Abstract
Curcumin derivatives have been shown to inhibit replication of human influenza A viruses (IAVs). However, it is not clear whether curcumin and its derivatives can inhibit neuraminidase (NA) of influenza virus. In this study, a meaningful 3D quantitative structure–activity relationship model (comparative molecular field analysis R2 = 0.997, q2 = 0.527, s = 0.064, F = 282.663) was built to understand the chemical–biological interactions between their activities and neuraminidase. Molecular docking was used to predict binding models between curcumin derivatives and neuraminidase. Real-time polymerase chain reactions showed that the five active curcumin derivatives might have direct effects on viral particle infectivity in H1N1-infected lung epithelial (MDCK) cells. Neuraminidase activation assay showed that five active curcumin derivatives decreased H1N1-induced neuraminidase activation in MDCK cells. Indirect immunofluorescence assay indicated that two active curcumin derivatives (tetramethylcurcumin and curcumin) down-regulated the nucleoprotein expression. Curcumin inhibited IAV in vivo. The therapeutic mechanism of curcumin in the treatment of influenza viral pneumonia is related to improving the immune function of infected mice and regulating secretion of tumor necrosis-α, interleukin-6, and interferon-γ. These results indicate that curcumin derivatives inhibit IAV by blocking neuraminidase in the cellular model and curcumin also has anti-IAV activity in the animal model.
Keywords: Curcumin derivatives, 3D-QSAR, Docking, Neuraminidise, Antiviral effects
Introduction
Influenza A virus (IAV), a type of orthomyxoviridae virus responsible for frequent seasonal epidemics, is closely related to human health (Taubenberger and Morens 2008; Scalera and Mossad 2009). It causes acute respiratory disease by infection of the upper respiratory tract mucosa (Taubenberger and Morens 2008). The glycoproteins hemagglutinin (HA) and neuraminidase (NA) are embedded in the envelope of the influenza virus, and play an important role in the invasion of host cells and viral transmission (Smith et al. 2009). In the procession of viral shedding, neuraminidase cuts the sialic acid linkage formed between the sialic acid receptor on the surface of the host cell and the hemagglutinin to promote viral replication (Nayak and Jabbar 1989; Ohuchi et al. 2006). Hence, neuraminidase is considered to be an important target for treatment of influenza (Ohuchi et al. 2006).
Currently, amantadine, oseltamivir, and zanamivir are used for treatment of influenza A (Pica and Palese 2013), including inhibition of the matrix protein M2 (amantadine) and NA (oseltamivir and zanamivir). M2 ion channel blockers only inhibit IAV and have potential neurotoxicity (Zhao et al. 2012). Neuraminidase inhibitors (NAIs) prevent infected cells from releasing and transmitting progeny virions during the replication cycle by binding to the active site of viral neuraminidase (Belser et al. 2015). However, in recent years, because of the widespread use of these drugs, resistant strains of IAV have emerged (Hurt et al. 2009; Moscona 2009; Cho et al. 2013). Therefore, it is urgent to identify new antiviral drugs that are effective against influenza viruses.
Due to its versatile pharmacological properties, including anti-inflammatory, anti-tumor, and anti-oxidant activities (Maheshwari et al. 2006; Anand et al. 2007; Goel et al. 2007), curcumin has attracted considerable interest. In particular, curcumin is reported to have anti-influenza virus activity by interfering with the cellular signaling pathways (such as Toll-like receptor2/4, p38/c-Jun N-terminal kinase, mitogen-activated protein kinase, and nuclear factor-κB pathways) or with proteins required for influenza virus replication (Dai et al. 2018; Richart et al. 2018). Ou et al. (2013) reported that the presence of the double bonds in the central seven-carbon chain enhances the curcumin-dependent anti-IAV activity and curcumin might interfere with IAV entry by its interaction with the receptor binding region of hemagglutinin. Chen et al. (2013) also reported that curcumin can inhibit influenza virus by disrupting integrity of the viral envelope and liposomal membranes. However, it is not clear whether curcumin and its derivatives can inhibit neuraminidase of influenza virus.
In the present study, we evaluated the antiviral effects of curcumin derivatives as potent inhibitors of influenza H1N1 neuraminidase based on 3D quantitative structure–activity relationship (QSAR) (Duarte et al. 2019), molecular docking (Nilakantan et al. 1993) and biological activity evaluation both in vitro and in vivo.
Materials and methods
Virus stock
Influenza virus strain, A/Font Monmouth/47(H1N1, FM1), adapted to mice, was originally obtained from Chinese Center for Disease Control and Prevention. The virus was adapted to replication in the lungs of BALB/c mice by eight sequential passages through mouse lungs. The inoculated virus was plaque-purified in MDCK cells, then replicated in 9-day-old chicken embryos and stored at− 80 °C. The virus titer was determined as 50% tissue culture infectious dose (TCID50/mL) in confluent cells of 96-well microtiter plates (Greiner Bio-One, Frickenhausen, Germany).
Cells and animals
Madin–Darby canine kidney (MDCK) cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 IU/mL penicillin and 100 mg/mL streptomycin in a 5% CO2 incubator. Specific-pathogen-free female NIH mice weighing 13–15 g (Medical Experiment Center of Guangdong Province, Guangzhou, China) were used after 24 h quarantine, and were fed standard mouse chow and tap water ad libitum for the duration of the studies. The animals were housed in the Guangdong Association for the Accreditation of Laboratory Animal Care-accredited Guangzhou University of Chinese Medicine (GZUCM) Laboratory Animal Research Center. All experimental protocols were approved by the GZUCM Animal Care and Welfare Committee.
Compounds
Dihydrocurcumin, tetramethylcurcumin, bisdemethoxycurcumin, curcumin-d6, demethoxycurcumin, demethylcurcumin, dimethylcurcumin, curcumin, tetrahydrocurcumin, octahydrocurcumin, hexahydrocurcumin and oseltamivir carboxylate were acquired from Shanghai yuanye Bio-Technology Co. Ltd. (Shanghai, China). Purity was ≥ 99% by high-performance liquid chromatography (HPLC).
Cell viability assay
Confluent MDCK cells were treated with a series of graded concentrations of curcumin derivatives for 24 h. Confluent MDCK cells were treated with curcumin derivatives for 24 h. Cell viability was determined using the 3-(4,5-dimethylthiazol-2-yl)-2–5-diphenyltetrazolium bromide (MTT) method. The median cytotoxic concentration (CC50) was calculated as the concentration of the compound that reduced the number of viable cells to 50% of the viability of the untreated controls. The maximal non-cytotoxic concentration was defined as the maximal concentration of the sample that did not exert a cytotoxic effect and resulted in > 90% viable cells.
Neuraminidase inhibition screening assay
Influenza virus NA activity was measured by Neuraminidase Inhibitors Screening Kit. Neuraminidase Inhibitors Screen Kit (NO. P0309) was purchased from Beyotime Institute of Biotechnology Co. Ltd. (Shanghai, China), which contained 10 mL buffer, 1 mL NA, 1 mL fluorescent substrate and 1.2 mL Milli-Q water.
