Summary
Mammalian skeletal muscle possesses a unique ability to regenerate, which is primarily mediated by a population of resident muscle stem cells (MuSCs) and requires a concerted response from other supporting cell populations. Previous targeted analysis has described the involvement of various specific populations in regeneration, but an unbiased and simultaneous evaluation of all cell populations has been limited. Therefore, we used single-cell RNA-sequencing to uncover gene expression signatures of over 53,000 individual cells during skeletal muscle regeneration. Cells clustered into 25 populations and subpopulations, including a subpopulation of immune gene enriched myoblasts (immunomyoblasts) and subpopulations of fibro-adipogenic progenitors. Our analyses also uncovered striking spatiotemporal dynamics in gene expression, population composition, and cell-cell interaction during muscle regeneration. These findings provide insights into the cellular and molecular underpinning of skeletal muscle regeneration.
Subject Areas: Stem Cells Research, Developmental Biology, Bioinformatics
Graphical Abstract

Highlights
-
•
scRNA-seq of 53,000 cells reveals known and new cell subsets in regenerating muscle
-
•
Immune cells exhibit early infiltration, pro- and anti-inflammation and slow resolution
-
•
Linear trajectory of activated FAPs to Dpp4+ and Cxcl14+ cells in non-injured muscle
-
•
A subset of MuSCs enriched for immune gene expression in regenerating muscle
Stem Cells Research; Developmental Biology; Bioinformatics
Introduction
Tissue regeneration is a necessary process that allows damaged tissues to repair and remodel upon injury. Mammalian tissue regeneration is restricted to a subset of tissues, and incomplete repair can lead to scar formation or fibrotic deposition (Cordero-Espinoza and Huch, 2018, Larouche et al., 2018, Moyer and Wagner, 2011, Wynn and Vannella, 2016). Therefore, it is critical to understand the cell types and processes that mediate tissue healing in order to improve regenerative efficiency while limiting scar formation. Although regeneration is a complex and regulated process, skeletal muscle harbors a well-studied population of stem cells that supports its regenerative capacity (Yin et al., 2013). As such, skeletal muscle is an ideal tissue to investigate mechanisms underlying successful regeneration toward improving stem-cell-based therapies.
Skeletal muscle is composed of multinucleated mature muscle cells (myofibers), a resident pool of muscle stem cells (MuSCs, also called muscle satellite cells), and other populations such as fibro-adipogenic progenitors (FAPs), endothelial cells, tenocytes, and resident immune cells (Paylor et al., 2011). Upon injury, MuSCs activate, proliferate, differentiate, and fuse together to repair damaged myofibers. However, their appropriate responses are mediated by both resident and infiltrating cells (Paylor et al., 2011, Relaix and Zammit, 2012). Immediately after injury, neutrophils and macrophages invade the damaged tissue and sustain a pro-inflammatory environment to help clear necrotic tissue (Juhas et al., 2018, Lu et al., 2011). The pro-inflammatory environment sustains MuSC proliferation, whereas the anti-inflammatory environment allows for MuSC differentiation, providing synergism between the immune and stem cell responses (Arnold et al., 2007, Deng et al., 2012, Villalta et al., 2011). For example, anti-inflammatory macrophages couple angiogenesis with MuSC differentiation through the production of oncostatin M, underscoring the interdependence of various cellular responses (Christov et al., 2007, Latroche et al., 2017, Arnold et al., 2007, Burzyn et al., 2013, Hardy et al., 2016, Heredia et al., 2013, Joe et al., 2010, Segawa et al., 2008, Tidball and Wehling-Henricks, 2007). These and many other studies emphasize the importance of MuSCs, supporting cell types and intercellular communication networks for successful muscle regeneration. However, conventional research has relied on targeted approaches to evaluate population-specific characteristics and thus has not provided a full picture of the events and dynamics.
Single-cell RNA-sequencing (scRNA-seq) provides the opportunity to deconvolute heterogeneous tissue into individual cells based on their transcriptomic profiles (Hwang et al., 2018). In combination with various computational techniques, scRNA-seq has revolutionized our understanding of tissue function and exposed a tremendous amount of heterogeneity in homeostatic tissues (Zeng and Dai, 2019, Zilionis et al., 2019). For example, scRNA-seq has provided insights about the regeneration program in axolotl, identified an interstitial fat progenitor population in both mouse and human adipose tissue, and demonstrated the similarity of infiltrating myeloid cells in human and mouse lung cancer (Gerber et al., 2018, Merrick et al., 2019, Zilionis et al., 2019). scRNA-seq has been used to describe cell-cell communication networks within tumor microenvironments, small intestinal crypts, mouse bone marrow, and across liver endothelial cells, emphasizing the breadth of information that can be gained to understand the cellular and molecular regulation of tissue homeostasis and diseases (Boisset et al., 2018, Halpern et al., 2018, Kumar et al., 2018).
Recent scRNA-seq studies on mouse hindlimb muscle identified a new population of interstitial tenocytes that function during muscle repair and defined distinct transcriptional programs in quiescent and activated MuSCs (Dell’orso et al., 2019, Giordani et al., 2019). scRNA-seq on muscle FAPs during mouse development, regeneration, and from a Duchenne's muscular dystrophy model suggest that a Vcam1-positive FAP population underlies fibrotic persistence (Malecova et al., 2018). A recent publication highlighted the transcriptional diversity of cells from muscle organoids and outlined growth factors required for MuSC differentiation in organoid culture (Wang et al., 2018), whereas two recent pre-prints employed scRNA-seq to study cell-type-specific responses to muscle injury at 2, 4, 5, and 7 days post injury (Micheli et al., 2019, Pawlikowski et al., 2019). However, a comprehensive characterization from the immediate response through to near pre-injury levels has not been described at the single-cell transcriptomic level.
We employed scRNA-seq of skeletal muscle to understand the transcriptional dynamics that underpin muscle regeneration at six key regenerative time-points and in non-injured muscle. We selected early time points to capture the immediate cellular responses and later time points during muscle maturation and recovery to provide the most comprehensive scRNA-seq analysis of muscle regeneration to date. Our findings highlight the continuous transition of immune populations and suggest that two unique FAP populations are present in resting, non-injured muscle that adopt distinct transcriptional features immediately upon injury. We also identify a unique subpopulation of MuSCs enriched for immune-related transcripts and outline potential receptor-ligand pairs to identify key players in cell-communication networks during muscle regeneration. Our findings provide insights into muscle regeneration and serve as a foundation for future exploration of the potentially critical role of various cellular populations and subpopulations in effective skeletal muscle repair.
Results
scRNA-Seq Reveals Transcriptional Dynamics during Muscle Regeneration
To understand cellular dynamics and interactions during muscle regeneration, we performed scRNA-seq of single cell suspensions collected at various stages of muscle regeneration (summarized in Figure S1A). Time points were selected based on the published literature in an effort to capture cell-type heterogeneity. Specifically, we chose 0.5, 2, 3.5, and 5 days post injury (DPI) as these stages are highly dynamic, involve an immediate immune response, and yield progressive changes that support MuSC activation, proliferation, and differentiation into newly regenerated fibers (Arnold et al., 2007, Deng et al., 2012, Garry et al., 2016, Lu et al., 2011, Relaix and Zammit, 2012). We also analyzed muscles at 10 DPI, when the degenerated myofibers are largely regenerated, and 21 DPI, when regeneration is assumed to be nearly complete and muscle function recovered (Baghdadi and Tajbakhsh, 2018). We also evaluated the morphological features of these time points by histological sectioning (Figure S1B), to confirm that these time points would best capture regeneration dynamics using scRNA-seq.
To generate single-cell suspensions for scRNA-seq, tibialis anterior (TA) muscles were isolated from non-injured and injured mice at six regenerative time points (n = 3 mice per time point, pooled into one sample) for cell isolation. Fluorescence-activated cell sorting (FACS) was then used to select for single cells and exclude aggregated cells, dead cells, and debris (Figure S1C; detailed in Transparent Methods). The 10X Genomics Chromium Platform was used to generate single cell libraries, which were processed according to the manufacturer's instructions and sequenced on an Illumina NovaSeq platform. Individual cells were filtered based on mitochondrial RNA content, features, and reads/cell to yield a total of 53,193 cells across the seven samples with an average of 7,599 cells/sample (Figure S2). Since the seven samples were processed in four batches, we used principal-component analysis (PCA) to determine if batch effects contributed to the variance across samples (Figure S3). As these results did not suggest that samples were purely separated by batch, we combined all cells from the seven samples and performed unsupervised clustering and UMAP embedding using the Seurat R package (Butler et al., 2018).
