Abstract
Background
Klebsiella pneumoniae is an important cause of healthcare-associated infection. Carbapenemases have increasingly been reported in Enterobacteriaceae, especially in K. pneumoniae.
Propose
The objective of this study was to determine antibiotic resistance patterns, and the molecular epidemiology of multidrug resistant K. pneumoniae isolates, obtained from hospitalized patients in Shiraz, Iran.
Methods
In this study, 60 K. pneumoniaeisolates were collected from Nemazee and Faghihi referral hospitals. Antibiotic susceptibility testing and MIC were performed by disk diffusion test and Epsilometer (E)-test strips, respectively. Carbapenemase genes were identified by polymerase chain reaction and sequencing. Then, clonal relationships were analyzed, using PFGE.
Results
Thirty-three out of 60 K. pneumoniae isolates were resistant to carbapenems. Among the isolates, 86.6% were multidrug resistant (MDR). Polymyxin B (18.3%) and tigecycline (23.3%) were shown to be the most active agents against K. pneumoniae isolates. In our study, the high prevalence of blaNDM (45%) and blaOXA-48 (10%) was detected.
Conclusion
The results of this study revealed the widespread carbapenemase gene between different wards in hospitals as a risk factor for treatment options. PFGE analysis showed 11 clusters and 3 singletons based on an 80% similarity level. Also, PFGE analysis showed that there were similar genetic patterns among K. pneumoniae isolates and these patterns were responsible for the distribution of infection in hospitals.
Keywords: Klebsiella pneumoniae, pulsed-field gel electrophoresis, PFGE, carbapenemases
Introduction
Klebsiella pneumoniae is a known cause of nosocomial infections, especially bloodstream-associated infections (BSIs), respiratory tract infections (RTIs), and urinary tract infections (UTIs).1
This bacterium is the cause of hospital-acquired infections such as bloodstream infections, pneumonia, meningitis, sepsis, infections in newborns, and intensive care unit patients.2 In the last decade, the rate of morbidity and mortality related to these infections has increased.3 The incidence and mortality rates were reported as 7.1 per 100,000 per year and 1.3 per 100,000 per year, respectively.4 In recent years, one of the biggest concerns for public health has been resistance to carbapenems, known as carbapenem resistance Enterobacteriaceae (CRE).
Carbapenemases have three crucial classes including Ambler’s class A (KPC enzymes), class B metallo-beta-lactamase (NDM, VIM, and IMP enzymes), and class D oxacillinase (OXA-48-like enzymes).5,6 These classes are unique to some special microorganisms and geographical regions in the world; for instance, KPC producers are found all over the world, with reports from Greece, Israel, France, China, and South America.7,8 Amongst these beta-lactam genes, blaNDM and blaOXA have the most active role in creating microbial resistance.9
Given the increasing trend of antibiotic resistance in K. pneumoniae, which has become a global concern, the use of an efficient genotyping method in order to monitor and control the wide spread of epidemic clones between hospitals is essential.10,11 Among different typing methods, pulsed-field gel electrophoresis (PFGE) is considered the gold standard for typing bacteria.12 PFGE is a kind of molecular typing method that cuts DNA into smaller fragments by a specific enzyme, which helps to recognize the source and the outbreak of various species in the hospital.13 In this method, the change in banding patterns can explain the main genetic events, and determine the spread of carbapenemase resistance strains of K. pneumoniae in different regions. Surveying the molecular typing of carbapenemase-producing Enterobacteriaceae (CPE) isolates can provide a better perspective of bacterial dissemination.14,15
Due to the importance of the aforementioned variants of carbapenemase genes and the increase in the rate of these genes in hospitalized patients, especially in K. pneumoniae, finding their source is important. Hence, we investigated the antibiotic resistance and pattern of carbapenemase gene spread by PFGE among K. pneumoniae isolates obtained from hospitalized patients in Shiraz, southwest Iran.
Materials and Methods
Ethics
This study was approved by the Ethics Committee of Shiraz University of Medical Sciences, Shiraz, Iran (Approval No. IR.SUMS.REC.1395.S252). The study was carried out in accordance with the principles of the Declaration of Helsinki. However, there was no need to obtain informed consent as only leftovers from clinical specimens were used and personal details of all hospitalized patients were kept strictly confidential.
Bacterial Samples
Over an 8-month period, a total of 60 K. pneumoniae isolates were collected from Nemazee and Faghihi hospitals in Shiraz, Iran. These hospitals are two of the largest hospitals in southwestern Iran. The bacterial isolates were identified as K. pneumoniae via biochemical methods including oxidase, sugar fermentation, IMViC, Kliger’s iron agar, nitrate reduction, and motility tests.16
Antimicrobial Susceptibility Testing
Antimicrobial susceptibility testing was done by the standard disk diffusion method and European Committee on Antimicrobial Susceptibility Testing (EUCAST) (http://www.eucast.org/clinical_breakpoints/). Antimicrobial agents that were tested are as follows: ciprofloxacin (5 μg), cefepime (30 μg), gentamcin (10 μg), imipenem (10 μg), meropenem (10 μg), ceftazidime (30 μg), piperacillin (100 μg), ampicillin (25 μg), piperacillin/tazobactam (110/10 μg), aztreonam (30 μg), amikacin (30 μg), amoxicillin/clavulanic acid (20/10 μg), polymyxin B(300 U), and tigecycline (15 μg).
