Abstract
Purpose
Approximately 1% of individuals who carry a balanced reciprocal translocation (BRT) are subfertile. Current karyotyping does not have the resolution to determine whether the breakpoints of the involved chromosomes perturb genes important for fertility. The aim of this study was to apply single-molecule optical mapping (SMOM) to patients presenting for IVF (in vitro fertilization) to ascertain whether the BRT disrupted any genes associated with normal fertility.
Methods
Nine subfertile patients with different BRTs were recruited for the study. Methyltransferase enzyme DLE1 was used to fluorescently label their genomic DNA samples at the recognition motif CTTAAG. The SMOM was performed on the Bionano platform, and long molecules aligned against the reference genome hg19 to identify the breakpoint regions. Mate-pair and PCR-Sanger sequencing were used to confirm the precise breakpoint sequences.
Results
Both breakpoint regions in each of the nine BRTs were finely mapped to small regions of approximately 10 Kb, and their positions were consistent with original cytogenetic banding patterns determined by karyotyping. In three BRTs, breakpoints disrupted genes known to be associated with male infertility, namely NUP155 and FNDC3A [46,XY,t(5;13)(p15;q22)], DPY19L1 [46,XY,t(1;7)(p36.3;p15), and BAI3 [46,XY,t(3;6)(p21;q16)].
Conclusions
The SMOM has potential clinical application as a rapid tool to screen patients with BRTs for underlying genetic causes of infertility and other diseases.
Electronic supplementary material
The online version of this article (10.1007/s10815-020-01702-z) contains supplementary material, which is available to authorized users.
Keywords: Balanced reciprocal translocation (BRT), Infertility, Single-molecule optical mapping, Mate-pair sequencing
Introduction
Balanced reciprocal translocations (BRTs) are common chromosomal structural rearrangements formed by random de novo breakage and rejoining of two or more chromosomes and occur in approximately 0.2% of newborns [1, 2]. While most translocations are de novo in origin, some individuals inherit familial forms. Reciprocal translocation carriers generally do not display any apparent phenotypic disease because the vast majority of breakpoints (BPs) occur in non-repetitive regions, and thus, normal gene expression of the rearranged chromosomes is thought to be maintained [3, 4]. However, in about 5% of cases, BPs disrupt haploinsufficient genes or their regulatory elements, leading to clinical diseases [5, 6]. Male carriers of translocations can have reduced fertility due to misparing disrupting the sex vesicle, leading to spermatogenic arrest [7]. In contrast, female carriers of a BRT are generally not infertile, but instead are prone to recurrent miscarraige. For BRTs with large translocated segments, apparent infertility in the female may actually represent very early pregnancy loss due to malsegragation of the translocation.
The main issues that plague couples where one partner is a translocation carrier is a high probability of reproductive failure due to biased formation of unbalanced gametes, which typically result in either implantation failure or miscarriage of an established fetus [2]. In some cases, if the unbalanced translocation is compatible with fetal development, a child can be born with a chromosome disease syndrome. Couples who have suffered repeated miscarriage often now resort to assisted reproductive treatment (ART) and, for some, preimplantation genetic testing (PGT) to select embryos with a normal or balanced chromosome complement for transfer [8, 9]. In some cases, more commonly now, couples have additionally requested selection of, or at least identification of, non-carrier embryos as options for transfer to avoid passing on the parental translocation with its associated problems to their offspring [10, 11], thus removing the genetic abnormality from the future family germline.
Since the early 1970s, conventional karyotyping has been used as the mainstream cytogenetic technique to detect the presence of chromosome structures in prenatal and postnatal samples. Recently, it has been considered possibly an essential tool in investigating either miscarriage and subfertility in any couple seeking to start or expand a family. While it is efficient in identifying large rearrangements, the technique has limitations. Principally, while it can readily identify the gross chromosome rearrangements, depending on genome location and the skill of the cytogeneticist, the resolution of cytogenetic G banding is only 3–10 Mb [12, 13] which is a distance that limits the implications for understanding potential gene effects. In contrast, while fluorescence in situ hybridization (FISH) using targeted probes can achieve much higher resolution than karyotyping, specific probe design to investigate finer details of the rearrangement is challenging and, the technique is difficult to perform and interpretation of results can be ambiguous [14]. More recently, to better identify the exact point of the translocation, next-generation sequencing (NGS) approaches including mate-pair sequencing (MPS) [10] and whole genome sequencing (WGS) [5, 15, 16] have been applied to mapping chromosome breakpoints; however, these techniques have some limitations since the short reads can be difficult to accurately map in some regions that are often involved in the translocation breakpoints.