UMAP embedding of all seven samples colored by time point highlighted the progressive nature of muscle regeneration (Figure 1A). Unsupervised clustering identified a total of 25 clusters across the seven time-points (Figures S4 and S5), which we manually grouped into meta-clusters based on marker gene expression to simplify visualization (Figures 1B and 1C). MuSCs, pericytes, endothelial cells, and myonuclei all formed discrete clusters (Figure 1B). Immune cells formed several discrete clusters including neutrophils, T-cells, and a large cluster of cells containing eleven subpopulations (Figure S4). Similarly, a population of mesenchymal cells containing fibroblasts, tenocytes, and fibro-adipogenic progenitors (FAPs) formed eight clusters (Figure S4), which we henceforth refer to as FAPs for simplicity. The cell populations identified are consistent with the previously published literature (Baghdadi and Tajbakhsh, 2018), suggesting our data recapitulated key events involved in regeneration.
Figure 1.
Single-Cell RNA Profiling of Over 53,000 Cells during Skeletal Muscle Regeneration
(A) Single-cell suspensions were generated from non-injured whole tibialis anterior muscle and six time points following injury with cardiotoxin (CTX). UMAP embedding of scRNA-seq data colored by time point highlights the progressive nature of skeletal muscle regeneration. The regeneration time point color correlates with the timeline scheme.
(B) UMAP embedding of scRNA-seq data colored by meta-clusters to simplify visualization.
(C) Violin plots grouped by meta-clusters demonstrate cell-type marker gene expression, which was used to classify meta-clusters.
(D) Relative proportion of cell types at each time point. Shows initial abundance of inflammatory immune cells and gradual decrease of immune cell abundance with a concurrent increase in FAP populations. Regeneration time point plotted along x axis, relative abundance as a % of total cells along the y axis. Abbreviations: APC, antigen-presenting cells; CTX, cardiotoxin; DCs, dendritic cells; Dividing IC, dividing immune cells; DPI, days post injury; ECs, endothelial cells; FAPs, fibro-adipogenic progenitors; MΦ, macrophages; MuSCs, muscle satellite cells; NI, non-injured.
In non-injured muscle, the primary cell types identified were FAPs and myonuclei (which were presumably released during tissue mincing and digestion and isolated by FACS as single cells), accounting for 62% and 24% of the total population, respectively (Figure 1D). MuSCs comprised 3.1% of the total cell populations, and immune cells made up 7% (Figure 1D). Immediately after injury, immune cells accounted for 87% of the total fraction of cells, whereas the abundance of FAPs dropped to 10% (Figure 1D), highlighting the immediate cell-type response to muscle injury. Few MuSCs were detected by scRNA-seq at 0.5 and 2 DPI, although they were readily detected in cross sections (Figure S6). This may be due to inefficient digestion/isolation of highly inflammatory muscle tissue at these early time points (Goodyear et al., 2014, Trapecar et al., 2017). Nonetheless, MuSCs were readily detectable (3.8%) by 3.5 DPI, along with an increased abundance of FAPs and shifts in the immune cell types (Figure 1D). Consistent with the ongoing resolution of muscle injury, the relative fraction of immune cells began to decline by 5 DPI, whereas the relative fraction of FAPs increased through 21 DPI (Figure 1D). A simple comparison of the transcriptional features from non-injured and 21 DPI also suggested that muscle-specific transcripts were enriched in non-injured compared with 21 DPI, whereas Col1a1 appeared to be enriched at 21 DPI compared with non-injured (Figure S7). This may indicate on-going fibrosis and tissue remodeling at 21 DPI that has not resolved to pre-injury levels. In summary, these data highlight the progressive nature of muscle regeneration and represent the largest scRNA-seq profile of muscle regeneration to date.
Profiling of Immune Cells Reveals a Dynamic and Progressive Immune Response
Immune cells comprised the largest population in our data and displayed the most dynamic, transient, and time-dependent transcriptional features compared with other cell populations. In non-injured muscle, we detected a small population of resident Cd3+ and Cd4+ T cells, as well as small populations of dendritic cells, monocytes, and neutrophils (Figure 2A, vii-viii). Immediately upon injury, leukocytes, M1 macrophages, and neutrophils were the primary cell types detected (Figure 2A, i-ii). Leukocytes were enriched for Vcan, Cxcl3, and Chil3; M1 macrophages expressed Cd36, Arg1, Spp1 Fabp4, and Fabp5; and neutrophils specifically expressed S100a8 and S100a9 (Figure S5). Neutrophils also expressed Csf1 at these early regeneration stages, which has been shown to modulate the tissue-resident macrophages' response and thus outlines the progressive inter-cellular communication network (Braza et al., 2018). Nonetheless, these early-stage immune populations were transient and not detected at the subsequent time points.
Figure 2.
Immune Cell Dynamics during Muscle Regeneration
(A) UMAP embedding of immune cell populations during muscle regeneration, colored by cluster. Panels (i)–(vii) time-point-specific immune cell populations. Arrows drawn to highlight the progressive nature of the immune response and subsequent resolution to near non-injured levels by 21 DPI. Panel (viii) UMAP embedding colored to show all immune cell populations and their cluster identity.
(B) Violin plots showing gene-specific expression trends as a function of regeneration time point. Top panel of violin plots highlights pro-inflammatory gene signatures that are enriched at 0.5 and 2 DPI, whereas the bottom panel shows later-stage anti-inflammatory and antigen presentation gene signatures.
(C) Density plot to demonstrate immune population dynamics throughout the course of regeneration. Immediate response is primarily mediated by pro-inflammatory immune cells such as neutrophils and M1 MΦ’s and subsequently followed by anti-inflammatory immune populations. X axis is hours post injury (with 0 being non-injured); along the y axis is fraction of the immune cell population out of total cells per time point, expressed as %. Abbreviations: DPI, days post injury; MΦ, macrophages.
At 3.5 and 5 DPI, we detected a population of Il7r+ macrophages, M2 macrophages, and Ly6c+ monocytes (Figure 2A and iii-iv). The Il7r+ macrophages expressed Gpnmb, Msrb1, and Pld3; M2 macrophages were enriched for C1qa, C1qb, C1qc, Ms4a6, and Ms4a7; and Ly6c+ monocytes expressed Cd52, Ccr2, and Tlr2. We also detected a late-stage population of macrophages with M2-like characteristics, which we labeled Mrc1+ macrophages. These macrophages were detected at 5 and 10 DPI and were enriched for H2-Aa, H2-Eb1, and H2-Ab1, as well as markers of immature dendritic cells such as Tmem176a, Tmem176b, and Cd81 (Figures 2A and iv-v and S5). T cells were most abundant at 10 and 21 DPI when most myofibers are fully regenerated, suggesting they may play a role in muscle remodeling (Figure 2A and v-vi).
To highlight the gene expression characteristics of the immune response, we analyzed time-point-specific gene expression. These data suggest that the immediate response to muscle injury is governed by a pro-inflammatory phenotype, which subsequently switches to an anti-inflammatory phenotype that yields a gradual resolution (Figures 2B and S8). Chil3, Tnf, Ptgs2, Ccl2, and Cxcl3 have known pro-inflammatory roles and were markedly enriched and specific to 0.5 and 2 DPI (Figure 2B) (Yang and Hu, 2018). Later time-point-specific gene expression characteristics included Tmem176b, Cd74, H2-Eb1, H2-Aa, and Ms4a7 (Figure 2B), which are markers for anti-inflammatory macrophages and dendritic cells in our dataset. The clear switch in gene expression signatures from 2 to 3.5 DPI is consistent with the switch from a pro- to anti-inflammatory immune environment (Figures S8 and 2C) and is nicely recapitulated by the clockwise shift of immune cell types during regeneration. Thus, these data will further serve the community as a tool to explore the immune-cell-specific transcriptional characteristics during muscle regeneration.
