Abstract
Embryos containing distinct cell lines are referred to as mosaic embryos, which are considered to be caused by mitotic errors in chromosome segregation during preimplantation development. As the accuracy and resolution of detection techniques improve, more and more mosaic embryos were identified recently. The impacts of mosaic embryos on survival and potential pregnancy outcome have been reported to be diverse in different studies. Because of the universality and clinical significance of mosaicism, it is essential to unravel the mechanisms and consequences with regard to this phenomenon in human pre- and post-implantation embryos. The purpose of this review is to explore the mechanisms, causes of mosaicism, and the development of pre- and post-implantation mosaic embryos in the light of recent emerging data, with the aim of providing new references for clinical applications.
Keywords: Mosaicism, Preimplantation genetic testing, Trophectoderm, Inner cell mass, Chromosome, Non-invasion preimplantation genetic testing
Introduction
Preimplantation embryonic mosaicism is a prevalent phenomenon defined as the simultaneous presence of two or more different cell lines in an embryo. The general origin of the mosaicism is due to post-zygotic mitotic errors, as opposed to the view that aneuploidy results from meiotic errors. Although the first few divisions after fertilization are mitosis, they are not identical to the typical mitotic pattern of somatic cells. Before the activation of the human genome, most of the materials and energy necessary for embryo division rely on oocytes. This is why embryos are prone to chromosomal segregation errors in the first development stage. According to the cell lineage involved and when mitotic errors occur, it can be divided into four forms: total mosaicism, inner cell mass (ICM) mosaicism, trophectoderm (TE) mosaicism, and ICM/TE mosaicism. Aneuploid cells in the total mosaic embryos are equally distributed in TE and ICM, rather than preferentially distributed in any cell line. For the other three types termed confined mosaicism, aneuploid cells are partially or entirely localized to specific cell lines [1, 2]. Furthermore, confined mosaicism can be composed of confined placental mosaicism (CPM) or true fetal mosaicism (TFM), and the former occurs more frequently than the latter [3]. In human embryos, mosaicism is thought to be associated with recurrent implantation failure, spontaneous miscarriages, and stillbirths or live births with a wide spectrum of genomic abnormalities. The incidence of mosaicism in embryos varies greatly in different studies, which is related not only to multiple external technical factors but also to complex internal factors. Here, we will elaborate on the occurrence and development of mosaicism and its clinical application.
The association between gametogenesis and mitosis after fertilization
It is well known that uniform aneuploidy, which is derived from meiotic chromosomal malsegregation, is critically dependent on maternal age [4]. The current mainstream thinking believes that errors in maternal meiosis, especially the MI, are the main factors leading to aneuploidy [5–7], while the paternal meiosis errors only account for 1% [8]. The reason can be attributed to the following: there is a rigorous checkpoint in the process of male spermatogenesis, which can effectively avoid the development of abnormal chromosomes while in the process of oogenesis, such checkpoints are often lacking or not strict enough [9]. Further, in the process of oogenesis, oocytes are stored in follicular pools since the fetal period and arrested in the prophase of meiosis I until ovulation occurs years later. During this long-term period, cohesion of sister chromatids is deteriorated with maternal age [7, 10]. Thus, oogenesis is more prone to errors than spermatogenesis.
On the contrary, some previous studies revealed that chromosomal mosaicism from mitotic errors is independent of maternal age [6, 11]. Although meiosis and mitosis possess very different mechanisms and consequences, they are nearly related to each other. Kuliev and Verlinsky indicated that errors from meiosis I could be corrected during meiosis II, whereby so-called “aneuploidy rescue,” but the corrected oocytes are more predisposed to generating aneuploidy affecting uninvolved chromosome during mitosis after fertilization, alluding it might damage meiotic apparatus instead of specific chromosome only [5]. Griffin and Ogur suggested that abnormal meiosis may be a catalyst for the subsequent presence of mosaicism [12]. Previous studies have observed that there is a gradual transition from acentriolar spindle to centrosome-driven spindle between meiosis and mitosis in rodents [13]. A recent published study utilized kinesin-14 HSET as a tool for transforming mouse oocytes from meiotic multiple acentriolar microtubule-organizing centers (aMTOCs) to a mitotic-like focused mode. HSET is a microtubule cross-linked motor, which plays an important role in regulating spindle assembly, the length of spindle, and organization of the pole. HSET can separate anti-parallel microtubules and arrange them into parallel bundles during mitosis. This study confirmed that meiotic barrel-shaped spindles are indispensable to the circumvention of chromosomal misalignment and malsegregation. Therefore, we presumed that the shift from meiotic toward mitotic mode might disturb the stability of genome during early preimplantation embryonic development [14]. Moreover, the first mitotic divisions after fertilization require the integrity of centrosome originating from spermatozoon. It was found that the incidence of embryo mosaicism from ejaculated sperm was significantly lower than that of testicular sperm extraction (TESE) [15]. Defects in the centrosome and the physiological transformation of apparatus may be factors in the fallibility of early stages after fertilization.
On the other hand, human oocytes play an important role in providing proteins and mRNA for cleavage and development of early embryos until the embryo genome is fully activated 3 days after fertilization [6, 16]. Telomere attrition in oocyte could promote genomic instability via non-homologous end joining (NHEJ), and the inefficient telomere reconstitution could give rise to the recombination of chromothripsis and subsequent mosaicism [17]. With the proliferation and consumption of the cells in embryo, the maternal transcripts gradually diminished, which provides a possible explanation that the second or third cleavage is more prone to mitotic errors comparing with the first [18–21]. Cell-cell interaction acts as a mediator either taking part in the cell fate decisions or inducing the demise of highly mosaic embryos [22, 23].
Mechanisms and molecular biological events in mosaicism formation
Mechanisms of mosaicism formation
There are various theories that can elucidate the mechanism of mosaicism formation such as anaphase lagging, endoreplication, non-disjunction, demolition, cytokinesis errors, or cell fusion. Herein, we will discuss the most common theories under the light of the recent data.
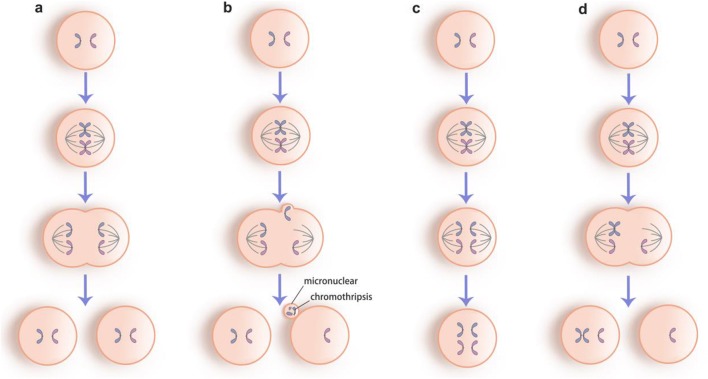
Some studies indicated that more than 50% of all mosaic embryos are caused by anaphase lagging [24]. The lagging chromosomes separated from the main group of chromosomes during anaphase result in a monosomy and a disomy of this chromosome in each daughter cell, respectively (Fig. 1b). Recent time-lapse imaging for abnormal chromosomal segregation in human embryos intuitively supported this theory as it showed a presence of micronuclear containing one or a few number of chromosome(s) during anaphase [25]. This extra structure can destroy the integrity of chromosomes leading to a detrimental rearrangement termed chromothripsis, concomitant with the dysfunction of kinetochore to attach to the spindle adequately [26]. As a matter of fact, some clinical embryologists have proposed that the appearance of micronucleus encapsulating fractured chromosomes is often associated with the increase of chromosome aneuploidy and is negatively correlated with the implantation rate [27]. Further, previous studies found that chromosome loss is more common than gain in human preimplantation embryos [8, 24, 28–32]. These observations supported anaphase lagging as the primary mechanism in mosaicism formation.
Fig. 1.
The most common mechanisms of mosaicism formation. a normal mitotic process. b Anaphase lagging with a micronuclear containing a fragmented chromosome termed chromothripsis. c Endoreplication of chromosomes without cell division. d Non-disjunction of the sister chromatids
Duplication of chromosome(s) without cell division can result in endoreplication, in which a trisomy or polyploidy would be generated (Fig. 1c). Polyploidy is thought to be of relevance for multipolar spindle in mitosis giving rise to multinucleation and chaotic embryos [18, 25, 33] and is supposed as a prevalent phenomenon in tumor invasion as well. Given that tetraploidization is a characteristic in human placenta and a motivator in the invasion of extra villous trophoblasts into decidua [34], we speculate that endoreplication is more common in trophoblast than embryo proper.
The failure of the sister chromatids to separate properly is referred as non-disjunction, resulting in a pair of daughter cells with reciprocal chromosomal complement (Fig. 1d). This was thought to be the most common mechanism in preimplantation development, but current views suggested that it is a rare event during mosaicism formation. Munne et al. found that only 2 out of 28 embryos showed reciprocal aneuploidy for the same chromosome [11]. This observation was supported by Mertzanidou et al. who showed no concurrence of the monosomy and trisomy for any chromosome in all embryos [28]. Non-disjunction is considered not only a mechanism of mitotic mosaicism but also one of the basic mechanisms leading to aneuploidy in meiosis and is influenced by chromosome-specific effects in different stages; thus, chromosomes are prone to non-disjunction in either MI or MII depending on different types [10, 35, 36]. Moreover, the incidence of this event is supposed to increase with old maternal age mostly because of the age-related malfunctioning of the female mitosis apparatus at early embryonic cleavage development stage [4].