Single-molecule optimal mapping (SMOM) on the Bionano platform is a newly developed technology which uses a non-destructive chemistry for sequence motif labeling of long DNA molecules, enabling genome assembly and construction of high-resolution karyotypes. By imaging extremely long genomic molecules megabases in size, the structural variation (SV) and copy number of complex regions of the genome including interspersed and long tandem repeats can be more easily elucidated [17]. In other studies, reanalysis of genomes from the 1000 Genome project by the SMOM found that such analysis alone correlated well with the original SNP data to phylogenetically classify the different subpopulations [18]. More recently, the SMOM has been used to unravel the structure of complex disease loci such as FSHD1 and correctly predict genotypes [19]. Thus, we postulated that the SMOM could also be used successfully as a rapid tool for fine examination of structural variations associated with chromosome rearrangements such as translocations. In this study, we investigated the potential of the SMOM to elucidate the breakpoint regions of nine translocation patients with subfertility and shed light on possible secondary gene disturbances that may have contributed to their fertility issues.
Materials and methods
Subject enrollment
A total of nine patients (7 males and 2 female) who carried reciprocal translocations and were seeking fertility assistance were recruited to this study. The experimental protocol was approved by the Ethics Committee of the Chinese PLA General Hospital (Approval number S2013-092-02). Written informed consent was obtained from each of the nine patients for the laboratory to conduct further genetic testing of their BRT.
Bionano genomic DNA labeling and data collection
High molecular weight (HMW) genomic DNA was isolated using the Bionano Prep™ Blood and Cell Culture DNA Isolation Kit (Bionano Genomics) from fresh blood samples collected in EDTA tubes. DNA quantification was performed using the Qubit dsDNA BR assay kit (Thermo Fisher Scientific) with a Qubit 2.0 Fluorometer (Thermo Fisher Scientific). The integrity and size range of the HMW DNA was confirmed by pulsed-field gel electrophoresis. A total of 1 μg of high molecular weight DNA was then labeled using the DLS DNA Labeling Kit (Bionano Genomics), which uses the methyltransferase enzyme DLE1 to fluorescently label DNA at the recognition motif CTTAAG. The labeled DNA was loaded into the flow cell of the Saphyr Chip (Bionano Genomics) and analyzed by collecting 5–20 Gb of data per scan (total of 160–480 Gb of data per sample per flow cell).
Bionano data assembly, SV calling, and identification of breakpoint regions
All data was analyzed by the Bionano software freely available from the Bionano Genomics support website (https://bionanogenomics.com/support). Raw DNA molecules from the BNX files were filtered based on a minimum length of 150 Kb and a minimum of nine labeled sites per molecule. De novo assembly of single molecules into consensus genome maps was performed with Bionano Solve v3.2.1 using RefAligner module. For SV calling, we performed in silico DLE1 digestion of the human reference genome (GRCh37, hg19) to generate the reference map. Following alignment of consensus genome maps with the reference map, SVs were identified using the Bionano custom SV caller. The minimal breakpoint regions were defined using the boundaries of the DLE mark positions on each chromosome closest to the crossover points. The whole SMOM procedure, from a sample to a molecular karyotype, can be completed in a 3-day period.
Mate-pair sequencing
Mate-pair sequencing (MPS) was used to independently confirm translocation breakpoint regions detected by the Bionano analysis. In brief, genomic DNA was sheared to generate fragment sizes of ~ 5 Kb and then biotinylated with Tn5 transposase. DNA fragments were end-repaired and size-selected by agarose gel electrophoresis. Following selection of 4.5–5.5 Kb fragments, molecules were circularized via intramolecular ligation using T3 DNA ligase (New England Biolabs) and the remaining linear molecules were removed by DNA exonuclease treatment. The circularized DNA fragments were then sheared to generate fragments of 300–500 bp. Next, the biotinylated fragments were purified using M280 Dynabeads (Thermo Fisher Scientific), end-repaired, A-tailed, and ligated to Illumina pair-end adaptors. PCR amplification was finally used to generate the library for sequencing.
Approximately 20–50 million single molecules from the mate-pair libraries were sequenced on the HiSeq2500 platform (Illumina) as 200-bp or 250-bp paired-end (PE) reads at an overall 30- to 80-fold sequencing depth. Trimmed genome-specific reads were then mapped to the hg19 reference genome using the Burrows-Wheeler transform algorithm, paired-end sequencing reads were binned, and alignments with the hg19 reference genome were scrutinized to identify pairs with discordant sequences representing the two involved translocation chromosomes. As previously described [10], the actual breakpoint sequences were finally identified using a series of forward and reverse primers designed from the known sequences of the paired-ends and sequencing the PCR amplicons containing the breakpoint region by standard Sanger sequencing.