3D-QSAR studies
We selected compounds with quantitative IC50 values as data set, and divided the 11 compounds randomly into a training set (nine compounds) and a testing set (three compounds). We converted the IC50 values to pIC50 values with the formula pIC50 = Δlog IC50. We used the pIC50 values to develop the 3D-QSAR model by combining the activity data with the generated hypothesis. The chemical structures and biological activities of the 11 compounds are listed in Table 1. We used the Sybyl-X2.1.1 molecular model package to perform the modeling studies.
Table 1.
Structure and activity data of curcumin derivatives as influenza H1N1 virus neuraminidase inhibitors
| No | Compd name | Structure | IC50 (μM) | pIC50 (expt.) | pIC50 (pred.) | Residual |
|---|---|---|---|---|---|---|
| 1 | Dihydrocurcumin | 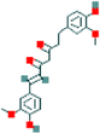 |
219.15 | 3.66 | 3.65 | 0.01 |
| 2 | Tetramethylcurcumin | 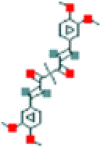 |
225.8 | 3.65 | 3.64 | 0.01 |
| 3 | Bisdemethoxycurcumin | 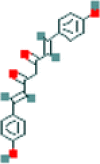 |
70.12 | 4.15 | 4.17 | − 0.02 |
| 4 | Curcumin-d6 | 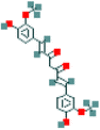 |
18,769.02 | 1.73 | 1.71 | 0.02 |
| 5 | Demethoxycurcumin | 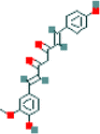 |
199.64 | 3.70 | 3.77 | − 0.07 |
| 6 | Demethylcurcumin | 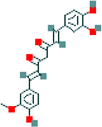 |
62.77 | 4.20 | 4.12 | 0.08 |
| 7 | Dimethylcurcumin | 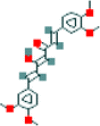 |
1610.82 | 2.79 | 2.80 | − 0.01 |
| 8 | Curcumin | 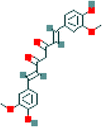 |
450.27 | 3.35 | 3.35 | 0 |
| 9a | Tetrahydrocurcumin | 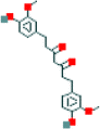 |
203.04 | 3.69 | 3.63 | 0.06 |
| 10a | Octahydrocurcumin | 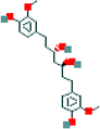 |
1534.48 | 2.81 | 3.65 | − 0.84 |
| 11a | Hexahydrocurcumin |  |
33,695.47 | 1.47 | 2.82 | − 1.35 |
Oseltamivir carboxylate was chosen as a positive control in the NA activity inhibition assay, the IC50 of oseltamivir carboxylate was 225.42 μM
aMolecules in the testing set
To derive the comparative molecular field analysis (CoMFA) descriptor fields, we generated a 3D cubic lattice from the space covering the nine aligned compounds. The standard Tripos force fields were used to calculate the space and electrostatic potential fields of the CoMFA. To establish quantitative relationships between biological activities and molecular descriptors in CoMFA, we used partial least squares regression analysis (Golbraikh and Tropsha 2002). The leave one method was used for cross-validation analysis (Golubović et al. 2018). Then, non-cross-validation was performed to obtain a conventional correlation coefficient r2 and estimated standard errors. To enhance the predictive ability, the region focusing method was used in the CoMFA model (Xu et al. 2016).
To predict the power of the 3D-QSAR model, we verified by the test set compounds (compounds marked * in Table 1). The QSAR model was validated by the leave-n-out techniques, which helped to assess the stability of the model to changes in the training set. We performed an external validation using a testing set whose structures and activities were not considered in the model building (Patel et al. 2008). Gao et al. (2011) suggested that the 3D-QSAR model performed well with Rtest2 > 0.6 or qtest2 > 0.5, where Rtest2 is the normalized correlation coefficient of the testing set, and qtest2 is the cross-validation correlation coefficient of the testing set. To validate the predictive power of the 3D-QSAR model, the activity of a three-compound testing set was predicted (Table 1). The CoMFA model was used to predict the pIC50 for each aligned conformation of the compound.
Docking studies
To investigate further the binding patterns and the SAR between curcumin derivatives and neuraminidase, we performed molecular docking. The docking procedure was performed using the Surflex-dock module of Sybyl-X2.1.1, and the receptor–ligand interactions were carried out by the Discovery Studio 2016 software. The curcumin derivatives were processed by adding polar hydrogen atoms, giving the Gasteiger–Huckel charge, and using Maxim’s genetic algorithm to perform energy minimization in the Tripos force field. The X-ray structure of influenza virus neuraminidase (code: 4HZZ) was taken from the RSCB Protein Data Bank (Fang et al. 2011). The ligand-binding site of neuraminidase was defined by the reference ligand 39G. The semi-flexible method was used for docking, and the crystal structure of the original ligand as a reference for docking. The parameters in the docking process were set as follows: expansion coefficient 1, threshold parameter 0.5, and other parameters were system default values. The Total Score function was used to score the situation of molecular docking, which is an empirical scoring function derived from the binding energies of protein–ligand complexes and their X-ray structures. This function takes into account factors such as polarity, hydrophobicity, enthalpy, and solvation. The larger the value, the more stable the docking compound, which means better matching and binding of small molecules and large proteins (Li et al 2013). It is generally accepted that Total Score is > 6 for better activity, and ≥ 9 for very good activity.
Real-time reverse transcriptase-polymerase chain reaction (RT-PCR)
RT-PCR was used to assess the ability of curcumin derivatives and oseltamivir carboxylate to inhibit H1N1 virus. Confluent MDCK cell layers infected with 100 μL 100 TCID50 IAV (H1N1) were treated with curcumin derivatives or oseltamivir carboxylate (20 mM, non-toxic concentration, data not shown) for 24 h. Then, cells were collected to detect the mRNA expression level of H1N1 M gene by using IAV nucleic acid test kit (RT-PCR). The primer sequences used for the detection of viral RNA were 5′-TTC TAA CCG AGG TCG AAA CG-3′ and 5′-ACA AAG CGT CTA CGC TGC AG-3′.
Neuraminidase inhibition activity assay
Cell culture supernatants were collected and treated for neuraminidase activity [expressed as relative luminescence units (mU/mL)] using the Neuraminidase Activity Fluorometric Assay Kit (K732-100, BioVision, Milpitas, CA, USA). Fluorometry was measured using POLARstar OPTIMA Multifunction Fluorescence Microplate Reader (BMG, Germany).
Indirect immunofluorescence microscopy
Confluent MDCK cell layers were infected with 100 μL 100 TCID50 IAV (H1N1) for 2 h at 37 °C. After removing the virus-containing medium, the cells were treated with curcumin derivatives at maximum non-toxic doses (MNTDs) in DMEM. Cells were fixed with ice-cold acetone/methanol (40:60, Mallinckrodt Baker B.V., Deventer, The Netherlands) for 15 min after 8 h infection. Then, cells were stained using the mouse monoclonal antibody (1 h incubation, 1:100 in phosphate-buffered saline; PBS) directed against IAV nucleoprotein (NP) (Millipore, Molsheim, France). The Alexa Fluor 488 goat anti-mouse IgG (H&L) (Invitrogen, Eugene, OR, USA) was used (1 h incubation, 1:100 in PBS) as secondary antibody. Nuclei were stained with 40,6-diamidino-2-phenylindole (DAPI) (Sigma–Aldrich Chemie GmbH, Munich, Germany). An Olympus IX 1 fluorescence microscope (Olympus, Planegg, Germany) was used to visualize fluorescence.
