Journal of Medical Microbiology. 2014;304:1100–1108. doi: 10.1016/j.ijmm.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 14.Vublik D, et al. Outbreak of KPC-3-producing ST15 and ST348 Klebsiella pneumoniae in a Portuguese hospital. Epidemiology &. Infection. 2017;145:595–599. doi: 10.1017/S0950268816002442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matheeussen V, et al. Emergence of colistin resistance during treatment of recurrent pneumonia caused by carbapenemase producing Klebsiella pneumoniae. Diagnostic Microbiology and Infectious Disease. 2019;94:407–409. doi: 10.1016/j.diagmicrobio.2019.02.014. [DOI] [PubMed] [Google Scholar]
- 16.Markovska R, et al. Dissemintaion of successful international clone ST15 and clonal complex 17 among Bulgarian CTX-M-15 producing K. pneumoniae isolates. Diagnostic Microbiology and Infectious Disease. 2017;89:310–313. doi: 10.1016/j.diagmicrobio.2017.08.012. [DOI] [PubMed] [Google Scholar]
- 17.Brisse S, et al. wzi gene sequencing, a rapid method for determination of capsular type for Klebsiella strains. Journal of Clinical Microbiology. 2013;51:4073–4078. doi: 10.1128/JCM.01924-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou K, et al. Use of whole-genome sequencing to trace, control and characterize the regional expansion of extended-spectrum β-lactamase producing ST15 Klebsiella pneumoniae. Scientific Reports. 2016;6:20840. doi: 10.1038/srep20840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Withey S, Cartmell E, Avery LM, Stephenson T. Bacteriophages – potential for application in wastewater treatment process. Science of the Total Environment. 2005;339:1–18. doi: 10.1016/j.scitotenv.2004.09.021. [DOI] [PubMed] [Google Scholar]
- 20.Golkar Z, Bagasra O, Pace DG. Bacteriophage therapy: a potential solution for the antibiotic resistance crisis. The Journal of Infection in Developing Countries. 2014;8:129–136. doi: 10.3855/jidc.3573. [DOI] [PubMed] [Google Scholar]
- 21.Tabassum R, et al. Complete genome analysis of a Siphoviridae phage TSK1 showing biofilm removal potential against Klebsiella pneumoniae. Scientific Reports. 2018;8:17904. doi: 10.1038/s41598-018-36229-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kubler-Kielb J, et al. The capsular polysaccharide and lipopolysaccharide structures of two carbapenem resistant Klebsiella pneumoniae outbreak isolates. Carbohydrate Research. 2013;369:6–9. doi: 10.1016/j.carres.2012.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bellich B, et al. Structure of the capsular polysaccharide of the KPC-2-producing Klbesiella pneumoniae strain KK207-2 and assignment of the glycosyltransferases functions. International Journal of Biological Macromolecules. 2019;130:536–544. doi: 10.1016/j.ijbiomac.2019.02.128. [DOI] [PubMed] [Google Scholar]
- 24.Pan Y-J, et al. Identification of three podoviruses infecting Klebsiella enconing capsule depolymerases that digest specific capsular types. Microbial. Biotechnology. 2019;12:472–486. doi: 10.1111/1751-7915.13370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pan Y-J, et al. Identification of capsular types in carbapenem-resistant Klebsiella pneumoniae strains by wzc sequencing and implications for capsule depolymerase treatment. Antimicrobial Agents and Chemotherapy. 2015;59:1038–1047. doi: 10.1128/AAC.03560-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ackermann HW. Frequency of morphological phage descriptions in the year 2000. Archives of Virology. 2001;146:843–857. doi: 10.1007/s007050170120. [DOI] [PubMed] [Google Scholar]
- 27.Vuotto C, et al. Biofilm formation and antibiotic resistance in Klebsiella pneumoniae urinary strains. Journal of Applied Microbiology. 2017;123:1003–1018. doi: 10.1111/jam.13533. [DOI] [PubMed] [Google Scholar]
- 28.Bull JJ, Vegge CS, Schmerer M, Chaudhry WN, Levin BR. Phenotypic resistance and the dynamics of bacterial escape from phage control. PLoS ONE. 2014;9:e94690. doi: 10.1371/journal.pone.0094690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seed KD. Battling phages: How bacteria defend against viral attack. PLoS Pathogens. 2015;11:e1004847. doi: 10.1371/journal.ppat.1004847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schroll D, Rogers S, Adhya S, Merril CR. Bacteriophage K1-5 encodes two different tail fiber proteins, allowing it to infect and replicate on both K1 and K5 strains of Escherichia coli. Journal of Virology. 2001;75:2509–2515. doi: 10.1128/JVI.75.6.2509-2515.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Drulis-Kawa Z, et al. Isolation and characterization of KP34 – a novel φKMV-like bacteriophage for Klebsiella pneumoniae. Applied Genetics and Molecular Biotechnology. 2011;90:1333–1345. doi: 10.1007/s00253-011-3149-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cao, F., et al Evaluation of the efficacy of a becteriophage in the treatment of pneumonia induced by multidrug resistance Klebsiella pneumoniae in mice. Hindawi Article ID 752930, 10.1155/2015/752930 (2015). [DOI] [PMC free article] [PubMed]