Results
Patient demographics
We recruited seven male infertility patients (BRT01-07) and two female infertility patients (BRT08 and 09) who had a positive diagnosis through karyotyping analysis of a reciprocal translocation (Table 1). Six of the seven male patients (BRT01-03 and BRT 05-07) were oligospermic, and one was normospermic (BRT04). Seven patients (BRT01-06 and BRT08) and their partners had experienced at least one period of infertility prior to conceiving naturally. However, all these natural conceptions resulted in an early miscarriage. After an extended period of trying to conceive with their partners, the remaining 2 patients (BRT07 and BRT09) were unable to achieve a pregnancy.
Table 1.
Patient reproductive history
| BRT carrier | Karyotype | Patient age in years | Partner age in years | Male infertility diagnosisa | Period(s) of infertilityb (length) | Natural pregnancies | Miscarriagesd |
|---|---|---|---|---|---|---|---|
| 01 | 46,XY,t(5;13)(p15;q22) | 34 | 33 | Oligospermia | 1 (2 years) | 1 | 1 |
| 02 | 46,XY,t(1;18)(p31;q22) | 34 | 34 | Oligospermia | 0 | 2(1c) | 2 |
| 03 | 46,XY,t(1;7)(q25;p15) | 34 | 33 | Oligospermia | 1 (2 years) + 1 (4 years) | 1 | 1 |
| 04 | 46,XY,t(2;13)(p11.2;q32) | 48 | 39 | Normospermia | 1 (2 years) | 2 | 2 |
| 05 | 46,XY,t(3;8)(q27;q11.2) | 31 | 28 | Oligospermia | 1 (2 years) | 2 | 2 |
| 06 | 46,XY,t(1;7)(p36.3;p15) | 39 | 40 | Oligospermia | 1 (1 year) | 3 | 3 |
| 07 | 46,XY,t(3;6)(p21;q16) | 33 | 34 | Oligospermia | 1 (> 5 years) | 0 | 0 |
| 08 | 46,XX,t(1;15)(q12;q21) | 29 | 27 | Not applicable | 1 (3 years) | 3 | 3 |
| 09 | 46,XX,t(5;14)(q11;q32) | 37 | 38 | Not applicable | 1 (5 years) | 0 | 0 |
BRT, balanced reciprocal translocation
aDiagnosis based on semen parameters of sperm count, motility, and morphology
bDefined as 1 year of trying to conceive with unprotected sex
cBiochemical pregnancy
dMiscarriages included biochemical pregnancies and those that were lost in the early first trimester
Single-molecule optical mapping analysis of BRTs
Genomic DNA from each translocation carrier was subjected to the SMOM analysis, and all nine genomes were assembled from the overlapping long reads. The Bionano QC parameters for each sample are summarized in Supplementary Table 1. On average, 407 Gb (range 321–515 Gb) of data was collected per sample with a DNA molecule size of 245 Kb (range 211–280 Kb). The average labeling rate was 14.5 labels (range 12.0–16.0) per 100 Kb, and the average molecule map rate was 73.8% (range 59.5–91.6%), equating to an average effective coverage depth of 80 × (range 56–108 ×).
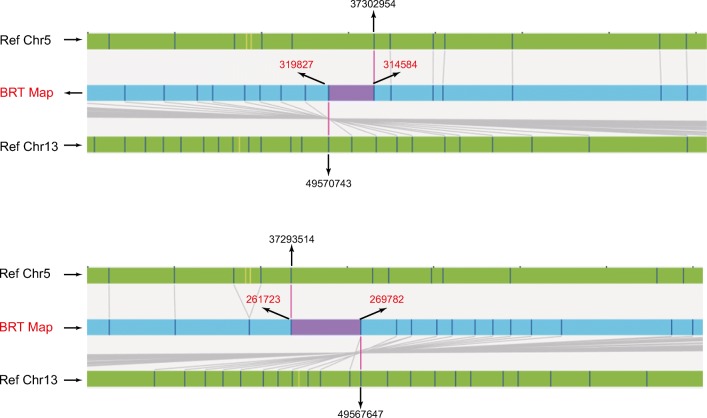
The assembled genome maps for each translocation pair were aligned to the in silico DLE1-digested reference sequence to identify the breakpoint junction regions. For all carriers, we were able to successfully map the two respective chromosome regions containing the breakpoints (Table 2, Fig. 1). The average resolution of the 18 junction regions identified by the Bionano was 9.1 Kb (range 2.8–38.6 Kb). All finely mapped breakpoints were essentially consistent with the broad cytogenetic locations determined by standard karyotyping. Within these defined regions, we identified the presence of potentially disrupted gene(s), including NUP155 and FNDC3A in the 5p and 13q BP regions of patient BRT01, DPY19L1 in the 7p region of BRT06, BAI3 in the 6q region of patient BRT07, and SPPL2A in the 15q region of patient BRT08. Of these genes, FNDC3A [20], NUP155 [21], and DPY19L1 [22, 23] and BAI3 [24] have been previously associated with male infertility.