These antibiotics were purchased from MAST Chemical Company, UK. Escherichia coli ATCC 25922 was used as the standard strain in the antibacterial susceptibility testing. The MIC (minimum inhibitory concentration) by Epsilometer (E)-test strips (HiMedia, Co., India) was determined for isolates that were not susceptible to imipenem and meropenem in the disk diffusion for confirmation.17 In the current study, multidrug resistant (MDR) isolates were determined as acquired resistance to at least one agent in three or more antimicrobial groups.18
Carbapenemase Screening Assays by Phenotypic Test
The Modified Hodge test (MHT) was performed to identify the carbapenemase, according to CLSI guidelines.17 To detect MBL (metallo-beta-lactamase) producing isolates phenotypically, we performed a double-disk synergy test (DDST). Briefly, 0.5 m EDTA solution was prepared by dissolving 186.1 g of EDTA in 100 mL of distilled water, and the pH was adjusted to 8 by adding NaOH. Then, an IPM (10 µg) disk was placed 20 mm apart from a blank disk containing 10 µL of 0.5 m EDTA (750 µg) in a plate containing Muller–Hinton agar with cultured K. pneumoniae. The result was considered after 16–18 h of incubation at 37 °C. In the DDST, zone inhibition enhancement between IPM and EDTA disks was considered MBL positive.19
DNA Extraction and Detection of Carbapenemase Genes
The boiling method was applied to extract the template DNA to detect carbapenemase genes. The primers were used to detect blaKPC, blaOXA-48-like, blaIMP, blaVIM, and blaNDM as described by Doyle et al.5
The amplification reaction was carried out in a final volume of 25 μL containing 0.5 μL of each primer (10 pM), 12.5 μL of DNA Polymerase Master Mix RED (Ampliqon, Co., Denmark), 1 μL of DNA, and 10.5 μL of DNase and RNase-free water in a FlexCycler PCR. The Thermal Cycler (Analytik Jena, Germany) was performed through the following cycling conditions: an initial denaturation step at 95 °C for 5 min, followed by 35 cycles of DNA denaturation at 95 °C for 45 s, annealing at 45 s, and extension at 72 °C for 1 min, and a final extension at 72 °C for 8 min.
The PCR products were analyzed with gel electrophoresis on a 1.5% agarose gel in 0.5 Tris, EDTA, and Boric acid (TBE) buffer, followed by staining with KBC stain (CinnaGen, Iran) and visualized using a UV transilluminator. Positive controls for these genes were obtained from Pasteur Institute of Iran, Tehran.
DNA Sequencing
The purified PCR products of blaNDM and blaOXA-48 positive isolates were sequenced (Macrogen, South Korea). Then, the obtained nucleotide sequences were compared using online BLAST software (www.ncbi.nih.gov/BLASTprogram), and confirmed as NDM-1 variant.
Pulsed-Field Gel Electrophoresis Procedure
All isolates were typed using a PFGE technique based on the PulseNet One-Day (24–28 h) Standardized Laboratory Protocol.20 Briefly, the genomic DNA of E. coli isolates and Salmonellaser. Braenderup H9812 (standard marker) were embedded within agarose plugs. XbaI restriction endonuclease (10 U/μL) was used to digest genomic DNA according to the manufacturer’s recommendations (Thermo Scientific, Waltham, MA, USA). The obtained restriction fragments were separated by the CHEF-DR III System (Bio-Rad Laboratories, Hercules, CA, USA), using 1% X174 Agarose LFTM (Amresco, Solon, OH, USA), with switch times of 6–36 s, a voltage of 6 V, and an involved angle of 120° at 14 °C for 18–19 h. Gels were stained with GelRed™ (Biotium Inc., UK) and imaged with the UVItec Gel Documentation System (UVItec Ltd., Cambridge, UK). PFGE profiles were analyzed by Bionumerics software, version 7.1 (Applied Maths, Sint-Martens-Latem, Belgium). The Dice similarity coefficient with a UPGMA dendrogram was produced based on 1.5% tolerance windows and 1.5% optimization. An 80% cutoff line was considered to analyze genetic relatedness.
Statistical Analysis
Statistical analysis was performed using the chi-square test through SPSS 20 software; P values <0.05 were considered to be statistically significant.
