Abstract
The presence of Giardia and Cryptosporidium was investigated in 274 faecal samples of alpacas (Vicugna pacos) from 12 herds from Peru by immunofluorescence microscopy and PCR amplification and sequencing of fragments of the ssu-rRNA and β-giardin genes from Giardia spp., as well as the ssu-rRNA gene from Cryptosporidium spp. A total of 137 samples (50.0%) were positive for Giardia spp., and 12 samples (4.4%) for Cryptosporidium spp. In ten samples (3.6%), co-infection by both pathogens was found. Herd prevalence was found to be 91.7% (11/12 herds) for Giardia and 58.3% (7/12 herds) for Cryptosporidium. Regarding the age of the animals, although Giardia was detected in animals as young as 1 week, the prevalence increased with age, reaching 80% by 8 weeks. Similarly, the highest percentage of Cryptosporidium detection (20%) was also found in the 8 week-old group. By PCR, 92 of the 274 analysed samples were positive for Giardia. Sequencing of the amplicons showed the existence of Giardia duodenalis assemblage A in 67 samples; G. duodenalis assemblage E in 24 samples; and inconsistent results between the two molecular markers used in a further sample. Cryptosporidium was only detected by PCR in 3 of the 274 samples; Cryptosporidium parvum was identified in two samples and Cryptosporidium ubiquitum in one sample. This study is the first performing molecular characterisation of both parasites in Peruvian alpacas, and the first report of C. ubiquitum in this host. The identification of G. duodenalis assemblage A, C. parvum and C. ubiquitum, suggests that zoonotic transmission of these enteropathogens between alpacas and humans is possible.
Keywords: Vicugna pacos, Giardia, Cryptosporidium, Immunofluorescence microscopy, Molecular characterisation, Peru
1. Introduction
The South American camelids (SACs) are a group of artiodactyl mammals of the family Camelidae which includes alpacas (Vicugna pacos), guanacos (Lama guanicoe), llamas (Lama glama), and vicuñas (Vicugna vicugna). Alpacas, the smallest domestic SAC, are primarily distributed in Peru, with smaller populations in Bolivia, Chile and Ecuador, where they are a major source of fiber and meat. There are also numerous alpaca farms in the USA, Australia, New Zealand and different European countries as consequence of their exportation from Peru. As with most other ruminant species, they are at increased risk of acquiring infections of diverse aetiology during the first months of life, and diarrhoea is an important cause of morbidity and mortality in neonatal animals. The most common pathogens causing diarrhoea in neonatal SACs are coronavirus, Escherichia coli, Cryptosporidium spp., Giardia duodenalis and coccidia (Cebra et al., 2003, Whitehead and Anderson, 2006).
Giardia and Cryptosporidium are two genera of protozoan parasites that infect humans, domesticated livestock, companion animals and wildlife worldwide (Xiao and Fayer, 2008). G. duodenalis is considered a multispecies complex infecting humans and many other species of mammals. Using a variety of genetic loci, eight genetic groups or assemblages (A → H) of G. duodenalis have been recognised: assemblages A and B from humans and many other mammals; assemblages C and D from dogs; assemblage E from artiodactyls; assemblage F from cats; assemblage G from rodents; and assemblage H from seals (Feng and Xiao, 2011).
The genus Cryptosporidium currently comprises 19 valid species and over 40 genotypes (Fayer, 2010). Ruminants can be infected by several Cryptosporidium species, being Cryptosporidium parvum the most commonly found. Together with Cryptosporidium hominis, this species is responsible for the majority of human infections. However, in some areas, such as Peru, Cryptosporidium meleagridis infection rates are as high as those of C. parvum (Cama et al., 2008, Xiao, 2010).
These parasites have been previously reported in SAC, including alpacas (Trout et al., 2008, Twomey et al., 2008). However, despite their economic importance, little information is available, and few studies have used molecular methods to characterise the Giardia and Cryptosporidium involved in the infections. The aim of the present study was to investigate the presence of Giardia and Cryptosporidium in alpacas from the Peruvian altiplano, applying molecular tests to characterise the isolates.
2. Materials and methods
2.1. Faecal samples
Between the years 2009 and 2010, a total of 274 faecal samples were collected directly from the rectums of suri (silky-haired) and huacaya (crimped-haired) breed alpacas (V. pacos), selected at random from 12 herds located in the Puno and Cusco regions of the Peruvian altiplano (Table 1 ).
Table 1.