Divergence and Bilineage Trajectory of FAP Populations
FAPs reside in the muscle interstitium and play a role in mediating the immune response and ECM remodeling to support skeletal muscle regeneration (Biferali et al., 2019). Based on the expression of Pdgfra, Sca1, and Cd34, we identified a heterogeneous and dynamic cluster of FAPs throughout regeneration (Figures 3A and 3B). Tenocytes marked by the expression of Tnmd and Scx clustered closely with FAPs, all within the larger mesenchymal cell population (Figure 3B). Unlike the immune population which exhibited a progressive nature to yield a nearly resolved state by 21 DPI, FAP populations followed a linear trajectory emerging from 0.5 DPI. Upon muscle damage, we detected a FAP population with distinct transcriptional characteristics compared with the non-injured populations. We labeled this subpopulation as activated FAPs as they were marked by the expression of Cxcl5, Cxcl3, Ccl7, and Ccl2 (Figure 3B). Activated FAPs were detected at early regeneration stages (0.5 and 2 DPI) and transitioned into a Wisp1+ FAP subpopulation at 3.5 and 5 DPI (Figure 3B). Wisp1+ FAPs were enriched for ECM-remodeling factors such as Col8a1, Col12a1, Col16a1, Col11a1, Tnc, Fbn2, and Adam12. Unique to 10 DPI, we detected a population of Dlk1+ FAPs (Figure 3C) enriched for genes that show complex imprinting patterns such as B830012L14Rik, Meg3, Airn, Peg3, Zim1, H19, and Igf2 (Ye et al., 2014). Osr1+ FAPs and a fibroblast population (enriched for genes encoding type I collagen) were the primary populations at 21 DPI. Osr1+ FAPs expressed cell-signaling-related genes such as Ccl1, Bmp4, Bmp5, and Wnt5a, and interestingly, some of the Osr1+ FAPs diverged into the two populations identified in the non-injured muscle: a Dpp4+ FAP population and a Cxcl14+ FAP population (Figures 3A and 3C). The Dpp4+ FAPs also expressed Pi16 and Wnt2, thus representing the muscle analogue of the reticulum interstitial adipose progenitors recently identified in the adipose tissues (Merrick et al., 2019). The Cxcl14+ FAPs expressed genes encoding secreted enzymes, such as Enpp2, Crispld2, and Hsd11b1. Furthermore, recent scRNA-seq of FAP in muscle identified two similar subpopulations (Scott et al., 2019), further corroborating our data and analysis. To highlight the stage-specific gene expression characteristics, we evaluated markers enriched in the FAP populations by regeneration time point (Figure 3C). Early response genes were implicated in cytokine interactions, whereas the later time points were enriched for ECM factors (Figure 3C), suggesting FAPs may function to mediate immune infiltration and muscle remodeling at early and late regenerative stages, respectively. A gene signature list for the top 50 enriched genes for each subpopulation is provided in Table S2.
Figure 3.
Fibro-Adipogenic Progenitor Population Dynamics during Muscle Regeneration
(A) UMAP embedding of all cells profiled by scRNA-seq during regeneration. Left panel colored by time point, right panel colored by FAP cluster identity. Dotted line circles the FAP population.
(B) Feature plots showing the normalized expression of cluster-specific genes. Pdgfra is expressed by most cells, whereas Cxcl5 is restricted to the activated FAP population. Wisp1 is expressed mostly in FAPs from 3.5 to 5 DPI, whereas Cxcl14 and Dpp4 may represent two divergent FAP subpopulations present in resting, non-injured muscle.
(C) Violin plots of genes exhibiting time-point-specific expression dynamics. The top panel highlights enrichment of immune-modulatory factors at early time points, the middle panel highlights genes enriched at 3.5–10 DPI, whereas the bottom highlights the expression of genes enriched at 21 DPI and in non-injured muscle.
(D) Pseudotime trajectory inference using Monocle of the FAP populations (not including tenocytes). Activated FAPs were selected as the start, and Monocle arranged cells accordingly. UMAP embedding and trajectory inference exhibit a similar pattern in which activated FAPs diverge into two subpopulations present in resting muscle.
(E) Plot to highlight the putative interactome of FAPs, highlighting their diverse role in response to muscle injury. Cell types are grouped by meta-clusters to simplify visualization. Each connection is the sum of interactions across all of the time points. Abbreviations: APC, antigen-presenting cells; DCs, dendritic cells; Dividing IC, dividing immune cells; DPI, days post injury; ECs, endothelial cells; FAPs, fibro-adipogenic progenitors; MΦ, macrophages; MuSCs, muscle satellite cells; NI, non-injured.
The directional progression and divergence of the FAP populations from 0.5 DPI to the two subpopulations detected in non-injured muscle was a unique attribute to this cluster of cells. To determine if these populations may represent transitional states, we employed Monocle for trajectory inference and selected activated FAPs as the start point. Monocle is an unsupervised algorithm that aligns cells along an inferred trajectory and can robustly recapitulate differentiation programs and other biological processes (Trapnell et al., 2014). Monocle arranged FAPs along a common trajectory that diverged into two distinct branches, which coincided with the two subpopulations (Cxcl14+ and Dpp4+) detected in non-injured muscle (Figures 3D and S9A–S9C). Activated and Wisp1+ FAPs localized toward the start of pseudotime, whereas the fibroblasts did not appear to have any spatial bias. Osr1+ and Dlk1+ FAPs were distributed along the two major branches (Figure 3D). Gene expression plots of Dpp4 and Cxcl14 highlight the divergent fates the two FAP subpopulations occupy in pseudotime, confirming that Cxcl14+ and Dpp4+ FAPs represent two distinct subpopulations in non-injured muscle (Figure S9C). As the tenocytes represent a relatively well-characterized population distinct from FAPs (Subramanian and Schilling, 2015), they were not included in trajectory analysis. These results were further corroborated by Slingshot, a semi-unsupervised clustering algorithm that uses scRNA-seq data to construct cell lineages and scored highest for accuracy and stability compared with all tree-based inference methods (Saelens et al., 2019, Street et al., 2018). Lineage inference with Slingshot on subclustered and UMAP embedded FAPs similarly produced a trajectory that diverged to yield the Dpp4+ and Cxcl14+ FAPs, consistent with Monocle and the global UMAP structure (Figures S9D–S9F). These results suggest that the FAP populations represent a continuous state during regeneration and may diverge into the Dpp4+ and Cxcl14+ FAP subpopulations present in non-injured muscle.
To better understand the requirement of FAPs, we evaluated the co-enrichment of receptor-ligand pairs across all cells per time point using a published receptor-ligand dataset (Ramilowski et al., 2015). For each time point, we evaluated the expression of receptor-ligand pairs enriched across all cell types and plotted the sum of these interactions to identify which cells may mediate cell-cell communication. Given that tenocytes express high levels of collagen-related genes, we were not surprised to identify a high enrichment of putative interactions among FAPs and tenocytes (Figure 3E). Nonetheless, FAPs were also enriched for putative receptor-ligand pairs across various immune cell types, consistent with their immunomodulatory and ECM remodeling functions (Figure 3E and Table S3). In sum, these data highlight the diverse and perhaps supportive role for FAPs in muscle regeneration.
