Abstract
Purpose
The aim of this study was to identify a co-existing hydatidiform mole (HM) in twin pregnancy from the abnormal mixed-genomic products of conception (POC) after assisted reproduction by histopathological review, evaluation of p57kip2 immunostaining and short tandem repeat genotyping.
Methods
Thirty-seven patients were collected with suspicion for HM by pathological morphology. They had two embryos individually transferred to their uterus after in vitro fertilization and presented two gestational sacs with undeveloped embryos or one sac with an abnormal area by ultrasonography.
Results
Thirty patients were diagnosed as singleton pregnancy, including twenty-two non-molar gestations, six trisomy gestations, one homozygous complete mole and one heterozygous partial mole. Although six patients had ultrasonic imaging of two gestational sacs, the embryonic components in the vacant sac might fade away after transferring. Other seven patients were considered as twin pregnancy by the allelic genotype from two individual conceptions. For the patients with uniform p57kip2 positivity, excessive paternal alleles indicated the potential partial HM in the twin pregnancy. For the patients demonstrated divergent and/or discordant p57kip2 immunostaining, twin pregnancy with co-existing complete HM or mosaic conception were confirmed by genotyping of different villi population respectively. These patients were monitored by serum β-HCG, while one twin pregnancy with complete mole suffered invasive mole and received chemotherapy.
Conclusions
A strategy composed of selective clinicopathological screening, immunohistochemical interpretation and accurate genotyping is recommended for diagnostically challenging mixed-genomic POC of potential twin pregnancy with HM, especially to differentiate a non-molar mosaic conception from a partial mole.
Electronic supplementary material
The online version of this article (10.1007/s10815-019-01650-3) contains supplementary material, which is available to authorized users.
Keywords: Hydatidiform mole, p57kip2, Short tandem repeat, Twin pregnancy, Assisted, Reproduction
Introduction
Hydatidiform mole (HM) is a pre-malignant lesion of persistent gestational trophoblastic disease (PGTD), associated with aberrant fertilization and excessive paternal genome [1]. Complete HMs are usually diploid androgenetic in nature, and most partial HMs develop as androgenetic triploids [2]. It is suggested that HM developed due to a potential oocyte defect, such as dispermic fertilization or fertilization of an empty ovum (null-genome). However, it is not eliminated with the use of assisted reproductive techniques (ART) [3]. A hydatidiform mole co-existing with a fetus, namely a twin pregnancy with hydatidiform mole (TPHM), is a rare entity with an estimated incidence of 1 in 22,000–100,000 natural pregnancies, and this number may be increasing because of the greater use of ART. Although the risk of PGTD varies greatly (14–62.5%) in various publications, the counseling and management of HM are suggested to TPHM patients [4–7].
A histological section of the molar placenta is characterized by hydatidiform swelling of the chorionic villi and trophoblastic hyperplasia. However, early complete HM, partial HM, digynic gestation and non-molar hydropic abortion (NMA) can be easily misinterpreted [8]. Immunohistochemical analysis of p57kip2 is a useful approach in the vast majority (> 95%) of samples through the distinct expression patterns of complete HM versus partial HM and NMA [9, 10]. Short tandem repeat (STR) genotyping allows for determination of both ploidy and the maternal/paternal contributions of chromosome complements. Therefore, STR can differentially diagnose complete HM, partial HM, and NMA by discerning androgenetic diploidy, diandric triploidy, and biparental diploidy, respectively. However, aberrant p57kip2 expression patterns can be encountered as well as complex genotyping results. In addition to mosaicism or chimerism, a potential TPHM is an important differential diagnosis, especially following ART.
After in vitro fertilization (IVF) and two-embryo transferring, clinical pathologists might be confused by the abnormal mixed-genomic products of conception (POC) from two zygotes, which could lead to misdiagnosis of TPHM [11]. We propose that appropriate integration of molecular genotyping and p57kip2 immunohistochemistry will assist in the recognition and better classification of genetically complex cases, particularly suspicious HM in twin pregnancy. Furthermore, integration will contribute to more complete knowledge in this field and will help ensure appropriate patient care and treatment.
Materials and methods
Case selection
All potentially molar POC specimens were collected by the Department of Pathology of Peking University Third Hospital and subjected to an algorithmic diagnostic process using p57kip2 immunohistochemical staining and STR genotyping because of clinical or pathological concerns. Thirty-seven patients after two-embryo transferring were identified with certain degrees of suspicion for molar gestation and reviewed by two independent pathologists. Maternal age, clinical gestational age, and the relevant clinical history were collected from the medical records. The Peking University Institution Review Board approved the study. A written consent form was signed by each patient.
Immunohistochemical staining of p57kip2
Immunohistochemical staining was performed by heat-induced antigen retrieval (10-mM citrate buffer, pH 6.0) followed by the polymer method using a mouse monoclonal antibody against human p57kip2 protein (clone 57P06; Maixin Biotech Co., Ltd., Fuzhou, China). Decidual stromal cells and intermediate trophoblastic cells were positive in all cases and provided a reliable internal control. Absence of nuclear staining and < 10% nuclear staining of cytotrophoblast and villous stromal cells were considered negative results; this was consistent with the diagnosis of complete mole. Diffuse nuclear staining of cytotrophoblast and villous stromal cells was considered a positive result, consistent with all forms of non-molar specimens as well as partial moles; therefore, we could not distinguish these entities.