Prevalence of Giardia and Cryptosporidium in alpacas from Peru and assemblages or species identified.
| Analysed samples (n) |
Giardia |
Cryptosporidium |
|||||
|---|---|---|---|---|---|---|---|
| Positive samples (%) | Intensity of infectiona | Molecular characterisation (A/E)b (%A)c | Positive samples (%) | Intensity of infectiona | Molecular characterisation (species) | ||
| Herd | |||||||
| 1 | 21 | 7 (33.3) | 1.9 | 0/3 (0.0) | 1 (4.8) | 1 | C. ubiquitum |
| 2 | 30 | 21 (70.0) | 2.4 | 5/9 (35.7) | 1 (3.3) | 1 | – |
| 3 | 22 | 1 (4.5) | 1.0 | 1/0 (100.0) | 0 (0.0) | – | – |
| 4 | 14 | 9 (64.3) | 1.4 | 1/4 (20.0) | 1 (7.1) | 1 | – |
| 5 | 38 | 16 (42.1) | 1.6 | 6/1 (85.7) | 0 (0.0) | – | – |
| 6 | 8 | 0 (0.0) | – | – | 1 (12.5) | 1 | – |
| 7 | 2 | 1 (50.0) | 1.0 | 1/0 (100) | 0 (0.0) | – | – |
| 8 | 43 | 26 (60.5) | 2.4 | 17/3 (85.0) | 3 (7.0) | 1 | – |
| 9 | 11 | 2 (18.2) | 1.5 | 1/0 (100) | 0 (0.0) | – | – |
| 10 | 22 | 15 (68.2) | 1.6 | 12/0 (100) | 3 (13.6) | 1 | C. parvum |
| 11 | 35 | 24 (68.6) | 2.7 | 16/2 (88.9) | 1 (2.9) | 1 | – |
| 12 | 8 | 6 (75.0) | 2.5 | 2/0 (100) | 0 (0.0) | – | – |
| No data | 20 | 9 (45.0) | 2.1 | 5/2 (71.4) | 1 (5.0) | 1 | C. parvum |
| Age (weeks) | |||||||
| 1 | 7 | 3 (42.9) | 1.7 | 2/0 (100) | 0 (0.0) | – | – |
| 2 | 9 | 3 (33.3) | 2.0 | 0/2 (0.0) | 0 (0.0) | – | – |
| 3 | 14 | 2 (14.3) | 2.0 | 1/0 (100) | 0 (0.0) | – | – |
| 4 | 45 | 20 (44.4) | 2.2 | 11/3 (78.6) | 0 (0.0) | – | – |
| 5 | 33 | 17 (51.5) | 2.7 | 10/2 (83.3) | 3 (9.1) | 1 | – |
| 6 | 76 | 47 (61.8) | 2.0 | 28/4 (87.5) | 5 (6.6) | 1 | C. parvum |
| 7 | 8 | 5 (62.5) | 2.8 | 2/0 (100) | 0 (0.0) | – | – |
| 8 | 5 | 4 (80.0) | 2.8 | 1/2 (33.3) | 1 (20.0) | 1 | – |
| 9 | 2 | 0 (0.0) | – | – | 0 (0.0) | – | – |
| 10 | 8 | 4 (50.0) | 1.3 | 1/1 (50.0) | 0 (0.0) | – | – |
| >10 | 35 | 7 (20.0) | 1.9 | 0/3 (0.0) | 2 (5.7) | 1 | C. ubiquitum |
| No data | 32 | 25 (78.1) | 2.0 | 11/7 (61.1) | 1 (3.1) | 1 | C. parvum |
| Breed | |||||||
| Huacaya | 140 | 78 (55.7) | 2.0 | 41/14 (74.5) | 6 (4.3) | 1 | C. parvum |
| Suri | 75 | 35 (46.7) | 2.3 | 20/3 (87.0) | 3 (4.0) | 1 | – |
| No data | 59 | 24 (40.7) | 2.2 | 6/7 (46.2) | 3 (5.1) | 1 | C. parvum/C. ubiquitum |
| Sex | |||||||
| Female | 121 | 52 (43.0) | 2.1 | 22/14 (61.1) | 6 (5.0) | 1 | C. ubiquitum |
| Male | 93 | 45 (48.4) | 2.4 | 25/5 (83.3) | 2 (2.2) | 1 | – |
| No data | 60 | 40 (66.7) | 2.1 | 20/5 (80.0) | 4 (6.7) | 1 | C. parvum |
| Diarrhoeic processes | |||||||
| Absence | 184 | 84 (45.7) | 2.4 | 38/13 (74.5) | 7 (3.8) | 1 | – |
| Presence | 76 | 44 (57.9) | 1.7 | 24/9 (72.7) | 4 (5.3) | 1 | C. parvum |
| No data | 14 | 9 (64.3) | 2.1 | 5/2 (71.4) | 1 (7.1) | 1 | C. parvum/C. ubiquitum |
| Total | 274 | 137 (50.0) | 2.1 | 67/24 (73.6)d | 12 (4.4) | 1 | |
Determined semiquantitatively according to the average number of parasitic forms in 10 randomly selected microscopic fields at 400× magnification: 0 (no cysts/oocysts), 1 (≤1 cyst/oocyst), 2 (2–5 cysts/oocysts), 3 (6–10 cysts/oocysts) and 4 (≥10 cysts/oocysts).
