Artificial intelligence (AI) within GI endoscopy is an area of growing interest and ongoing innovation. The use of computer-aided diagnosis (CADx) and computer-aided detection (CADe) technologies based on AI algorithms may help augment endoscopists’ performance if these tools can be effectively incorporated into the endoscopy suite.1
Here we demonstrate the use of a previously validated2 CADe system for colon polyp detection, used in real time during screening colonoscopy, and we discuss several practical considerations when CADe technologies are used in the endoscopy suite.
Description of the technology
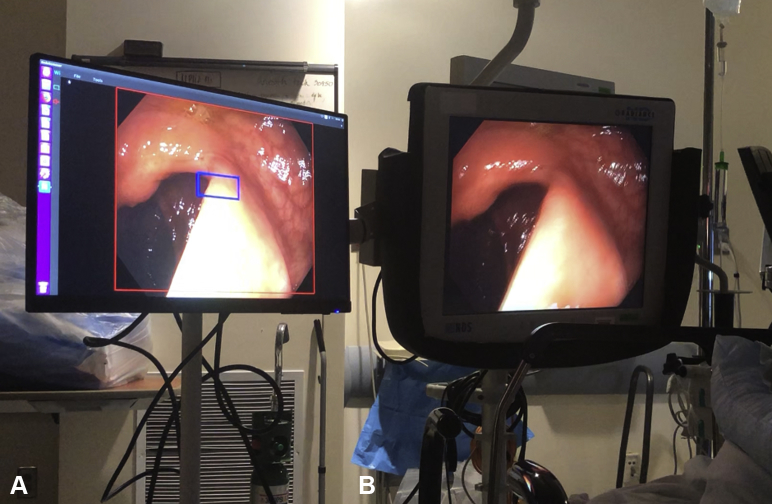
The automatic polyp detection system used in this video (Video 1, available online at www.VideoGIE.org) was developed by Shanghai Wision AI Co, Ltd, Shanghai, China. It is a convolutional neural network based on SegNet architecture that has been prospectively studied in a prior randomized clinical trial with use of a dual-monitor setup.3 For the purposes of this video study, a similar dual-monitor setup was used (Fig. 1). The research software uses a deep learning algorithm to detect polyps in nearly real time, with a current latency of approximately 46.56 ± 2.79 ms, and the indicator, or “visual alarm,” can be configured to display as a hollow blue box or as a paint-filled visual indicator (Fig. 2). A prior validation study showed a per-image sensitivity of 94.38%, a per-image specificity of 95.92%, and an area under the receiver operating characteristic curve of 0.984 for the detection of polyps.2 For this video study, standard Olympus 190-series high-definition white-light colonoscopes (CF-HQ190L/I; Olympus Corp, Tokyo, Japan) were used. The current system is still being evaluated in an experimental setting, but this and similar systems may be available for commercial use in the future.
Figure 1.
Another example of the dual-monitor setup, in which the computer-aided detection output lies adjacent and parallel to the primary monitor. (A) Computer aided detection output on a supplementary monitor. (B) Primary monitor displaying high-definition white light output.
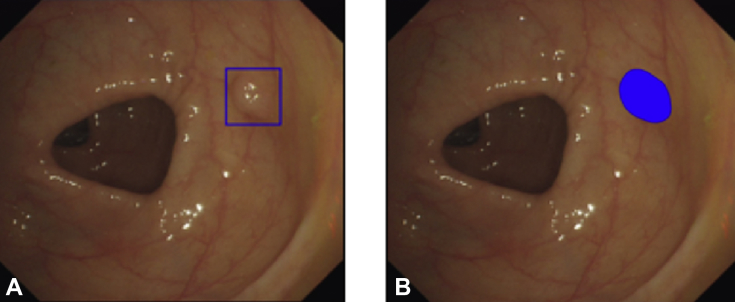
Figure 2.
Example output of the computer-aided detection system. The indicator can be programmed to display a variety of visual indicators. A hollow box drawn around the suspected polyp (A) is currently the preferred approach in most systems. (B) Shows a paint-fill visual indicator.
Considerations
CADe can be used with a single- or dual-monitor configuration. In the single-monitor configuration, the CADe output is overlaid directly on the primary endoscopy screen (Fig. 3). In the dual-monitor setup, such as that displayed in Video 1, the endoscopist uses the standard primary endoscopy monitor, along with a second monitor that is adjacent to the primary monitor. The second monitor displays the standard colonoscopy video with AI overlay for CADe polyp detection (Fig. 3). Although a single-monitor setup may be preferred in the future so that the endoscopist can concentrate on just 1 screen, a dual-monitor setup may be preferred for certain study designs and may be less burdensome if latency is greater. Studies show that screen latency of more than 50 to 100 milliseconds may have a negative impact on performance in certain virtual environments.4 Prior experience with multiscreen configurations, such as with the full-spectrum endoscopy technology,5 suggests that physicians can adapt to using multiple screens, but this may affect gaze patterns in an adverse way.6
Figure 3.