According to Lewis et al, the abnormal p57kip2 expression pattern was divided into two types [12]. One pattern was interpreted as “discordant” when any combination/admixture of negative and positive results for villous stromal cells and cytotrophoblast within individual villi was observed. The other pattern was interpreted as “divergent” when two populations of villi, each with different morphologies, exhibited two different staining patterns (positive in one set and negative in the other set). Discordant and divergent p57kip2 expression patterns have been reported in androgenetic/biparental mosaic/chimeric conceptions, as well as in twin pregnancies [13].
Molecular genotyping using STR markers
Ten 5-μm-thick sections were cut serially from formalin-fixed paraffin-embedded tissue blocks, and the middle sections were stained with hematoxylin and eosin (HE) and p57kip2 immunostaining. According to the HE slides, areas of well-separated maternal deciduas and villous tissue were removed from unstained slides. The p57KIP2 immunostaining slide was used to guide the selection of discrete regions in villous tissue. DNA was extracted using the hydrothermal pressure method of simultaneous deparaffinization and lysis of formalin-fixed paraffin-embedded tissue followed by conventional column purification to obtain high-quality DNA [14].
The PowerPlex 16 System (Promega Corporation, Madison, WI, USA) was performed for molecular genotyping according to methods described elsewhere [15]. Tissue genotyping was performed by multiplex polymerase chain reaction (PCR) at 15 autosomal STR loci (Penta E, D18S51, D21S11, TH01, D3S1358, FGA, TPOX, D8S1179, vWA, Penta D, CSF1PO, D16S539, D7S820, D13S317, D5S818) and the sex-determining marker (Amelogenin). PCR amplification and capillary electrophoresis were performed according to the manufacturer’s instructions. Molecular diagnostic criteria of complete mole, partial mole, digynic triploid gestation, and NMA have been described in previous literature [15, 16].
Results
Clinical findings
All of the patients ranged in age from 23 to 43 years (mean and median, 33.4 and 34 years) and had two embryos individually transferred to their uterus after IVF. Twenty-seven patients showed only one sac with or without an abnormal echogenic area by ultrasonography examination. Other ten patients demonstrated two gestational sacs, and their pregnancies had to be terminated because there was no evidence of fetal development or a fetal heartbeat. Mean ± SD gestation weeks at the end of pregnancy were 8 ± 1.6 weeks (range 5–11.3 weeks). All these patients had no familial history of HM.
Diagnosis of suspicious HM in patients with one gestational sac
As shown in Table 1, in the five patients with one sac and adjacent abnormal echogenicity, two were diagnosed with NMA according to positive p57kip2 immunostaining and balanced biparental allelic genotyping. Patient 22 had an empty sac and high level serum β-HCG (> 200,000 mIU/mL). The cauliflower-like villi with trophoblastic hyperplasia and negative p57kip2 immunostaining indicated a complete mole, which were confirmed by monospermic androgenetic diploid genotyping (Table S1). Patient 31 presented at 10 weeks with an undeveloped fetus in sac and an anomalous echogenic area. Irregularly shaped villi have scalloped contours, mild trophoblastic hyperplasia, and positive p57kip2 immunostaining. STR genotyping showed diandric monogynic triploid alleles, which supported a dispermic partial mole (Table S1). They did not reveal any signs of PGTD at 1-year follow-up.
Table 1.
Diagnosis of 37 patients following assisted reproduction
| Diagnosis | One sac | Two sacs | Total | |
|---|---|---|---|---|
| Without abnormal echogenicity | With abnormal echogenicity | |||
| Singleton pregnancy | ||||
| Non-molar gestation | 15 | 2 | 5 | 22 |
| Trisomy | 5 | 0 | 1 | 6 |
| Hydatidiform mole | 0 | 2 | 0 | 2 |
| Twin pregnancy | ||||
| Both non-molar gestation | 0 | 0 | 1 | 1 |
| Including one complete mole | 1 | 1 | 0 | 2 |
| Including one suspected partial mole | 0 | 0 | 3 | 3 |
| Including one mosaic pregnancy | 1 | 0 | 0 | 1 |
| Total | 22 | 5 | 10 | 37 |
According to high-level serum β-HCG and aberrant ultrasonography findings showing one fetus surrounded by a “snowstorm” area (70 × 41 mm), the pregnancy of patient 3 was terminated at 11 + 2/7 weeks. The placenta was composed of grossly normal tissue and a cystic mole. Divergent p57kip2 expression was found in two different populations of villi: no p57kip2 immunostaining signals were observed in the large hydropic villi and the small villi with mild stromal fibrosis (Fig. 1 a and b) demonstrated normal p57kip2 staining. The fetus showed balanced biallelic profiles of biparental origins, whereas the hydropic villi demonstrated exclusively monospermic paternal alleles (Fig. 1c). This patient was diagnosed with a twin pregnancy with homozygous complete mole and a co-existent fetus (Table S1). One month later, an invasive mole was unfortunately suspected because of an abnormal increase of serum β-HCG (8442 mIU/μL), presence of an irregular intrauterine mass, and multiple pulmonary nodules revealed by imaging. A clinical diagnosis of PGTD (FIGO III) was made, and chemotherapy (etoposide and floxuridine) was performed. After four cycles (each course lasted 5 days and had 18-day intervals) of chemotherapy, the serum β-HCG level decreased to 4.91 mIU/mL. She continued follow-up in the gynecology–oncology clinic, and her serum β-HCG level returned to normal (0.1 mIU/mL).