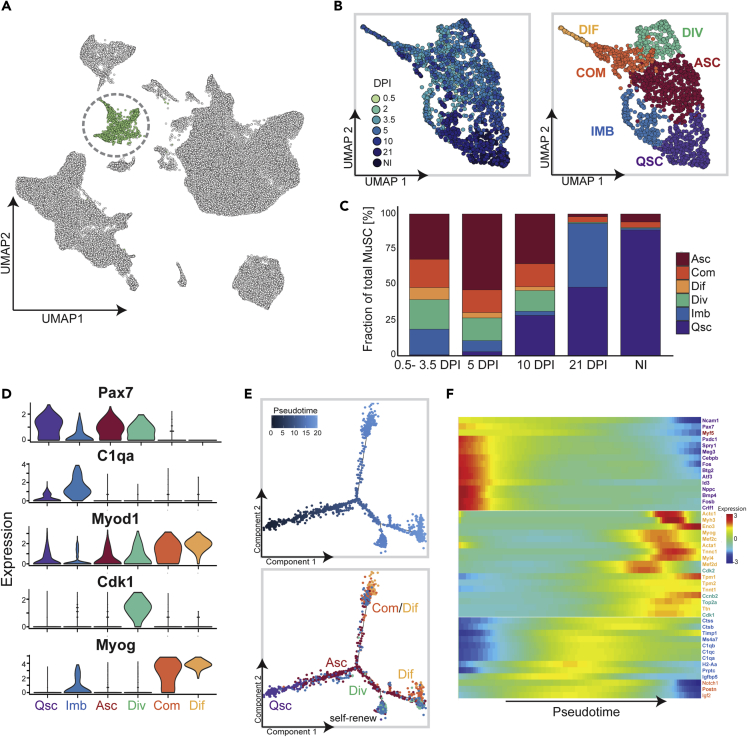
MuSC Sub-clustering Reveals a Subpopulation of Myoblasts with Immune Gene Characteristics
MuSCs from all regeneration time points formed a single population distinct from other clusters (Figures 1B and 4A). To better understand the transcriptional features within the MuSC population, we performed unsupervised clustering on the MuSCs containing the transcripts of 1,727 cells. This identified a total of six unique subclusters that we labeled according to their gene expression profiles (Figures 4A and 4B). Specifically, we identified a subpopulation of quiescent MuSCs that expressed Pax7, Sdc4, Col3a1, Pten, and Spry1 (Fukada et al., 2007, Pietrosemoli et al., 2017, Shea et al., 2010, van Velthoven et al., 2017, Yue et al., 2017), which comprised MuSCs mostly from non-injured muscle (Figures 4B, 4C, and S10). Activated MuSCs expressed Pax7, Myod1, Islr, and Itm2a, whereas dividing MuSCs also expressed Pax7 but were enriched for cell-cycle-related genes Mik67, Top2a, and Cdk1 (Lagha et al., 2013). The committed and differentiated subpopulations expressed differentiation and mature muscle markers such as Myogenin, Ttn, Myh3, and Myl4, with higher expression levels in the differentiated subpopulation. MuSCs from 0.5 to 5 DPI were mostly composed of activated, dividing, committed, and differentiated MuSCs, respectively (Figure S10), consistent with known stages of MuSC-mediated myogenesis in response to injury.
Figure 4.
MuSC Sub-cluster Analysis Reveals Subpopulation Enriched for Immune Gene Expression
(A) UMAP embedding of all cells profiled during muscle regeneration. MuSCs are colored in green and circled; all other cells are colored in gray.
(B) Subclustering of MuSCs. Left panel colored by regeneration time point; right panel colored by cluster identity.
(C) Bar graphs to represent the proportion of MuSC subcluster for each time point; 0.5, 2, and 3.5 DPI were combined owing to low number of MuSCs detected at 0.5 and 2 DPI.
(D) Violin plots to show subcluster-specific gene expression and enrichment of C1qa in the immuno-myoblast sub-cluster. Colored by cluster identity.
(E) Pseudotime trajectory inference using Monocle, cells are plotted along the inferred trajectory. Quiescent MuSCs were selected as the start of pseudotime, which is indicated in the top panel. The bottom panel is colored by the cluster identities.
(F) Heatmap of selected genes to show their dynamic expression along pseudotime. Genes enriched in quiescent MuSCs are enriched at the start of pseudotime, whereas differentiation-specific genes are expressed at the end of pseudotime. Gene names are colored by subcluster identity in which they are enriched. Expression values are the log(expression value +0.1). Abbreviations: MuSCs, muscle satellite cells; QSC, quiescent MuSCs; ASC, activated MuSCs; IMB, immuno-myoblasts; DIV, dividing MuSCs; DIF, differentiated MuSCs; COM, committed MuSCs; DPI, days post injury.
Surprisingly, we also identified an MuSC subpopulation that has not been previously described. This population was enriched for genes involved in immune cell complement activation (C1qa, C1qb, C1qc), major histocompatibility class II antigens (H2-Eb1, H2-Aa, H2-Ab1) and members of the cathepsin family (Ctsb, Ctss), in addition to the expression of myogenic genes (Figure S10 and Table S4). We therefore coined this subpopulation of MuSCs as immunomyoblasts (IMBs). A small proportion of MuSCs at early regeneration stages fell within this sub-cluster, whereas at 21 DPI nearly 50% of detected MuSCs were immunomyoblasts (Figure 4C). Although this subpopulation had a similar gene expression profile to quiescent, activated, and dividing MuSCs, the expression of immune-related genes was restricted to this population (Figure S10). In summary, a progressive change in gene expression mediates the transition from quiescent to differentiated MuSCs, and our results highlight the robustness of scRNA-seq in recapitulating this dynamic and capture a unique subpopulation of MuSCs that have not been described to date.
Given that our data recapitulated known stages of MuSC transition from quiescence to differentiation, we used Monocle to assess if the IMB subpopulation represents a previously undescribed MuSC fate (Trapnell et al., 2014). Monocle arranged the majority of cells along two trajectories and one smaller trajectory (Figure 4E). Quiescent MuSCs plotted tightly together at the start of pseudotime, whereas activated MuSCs did not exhibit a spatial bias, suggesting that the activation state of MuSCs is continuous. Committed and differentiated MuSCs localized at the two ends of the major branchpoint, whereas dividing cells were restricted to the lower half of the pseudotime space (Figure 4E). This may suggest that MuSCs go from quiescence to activation and then can be fated to (1) differentiate (top right branch) or (2) proliferate and then differentiate (bottom branch furthest to right) or self-renew (bottom branchpoint toward the left) (Figure 4E). Consistent with this notion, Pax7 and Cdk1 expression was enriched in the lower half of pseudotime space, whereas Myog was restricted to the end of the major branchpoint (top and bottom right branches, Figure S11A). We next plotted the gene expression of markers associated with quiescent and differentiated MuSCs. The expression of quiescent markers was enriched at the start of pseudotime, whereas differentiation markers were enriched at the end (Figure 4F). Also enriched toward the end of pseudotime were cell-cycle-related genes (Figure 4F), which may represent gene expression characteristics of a self-renewing MuSC population. Although immunomyoblasts did not occupy a specific pseudotime space, gene signatures of this population (i.e., Ctss, Ctsb, Ms4a7, H2-Aa, C1qa, C1qb, C1qc) were transiently expressed along the progression of pseudotime (Figure 4F), suggesting that this subpopulation may represent a transitional MuSC state. We also used Slingshot to corroborate these findings and selected quiescent MuSCs as the start point and differentiated MuSCs as the endpoint (Figure S11B). Based on the constructed minimum spanning tree (MST), Slingshot identified three distinct lineages in the MuSC cluster that highlight the (1) differentiation trajectory, (2) the dividing trajectory, and (3) the immunomyoblast trajectory (Figure S11B). Genes that defined the immunomyoblast fate highlighted the distinct transcriptional features of this subpopulation and promoted us to further evaluate their expression in MuSCs.
