Summary
The open reading frame (ORF) 7a of the SARS-associated coronavirus (SARS-CoV) encodes a unique type I transmembrane protein of unknown function. We have determined the 1.8 Å resolution crystal structure of the N-terminal ectodomain of orf7a, revealing a compact seven-stranded β sandwich unexpectedly similar in fold and topology to members of the Ig superfamily. We also demonstrate that, in SARS-CoV- infected cells, the orf7a protein is expressed and retained intracellularly. Confocal microscopy studies using orf7a and orf7a/CD4 chimeras implicate the short cytoplasmic tail and transmembrane domain in trafficking of the protein within the endoplasmic reticulum and Golgi network. Taken together, our findings provide a structural and cellular framework in which to explore the role of orf7a in SARS-CoV pathogenesis.
Introduction
Severe acute respiratory syndrome (SARS) is an atypical pneumonia displaying unusually high rates of morbidity and mortality (Stadler et al., 2003). The illness is a direct result of infection by a coronavirus (SARS-CoV) that was first identified in March of 2003. SARS-CoV is sufficiently divergent from all previously identified coronaviruses that it may represent a distinct lineage (Marra et al., 2003, Rota et al., 2003). Current evidence suggests the virus emerged from nonhuman sources (Guan et al., 2003, Yu et al., 2003), possibly as a recombination event between mammalian-like and avian-like parent viruses (Rest and Mindell, 2003, Stanhope et al., 2004, Stavrinides and Guttman, 2004).
The genomic sequences of numerous SARS-CoV isolates have been determined (http://www.ncbi.nlm.nih. gov/genomes/SARS/SARS.html). The principal “conserved” open reading frames occur in the same order and are of similar size as those found in other coronaviruses. These include, from 5′ to 3′, genes for the replicase (rep), spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins. In addition to the conserved genes, six or more novel open reading frames are predicted at the 3′ end of the SARS-CoV genome (ORFs 3a, 3b, 7a, 7b, 8ab, and 9b) (Snijder et al., 2003). So far, the functions of these genes remain unknown. Their absence from other genomes suggests that they might carry out unique functions in SARS-CoV replication, assembly, or virulence. Similarly positioned “accessory genes” have proven dispensable for coronavirus viability in vitro, although their deletion often leads to viral attenuation in vivo (de Haan et al., 2002). These genes are therefore particularly interesting, considering that nonessential accessory genes from a wide array of viruses function to circumvent host innate and adaptive immune responses (Alcami and Koszinowski, 2000, Ploegh, 1998).
In this study, we examine the product of the SARS-CoV accessory gene ORF 7a (Snijder et al., 2003) (also known as ORF 8 or X4) (Marra et al., 2003, Rota et al., 2003). Sequence analysis predicts that ORF 7a encodes a type I transmembrane protein, 122 amino acids in length, consisting of a 15 residue N-terminal signal peptide, an 81 residue luminal domain, a 21 residue transmembrane segment, and a 5 residue cytoplasmic tail. Although the orf7a sequence has been identified in all isolates of SARS-CoV collected from both human and animal sources, it appears to be unique to SARS, displaying no significant similarity to any other viral or nonviral protein. Here, we examine the orf7a accessory protein in an attempt to clarify its biological significance and evaluate its potential as a therapeutic target. Using an E. coli expression system, we have successfully produced the luminal domain of orf7a as soluble protein by oxidative refolding. We have determined the crystal structure of orf7a to 1.8 Å resolution, revealing a compact Ig-like domain. In addition, monoclonal antibodies specific for both the native and denatured forms of orf7a have been produced, allowing for the analysis of orf7a expression in SARS-CoV-infected cells. Further, we examined orf7a cellular trafficking by immunofluorescence microscopy, revealing predominant in tracellular retention within the Golgi network that is mediated by the transmembrane and short cytoplasmic tail of the protein.
Results
Production of Soluble Refolded Orf7a Protein
We initiated our studies of orf7a by cloning a cDNA fragment encoding the mature N-terminal ectodomain into a bacterial expression vector. Based on the presence of four cysteine residues and a predicted secretory signal peptide, we expected that two disulfide bonds would be required for proper folding of the orf7a ectodomain. Indeed, the bacterially expressed recombinant protein proved insoluble. It was therefore re cov ered from inclusion bodies, denatured in guanidine hydrochloride, and then oxidatively refolded by rapid dilution. The resulting soluble protein was purified on size exclusion chromatography eluting at ∼9 kDa, the correct calibrated molecular weight expected for the compactly folded monomer. We verified the identity of the orf7a ectodomain fragment by electrospray mass spectrometry (see Experimental Procedures). The observed mass is consistent with two disulfide bonds in the refolded molecule. The protein runs as a single spe cies on native PAGE and is stable in solution at 10 mg/ml over a period of several weeks.
Structure of the Orf7a Luminal Domain
We next initiated a structural-genomics-type examination of orf7a. We hoped to gain insight into the potential function of orf7a by investigating the structural relationships between orf7a and other well-characterized proteins. Crystallization screening of the refolded orf7a protein yielded diffraction-quality hexagonal crystals belonging to space group P31 (a = b = 37.10 Å, c = 55.33 Å), which grew over a 3 day period in hanging drops to an approximate size of 0.2 × 0.1 × 0.1 mm. Initial phasing was accomplished by multiple isomor phous replacement (MIR) using data collected from three different Pt heavy atom derivatives (Table 1). The resulting electron density maps were readily interpretable (Figure 1A). An initial model, spanning residues 1–67 without a main chain break, was obtained directly from experimental phase using the autobuild feature of ARP/wARP (Morris et al., 2003). Further refinement was carried out in CNS with only minor model building required in O (Jones et al., 1991). The final model has excellent geometry, with 94.8% of all residues residing in the most favored region of the Ramachandran plot and the remaining 5.2% in the additionally allowed region as defined by PROCHECK (Laskowski et al., 1993). There are no residues in the disallowed or generously allowed regions. The final model has an Rwork of 22.3% to 1.8 Å resolution, with an Rfree of 27.5%.
Table 1.
Summary of Data Collection, Phasing, and Refinement for SARS Orf7a
| Data Collectiona | ||||
|---|---|---|---|---|
| Space group and unit cell (Å) | P31 a = b = 37.10 Å, c = 55.33 Å | |||
| Data set | native | K2Pt(CN)4 | K2Pt(NO2)4 | K2PtCl4 |
| Wavelength (Å) | 0.90 | 1.5418 | 0.90 | 0.90 |
| X-ray source | APS 14BMb | Rigaku | APS 14BM | APS 14BM |
| Resolution (Å) (outer shell) | 20–1.8 (1.88–1.8) | 20–2.0 (2.09–2.0) | 20–2.0 (2.09–2.0) | 20–2.0 (2.09–2.0) |
| Observations/unique | 55,822/7,744 | 41,928/5,770 | 25,115/5,656 | 101,513/5,759 |
| Completeness (%) | 98.7 (100) | 99.3 (98.4) | 99.2 (100) | 99.1 (98.7) |
| Rsym (%) | 4.7 (29.7) | 7.3 (74.5) | 2.9 (14.8) | 5.1 (32.1) |
| I/σ | 34.9 (5.2) | 25.5 (1.9) | 47.7 (10.3) | 25.0 (4.0) |
| MIR Phasing Statisticsc | ||||
|---|---|---|---|---|
| Heavy atom sites | 1 | 1 | 2 | |
| Rcullis isomorphous/anomalous | 0.77/0.98 | 0.85/0.95 | 0.86/0.99 | |
| Phasing power isomorphous/anomalous | 0.98/0.35 | 0.58/0.55 | 0.55/0.28 | |
| Figure of merit | 0.38 | |||
| Refinement Summaryd | ||||
|---|---|---|---|---|
| Resolution (Å) | 20–1.8 (1.88–1.8) | |||
| Reflections Rwork/Rfree | 7,741 (422) | |||
| No. protein atoms/solvent | 534/142 | |||
| Rwork overall (outer shell) (%) | 22.3 (30.1) | |||
| Rfree overall (outer shell) (%) | 27.5 (31.8) | |||
| Rmsd bond length (Å)/angles (°) | 0.005/1.3 | |||
| Rmsd dihedral/improper (°) | 25.8/0.69 | |||
| Ramachandran plot | ||||
| Most favored/additional (%) | 94.8/5.2 | |||
| Est. coordinate error (Å) | 0.24 | |||
Values as defined in SCALEPACK (Otwinowski and Minor, 1997).