Results
A total of 60 K. pneumoniae isolates were isolated from male (31, 51.7%), and female (29, 48.3%) patients at Nemazee and Faghihi hospitals. Collected samples are as follows: urine (30, 50%), sputum (8, 13.3%), throat (5, 8.3%), blood (8, 13.3%), wound (4, 6.7%), nasal/c (2, 3.3%), abscess (2, 3.3%), and plural (1, 1.7%). The samples were collected from hospitalized patients in the following wards: the ICU (25, 41.6%), internal (15, 25%), transplant (6, 10%), neonatal (5, 8.3%), surgery (4, 4.6%), nephrology (4, 4.6%), and OPD (1, 1.7%) (Table 1).
Table 1.
Characteristics of 60 Klebsiella pneumoniae Isolates from Nemazee and Faghihi referral hospitals
| Case Number | Age | Gender | Specimen | Ward | Hospital | PFGE Clusters | Pulsotype |
|---|---|---|---|---|---|---|---|
| 3 | 86 | F | Urine | Internal | Nemazee | C3 | 39 |
| 7 | 5 | M | Urine | Neonatal | Nemazee | B3 | 20 |
| 8 | 30 | F | Abscess | Nephrology | Nemazee | Singleton | 19 |
| 11 | 12 | M | Sputum | ICU | Faghihi | A3 | 8 |
| 13 | 28 | F | Urine | Internal | Nemazee | B2 | 14 |
| 14 | 71 | F | Urine | Internal | Nemazee | B2 | 15 |
| 15 | 60 | F | Urine | ICU | Nemazee | C3 | 38 |
| 16 | 70 | M | Blood | ICU | Nemazee | A3 | 9 |
| 17 | 60 | F | Blood | ICU | Nemazee | C3 | 38 |
| 19 | 60 | F | Plural | ICU | Nemazee | C3 | 36 |
| 20 | 25 | F | Urine | ICU | Nemazee | A3 | 7 |
| 21 | 30 | M | Urine | ICU | Faghihi | A3 | 10 |
| 23 | 43 | M | Sputum | ICU | Nemazee | C3 | 36 |
| 24 | 79 | M | Blood | ICU | Nemazee | A2 | 5 |
| 26 | 57 | M | Urine | Internal | Nemazee | B2 | 17 |
| 27 | 48 | F | Urine | Internal | Nemazee | C3 | 37 |
| 28 | 7 | F | Urine | Neonatal | Nemazee | A1 | 2 |
| 30 | 43 | M | Sputum | ICU | Nemazee | C1 | 22 |
| 32 | 36 | F | Urine | Neonatal | Nemazee | D2 | 44 |
| 33 | 16 | F | Urine | Neonatal | Nemazee | D2 | 44 |
| 36 | 40 | F | Urine | ICU | Faghihi | C1 | 25 |
| 38 | 63 | M | Urine | Transplant | Nemazee | C1 | 24 |
| 39 | 63 | F | Urine | ICU | Nemazee | C1 | 23 |
| 41 | 27 | F | Sputum | ICU | Nemazee | C2 | 28 |
| 42 | 3 | M | Urine | ICU | Nemazee | A3 | 7 |
| 45 | 63 | F | Abscess | OPD | Nemazee | Singleton | 45 |
| 48 | 79 | M | Urine | Transplant | Nemazee | A3 | 10 |
| 50 | 99 | F | Urine | Surgery | Nemazee | C2 | 26 |
| 53 | 3 | M | Sputum | Internal | Nemazee | B2 | 18 |
| 54 | 56 | F | Urine | Transplant | Nemazee | B1 | 11 |
| 55 | 17 | M | Urine | Transplant | Nemazee | C2 | 26 |
| 56 | 36 | M | Urine | Transplant | Nemazee | B1 | 12 |
| 61 | 9 | M | Blood | Internal | Nemazee | C2 | 27 |
| 62 | 6 | M | Sputum | Internal | Nemazee | C2 | 28 |
| 78 | 37 | F | Throat | ICU | Nemazee | D1 | 41 |
| 79 | 2 | M | Sputum | Nephrology | Nemazee | Singleton | 13 |
| 90 | 12 | F | Blood | ICU | Nemazee | A2 | 6 |
| 96 | 54 | M | Urine | Internal | Nemazee | C2 | 28 |
| 98 | 30 | F | Nasal/c | ICU | Nemazee | D1 | 43 |
| 101 | 59 | M | Nasal/c | Internal | Nemazee | B2 | 16 |
| 105 | 5 | F | Urine | Internal | Nemazee | C2 | 28 |
| 113 | 62 | F | Sputum | ICU | Nemazee | C2 | 32 |
| 121 | 10 | M | Urine | Internal | Nemazee | C2 | 35 |
| 124 | 53 | M | Wound | Internal | Nemazee | B3 | 20 |
| 136 | 1 | M | Urine | Surgery | Nemazee | C2 | 34 |
| 138 | 1 | M | Throat | ICU | Nemazee | A1 | 1 |
| 143 | 5 | M | Wound | Surgery | Nemazee | C2 | 32 |
| 144 | 4 | M | Throat | ICU | Nemazee | A1 | 3 |
| 147 | 2 | M | Blood | Nephrology | Nemazee | C2 | 31 |
| 154 | 1 | F | Urine | ICU | Nemazee | C2 | 29 |
| 157 | 4 | F | Urine | Internal | Nemazee | C2 | 32 |
| 180 | 26 | F | Blood | Internal | Faghihi | C1 | 22 |
| 184 | 58 | F | Wound | ICU | Nemazee | D1 | 42 |
| 185 | 3 | M | Blood | Neonatal | Nemazee | B3 | 21 |
| 188 | 27 | M | Wound | ICU | Nemazee | D1 | 40 |
| 196 | 71 | F | Urine | Nephrology | Nemazee | C1 | 24 |
| 197 | 35 | F | Throat | ICU | Nemazee | A1 | 4 |
| 202 | 4 | M | Urine | Surgery | Nemazee | C2 | 30 |
| 211 | 30 | M | Urine | Transplant | Nemazee | C2 | 33 |
| 217 | 41 | M | Throat | ICU | Nemazee | C2 | 29 |
| □P value | 0.00 | 0.01 | 0.001 | 0.20 |
Notes: □P values for statistical differences between PFGE patterns by the different variables.