Table 2.
Breakpoint definition of BRTs
| BRT carrier | Karyotype | SMOM | MPS + Sanger sequencing | ||
|---|---|---|---|---|---|
| Minimum coordinates of BP regions (size in Kb)# | Gene mapping in BP regions | BP coordinate | Gene disrupted (position) | ||
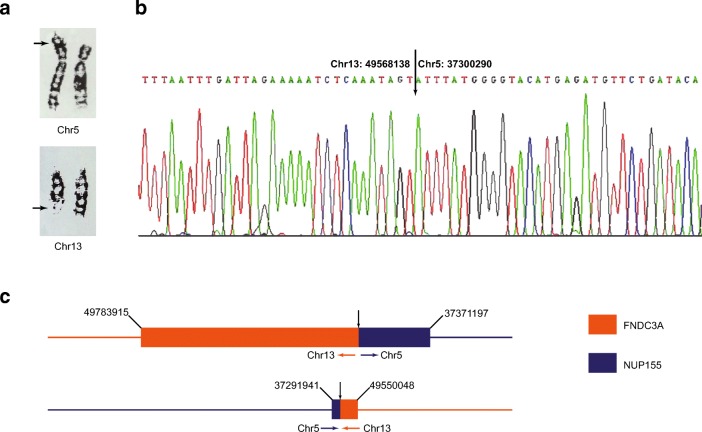
| 01 | 46,XY,t(5;13)(p15;q22) | chr5: 37293514-37302954 (9.4) | NUP155 | chr5:37300290 | NUP155 |
| chr13: 49567647-49570743 (3.1) | FNDC3A | chr13:49568138 | FNDC3A | ||
| 02 | 46,XY,t(1;18)(p31;q22) | chr1: 106973482-106979317 (5.8) | - | - | - |
| chr18: 31937692-31948951 (11.3) | - | - | - | ||
| 03 | 46,XY,t(1;7)(q25;p15) | chr1: 195806319-195819568 (13.3) | - | chr1:195810432 | - |
| chr7: 17194683-17225513 (30.8) | - | chr7:17202931 | - | ||
| 04 | 46,XY,t(2;13)(p11.2;q32) | chr2: 82728043-82733890 (5.8) | - | chr2:82730517 | - |
| chr13: 106887575-106891812 (4.2) | - | chr13:106891119 | - | ||
| 05 | 46,XY,t(3;8)(q27;q11.2) | chr3: 192940440-192944347 (3.9) | - | chr3:192943480 | - |
| chr8: 59623611-68028053 (11.9) | - | chr8:59630115 | - | ||
| 06 | 46,XY,t(1;7)(p36.3;p15) | chr1: 5268734-5280394 (11.7) | - | chr1:5269661 | - |
| chr7: 34970070-34982696 (12.6) | DPY19L1 | chr7:34980819 | DPY19L1 | ||
| 07 | 46,XY,t(3;6)(p21;q16) | chr3: 84963340-84976902 (13.6) | - | chr3:84971891 | - |
| chr6: 69858438-69868358 (9.9) | BAI3 | chr6:69858707 | BAI3 | ||
| 08 | 46,XX,t(1;15)(q12;q21) | chr1: 162955731-162960008 (4.3) | - | chr1:162959425 | - |
| chr15: 51020174-51029746 (9.6) | SPPL2A | chr15:51025649 | SPPL2A | ||
| 09 | 46,XX,t(5;14)(q11;q32) | chr5: 50217079-50228607 (11.5) | - | chr5:50217922 | - |
| chr14: 90511140-90522191 (11.1) | - | chr14:90512839 | - | ||
BRT, balanced reciprocal translocation; SMOM, single-molecule optimal mapping; MPS, mate-pair sequencing
#Minimum breakpoints were defined as the distance between opposing CTTAAG labeling sites either side of the breakpoint of the two chromosomes involved in the translocation
Fig. 1.