Advanced Photon Source, Beamline 14BM.
Values as defined in SHARP (Morris et al., 2003).
Values as defined in CNS (Brunger et al., 1998).
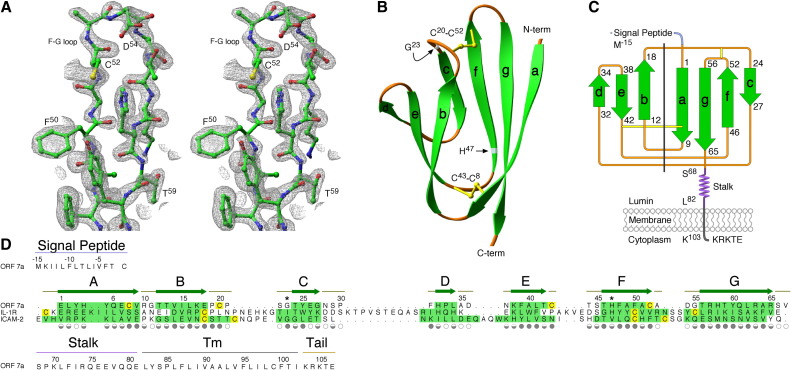
Figure 1.
Three-Dimensional Structure of the Orf7a Luminal Domain
(A) Stereoview of the 2Fo − Fc electron density composite omit map shown as gray mesh with orf7a residues depicted as a ball-and-stick model. The view is of the F-G loop drawn at a contour level of 2σ.
(B) Ribbon trace of the orf7a luminal domain showing the two sheets of the Ig-like β sandwich. The disulfides are labeled by their Cys positions. Two reported polymorphisms (Gly23 and His47) are indicated in silver.
(C) Topological diagram of the orf7a fold. A dashed line separates the BED and AGFC sheets. The β strands are labeled A–G and displayed in green.
(D) Structure-based alignment of orf7a with the N-terminal domains of IL-1R and ICAM-2. The β strands in each are highlighted in green, and Cys residues are in yellow. Symbols indicating the solvent accessibility of the orf7a side chains are shown below each position. Filled circles represent greater than 60% solvent inaccessible, half-filled between 30% and 60% inaccessible, and empty circles less than 30% inaccessible. Regions within the orf7a sequence are labeled: signal peptide, stalk, transmembrane (Tm), and cytoplasmic tail. Positions of polymorphism are indicated with asterisks.
Our model for the orf7a luminal domain consists of seven β strands which form two β sheets, compactly arranged in an Ig-like β sandwich fold (Figure 1B). However, the precise topology of orf7a is distinctive from that of typical Ig-superfamily members (Figure 1C). Strand A, instead of running antiparallel to strand B, has switched sheets and lies parallel to strand G. Like many C1-type Ig domains, both the C′ and C″ strands are absent. In addition, the C and D strands are both very short: the C strand is only 4 amino acids in length, while the D strand is just 3 residues long. The structure of orf7a also contains two unusual disulfide bonds connecting the BED and AGFC sheets. Neither occurs in the typical position occupied by the Ig-superfamily canonical disulfide (i.e., connecting the B and F strands). The first disulfide of orf7a connects the end of strand A to the E-F loop, while the second disulfide connects the short B-C and F-G loops.
We sought to identify proteins of similar topology by performing a Dali search (Holm and Sander, 1995). The two most similar structures identified were the N-terminal domain of the human intercellular adhesion molecule-2 (ICAM-2) (Protein Data Bank [PDB] ID 1ZXQ, fragment 1–85, rmsd of 2.3 Å for 55 aligned residues, with 6% sequence identity) and the N-terminal domain of the human interleukin-1 receptor (IL-1R) (PDB ID 1IRA, fragment 1–94, rmsd of 2.0 Å for 46 aligned residues, with 11% sequence identity). Both structures are considered examples of I-set Ig domains as defined by SCOP (Murzin et al., 1995). Comparisons of orf7a with 55 additional members of the Ig superfamily identified by Dali revealed sequence identities ranging from 2% to 16% for aligned core residues, consistent with our failure to predict an Ig-like fold from the primary sequence alone. Also, compared with most Ig folds, the orf7a luminal domain is extremely small, consisting of only 65 amino acids. For comparison, the average length of an I-set domain in HOMSTRAD (Mizuguchi et al., 1998) is 98 amino acids. In addition to the short C and D strands, the membrane-distal B-C and F-G loops are also comparatively short. These differences are best seen in the structure-based alignment of the I-set domains with orf7a (Figure 1D).
Localization of Orf7a in SARS-CoV-Infected Cells
In order to characterize the expression and intracellular trafficking of orf7a, we generated monoclonal antibodies (mAbs) specific for the luminal domain. Recombinant soluble-refolded orf7a protein was used to immunize mice. Solid-phase ELISA identified 24 hybridomas producing mAb specific for the refolded protein. These were characterized in cell staining, Western blotting, and immunoprecipitation assays (Table 2A). To prove that the IgG2b isotype clone 2E11 was specific for native orf7a, immunoprecipitation studies were performed using lysate from SARS-CoV-infected cells. Vero cells were infected with SARS-CoV at a multiplicity of infection (moi) of 0.01 for 48 hr in accordance with the rules and regulations of Washington University and the “Laboratory Biosafety Guidelines for Handling and Processing Specimens Associated with SARS-CoV” as put forward by the Department of Health and Human Services Centers for Disease Control and Prevention (CDC). All manipulation of infectious samples took place in a BSL-3 biological safety facility. The infected cells were washed, solubilized in 1.0% Triton X-100, and immune complexes were recovered on protein A Sepharose. As expected, the 2E11 clone immunoprecipitated a single band of ∼12 kDa. This protein was identified as orf7a by N-terminal sequencing (Table 2B). An identical sequence was obtained using lysate from 293T cells transfected with an orf7a cDNA. In both cases, the predicted 15 amino acid signal peptide (MKIILFLTLIVFTSC) of orf7a had been removed, presumably by signal peptidase cleavage. These results and the inability of 2E11 to recognize denatured (boiled-reduced) recombinant orf7a protein on Western blot support the conclusion that 2E11 is specific for orf7a in its natively expressed form. The ability of the conformation-dependent 2E11 mAb (raised against recombinant, refolded orf7a) to immunoprecipitate orf7a protein from SARS-CoV-infected cells strongly suggests that the conformation of the refolded and natively expressed proteins is similar if not identical. Final confirmation of this awaits the development of assays for the as yet unknown function of orf7a.