Molecular biological events in mosaicism formation
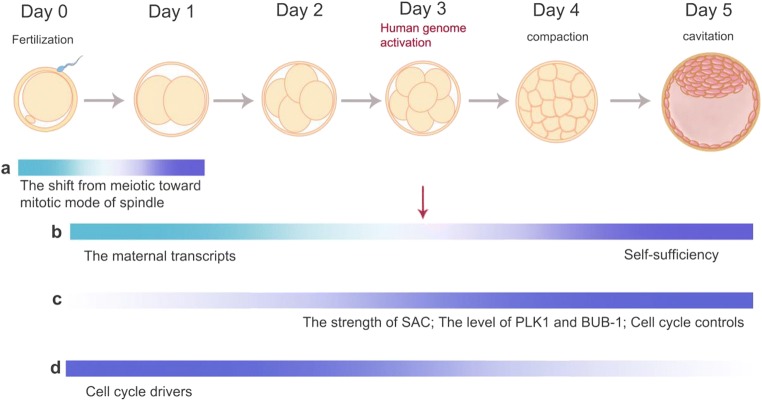
Above and beyond these essential mechanisms, there are some specific biological events in early human embryos; those could correct mitotic errors to avoid mosaicism formation (Fig. 2). The spindle assembly checkpoint (SAC) is responsible for arresting mitotic cells at the onset of the anaphase until all kinetochores of chromosomes are attached to the spindle properly. Chromosomal misalignment and spindle dysfunction in human preimplantation embryos can activate SAC, but it is insufficient to delay the onset of anaphase and induce the apoptosis of aneuploid cells [16, 21, 37]. In rapidly dividing cells, SAC could not effectively inhibit anaphase-promoting complex/cyclosome (APC/C) activity in a relaxed state. Experiments in early mouse embryos showed that SAC lost the ability to correct errors effectively that give rise to aneuploidy and mosaicism [37, 38]. Study in the Caenorhabditis elegans embryo indicated that both the ratio of DNA or kinetochore to cytoplasm and lineage-specific factors are the important determinants of SAC strength. This means that SAC would strengthen as cell size decreases progressively [39]. However, it remains largely unknown whether this correlation also holds in human early embryos. BUB-1 is one of the essential components of SAC, which is located on the kinetochore and plays an accurate role in promoting chromosome biorientation [40]. It was found that the level of BUB-1 is low in preimplantation embryos and is related to the occurrence of spontaneous miscarriage [41, 42]. Previous study in mouse embryos demonstrated that Polo-like kinase 1 (PLK1), a key cytokinesis-related gene involved in regulation of spindle assembly, anaphase onset, and APC/C activation [13], showed lower expression before the activation of the embryo genome at the 8-cell stage compared with the subsequent blastocyst stage. Compromised level of PLK1 could contribute to chromosome malsegregation and aneuploid karyotypes [43].
Fig. 2.
The molecular biological mechanisms that mosaicism is more common in early cleavage-stage embryos. a The shift from meiotic toward mitotic mode of spindle might disturb the stability of genome. b The diminished maternal transcripts before the activation of the embryo genome at the 8-cell stage and the increased self-sufficiency after the activation of the human genome. c The spindle assembly checkpoint (SAC) could not effectively inhibit anaphase-promoting complex/cyclosome (APC/C) activity in early embryos and would strengthen as cell size decreases progressively. The Polo-like kinase 1 (PLK1) and BUB-1 show compromised level before the activation of the embryo genome at the 8-cell stage compared with the subsequent blastocyst stage. Cell cycle controls remain at a low-level until 8-cell stage and increase in later stage. d There is overexpression of cell cycle drivers, and in the blastocyst, the opposite is true
Apart from these crucial regulatory elements for mitosis mentioned above, cell cycle checkpoints remain at a low-level until 8-cell stage, while there is overexpression of cell cycle driver. Due to the shortening of the G1 phase and rapid proliferation [16], cells lack enough time and regulators to respond to DNA damage and maintain the genetic integrity of daughter cells [44]. All these can make early preimplantation embryos more sensitive to adverse external influences and thus to mosaicism.
The variation in frequencies in pre- and post-implantation of mosaic embryos
The frequencies of mosaicism in preimplantation embryos vary considerably in different previous studies. The incidence of mosaicism in cleavage-stage embryos was estimated to be 15–93% [4, 20, 28, 30, 31, 45–48] while the rate was reported to vary from 3 to 44% in later stages such as morula and blastocyst [11, 35, 48–54]. Diploid-aneuploid mosaicism is considered to be the most common in mosaic embryos [45]. Data showed visible heterogeneity in the rate of mosaicism among various laboratories. There may be several reasons for the discrepancies.
Artificial factors
One of the important reasons is the heterogeneity of the diagnostic criteria of mosaicism in different laboratories and centers. A high level of evidence for identifying mosaicism is a double biopsy and blinded analysis showing reciprocal aneuploidies [55]. Nonetheless, this standard is difficult to implement because the occurrence of reciprocal aneuploidies has scarcely been reported, which indicated that non-disjunction is a rare mechanism in the formation of mosaic embryos. Most investigators identified mosaicism as long as cells analyzed with different chromosome constitutions appeared in the same embryos, whereupon the stringency of methodology and platform is also a variable factor between different centers. In addition, because of ethical issues, most studies were based on discarded embryos, and only a small number of studies use euploid human embryos of single gene or X-linked disorders [46]. Thus, this may lead to conclusions that cannot be extrapolated to all types of embryos.
Technical/external factors
Another important factor that influence the frequency of mosaicism is the technical procedure used for diagnosis of mosaicism. Fluorescence in situ hybridization (FISH) is a classical and conventional technique initially applied in preimplantation genetic diagnosis (PGD), primarily for sexing in terms of X-linked disorders [56, 57]. Previous studies applied biopsy of only one or two blastomere(s) coupled with FISH in cleavage-stage embryos, which was proved to be poorly representative of the rest of the embryos. The limited number of probes that can be used to label the chromosomes in a cycle is another inherent limitation of FISH usage. Moreover, the occurrence of mosaicism is chromosome-specific while all of the chromosomes are potential to be implicated in different propensities. Thus, the number and type of chromosomes investigated resulted in the high heterogeneity in frequencies from different laboratories [58]. Early randomized controlled trials conducted with FISH technique found that the live birth rate after preimplantation genetic screening (PGS) did not improve but decreased compared with the control group, which fully exposed the limitations and inapplicability of FISH [59–61]. One of the determinant reasons may be that the biopsy process at cleavage stage has significant impacts on the embryo, especially when two blastomeres are aspirated. Because there are so limited amounts of cells in embryos at this stage, removal of a number of two could be detrimental to embryos [62]. By contrast, it is logical that the biopsy of TE in blastocyst combined with comprehensive chromosome screening (CCS) is more robust and reliable because multiple cells meet the requirement for detection of mosaicism upholding the principle of security and demonstrate higher accuracy with reducing the deviation.
The latest next-generation sequencing (NGS) protocol offers more advantages than the array CGH implemented in previous years: higher cost-effectiveness, increased resolution to detect segmental aneuploid, wider dynamic range for interpretation of low-rate aneuploidy, higher throughput, reduced hands-on time, and decreased artificial background noise. Recent studies have indicated that the sensitivity of NGS to detect aneuploidy is the same as array CGH, and the consistency can reach almost 100% [63]. Hitherto, the ability of NGS to detect mosaicism correctly was reported to contain as low as 17% aneuploidy in a sample while array CGH can measure a shift from normality only when the level of aneuploid cells in the sample was > 25% [64, 65]. Theoretically, increasing sensitivity can also reduce specificity leading to false positives and discard euploid embryos by mistake, which gives rise to adverse pregnancy outcomes as well. Nonetheless, studies have shown that the pregnancy outcomes of euploid embryo transplantation detected by NGS were better than those of array CGH [66]. Therefore, NGS has been widely accepted as an alternative to other methods because of its high accuracy and distinctive preponderance.