As the IMB subpopulation of MuSCs has not been described, we sought to confirm the expression of some of the features uniquely enriched in this population. To this end, we used FACS to isolate MuSCs from non-injured and 5 DPI hindlimb muscles of Pax7CreER:sfGFP reporter mice, which allowed the conditional labeling of Pax7-positive cells and all of the progeny with nuclear-membrane GFP upon administration of tamoxifen (Figure S12A) (Luo et al., 2017, Murphy et al., 2011). We isolated RNA, generated cDNA, and probed the expression of some of the aforementioned markers enriched in the IMB population identified in our scRNA-seq dataset by RT-PCR. This confirmed that MuSCs isolated from 5 DPI muscle indeed showed increased expression of these markers (Figure 5A). Specifically, Ctsb, C1qa, Ctqc, and Lyz2 were not detectable in MuSCs isolated from non-injured muscles but were expressed in MuSCs isolated from 5 DPI muscles. As mRNA does not always correlate to protein expression (Liu et al., 2016), we performed immunofluorescence (IF) with selected antibodies on muscle sections or myoblasts growing on cultured myofibers. We first probed the expression of C1q and Pax7 by IF from non-injured and 5, 10, and 21 DPI samples. Quantification of C1q and Pax7 co-expression suggested that only a subset of Pax7-positive cells was also positive for C1q and this was dynamic during the course of regeneration, corroborating the results from our scRNA-seq dataset (Figure 5B). Co-localization of Pax7 and C1q was highest at 10 DPI (61% of Pax7+ cells were also C1q+) and subsequently decreased by 21 DPI to 20% (Figure 5B). Since MHC II expression was also enriched in the in silico identified immunomyoblast subpopulation, we evaluated the co-expression of Pax7 and MHC II on muscle sections from non-injured and 5, 10 and 21 DPI (Figure 5C). Quantification of Pax7 and MHC II co-localization confirmed that a subset of Pax7+ cells are also positive for MHC II, and the co-expression of Pax7 and MHC II is enriched at 10 DPI (17%, Figure 5C). Given that MHC II is a surface protein primarily expressed in antigen-presenting cells (Rock et al., 2016), we used FACS and the Pax7CreER:sfGFP reporter mouse to better resolve MHC II expression in MuSCs. We digested hindlimb muscle from 5 DPI mice, stained with MHC II conjugated to PE or the respective isotype control, and subjected the samples to FACS to determine the presence of any GFP+/MHC II+ cells. Only a subset of GFP+ cells were also MHC II positive after injury, whereas the isotype control did not exhibit binding, suggesting that the MHC II antibody is specific for the target antigen (Figure S12B). Furthermore, imaging of 5 DPI MuSCs after FACS clearly demonstrated surface expression of MHC II (Figure 5D). Finally, we evaluated the expression of Ctsb and Apoe by IF using single myofiber culture, as this is commonly used to mimic the MuSC-myofiber interaction in vitro (Bischoff, 1986). Indeed, a subset of Myod+ cells were also positive for cytoplasmic Ctsb (28%, Figure S12C), whereas ApoE was detected in the cytoplasm 22% of Myod+ cells (Figure S12D). Overall, these data suggest that a subpopulation of MuSCs enriched in immune-gene expression may transiently exist after muscle injury, consistent with the identification of this population in silico in our scRNA-seq data.
Figure 5.
Evaluation of Immune Gene Expression in MuSCs
(A) RT-PCR to detect the expression of immune genes in MuSCs isolated from non-injured muscle (-CTX) or 5 DPI (+CTX). Genes are marked along the top.
(B) Immunofluorescence to detect Pax7 and C1q on sections from non-injured and 5, 10, and 21 DPI TA muscle. Left panel: C1q single channel, right panel: merged. Bar on right: quantification of Pax7+ cells (total number counted in black) that are also positive for C1q (% in green). Scale bar: 50 μm.
(C) Immunofluorescence to detect Pax7 and MHC-II on sections from non-injured and 5, 10 and 21 DPI TA muscle. Left panel: MHCII single channel, right panel: merged. Bar on right: quantification of Pax7+ cells (total number counted in black) also positive for MHC II (% in green). Scale bar: 50 μm.
(D) Images MuSCs isolated 5 DPI from Pax7CreER:sfGFP mice using FACS based on nuclear-membrane GFP (top) and MHCII (middle). The merged panels (bottom) highlight the surface localization of MHCII expression in Pax7-progeny cells (related to Figures S12A and S12B).
(E) Plot to highlight the potential interactions of immunomyoblasts with all other cells. Interactions are primarily enriched from IMB to DCs, FAPs, and monocytes. Abbreviations: APC, antigen-presenting cells; CTX, cardiotoxin; DCs, dendritic cells; Dividing IC, dividing immune cells; DPI, days post injury; ECs, endothelial cells; FAPs, fibro-adipogenic progenitors; MΦ, macrophages; MuSCs, muscle satellite cells; TA, tibialis anterior muscle.
To further explore the potential function of this subpopulation, we analyzed enrichment of receptor-ligand pairs between immunomyoblasts and all other meta-clusters (from Figure 1B). Given that immunomyoblasts were enriched for immune-related genes, it was not surprising that receptor-ligand pairs were also enriched between immunomyoblasts and immune cell populations (Figures 5E and Table S5). Although the functional impact of this subpopulation remains to be determined, our data suggest that MuSCs can activate an immunogenic transcriptional program that may play a role in muscle regeneration and/or immune cell modulation in response to injury.
Discussion
Our data containing the transcripts of over 53,000 cells during muscle regeneration highlights the complex nature of tissue regeneration and identifies a subpopulation of MuSCs with immune gene characteristics. Although batch effects remain a technical and analytic challenge for the present dataset, non-linear batch correction methods aim to minimize the differences observed across similar samples processed using different platforms or under different experimental conditions (Stuart et al., 2019). Given that each sample in our dataset is a unique time point, we did not perform batch correction as we did not have multiple samples from the same time point (although samples were processed as uniformly as possible). Therefore, our datasets were merged and subsequently analyzed in order to preserve the expected heterogeneity across regeneration time points. PCA and UMAP embedding highlight that samples cluster by regeneration time point and not by batch (Figure S3). However, future experiments to include cell tagging will clarify potential batch effects across regeneration time points (Gehring et al., 2020, Kong et al., 2020). Nonetheless, our dataset recapitulates key features of muscle regeneration. Specifically, the immune cell populations were the largest in number and displayed the most time-dependent dynamics. Although 21 DPI immune characteristics approached the non-injured muscle state, they were fairly distinct as we detected small populations of pro- and anti-inflammatory populations, dendritic cells, T cells, and leukocytes. These late-regeneration-stage immune cells may function to further model the regenerated muscle or may be a signature of injured muscle and thereby impart a “memory” on the injured muscle (Burzyn et al., 2013), which will be important to consider for clinical applications in which injury precedes therapy.
In contrast to the immediate and subsequent resolution of the immune response, the mesenchymal populations displayed a linear trajectory from activated FAPs through to the non-injured subpopulations. In non-injured muscle, we identified two subpopulations of FAPs. One, which we termed Cxcl14+ FAPs, and another Dpp4+ population with similar features to the reticular interstitial progenitors identified in mouse and human adipose tissue and recently described in muscle (Merrick et al., 2019, Scott et al., 2019). Upon injury, activated FAPs were transcriptionally distinct from the non-injured populations and progressed to Wisp1+ FAPs at 3.5 and 5 DPI. The transient expression of Wisp1 in the FAP population was consistent with a recently reported function for Wisp1 in FAPs, in which its expression is enriched in FAPs at 3 DPI and required for MuSC asymmetric expansion (Lukjanenko et al., 2019). Thus, this population of FAPs warrants further investigation as they may play a role in MuSC function and skeletal muscle regeneration as a whole. Our dataset also identified a subpopulation of Dlk1+ FAPs that appeared to be specific to 10 DPI. Interestingly, Dlk1 is a marker for pre-adipocytes and FAP accumulation can lead to intra-muscular adipocyte infiltration (Biferali et al., 2019, Hudak and Sul, 2013). Whether the Dlk1+ FAP population represents a pre-adipocyte state during muscle regeneration will also require future studies. At 21 DPI, we detected a unique population of fibroblasts enriched for collagen type 1 gene expression and therefore may represent persistent, unresolved fibrosis after injury. Osr1+ FAPs were the predominant cell type present at 21 DPI, and a small population of Osr1+ FAPs was also present in non-injured muscle. A recent report suggests that Osr1+ FAPs constitute the FAP progenitor pool in response to freeze injury (Stumm et al., 2018). However, our data did not imply any specific progenitor population and, more strikingly, did not identify a subset of FAPs enriched for cell-cycle-related genes. These data demonstrate the possibly diverse role of the FAP populations and will serve as a platform to further our understanding of the functional significance of these subpopulations.