Antiviral study in mice
NIH mice were randomly divided into six groups, 12 in each group: normal control group (NC); placebo control group (IAV-C + sterile saline, suspended in 0.5% Tween 80); curcumin groups (H1N1 virus + curcumin 25, 50 and 100 mg/kg); and positive control ribavirin (70 mg/kg). The challenge inoculation of ~ 105 50% cell culture infectious doses (CCID50)/mouse was equivalent to five 50% mouse lethal challenge doses (MLD50). Under mild anesthesia, mice were infected with 50 μL 15 MLD50 IAV (sterile in PBS, pH 7.2) intranasally (without the NC group). Then, mice were treated with intraperitoneal injection of curcumin (25, 50 and 100 mg/kg) or ribavirin (70 mg/kg) for 1 day prior to viral challenge and received concomitant treatment for 3 days after infection. On day 4 after infection, the mice were sacrificed to collect relevant samples, and the body weight, lung wet weight, and the degree of lung pathological changes were measured.
Survival experiments
NIH mice were anesthetized in a chamber containing absorbent cotton saturated with 30 mL diethyl ether for about 30 s and infected intranasally with 2 MLD50 of IAV in sterile PBS (pre-cooled). Then, mice were treated with oral gavage of curcumin (25, 50 and 100 mg/kg) or ribavirin (70 mg/kg) for 1 day before infection in the protective administration group, and mice in the therapeutic administration groups were treated 6 h after infection. Mice received these treatments for 12 days and were monitored daily for 15 days after infection. For the IAV-C group, the mice were given saline (0.5% Tween 80) only at the same intervals. Fifteen days after infection, the mice were continuously observed daily for weight loss, survival, and clinical symptoms of illness (e.g., wrinkled fur, inactivity, loss of appetite, hunchback posture, and rapid shallow breathing) to assess the survival rate and mean days to death. The lung index was calculated as follows: Lung index = A/B × 100, where A is the lung weight, and B is the body weight (Li et al. 2012).
Cytokine in lung homogenate in mice infected with IAV
After the mice were killed by exsanguination, the lungs were removed, homogenized, and centrifuged at 4000×g for 15 min at 4 °C. Before the analysis of cytokine levels, the supernatant of the lung homogenate was stored at − 80 °C. Anti-mouse enzyme-linked immunosorbent assay (ELISA) kits (eBioscience, San Diego, CA, USA) were used to measure the concentrations of interleukin (IL)-6, tumor necrosis factor (TNF)-α and interferon (IFN)-γ in the bronchoalveolar lavage fluid.
Results
Neuraminidase inhibitory activity
We selected 11 curcumin derivatives (Table 1) to perform the neuraminidase inhibition screening assay. Most of them had an inhibitory effect on the structural protein NA of H1N1 virus, with IC50 values ranging from 62.77 to 33,695.47 μM, while oseltamivir carboxylate (positive control) had an IC50 value of 225.42 μM. Eleven active compounds were selected for 3D-QSAR and the docking study.
CoMFA statistical results
The 11 curcumin derivatives selected in the neuraminidase inhibition screening assay were used to perform the 3D QSAR studies (CoMFA), and the CoMFA statistical coefficients are shown in Table 2. When the cross-validation correlation coefficient q2 is > 0.5, the prediction model is reliable (Chen et al. 2011). The larger q2 is, the stronger the prediction ability is. In the CoMFA model, the cross-validation coefficient q2 was 0.527 and the best composition score was 4. Partial least squares regression analysis yielded a model correlation coefficient R2 of 0.997 and an F statistic of 282.663. The cross-validation coefficient was used to evaluate the predictive ability of the fitting equation. The higher R2 value is, the higher the ratio of the two variables, and the better the fitting degree of the model and data; the F statistics are greater than the critical value K, that is, the effect of the regression analysis is significant.
Table 2.
Statistical results of the CoMFA model
| ONC | q2 | R2 | F | SEE |
|---|---|---|---|---|
| 4 | 0.527 | 0.997 | 282.663 | 0.064 |
ONC optimal number of the principal components, q2 cross-validated correlation coefficient of the training set, R2 conventional correlation coefficient of the training set, F statistical squared deviation ratio, SEE standard error of the estimate
Compared with the actual activities, the predicted activities of the CoMFA model were in general agreement with the original data (Fig. 1), indicating that the predictive ability of the model was credible. The results of the actual and predicted pIC50 values for the training set and testing set are shown in Fig. 1. It can be seen from the figure that the actual value was close to the predicted value, and the prediction of the pIC50 value by the 3D-QSAR model was reasonably accurate. The actual and predicted values were close, indicating that the model established in this study had statistical significance. The calculated and experimental values of the external verification test set were also similar (red triangle in Fig. 1), indicating that the model had a strong predictive ability and was successfully constructed. Five active compounds with better inhibition of NA activity in vitro were selected for further study.
Fig. 1.
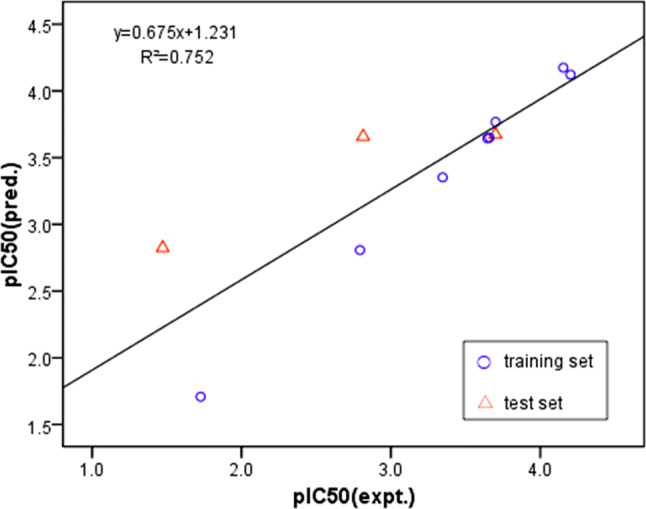
Fitness graphs between observed activity and predicted activity for the training set and the testing set compounds
CoMFA contour maps
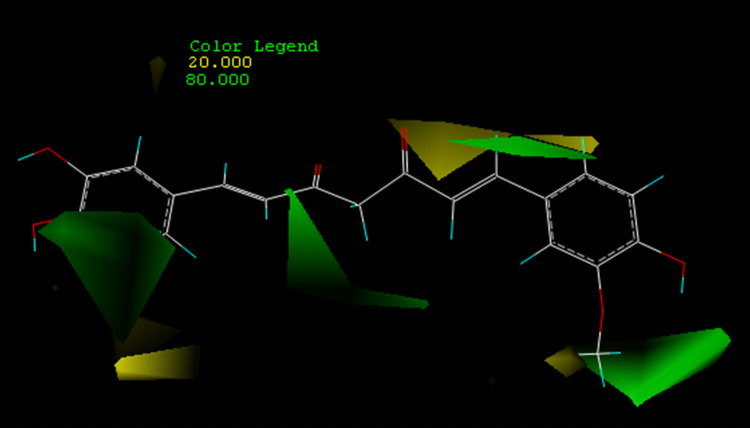
Different colors in the CoMFA model represent regions of reduced or increased activity due to spatial variations of different molecules. The CoMFA steric field is represented as a contour map in Fig. 2.