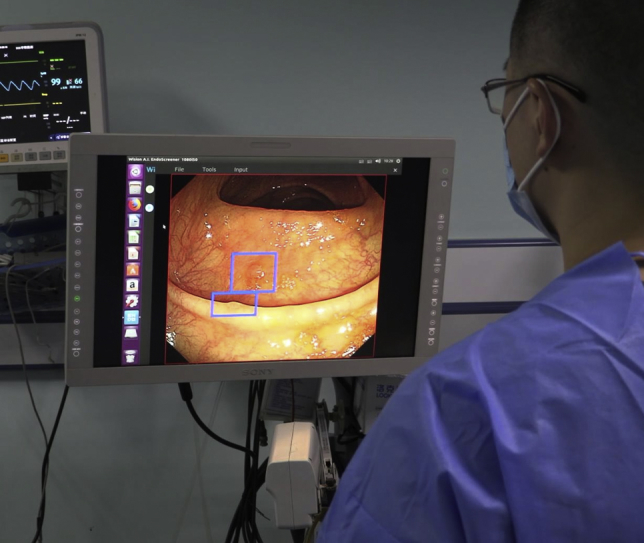
A single-monitor configuration, in which the computer-aided detection output is displayed directly on the primary monitor.
Many proposed solutions for CADe involve an additional processor or central processing unit, either on a mobile cart or as a horizontally placed processor, which can sit atop the endoscopy processor and light source. We expect that future iterations of CADe technology may be either cloud based or incorporated into the endoscopy processor, obviating the need for a separate CADe processing unit.
Intraprocedural considerations
As shown in Video 1, good mucosal inspection techniques, including careful suction of fluid and debris and full insufflation of the colonic lumen during withdrawal, are paramount to ensure optimal performance of the CADe system and to limit false-positive results (Fig. 4). CADe technologies are designed to detect polyps that are partially or fully in the visual field but that otherwise may be missed by the endoscopist. Careful mucosal visualization is still paramount, and in the future, devices that enhance mucosal visualization, such as the EndoCuff Vision distal endoscope attachment (Olympus Corp, Tokyo, Japan),7 may be paired with CADe technology to further improve adenoma detection.
Figure 4.
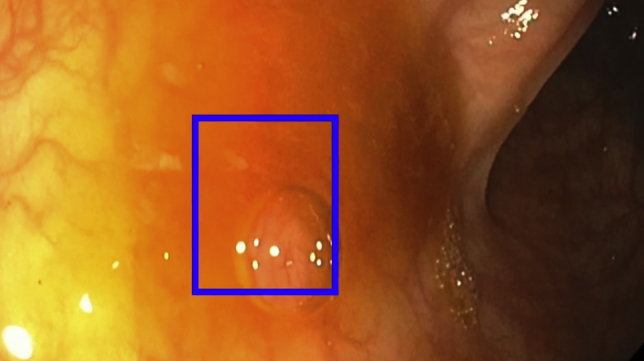
Example of a false-positive result. In this case, a bubble is identified as a possible polyp by the computer-aided detection system, before it is washed away (full sequence shown in Video 1).
Cases
Case 1 represents the detection of a flat lesion during colonoscopy by the CADe system. An ideal CADe technology should demonstrate high sensitivity for the detection of flat lesions such as sessile serrated adenomas, which often have subtle surface features and otherwise serve as a challenge for detection with the naked eye (Fig. 5).8
Figure 5.