The striking dynamics of the immune and mesenchymal populations prompted us to further explore MuSC transcriptional features, by which we identified a subpopulation of MuSCs enriched for immune-related genes. We determined by IF that a subpopulation of MuSCs indeed express immune-related genes upon injury and further evaluated the expression of MHC II on the surface of a subset of MuSCs via FACS. This subpopulation of MuSCs has not been defined as a discrete subpopulation but overlaps with the previously described literature. Early studies suggested that a specific subset of bone-marrow-derived stem cells contribute to muscle regeneration through fusion into the newly regenerated myofibers and that Cd45+ cells can be myogenic upon muscle injury (Doyonnas et al., 2004, Palermo et al., 2005, Polesskaya et al., 2003, Seale et al., 2004). MuSCs can also act as antigen-presenting cells upon viral transduction (Cao et al., 2004), suggesting an immune transcriptional program exists in MuSCs. A recent study further suggests that non-hematopoietic cells can express MHC II (Wosen et al., 2018). Various datasets analyzing the expression profiles of quiescent and activated MuSCs also show that inflammatory gene signatures are enriched in activated MuSCs (Pietrosemoli et al., 2017). However, these studies used bulk-RNA methods and thus were unable to assign the expression of inflammatory and complement-related transcripts to a specific subpopulation. Furthermore, a recent study identified immune-related gene expression networks by RNA-sequencing of Pax3-positive cells in mouse limb during fetal myogenesis (Singh et al., 2018), when a subset of embryonic myoblasts take the sublaminar position to become MuSCs. Similarly, our in silico analysis showed that IMBs were most abundant at 21 DPI, perhaps suggesting a synergism between the immune system and the homing of quiescent MuSCs upon completion of regeneration. Interestingly, at 21 DPI we detected a diverse range of immune cells including pro-inflammatory subsets, which may secret pro-inflammatory cytokines to promote the expansion of MuSCs including IMBs (Fu et al., 2015). Although further functional analyses will be required to confirm if the IMBs represent a true subpopulation of MuSCs with a specific function, our single cell transcriptional profiling and preliminary IF and FACS data provide evidence for the existence of this subpopulation of transitional myoblasts.
Limitations of the Study
Given that each time point was a single sample, there may be batch effects that cannot be teased out with the current dataset. Although PCA, clustering, and UMAP embedding did not suggest a strong batch effect, future experiments to include cell tagging or additional samples will be necessary to elucidate batch effects. In silico identification of a subpopulation of MuSCs enriched for immune-gene expression (immunomyoblasts) will require further functional validation to confirm that this subpopulation is not an artifact of scRNA-seq sample preparation. Although IF suggested that a subset of MuSCs express C1q and MHC-II upon muscle injury, future efforts to evaluate the transcriptional features and functional role of this MuSCs population will help determine the functional relevance of this subpopulation in muscle regeneration.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
We thank Dr. Matthew Olson for his help with immune cell identification, Dr. Timothy Ratliff and Dr. Gregory Cresswell for assistance with the 10X Genomics Platform, Dr. Phillip San Miguel and Purdue's Genomics Core for RNA-sequencing, Drs. Nadia Atallah and Jun Wan for cluster computing access and advice on data analysis, Purdue's Flow Cytometry and Cell Separation Facility for assistance with FACS data collection and analysis, and Jun Wu for technical support.
This work was supported by grants from the US National Institutes of Health (R01AR071649), the National Institute of Food and Agriculture (NC-1184) and Purdue University Center for Cancer Research (P30CA023168). Funding for open access charge: NIH R01AR071649.
Author Contributions
S.N.O. conceived and conducted the experiments, performed the data analysis, and wrote the paper; F.Y. helped with experimental design and performed experiments; J.Q. performed experiments; L.F.B. helped with data analysis; S.K. conceived and supervised the project, analyzed the data, and revised the manuscript. All authors read and provided feedback on the manuscript.
Declaration of Interests
The authors declare that they have no conflicts of interest with the contents of this article.
Published: April 24, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.100993.
Data and Code Availability
The accession number for the single-cell RNA-sequencing data reported in this paper is: GSE138826.
Supplemental Information
References
- Arnold L., Henry A., Poron F., Baba-Amer Y., van Rooijen N., Plonquet A., Gherardi R.K., Chazaud B. Inflammatory monocytes recruited after skeletal muscle injury switch into antiinflammatory macrophages to support myogenesis. J. Exp. Med. 2007;204:1057–1069. doi: 10.1084/jem.20070075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baghdadi M.B., Tajbakhsh S. Regulation and phylogeny of skeletal muscle regeneration. Dev. Biol. 2018;433:200–209. doi: 10.1016/j.ydbio.2017.07.026. [DOI] [PubMed] [Google Scholar]
- Biferali B., Proietti D., Mozzetta C., Madaro L. Fibro–adipogenic progenitors cross-talk in skeletal muscle: the social network. Front. Physiol. 2019;10:1074. doi: 10.3389/fphys.2019.01074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff R. Proliferation of muscle satellite cells on intact myofibers in culture. Dev. Biol. 1986;115:129–139. doi: 10.1016/0012-1606(86)90234-4. [DOI] [PubMed] [Google Scholar]
- Boisset J.-C., Vivié J., Grün D., Muraro M.J., Lyubimova A., van Oudenaarden A. Mapping the physical network of cellular interactions. Nat. Methods. 2018;15:547–553. doi: 10.1038/s41592-018-0009-z. [DOI] [PubMed] [Google Scholar]
- Braza M.S., Conde P., Garcia M., Cortegano I., Brahmachary M., Pothula V., Fay F., Boros P., Werner S.A., Ginhoux F. Neutrophil derived CSF1 induces macrophage polarization and promotes transplantation tolerance. Am. J. Transplant. 2018;18:1247–1255. doi: 10.1111/ajt.14645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burzyn D., Kuswanto W., Kolodin D., Shadrach J.L., Cerletti M., Jang Y., Sefik E., Tan T.G., Wagers A.J., Benoist C. A special population of regulatory T cells potentiates muscle repair. Cell. 2013;155:1282–1295. doi: 10.1016/j.cell.2013.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao B., Bruder J., Kovesdi I., Huard J. Muscle stem cells can act as antigen-presenting cells: implication for gene therapy. Gene Ther. 2004;11:1321–1330. doi: 10.1038/sj.gt.3302293. [DOI] [PubMed] [Google Scholar]
- Christov C., Chrétien F., Abou-Khalil R., Bassez G., Vallet G., Authier F.-J., Bassaglia Y., Shinin V., Tajbakhsh S., Chazaud B. Muscle satellite cells and endothelial cells: close neighbors and privileged partners. Mol. Biol. Cell. 2007;18:1397–1409. doi: 10.1091/mbc.E06-08-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero-Espinoza L., Huch M. The balancing act of the liver: tissue regeneration versus fibrosis. J. Clin. Invest. 2018;128:85–96. doi: 10.1172/JCI93562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell’orso S., Juan A.H., Ko K.-D., Naz F., Gutierrez-Cruz G., Feng X., Sartorelli V. Single-cell analysis of adult skeletal muscle stem cells in homeostatic and regenerative conditions. Development. 2019;146 doi: 10.1242/dev.174177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng B., Wehling-Henricks M., Villalta S.A., Wang Y., Tidball J.G. IL-10 triggers changes in macrophage phenotype that promote muscle growth and regeneration. J. Immunol. 2012;189:3669–3680. doi: 10.4049/jimmunol.1103180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyonnas R., LaBarge M.A., Sacco A., Charlton C., Blau H.M. Hematopoietic contribution to skeletal muscle regeneration by myelomonocytic precursors. Proc. Natl. Acad. Sci. U S A. 2004;101:13507–13512. doi: 10.1073/pnas.0405361101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X., Xiao J., Wei Y., Li S., Liu Y., Yin J., Sun K., Sun H., Wang H., Zhang Z. Combination of inflammation-related cytokines promotes long-term muscle stem cell expansion. Cell Res. 2015;25:655–673. doi: 10.1038/cr.2015.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukada S., Uezumi A., Ikemoto M., Masuda S., Segawa M., Tanimura N., Yamamoto H., Miyagoe-Suzuki Y., Takeda S. Molecular signature of quiescent satellite cells in adult skeletal muscle. Stem Cells. 2007;25:2448–2459. doi: 10.1634/stemcells.2007-0019. [DOI] [PubMed] [Google Scholar]