Bionano map showing breakpoint analysis of BRT01 [46,XY,t(5;13)(p15;q22)]. Chromosome 5 and 13 reference maps aligned to the BRT map are shown by arrows. The chromosome and genome map locations identified in the translocation regions are labeled. Coordinates in black are the hg19 locations; those in red are the genome map position for DLE labeling sites
Validation of breakpoint locations by mate-pair sequencing
To confirm the single-molecule mapping results, we performed the MPS analysis and then used the simple Sanger sequencing on breakpoint PCR products to accurately locate the breakpoints, on both chromosomes, down to the single nucleotide resolution (Table 2, Fig. 2). With the exception of BRT02, we were able to precisely map the nucleotide sequence position on both translocation chromosomes where de novo breakage and rejoining had occurred. However, in the case of BRT02, recombination points were located in highly repetitive sequence regions that were not able to be analyzed by the PCR and Sanger sequencing. Importantly, these sites physically disrupted those genes predicted by the SMOM analysis to exist in the breakpoint regions.
Fig. 2.
Breakpoint validation of BRT01 [46,XY,t(5;13)(p15;q22)]. a Karyogram of chromosomes 5 and 13. Arrows indicate the cytogenetic breakpoints of the translocation chromosomes. b Sanger sequencing. The actual BP positions on chromosomes 5 and 13 are defined. c Genome map showing disruption of two genes FNDC3A and NUP155 as a result of the translocation event. Numbers represent corresponding coordinates from the hg19 reference genome
Discussion
Infertility is a complex disease caused by physical, genetic, and environmental factors [25]. While numerous genetic causes have been identified in the male [26, 27], chromosome translocations represent a unique type of genetic abnormality that has been associated with both male and female infertilities or subfertilities, affecting approximately 1% of carriers [28]. We therefore postulated that translocation structural variations at the breakpoint regions may potentially perturb important genes required for normal fertility, reducing reproductive potential and the chance of achieving pregnancy. To test this hypothesis, we applied a novel genome mapping technology called the SMOM to examine genotype and phenotype correlations in nine BRT patients with subfertility. The SMOM analysis was able to successfully and finely map the breakpoint regions to small regions of around 10 Kb, allowing the identification of genes spanning the breakpoint interval(s). In four BRT cases, we found that the gene sequences of FNDC3A, NUP155, DPY19L1, SPPL2A, and BAI3 were disrupted and, with the exception of SPPL2A, which encodes an aspartic peptidase expressed in most tissues, the other four genes have reported known associations with male infertility.
In the first case (BRT01), analysis of the 46,XY,t(5;13)(p15;q22) translocation showed that both breakpoints interrupted two genes, FNDC3A and NUP155. The gene FNDC3A is a fibronectin domain protein involved in normal tissue development and regulation of cellular metabolism. In a mouse model, FNDC3A has been shown to be required for adhesion between spermatids and Sertoli cells and homozygous mutation causes sterility [20]. The other gene NUP155 is critical for nuclear pore formation and RNA transport, and in a Drosophila model, NUP155 and related family member NUP154 are essential for both male and female gametogenesis [21]. Based on these findings, we speculate that the combined gene disturbances could have potentially caused reduced sperm production and quality. In the second case (BRT06), the 46,XY,t(1;7)(p36.3;p15) translocation resulted in disruption of the gene DPY19L1 (homologue DPY19L2), which appears to be critical for spermiogenesis and its deficiency is linked to globozospermia [22, 23]. While there was no evidence of globozospermia, the patient was diagnosed with oligospermia. In the third case (BRT07), the 46,XY,t(3;6)(p21;q16) translocation interrupted the gene BAI3 which is associated with intellectual ability. However, in expression studies of human fertile and infertile spermatozoa, BAI3 expression was higher in fertile sperm [24]. However, there is no clear association of the gene defect with oligospermia. Thus, while the role of FNDC3A, NUP155, DY19L1, and BAI3 in male fertility still remains speculative, by association with subfertility, we propose that it is likely that the lower expression of these genes through haploinsufficiency may have played a potential role in reducing the reproductive potential of the male partner, leading to a history of infertility. For the remaining translocations, 46,XY,t(1;18)(p31;q22); 46,XY,t(1;7)(q25;p15); 46,XY,t(2;13)(p11.2;q32); 46,XY,t(3;8)(q27;q11.2); and 46,XX,t(5;14)(q11;q32), the breakpoint regions identified were devoid of genes. We speculate that the patients carrying these translocations may be subfertile simply due to very early pregnancy loss caused by malsegregation of the translocation [7].