Table 2.
Characterization of Monoclonal Antibodies and Orf7a N-Terminal Sequencing
| A. Representative Set of Anti-Orf7a Hybridomas | ||||||
|---|---|---|---|---|---|---|
| Hybridoma | Isotype | Permeabilized Cell Staininga | Boiled Reducedb | Nonboiled Nonreducedb | IP Triton X-100c | |
| 2E11 | IgG2b | ++++ | — | ++++ | ++++ | |
| 1H4 | IgG1 | +++ | — | ++++ | +++ | |
| 3B1 | IgG1 | ++ | — | ++ | + | |
| 1A2 | IgG1 | +++ | ++ | — | + | |
| 2F6 | IgG3 | + | ++++ | — | + | |
| 1D10 | IgG3 | ++ | ++++ | — | + | |
| B. N-Terminal Protein Sequencing of Orf7a Immunoprecipitated with the 2E11 Monoclonal Antibody | ||||||
|---|---|---|---|---|---|---|
| Source | Peptide Sequence | Positions | ||||
| SARS-CoV-infected Vero cellsd | ELYHYQE(X)fVRGTTV | 1–14 | ||||
| Orf7a-transfected 293T cellse | ELYHYQE(X)VRGTTV | 1–14 | ||||
Assessed by FACS using 1% saponin-treated cells.
Treatment of recombinant orf7a before Western blot.
Immunoprecipitation in 1% Triton X-100.
Cell lysates prepared from Vero cells at 48 hr postinfection.
Cell lysates prepared from 293T cells transfected with an orf7a cDNA at 72 hr.
The amino acid cysteine cannot be observed by this method, as indicated by (X).
We next examined the expression of ORF 7a in SARS-CoV-infected cells by confocal microscopy using the 2E11 antibody. Infected Vero cells were fixed and stained for either SARS-CoV S protein (Figures 2A and 2B) or orf7a (Figures 2C and 2D). While the S protein was readily detectable at the plasma membrane, little orf7a could be detected. Permeabilization of SARS-CoV-infected cells with saponin significantly increased the orf7a staining, demonstrating that the majority of orf7a remains intracellular (mainly in the perinuclear region; Figures 2E and 2F).
Figure 2.
Intracellular Retention of Orf7a
Vero cells were either mock infected ([A], [C], and [E]) or infected with SARS-CoV ([B], [D], and [F]) for 18 hr at an moi of 5. As expected, the spike protein was clearly present at the surface of the SARS-CoV-infected but not mock-infected cells ([A] and [B]). Incubation with the anti-orf7a monoclonal antibody 2E11 demonstrated only limited cell-surface expression of orf7a. An intracellular pool of orf7a was clearly evident after saponin permeabilization ([E] and [F]). The anti-SARS antibody is a mixture of hybridoma supernatants specific for SARS proteins and found to primarily recognize the S protein by Western blotting (a gift of Larry Anderson, CDC). Intracellular retention of orf7a does not require other viral proteins. When an orf7a cDNA is used to transiently transfect 293T cells, very little orf7a can be detected at the cell surface using 2E11 (G). Again, saponin pretreatment allowed staining of the intracellular orf7a (H). Addition of a C-terminal GFP tag did not significantly alter this intracellular distribution. A good colocalization was observed between orf7a staining (red) and GFP fluorescence (green) in this transfectant ([I] and [J]). The orf7a-GFP distribution was distinct from that seen for GFP alone (K).
Localization Studies of Orf7a in Cell Transfectants
Vero cells transfected with an ORF 7a cDNA showed a similar pattern; intact cells displayed little surface orf7a staining, while permeabilized cells displayed intense int racellular staining (Figures 2G and 2H). From this result, we conclude that orf7a does not require the expression of other viral proteins for its intracellular trafficking and retention. To facilitate analysis of the intracellular distribution, a green fluorescence protein (GFP) tag was fused in-frame to the carboxyl terminus. The fluorescent pattern was clearly distinct from GFP alone (Figure 2K). The fusion tag also did not alter the observed intracellular distribution of orf7a; the majority of the orf7a-GFP was still retained in a perinuclear location (Figures 2I and 2J).
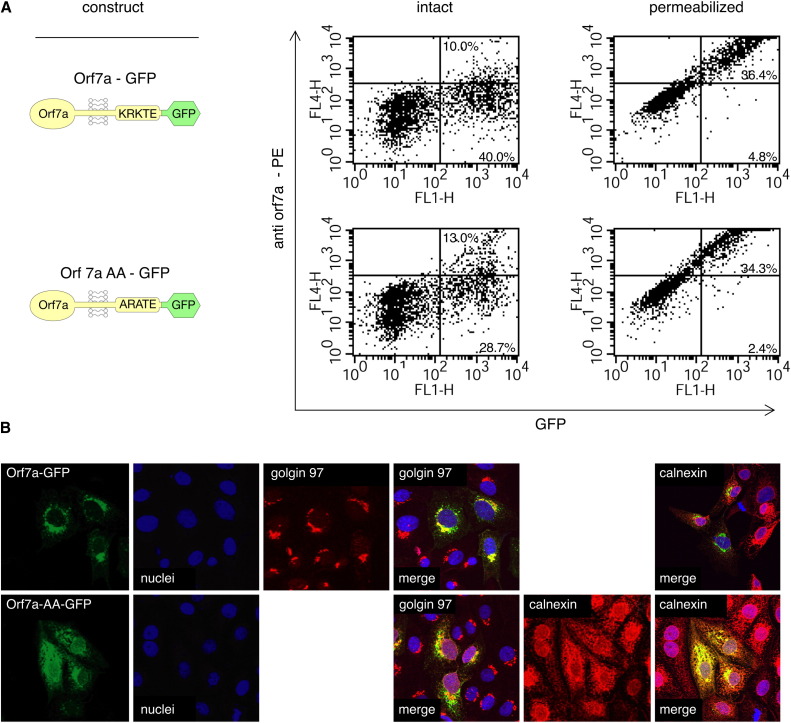
We confirmed the low level of cell-surface expression of orf7a by flow cytometry using 293T cells transfected with an orf7a-GFP cDNA. Intact cells displayed little orf7a-positive/GFP-positive staining, whereas saponin-permeabilized cells showed a significant increase in the double-positive population (Figure 3A, upper panels). Next, permeabilized Vero cell transfectants were stained using an antibody to Golgin 97, a protein known to localize in the trans-Golgi network (Griffith et al., 1997). The merged composite images (Figure 3B, top row) show a strong colocalization between orf7a-GFP and Golgin 97 staining. A much weaker association was seen with the ER marker calnexin (Galvin et al., 1992). Although differences in the fixation procedures and antibodies used make a direct comparison difficult, these results are in general agreement with that of Fielding et al., who resolved perinuclear localization of orf7a with the subcellular markers GRP94 and Sec 31 (Fielding et al., 2004), collectively placing orf7a in the ER and Golgi compartments.
Figure 3.
Intracellular Localization of Orf7a
(A) Flow cytometry analysis of protein expression levels on 293T cell transfectants. Direct comparison of orf7a-GFP with its cytoplasmic tail mutant, orf7aAA-GFP, in which Lys103 and Lys105 were replaced by Ala. The extent of transfection is revealed by GFP fluorescence. Both intact and saponin-permeabilized cells were stained with the anti-orf7a mAb 2E11. Significantly more anti-orf7a mAb staining was seen in the permeabilized cells, suggesting that the majority of the protein was intracellular.