Fig. 1.
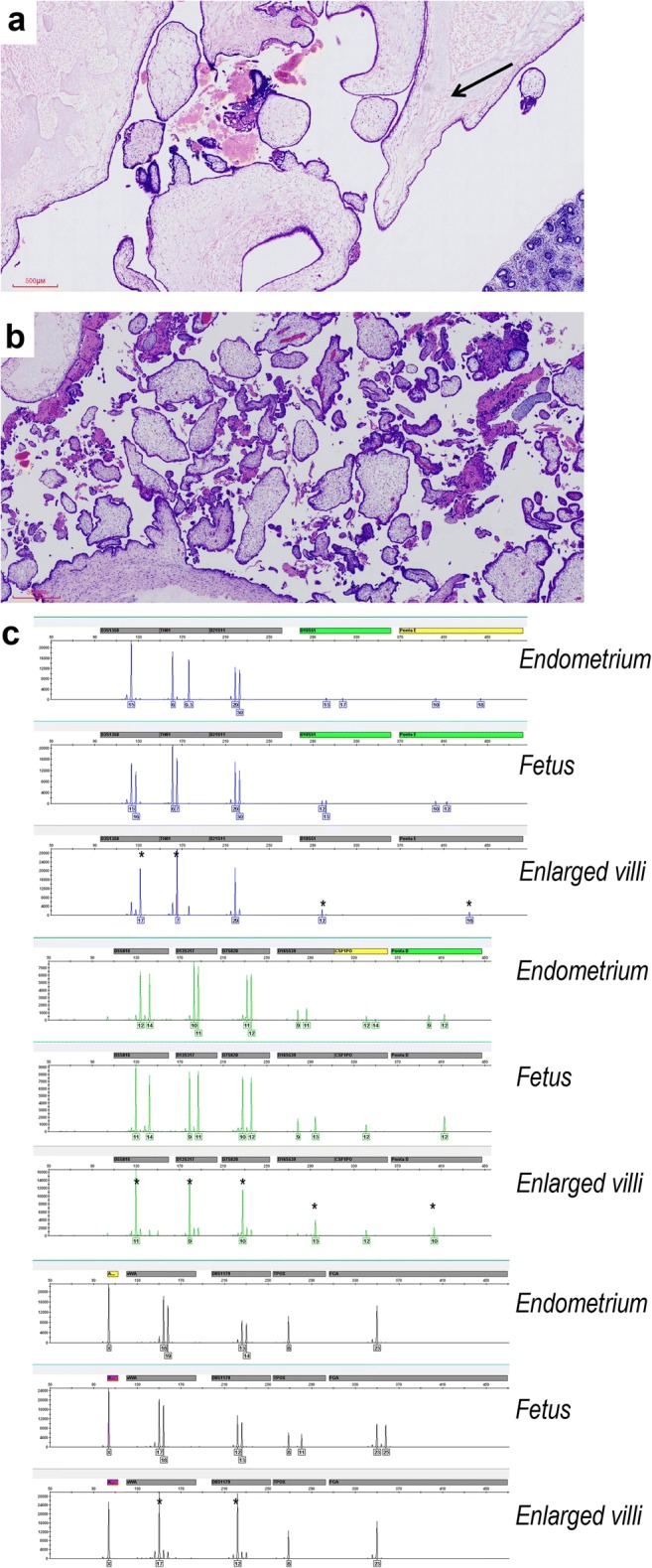
Twin pregnancy with monospermic homozygous complete mole and a co-existent fetus (patient 3 in Table S1). a Large hydropic villi with cistern formation near the fetal components (arrow) (× 40 magnification). b Small fibrotic villi in other areas (× 40 magnification). c Genotyping revealed complete mole with monospermic homozygous paternal allele at 11 STR loci (asterisk) in the hydropic chorionic villus and biparental fetal conception
Furthermore, there were 22 patients presenting one sac without abnormal sonographic area. In these patients, fifteen were diagnosed with NMA, including one with ectopic pregnancy in right ovary, six with empty sac, and eight with one undeveloped fetus. They had intermediate level of serum β-HCG (< 30,000 mIU/mL), hydropic villi with mild trophoblastic hyperplasia, positive p57kip2 staining, and biparental diploid genotyping. Other five patients manifested enlarged villi with cellular stroma and trophoblastic inclusion, which simulated the morphology of partial mole. Trisomy was the final diagnosis according to p57kip2 positivity and biparental diploid genome with three alleles in specific chromosome, such as chromosome 7, 15, or 16 (Table S1).