In the general population of infertile patients presenting for assisted conception, up to 4% of patients are identified with a translocation [29, 30], suggesting that BRTs may be a more common cause of infertility than previously recognized. Patients who have a BRT and underlying subfertility can seek genetic counseling to discuss their reproductive options. The dilemma for these couples is whether natural conception or treatment with IVF and PGT will give them the best chance of conceiving a child that is normal or balanced with respect to the translocation [31]. As part of reaching an informed decision, the genetic counselor may suggest exploring the option of the Bionano analysis to find out whether their translocation has disturbed genes important for normal fertility. If this indeed proves to be the case, patients may then decide that IVF to overcome subfertility combined with PGT to select embryos with a normal or balanced chromosome constitution is the best approach for them to pursue. Further, if they proceed along this pathway, the additional option of selecting a non-carrier embryo without the balanced translocation can be also be discussed.
Based on our findings, the SMOM analysis may also have other useful clinical applications for carriers of a reciprocal translocation. A small proportion of these patients have disease phenotypes [5, 6] and thus may wish to know whether there is genetic cause of their disease and potential therapies. Another useful application would be during prenatal diagnosis where occasionally incidental de novo BRTs are found [32, 33], but their pathogenicity related to the SV is unknown by karyotyping. In these scenarios, amniocyte DNA could be rapidly assessed by the SMOM in a 3-day turnaround time to identify whether the SV associated with the translocation interrupts disease-specific genes or interferes with their gene expression. If the gene has a dominant phenotype, genetic counseling could then be recommended for management of the pregnancy.
In conclusion, the SMOM on the Bionano instrument is a useful and rapid tool for defining the SV caused by BRTs. At a research level, the method could provide valuable information regarding the architecture of chromosomes around the breakpoints and potentially uncover interesting genes and their functions. For clinical utility, the SMOM has additional applications for translocation and other structural variation carriers, including investigating potential causes of infertility, identifying disease gene mutations in fetal de novo balanced translocations inadvertently found at prenatal diagnosis, and analyzing de novo translocations found in children with abnormal phenotypes to identify potential therapies to treat symptoms.
Electronic supplementary material
(DOCX 13.6 kb)
Acknowledgments
The authors thank the patients who participated in this research study.
Authors’ roles
H.W. and B.X.: study design and management and supervision of the study. A.M. and D.S.C.: study design, statistical analysis, and manuscript preparation. Z.J., A.M., H.Z., and T.Y.: experimental procedures and statistical analysis. X.Z. and T.M.: data analysis. S.W., L.W., and S.L.: supervision. M.X.: data analysis and manuscript preparation. Y.Y.: supervision and manuscript review.
Funding information
This work was supported by National Key Research and Development Program of China (2018YFC1003100).
Compliance with ethical standards
Conflict of interest
A.M., H.Z., X.Z., T.Y, T.M., M.X., and D.S.C. are employees of Berry Genomics Corporation. None of the authors holds any stocks or bonds in the company. The other authors declare that they have no conflict of interest.
Ethical approval
Ethical approval for performing this study was obtained from the Ethics Committee of the Chinese PLA General Hospital (Approval number S2013-092-02).
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Hui Wang, Zhengjun Jia and Aiping Mao contributed equally to this work.
Contributor Information
David S. Cram, Email: david.cram@berrygenomics.com
Yuanqing Yao, Email: yqyao@126.com.
References
- 1.Ogilvie CM, Braude P, Scriven PN. Successful pregnancy outcomes after preimplantation genetic diagnosis (PGD) for carriers of chromosome translocations. Hum Fertil (Camb) 2001;4(3):168–171. doi: 10.1080/1464727012000199252. [DOI] [PubMed] [Google Scholar]
- 2.Wilch ES, Morton CC. Historical and clinical perspectives on chromosomal translocations. Adv Exp Med Biol. 2018;1044:1–14. doi: 10.1007/978-981-13-0593-1_1. [DOI] [PubMed] [Google Scholar]