The prevalence of each antibiotic resistance was as follows: imipenem 33 (55%), meropenem 33 (55%), cefepime 49 (81.7%), ciprofloxacin 39 (65%), gentamcin 40 (66.7%), amikacin 36 (60%), aztreonam 49 (81.7%), ceftazidime 50 (83.3%), piperacillin 53 (88.3%), ampicillin 60 (100%), polymyxin B 11 (18.3%), piperacillin/tazobactam 42 (70%), tigecycline 14 (23.3%), and amoxicillin/clavulanic acid 50 (83.3%) (Figure 1). Our results showed that there were no statistically significant differences in the antibiotic resistance rate of isolates between different specimens except for amoxicillin/clavulanic acid (p=0.02). Also, no statistically significant differences were found between antibiotic resistances in different wards; but most antibiotic resistance was detected in ICU and internal wards. Based on MIC (MIC>4) results, 33 isolates were resistant to imipenem or meropenem.
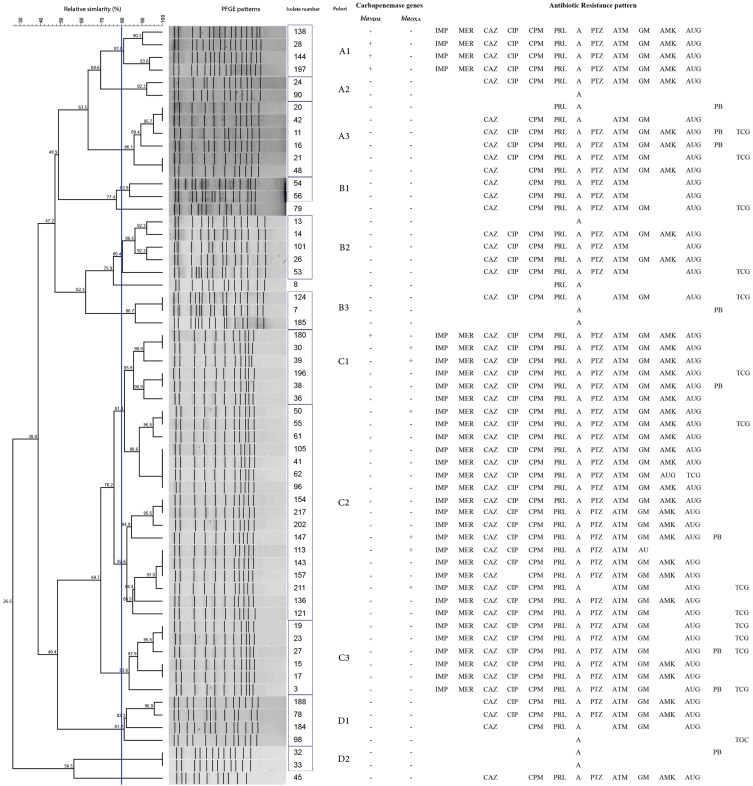
Figure 1.
Dendrogram of 60 K. pneumoniae isolates based on PFGE patterns after digestion with enzyme XbaІ associated with presence of carbapenemase genes and antibiotic resistance pattern; ceftazidime (CAZ), ciprofloxacin (CIP), cefepime (CPM), piperacillin (PRL), ampicillin (A), polymyxin B (PB), tigecycline (TGC) piperacillin/tazobactam (PTZ), aztreonam (ATM), gentamicin (GM), amikacin (AMK) amoxicillin/clavulanic acid (AUG), imipenem (IMP), meropenem (MEM).