- Garry G.A., Antony M.L., Garry D.J. Cardiotoxin induced injury and skeletal muscle regeneration. In: Clifton N.J., editor. Methods in Molecular Biology. Humana Press, Inc.; 2016. pp. 61–71. [DOI] [PubMed] [Google Scholar]
- Gehring J., Hwee Park J., Chen S., Thomson M., Pachter L. Highly multiplexed single-cell RNA-seq by DNA oligonucleotide tagging of cellular proteins. Nat. Biotechnol. 2020;38:35–38. doi: 10.1038/s41587-019-0372-z. [DOI] [PubMed] [Google Scholar]
- Gerber T., Gerber T., Murawala P., Knapp D., Masselink W., Schuez M., Gac-santel M., Nowoshilow S., Kageyama J., Khattak S. Single-cell analysis uncovers convergence of cell identities during axolotl limb regeneration. Science. 2018;362 doi: 10.1126/science.aaq0681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordani L., He G.J., Negroni E., Sakai H., Law J.Y.C., Siu M.M., Wan R., Corneau A., Tajbakhsh S., Cheung T.H. High-Dimensional single-cell cartography reveals novel skeletal muscle-resident cell populations. Mol. Cell. 2019;74:609–621.e6. doi: 10.1016/j.molcel.2019.02.026. [DOI] [PubMed] [Google Scholar]
- Goodyear A.W., Kumar A., Dow S., Ryan E.P. Optimization of murine small intestine leukocyte isolation for global immune phenotype analysis. J. Immunol. Methods. 2014;405:97–108. doi: 10.1016/j.jim.2014.01.014. [DOI] [PubMed] [Google Scholar]
- Halpern K.B., Shenhav R., Massalha H., Toth B., Egozi A., Massasa E.E., Medgalia C., David E., Giladi A., Moor A.E. Paired-cell sequencing enables spatial gene expression mapping of liver endothelial cells. Nat. Biotechnol. 2018;36:962. doi: 10.1038/nbt.4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy D., Besnard A., Latil M., Jouvion G., Briand D., Thépenier C., Pascal Q., Guguin A., Gayraud-Morel B., Cavaillon J.-M. Comparative study of injury models for studying muscle regeneration in mice. PLoS One. 2016;11:e0147198. doi: 10.1371/journal.pone.0147198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heredia J.E., Mukundan L., Chen F.M., Mueller A.A., Deo R.C., Locksley R.M., Rando T.A., Chawla A. Type 2 innate signals stimulate fibro/adipogenic progenitors to facilitate muscle regeneration. Cell. 2013;153:376–388. doi: 10.1016/j.cell.2013.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudak C.S., Sul H.S. Pref-1, a gatekeeper of adipogenesis. Front. Endocrinol. (Lausanne) 2013;4:79. doi: 10.3389/fendo.2013.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang B., Lee J.H., Bang D. Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp. Mol. Med. 2018;50:96. doi: 10.1038/s12276-018-0071-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joe A.W.B., Yi L., Natarajan A., Le Grand F., So L., Wang J., Rudnicki M.A., Rossi F.M.V. Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nat. Cell Biol. 2010;12:153–163. doi: 10.1038/ncb2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhas M., Abutaleb N., Wang J.T., Ye J., Shaikh Z., Sriworarat C., Qian Y., Bursac N. Incorporation of macrophages into engineered skeletal muscle enables enhanced muscle regeneration. Nat. Biomed. Eng. 2018;2:942–954. doi: 10.1038/s41551-018-0290-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong W., Biddy B.A., Kamimoto K., Amrute J.M., Butka E.G., Morris S.A. CellTagging: combinatorial indexing to simultaneously map lineage and identity at single-cell resolution. Nat. Protoc. 2020;15:1–23. doi: 10.1038/s41596-019-0247-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M.P., Du J., Lagoudas G., Jiao Y., Sawyer A., Drummond D.C., Lauffenburger D.A., Raue A. Analysis of single-cell RNA-seq identifies cell-cell communication associated with tumor characteristics. Cell Rep. 2018;25:1458–1468.e4. doi: 10.1016/j.celrep.2018.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagha M., Mayeuf-Louchart A., Chang T., Montarras D., Rocancourt D., Zalc A., Kormish J., Zaret K.S., Buckingham M.E., Relaix F. Itm2a is a Pax3 target gene, expressed at sites of skeletal muscle formation in vivo. PLoS One. 2013;8:e63143. doi: 10.1371/journal.pone.0063143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larouche J., Greising S.M., Corona B.T., Aguilar C.A. Robust inflammatory and fibrotic signaling following volumetric muscle loss: a barrier to muscle regeneration. Cell Death Dis. 2018;9:409. doi: 10.1038/s41419-018-0455-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latroche C., Weiss-Gayet M., Muller L., Gitiaux C., Leblanc P., Liot S., Ben-Larbi S., Abou-Khalil R., Verger N., Bardot P. Coupling between myogenesis and angiogenesis during skeletal muscle regeneration is stimulated by restorative macrophages. Stem Cell Reports. 2017;9:2018–2033. doi: 10.1016/j.stemcr.2017.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Beyer A., Aebersold R. On the dependency of cellular protein levels on mRNA abundance. Cell. 2016;165:535–550. doi: 10.1016/j.cell.2016.03.014. [DOI] [PubMed] [Google Scholar]
- Lu H., Huang D., Ransohoff R.M., Zhou L. Acute skeletal muscle injury: CCL2 expression by both monocytes and injured muscle is required for repair. FASEB J. 2011;25:3344–3355. doi: 10.1096/fj.10-178939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukjanenko L., Karaz S., Stuelsatz P., Gurriaran-Rodriguez U., Michaud J., Dammone G., Sizzano F., Mashinchian O., Ancel S., Migliavacca E. Aging disrupts muscle stem cell function by impairing matricellular WISP1 secretion from fibro-adipogenic progenitors. Cell Stem Cell. 2019;24:433–446.e7. doi: 10.1016/j.stem.2018.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C., Keown C.L., Kurihara L., Zhou J., He Y., Li J., Castanon R., Lucero J., Nery J.R., Sandoval J.P. Single-cell methylomes identify neuronal subtypes and regulatory elements in mammalian cortex. Science. 2017;357:600–604. doi: 10.1126/science.aan3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malecova B., Gatto S., Etxaniz U., Passafaro M., Cortez A., Nicoletti C., Giordani L., Torcinaro A., De Bardi M., Bicciato S. Dynamics of cellular states of fibro-adipogenic progenitors during myogenesis and muscular dystrophy. Nat. Commun. 2018;9:3670. doi: 10.1038/s41467-018-06068-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick D., Sakers A., Irgebay Z., Okada C., Calvert C., Morley M.P., Percec I., Seale P. Identification of a mesenchymal progenitor cell hierarchy in adipose tissue. Science. 2019;364:eaav2501. doi: 10.1126/science.aav2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheli A.J. De, Fraczek P., Soueid-Baumgarten S., Ravichandran H., Vlaminck I. De, Elemento O., Cosgrove B.D. Single-cell analysis of the muscle stem cell hierarchy identifies heterotypic communication signals involved in skeletal muscle regeneration. bioRxiv. 2019:671032. doi: 10.1016/j.celrep.2020.02.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer A.L., Wagner K.R. Regeneration versus fibrosis in skeletal muscle. Curr. Opin. Rheumatol. 2011;23:568–573. doi: 10.1097/BOR.0b013e32834bac92. [DOI] [PubMed] [Google Scholar]
- Murphy M.M., Lawson J.A., Mathew S.J., Hutcheson D.A., Kardon G. Satellite cells, connective tissue fibroblasts and their interactions are crucial for muscle regeneration. Development. 2011;138:3625–3637. doi: 10.1242/dev.064162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palermo A.T., LaBarge M.A., Doyonnas R., Pomerantz J., Blau H.M. Bone marrow contribution to skeletal muscle: a physiological response to stress. Dev. Biol. 2005;279:336–344. doi: 10.1016/j.ydbio.2004.12.024. [DOI] [PubMed] [Google Scholar]