(B) The same cDNA constructs were introduced into Vero cells and examined by confocal microscopy. Orf7a-GFP colocalizes best with the Golgi marker Golgin 97 (see Golgin 97 merge in upper row), while orf7a-AA-GFP colocalizes best with the ER resident protein calnexin (see calnexin merge in lower row). In all cases, the nuclei were counterstained blue with Topro, and the GFP fluorescence appears green. Optical slices were reconstructed into a three-dimensional image to show colocalization before compression into a two-dimensional representation.
Localization Studies of Orf7a Variants and Orf7a/CD4 Chimeras
The short orf7a cytoplasmic tail contains three positively charged residues proximal to the membrane (Lys103, Arg104, and Lys105 in Figure 1D). This triplet sequence ([Arg/Lys][X][Arg/Lys]) has been found in several Golgi resident proteins and appears to be required for recognition by the COPII vesicular system implicated in the transport of proteins from ER to Golgi compartments (Giraudo and Maccioni, 2003; Bickford et al., 2004). To examine the role of the orf7a cytoplasmic tail, we constructed a mutant orf7a-GFP fusion in which the two Lys tail residues were changed to Ala. The orf7a-AA-GFP tail mutant, like the wild-type, displayed only a low level of expression at the plasma membrane of transfected cells (Figure 3A, lower panels). The majority of the mutant protein colocalized with antibodies directed against the ER resident protein calnexin (Figure 3B, bottom row).
The extremely short length of the orf7a cytoplasmic tail (five amino acids) suggested that additional targeting information might exist elsewhere in the protein. Not surprisingly, transfer of the orf7a cytoplasmic tail was insufficient to confer intracellular retention on the cell-surface protein CD4 (Figure 4A, compare the first two panels). In contrast, transfer of both the orf7a transmembrane domain and cytoplasmic tail (26 amino acids) resulted in an increase in the amount of CD4 marker protein retained in the Golgi, as seen by comparing the colocalization of this chimeric construct with Golgin 97 (Figure 4B). However, localization of the CD4/orf7a TM tail construct was not as tight as that seen for orf7a alone, suggesting that residues in the orf7a stalk or luminal domain may also contribute to orf7a intracellular targeting. This situation, where residues outside the transmembrane domain help to fine-tune localization within the Golgi, has been observed for other membrane bound Golgi-resident proteins (Burke et al., 1994). Still, the dramatic difference between the CD4/orf7a TM tail and the CD4-GFP localizations indicates that the targeting signal exists primarily within the transmembrane and cytoplasmic tail.
Figure 4.
The Intracellular Retention Signal of Orf7a
(A) The majority of the CD4-GFP control protein is seen at the plasma membrane by FACS analysis using intact 293T cells (left). This distribution did not change when the cytoplasmic tail of CD4 was replaced with that of orf7a (middle). In contrast, significantly more protein was retained inside the cell when the entire CD4 transmembrane domain and tail were replaced by the orf7a transmembrane domain and tail (right).
(B) Vero cells expressing the CD4-fusion constructs were permeabilized 24 hr posttransfection and stained for Golgin 97 (red). Confocal microscopy was used to generate a three-dimensional reconstruction of the cells. Only the merged (Golgin 97 + GFP) images are shown (lower panels). In all three images, total fluorescence was normalized to the area of the cell displaying the most intense staining (cell surface for CD4 and CD4-tail, Golgi for CD4-Tm tail).
Discussion
Our structural studies of the orf7a luminal domain have established that it adopts an extremely compact Ig-like β sandwich fold topology, despite an absence of significant sequence similarity to other members of the Ig superfamily. This common structural fold occurs in a wide variety of proteins, where it performs a diverse set of functions. For example, the fold is found in proteins of the extracellular matrix, muscle proteins, proteins of the immune system, cell-surface receptors, enzymes, transcription factors, and a wide variety of viral proteins (Clarke et al., 1999). An automated comparison of the luminal domain against 189 active site templates revealed no obvious enzymatic sites (Watson et al., 2003). Further, comparison of the structure with Ig superfamily members did not reveal any obvious conserved functional regions. As a result, we find it difficult to draw conclusions about the function of orf7a from the structure alone. Still, some features are worth noting. Because of the unusual disulfide-bonding pattern, the two β sheets bow away from each other in the middle. This separation allows the formation of a deep hydrophobic pocket near the middle of the A strand. In most Ig folds, the B strand hydrogen bonds with the A strand. In orf7a, the peptide backbone of the B strand is free and passes close to the deep hydrophobic A pocket (Figure 5B). Therefore, it may be possible for a peptide strand from another protein to hydrogen bond with the B strand while inserting a side chain into the A pocket. Similar examples of donor-strand binding have been observed in the assembly of bacterial Ig-superfamily chaperones (Sauer et al., 2002). Second, there exists a deep groove on the backside of the orf7a molecule, formed between the C-D and E-F loops. Interestingly, one of the few reported sites of polymorphism (His47-Asn47) occurs along this groove and may represent an adaptation to accommodate a binding partner or ligand.
Figure 5.
Surface Features of the Orf7a Luminal Domain
(A) Backbone worm shown in the same orientation as Figure 1B (right), and also the reverse orientation (left).
(B) The molecular surface is shown color coded by curvature to highlight local topography. The B strand and A pocket are indicated. Positions of known polymorphism are labeled. The deep groove that runs along the base of the molecule is indicated.
(C) Electrostatic potential mapped onto the accessible molecular surface; blue denotes a net positive charge (+4 kT), and red denotes a negative (−4 kT). The membrane distal face (top) is primarily acidic.
Our fluorescence localization studies suggest that the intracellular retention of orf7a requires both the transmembrane and cytosolic tail regions. Transfer of the orf7a cytoplasmic tail alone onto CD4 (CD4/orf7a-tail-GFP fusion) was unable to prevent surface expression of the marker protein. We also found that residues Lys103 and Lys105 of the orf7a cytoplasmic tail are required for efficient exit from the ER. Several interpretations exist for the higher steady-state ER retention of the Lys minus mutant. First, the charged Lys residues could serve as a stop-transfer sequence for translocation of the transmembrane domain. Their substitution to smaller nonpolar Ala residues may disrupt the conformation of the transmembrane domain or shift its position within the lipid bilayer enough to destroy recognition of an export signal. At this time, it is unclear what causes cargo to collect in nascent COPII vesicles (LaPointe et al., 2004); therefore, it remains possible that the Lys-Arg-Lys sequence is recognized directly. Regardless, our data indicate that the charged Lys residues are important for exit of orf7a from the ER.
How does the trafficking signal in orf7a compare with other viral ER/Golgi trafficking signals? It is well established that the coronavirus E molecule is held in the Golgi by a signal located in its cytoplasmic tail (Corse and Machamer, 2002). Little primary sequence homology exists between the E proteins of the three coronavirus groups. Therefore, the exact trafficking determinant being recognized remains unclear. Still, because the E signal occurs outside the transmembrane region, it is most likely different from the orf7a signal. A second type of Golgi targeting signal occurs in the first transmembrane domain of the coronavirus M protein (Klum per man et al., 1994). Specifically, four polar residues that line up on one face of a predicted α helix appear to be critical for the retention of M in the Golgi complex (Machamer et al., 1993). Here, the lack of polar transmembrane residues and different membrane topology suggest that the orf7a signal operates differently. Several single-stranded RNA viruses bud into the Golgi. For many of these, the principle glycoprotein component of the envelope consists of a heterodimeric complex of two single-pass transmembrane proteins. In almost every case, the Golgi targeting signal maps to the transmembrane domain and adjacent carboxy-terminal residues of one chain of the heterodimer. Examples of this type include the rubella virus envelope protein E2 (Hobman et al., 1995) and the envelope proteins from probably all members of the Bunyaviridae family (Bupp et al., 1996, Shi and Elliott, 2002). No detectable sequence homology exists among these regions, and it is likely that they are conformational in character, making it difficult to determine their relationship to the orf7a signal.