Carbapenemase and MBL Screening
The DDST and MHT were performed on 33 carbapenem resistant isolates and the following results showed that 31 (93.9%) of isolates were MBL producing and 2 (6%) isolates were negative. Also, the results of MHT revealed that 30 out of 33 carbapenem resistant isolates were carbapenemase producing and 2 isolates were negative.
Molecular Analysis of Carbapenemase Genes
Of 60 K. pneumoniae isolates, 33 were resistant to carbapenems. The frequency of blaNDM (27, 45%) and blaOXA-48 (6, 10%) genes was detected amongst the isolates, which was confirmed by sequencing. However, in this study, blaKPC, blaVIM, and blaIMP were not detected.
There was a significant difference between blaNDM-1 positive and different specimens (p=0.01); the presence of blaNDM-1 was significantly higher in urine in comparison with the other samples.
Genetic Relatedness of Isolates
A total of 33 isolates were chosen due to resistance to carbapenems, and the other 27 isolates were opted to type by PFGE technique for the type of specimens and ward of the hospital. The PFGE dendrogram revealed 11 clusters and 3 singletons based on an 80% similarity level, with a high prevalence of cluster C2, being related to 17 isolates, followed by clusters C1, C3, and A3 linked to 6 strains. The least common clusters were A2 and D2 (2 isolates) (Figure 1). The 17 isolated strains of cluster C2 were from the internal, ICU, surgery, transplant, and central lab wards (Figure 1).
The genetic patterns of C2 (isolates no. 41, 96, 105, and 62), C3 (isolates no. 19, 23, 15, and 17), and C1 (isolates no. 198 and 38) of K. pneumoniae isolated had a high genetic similarity to the other groups. In the biggest cluster (C2), 4 isolates were similar (Figure 1). In this study, all 29 blaNDM-1 positive isolates belonged to 4 clusters (A1, C1, C2, and C3). The isolates in the A1 profile were not multidrug resistant and 7 clusters (A2, A3, B1, B2, B3, D1, and D2) were sensitive to imipenem and meropenem (Figure 1). The occurrence of different pulse types over the origin and ward is shown in Table 1.
Discussion
K. pneumoniae, an important cause of nosocomial infections, causes severe morbidity and mortality, especially in intensive care unit, pediatric, surgical, and transplant wards. Also, this bacterium is one of the most important causes of nosocomial infections with multidrug resistance.1 K. pneumoniae carbapenemases (KPCs) have been widely reported in the world, causing problems in the treatment of infections.6 The spread of K. pneumoniae with resistance to different antibiotics will limit effective treatment options.9 In most countries, the rates of resistance to carbapenems (imipenem, meropenem, and ertapenem) are on the rise in K. pneumoniae.9,21 Previous studies analyzed the use of broad-spectrum antibiotics (such as carbapenems), urinary catheters, and mechanical ventilation as the risk factors for acquiring CRE strains among hospitalized patients.22 In the current study, the rate of multidrug resistance among K. pneumoniae isolates was 86.6%.
Also, resistance to imipenem and meropenem was 55%, and the rate of imipenem and meropenem resistance was increasing. The rate of resistance to imipenem and meropenem in our strains is in line with other studies in Tehran, Egypt, and China.23–25 Carbapenemase such as blaNDM is a plasmid-borne gene;25 so, it may have readily been transmitted to other susceptible bacteria and make them resistant to carbapenemase, which might lead to failure in controlling spread of these bacteria in the hospital environment. This study investigated the prevalence of carbapenemase genes (blaNDM and blaOXA-48 were 45% and 10%, respectively); these genes were isolated from different wards, especially the ICU. In addition, KPC isolates (13/25, 52%) were obtained from patients in the ICU, which highlights the importance of this ward, and the length of hospitalization in this section. Our results were in line with studies from Turkey and Tehran.26,27 In this study, the blaOXA-48 gene was detected in 6 (10%) of the KPCs producing isolates. The OXA-48 gene is one of the most blaOXA-48-like common variants belonging to class D carbapenemase. The plasmid blaOXA-48 gene is located within a composite transposon, namely Tn1999, that flanks the carbapenemase gene and facilitates mobilizing an intervening DNA segment.28 The first report of the blaOXA-48 gene was reported in Turkey in 2001.29 Different distribution of the blaOXA-48 gene was reported in different parts of Iran, in Tehran in 2014 (96.4%) and Isfahan in 2018 (90.2%).29,30 Also, the prevalence of OXA-48 genes among K. pneumoniae strains was reported from several countries including China, France, Turkey, and Saudi Arabia,31–34 which raises serious concerns when treating patients in different parts of the world. In the current study, the results showed that a combination of phenotypic and molecular tests should be used simultaneously to detect carbapenemase.