- 3.Burrow AA, Williams LE, Pierce LC, Wang YH. Over half of breakpoints in gene pairs involved in cancer-specific recurrent translocations are mapped to human chromosomal fragile sites. BMC Genomics. 2009;10:59. doi: 10.1186/1471-2164-10-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weckselblatt B, Rudd MK. Human structural variation: mechanisms of chromosome rearrangements. Trends Genet. 2015;31(10):587–599. doi: 10.1016/j.tig.2015.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nilsson D, Pettersson M, Gustavsson P, Forster A, Hofmeister W, Wincent J, et al. Whole-genome sequencing of cytogenetically balanced chromosome translocations identifies potentially pathological gene disruptions and highlights the importance of microhomology in the mechanism of formation. Hum Mutat. 2017;38(2):180–192. doi: 10.1002/humu.23146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schluth-Bolard C, Labalme A, Cordier MP, Till M, Nadeau G, Tevissen H, Lesca G, Boutry-Kryza N, Rossignol S, Rocas D, Dubruc E, Edery P, Sanlaville D. Breakpoint mapping by next generation sequencing reveals causative gene disruption in patients carrying apparently balanced chromosome rearrangements with intellectual deficiency and/or congenital malformations. J Med Genet. 2013;50(3):144–150. doi: 10.1136/jmedgenet-2012-101351. [DOI] [PubMed] [Google Scholar]
- 7.Kurahashi H, Kogo H, Tsutsumi M, Inagaki H, Ohye T. Failure of homologous synapsis and sex-specifc reproduction problems. Front Genet. 2012;3:112. doi: 10.3389/fgene.2012.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fischer J, Colls P, Escudero T, Munne S. Preimplantation genetic diagnosis (PGD) improves pregnancy outcome for translocation carriers with a history of recurrent losses. Fertil Steril. 2010;94(1):283–289. doi: 10.1016/j.fertnstert.2009.02.060. [DOI] [PubMed] [Google Scholar]
- 9.Zhang W, Liu Y, Wang L, Wang H, Ma M, Xu M, Xu X, Gao Z, Duan J, Cram DS, Yao Y. Clinical application of next-generation sequencing in preimplantation genetic diagnosis cycles for Robertsonian and reciprocal translocations. J Assist Reprod Genet. 2016;33(7):899–906. doi: 10.1007/s10815-016-0724-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang L, Shen J, Cram DS, Ma M, Wang H, Zhang W, Fan J, Gao Z, Zhang L, Li Z, Xu M, Leigh DA, Trounson AO, Liu J, Yao Y. Preferential selection and transfer of euploid noncarrier embryos in preimplantation genetic diagnosis cycles for reciprocal translocations. Fertil Steril. 2017;108(4):620–627. doi: 10.1016/j.fertnstert.2017.07.010. [DOI] [PubMed] [Google Scholar]
- 11.Treff NR, Thompson K, Rafizadeh M, Chow M, Morrison L, Tao X, Garnsey H, Reda CV, Metzgar TL, Neal S, Jalas C, Scott RT Jr, Forman EJ. SNP array-based analyses of unbalanced embryos as a reference to distinguish between balanced translocation carrier and normal blastocysts. J Assist Reprod Genet. 2016;33(8):1115–1119. doi: 10.1007/s10815-016-0734-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trask BJ. Human cytogenetics: 46 chromosomes, 46 years and counting. Nat Rev Genet. 2002;3(10):769–778. doi: 10.1038/nrg905. [DOI] [PubMed] [Google Scholar]
- 13.Wapner RJ, Martin CL, Levy B, Ballif BC, Eng CM, Zachary JM, Savage M, Platt LD, Saltzman D, Grobman WA, Klugman S, Scholl T, Simpson JL, McCall K, Aggarwal VS, Bunke B, Nahum O, Patel A, Lamb AN, Thom EA, Beaudet AL, Ledbetter DH, Shaffer LG, Jackson L. Chromosomal microarray versus karyotyping for prenatal diagnosis. N Engl J Med. 2012;367(23):2175–2184. doi: 10.1056/NEJMoa1203382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carpenter NJ. Molecular cytogenetics. Semin Pediatr Neurol. 2001;8(3):135–146. doi: 10.1053/spen.2001.26447. [DOI] [PubMed] [Google Scholar]
- 15.Aristidou C, Koufaris C, Theodosiou A, Bak M, Mehrjouy MM, Behjati F, Tanteles G, Christophidou-Anastasiadou V, Tommerup N, Sismani C. Accurate breakpoint mapping in apparently balanced translocation families with discordant phenotypes using whole genome mate-pair sequencing. PLoS One. 2017;12(1):e0169935. doi: 10.1371/journal.pone.0169935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang D, Wang Y, Ji X, Hu H, Zhang J, Meng L, Lin Y, Ma D, Jiang T, Jiang H, Asan, Song L, Guo J, Hu P, Xu Z. Clinical application of whole-genome low-coverage next-generation sequencing to detect and characterize balanced chromosomal translocations. Clin Genet. 2017;91(4):605–610. doi: 10.1111/cge.12844. [DOI] [PubMed] [Google Scholar]