The evacuation specimens of patient 4 grossly appeared as two villous populations. The normal-appearing villi were normal p57kip2 staining, whereas the enlarged villi with cellular stroma were “discordant” p57kip2 staining pattern (Fig. 2 a and b). DNA genotyping demonstrated that the enlarged villi shared the same biparental alleles with the normal-appearing villi at each locus but had double-dose homozygous paternal allele at 10 of the 15 autosomal loci (Fig. 2c). It indicated that the enlarged villi were mosaic with two cellular populations, one androgenetic in the stroma and one diploid biparental in the cytotrophoblast, both originating from the same zygote and a single-sperm fertilization. The final diagnosis appeared to be a twin pregnancy with usual non-molar and mosaic non-molar components, rather than a monospermic partial mole (Table S1). The maternal blood total β-HCG decreased gradually to 1576 mIU/mL at 1 week and to the normal range at 10 weeks postpartum. Patient 18 also had two villous populations, in which the normal villi were positive p57kip2 staining and the enlarge villi were negative in both cytotrophoblast and stromal cells (Fig. 3 a and b). However, two villous populations were absolutely mixed and could not be distinguished by manual dissection. The genotyping assay of the mixed villi suggested polyploid genome fertilized by at least two sperms (dispermy) (Fig. 3c). On the basis of high-level β-HCG (300,000 mIU/mL), negative p57kip2 immunostaining, and excessive paternal alleles, we considered this patient with “twin pregnancy, favoring one co-existing complete mole” and advised her to enter the standard clinical follow-up (Table S1).
Fig. 2.
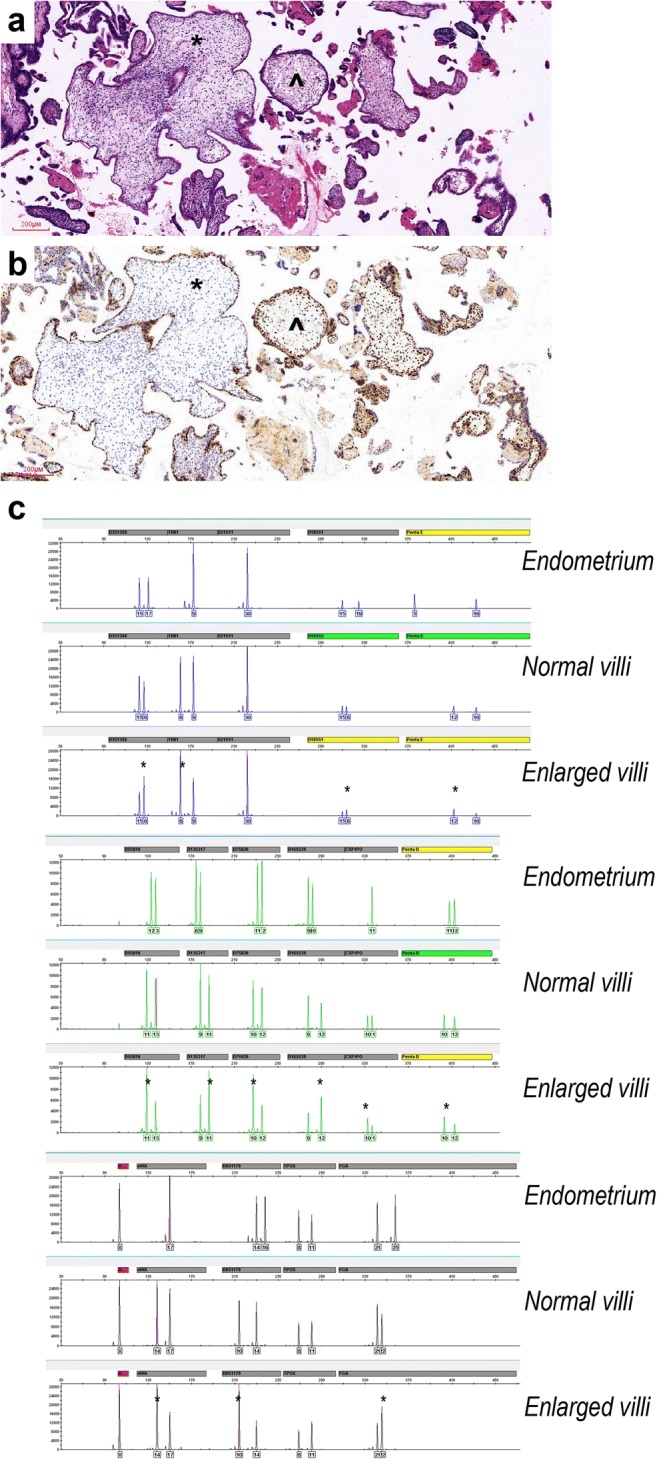
Twin pregnancy with non-molar and mosaic hydropic components (patient 4 in Table S1). a Normal villi (angle) and enlarged chorionic villi with cellular stroma (asterisk) (× 100 magnification). b Normal villi were uniform p57kip2-positive (angle), and enlarged villi were p57kip2-positive in cytotrophoblast and negative in stromal cells (asterisk) (immunohistochemistry; × 100 magnification). c The normal villi shared the same biparental alleles with the enlarged villi at each locus. The latter didn’t show two paternal alleles at any locus, and the present paternal allele at most loci was in double dose (asterisk), which suggested a possible androgenetic/biparental mosaic
Fig. 3.