In a study in Tehran, Iran, PFGE as the molecular typing method revealed a high genomic relatedness among K. pneumoniae isolates. Epidemiological investigation such as PFGE is used for the bacteria isolates outbreak and also isolates transmission among patients, and within hospital wards.23
In this study, PFGE analysis showed that K. pneumoniae was divided into 11 clusters and 3 singletons. The largest clusters had 4 similar isolates, and the other clusters had 2 or 3 identical isolates (Figure 1). Remarkably, clusters A1, C1, and C2 (isolates no. 138, 28, 144, and 197; 180, 30, and 39; and 105, 41, 61, 96, 154, and 217) had similar patterns of resistance and carbapenemase genes. Also, PFGE profiles showed the most prevalent carbapenemase-producing K. pneumoniae was found in cluster C1 in the ICU ward. In this cluster, all of the K. pneumoniae isolates had blaNDM or blaOXA. K. pneumoniae is one of the most important pathogens found in the intestinal tract and in the human nasopharynx. The carrier rate is 60–70% in the hospital, and related nosocomial infections are transmitted through person to person, environment, or contaminated water and food, infected individuals, contaminated healthcare personnel’s skin, or contact via shared items and surfaces.27 Due to similarity of these isolates, inter-hospital transfer between patients is likely to happen, for example in the ICU, transplant, and internal wards. In the current study, the isolate numbers 20 and 42 were similar, which were detected in the urine sample of patients admitted to the ICU. It seems that this might be due to transmission of isolates between patients in the ICU. Although the hospital environment such as door knobs, bed sheets, stethoscopes, etc. can also be considered as transmitters, the major transmission route has been said to be hospital staff, nurses, doctors, and colonies of patients which might cause cross-transmission. Also, medical staff and employees working outside the hospital cause the wide spread of strains in different hospital wards.25 Infection control precautions applying the basic rules of sanitation, efficient use of disinfectants, and careful handwashing would significantly decrease such transmissions.35
A study from Egypt reported that they had isolated clonally related strains which were from different sources including different patients in a hospital and environmental samples.24 In another study in Kurdistan, Iran, PFGE analysis of K. pneumoniae isolates was performed, and they identified 11 various profiles and one major clone, and transmission between patients and different wards was also proposed.36
According to a previous study, environmental reservoirs in hospitals can cause the transmission of different bacteria and antibiotic resistance in different wards.25 As previously reported, antimicrobial resistance genes can easily spread among different species and strains and distribute among the hospital wards. Transmission of microbial infections and resistant genes to carbapenems was reported in hospital wards.23,26,27
The role of infection control and careful surveillance is very important in hospitals in order to reduce infections and antibiotic resistance. We can use this strategy to prevent and manage antimicrobial resistance in nosocomial settings and reduce the outbreaks.
In general, the results of this study showed that there were similar genetic patterns among isolated K. pneumoniae isolates from Nemazee referral hospital, in Shiraz. These patterns are responsible for dissemination of infection in hospitals. However, we did not see any similar genetic patterns between the collected isolates from Faghihi hospital, which might be due to timing when the samples were collected; only four K. pneumoniae isolates were isolated from this hospital. Moreover, there is a genetic correlation between these patterns; also, they might have similar genetic origin. However, we understood that most of these types are resistant to most common antibiotics and 29 isolates had carbapenemase genes; hence, designing a protective program to control infections created in different parts of hospitals, especially the ICU, is vital.
Environmental samples could have been collected in this study, which limits better explanation of the spread of our studied resistant bacteria.
Conclusion
It is important to identify the antibiotic resistant genes in each hospital via proper molecular methods and to determine the antibiotic resistance pattern in order to prescribe proper antibiotics for an effective treatment. Genotypic methods for carbapenemase detection such as PCR as a rapid sensitive method and finding the source of infection in outbreaks using PFGE may help to develop infection control strategies and decrease the chance of spreading organisms in hospitals.