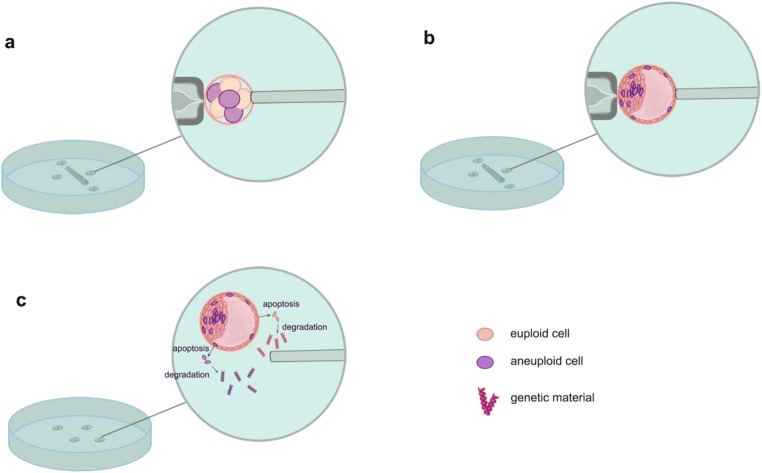
Both the biopsies in the cleavage and blastocyst stages are invasive processes, and the potential damage to embryos is still controversial. On the other hand, two mathematical models established by Gleicher et al. demonstrated that even in the ideal scenario, assuming uniform distribution of mosaicism in TE, a single biopsy of at least 27 TE cells is required to obtain minimal diagnostic predictability [67]. This raises more questions about the effectiveness and accuracy of invasive biopsies. Circulating cell-free DNA sampling from blastocoel fluid and spent culture media of blastocysts in conjunction with NGS has been sought as an attractive approach to obtain genetic information of preimplantation embryos, which is known as non-invasive preimplantation genetic testing (NIPGT). Intriguingly, encouraging findings are being made in vast majority of investigations. The concordance rates on genetic constitution of cell-free source compared with TE biopsies varied from 33 to 100% while whole embryo biopsies from 50 to 99%, which supports NIPGT as a reliable and promising source of embryonic genetic information [68–71]. However, Capalbo et al. argued that blastocoel fluid and spent culture media genetic analysis cannot provide sufficiently reliable results to be employed neither for gene-level abnormalities (PGT-M) nor for chromosomal aneuploidies (PGT-A) in clinical settings. They identified higher maternal contamination by the characterization of discordant genotypes due to the dominant paternal allele drop-out (ADO) [72]. Most of these studies obtained DNA information in spent culture media from day 3 to day 5, while one recent study collected culture media from thawed blastocysts after 24 h cultured. Obviously, this method avoided the maternal contamination to the greatest extent. The main conclusion they brought was that NIPGT-A in spent medium may be more reliable than TE biopsy; they assumed that DNA in spent culture medium might originate from both ICM and TE while the DNA in the TE biopsy might only represent the TE content [73]. This finding provided a new channel for the research and discussion of mosaicism. DNA materials of embryos from blastocoel fluid and spent culture media are supported to be marginalized from aneuploid or euploid cells in mosaic embryos by way of apoptosis during development (Fig. 3) [74]. Whether the cell-free DNA represent the corresponding embryos truly remains controversial. On the other hand, it is plausible that cell-free DNA may be better represented than TE biopsies since it originates from embryonic cells uniformly, and the challenge of mosaicism detection will also be readily solved. What is well known to us is that mosaicism is a major obstacle to NIPGT; however, if we can better understand the source of DNA in spent culture medium, it may be of great significance to non-invasive testing of mosaic embryos. As such, well-designed studies should be performed to explore the actual biological origin and mechanism of DNA materials in the extraembryonic environment. The practicability of NIPGT to diagnose mosaicism also requires other trials.
Fig. 3.
a The biopsy of only one or two blastomere(s) coupled with FISH in cleavage-stage mosaic embryos. b The biopsy of TE in mosaic blastocyst combined with comprehensive chromosome screening (CCS). Both a and b are misdiagnosed, and the potential damage to embryos is still controversial. c Circulating cell-free DNA sampling from blastocoel fluid and spent culture media of blastocysts in conjunction with NGS has been sought as an attractive approach to obtain genetic information of preimplantation embryos with the assumption that cell-free DNA materials should be marginalized from both aneuploid and euploid cells in mosaic embryos by way of apoptosis during development
Biological/internal factors
Apart from possible technical and artificial issues, the existence of self-selection and correction of abnormal cells in preimplantation embryos can be responsible for the reduced mosaic embryos from cleavage to blastocyst stage. A study indicated that the incidence of mosaic embryos decreased from 83 on day 4 to 42% on day 8 in vitro. It also illustrated the differentiation of embryos and formation of cavitation as a pivot to the negative selection of abnormal cells [48]. Further, the mosaicism rates were reported to be as low as 2% in chorionic villus sampling while reduced to be approximately 0.2% in amniocentesis, which supported a nature selection process during development [3, 75].
Embryos can reduce the magnitude of mosaicism and even eventually develop into completely euploid embryos via arrest/apoptosis of aneuploid cells or the preferential proliferation of euploid cells, which is referred as clonal depletion. Cell cycle checkpoints can arrest cells with chromosomal abnormalities in metaphase and delay the onset of anaphase until the DNA repair system corrects the errors; otherwise, it will initiate programmed cell apoptosis. In the earliest stage from fertilization to 8-cell embryo development, DNA repair is regulated by maternal transcripts in the absence of efficient checkpoints. It has been proposed that the ability to repair DNA damages in oocytes is less than 8–10% of the haploid genome and the occurrence of chromothripsis caused by abortive apoptosis in preimplantation embryos could give rise to multiple and chaotic chromosome rearrangements [76]. During the differentiation from morula to blastocyst stage, cell cycle checkpoints are reestablished to be more stringent and aneuploid cells can be targeted for clearance [21, 77]. The level of transcripts of apoptotic genes, such as BAX and BIX, is very low in the zygotes and increased rapidly in the morula whereas the anti-apoptotic gene BCL shows the opposite [78]. Bolton et al. revealed that aneuploid cells in ICM are eliminated by apoptosis while those in the TE show restriction on proliferation in a mouse model [79]. This suggests a lack of apoptotic pathways during early stage and provides an explanation for the lower rate of mosaicism at the blastocyst stage compared with the cleavage stage.
In addition to the clonal depletion mechanisms that positively or negatively eliminate abnormal blastomeres as mentioned above, another hypothesis is that aneuploid cells in mosaic embryos can be cleared by self-correction such as “trisomy rescue.” Trisomic cells can eliminate excess chromosome(s) through anaphase lagging or demolition. Similarly, monosomic cells can also self-correct by endoreplication of the lost chromosome. There is currently no direct evidence for this mechanism; however, since self-correction is considered a prevalence mechanism, the incidence of subsequent uniparental disomy (UPD) should theoretically be at least one-third, but studies showed it was as low as 0~2.1% in both preimplantation embryos and newborns [3, 31, 80, 81]. This fact suggests that trisomy rescue may be a rare mechanism in the process of embryonic development and progression.
Preferential allocation means that aneuploid cells in embryos are prone to distribute to trophectoderm (TE) whereas euploid cells tend to be placed in inner cell mass (ICM). In the blastocyst stage, multiple studies have shown that the extent of mosaicism in TE is consistent with ICM, and there is no tendency of euploid cell distribution in ICM. Therefore, TE biopsy exhibits a good representative of the chromosomal constitution of the ICM and even the whole embryo [22, 49, 50, 53, 54, 79, 82–84]. Nevertheless, TE biopsy is not able to predict pregnancy outcomes, such as ongoing pregnancy and miscarriage [85]. Confined placental mosaicism (CPM) is more common than true fetal mosaicism (TFM) in the gestation period. The incidence of mosaicism in chorionic villus sampling (CVS) was reported to be 1.8~2.2% [3, 86], of which only 13–28% in the followed amniocentesis showed TFM involvement [3, 75]. Both fetal and mesenchymal cells are derived from the epiblast, and the cytotrophoblast is derived from the trophoblast, the former shows a closer relative to the fetus. The possibility of fetal involvement in the mosaicism of mesenchymal cells was significantly higher than that of cytotrophoblast [75, 87]. This finding indicated that the preferential allocation of aneuploidy to non-fetal tissue may occur in the post-implantation stage. However, the possibility that apoptosis of aneuploid cells in fetal tissue increases while aneuploid cells in placenta are compatible with live birth cannot be ruled out.
Embryo selection for transfer
The fate and viability of mosaic embryos pre- and post-implantation are determined by multiple factors such as the type of chromosome involved, the location of aneuploid cells, and the degree of mosaicism. Although mosaicism can occur on each chromosome, the likelihood of involvement is not the same but varies widely with the time of occurrence of the mosaicism (meiosis or mitosis), the source (maternal or paternal), and its own characteristics (size and type). Accordingly, the aneuploidy of different chromosomes will lead to different developmental destinies and clinical manifestation. Here, we discuss the various factors affecting the viability of mosaic embryo as follows.
The type of chromosome involved
Mosaicism can impact single or multiple chromosomes with copy number variation of whole chromosome. Complex mosaic blastocysts present more compromised embryonic development potential and pregnancy outcome compared with other types, such as single or double mosaic embryos [11]. It is noteworthy that the majority of monosomy originate from mitotic errors, while most trisomy originate from maternal meiotic errors [31, 49]. It was previously thought that mosaic embryos derived from early meiotic errors showed a higher degree of abnormalities, compared with mosaic embryos occurred after fertilization, especially mitotic errors at the last phase of in vitro development that had a lower mosaic extent. In addition, the viability of monosomic cells is lower than trisomic cells; thus, the majority of monosomic cells will be cleared in the post-implantation phase [88, 89]. Preimplantation Genetic Diagnosis International Society (PGDIS) recommended that mosaic euploid-monosomy transfer should be preferred over euploid-trisomy [90]. However, Munne et al. found no significant discrepancy in pregnancy outcomes between monosomic and trisomic mosaic embryos [11]. This suggests that ploidy is not the only factor determining the survival potential of embryos. Certain types of chromosomes can pose a fatal threat on the development of embryos, while others can coexist with embryos or even live births. Moreover, different types of chromosomes are not equally affected. Aneuploidy 16 is the most frequently affected chromosome in preimplantation embryos and miscarriage products of conception, which is strongly associated with potential intrauterine growth restriction. Further, chromosomes X, 21, and 22 were also reported to be the most susceptible to whole chromosome errors [4, 8, 49, 91–93]. Different types of chromosomes show different copy number tendencies. Chromosomes 11 and 15 are more prone to be trisomy, while chromosomes 20 and 18 are more prone to be monosomy [24, 29]. This suggests that the origin of different chromosomes may also be diverse. Chromosome length is positively correlated with chromosome-specific rates of mitotic error but negatively with meiotic error in preimplantation embryo [8].