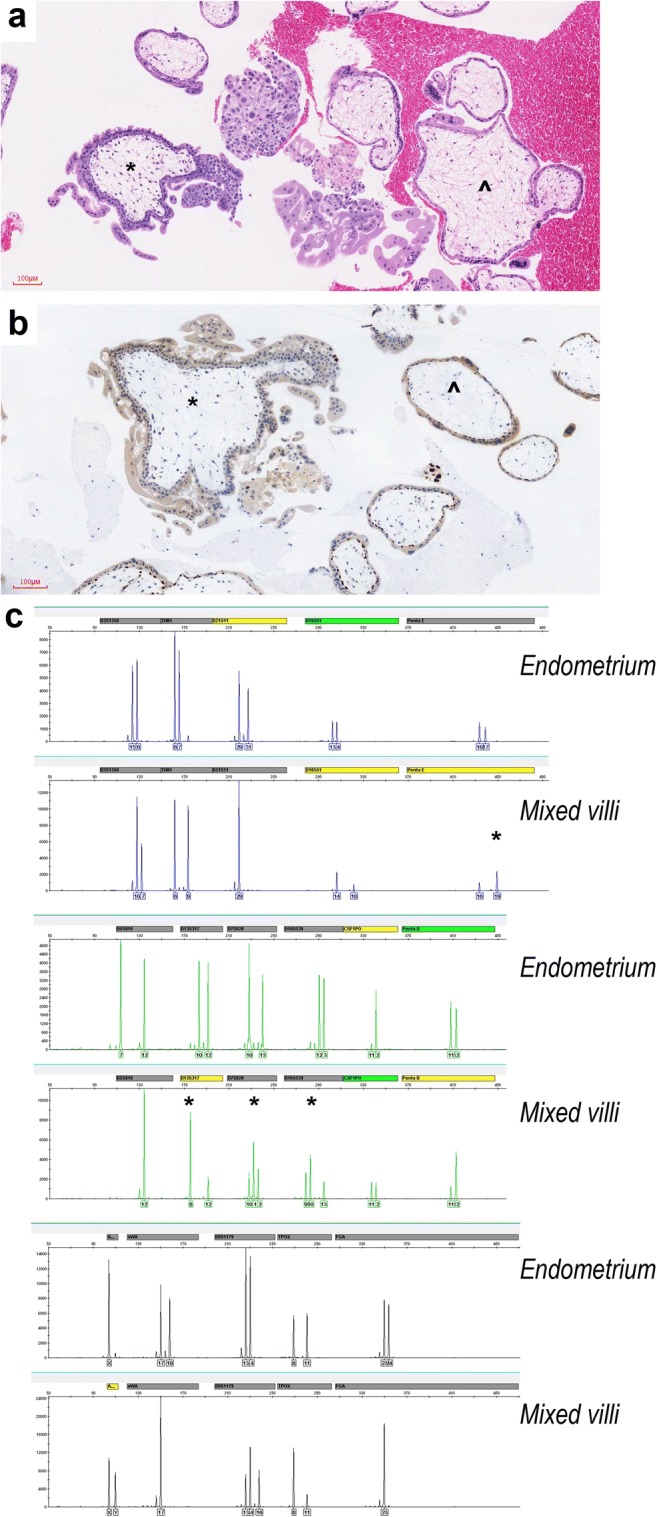
Twin pregnancy, favoring one co-existing complete mole (patient 18 in Table S1). a Hydropic villi without trophoblastic hyperplasia (angle) and cauliflower-like villi with karyorrhectic nuclear debris and trophoblastic hyperplasia (asterisk) (× 200 magnification). b Hydropic villi were p57kip2-positive (angle) and cauliflower-like villi were p57kip2-negative (asterisk) in cytotrophoblast (Immunohistochemistry; × 200 magnification). c Genotyping of the mixed villi suggested polyploid genome fertilized by at least two sperms according to the excessive paternal alleles at four of eleven informative loci (Penta E, D13S317, D7S820 and D16S539) (asterisk)
Diagnosis of suspicious HM in patients with two gestational sacs
Ten patients had two gestational sacs, mostly including one vacant sac and another with an undeveloped embryo. The hydropic villi exhibited uniformly positive p57kip2 immunostaining, which excluded the possibility of complete mole. Five of them showed balanced biparental origins in all STR loci and were diagnosed as non-molar singleton gestations. As shown in Table S1, patient 21 demonstrated some morphological features of partial mole, but she was diagnosed as trisomy 21 by three alleles at D21S11 locus in genotyping profile. For other four patients, the mixed gestational villi from different sacs demonstrated complicated DNA genotyping in most STR loci, indicating different genetic components from two zygotes. However, the concordant morphology and uniform p57kip2 positivity of villi made it difficult to discriminate one POC from another. Patient 6 was considered with “twin pregnancy, favoring non-molar gestation” based on the excessive maternal alleles at all STR loci. On the contrary, patients 10, 12, and 17 demonstrated excessive paternal alleles at four to six informative loci. An indeterminate diagnosis of “twin pregnancy, PHM cannot be excluded” was rendered and potentially led to clinical management for some abbreviated time frame.
Discussion
The use of ART is increasing; however, reports of molar pregnancy following ART remain scarce. A study using the Human Fertility and Embryology Authority data reported that the incidence of molar pregnancy was considerably lower in ART pregnancies than in spontaneous conceptions [3]. Serial embryo observation and morphology-based selection in standard IVF may be relevant to this phenomenon. Partial/complete HM with coexisting fetus is a rare condition [17]. But the increased incidence of iatrogenic multiple gestations may cause a concomitant increase of multiple gestations with complete mole and coexisting fetus (CHMCF) [18–20]. About 24–30% of CHMCF involved ovulation induction treatment, which might increase the rate of a nuclear “empty” egg supporting development of complete mole [20–22]. The counseling and management of CHMCF remained controversial because their risk of PGTD varies greatly in publications [4–7, 17]. At all events, the potential risk of recurrence if a patient has a mole through IVF may also suggest that pre-implantation genetic diagnosis should be considered in the next cycle.