Acknowledgments
The authors would like to thank Basic Sciences in Infectious Diseases Research Center, Shiraz University of Medical Sciences, Shiraz, Iran (Grant number 94-9600). The authors alone are responsible for the content and writing of the paper and declare no conflicts of interest. The authors wish to thank Mr H. Argasi at the Research Consultation Center (RCC) of Shiraz University of Medical Sciences for his invaluable assistance in editing this manuscript.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Pitout JD, Nordmann P, Poirel L. Carbapenemase-producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob Agents Chemother. 2015;59(10):5873–5884. doi: 10.1128/AAC.01019-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guo Y, Zhou H, Qin L, et al. Frequency, antimicrobial resistance and genetic diversity of Klebsiella pneumoniae in food samples. PLoS One. 2016;11(4):e0153561. doi: 10.1371/journal.pone.0153561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giani T, Pini B, Arena F, et al. Epidemic diffusion of KPC carbapenemase-producing Klebsiella pneumoniae in Italy: results of the first countrywide survey, 15 May to 30 June 2011. Euro Surveill. 2013;18(22):1–7. [PubMed] [Google Scholar]
- 4.Meatherall BL, Gregson D, Ross T, Pitout JD, Laupland KB. Incidence, risk factors, and outcomes of Klebsiella pneumoniae bacteremia. Am J Med. 2009;122(9):866–873. doi: 10.1016/j.amjmed.2009.03.034 [DOI] [PubMed] [Google Scholar]
- 5.Doyle D, Peirano G, Lascols C, Lloyd T, Church DL, Pitout JD. Laboratory detection of enterobacteriaceae that produce carbapenemases. J Clin Microbiol. 2012;50(12):3877–3880. doi: 10.1128/JCM.02117-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonnin RA, Jousset AB, Urvoy N, et al. Detection of GES-5 carbapenemase in Klebsiella pneumoniae, a newcomer in France. Antimicrob Agents Chemother. 2017;61(3):1–4. doi: 10.1128/AAC.02263-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Correa L, Martino MD, Siqueira I, et al. A hospital-based matched case-control study to identify clinical outcome and risk factors associated with carbapenem-resistant Klebsiella pneumoniae infection. BMC Infect Dis. 2013;13:80. doi: 10.1186/1471-2334-13-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cury AP, Andreazzi D, Maffucci M, Caiaffa HH Jr, Rossi F. The modified Hodge test is a useful tool for ruling out Klebsiella pneumoniae carbapenemase. Clinics (Sao Paulo). 2012;67(12):1427–1431. doi: 10.6061/clinics [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xie L, Dou Y, Zhou K, et al. Coexistence of blaOXA-48 and truncated blaNDM-1 on different plasmids in a Klebsiella pneumoniae isolate in China. Front Microbiol. 2017;8:133. doi: 10.3389/fmicb.2017.00133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paterson DL, Bonomo RA. Extended-spectrum beta-lactamases: a clinical update. Clin Microbiol Rev. 2005;18(4):657–686. doi: 10.1128/CMR.18.4.657-686.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li W, Raoult D, Fournier PE. Bacterial strain typing in the genomic era. FEMS Microbiol Rev. 2009;33(5):892–916. doi: 10.1111/j.1574-6976.2009.00182.x [DOI] [PubMed] [Google Scholar]
- 12.Ripabelli G, Sammarco M, Flocco R, Scutellà M, Recchia L, Grasso G. Klebsiella pneumoniae isolated from intensive care unit patients with respiratory tract infections: characterization by pulsed-field gel electrophoresis, antimicrobial resistance and pcrs for carbapenemase genes detection. J Respir Med Lung Dis. 2017;2(1):1008. [Google Scholar]
- 13.Gouby A, Neuwirth C, Bourg G, et al. Epidemiological study by pulsed-field gel electrophoresis of an outbreak of extended-spectrum beta-lactamase-producing Klebsiella pneumoniae in a geriatric hospital. J Clin Microbiol. 1994;32(2):301–305. doi: 10.1128/JCM.32.2.301-305.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tenover FC, Arbeit RD, Goering RV, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33(9):2233–2239. doi: 10.1128/JCM.33.9.2233-2239.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ripabelli G, Tamburro M, Guerrizio G, et al. Tracking multidrug-resistant Klebsiella pneumoniae from an Italian hospital: molecular epidemiology and surveillance by PFGE, RAPD and PCR-based resistance genes prevalence. Curr Microbiol. 2018;75(8):977–987. doi: 10.1007/s00284-018-1475-3 [DOI] [PubMed] [Google Scholar]
- 16.Mahon CR, Lehman DC, Manuselis G. Textbook of Diagnostic Microbiology-E-Book. Elsevier Health Sciences; 2014. [Google Scholar]
- 17.CLSI. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fourth Informational Supplement. M100-S24 January; 2017. [Google Scholar]
- 18.Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. doi: 10.1111/j.1469-0691.2011.03570.x [DOI] [PubMed] [Google Scholar]