Why does orf7a contain an intracellular targeting signal? Coronaviruses acquire their membrane envelope by budding into the lumen of an ER to Golgi intermediate compartment (ERGIC) (Klumperman et al., 1994, Krijnse-Locker et al., 1994). Three or four viral proteins incorporate into this envelope, by far the most abundant being the M protein. In cells forced to express M by cDNA transfection, a small amount of E is sufficient to trigger the formation of virus-like particles (Bos et al., 1996, Vennema et al., 1996). It is still unclear what makes the M and E proteins gather in the ERGIC during infection (Lontok et al., 2004). When expressed individually, both move past the virus-assembly site (Corse and Machamer, 2002, Swift and Machamer, 1991). Two other coronavirus proteins integrate into the budding membrane: the S protein and, in the subset of coronaviruses that express it, the hemagglutinin–esterase (HE) protein. Both localize to the pre-Golgi by forming specific interactions with M (Nguyen and Hogue, 1998). If SARS-CoV buds into the ERGIC as other coronaviruses do, then our data indicate that orf7a traffics through the budding compartment. It is conceivable that orf7a may play a role in viral assembly or budding events unique to SARS-CoV. In support of this idea, Tan et al. have presented coimmunoprecipitation data suggesting an interaction between orf7a and the product of another SARS-CoV accessory gene, ORF3a, a protein which in turn interacts with the structural proteins M, E, and S (Tan et al., 2004). Alternatively, orf7a may itself be packaged into virions, possibly to serve as a secondary attachment protein in a manner analogous to HE. So far, studies aimed at identifying the structural proteins of the SARS-CoV virion have failed to detect orf7a (Krokhin et al., 2003, Ying et al., 2004), although it is worth noting that these methods also have failed to detect the virion-associated E protein. We tested whether our anti-orf7a monoclonal antibodies could block the production of virus or its cytopathic effects in SARS-CoV-infected Vero cells. However, no neutralization effect was observed (data not shown).
What other functions could orf7a serve within the context of the ER/Golgi network? A significant number of viral accessory proteins have been found to be critical for the evasion of host-mediated immunity, interfering with diverse processes, including apoptosis, complement activation, cytokine signaling, and innate and adaptive immune surveillance (Alcami and Koszinowski, 2000, Ploegh, 1998). While many of these proteins act at the plasma membrane or are secreted from infected cells, others have been found to operate within the secretory pathway, where they downregulate a variety of cell surface receptors, including signaling, co stimulatory, and adhesion molecules. For example, several viral accessory proteins have evolved to specifically prevent activation of NK or CD8 T cells by interfering with classical and nonclassical MHC function within ER and/or Golgi compartments (Orange et al., 2002). The murine cytomegalovirus m152 protein blocks surface expression of MHC class I as well as ligands of NKG2D by sequestering them in the ERGIC (Lodoen et al., 2003, Ziegler et al., 1997). The human cytomegalovirus US2 and US11 proteins, which are both like orf7a (type I transmembrane proteins that adopt Ig-like folds [Gewurz et al., 2001] ), catalyze the dislocation of MHC class I molecules from ER to cytosol; this dislocation results in its rapid degradation (Lilley and Ploegh, 2004, Ye et al., 2004 ). The bovine papillomavirus E5 protein retains MHC class I molecules in the Golgi and prevents their transport to the cell surface (Marchetti et al., 2002 ). RNA viruses can also en code MHC subversion proteins. For example, HIV-1 uses Nef to selectively downregulate HLA-A and HLA-B (Cohen et al., 1999 ). To see whether orf7a might have an analogous immunomodulatory function, we examined 293T cells transiently transfected with orf7a by flow cytometry. Our preliminary experiments indicated that orf7a expression does not result in significant loss of HLA-A expression (data not shown). The possible interference of orf7a with other immune surveillance mechanisms is currently under investigation.
To date, significant progress has been made in understanding those genes common to all coronaviruses encoding “essential” replication or structural functions. However, comparatively little is known about the coronavirus group-specific “accessory” genes. To help elucidate the significance of one of these, we have determined the structure and subcellular localization of SARS-CoV orf7a. Our results establish a structural and cellular framework for experiments directed at understanding the function of orf7a within the SARS-CoV life cycle. Toward this end, we are pursuing both biochemical and genetic approaches to identify potential orf7a molecular interactions, as these may prove suitable as targets for antiviral intervention.
Experimental Procedures
Protein Expression and Refolding
A fragment of ORF 7a cDNA encoding amino acids −2 through 81 was inserted between the NcoI and BamHI sites of the bacterial expression vector pET-21a. This plasmid was introduced into BL21(DE3)-RIL codon (+) E. coli cells for expression. Recombinant protein was recovered as an inclusion body pellet, denatured, reduced, and then renatured by rapid dilution in refolding buffer consisting of 1 M arginine, 100 mM Tris-HCl, 2 mM EDTA, 200 µM PMSF, 5 mM reduced glutathione, and 500 µM oxidized glutathione at a final pH of 8.3. After 24 hr, the soluble-refolded protein was collected over YM10 membrane in a stirred cell concentrator, passed through a 0.45 μm filter, and subjected to sizing on Superdex 200 using an AKTA-FPLC. The original cDNA clone differed by a single nucleotide from the published sequence (C→A at position 27,360 as numbered in NCBI Accession No. AY278741.1). All of our constructs maintain the resulting Leu15 to Ile15 replacement. The final recombinant fragment of orf7a spanned amino acid residues M(−3)SC/ELY···EEVQQE81* and contained no extraneous tags.
Crystal Growth and Preparation
Purified orf7a protein (8 mg/ml in 20 mM HEPES [pH 7.4], 20 mM NaCl) was mixed with an equal volume of reservoir solution containing 16% polypropylene glycol 400 and 100 mM NaOAc/HCl at pH 5.35, then left to equilibrate in hanging drops. Hexagonal crystals belonging to space group P31 (a = b = 37.10 Å, c = 55.33 Å) grew as triangular rods. These crystals were prepared for flash cooling at 100 K by transfer into reservoir solution containing 20% ethylene glycol for about 20 s. Heavy-atom Pt derivatives were used to determine the initial phasing by multiple isomorphous replacement (MIR). Fresh 10 mM stock solutions of potassium tetracyanoplatinate (II) [K2Pt(CN)4], potassium tetrachloroplatinate (II) (K2PtCl4), and potassium tetranitroplatinate (II) [K2Pt(NO2)4] were prepared in well solution. Hanging drops containing crystals were supplemented with one-tenth volume of heavy-atom stock solution and held at room temperature for various times.