During the first trimester, the ultrasound detection rate of TPHM is only approximately 68% for the lack of typical “storm-like” echo pattern [23]. In present study, three patients with twin pregnancy demonstrated only one gestational sac. On the other hand, TPHM always displays some normal villi and numerous edematous villi in the mixed POC, simulating an early NMA or a partial mole. Furthermore, TPHM should be excluded from other rare confusing conditions without risk of PGTD, such as androgenetic/biparental mosaic conceptions or placenta mesenchymal dysplasia [24].
Based on the unique genetic features of complete moles (purely androgenetic conceptions), partial moles (diandric triploid conceptions), and NMAs (balanced biparental conceptions), p57 immunostaining (a paternally imprinted, maternally expressed gene), and DNA genotyping could refine the diagnosis and subtyping of HM [24–26]. In thirty-seven patients after two-embryo transferring of present study, 22 NMAs, 6 trisomy syndromes, and 2 HMs were confirmed as singleton pregnancies by typical p57kip2 immunostaining and STR genotyping. Although six patients had two gestational sacs in ultrasonic findings, the embryonic components in the vacant sac might fade away after transferring.
However, after an anembryonic gestation, mixed mole-like hydropic villi originating from two independent gestational sacs could be the only remnants of a twin pregnancy, which makes DNA genotyping more difficult. Therefore, strategies integrating p57 kip2 staining with genotyping could triage patients with better cost efficiency and shorter turnaround times. The correct interpretation of discordant and/or divergent p57kip2 staining patterns and precise separation of different components to assure accurate molecular genotyping are critical [27, 28].
Firstly, uniform p57kip2 positivity and unbalanced biparental genotyping with excessive maternal alleles in all loci suggested that a diagnosis of “twin pregnancy, favoring non-molar gestation” was sufficient for the unnecessity of follow-up. On the contrary, because overrepresentation of the paternal genome was a fundamental genetic event of molar pregnancy, excessive paternal alleles in at least four informative loci should point to an equivocal diagnosis of “twin pregnancy, partial HM cannot be excluded”, even if the paternal:maternal ratio of suspicious loci was less than 2:1 (the standard situation of partial mole). Abbreviated follow-up might be recommended in view of the low risk of PGTD in partial mole. Secondly, some patients showed divergent or discordant p57kip2 staining patterns and complicated tetraploid or pentaploid genotyping of mixed POC. If the different villi populations could be separated manually, STR genotyping would give accurate diagnosis, such as TPHM. If the two different villi populations could not to be absolutely separated, “twin pregnancy, favoring one co-existing complete mole” would be appropriate according to high level β-HCG, the negative p57kip2 immunotype, and excessive paternal alleles in at least four informative STR loci. CHMCF after ART should be monitored by serum β-HCG and given chemotherapy timely because their incidence of PGTD was 53.3% in previous reports and 25% (1 of 4) in present study [7, 29]. Uncommonly, the divergent and discordant p57kip2 immunostaining villi populations were presented at the same patient. By genotyping, the normal villi with uniform p57kip2 positivity were from a biparental diploid NMA, and the enlarge villi with discordant p57kip2 staining indicated a rare androgenetic/biparental mosaic conception rather than a partial mole. The mechanism and risk of PGTD in such mosaic conceptions were not ascertained, and follow-up as HM was warranted in view of harboring androgenetic cell lines [12, 30].
So far, there are only few case reports of CHMCF following ART in the literature [29]. The strengths of this study are the sample size and the availability of complete information on morphological, immunohistochemical, and genetic analyses. One of the limitations of this study was that laser microdissection was not used to precisely identify the genotypes of abnormal villi from the closely intermixed villi. Otherwise, patients could have been specifically interpreted at the genetic level instead of an “indeterminate” diagnosis. For another, due to the short follow-up duration, it is difficult to conclude the risk of PGTD of the “indeterminate” patients.
In conclusion, for diagnostically challenging cases of possible TPHM, a strategy composed of selective clinicopathological screening, immunohistochemical interpretation, and accurate genotyping is recommended. An evaluation of p57kip2 staining patterns should be used as an initial assay to distinguish potential HM after IVF and two-embryo transferring. Independent DNA genotyping of different villi populations with abnormal p57kip2 expression can usually contribute a definitive diagnosis. Rational integration of ancillary techniques into routine practice will provide refined diagnosis, facilitate accurate assessment of the risk of PGTD, and guide appropriate management of HM in twin pregnancies following ART.
Electronic supplementary material
(XLSX 11 kb)
Contributions
Yan Liu played a major role in data collection and overview, interpreted the data of genotyping and immunochemistry, edited and revised the manuscript for intellectual content.
Xingzheng Zheng played a major role in data collection, interpreted the data of genotyping, and revised the manuscript for intellectual content.
Yuxiang Wang played a major role in pathological review, interpreted the data of immunochemistry, and revised the manuscript for intellectual content.
Yan Li played a major role in the molecular genotyping and immunochemistry experiments.
Congrong Liu played a major role in pathological review, interpreted the data, and revised the manuscript for intellectual content.