- 19.Behera B, Mathur P, Das A, Kapil A, Sharma V. An evaluation of four different phenotypic techniques for detection of metallo-β-lactamase producing pseudomonas aeruginosa. Indian J Med Microbiol. 2008;26(3):233. doi: 10.4103/0255-0857.39587 [DOI] [PubMed] [Google Scholar]
- 20.Control CfD, Prevention. Standard Operating Procedure for PulseNet PFGE of Escherichia Coli O157: H7, Escherichia Coli Non-O157 (STEC), Salmonella Serotypes, Shigella Sonnei and Shigella Flexneri. Atlanta: Centers for Disease Control and Prevention; 2013. [Google Scholar]
- 21.Queenan AM, Bush K. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev. 2007;20(3):440–458. doi: 10.1128/CMR.00001-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu J, Ding B, Xu X, et al. Klebsiella pneumoniae: development of carbapenem resistance due to acquisition of blaNDM-1 during antimicrobial therapy in twin infants with pneumonia. Front Microbiol. 2015;6:1399. doi: 10.3389/fmicb.2015.01399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shahcheraghi F, Aslani MM, Mahmoudi H, et al. Molecular study of carbapenemase genes in clinical isolates of enterobacteriaceae resistant to carbapenems and determining their clonal relationship using pulsed-field gel electrophoresis. J Med Microbiol. 2017;66(5):570–576. doi: 10.1099/jmm.0.000467 [DOI] [PubMed] [Google Scholar]
- 24.Mohamed ER, Aly SA, Halby HM, Ahmed SH, Zakaria AM, El-Asheer OM. Epidemiological typing of multidrug-resistant Klebsiella pneumoniae, which causes paediatric ventilator-associated pneumonia in Egypt. J Med Microbiol. 2017;66(5):628–634. doi: 10.1099/jmm.0.000473 [DOI] [PubMed] [Google Scholar]
- 25.Zheng R, Zhang Q, Guo Y, et al. Outbreak of plasmid-mediated NDM-1-producing Klebsiella pneumoniae ST105 among neonatal patients in Yunnan, China. Ann Clin Microbiol Antimicrob. 2016;15(1):10,1–8. doi: 10.1186/s12941-016-0124-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Celikbilek N, Unaldi O, Kirca F, Gozalan A, Acikgoz ZC, Durmaz R. Molecular characterization of carbapenem-resistant Klebsiella pneumoniae species isolated from a Tertiary Hospital, Ankara, Turkey. Jundishapur J Microbiol. 2017;10(10):1–7. doi: 10.5812/jjm [DOI] [Google Scholar]
- 27.Solgi H, Badmasti F, Aminzadeh Z, et al. Molecular characterization of intestinal carriage of carbapenem-resistant enterobacteriaceae among inpatients at two Iranian university hospitals: first report of co-production of bla NDM-7 and bla OXA-48. Eur J Clin Microbiol. 2017;36(11):2127–2135. doi: 10.1007/s10096-017-3035-3 [DOI] [PubMed] [Google Scholar]
- 28.Berger S, Alauzet C, Aissa N, et al. Characterization of a new blaOXA-48-carrying plasmid in enterobacteriaceae. Antimicrob Agents Chemother. 2013;57(8):4064–4067. doi: 10.1128/AAC.02550-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moghadampour M, Rezaei A, Faghri J. The emergence of blaOXA-48 and blaNDM among ESBL-producing Klebsiella pneumoniae in clinical isolates of a tertiary hospital in Iran. Acta Microbiol Immunol Hung. 2018;65(3):335–344. doi: 10.1556/030.65.2018.034 [DOI] [PubMed] [Google Scholar]
- 30.Azimi L, Nordmann P, Lari AR, Bonnin RA. First report of OXA-48-producing Klebsiella pneumoniae strains in Iran. GMS Hyg Infect Control. 2014;9(1):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo L, An J, Ma Y, et al. Nosocomial outbreak of OXA-48-producing Klebsiella pneumoniae in a Chinese hospital: clonal transmission of ST147 and ST383. PLoS One. 2016;11(8):e0160754. doi: 10.1371/journal.pone.0160754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cuzon G, Ouanich J, Gondret R, Naas T, Nordmann P. Outbreak of OXA-48-positive carbapenem-resistant Klebsiella pneumoniae isolates in France. Antimicrob Agents Chemother. 2011;55(5):2420–2423. doi: 10.1128/AAC.01452-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nazik H, Ongen B, Ilktac M, et al. Carbapenem resistance due to Bla^ sub OXA-48^ among ESBL-producing Esherichia Coli And Klebsiella Pneumoniae isolates in a Univesity Hospital, Turkey. Southeast Asian J Trop Med Public Health. 2012;43(5):1178. [PubMed] [Google Scholar]
- 34.Shibl A, Al-Agamy M, Memish Z, Senok A, Khader SA, Assiri A. The emergence of OXA-48-and NDM-1-positive Klebsiella pneumoniae in Riyadh, Saudi Arabia. Int J Infect Dis. 2013;17(12):e1130–e1133. doi: 10.1016/j.ijid.2013.06.016 [DOI] [PubMed] [Google Scholar]
- 35.World Health Organization. WHO Guidelines on Hand Hygiene in Health Care (Advanced Draft): Global Safety Challenge 2005–2006: Clean Care Is Safer Care. Geneva: World Health Organization; 2006. [Google Scholar]
- 36.Peerayeh SN, Derakhshan S, Fallah F, Bakhshi B. Strain typing and molecular characterization of CTX-M-1 group ESBL in clinical Klebsiella pneumoniae isolated from children. Arch Pediatr Infect Dis. 2017;5(2):1–7. [Google Scholar]