Data Collection and Model Refinement
Single-wavelength diffraction data were collected on our home source and at the Advance Photon Source Beamline ID-14BM (Argonne National Laboratory) (Table 1). Data were integrated and scaled with DENZO, SCALEPACK, and HKL2000 (Otwinowski and Minor, 1997 ). Heavy-atom sites were located in CNS (Brunger et al., 1998 ) and refined with SOLVE (Terwilliger and Berendzen, 1999 ). A random set of reflections containing 5% of the data was excluded from the refinement for calculation of Rfree. The experimental electron density map was subjected to density modification with DM (Cowtan, 1994 ). The experimental phase maps are of exceptional quality at 2.2 Å. There is no electron density for the first 2 and the last 14 amino acid residues of the 84 encoded by the construct. Solvent accessibilities were calculated with NACCESS (probe radius 1.4 Å) (Hubbard et al., 1991 ). Molecular diagrams were drawn using the programs GRASP (Nicholls et al., 1993 ) and RIBBONS (Carson, 1991 ).
Antibody Generation
BALB/c mice were immunized intraperitoneally with 25 μg of recombinant orf7a protein in 0.2 ml of Ribi adjuvant (Corixa) and boosted twice at 21 day intervals using the same formulation. Test bleeds were used to select a single mouse for fusion. This mouse was boosted intravenously with 5 μg protein diluted in PBS 14 days after its last injection. Three days later, the animal was sacrificed and the spleen was removed. The murine nonsecreting myeloma cell line P3x63Ag8.653 was used as the partner in a standard PEG 1500 fusion. Hybridomas were seeded directly into 96-well plates containing peritoneal macrophages as feeder cells. After 10 days of selection in hypoxanthine, aminopterin, and thymidine solutions (HAT), supernatants were screened by ELISA for anti-orf7a mAbs using solid-phase orf7a protein. Positive hybridomas were expanded in Iscove’s modified Dulbecco’s medium containing HT, 20% FBS (low IgG fetal bovine serum, Hyclone), 3% hybridoma cloning growth factor (IGEN), 4 mM L-glutamine, and antibiotics, then cloned by limiting dilution.
Immunoprecipitations and N-Terminal Sequencing
Vero cells were infected with SARS-CoV (Urbani) at an moi of 0.01. At 48 hr postinfection, the cells were washed in 50 mM HEPES (pH 7.5), 1 mM EGTA, 1500 µM MgCl2, 150 mM NaCl, and 1× complete protease inhibitors (Roche), then suspended at 1 × 107 cells/ml in the same buffer. The cells were lysed with an equal volume of 2.0% Triton X-100 in 150 mM NaCl. After centrifugation, supernatants were precleared with protein A Sepharose. The anti-orf7a mAb 2E11 was added to 10 μg/ml and held 60 min at 4°C. Immune complexes were recovered on protein A Sepharose and washed in cold lysis buffer without protease inhibitors before being heated to 90°C in loading buffer (62.5 mM Tris-HCl [pH 6.0], 2% SDS, 5% 2-mercaptoethanol, 500 mM sucrose) for 5 min. After separation on SDS-PAGE, the proteins were transferred to polyvinylidene difluoride membrane. The blot was rinsed in distilled water and then methanol before soaking 2 min in 1.1% w/v Coomassie blue, 40% v/v methanol/water, and 1% acetic acid. Several changes of 50% methanol were required to destain the blot. The orf7a region was excised for N-terminal sequencing (Table 2B).
Orf7a and CD4 Fusion Constructs
ORF 7a was also cloned into the mammalian expression vector pEGFP-N1 (BD Biosciences) downstream of the CMV promoter and in-frame with the GFP fusion tag. NheI and BamHI restriction sites were engineered to flank the insert. The resulting orf7a/linker/GFP fusion gene encodes MAIILF···IKRKTE/DPPVAT/MVSKG···DELYK*. Similarly, a full-length cDNA encoding human CD4 (nucleotides 153–1529 in accession number NM_000616) was inserted in pEGFP-N1 in-frame with the GFP tag. The resulting CD4/linker/GFP fusion protein spanned MARGVP···CSPI/EDPPVAT/MVSKG···DELYK*. Two chimeric fusion genes were made from these constructs, a CD4/orf7a tail-GFP fusion having the amino acid sequence MARGVP···IFFCV/KRKTE/DPPVAT/MVSKG···DELYK* and a CD4/orf7a TM tail-GFP fusion having the sequence MARGVP···TWSTPVD/LYSPL···KRKTE/ DPPVAT/MVSKG···DELYK*. Orf7aAA-GFP was made by changing the orf7a cytoplasmic tail sequence from KRKTE to ARATE.
Immunofluorescence Localizations
Vero E6 (ATCC CRL-1586) and 293T/17 (CRL-11268) cells were cultured in DMEM supplemented with 10% heat-inactivated fetal bovine serum, 1 mM glutamine, and 100 U/ml penicillin/streptomycin. The cells were incubated in a 95% air, 5% CO2 humidified incubator at 37°C. Vero cells were plated onto glass cover slips in 3.5 cm diameter tissue culture dishes and incubated overnight at 37°C. The cells were transfected using the LT-1 (Mirrus) transfection reagent (1 μg plasmid DNA and 4 μl of transfection reagent per dish) as previously described (Pekosz and Lamb, 1999 ). Eighteen hours posttransfection, the cells were washed in PBS and fixed in 1% methanol-free formaldehyde for 10 min at room temperature. After extensive washing in PBS, the cells were incubated with antibody diluted in PBS containing 3% normal goat sera for 1 hr. The cells were washed three times with PBS and then incubated with a secondary antibody for 30 min as appropriate. The cover slips were mounted onto microscope slides using Prolong (Molecular Probes) and visualized on a Zeiss LSM 510 confocal microscope. Primary antibodies used were 2E11 (anti-orf7a, 1:100 dilution mouse mono clonal), anti Golgin 97 (Sigma, 1:500 dilution, mouse monoclonal), or anti-calnexin (Sigma, 1:500 dilution, rabbit polyclonal). Secondary antibodies include goat anti-mouse IgG or goat anti-rabbit IgG conjugated to Alexafluor 594 (Molecular Probes; 1:500 dilution). When appropriate, cells were permeabilized by the addition of 0.1% saponin to all buffers postfixation. Nuclei were counter stained with Topro included with the secondary antibody.
Fluorescence-Activated Cell Sorting
293T or Vero cells were plated overnight onto 3.5 cm dishes. The cells were transfected with the indicated plasmids (1 μg plasmid per dish, 2 or 4 μl of transfection reagent for 293T or Vero cells, respectively) and stained for flow cytometry as previously described (Pekosz and Lamb, 1999 ). Primary antibody 2E11 (dilution 1:100 of 2.4 mg/ml stock) was followed by goat anti-mouse IgG conjugated to Alexafluor 647 (Molecular Probes; 1:1000 dilution) or anti-human CD4 conjugated to Tri-color (Caltag; 1:500 dilution). The cells were analyzed on a FACSCalibur flow cytometer using CellQuest software.