All authors approved the submitted manuscript and agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Funding information
This project was funded by the following sources:
1. National Key Research and Development Program of China, Contract grant number: 2018YFC1004000.
2. Leading Academic Discipline Project of Beijing Education Bureau, Contract grant number: BMU20110254.
Compliance with ethical standards
"The use of patient data for the present study has been approved by the local ethical committee. A written consent form was signed by each patient.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yan Liu, Email: laylaly@bjmu.edu.cn.
Congrong Liu, Email: congrong_liu@163.com.
References
- 1.Berkowitz RS, Goldstein DP. Clinical practice. Molar pregnancy. N. Engl. J. Med. 2009;360:1639–1645. doi: 10.1056/NEJMcp0900696. [DOI] [PubMed] [Google Scholar]
- 2.Slim R, Mehio A. The genetics of hydatidiform moles: new lights on an ancient disease. Clin. Genet. 2007;71:25–34. doi: 10.1111/j.1399-0004.2006.00697.x. [DOI] [PubMed] [Google Scholar]
- 3.Nickkho-Amiry M, Horne G, Akhtar M, Mathur R, Brison DR. Hydatidiform molar pregnancy following assisted reproduction. J. Assist. Reprod. Genet. 2019;36:667–671. doi: 10.1007/s10815-018-1389-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Albayrak M, Ozer A, Demir OF. Ozer S, Erkaya S. Complete mole coexistent with a twin fetus. Arch. Gynecol. Obstet. 2010;281:119–122. doi: 10.1007/s00404-009-1076-2. [DOI] [PubMed] [Google Scholar]
- 5.Kihara M, Usui H, Tanaka H, Inoue H, Matsui H, Shozu M. Complicating preeclampsia as a predictor of poor survival of the fetus in complete hydatidiform mole coexistent with twin fetus. J Reprod Med. 2012;57:325–328. [PubMed] [Google Scholar]
- 6.Lin LH, Maestá I, Braga A, Sun SY, Fushida K, Francisco RPV, Elias KM, Horowitz N, Goldstein DP, Berkowitz RS. Multiple pregnancies with complete mole and coexisting normal fetus in North and South America: a retrospective multicenter cohort and literature review. Gynecol. Oncol. 2017;145:88–95. doi: 10.1016/j.ygyno.2017.01.021. [DOI] [PubMed] [Google Scholar]
- 7.Niemann I, Sunde L, Petersen LK. Evaluation of the risk of persistent trophoblastic disease after twin pregnancy with diploid hydatidiform mole and coexisting normal fetus. Am. J. Obstet. Gynecol. 2007;197(45):e1–e5. doi: 10.1016/j.ajog.2007.02.038. [DOI] [PubMed] [Google Scholar]
- 8.Hui P, Martel M. Parkash V (2005) Gestational trophoblastic diseases: recent advances in histopathologic diagnosis and related genetic aspects. Adv. Anat. Pathol. 2005;12:116–125. doi: 10.1097/01.pap.0000163960.11107.73. [DOI] [PubMed] [Google Scholar]
- 9.McConnell TG, Murphy KM, Hafez M, Vang R, Ronnett BM. Diagnosis and subclassification of hydatidiform moles using p57 immunohistochemistry and molecular genotyping: validation and prospective analysis in routine and consultation practice settings with development of an algorithmic approach. Am. J. Surg. Pathol. 2009;33:805–817. doi: 10.1097/PAS.0b013e318191f309. [DOI] [PubMed] [Google Scholar]
- 10.Ronnett BM, DeScipio C, Murphy KM. Hydatidiform moles: ancillary techniques to refine diagnosis. Int. J. Gynecol. Pathol. 2011;30:101–116. doi: 10.1097/PGP.0b013e3181f4de77. [DOI] [PubMed] [Google Scholar]
- 11.Ronnett BM. Hydatidiform moles: ancillary techniques to refine diagnosis. Arch. Pathol. Lab. Med. 2018;142:1485–1502. doi: 10.5858/arpa.2018-0226-RA. [DOI] [PubMed] [Google Scholar]
- 12.Lewis GH, DeScipio C, Murphy KM, Haley L, Beierl K, Mosier S, Tandy S, Cohen DS, Lytwyn A, Elit L, Vang R, Ronnett BM. Characterization of androgenetic/biparental mosaic/chimeric conceptions, including those with a molar component: morphology, p57 immnohistochemistry, molecular genotyping, and risk of persistent gestational trophoblastic disease. Int. J. Gynecol. Pathol. 2013;32:199–214. doi: 10.1097/PGP.0b013e3182630d8c. [DOI] [PubMed] [Google Scholar]
- 13.Hoffner L, Dunn J, Esposito N, Macpherson T, Surti U. P57KIP2 immunostaining and molecular cytogenetics: combined approach aids in diagnosis of morphologically challenging cases with molar phenotype and in detecting androgenetic cell lines in mosaic/chimeric conceptions. Hum. Pathol. 2008;39:63–72. doi: 10.1016/j.humpath.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 14.Zhong H, Liu Y, Talmor M, Wu B, Hui P. Deparaffinization and lysis by hydrothermal pressure (pressure cooking) coupled with chaotropic salt column purification: a rapid and efficient method of dna extraction from formalin-fixed paraffin-embedded tissue. Diagn. Mol. Pathol. 2013;22:52–58. doi: 10.1097/PDM.0b013e318263f092. [DOI] [PubMed] [Google Scholar]