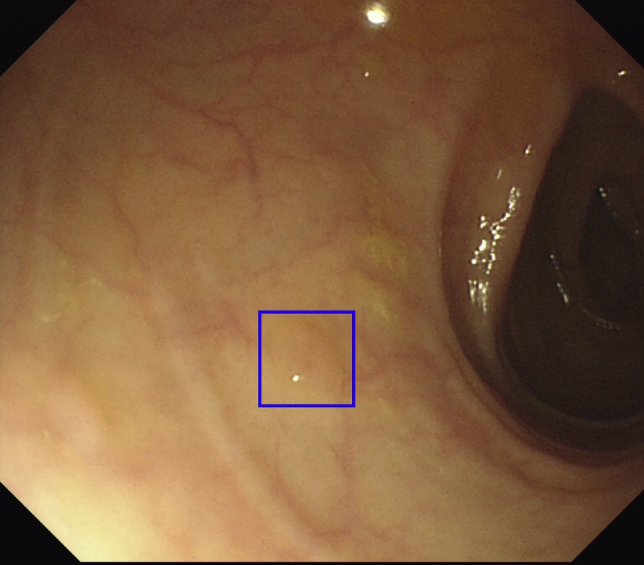
A sessile serrated lesion identified by the computer-aided detection system. Flat lesions may serve as a particular challenge to the endoscopist and serve as an important target for any computer-aided detection system.
Case 2 represents the detection of small polyps during colonoscopy, which otherwise may be missed by the endoscopist.
Case 3 represents the delayed detection of a polyp. In some cases, with the current iteration of the technology, the endoscopist may detect the polyp before the deep learning algorithm. These cases serve as an opportunity to iteratively improve CADe software performance, because earlier detection, as soon as the polyp is on screen, is preferred.
Case 4 represents the detection of a sessile serrated polyp with a mucous cap by the CADe system.
Case 5 represents the detection of a laterally spreading tumor by the CADe system.
Conclusions
Multiple CADe technologies have already shown great promise in detecting colon polyps. More importantly, ongoing prospective clinical trials are already beginning to report data on the impact of CADe on adenoma detection. Similar approaches are being applied to other GI lesions as well, ranging from Barrett’s esophagus to dysplasia detection in inflammatory bowel disease. GI endoscopists will need to familiarize themselves with how AI systems will be incorporated into the endoscopy suite, and we believe that CADe systems for colon polyp detection represent the starting point for this process.
Acknowledgment
Research support for this work was provided by Shanghai Wision AI.
Disclosure
Dr Berzin is a consultant for Wision AI, Fujifilm, and Medtronic. All other authors disclosed no financial relationships relevant to this publication.
Footnotes
If you would like to chat with an author of this article, you may contact Dr Glissen Brown at jglissen@bidmc.harvard.edu.
Supplementary data
A video primer outlining a description of the computer aided detection technology as well as intraprocedural considerations and 5 video cases.
References
- 1.Vinsard D.G., Mori Y., Misawa M. Quality assurance of computer-aided detection and diagnosis in colonoscopy. Gastrointest Endosc. 2019;90:55–63. doi: 10.1016/j.gie.2019.03.019. [DOI] [PubMed] [Google Scholar]
- 2.Wang P., Xiao X., Glissen Brown J.R. Development and validation of a deep-learning algorithm for the detection of polyps during colonoscopy. Nat Biomed Eng. 2018;2:1–748. doi: 10.1038/s41551-018-0301-3. [DOI] [PubMed] [Google Scholar]
- 3.Wang P., Berzin T.M., Glissen Brown J.R. Real-time automatic detection system increases colonoscopic polyp and adenoma detection rates: a prospective randomised controlled study. Gut. 2019;68:1813–1819. doi: 10.1136/gutjnl-2018-317500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buker T.J., Vincenzi D.A., Deaton J.E. The effect of apparent latency on simulator sickness while using a see-through helmet-mounted display: reducing apparent latency with predictive compensation. Hum Factors. 2012;54:235–249. doi: 10.1177/0018720811428734. [DOI] [PubMed] [Google Scholar]
- 5.Gralnek I.M., Siersema P.D., Halpern Z. Standard forward-viewing colonoscopy versus full-spectrum endoscopy: an international, multicentre, randomised, tandem colonoscopy trial. Lancet Oncol. 2014;15:353–360. doi: 10.1016/S1470-2045(14)70020-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lami M., Singh H., Dilley J.H. Gaze patterns hold key to unlocking successful search strategies and increasing polyp detection rate in colonoscopy. Endoscopy. 2018;50:701–707. doi: 10.1055/s-0044-101026. [DOI] [PubMed] [Google Scholar]
- 7.Ngu W.S., Bevan R., Tsiamoulos Z.P. Improved adenoma detection with Endocuff Vision: the ADENOMA randomised controlled trial. Gut. 2019;68:280–288. doi: 10.1136/gutjnl-2017-314889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma M.X., Bourke M.J. Sessile serrated adenomas: how to detect, characterize and resect. Gut Liver. 2017;11:747–760. doi: 10.5009/gnl16523. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A video primer outlining a description of the computer aided detection technology as well as intraprocedural considerations and 5 video cases.