Uniparental disomy (UPD) is defined as a phenomenon that two homologous chromosomes are inherited from one parent rather than each of the two parents, respectively. UPD can result in imprinting disorders and serious recessive disorders. Well-known or proposed chromosomes involved in clinical significant UPD are chromosomes 6, 7, 11, 14, 15, 16, and 20 [94, 95]. Although UPD has proven to be a clinically rare phenomenon, its occurrence can lead to complex and intractable diseases such as Angelman syndrome and Prader-Willi syndrome [1]. Furthermore, UPD affects only a part of chromosome, called segmental UPD, which often occurs in Beckwith-Wiedemann syndrome. Therefore, detailed discussion is warranted when mosaic embryos involving these chromosomes are considered for transfer.
The location of aneuploid cells
The distribution of chromosome aneuploidy in different cell lines is also heterogeneous. Different chromosomal abnormalities showed different distribution patterns in fetal tissue, cytotrophoblast, and mesenchyme. Malvestiti et al. found that mosaic aneuploidy can involve all chromosomes, but chromosomes 1, 6, and 17 are commonly observed in CPM, while chromosomes 4, 8, 12, 13, 16, 18, 20, and 21 are preferentially observed in fetal tissue [3]. Mesenchymal cells are more closely related to fetal tissue than cytotrophoblast, so the mosaicism of chromosomes in mesenchymal cells is more likely to be involved in fetal tissue [75, 87]. Although some chromosomes are confined to extra fetal tissues, abnormalities in certain chromosomes (particularly chromosomes 2, 7, 16, and possibly 22) in CPM may also increase the risk of intrauterine growth restriction or other complications [19, 91], which results in stillbirth or congenital defect particularly when both the mesenchyme and the cytotrophoblast are affected [3].
Segmental mosaicism
With the development and progress of detection technology, more and more segmental aneuploidy has been discovered gradually with a majority being mitotic in origin. So far, the frequencies of segmental aneuploidy have been reported ranging from 1.1 to 70% in different literatures [31, 46, 96, 97]. It should be noted that during chromosome separation, large chromosomes are more likely to break and result in segmental mosaicism [11, 51, 97, 98]. Conversely, small chromosomes and acrocentric chromosomes are more prone to copy number errors [99, 100]. In addition to the size, fragile sites of chromosomes may be an important factor leading to segmental mosaicism [22, 101]. Studies demonstrated that chromosomal breaks appear to concentrate at fixed loci [102, 103]. Chromosome 19 is common in segmental abnormalities in virtue of the highest GC content even its relative small size [103]. Chromosomal abnormalities affecting centromeres (such as whole chromosome aneuploidy or entire chromosome arm breakage) are more likely to result from meiosis and inclined to be passed on to the next generation, so the impact on pregnancy may be more pronounced. However, segmental mosaicism is usually accompanied by the formation of micronucleus and chromothripsis, in which the DNA fragmentation followed by cell cycle arrest is inevitable [27, 82].
From the above, segmental mosaic embryos should be given priority to be transferred compared with whole chromosomal mosaic embryos. It is worth noting that the true incidence of segmental mosaicism may be overestimated due to technical and biological errors. Several studies suggested that NGS is more sensitive, and its ability to detect low-level and segmental aneuploidy is better than array CGH [66, 96, 104]. It was reported that the minimum segmental abnormality the high-resolution NGS can detect is as small as 1.8 Mb [105]. In comparison with array CGH, it was proved that NGS can detect more segmental abnormalities [96]. In addition to artificial factors, biological factors such as cell cycle S phase interference are also a major issue. The S phase of the cell cycle may contain DNA replication domains with different copy numbers, leading to false positives [106]. Furthermore, rapidly dividing cells such as tumor cells or blastomere contain a longer S phase and a shorter G1 phase causing a larger proportion of cells remaining in S phase [102, 107]. This increases the risk of misdiagnosis especially the single cell biopsy, and we must manage the diagnosis of segmental mosaicism with prudence.
The degree of mosaicism
Grati et al. proposed a set of scoring system for prioritizing mosaic aneuploid embryos. They indicated that mosaic trisomies 1, 3, 10, 12, and 19 have the highest priority for transfer due to the very low risk of any adverse outcomes while mosaic trisomies 13, 14, 16, 18, 21, 45, and X should be avoided because of the high risk of compatibility with live birth [108]. However, this scoring system was based solely on chorionic villus samples and products of conception and could not evaluate the degree of mosaicism in preimplantation embryos so that the scoring criterion failed to use the proportion of aneuploid cells in embryos for prioritization. There is no general consensus and unified standard about the degree of mosaicism. Studies in mice suggested that the implantation rates of mosaic embryos are the same as those of the control group, provided the existence of a sufficient proportion of normal cells [79]. Theoretically, any mosaic embryo can survive in the presence of euploid cells. PGDIS suggested that the proportion of aneuploid cells in mosaic embryos should be considered, but the level of mosaicism in biopsy samples cannot represent the state of rest embryos [90]. CoGEN indicated that even though biopsy samples may be less representative, when embryos contain higher levels of mosaicism in the trophectoderm, a higher probability with aneuploidy in the inner cell mass is logical [109]. Previous studies have revealed that the grade and the distribution of mosaic abnormal cells were correlated with the proportion of aneuploid cells and the likelihood of being diagnosed on clinical TE biopsies [50]. Remarkably, a recent study demonstrated an overall high sensitivity but relatively low specificity of only 67% in the context of PGT-A, indicating a dramatic number of false positive diagnosis based on TE biopsy result [110]. As a consequence of this observation, we should acknowledge that the interpretation of the results warrants further consideration, and more efficient methods and platforms are required as the degree of mosaicism is an important factor in considering the transfer of mosaic embryos.
Clinical outcome of mosaic embryo transfer
The clinical outcome of mosaic embryo transfer has been reported in many publications, but the debate on mosaic embryos remains intensive. The reported rate of ongoing pregnancy after mosaic embryo transfer ranged from 15 to 41%, showing significant decrease compared with the control group [11, 52, 82, 111]. Mosaic in placenta may have a higher risk of prenatal and perinatal complications [112]. A recent worldwide web-based survey reported that 20% of respondent IVF units performed transfers of chromosomally “abnormal” embryos, of which 49.3% cycles resulted in ongoing pregnancies or live births with a low miscarriage rate (9.3%) [113]. These results suggest that mosaic embryos also have the potential to develop into normal karyotype offspring and should be classified as viable rather than discarded [114].
Based on the proposed criteria for mosaic embryo transfer, it is necessary to thoroughly review the risks and limitations in appropriate genetic counseling for patients to make prudent decisions in the context of clinical management. Patients should be fully informed of some factors leading to misdiagnosis, for instance, the false negative resulting from the inability of NGS to effectively identify reciprocal chromosomal aneuploidy and euploidy/aneuploid mosaic embryos and the false positive results due to heterogeneity at the cell cycle stage [115]. These issues suggest that normal or low-level mosaic embryos diagnosed by preimplantation genetic screening may actually be complete aneuploidy, while embryos diagnosed as abnormal may also be completely normal and mistakenly discarded. Non-invasive PGT of mosaic embryos is also confronted with new opportunities and challenges. Once a pregnancy has been established after a transfer of mosaic embryo, periodic prenatal diagnosis should be strongly recommended thereafter to track the possible outcomes. The benefits and limitations of prenatal diagnosis should also be provided by a genetic counselor. At present, the application value of mosaic embryos has been affirmed to some degree. Nevertheless, how to make the best use of this special embryo type needs more rigorous clinical controlled studies.
Funding information
The work was funded by the Central Guiding the Science and Technology Devlopment of the Local (2018080802D0081) (Zhiguo Zhang) and College Natural Science Project of Anhui Province (KJ2019A0287) (Yan Hao).