- 15.Zheng XZ, Hui P, Chang B, Gao ZB, Li Y, Wu BQ, Zhang B. STR DNA genotyping of hydatidiform moles in south China. Int. J. Clin. Exp. Pathol. 2014;7:4704–4719. [PMC free article] [PubMed] [Google Scholar]
- 16.Hui P. Molecular diagnosis of gestational trophoblastic disease. Expert. Rev. Mol. Diagn. 2010;10:1023–1034. doi: 10.1586/erm.10.93. [DOI] [PubMed] [Google Scholar]
- 17.Rao AR, Dafle K, Padmashri G, Rao DR, Sivakumar NC. Pregnancy outcome with coexisting mole after intracytoplasmic sperm injection: a case series. J Hum Reprod Sci. 2015;8:178–181. doi: 10.4103/0974-1208.165149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wee L, Jauniaux E. Prenatal diagnosis and management of twin pregnancies complicated by a co-existing molar pregnancy. Prenat. Diagn. 2005;25:772–776. doi: 10.1002/pd.1272. [DOI] [PubMed] [Google Scholar]
- 19.Massardier J, Golfier F, Journet D, Frappart L, Zalaquett M, Schott AM, Lenoir VT, Dupuis O, Hajri T, Raudrant D. Twin pregnancy with complete hydatidiform mole and coexistent fetus: obstetrical and oncological outcomes in a series of 14 cases. Eur. J. Obstet. Gynecol. Reprod. Biol. 2009;143:84–87. doi: 10.1016/j.ejogrb.2008.12.006. [DOI] [PubMed] [Google Scholar]
- 20.Piura B, Rabinovich A, Hershkovitz R, Maor E, Mazor M. Twin pregnancy with a complete hydatidiform mole and surviving co-existent fetus. Arch. Gynecol. Obstet. 2008;278:377–382. doi: 10.1007/s00404-008-0591-x. [DOI] [PubMed] [Google Scholar]
- 21.Dolapcioglu K, Gungoren A, Hakverdi S, Hakverdi AU, Egilmez E. Twin pregnancy with a complete hydatidiform mole and co-existent live fetus: two case reports and review of the literature. Arch. Gynecol. Obstet. 2009;279:431–436. doi: 10.1007/s00404-008-0737-x. [DOI] [PubMed] [Google Scholar]
- 22.Bruchim I, Kidron D, Amiel A, Altaras M, Fejgin MD. Complete hydatidiform mole and a coexistent viable fetus: report of two cases and review of the literature. Gynecol. Oncol. 2000;77:197–202. doi: 10.1006/gyno.2000.5733. [DOI] [PubMed] [Google Scholar]
- 23.Vaisbuch E, Ben-Arie A, Dgani R, Perlman S, Sokolovsky N, Hagay Z. Twin pregnancy consisting of a complete hydatidiform mole and co-existent fetus: report of two cases and review of literature. Gynecol. Oncol. 2005;98:19–23. doi: 10.1016/j.ygyno.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 24.Buza N, Hui P. Partial hydatidiform mole: histologic parameters in correlation with DNA genotyping. Int. J. Gynecol. Pathol. 2013;32:307–315. doi: 10.1097/PGP.0b013e3182626011. [DOI] [PubMed] [Google Scholar]
- 25.Madi JM, Braga AR, Paganella MP, Litvin IE, Da Ros Wendland EM. Accuracy of p57KIP2 compared with genotyping for the diagnosis of complete hydatidiform mole: protocol for a systematic review and meta-analysis. Syst Rev. 2016;5:169. doi: 10.1186/s13643-016-0349-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hui P, Buza N, Murphy KM, Ronnett BM. Hydatidiform moles: genetic basis and precision diagnosis. Annu. Rev. Pathol. 2017;12:449–485. doi: 10.1146/annurev-pathol-052016-100237. [DOI] [PubMed] [Google Scholar]
- 27.Landolsi H, Missaoui N, Brahem S, Hmissa S, Gribaa M, Yacoubi MT. The usefulness of p57(KIP2) immunohistochemical staining and genotyping test in the diagnosis of the hydatidiform mole. Pathol. Res. Pract. 2011;207:498–504. doi: 10.1016/j.prp.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 28.Banet N, DeScipio C, Murphy KM, Beierl K, Adams E, Vang R, Ronnett BM. Characteristics of hydatidiform moles: analysis of a prospective series with p57 immunohistochemistry and molecular genotyping. Mod. Pathol. 2014;27:238–254. doi: 10.1038/modpathol.2013.143. [DOI] [PubMed] [Google Scholar]
- 29.Nobuhara I, Harada N, Haruta N, Higashiura Y, Watanabe H, Watanabe S, Hisanaga H, Sado T. Multiple metastatic gestational trophoblastic disease after a twin pregnancy with complete hydatidiform mole and coexisting fetus, following assisted reproductive technology: case report and literature review. Taiwan J Obstet Gynecol. 2018;57:588–593. doi: 10.1016/j.tjog.2018.06.020. [DOI] [PubMed] [Google Scholar]
- 30.Surti U, Hoffner L, Kolthoff M, Dunn J, Hunt J, Sniezek L, Macpherson T. Persistent gestational trophoblastic disease after an androgenetic/biparental fetal chimera: a case report and review. Int. J. Gynecol. Pathol. 2006;25:366–372. doi: 10.1097/01.pgp.0000215295.45738.ed. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX 11 kb)