- Pawlikowski B., Betta N.D., Elston T., O’Rourke R., Jones K., Olwin B.B. A cellular atlas of skeletal muscle regeneration and aging. bioRxiv. 2019:635805. [Google Scholar]
- Paylor B., Natarajan A., Zhang R.-H., Rossi F. Nonmyogenic cells in skeletal muscle regeneration. Curr. Top. Dev. Biol. 2011;96:139–165. doi: 10.1016/B978-0-12-385940-2.00006-1. [DOI] [PubMed] [Google Scholar]
- Pietrosemoli N., Mella S., Yennek S., Baghdadi M.B., Sakai H., Sambasivan R., Pala F., Di Girolamo D., Tajbakhsh S. Comparison of multiple transcriptomes exposes unified and divergent features of quiescent and activated skeletal muscle stem cells. Skelet. Muscle. 2017;7:28. doi: 10.1186/s13395-017-0144-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polesskaya A., Seale P., Rudnicki M.A. Wnt signaling induces the myogenic specification of resident CD45+ adult stem cells during muscle regeneration. Cell. 2003;113:841–852. doi: 10.1016/s0092-8674(03)00437-9. [DOI] [PubMed] [Google Scholar]
- Ramilowski J.A., Goldberg T., Harshbarger J., Kloppmann E., Lizio M., Satagopam V.P., Itoh M., Kawaji H., Carninci P., Rost B. A draft network of ligand–receptor-mediated multicellular signalling in human. Nat. Commun. 2015;6:7866. doi: 10.1038/ncomms8866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relaix F., Zammit P.S. Satellite cells are essential for skeletal muscle regeneration: the cell on the edge returns centre stage. Development. 2012;139:2845–2856. doi: 10.1242/dev.069088. [DOI] [PubMed] [Google Scholar]
- Rock K.L., Reits E., Neefjes J. Present yourself! By MHC class I and MHC class II molecules. Trends Immunol. 2016;37:724–737. doi: 10.1016/j.it.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saelens W., Cannoodt R., Todorov H., Saeys Y. A comparison of single-cell trajectory inference methods. Nat. Biotechnol. 2019;37:547–554. doi: 10.1038/s41587-019-0071-9. [DOI] [PubMed] [Google Scholar]
- Seale P., Ishibashi J., Scimè A., Rudnicki M.A. Pax7 is necessary and sufficient for the myogenic specification of CD45+:sca1+ stem cells from injured muscle. PLoS Biol. 2004;2:e130. doi: 10.1371/journal.pbio.0020130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segawa M., Fukada S., Yamamoto Y., Yahagi H., Kanematsu M., Sato M., Ito T., Uezumi A., Hayashi S., Miyagoe-Suzuki Y. Suppression of macrophage functions impairs skeletal muscle regeneration with severe fibrosis. Exp. Cell Res. 2008;314:3232–3244. doi: 10.1016/j.yexcr.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Shea K.L., Xiang W., LaPorta V.S., Licht J.D., Keller C., Basson M.A., Brack A.S. Sprouty1 regulates reversible quiescence of a self-renewing adult muscle stem cell pool during regeneration. Cell Stem Cell. 2010;6:117–129. doi: 10.1016/j.stem.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A.J., Chang C.-N., Ma H.-Y., Ramsey S.A., Filtz T.M., Kioussi C. FACS-Seq analysis of Pax3-derived cells identifies non-myogenic lineages in the embryonic forelimb. Sci. Rep. 2018;8:7670. doi: 10.1038/s41598-018-25998-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott R.W., Arostegui M., Schweitzer R., Rossi F.M., Underhill T.M. Hic1 defines quiescent mesenchymal progenitor subpopulations with distinct functions and fates in skeletal muscle regeneration. Cell Stem Cell. 2019;25:797–813.e9. doi: 10.1016/j.stem.2019.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Street K., Risso D., Fletcher R.B., Das D., Ngai J., Yosef N., Purdom E., Dudoit S. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics. 2018;19:477. doi: 10.1186/s12864-018-4772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Stoeckius M., Smibert P., Satija R., Hafemeister C., Papalexi E., Mauck W.M., III, Hao Y. Comprehensive integration of single-cell data resource comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumm J., Vallecillo-García P., Vom Hofe-Schneider S., Ollitrault D., Schrewe H., Economides A.N., Marazzi G., Sassoon D.A., Stricker S. Odd skipped-related 1 (Osr1) identifies muscle-interstitial fibro-adipogenic progenitors (FAPs) activated by acute injury. Stem Cell Res. 2018;32:8–16. doi: 10.1016/j.scr.2018.08.010. [DOI] [PubMed] [Google Scholar]
- Subramanian A., Schilling T.F. Tendon development and musculoskeletal assembly: emerging roles for the extracellular matrix. Development. 2015;142:4191–4204. doi: 10.1242/dev.114777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tidball J.G., Wehling-Henricks M. Macrophages promote muscle membrane repair and muscle fibre growth and regeneration during modified muscle loading in mice in vivo. J. Physiol. 2007;578:327–336. doi: 10.1113/jphysiol.2006.118265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapecar M., Khan S., Roan N.R., Chen T.-H., Telwatte S., Deswal M., Pao M., Somsouk M., Deeks S.G., Hunt P.W. An optimized and validated method for isolation and characterization of lymphocytes from HIV+ human gut biopsies. AIDS Res. Hum. Retroviruses. 2017;33:S31–S39. doi: 10.1089/aid.2017.0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M., Lennon N.J., Livak K.J., Mikkelsen T.S., Rinn J.L. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Velthoven C.T.J., de Morree A., Egner I.M., Brett J.O., Rando T.A. Transcriptional profiling of quiescent muscle stem cells in vivo. Cell Rep. 2017;21:1994–2004. doi: 10.1016/j.celrep.2017.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villalta S.A., Rinaldi C., Deng B., Liu G., Fedor B., Tidball J.G. Interleukin-10 reduces the pathology of mdx muscular dystrophy by deactivating M1 macrophages and modulating macrophage phenotype. Hum. Mol. Genet. 2011;20:790–805. doi: 10.1093/hmg/ddq523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Allen W.E., Wright M.A., Sylwestrak E.L., Samusik N., Vesuna S., Evans K., Liu C., Ramakrishnan C., Liu J. Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science. 2018;361 doi: 10.1126/science.aat5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wosen J.E., Mukhopadhyay D., MacAubas C., Mellins E.D. Epithelial MHC class II expression and its role in antigen presentation in the gastrointestinal and respiratory tracts. Front. Immunol. 2018;9:2144. doi: 10.3389/fimmu.2018.02144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynn T.A., Vannella K.M. Macrophages in tissue repair, regeneration, and fibrosis. Immunity. 2016;44:450–462. doi: 10.1016/j.immuni.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Hu P. Skeletal muscle regeneration is modulated by inflammation. J. Orthop. Transl. 2018;13:25–32. doi: 10.1016/j.jot.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye A., He H., Kim J. Paternally expressed Peg3 controls maternally expressed Zim1 as a trans factor. PLoS One. 2014;9:e108596. doi: 10.1371/journal.pone.0108596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H., Price F., Rudnicki M.A. Satellite cells and the muscle stem cell niche. Physiol. Rev. 2013;93:23–67. doi: 10.1152/physrev.00043.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue F., Bi P., Wang C., Shan T., Nie Y., Ratliff T.L., Gavin T.P., Kuang S. Pten is necessary for the quiescence and maintenance of adult muscle stem cells. Nat. Commun. 2017;8:14328. doi: 10.1038/ncomms14328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng T., Dai H. Single-cell RNA sequencing-based computational analysis to describe disease heterogeneity. Front. Genet. 2019;10:629. doi: 10.3389/fgene.2019.00629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilionis R., Engblom C., Pfirschke C., Savova V., Zemmour D., Saatcioglu H.D., Krishnan I., Maroni G., Meyerovitz C.V., Kerwin C.M. Single-cell transcriptomics of human and mouse lung cancers reveals conserved myeloid populations across individuals and species. Immunity. 2019;50:1317–1334.e10. doi: 10.1016/j.immuni.2019.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the single-cell RNA-sequencing data reported in this paper is: GSE138826.