G. duodenalis assemblage A or E.
Percentage of samples in which the assemblage A of G. duodenalis was identified respect to the number of amplified samples.
There was an incongruence between the two molecular markers used in one sample.
After collection, aliquots of the faecal samples were resuspended in distilled water, put into screw top specimen cups, labelled with the animal identification and sent to the laboratory of Animal Health and Zoonoses (SALUVET) research group (Madrid, Spain). Data regarding sampling location, age, breed, sex and presence or absence of diarrhoeic processes were also recorded when possible.
2.2. Detection of Giardia cysts and Cryptosporidium oocysts by epifluorescence microscopy
Immunofluorescence staining was performed using the Crypto/Giardia Cel IF Test (Cellabs Pty Ltd., Brookvale, Australia) according to the manufacturer's instructions. The cysts/oocysts were identified by epifluorescence microscopy at 400× magnification on the basis of their size, shape and the pattern and intensity of immunofluorescence staining.
For detection, the whole slide was examined and the intensity of infection was determined semi-quantitatively according to the average number of parasitic forms in 10 randomly selected microscopic fields at 400× magnification: 0 (no cysts/oocysts), 1 (1 cyst/oocyst), 2 (2–5 cysts/oocysts), 3 (6–10 cysts/oocysts) and 4 (≥10 cysts/oocysts).
2.3. Molecular characterisation of Giardia spp. and Cryptosporidium spp.
Nucleic acids were extracted from the faecal samples as described by McLauchlin et al. (1999). Briefly, 200 μl of the sample was mechanically disrupted in the presence of guanidinium thiocyanate with zirconia beads using a Bead-Beater-8™ (BioSpec Products, Inc., Batlesville, OK, USA) and the DNA released from disrupted cysts/oocysts was purified using coarse activated silica and stored at −20 °C until use.
For the molecular characterisation of Giardia isolates, two nested PCR protocols were used to amplify fragments of 292 bp and 510 bp of the ssu-rRNA and β-giardin genes, respectively (Appelbee et al., 2003, Lalle et al., 2005). For Cryptosporidium, a two-step nested PCR protocol amplifying a fragment of 819–825 bp of the ssu-rRNA was employed (Xiao et al., 2000). Negative and positive controls were included for all PCR runs. The PCR products were subjected to electrophoresis on 1% agarose/ethidium bromide gels.
PCR products were purified using GeneClean® Turbo Kit (Qbiogene, Inc., Carlsbad, CA, USA) and sequenced in both directions using the forward and reverse inner primers with the Big Dye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) and a 3730 DNA analyser (Applied Biosystems) at the Unidad Genómica del Parque Científico de Madrid. Sequence data was examined using SeqMan® 7.0 (DNASTAR®, Madison, WI, USA) and BioEdit 7.0.5.3 (®1997–2005 Tom Hall, Ibis Therapeutics, Carlsbad, CA, USA). The resulting sequences were compared with those of Giardia spp. and Cryptosporidium spp. deposited in GenBank (National Institutes of Health, Bethesda, MD, USA) using the program BLAST v.2.2.12 (http://www.ncbi.nlm.nih.gov/BLAST/, National Center for Biotechnology Information, Bethesda, MD, USA). Phylogenetic and molecular evolutionary analyses were made using MEGA 5 (Tamura et al., 2011) on the basis of genetic distances calculated by the Kimura two-parameter model (Kimura, 1980).
2.4. Statistical analysis
Prevalence data with respect to breed, age, sex and presence or absence of diarrhoeic processes were analysed using the Chi-square or Fisher's exact test. Intensity of infection was analysed by the Kruskal–Wallis test (non-parametric anova) using the GraphPad Instat®, version 3.05, statistical software (©1992–2000 GraphPad Software, La Jolla, CA, USA). Differences were considered significant at a probability level of P < 0.05.
2.5. Nucleotide sequence accession numbers
The nucleotide sequences of the isolates analysed in this study have been deposited in the GenBank database under accession numbers JN812214–JN812336.
3. Results
The application of immunofluorescence microscopy and PCR techniques on 274 alpaca faecal samples showed that 137 animals (50.0%) were infected by Giardia spp. and 12 animals by Cryptosporidium spp. (4.4%) (Table 1). In 10 of these samples (3.6%), co-infection by both pathogens was found.