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Taylor TH, Gitlin SA, Patrick JL, Crain JL, Wilson JM, Griffin DK. The origin, mechanisms, incidence and clinical consequences of chromosomal mosaicism in humans. Hum Reprod Update. 2014;20(4):571–581. doi: 10.1093/humupd/dmu016. [DOI] [PubMed] [Google Scholar]
- 2.Vera-Rodriguez M, Rubio C. Assessing the true incidence of mosaicism in preimplantation embryos. Fertil Steril. 2017;107(5):1107–1112. doi: 10.1016/j.fertnstert.2017.03.019. [DOI] [PubMed] [Google Scholar]
- 3.Malvestiti F, Agrati C, Grimi B, Pompilii E, Izzi C, Martinoni L, Gaetani E, Liuti MR, Trotta A, Maggi F, Simoni G, Grati FR. Interpreting mosaicism in chorionic villi: results of a monocentric series of 1001 mosaics in chorionic villi with follow-up amniocentesis. Prenat Diagn. 2015;35(11):1117–1127. doi: 10.1002/pd.4656. [DOI] [PubMed] [Google Scholar]
- 4.Rius M, Daina G, Obradors A, Ramos L, Velilla E, Fernandez S, et al. Comprehensive embryo analysis of advanced maternal age-related aneuploidies and mosaicism by short comparative genomic hybridization. Fertil Steril. 2011;95(1):413–416. doi: 10.1016/j.fertnstert.2010.07.1051. [DOI] [PubMed] [Google Scholar]
- 5.Kuliev A, Verlinsky Y. Meiotic and mitotic nondisjunction: lessons from preimplantation genetic diagnosis. Hum Reprod Update. 2004;10(5):401–407. doi: 10.1093/humupd/dmh036. [DOI] [PubMed] [Google Scholar]
- 6.Frumkin T, Malcov M, Yaron Y, Ben-Yosef D. Elucidating the origin of chromosomal aberrations in IVF embryos by preimplantation genetic analysis. Mol Cell Endocrinol. 2008;282(1–2):112–119. doi: 10.1016/j.mce.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 7.Chiang T, Schultz RM, Lampson MA. Meiotic origins of maternal age-related aneuploidy. Biol Reprod. 2012;86(1):1–7. doi: 10.1095/biolreprod.111.094367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCoy RC, Demko ZP, Ryan A, Banjevic M, Hill M, Sigurjonsson S, Rabinowitz M, Petrov DA. Evidence of selection against complex mitotic-origin aneuploidy during preimplantation development. PLoS Genet. 2015;11(10):e1005601. doi: 10.1371/journal.pgen.1005601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pujol A, Benet J, Staessen C, Van Assche E, Campillo M, Egozcue J, et al. The importance of aneuploidy screening in reciprocal translocation carriers. Reproduction. 2006;131(6):1025–1035. doi: 10.1530/rep.1.01063. [DOI] [PubMed] [Google Scholar]
- 10.Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet. 2001;2(4):280–291. doi: 10.1038/35066065. [DOI] [PubMed] [Google Scholar]
- 11.Munne S, Blazek J, Large M, Martinez-Ortiz PA, Nisson H, Liu E, et al. Detailed investigation into the cytogenetic constitution and pregnancy outcome of replacing mosaic blastocysts detected with the use of high-resolution next-generation sequencing. Fertil Steril. 2017;108(1):62–71. doi: 10.1016/j.fertnstert.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 12.Griffin DK, Ogur C. Chromosomal analysis in IVF: just how useful is it? Reproduction. 2018;156(1):F29–F50. doi: 10.1530/REP-17-0683. [DOI] [PubMed] [Google Scholar]
- 13.Baran V, Brzakova A, Rehak P, Kovarikova V, Solc P. PLK1 regulates spindle formation kinetics and APC/C activation in mouse zygote. Zygote. 2016;24(3):338–345. doi: 10.1017/S0967199415000246. [DOI] [PubMed] [Google Scholar]
- 14.Bennabi I, Queguiner I, Kolano A, Boudier T, Mailly P, Verlhac MH, et al. Shifting meiotic to mitotic spindle assembly in oocytes disrupts chromosome alignment. EMBO Rep. 2018;19(2):368–381. doi: 10.15252/embr.201745225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Silber S, Escudero T, Lenahan K, Abdelhadi I, Kilani Z, Munne S. Chromosomal abnormalities in embryos derived from testicular sperm extraction. Fertil Steril. 2003;79(1):30–38. doi: 10.1016/s0015-0282(02)04407-2. [DOI] [PubMed] [Google Scholar]
- 16.Ambartsumyan G, Clark AT. Aneuploidy and early human embryo development. Hum Mol Genet. 2008;17(R1):R10–R15. doi: 10.1093/hmg/ddn170. [DOI] [PubMed] [Google Scholar]
- 17.Keefe DL. Telomeres and genomic instability during early development. Eur J Med Genet. 2019;103638. 10.1016/j.ejmg.2019.03.002. [DOI] [PubMed]
- 18.Mertzanidou A, Spits C, Nguyen HT, Van de Velde H, Sermon K. Evolution of aneuploidy up to day 4 of human preimplantation development. Hum Reprod. 2013;28(6):1716–1724. doi: 10.1093/humrep/det079. [DOI] [PubMed] [Google Scholar]
- 19.Sachdev NM, Maxwell SM, Besser AG, Grifo JA. Diagnosis and clinical management of embryonic mosaicism. Fertil Steril. 2017;107(1):6–11. doi: 10.1016/j.fertnstert.2016.10.006. [DOI] [PubMed] [Google Scholar]
- 20.Munne S, Weier HU, Grifo J, Cohen J. Chromosome mosaicism in human embryos. Biol Reprod. 1994;51(3):373–379. doi: 10.1095/biolreprod51.3.373. [DOI] [PubMed] [Google Scholar]
- 21.Jacobs K, Van de Velde H, De Paepe C, Sermon K, Spits C. Mitotic spindle disruption in human preimplantation embryos activates the spindle assembly checkpoint but not apoptosis until day 5 of development. Mol Hum Reprod. 2017;23(5):321–329. doi: 10.1093/molehr/gax007. [DOI] [PubMed] [Google Scholar]
- 22.Fragouli E, Lenzi M, Ross R, Katz-Jaffe M, Schoolcraft WB, Wells D. Comprehensive molecular cytogenetic analysis of the human blastocyst stage. Hum Reprod. 2008;23(11):2596–2608. doi: 10.1093/humrep/den287. [DOI] [PubMed] [Google Scholar]
- 23.Capalbo A, Rienzi L. Mosaicism between trophectoderm and inner cell mass. Fertil Steril. 2017;107(5):1098–1106. doi: 10.1016/j.fertnstert.2017.03.023. [DOI] [PubMed] [Google Scholar]
- 24.Coonen E, Derhaag JG, Dumoulin JC, van Wissen LC, Bras M, Janssen M, Evers JL, Geraedts JP. Anaphase lagging mainly explains chromosomal mosaicism in human preimplantation embryos. Hum Reprod. 2004;19(2):316–324. doi: 10.1093/humrep/deh077. [DOI] [PubMed] [Google Scholar]
- 25.Daughtry BL, Chavez SL. Time-lapse imaging for the detection of chromosomal abnormalities in primate preimplantation embryos. Methods Mol Biol. 1769;2018:293–317. doi: 10.1007/978-1-4939-7780-2_19. [DOI] [PubMed] [Google Scholar]
- 26.Vazquez-Diez C, Yamagata K, Trivedi S, Haverfield J, FitzHarris G. Micronucleus formation causes perpetual unilateral chromosome inheritance in mouse embryos. Proc Natl Acad Sci U S A. 2016;113(3):626–631. doi: 10.1073/pnas.1517628112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kort DH, Chia G, Treff NR, Tanaka AJ, Xing T, Vensand LB, Micucci S, Prosser R, Lobo RA, Sauer MV, Egli D. Human embryos commonly form abnormal nuclei during development: a mechanism of DNA damage, embryonic aneuploidy, and developmental arrest. Hum Reprod. 2016;31(2):312–323. doi: 10.1093/humrep/dev281. [DOI] [PubMed] [Google Scholar]
- 28.Mertzanidou A, Wilton L, Cheng J, Spits C, Vanneste E, Moreau Y, et al. Microarray analysis reveals abnormal chromosomal complements in over 70% of 14 normally developing human embryos. Hum Reprod. 2013;28(1):256–264. doi: 10.1093/humrep/des362. [DOI] [PubMed] [Google Scholar]
- 29.Ioannou D, Fonseka KG, Meershoek EJ, Thornhill AR, Abogrein A, Ellis M, Griffin DK. Twenty-four chromosome FISH in human IVF embryos reveals patterns of post-zygotic chromosome segregation and nuclear organisation. Chromosom Res. 2012;20(4):447–460. doi: 10.1007/s10577-012-9294-z. [DOI] [PubMed] [Google Scholar]
- 30.Daphnis DD, Delhanty JD, Jerkovic S, Geyer J, Craft I, Harper JC. Detailed FISH analysis of day 5 human embryos reveals the mechanisms leading to mosaic aneuploidy. Hum Reprod. 2005;20(1):129–137. doi: 10.1093/humrep/deh554. [DOI] [PubMed] [Google Scholar]
- 31.Johnson DS, Gemelos G, Baner J, Ryan A, Cinnioglu C, Banjevic M, Ross R, Alper M, Barrett B, Frederick J, Potter D, Behr B, Rabinowitz M. Preclinical validation of a microarray method for full molecular karyotyping of blastomeres in a 24-h protocol. Hum Reprod. 2010;25(4):1066–1075. doi: 10.1093/humrep/dep452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Capalbo A, Bono S, Spizzichino L, Biricik A, Baldi M, Colamaria S, Ubaldi FM, Rienzi L, Fiorentino F. Sequential comprehensive chromosome analysis on polar bodies, blastomeres and trophoblast: insights into female meiotic errors and chromosomal segregation in the preimplantation window of embryo development. Hum Reprod. 2013;28(2):509–518. doi: 10.1093/humrep/des394. [DOI] [PubMed] [Google Scholar]