Mass Spectrometry
The sequence of the orf7a luminal domain construct (MSCEL.... EVQQE*) predicts a molecular weight of 9412.55 Amu without the N-terminal Met (usually removed by endogenous amino-peptidase activity). To prepare a sample for electrospray mass spectrometry (ESMS), 10 μg of protein was brought to a volume of 100 μl in water and mixed with 100 μl of 20% trichloroacetic acid. This mixture was held on ice for 30 min. The sample was spun in a microfuge at 16,000 × g at 4°C for 20 min. The precipitate was washed with 300 μl of cold acetone and spun again at 16,000 × g at 4°C for 5 min. The protein pellet was air dried and resuspended in 20 μl of 60% acetonitrile with 0.1% formic acid for analysis. ESMS yielded the expected molecular weight but also a smaller fragment corresponding to a loss of nine amino acids from the stalk at the C terminus. The protein used for the crystallization trials contained a mixture of the full-length and truncated forms. The observed weights also indicate two disulfide bonds per monomer, with the extra cysteine capped by glutathione (briefly, 9713.81 Amu = 9412.55 fragment − 5.04 Amu for 5 Hs + 306.3 Amu for oxidized glutathione, and similarly 8573.59 Amu = 8272.33 Amu fragment − 5.04 Amu for 5 Hs + 306.3 Amu for oxidized glutathione).
Acknowledgments
We thank Drs. Stuart Kornfeld, Linton Traub, Kenneth Murphy, Marcel Fremont, and Skip Virgin for comments on the manuscript. We also thank Dr. Vincent Magrini of the Washington University School of Medicine Genome Sequencing Center for the ORF 7a cDNA clone 79A01. The National Institutes of Health Grants GM62414-04 (D.H.F.) and R21AI059328 (A.P., M.S.D., and D.H.F.) supported this work.
The atomic coordinates and structure factors (accession code 1XAK) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org). Orf7a appears as APC35380 in the target list of the Midwest Center for Structural Genomics (http://www.mcsg.anl.gov).
Published: January 11, 2005
References
- Alcami A., Koszinowski U.H. Viral mechanisms of immune evasion. Immunol. Today. 2000;21:447–455. doi: 10.1016/S0167-5699(00)01699-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickford L.C., Mossessova E., Goldberg J. A structural view of the COPII vesicle coat. Curr. Opin. Struct. Biol. 2004;14:147–153. doi: 10.1016/j.sbi.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Bos E.C., Luytjes W., van der Meulen H.V., Koerten H.K., Spaan W.J. The production of recombinant infectious DI-particles of a murine coronavirus in the absence of helper virus. Virology. 1996;218:52–60. doi: 10.1006/viro.1996.0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger A.T., Adams P.D., Clore G.M., DeLano W.L., Gros P., Grosse-Kunstleve R.W., Jiang J.S., Kuszewski J., Nilges M., Pannu N.S. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Bupp K., Stillmock K., Gonzalez-Scarano F. Analysis of the intracellular transport properties of recombinant La Crosse virus glycoproteins. Virology. 1996;220:485–490. doi: 10.1006/viro.1996.0336. [DOI] [PubMed] [Google Scholar]
- Burke J., Pettitt J.M., Humphris D., Gleeson P.A. Medial-Golgi retention of N-acetylglucosaminyltransferase I. Contribution from all domains of the enzyme. J. Biol. Chem. 1994;269:12049–12059. [PubMed] [Google Scholar]
- Carson M. Ribbons 2.0. J. Appl. Crystallogr. 1991;24:958–961. [Google Scholar]
- Clarke J., Cota E., Fowler S.B., Hamill S.J. Folding studies of immunoglobulin-like beta-sandwich proteins suggest that they share a common folding pathway. Struct. Fold. Des. 1999;7:1145–1153. doi: 10.1016/s0969-2126(99)80181-6. [DOI] [PubMed] [Google Scholar]
- Cohen G.B., Gandhi R.T., Davis D.M., Mandelboim O., Chen B.K., Strominger J.L., Baltimore D. The selective downregulation of class I major histocompatibility complex proteins by HIV-1 protects HIV-infected cells from NK cells. Immunity. 1999;10:661–671. doi: 10.1016/s1074-7613(00)80065-5. [DOI] [PubMed] [Google Scholar]
- Corse E., Machamer C.E. The cytoplasmic tail of infectious bronchitis virus E protein directs Golgi targeting. J. Virol. 2002;76:1273–1284. doi: 10.1128/JVI.76.3.1273-1284.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowtan K.D. DM: an automated procedure for phase improvement by density modification. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography. 1994;31:34–38. [Google Scholar]
- de Haan C.A., Masters P.S., Shen X., Weiss S., Rottier P.J. The group-specific murine coronavirus genes are not essential, but their deletion, by reverse genetics, is attenuating in the natural host. Virology. 2002;296:177–189. doi: 10.1006/viro.2002.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielding B.C., Tan Y.J., Shuo S., Tan T.H., Ooi E.E., Lim S.G., Hong W., Goh P.Y. Characterization of a unique group-specific protein (U122) of the severe acute respiratory syndrome coronavirus. J. Virol. 2004;78:7311–7318. doi: 10.1128/JVI.78.14.7311-7318.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvin K., Krishna S., Ponchel F., Frohlich M., Cummings D.E., Carlson R., Wands J.R., Isselbacher K.J., Pillai S., Ozturk M. The major histocompatibility complex class I antigen-binding protein p88 is the product of the calnexin gene. Proc. Natl. Acad. Sci. USA. 1992;89:8452–8456. doi: 10.1073/pnas.89.18.8452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gewurz B.E., Gaudet R., Tortorella D., Wang E.W., Ploegh H.L., Wiley D.C. Antigen presentation subverted: Structure of the human cytomegalovirus protein US2 bound to the class I molecule HLA-A2. Proc. Natl. Acad. Sci. USA. 2001;98:6794–6799. doi: 10.1073/pnas.121172898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudo C.G., Maccioni H.J. Endoplasmic reticulum export of glycosyltransferases depends on interaction of a cytoplasmic dibasic motif with Sar1. Mol. Biol. Cell. 2003;14:3753–3766. doi: 10.1091/mbc.E03-02-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith K.J., Chan E.K., Lung C.C., Hamel J.C., Guo X., Miyachi K., Fritzler M.J. Molecular cloning of a novel 97-kd Golgi complex autoantigen associated with Sjogren's syndrome. Arthritis Rheum. 1997;40:1693–1702. doi: 10.1002/art.1780400920. [DOI] [PubMed] [Google Scholar]
- Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L., Luo S.W., Li P.H., Zhang L.J., Guan Y.J. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- Hobman T.C., Woodward L., Farquhar M.G. Targeting of a heterodimeric membrane protein complex to the Golgi: rubella virus E2 glycoprotein contains a transmembrane Golgi retention signal. Mol. Biol. Cell. 1995;6:7–20. doi: 10.1091/mbc.6.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L., Sander C. Dali: a network tool for protein structure comparison. Trends Biochem. Sci. 1995;20:478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- Hubbard S.J., Campbell S.F., Thornton J.M. Molecular recognition. Conformational analysis of limited proteolytic sites and serine proteinase protein inhibitors. J. Mol. Biol. 1991;220:507–530. doi: 10.1016/0022-2836(91)90027-4. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou J.Y., Cowan S.W. Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Klumperman J., Locker J.K., Meijer A., Horzinek M.C., Geuze H.J., Rottier P.J. Coronavirus M proteins accumulate in the Golgi complex beyond the site of virion budding. J. Virol. 1994;68:6523–6534. doi: 10.1128/jvi.68.10.6523-6534.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krijnse-Locker J., Ericsson M., Rottier P.J., Griffiths G. Characterization of the budding compartment of mouse hepatitis virus: evidence that transport from the RER to the Golgi complex requires only one vesicular transport step. J. Cell Biol. 1994;124:55–70. doi: 10.1083/jcb.124.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krokhin O., Li Y., Andonov A., Feldmann H., Flick R., Jones S., Stroeher U., Bastien N., Dasuri K.V., Cheng K. Mass spectrometric characterization of proteins from the SARS virus: a preliminary report. Mol. Cell. Proteomics. 2003;2:346–356. doi: 10.1074/mcp.M300048-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaPointe P., Gurkan C., Balch W.E. Mise en place—this bud’s for the Golgi. Mol. Cell. 2004;14:413–414. doi: 10.1016/s1097-2765(04)00267-9. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- Lilley B.N., Ploegh H.L. A membrane protein required for dislocation of misfolded proteins from the ER. Nature. 2004;429:834–840. doi: 10.1038/nature02592. [DOI] [PubMed] [Google Scholar]