Fig. 2.
CoMFAsteric contour map. Green contours indicate regions where bulky groups increase activity, whereas yellow contours indicate regions where bulky groups decrease activity
In the stereo contour map, the yellow and green regions represent areas where small and large volume groups enhance activity, respectively. We selected the most potent inhibitor, demethylcurcumin (pIC50 = 4.20), as a reference for assisted visualization. There were four green areas and five yellow areas around the composite zones. The green outline around the meta-hydroxyl group of the phenyl ring indicated where it favored the space volume, such as a meta-methoxy group of the other benzene ring. Large volume groups at these positions may facilitate interactions between the ligand and its receptor, which accounted for why demethoxycurcumin activity (pIC50 = 3.70) was lower than demethylcurcumin activity (pIC50) = 4.20. The difference in activity between bisdemethoxycurcumin (pIC50 = 4.15) and demethylcurcumin (pIC50 = 4.20) was also reasonably explained. In addition, the green contour around the central seven carbon chain showed that the double bonds in the central seven-carbon chain may be beneficial for the interaction between the ligand and its receptor. For example, the activity of dihydrocurcumin (pIC50 = 3.66) and tetrahydrocurcumin (pIC50 = 3.69) was higher than that of hexahydrocurcumin (pIC50 = 1.47) and octahydrocurcumin (pIC50 = 2.81).
Docking study
In order to explore the binding patterns between curcumin derivatives and neuraminidase, molecular docking was performed to help understand the SARs between molecules and proteins. Sybyl-X2.1.1 was applied to carry out the docking studies. Oseltamivir carboxylate was used as a positive control to assess the ability of other molecules to bind to NA. The Total Score function was used to comprehensively score the situation of molecular docking, which is an empirical scoring function derived from the binding energies of protein–ligand complexes and their X-ray structures. It is generally accepted that Total Score is > 6 for better activity, and ≥ 9 for very good activity (Golbraikh and Tropsha 2002). As shown in Fig. 3, the Total Scores of 11 curcumin derivatives and oseltamivir carboxylate were all > 6, indicating that these compounds were likely to have inhibited NA. Thus, according to NA inhibitory activity assays, the binding mode between demethylcurcumin (IC50 = 62.77 μM) and oseltamivir carboxylate (IC50 = 225.42 μM) was further investigated in the docking study, and the top five compounds with higher IC50 were selected for in vitro activity verification.
Fig. 3.
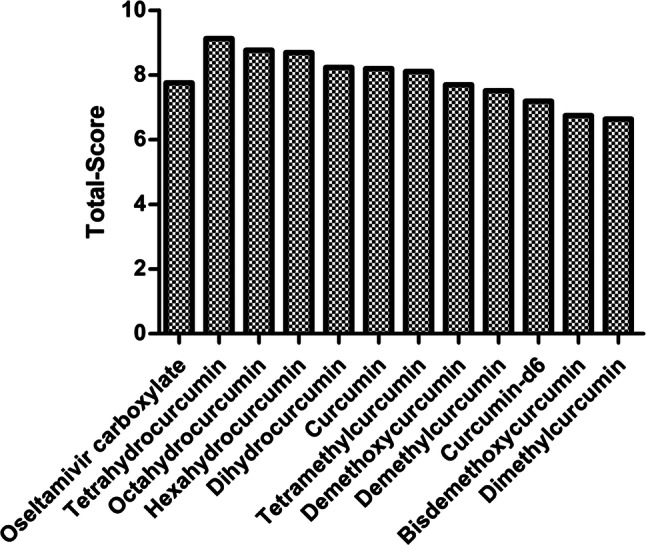
Total-scores of curcumin derivatives and neuraminidase, while oseltamivir carboxylate served as the control
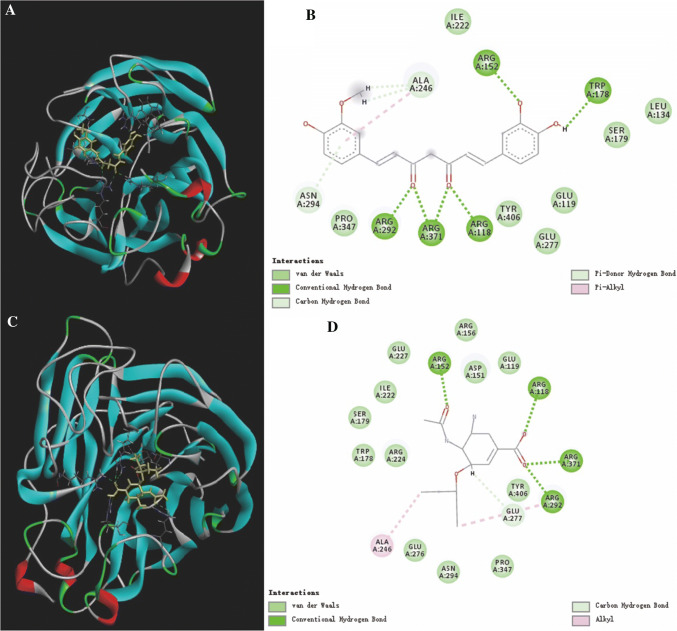
The 3D and 2D binding models between neuraminidase and demethylcurcumin (A and B), and neuraminidase and oseltamivir carboxylate (C and D) were analyzed (Fig. 4). Key residues, H bonds, van der Waals forces, π-alkyl and alkyl groups were analytically labeled and visualized to understand further the details of neuraminidase binding to molecules. The chemical bonds corresponding to the residues are listed in Table 3.
Fig. 4.
3D and 2D binding models between neuraminidase and demethylcurcumin (A, B), neuraminidase and oseltamivir carboxylate (C, D)
Table 3.
The residues for the chemical bonds between small molecules and neuraminidase
| Molecules | Conventional hydrogen bond residues | Pi-donor hydrogen bond residues | Pi-alkyl residues | Alkyl residues |
|---|---|---|---|---|
| Demethylcurcumin | ARGA152, ARGA118, ARGA371, ARGA292, TRPA178 | ASNA294, ALAA246 | ALAA246 | – |
| Oseltamivir carboxylate | ARGA152, ARGA118, ARGA371, ARGA292 | – | – | ALAA246 |
There were four conventional H bonds between the oseltamivir carboxylate and five residues of neuraminidase, which indicated that the binding region of this portion of the H-bond was important for the stable binding between oseltamivir carboxylate and NA (Fig. 4D). The combination of demethylcurcumin with NA produces five conventional H bonds (Fig. 4B), indicating that demethylcurcumin can be considered as a potential NA inhibitor. In addition, there were two π-donor H bonds and one π-alkyl bond between demethylcurcumin and NA, but only one alkyl bond between oseltamivir carboxylate.