- 33.Vazquez-Diez C, FitzHarris G. Causes and consequences of chromosome segregation error in preimplantation embryos. Reproduction. 2018;155(1):R63–R76. doi: 10.1530/REP-17-0569. [DOI] [PubMed] [Google Scholar]
- 34.Velicky P, Meinhardt G, Plessl K, Vondra S, Weiss T, Haslinger P, Lendl T, Aumayr K, Mairhofer M, Zhu X, Schütz B, Hannibal RL, Lindau R, Weil B, Ernerudh J, Neesen J, Egger G, Mikula M, Röhrl C, Urban AE, Baker J, Knöfler M, Pollheimer J. Genome amplification and cellular senescence are hallmarks of human placenta development. PLoS Genet. 2018;14(10):e1007698. doi: 10.1371/journal.pgen.1007698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Munne S, Grifo J, Wells D. Mosaicism: “survival of the fittest” versus “no embryo left behind”. Fertil Steril. 2016;105(5):1146–1149. doi: 10.1016/j.fertnstert.2016.01.016. [DOI] [PubMed] [Google Scholar]
- 36.Bazrgar M, Gourabi H, Valojerdi MR, Yazdi PE, Baharvand H. Self-correction of chromosomal abnormalities in human preimplantation embryos and embryonic stem cells. Stem Cells Dev. 2013;22(17):2449–2456. doi: 10.1089/scd.2013.0053. [DOI] [PubMed] [Google Scholar]
- 37.Vazquez-Diez C, Paim L, FitzHarris G. Cell-size-independent spindle checkpoint failure underlies chromosome segregation error in mouse embryos. Curr Biol. 2019;29(5):865–873. doi: 10.1016/j.cub.2018.12.042. [DOI] [PubMed] [Google Scholar]
- 38.Zhang M, Kothari P, Lampson MA. Spindle assembly checkpoint acquisition at the mid-blastula transition. PLoS One. 2015;10(3):e119285. doi: 10.1371/journal.pone.0119285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gerhold AR, Poupart V, Labbe JC, Maddox PS. Spindle assembly checkpoint strength is linked to cell fate in the Caenorhabditis elegans embryo. Mol Biol Cell. 2018;29(12):1435–1448. doi: 10.1091/mbc.E18-04-0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edwards F, Maton G, Gareil N, Canman JC, Dumont J. BUB-1 promotes amphitelic chromosome biorientation via multiple activities at the kinetochore. Elife. 2018;7. 10.7554/eLife.40690. [DOI] [PMC free article] [PubMed]
- 41.Wells D, Bermudez MG, Steuerwald N, Thornhill AR, Walker DL, Malter H, Delhanty JD, Cohen J. Expression of genes regulating chromosome segregation, the cell cycle and apoptosis during human preimplantation development. Hum Reprod. 2005;20(5):1339–1348. doi: 10.1093/humrep/deh778. [DOI] [PubMed] [Google Scholar]
- 42.Shi Q, Hu M, Luo M, Liu Q, Jiang F, Zhang Y, Wang S, Yan C, Weng Y. Reduced expression of Mad2 and Bub1 proteins is associated with spontaneous miscarriages. Mol Hum Reprod. 2011;17(1):14–21. doi: 10.1093/molehr/gaq065. [DOI] [PubMed] [Google Scholar]
- 43.Fan Y, Zhao HC, Liu J, Tan T, Ding T, Li R, et al. Aberrant expression of maternal Plk1 and Dctn3 results in the developmental failure of human in-vivo- and in-vitro-matured oocytes. Sci Rep. 2015;5:8192. doi: 10.1038/srep08192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kiessling AA. Timing is everything in the human embryo. Nat Biotechnol. 2010;28(10):1025–1026. doi: 10.1038/nbt1010-1025. [DOI] [PubMed] [Google Scholar]
- 45.van Echten-Arends J, Mastenbroek S, Sikkema-Raddatz B, Korevaar JC, Heineman MJ, van der Veen F, Repping S. Chromosomal mosaicism in human preimplantation embryos: a systematic review. Hum Reprod Update. 2011;17(5):620–627. doi: 10.1093/humupd/dmr014. [DOI] [PubMed] [Google Scholar]
- 46.Vanneste E, Voet T, Le Caignec C, Ampe M, Konings P, Melotte C, et al. Chromosome instability is common in human cleavage-stage embryos. Nat Med. 2009;15(5):577–583. doi: 10.1038/nm.1924. [DOI] [PubMed] [Google Scholar]
- 47.Harper JC, Coonen E, Handyside AH, Winston RM, Hopman AH, Delhanty JD. Mosaicism of autosomes and sex chromosomes in morphologically normal, monospermic preimplantation human embryos. Prenat Diagn. 1995;15(1):41–49. doi: 10.1002/pd.1970150109. [DOI] [PubMed] [Google Scholar]
- 48.Santos MA, Teklenburg G, Macklon NS, Van Opstal D, Schuring-Blom GH, Krijtenburg PJ, et al. The fate of the mosaic embryo: chromosomal constitution and development of day 4, 5 and 8 human embryos. Hum Reprod. 2010;25(8):1916–1926. doi: 10.1093/humrep/deq139. [DOI] [PubMed] [Google Scholar]
- 49.Johnson DS, Cinnioglu C, Ross R, Filby A, Gemelos G, Hill M, Ryan A, Smotrich D, Rabinowitz M, Murray MJ. Comprehensive analysis of karyotypic mosaicism between trophectoderm and inner cell mass. Mol Hum Reprod. 2010;16(12):944–949. doi: 10.1093/molehr/gaq062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Capalbo A, Wright G, Elliott T, Ubaldi FM, Rienzi L, Nagy ZP. FISH reanalysis of inner cell mass and trophectoderm samples of previously array-CGH screened blastocysts shows high accuracy of diagnosis and no major diagnostic impact of mosaicism at the blastocyst stage. Hum Reprod. 2013;28(8):2298–2307. doi: 10.1093/humrep/det245. [DOI] [PubMed] [Google Scholar]
- 51.Nakhuda G, Jing C, Butler R, Guimond C, Hitkari J, Taylor E, et al. Frequencies of chromosome-specific mosaicisms in trophoectoderm biopsies detected by next-generation sequencing. Fertil Steril. 2018;109(5):857–865. doi: 10.1016/j.fertnstert.2018.01.011. [DOI] [PubMed] [Google Scholar]
- 52.Greco E, Minasi MG, Fiorentino F. Healthy babies after intrauterine transfer of mosaic aneuploid blastocysts. N Engl J Med. 2015;373(21):2089–2090. doi: 10.1056/NEJMc1500421. [DOI] [PubMed] [Google Scholar]
- 53.Tortoriello DV, Dayal M, Beyhan Z, Yakut T, Keskintepe L. Reanalysis of human blastocysts with different molecular genetic screening platforms reveals significant discordance in ploidy status. J Assist Reprod Genet. 2016;33(11):1467–1471. doi: 10.1007/s10815-016-0766-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Orvieto R, Shuly Y, Brengauz M, Feldman B. Should pre-implantation genetic screening be implemented to routine clinical practice? Gynecol Endocrinol. 2016;32(6):506–508. doi: 10.3109/09513590.2016.1142962. [DOI] [PubMed] [Google Scholar]
- 55.Capalbo A, Ubaldi FM, Rienzi L, Scott R, Treff N. Detecting mosaicism in trophectoderm biopsies: current challenges and future possibilities. Hum Reprod. 2017;32(3):492–498. doi: 10.1093/humrep/dew250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Munne S, Lee A, Rosenwaks Z, Grifo J, Cohen J. Diagnosis of major chromosome aneuploidies in human preimplantation embryos. Hum Reprod. 1993;8(12):2185–2191. doi: 10.1093/oxfordjournals.humrep.a138001. [DOI] [PubMed] [Google Scholar]
- 57.Griffin DK, Handyside AH, Penketh RJ, Winston RM, Delhanty JD. Fluorescent in-situ hybridization to interphase nuclei of human preimplantation embryos with X and Y chromosome specific probes. Hum Reprod. 1991;6(1):101–105. doi: 10.1093/oxfordjournals.humrep.a137241. [DOI] [PubMed] [Google Scholar]
- 58.Wilton L, Thornhill A, Traeger-Synodinos J, Sermon KD, Harper JC. The causes of misdiagnosis and adverse outcomes in PGD. Hum Reprod. 2009;24(5):1221–1228. doi: 10.1093/humrep/den488. [DOI] [PubMed] [Google Scholar]
- 59.Staessen C, Platteau P, Van Assche E, Michiels A, Tournaye H, Camus M, et al. Comparison of blastocyst transfer with or without preimplantation genetic diagnosis for aneuploidy screening in couples with advanced maternal age: a prospective randomized controlled trial. Hum Reprod. 2004;19(12):2849–2858. doi: 10.1093/humrep/deh536. [DOI] [PubMed] [Google Scholar]
- 60.Platteau P, Staessen C, Michiels A, Van Steirteghem A, Liebaers I, Devroey P. Preimplantation genetic diagnosis for aneuploidy screening in patients with unexplained recurrent miscarriages. Fertil Steril. 2005;83(2):393–397. doi: 10.1016/j.fertnstert.2004.06.071. [DOI] [PubMed] [Google Scholar]
- 61.Hardarson T, Hanson C, Lundin K, Hillensjo T, Nilsson L, Stevic J, et al. Preimplantation genetic screening in women of advanced maternal age caused a decrease in clinical pregnancy rate: a randomized controlled trial. Hum Reprod. 2008;23(12):2806–12. doi: 10.1093/humrep/den217. [DOI] [PubMed] [Google Scholar]