- Lodoen M., Ogasawara K., Hamerman J.A., Arase H., Houchins J.P., Mocarski E.S., Lanier L.L. NKG2D-mediated natural killer cell protection against cytomegalovirus is impaired by viral gp40 modulation of retinoic acid early inducible 1 gene molecules. J. Exp. Med. 2003;197:1245–1253. doi: 10.1084/jem.20021973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lontok E., Corse E., Machamer C.E. Intracellular targeting signals contribute to localization of coronavirus spike proteins near the virus assembly site. J. Virol. 2004;78:5913–5922. doi: 10.1128/JVI.78.11.5913-5922.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machamer C.E., Grim M.G., Esquela A., Chung S.W., Rolls M., Ryan K., Swift A.M. Retention of a cis Golgi protein requires polar residues on one face of a predicted alpha-helix in the transmembrane domain. Mol. Biol. Cell. 1993;4:695–704. doi: 10.1091/mbc.4.7.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetti B., Ashrafi G.H., Tsirimonaki E., O'Brien P.M., Campo M.S. The bovine papillomavirus oncoprotein E5 retains MHC class I molecules in the Golgi apparatus and prevents their transport to the cell surface. Oncogene. 2002;21:7808–7816. doi: 10.1038/sj.onc.1205885. [DOI] [PubMed] [Google Scholar]
- Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- Mizuguchi K., Deane C.M., Blundell T.L., Overington J.P. HOMSTRAD: a database of protein structure alignments for homologous families. Protein Sci. 1998;7:2469–2471. doi: 10.1002/pro.5560071126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris R.J., Perrakis A., Lamzin V.S. ARP/wARP and automatic interpretation of protein electron density maps. Methods Enzymol. 2003;374:229–244. doi: 10.1016/S0076-6879(03)74011-7. [DOI] [PubMed] [Google Scholar]
- Murzin A.G., Brenner S.E., Hubbard T., Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- Nguyen V.P., Hogue B.G. Coronavirus envelope glycoprotein assembly complexes. Adv. Exp. Med. Biol. 1998;440:361–365. doi: 10.1007/978-1-4615-5331-1_47. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Sharp K.A., Honig B. The program Grasp. Biophys. J. 1993;64:166–170. [Google Scholar]
- Orange J.S., Fassett M.S., Koopman L.A., Boyson J.E., Strominger J.L. Viral evasion of natural killer cells. Nat. Immunol. 2002;3:1006–1012. doi: 10.1038/ni1102-1006. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pekosz A., Lamb R.A. Cell surface expression of biologically active influenza C virus HEF glycoprotein expressed from cDNA. J. Virol. 1999;73:8808–8812. doi: 10.1128/jvi.73.10.8808-8812.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploegh H.L. Viral strategies of immune evasion. Science. 1998;280:248–253. doi: 10.1126/science.280.5361.248. [DOI] [PubMed] [Google Scholar]
- Rest J.S., Mindell D.P. SARS associated coronavirus has a recombinant polymerase and coronaviruses have a history of host-shifting. Infect. Genet. Evol. 2003;3:219–225. doi: 10.1016/j.meegid.2003.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- Sauer F.G., Pinkner J.S., Waksman G., Hultgren S.J. Chaperone priming of pilus subunits facilitates a topological transition that drives fiber formation. Cell. 2002;111:543–551. doi: 10.1016/s0092-8674(02)01050-4. [DOI] [PubMed] [Google Scholar]
- Shi X., Elliott R.M. Golgi localization of Hantaan virus glycoproteins requires coexpression of G1 and G2. Virology. 2002;300:31–38. doi: 10.1006/viro.2002.1414. [DOI] [PubMed] [Google Scholar]
- Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J., Poon L.L., Guan Y., Rozanov M., Spaan W.J., Gorbalenya A.E. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler K., Masignani V., Eickmann M., Becker S., Abrignani S., Klenk H.D., Rappuoli R. SARS-Beginning to understand a new virus. Nat. Rev. Microbiol. 2003;1:209–218. doi: 10.1038/nrmicro775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanhope M.J., Brown J.R., Amrine-Madsen H. Evidence from the evolutionary analysis of nucleotide sequences for a recombinant history of SARS-CoV. Infect. Genet. Evol. 2004;4:15–19. doi: 10.1016/j.meegid.2003.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavrinides J., Guttman D.S. Mosaic evolution of the severe acute respiratory syndrome coronavirus. J. Virol. 2004;78:76–82. doi: 10.1128/JVI.78.1.76-82.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift A.M., Machamer C.E. A Golgi retention signal in a membrane-spanning domain of coronavirus E1 protein. J. Cell Biol. 1991;115:19–30. doi: 10.1083/jcb.115.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y.J., Teng E., Shen S., Tan T.H., Goh P.Y., Fielding B.C., Ooi E.E., Tan H.C., Lim S.G., Hong W. A novel severe acute respiratory syndrome coronavirus protein, U274, is transported to the cell surface and undergoes endocytosis. J. Virol. 2004;78:6723–6734. doi: 10.1128/JVI.78.13.6723-6734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T.C., Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vennema H., Godeke G.J., Rossen J.W., Voorhout W.F., Horzinek M.C., Opstelten D.J., Rottier P.J. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 1996;15:2020–2028. doi: 10.1002/j.1460-2075.1996.tb00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson J.D., Todd A.E., Bray J., Laskowski R.A., Edwards A., Joachimiak A., Orengo C.A., Thornton J.M. Target selection and determination of function in structural genomics. IUBMB Life. 2003;55:249–255. doi: 10.1080/1521654031000123385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Shibata Y., Yun C., Ron D., Rapoport T.A. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature. 2004;429:841–847. doi: 10.1038/nature02656. [DOI] [PubMed] [Google Scholar]
- Ying W., Hao Y., Zhang Y., Peng W., Qin E., Cai Y., Wei K., Wang J., Chang G., Sun W. Proteomic analysis on structural proteins of Severe Acute Respiratory Syndrome coronavirus. Proteomics. 2004;4:492–504. doi: 10.1002/pmic.200300676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D., Li H., Xu R., He J., Lin J., Li L., Li W., Xu X., Huang S., Huang J. Prevalence of IgG antibody to SARS-associated coronavirus in animal traders—Guangdong Province, China, 2003. MMWR Morb. Mortal. Wkly. Rep. 2003;52:986–987. [PubMed] [Google Scholar]
- Ziegler H., Thale R., Lucin P., Muranyi W., Flohr T., Hengel H., Farrell H., Rawlinson W., Koszinowski U.H. A mouse cytomegalovirus glycoprotein retains MHC class I complexes in the ERGIC/cis-Golgi compartments. Immunity. 1997;6:57–66. doi: 10.1016/s1074-7613(00)80242-3. [DOI] [PubMed] [Google Scholar]