- 33.Thurow H, Niemann H, Rudolph C, Stirm S. Host capsule depolymerase activity of bacteriophage particles active on Klebsiella K20 and K24 strains. Virology. 1974;58:306–309. doi: 10.1016/0042-6822(74)90166-4. [DOI] [PubMed] [Google Scholar]
- 34.Hsieh P-F, Lin H-H, Lin T-L, Chen Y-Y, Wang J-T. Two T7-like bacteriophages, K5-2 and K5-4, each encodes two capsule depolymerases: isolation and functional characterization. Scientific Reports. 2017;7:4624. doi: 10.1038/s41598-017-04644-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.D’Andrea MM, et al. φBO1E, a newly discovered lytic bacteriophage targeting carbapenemase-producing Klebsiella pneumoniae of the pandemic Clonal Group 258 clade II lineage. Scientific Reports. 2017;7:2614. doi: 10.1038/s41598-017-02788-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jamal M, Hussain T, Das CR, Andleeb S. Characterization of Siphoviridae phage Z and studying its efficacy against multidrug-resistant Klebsiella pneumoniae planktonic cells and biofilm. Journal in Medical Microbiology. 2015;64:454–462. doi: 10.1099/jmm.0.000040. [DOI] [PubMed] [Google Scholar]
- 37.Seema K, Harjai K, Chhibber S. Evidence to support the therapeutic potential of bacteriophage Kpn5 in burn wound infection caused by Klebsiella pneumoniae in BALB/c mice. Journal of Microbiology and Biotechnology. 2010;20:935–941. doi: 10.4014/jmb.0909.09010. [DOI] [PubMed] [Google Scholar]
- 38.Chhibber S, Kaur S, Kumari S. Therapeutic potential of bacteriophage in treating Klebsiella pneumoniae B5055-mediated lobar pneumonia in mice. Journal of Medical Microbiology. 2008;57:1508–1513. doi: 10.1099/jmm.0.2008/002873-0. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook, J., Fritsch, E. F. & Maniatis, T. Molecular cloning: a laboratory manual. No. Ed. 2 pp.xxxviii + 1546 pp., University of Texas South Western Medical Center, USA (ISBN 0879693096) (1989).
- 40.King, A. M. Q., Adams, M. J., Carstens, E. B. & Lefkowitz, E. J. Virus taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, California, USA (ISBN 978-0-12-384684-6) (2011).
- 41.Mirzaei MK, Nilsson AS. Isolation of phages for phage therapy: A comparison of spot tests and efficiency of plating analyses for determination of host range and efficacy. PLoS ONE. 2015;10:e0118557. doi: 10.1371/journal.pone.0118557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adams, M. H. Bacteriophage, Inter-science Publishers. New York, USA, pp.450–456 (Record Number: 19602204111) (1959).
- 43.Di Lallo, G. et al. Isolation and partial characterization of bacteriophages infecting Pseudomonas syringae pv. actinidiae, causal agent of kiwifruit bacterial canker. Journal of Basic Microbiology54, 1210–1221, 10.1002/jobm.201300951 (2014). [DOI] [PubMed]
- 44.Dömötör D, et al. Comparative analysis of two bacteriophages of Xantomonas arboricola pv. juglandis. Infection, Genetics and Evolution. 2016;43:371–377. doi: 10.1016/j.meegid.2016.06.011. [DOI] [PubMed] [Google Scholar]
- 45.Pettit RK, et al. Microplate alamar blue assay for Staplylococcus epidermidis biofilm susceptibility testing. Antimicrobial Agents and Chemotherapy. 2005;49:2612–2617. doi: 10.1128/AAC.49.7.2612-2617.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O’Toole GA, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signaling pathways: a genetic analysis. Molecular Microbiology. 1998;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 47.Gowrishankar, S., Mosioma, N. D. & Pandian, S. K. Coral-assosciated bacteria as a promising antibiofilm agent against methicillin-resistant and –susceptible Staphylococcus aureus biofilms. Evidence-Based Complementary and Alternative Medicine Article ID 862374, 10.1155/2012/862374 (2012). [DOI] [PMC free article] [PubMed]
- 48.Liao Y-C, Lin H-H, Sabharwal A, Haase EM, Scannapieco FA. MyPro: A seamless pipeline for automated prokaryotic genome assembly and annotation. Journal of Microbiological Methods. 2015;113:72–74. doi: 10.1016/j.mimet.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Overbeek R, et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST) Nucleic Acids Research. 2014;42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lukashin AV, Borodovsky M. GeneMark.hmm: New solutions for gene finding. Nucleic Acids Research. 1998;265:1107–1115. doi: 10.1093/nar/26.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meier-Kolthoff JP, Göker M. VICTOR: genome-based phylogeny and classification of prokaryotic viruses. Bioinformatics. 2017;33:3396–3404. doi: 10.1093/bioinformatics/btx440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rambaut, A. FigTree 1.4.3 - a graphical viewer of phylogenetic trees and a program for producing publication-ready figures. Molecular evolution, phylogenetics and epidemiology, http://tree.bio.ed.ac.uk/software/figtree (2006).
- 53.Verma V, Harjai K, Chhibber S. Characterization of a T7-like lytic bacteriophage of Klebsiella pneumoniae B5055: A potential therapeutic agent. Current Microbiology. 2009;59:274–281. doi: 10.1007/s00284-009-9430-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.