- 17.Chan EKF, Cameron DL, Petersen DC, Lyons RJ, Baldi BF, Papenfuss AT, Thomas DM, Hayes VM. Optical mapping reveals a higher level of genomic architecture of chained fusions in cancer. Genome Res. 2018;28(5):726–738. doi: 10.1101/gr.227975.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levy-Sakin M, Pastor S, Mostovoy Y, Li L, Leung AKY, McCaffrey J, Young E, Lam ET, Hastie AR, Wong KHY, Chung CYL, Ma W, Sibert J, Rajagopalan R, Jin N, Chow EYC, Chu C, Poon A, Lin C, Naguib A, Wang WP, Cao H, Chan TF, Yip KY, Xiao M, Kwok PY. Genome maps across 26 human populations reveal population-specific patterns of structural variation. Nat Commun. 2019;10(1):1025. doi: 10.1038/s41467-019-08992-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Q, Xu X, Ding L, Li H, Xu C, Gong Y, Liu Y, Mu T, Leigh D, Cram DS, Tang S. Clinical application of single-molecule optical mapping to a multigeneration FSHD1 pedigree. Mol Genet Genomic Med. 2019;7(3):e565. doi: 10.1002/mgg3.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Obholz KL, Akopyan A, Waymire KG, MacGregor GR. FNDC3A is required for adhesion between spermatids and Sertoli cells. Dev Biol. 2006;298(2):498–513. doi: 10.1016/j.ydbio.2006.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gigliotti S, Callaini G, Andone S, Riparbelli MG, Pernas-Alonso R, Hoffmann G, Graziani F, Malva C. Nup154, a new Drosophila gene essential for male and female gametogenesis is related to the nup155 vertebrate nucleoporin gene. J Cell Biol. 1998;142(5):1195–1207. doi: 10.1083/jcb.142.5.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ray PF, Coutton C, Arnoult C. Sun proteins and Dpy19l2 forming LINC-like links are critical for spermiogenesis. Biol Open. 2016;5(5):535–536. doi: 10.1242/bio.016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koscinski I, Elinati E, Fossard C, Redin C, Muller J, Velez de la Calle J, et al. DPY19L2 deletion as a major cause of globozoospermia. Am J Hum Genet. 2011;88(3):344–350. doi: 10.1016/j.ajhg.2011.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bansal SK, Gupta N, Sankhwar SN, Rajender S. Differential genes expression between fertile and infertile spermatozoa revealed by transcriptome analysis. PLoS One. 2015;10(5):e0127007. doi: 10.1371/journal.pone.0127007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cram DS, O’Bryan MK, de Kretser DM. Male infertility genetics--the future. J Androl. 2001;22(5):738–746. [PubMed] [Google Scholar]
- 26.Cram D, Lynch M, O’Bryan MK, Salvado C, McLachlan RI, de Kretser DM. Genetic screening of infertile men. Reprod Fertil Dev. 2004;16(5):573–580. doi: 10.10371/RD03097. [DOI] [PubMed] [Google Scholar]
- 27.Harton GL, Tempest HG. Chromosomal disorders and male infertility. Asian J Androl. 2012;14(1):32–39. doi: 10.1038/aja.2011.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chantot-Bastaraud S, Ravel C, Siffroi JP. Underlying karyotype abnormalities in IVF/ICSI patients. Reprod BioMed Online. 2008;16(4):514–522. doi: 10.1016/S1472-6483(10)60458-0. [DOI] [PubMed] [Google Scholar]
- 29.Stern C, Pertile M, Norris H, Hale L, Baker HW. Chromosome translocations in couples with in-vitro fertilization implantation failure. Hum Reprod. 1999;14(8):2097–2101. doi: 10.1093/humrep/14.8.2097. [DOI] [PubMed] [Google Scholar]
- 30.Schreurs A, Legius E, Meuleman C, Fryns JP, D’Hooghe TM. Increased frequency of chromosomal abnormalities in female partners of couples undergoing in vitro fertilization or intracytoplasmic sperm injection. Fertil Steril. 2000;74(1):94–96. doi: 10.1016/s0015-0282(00)00558-6. [DOI] [PubMed] [Google Scholar]
- 31.Scriven PN, Flinter FA, Khalaf Y, Lashwood A, Ogilvie CM. Benefits and drawbacks of preimplantation genetic diagnosis (PGD) for reciprocal translocations: lessons from a prospective cohort study. Eur J Hum Genet. 2013;21(10):1035–1041. doi: 10.1038/ejhg.2013.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Warburton D. De novo balanced chromosome rearrangements and extra marker chromosomes identified at prenatal diagnosis: clinical significance and distribution of breakpoints. Am J Hum Genet. 1991;49(5):995–1013. [PMC free article] [PubMed] [Google Scholar]
- 33.Vasilevska M, Ivanovska E, Kubelka Sabit K, Sukarova-Angelovska E, Dimeska G. The incidence and type of chromosomal translocations from prenatal diagnosis of 3800 patients in the republic of Macedonia. Balkan J Med Genet. 2013;16(2):23–28. doi: 10.2478/bjmg-2013-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 13.6 kb)