Validation of neuraminidase inhibition ability for curcumin derivatives
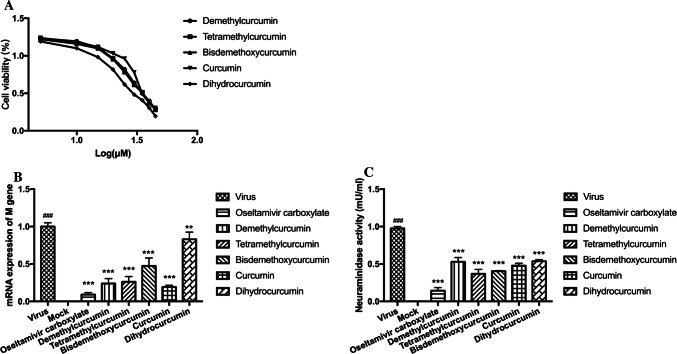
To validate the neuraminidase inhibition ability for the top five compounds that showed higher IC50 in the neuraminidase inhibition screening experiments, experiments including viral replication inhibition assay and neuraminidase inhibition assay in vitro were carried out. First, MTT cell proliferation assay was performed to measure the MNTD of the compounds. The MNTDs for tetramethylcurcumin, demethylcurcumin, bisdemethoxycurcumin, dihydrocurcumin and curcumin were 15, 20, 20, 20 and 25 μM, respectively (Fig. 5A). The MNTDs of the compounds were used in the viral replication inhibition assay and neuraminidase inhibition assay. The five curcumin derivatives showed different degrees of inhibition of H1N1 M gene mRNA expression (Fig. 5B). Demethylcurcumin and tetramethylcurcumin significantly decreased the mRNA expression levels of M gene about 4.17-fold (0.24 ± 0.07 μM) and about 3.85-fold (0.26 ± 0.07 μM) compared to mock-treated virus controls. Treatment with curcumin reduced expression about 5.26-fold (0.19 ± 0.03 μM). Bisdemethoxycurcumin and dihydrocurcumin decreased the M gene mRNA expression about 2.13-fold (0.47 ± 0.11 μM) and 1.20-fold (0.83 ± 0.09 μM), respectively. In addition, the positive drug group (oseltamivir carboxylate) almost inhibited the mRNA expression of M gene, which was about 11.11-fold compared to mock-treated virus controls.
Fig. 5.
A Cellular toxicity of five curcumin derivatives. (B) Influence of five curcumin derivatives treatment on the mRNA expression of M gene in H1N1-infected MDCK cells. MDCK cells were infected with 100 μL of 100TCID50 influenza virus A/Font Monmouth/47(H1N1, FM1) for 2 h at 37 °C. After removing the virus-containing medium, the cells were treated with curcumin derivatives at MNLDs in DMEM respectively. Cells were harvested 24 h after infection and mRNA expression levels of M genes were detected by real-time RT-PCR. Data represent the mean ± SD of 3 separate experiments. *P < 0.05, **P < 0.01, ***P < 0.001 relative to virus group. #P < 0.05, ##P < 0.01, ###P < 0.001 relative to mock control group. (C) Influence of curcumin derivatives treatment on neuraminidase activation in H1N1-infected MDCK cells. MDCK cells were infected with 100 μL of 100TCID50 influenza virus A/Font Monmouth/47(H1N1, FM1) for 2 h at 37 °C. After removing the virus-containing medium, the cells were treated with curcumin derivatives at MNLDs in DMEM respectively. Twenty-four hours post-infection the cell supernatant was analyzed for neuraminidase activity (expressed as mU/mL units) using Neuraminidase Activity Assay Kit. Data represent the mean ± SD of 3 separate experiments. *P < 0.05, **P < 0.01, ***P < 0.001 relative to virus group. #P < 0.05, ##P < 0.01, ###P < 0.001 relative to mock control group
Furthermore, the influence of curcumin derivatives on H1N1-induced neuraminidase activation in MDCK cells was analyzed by the supernatant. H1N1 infection decreased the neuraminidase activity relative to that in the mock-infected cells (Fig. 5C). Compared to virus controls, tetramethylcurcumin and bisdemethoxycurcumin decreased NA activity by 2.65-fold (0.37 ± 0.06 mU/mL) and 2.45-fold (0.40 ± 0.01 mU/mL), respectively, whose inhibitory effects were all better than that of the oseltamivir carboxylate group (0.44 ± 0.04 mU/mL). The other three derivatives also showed an inhibitory effect, in which curcumin, demethylcurcumin and dihydrocurcumin respectively decreased NA activity by about 2.04-fold (0.48 ± 0.03 mU/mL), 1.85-fold (0.53 ± 0.06 mU/mL) and 1.81-fold (0.54 ± 0.02 mU/mL).
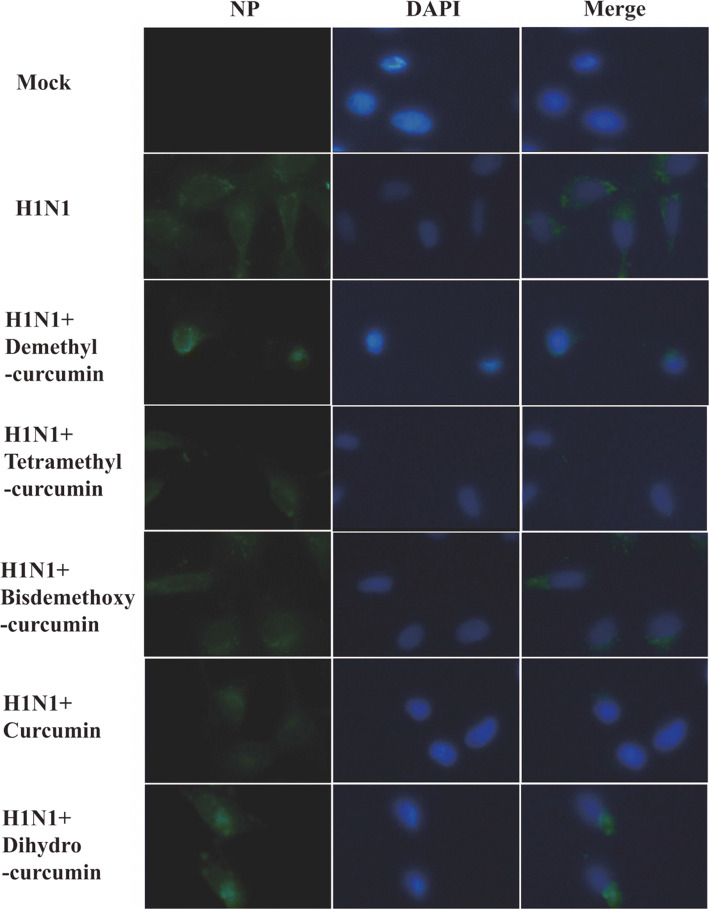
Influence of curcumin derivatives on the nuclear export of viral NP
To investigate the influence of curcumin derivatives on the reduction of the viral NP, MDCK cells were infected with H1N1 (100 TCID50) for 2 h at 37 °C and then were treated with curcumin derivatives at MNTDs. NP localization was detected 8 h after infection, at which time viral replication and transcription were ongoing, and newly formed viral particles began to be released and spread from infected cells. No immunofluorescence of viral NP was observed in the control group, while green fluorescent signal was observed in cells infected with virus (Fig. 6). Viral NP was mainly localized in the cytoplasm, with lesser amounts in the nucleus. Compared to the virus group, less NP was observed in the cytoplasm in cells treated with tetramethylcurcumin and curcumin. In contrast, in cells treated with demethylcurcumin, bisdemethoxycurcumin, and dihydrocurcumin, a strong green fluorescent signal was observed in the cytoplasm. These results suggested that tetramethylcurcumin and curcumin inhibited the nucleation of viral NP, leading to nuclear retention of NP, preventing viral assembly.