- 62.Coulam CB, Jeyendran RS, Fiddler M, Pergament E. Discordance among blastomeres renders preimplantation genetic diagnosis for aneuploidy ineffective. J Assist Reprod Genet. 2007;24(1):37–41. doi: 10.1007/s10815-006-9073-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fiorentino F, Bono S, Biricik A, Nuccitelli A, Cotroneo E, Cottone G, Kokocinski F, Michel CE, Minasi MG, Greco E. Application of next-generation sequencing technology for comprehensive aneuploidy screening of blastocysts in clinical preimplantation genetic screening cycles. Hum Reprod. 2014;29(12):2802–2813. doi: 10.1093/humrep/deu277. [DOI] [PubMed] [Google Scholar]
- 64.Goodrich D, Tao X, Bohrer C, Lonczak A, Xing T, Zimmerman R, et al. A randomized and blinded comparison of qPCR and NGS-based detection of aneuploidy in a cell line mixture model of blastocyst biopsy mosaicism. J Assist Reprod Genet. 2016;33(11):1473–1480. doi: 10.1007/s10815-016-0784-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mamas T, Gordon A, Brown A, Harper J, Sengupta S. Detection of aneuploidy by array comparative genomic hybridization using cell lines to mimic a mosaic trophectoderm biopsy. Fertil Steril. 2012;97(4):943–947. doi: 10.1016/j.fertnstert.2011.12.048. [DOI] [PubMed] [Google Scholar]
- 66.Maxwell SM, Colls P, Hodes-Wertz B, McCulloh DH, McCaffrey C, Wells D, Munné S, Grifo JA. Why do euploid embryos miscarry? A case-control study comparing the rate of aneuploidy within presumed euploid embryos that resulted in miscarriage or live birth using next-generation sequencing. Fertil Steril. 2016;106(6):1414–1419. doi: 10.1016/j.fertnstert.2016.08.017. [DOI] [PubMed] [Google Scholar]
- 67.Gleicher N, Metzger J, Croft G, Kushnir VA, Albertini DF, Barad DH. A single trophectoderm biopsy at blastocyst stage is mathematically unable to determine embryo ploidy accurately enough for clinical use. Reprod Biol Endocrinol. 2017;15(1):33. doi: 10.1186/s12958-017-0251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kuznyetsov V, Madjunkova S, Antes R, Abramov R, Motamedi G, Ibarrientos Z, et al. Evaluation of a novel non-invasive preimplantation genetic screening approach. PLoS One. 2018;13(5):e197262. doi: 10.1371/journal.pone.0197262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li P, Song Z, Yao Y, Huang T, Mao R, Huang J, Ma Y, Dong X, Huang W, Huang J, Chen T, Qu T, Li L, Zhong Y, Gu J. Preimplantation genetic screening with spent culture medium/blastocoel fluid for in vitro fertilization. Sci Rep. 2018;8(1):9275. doi: 10.1038/s41598-018-27367-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Magli MC, Pomante A, Cafueri G, Valerio M, Crippa A, Ferraretti AP, Gianaroli L. Preimplantation genetic testing: polar bodies, blastomeres, trophectoderm cells, or blastocoelic fluid? Fertil Steril. 2016;105(3):676–683. doi: 10.1016/j.fertnstert.2015.11.018. [DOI] [PubMed] [Google Scholar]
- 71.Vera-Rodriguez M, Diez-Juan A, Jimenez-Almazan J, Martinez S, Navarro R, Peinado V, Mercader A, Meseguer M, Blesa D, Moreno I, Valbuena D, Rubio C, Simon C. Origin and composition of cell-free DNA in spent medium from human embryo culture during preimplantation development. Hum Reprod. 2018;33(4):745–756. doi: 10.1093/humrep/dey028. [DOI] [PubMed] [Google Scholar]
- 72.Capalbo A, Romanelli V, Patassini C, Poli M, Girardi L, Giancani A, Stoppa M, Cimadomo D, Ubaldi FM, Rienzi L. Diagnostic efficacy of blastocoel fluid and spent media as sources of DNA for preimplantation genetic testing in standard clinical conditions. Fertil Steril. 2018;110(5):870–879. doi: 10.1016/j.fertnstert.2018.05.031. [DOI] [PubMed] [Google Scholar]
- 73.Huang L, Bogale B, Tang Y, Lu S, Xie XS, Racowsky C. Noninvasive preimplantation genetic testing for aneuploidy in spent medium may be more reliable than trophectoderm biopsy. Proc Natl Acad Sci U S A. 2019;116(28):14105–14112. doi: 10.1073/pnas.1907472116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Farra C, Choucair F, Awwad J. Non-invasive pre-implantation genetic testing of human embryos: an emerging concept. Hum Reprod. 2018;33(12):2162–2167. doi: 10.1093/humrep/dey314. [DOI] [PubMed] [Google Scholar]
- 75.Battaglia P, Baroncini A, Mattarozzi A, Baccolini I, Capucci A, Spada F, Pompilii E, Pittalis MC. Cytogenetic follow-up of chromosomal mosaicism detected in first-trimester prenatal diagnosis. Prenat Diagn. 2014;34(8):739–747. doi: 10.1002/pd.4358. [DOI] [PubMed] [Google Scholar]
- 76.Pellestor F, Gatinois V. Potential role of chromothripsis in the genesis of complex chromosomal rearrangements in human gametes and preimplantation embryo. Methods Mol Biol. 1769;2018:35–41. doi: 10.1007/978-1-4939-7780-2_3. [DOI] [PubMed] [Google Scholar]
- 77.Braude P, Bolton V, Moore S. Human gene expression first occurs between the four- and eight-cell stages of preimplantation development. Nature. 1988;332(6163):459–461. doi: 10.1038/332459a0. [DOI] [PubMed] [Google Scholar]
- 78.Yan L, Yang M, Guo H, Yang L, Wu J, Li R, et al. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat Struct Mol Biol. 2013;20(9):1131–9. doi: 10.1038/nsmb.2660. [DOI] [PubMed] [Google Scholar]
- 79.Bolton H, Graham S, Van der Aa N, Kumar P, Theunis K, Fernandez GE, et al. Mouse model of chromosome mosaicism reveals lineage-specific depletion of aneuploid cells and normal developmental potential. Nat Commun. 2016;7:11165. doi: 10.1038/ncomms11165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gueye NA, Devkota B, Taylor D, Pfundt R, Scott RJ, Treff NR. Uniparental disomy in the human blastocyst is exceedingly rare. Fertil Steril. 2014;101(1):232–236. doi: 10.1016/j.fertnstert.2013.08.051. [DOI] [PubMed] [Google Scholar]
- 81.Munne S. Preimplantation genetic diagnosis for aneuploidy and translocations using array comparative genomic hybridization. Curr Genomics. 2012;13(6):463–470. doi: 10.2174/138920212802510457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fragouli E, Alfarawati S, Spath K, Babariya D, Tarozzi N, Borini A, Wells D. Analysis of implantation and ongoing pregnancy rates following the transfer of mosaic diploid-aneuploid blastocysts. Hum Genet. 2017;136(7):805–819. doi: 10.1007/s00439-017-1797-4. [DOI] [PubMed] [Google Scholar]
- 83.Vera-Rodriguez M, Michel CE, Mercader A, Bladon AJ, Rodrigo L, Kokocinski F, et al. Distribution patterns of segmental aneuploidies in human blastocysts identified by next-generation sequencing. Fertil Steril. 2016;105(4):1047–1055. doi: 10.1016/j.fertnstert.2015.12.022. [DOI] [PubMed] [Google Scholar]
- 84.Chuang TH, Hsieh JY, Lee MJ, Lai HH, Hsieh CL, Wang HL, et al. Concordance between different trophectoderm biopsy sites and the inner cell mass of chromosomal composition measured with a next-generation sequencing platform. Mol Hum Reprod. 2018;24(12):593–601. doi: 10.1093/molehr/gay043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kushnir VA, Darmon SK, Barad DH, Gleicher N. Degree of mosaicism in trophectoderm does not predict pregnancy potential: a corrected analysis of pregnancy outcomes following transfer of mosaic embryos. Reprod Biol Endocrinol. 2018;16(1):6. doi: 10.1186/s12958-018-0322-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Carey L, Scott F, Murphy K, Mansfield N, Barahona P, Leigh D, Robertson R, McLennan A. Prenatal diagnosis of chromosomal mosaicism in over 1600 cases using array comparative genomic hybridization as a first line test. Prenat Diagn. 2014;34(5):478–486. doi: 10.1002/pd.4332. [DOI] [PubMed] [Google Scholar]
- 87.Lebedev I. Mosaic aneuploidy in early fetal losses. Cytogenet Genome Res. 2011;133(2–4):169–183. doi: 10.1159/000324120. [DOI] [PubMed] [Google Scholar]
- 88.Rubio C, Rodrigo L, Mercader A, Mateu E, Buendia P, Pehlivan T, et al. Impact of chromosomal abnormalities on preimplantation embryo development. Prenat Diagn. 2007;27(8):748–756. doi: 10.1002/pd.1773. [DOI] [PubMed] [Google Scholar]
- 89.Barbash-Hazan S, Frumkin T, Malcov M, Yaron Y, Cohen T, Azem F, Amit A, Ben-Yosef D. Preimplantation aneuploid embryos undergo self-correction in correlation with their developmental potential. Fertil Steril. 2009;92(3):890–896. doi: 10.1016/j.fertnstert.2008.07.1761. [DOI] [PubMed] [Google Scholar]
- 90.PGDIS. PGDIS position statement on chromosome mosaicism and preimplantation aneuploidy testing at the blastocyst stage. 15th International Conference on Preimplantation Genetic Diagnosis, Bologna, Italy, Reprod Biomed Online. 2016. http://www.pgdis.org/docs/newsletter_071816.html. Accessed 1 Sept 2019.