Fig. 6.
Influence of curcumin derivatives treatment on nuclear export of viral NP in H1N1-infected MDCK cells. MDCK cells were infected with A/Font Monmouth/47(H1N1, FM1) at 100TCID50. 5 curcumin derivatives treatments were performed continuously starting after infection. Eight hours post-infection NP localisation was visualised using specific antibodies by immunfluorescence. NP staining is shown in green. Nuclei are stained by DAPI (shown in blue). Photographs are
taken from one representative experiment. In total, three independent experiments were performed with similar results
Inhibitory effects of curcumin on IAV in vivo
Mice treated with curcumin at 100 mg/kg/day remained stable in weight and showed no significant clinical symptoms, indicating dose safety. To evaluate the therapeutic efficacy of curcumin against influenza A/Font Monmouth/47(H1N1, FM1) virus, a lethal murine infection model was used and all the mice were killed a few days after infection. The degree of lung pathological change of the placebo group was more serious than that of the normal control group, and the difference in lung index was significant (P < 0.01) (Table 4). Compared with the placebo control, the lung index of the ribavirin group was significantly improved, and the lung index inhibition rate was 43% (P < 0.01). However, the administration of curcumin at 100, 50, and 25 mg/kg/day improved the lung index inhibition rate at 22.42% (P < 0.05), 35.75% (P < 0.1), and 23.6% (P < 0.05), respectively.
Table 4.
Inhibition of the influenza activity of curcumin in H1N1-infected mice
| Treatment | Dose/(mg/kg/day) | Number/(n) | Lung index | Lung index inhibition (%) | Lung pathological change degree |
|---|---|---|---|---|---|
| Normal | – | 12 | 0.82 ± 0.12 | – | – |
| Placebo | – | 12 | 1.65 ± 0.41## | – | 3.29 ± 0.46 |
| Ribavirin | 70 | 12 | 0.94 ± 0.29** | 43** | 0.87 ± 0.52 |
| Curcumin | 100 | 12 | 1.28 ± 0.39* | 22.42* | 1.10 ± 0.39 |
| 50 | 12 | 1.06 ± 0.24** | 35.75** | 0.92 ± 0.47 | |
| 25 | 12 | 1.26 ± 0.42* | 23.6* | 1.02 ± 0.55 |
The mice were killed at 4 dpi. The lungs were removed and rinsed with sterile PBS. The lung index of mice and lung index inhibition were detected (n = 12). #P < 0.05, ##P < 0.01, ###P < 0.001, the values were compared with normal controls. *P < 0.05, **P < 0.01, ***P < 0.001, the values were compared with placebo controls
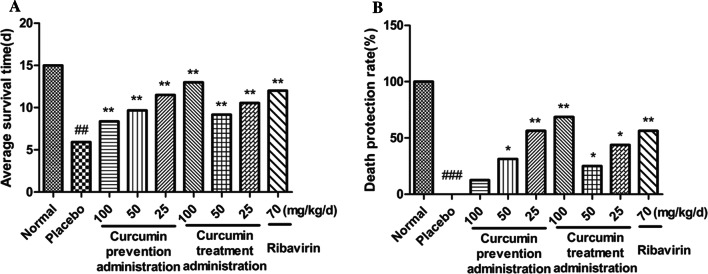
To investigate further the protective and therapeutic efficacy of curcumin against H1N1, mice were treated with curcumin at 100, 50, and 25 mg/kg/day before and after inoculation with IAV. Treatment with curcumin pre-infection and post-infection at all doses prolonged the average survival time and promote the death protection rate significantly, compared with placebo group (Fig. 7). It was observed that the death protection rate with pre-infection administration was dose dependent, while the death protection rate with post-infection administration at 100 mg/kg/day was the most obvious. These results showed that curcumin had a good death protection effect against IAV-induced pneumonia in mice.
Fig. 7.
A Average survival time. (B) Death protection rate (%). Mice were infected with 2 LD50 of the influenza A/Font Monmouth/47(H1N1, FM1) virus and treated with curcumin once a day for 12 days (n = 16). Clinical signs were observed for 15 days (n = 16). Data represent the mean ± SD of 3 separate experiments. #P < 0.05, ##P < 0.01, ###P < 0.001 relative to normal controls. *P < 0.05, **P < 0.01, ***P < 0.001 relative to placebo controls
Effects of curcumin on cytokine production in lung homogenates
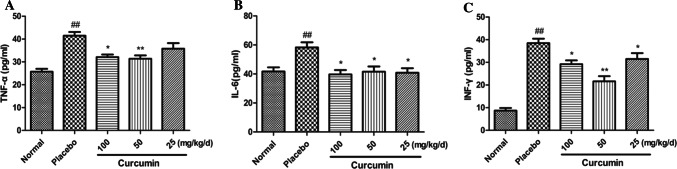
IAV infection causes an increase in pro-inflammatory cytokines, so we examined the effect of curcumin on the release of virus-induced proinflammatory cytokines. Lung tissues were harvested from IAV-infected mice on day 5 after infection, and several cytokines (TNF-α, IL-6, and IFN-γ) were analyzed by ELISA.
Elevation of proinflammatory cytokines is a typical response to IAV infection, so we established whether curcumin affected the virus-induced release of pro-inflammatory cytokines. Lung tissues were harvested from IAV-infected mice on day 5 after infection, and several cytokines (TNF-α, IL-6, and IFN-γ) were analyzed by ELISA. Compared to the control group, the levels of TNF-α, IL-6, and IFN-γ in the lung homogenate were significantly increased (Fig. 8). However, curcumin at different doses significantly attenuated the increases in TNF-α, IL-6 and IFN-γ levels in similar lung homogenates, except for TNF-α level in mice treated with 25 mg/kg/day curcumin.
Fig. 8.
Effects of curcumin on cytokine levels in lung homogenates from mice infected with IAV. Pro-inflammatory cytokine levels were analyzed in the lung homogenates of mice on day 5 post infection, by ELISA (n = 10). (A) The level of TNF-alpha in lung homogenates. (B) The level of INF-gamma in lung homogenates. (C) The level of IL-6 in lung homogenates. Data represent the mean ± SD of 3 separate experiments. #P < 0.05, ##P < 0.01, ###P < 0.001 relative to normal controls. *P < 0.05, **P < 0.01, ***P < 0.001 relative to placebo controls
Discussion
Influenza is a common respiratory infectious disease with high morbidity and mortality during epidemics. The emergence of adverse effects and resistance to antiviral drugs limits their clinical application (Fiers et al. 2004; Hurt et al. 2009; Pica and Palese 2013). Therefore, exploring new anti-influenza drugs is urgently needed (Suzuki et al. 2003). Curcumin, a coloring agent and spice commonly used in food (Goel et al. 2007), inhibits the replication of IAV in vitro and in vivo (Kannan and Kolandaivel 2017; Dai et al. 2018).
In this study, we used a 3D-QSAR model and docking model to investigate the inhibitory effects of curcumin derivatives against neuraminidase and influenza A/Font Monmouth/47(H1N1, FM1) in MDCK cells in vitro. Then, we detected the anti-influenza virus effects of curcumin in mice and revealed that it could significantly inhibit IAV and decreased TNF-α, IL-6 and IFN-γ pro-inflammatory cytokines in vivo.
In the present study, most of the curcumin derivatives exhibited significant inhibition of H1N1 neuraminidase. We established a meaningful 3D-QSAR model (CoMFA) to study the SAR between curcumin derivatives and neuraminidase, and predicted the activity of the ligand in the test set. In addition, establishment of the CoMFA model showed that the hydroxyl group at the meta-position of the benzene ring and the double bonds in the central seven-carbon chain in the curcumin derivatives may be essential for neuraminidase inhibitory activity. The docking model was also consistent with the predicted activity of QSAR.
Curcumin inhibits IAV infection (Mazur et al. 2007; Kannan and Kolandaivel 2017) by interfering with hemagglutinin activity, while tetrahydrocurcumin is less effective in inhibiting IAV infection (Ou et al. 2013). Further SAR analysis showed that the two-tone functional group as a receptor for Michael addition conjugate contributes to the differential inhibition of curcuminoid. Simulated docking of curcumin with the hemagglutinin structure indicates that curcumin binds to the region that constitutes the sialic acid anchoring residue, supporting the results obtained by inhibition of hemagglutinin activity. These studies indicate that the presence of the double bond in the central seven-carbon chain enhances curcumin-dependent anti-IAV activity and interferes with IAV entry through interaction of curcumin with the receptor-binding region of the viral hemagglutinin protein.
In this study, we reported the inhibitory effects of the curcumin derivatives on IAV neuraminidase. The addition of curcumin derivative after virus adsorption showed anti-influenza activity in MDCK cells. Therefore, we hypothesized that the curcumin derivative inhibits the activity of NA, thereby preventing the release of virions from infected cells. Indeed, the inhibitory effect of curcumin derivatives on IAV replication was not the same as the inhibitory effect of NA expression. It can be seen that curcumin showed the best inhibitory effect on viral replication, followed by demethylcurcumin, tetramethylcurcumin, and bisdemethoxycurcumin, and dihydrocurcumin showed a weak inhibitory effect, which is consistent with the results of Ou et al. (2013). However, tetramethylcurcumin and bisdemethoxycurcumin showed a similar effect on inhibition of NA, followed by curcumin, dihydrocurcumin and demethylcurcumin. These results indicated that curcumin derivatives may inhibit IAV through inhibiting many other proteins or signaling pathways, and not only neuraminidase protein. In addition, curcumin derivatives had different inhibitory effects on IAV neuraminidase protein, which was relative to their structures and binding models.
Finally, we investigated the influence of curcumin derivatives on the reduction of viral NP. Two curcumin derivatives (tetramethylcurcumin and curcumin) were shown to decrease expression of NP induced by IAV infection.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (No 81470186). We thank International Science Editing (https://www.internationalscienceediting.com) for editing this manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Anand P, Kunnumakkara AB, Newman RA, Aggarwal BB. Bioavailability of curcumin: problems and promises. Mol Pharm. 2007;4(6):807–818. doi: 10.1021/mp700113r. [DOI] [PubMed] [Google Scholar]
- Belser JA, Maines TR, Creager HM, Katz JM, Tumpey TM. Oseltamivir inhibits influenza virus replication and transmission following ocular-only aerosol inoculation of ferrets. Virology. 2015;484:305–312. doi: 10.1016/j.virol.2015.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LL, Dou J, Su ZZ, Zhou HM, Wang H, Zhou WD, Guo QL, Zhou CL. Synergistic activity of baicalein with ribavirin against influenza A (H1N1) virus infections in cell culture and in mice. Antivir Res. 2011;91(3):314–320. doi: 10.1016/j.antiviral.2011.07.008. [DOI] [PubMed] [Google Scholar]
- Chen TY, Chen DY, Wen HW, Ou JL, Chiou SS, Chen JM, Wong ML, Hsu WL. Inhibition of enveloped viruses infectivity by curcumin. PLoS ONE. 2013;8(5):e62482. doi: 10.1371/journal.pone.0062482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HG, Choi JH, Lee HK, Mun SK, Lee JB, Jho EH, Kang C, Lin YH. Oseltamivir-resistant influenza viruses isolated in South Korea from 2005 to 2010. Arch Virol. 2013;158(11):2365–2370. doi: 10.1007/s00705-013-1734-3. [DOI] [PubMed] [Google Scholar]
- Dai JP, Gu LM, Su Y, Wang QW, Zhao Y, Chen XX, Deng HX, Li WZ, Wang GF, Li KS. Inhibition of curcumin on influenza A virus infection and influenzal pneumonia via oxidative stress, TLR2/4, p38/JNK MAPK and NF-κB pathways. Int Immunopharmacol. 2018;54:177–187. doi: 10.1016/j.intimp.2017.11.009. [DOI] [PubMed] [Google Scholar]
- Duarte Y, Márquez-Miranda V, Miossec MJ. Integration of target discovery, drug discovery and drug delivery: a review on computational strategies. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2019;11(4):e1554. doi: 10.1002/wnan.1554. [DOI] [PubMed] [Google Scholar]