- 91.Harton GL, Cinnioglu C, Fiorentino F. Current experience concerning mosaic embryos diagnosed during preimplantation genetic screening. Fertil Steril. 2017;107(5):1113–1119. doi: 10.1016/j.fertnstert.2017.03.016. [DOI] [PubMed] [Google Scholar]
- 92.Menasha J, Levy B, Hirschhorn K, Kardon NB. Incidence and spectrum of chromosome abnormalities in spontaneous abortions: new insights from a 12-year study. Genet Med. 2005;7(4):251–263. doi: 10.1097/01.gim.0000160075.96707.04. [DOI] [PubMed] [Google Scholar]
- 93.Langlois S, Yong PJ, Yong SL, Barrett I, Kalousek DK, Miny P, Exeler R, Morris K, Robinson WP. Postnatal follow-up of prenatally diagnosed trisomy 16 mosaicism. Prenat Diagn. 2006;26(6):548–558. doi: 10.1002/pd.1457. [DOI] [PubMed] [Google Scholar]
- 94.Dawson AJ, Chernos J, McGowan-Jordan J, Lavoie J, Shetty S, Steinraths M, Wang JC, Xu J, Canadian College of Medical Geneticists committees CCMG guidelines: prenatal and postnatal diagnostic testing for uniparental disomy. Clin Genet. 2011;79(2):118–124. doi: 10.1111/j.1399-0004.2010.01547.x. [DOI] [PubMed] [Google Scholar]
- 95.Kearney HM, Kearney JB, Conlin LK. Diagnostic implications of excessive homozygosity detected by SNP-based microarrays: consanguinity, uniparental disomy, and recessive single-gene mutations. Clin Lab Med. 2011;31(4):595–613. doi: 10.1016/j.cll.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 96.Lai HH, Chuang TH, Wong LK, Lee MJ, Hsieh CL, Wang HL, et al. Identification of mosaic and segmental aneuploidies by next-generation sequencing in preimplantation genetic screening can improve clinical outcomes compared to array-comparative genomic hybridization. Mol Cytogenet. 2017;10:14. doi: 10.1186/s13039-017-0315-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rubio C, Bellver J, Rodrigo L, Castillon G, Guillen A, Vidal C, et al. In vitro fertilization with preimplantation genetic diagnosis for aneuploidies in advanced maternal age: a randomized, controlled study. Fertil Steril. 2017;107(5):1122–1129. doi: 10.1016/j.fertnstert.2017.03.011. [DOI] [PubMed] [Google Scholar]
- 98.Capalbo A, Rienzi L, Cimadomo D, Maggiulli R, Elliott T, Wright G, Nagy ZP, Ubaldi FM. Correlation between standard blastocyst morphology, euploidy and implantation: an observational study in two centers involving 956 screened blastocysts. Hum Reprod. 2014;29(6):1173–1181. doi: 10.1093/humrep/deu033. [DOI] [PubMed] [Google Scholar]
- 99.Franasiak JM, Forman EJ, Hong KH, Werner MD, Upham KM, Treff NR, Scott RT. Aneuploidy across individual chromosomes at the embryonic level in trophectoderm biopsies: changes with patient age and chromosome structure. J Assist Reprod Genet. 2014;31(11):1501–1509. doi: 10.1007/s10815-014-0333-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Iwarsson E, Malmgren H, Inzunza J, Ahrlund-Richter L, Sjoblom P, Rosenlund B, et al. Highly abnormal cleavage divisions in preimplantation embryos from translocation carriers. Prenat Diagn. 2000;20(13):1038–1047. [PubMed] [Google Scholar]
- 101.Mantikou E, Wong KM, Repping S, Mastenbroek S. Molecular origin of mitotic aneuploidies in preimplantation embryos. Biochim Biophys Acta. 2012;1822(12):1921–1930. doi: 10.1016/j.bbadis.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 102.Ramos L, Del RJ, Daina G, Martinez-Passarell O, Rius M, Tunon D, et al. Does the S phase have an impact on the accuracy of comparative genomic hybridization profiles in single fibroblasts and human blastomeres? Fertil Steril. 2014;101(2):488–495. doi: 10.1016/j.fertnstert.2013.10.031. [DOI] [PubMed] [Google Scholar]
- 103.Babariya D, Fragouli E, Alfarawati S, Spath K, Wells D. The incidence and origin of segmental aneuploidy in human oocytes and preimplantation embryos. Hum Reprod. 2017;32(12):2549–2560. doi: 10.1093/humrep/dex324. [DOI] [PubMed] [Google Scholar]
- 104.Friedenthal J, Maxwell SM, Munne S, Kramer Y, McCulloh DH, McCaffrey C, et al. Next generation sequencing for preimplantation genetic screening improves pregnancy outcomes compared with array comparative genomic hybridization in single thawed euploid embryo transfer cycles. Fertil Steril. 2018;109(4):627–632. doi: 10.1016/j.fertnstert.2017.12.017. [DOI] [PubMed] [Google Scholar]
- 105.Zheng H, Jin H, Liu L, Liu J, Wang WH. Application of next-generation sequencing for 24-chromosome aneuploidy screening of human preimplantation embryos. Mol Cytogenet. 2015;8:38. doi: 10.1186/s13039-015-0143-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dimitriadou E, Van der Aa N, Cheng J, Voet T, Vermeesch JR. Single cell segmental aneuploidy detection is compromised by S phase. Mol Cytogenet. 2014;7:46. doi: 10.1186/1755-8166-7-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Van der Aa N, Cheng J, Mateiu L, Zamani EM, Kumar P, Dimitriadou E, et al. Genome-wide copy number profiling of single cells in S-phase reveals DNA-replication domains. Nucleic Acids Res. 2013;41(6):e66. doi: 10.1093/nar/gks1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Grati FR, Gallazzi G, Branca L, Maggi F, Simoni G, Yaron Y. An evidence-based scoring system for prioritizing mosaic aneuploid embryos following preimplantation genetic screening. Reprod BioMed Online. 2018;36(4):442–449. doi: 10.1016/j.rbmo.2018.01.005. [DOI] [PubMed] [Google Scholar]
- 109.CoGEN Statement. COGEN position statement on chromosomal mosaicism detected in preimplantation blastocyst biopsies. 2017. https://www.ivf-worldwide.com/index.php?option=com_content&view=article&id=733&Itemid=464. Accessed 1 Sept 2019.
- 110.Popovic M, Dheedene A, Christodoulou C, Taelman J, Dhaenens L, Van Nieuwerburgh F, et al. Chromosomal mosaicism in human blastocysts: the ultimate challenge of preimplantation genetic testing? Hum Reprod. 2018;33(7):1342–1354. doi: 10.1093/humrep/dey106. [DOI] [PubMed] [Google Scholar]
- 111.Munné S, Spinella F, Grifo J, Zhang J, Beltran MP, Fragouli E, et al. Clinical outcomes after the transfer of blastocysts characterized as mosaic by high resolution next generation sequencing- further insights. Eur J Med Genet. 2019;103741. 10.1016/j.ejmg.2019.103741. [DOI] [PubMed]
- 112.Besser AG, Mounts EL. Counselling considerations for chromosomal mosaicism detected by preimplantation genetic screening. Reprod BioMed Online. 2017;34(4):369–374. doi: 10.1016/j.rbmo.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 113.Patrizio P, Shoham G, Shoham Z, Leong M, Barad DH, Gleicher N. Worldwide live births following the transfer of chromosomally “abnormal” embryos after PGT/A: results of a worldwide web-based survey. J Assist Reprod Genet. 2019;36(8):1599–1607. doi: 10.1007/s10815-019-01510-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Munne S. Status of preimplantation genetic testing and embryo selection. Reprod BioMed Online. 2018;37(4):393–396. doi: 10.1016/j.rbmo.2018.08.001. [DOI] [PubMed] [Google Scholar]
- 115.Scott RJ, Galliano D. The challenge of embryonic mosaicism in preimplantation genetic screening. Fertil Steril. 2016;105(5):1150–1152. doi: 10.1016/j.fertnstert.2016.01.007. [DOI] [PubMed] [Google Scholar]