- Fiers W, De Filette M, Birkett A, Neirynck S, Min Jou W. A “universal” human influenza A vaccine. Virus Res. 2004;103(1–2):173–176. doi: 10.1016/j.virusres.2004.02.030. [DOI] [PubMed] [Google Scholar]
- Fang JS, Huang DN, Zhao WX, Ge H, Luo HB, Xu J. A new protocol for predicting novel GSK-3beta ATP competitive inhibitors. J Chem Inf Model. 2011;51(6):1431–1438. doi: 10.1021/ci2001154. [DOI] [PubMed] [Google Scholar]
- Golbraikh A, Tropsha A. Beware of q2! J Mol Graph Model. 2002;20(4):269–276. doi: 10.1016/S1093-3263(01)00123-1. [DOI] [PubMed] [Google Scholar]
- Goel A, Kunnumakkara AB, Aggarwal BB. Curcumin as “Curecumin”: from kitchen to clinic. Biochem Pharmacol. 2007;75(4):787–809. doi: 10.1016/j.bcp.2007.08.016. [DOI] [PubMed] [Google Scholar]
- Gao L, Zu M, Wu S, Liu AL, Du GH. 3D QSAR and docking study of flavone derivatives as potent inhibitors of influenza H1N1 virus neuraminidase. Bioorg Med Chem Lett. 2011;21(19):5964–5970. doi: 10.1016/j.bmcl.2011.07.071. [DOI] [PubMed] [Google Scholar]
- Golubović M, Lazarević M, Zlatanović D, Golubović M, Lazarević M, Zlatanović D, Krtinić D, Stoičkov V, Mladenović B, Milić DJ, Sokolović D, Veselinović AM. The anesthetic action of some polyhalogenated ethers-Monte Carlo method based QSAR study. Comput Biol Chem. 2018;75:32–38. doi: 10.1016/j.compbiolchem.2018.04.009. [DOI] [PubMed] [Google Scholar]
- Hurt AC, Holien JK, Parker M, Kelso A, Barr IG. Zanamivir-resistant influenza viruses with a novel neuraminidase mutation. J Virol. 2009;83(20):10366–10373. doi: 10.1128/JVI.01200-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan S, Kolandaivel P. Antiviral potential of natural compounds against influenza virus hemagglutinin. Comput Biol Chem. 2017;71:207–218. doi: 10.1016/j.compbiolchem.2017.11.001. [DOI] [PubMed] [Google Scholar]
- Li YC, Peng SZ, Chen HM, Zhang FX, Xu PP, Xie JH, He JJ, Chen JN, Lai XP, Su ZR. Oral administration of patchouli alcohol isolated from Pogostemonis Herba augments protection against influenza viral infection in mice. Int Immunopharmacol. 2012;12(1):294–301. doi: 10.1016/j.intimp.2011.12.007. [DOI] [PubMed] [Google Scholar]
- Li Q, Qi JX, Wu Y, Kiyota H, Tanaka K, Suhara Y, Ohrui H, Suzuki Y, Vavricka CJ, Gao GF. Functional and structural analysis of influenza virus neuraminidase N3 offers further insight into the mechanisms of oseltamivir resistance. J Virol. 2013;87(18):10016–10024. doi: 10.1128/JVI.01129-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheshwari RK, Singh AK, Gaddipati J, Srimal RC. Multiple biological activities of curcumin: a short review. Life Sci. 2006;78(18):2081–2087. doi: 10.1016/j.lfs.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Mazur I, Wurzer WJ, Ehrhardt C, Pleschka S, Puthavathana P, Silberzahn T, Wolff T, Planz O, Ludwig S. Acetylsalicylic acid (ASA) blocks influenza virus propagation via its NF-kappaB-inhibiting activity. Cell Microbiol. 2007;9(7):1683–1694. doi: 10.1111/j.1462-5822.2007.00902.x. [DOI] [PubMed] [Google Scholar]
- Moscona A. Global transmission of oseltamivir-resistant influenza. N Engl J Med. 2009;360(10):953–956. doi: 10.1056/NEJMp0900648. [DOI] [PubMed] [Google Scholar]
- Nayak DP, Jabbar MA. Structural domains and organizational conformation involved in the sorting and transport of influenza virus transmembrane proteins. Annu Rev Microbiol. 1989;43:465–501. doi: 10.1146/annurev.mi.43.100189.002341. [DOI] [PubMed] [Google Scholar]
- Nilakantan R, Bauman N, Venkataraghavan R. New method for rapid characterization of molecular shapes: applications in drug design. J Chem Inf Comput Sci. 1993;33(1):79–85. doi: 10.1021/ci00011a012. [DOI] [PubMed] [Google Scholar]
- Ohuchi M, Asaoka N, Sakai T, Ohuchi R. Roles of neuraminidase in the initial stage of influenza virus infection. Microbes Infect. 2006;8(5):1287–1293. doi: 10.1016/j.micinf.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Ou JL, Mizushina Y, Wang SY, Chuang DY, Nadar M, Hsu WL. Structure–activity relationship analysis of curcumin analogues on anti-influenza virus activity. FEBS J. 2013;280(22):5829–5840. doi: 10.1111/febs.12503. [DOI] [PubMed] [Google Scholar]
- Patel PD, Patel MR, Kaushik-Basu N, Talele TT. 3D QSAR and molecular docking studies of benzimidazole derivatives as hepatitis C virus NS5B polymerase inhibitors. J Chem Inf Model. 2008;48(1):42–55. doi: 10.1021/ci700266z. [DOI] [PubMed] [Google Scholar]
- Pica N, Palese P. Toward a universal influenza virus vaccine: prospects and challenges. Annu Rev Med. 2013;64:189–202. doi: 10.1146/annurev-med-120611-145115. [DOI] [PubMed] [Google Scholar]
- Richart SM, Li YL, Mizushina Y, Chang YY, Chung TY, Chen GH, Tzen JT, Shia KS, Hsu WL. Synergic effect of curcumin and its structural analogue (monoacetylcurcumin) on anti-influenza virus infection. J Food Drug Anal. 2018;26(3):1015–1023. doi: 10.1016/j.jfda.2017.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Saito R, Masuda H, Oshitani H, Sato M, Sato I. Emergence of amantadine-resistant influenza A viruses: epidemiological study. J Infect Chemother. 2003;9(3):195–200. doi: 10.1007/s10156-003-0262-6. [DOI] [PubMed] [Google Scholar]
- Scalera NM, Mossad SB. The first pandemic of the 21st century: a review of the 2009 pandemic variant influenza A (H1N1) virus. Postgrad Med. 2009;121(5):43–47. doi: 10.3810/pgm.2009.09.2051. [DOI] [PubMed] [Google Scholar]
- Smith GJ, Vijaykrishna D, Bahl J, Lycett SJ, Worobey M, Pybus OG, Ma SK, Cheung CL, Raghwani J, Bhatt S, Peiris JS, Guan Y, Rambaut A. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature. 2009;459(7250):1122–1125. doi: 10.1038/nature08182. [DOI] [PubMed] [Google Scholar]
- Taubenberger JK, Morens DM. The pathology of influenza virus infections. Annu Rev Pathol. 2008;3(1):499–522. doi: 10.1146/annurev.pathmechdis.3.121806.154316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu HR, Fu L, Zhan P, Liu XY. 3D-QSAR analysis of a series of S-DABO derivatives as anti-HIV agents by CoMFA and CoMSIA. SAR QSAR Environ Res. 2016;27(12):999–1014. doi: 10.1080/1062936X.2016.1233580. [DOI] [PubMed] [Google Scholar]
- Zhao X, Jie YL, Rosenberg MR, Wan JT, Zeng SG, Cui W, Xiao YP, Li ZY, Tu ZC, Casarotto MG, Hu WH. Design and synthesis of pinanamine derivatives as anti-influenza A M2 ion channel inhibitors. Antiviral Res. 2012;96(2):91–99. doi: 10.1016/j.antiviral.2012.09.001. [DOI] [PubMed] [Google Scholar]