Giardia was detected in 11/12 herds examined (91.7%), with intra-herd prevalence ranging from 18.2% to 75%. The intensity of infection varied between 1.0 and 2.7. Although Giardia was detected in animals as young as 1 week, the prevalence and the intensity of infection increased with age reaching 80% and 2.8, respectively, at 8 weeks. Prevalence values of 50% and 20% were observed in animals 10 weeks and older than 10 weeks of age, respectively, but at lower intensities of infection. No statistically significant differences in prevalence and intensity of infection were observed in relation to breed, sex, or the existence of diarrhoeic process.
By PCR amplification of fragments of the ssu-rRNA and β-giardin genes, 92 of the 274 analysed samples (33.6%) were positive for Giardia. Sequencing of amplicons showed the existence of G. duodenalis assemblage A in 67 samples; G. duodenalis assemblage E in 24 samples; and in one sample there was an inconsistency between the two molecular markers used. For the amplified fragment of the ssu-rRNA gene, 6 sequences were observed, two matched others deposited in GenBank and 4 were new sequences (one close to G. duodenalis assemblage A and 3 close to G. duodenalis assemblage E) (Fig. 1 ). For the amplified fragment of the β-giardin gene, 3 sequences were observed, one corresponding to G. duodenalis assemblage A and two to G. duodenalis assemblage E. All sequences showed several SNPs with respect to the sequences deposited in GenBank (Fig. 2 ).
Fig. 1.
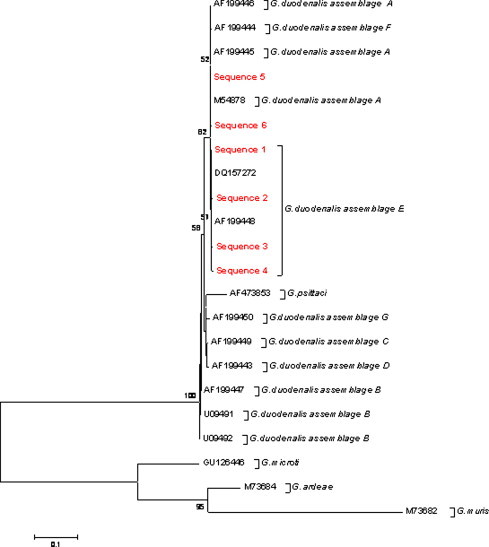
Phylogenetic relationships among Giardia species and assemblages inferred by a neighbour-joining analysis of a fragment of the ssu-rRNA gene sequence, based on genetic distances calculated by the Kimura two-parameter model. Numbers on branches are percentage bootstrap values (>50%) from 1000 replicates.
Fig. 2.
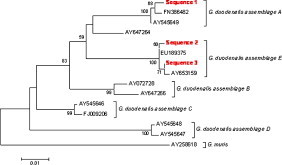
Phylogenetic relationships among G. duodenalis assemblages inferred by a neighbour-joining analysis of a fragment of the β-giardin gene sequence, based on genetic distances calculated by the Kimura two-parameter model. Numbers on branches are percentage bootstrap values (>50%) from 1000 replicates.
Faecal samples containing both G. duodenalis assemblages A and E were detected in 5 herds, whereas 5 herds contained only assemblage A and one herd had only assemblage E. Overall, G. duodenalis assemblage A was identified in a higher percentage of samples (73.6%). No significant differences were observed in relation to breed, sex or the presence of diarrhoeic processes.
Cryptosporidium spp. oocysts were observed by immunofluorescence microscopy in 12 faecal samples (4.4%) from animals aged ≥5 weeks belonging to 7 herds (58.3%) (Table 1). Intra-herd prevalence ranged from 2.9% to 13.6%, and the intensity of infection was very low (<1 oocyst per field). By amplification of a fragment of the ssu-rRNA, C. parvum was identified in two samples and C. ubiquitum in one sample (Fig. 3 ).
Fig. 3.
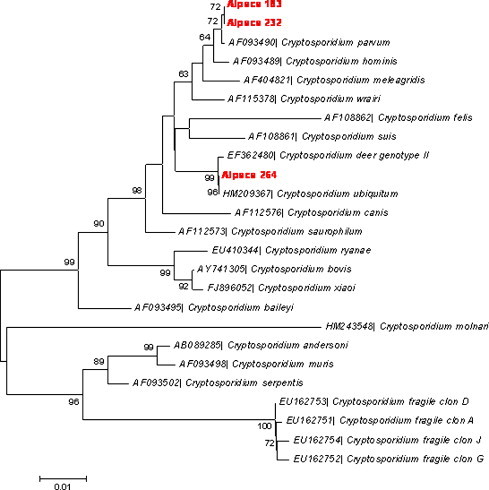
Phylogenetic relationships among Cryptosporidium species and genotypes inferred by a neighbour-joining analysis of a fragment of the ssu-rRNA gene sequence, based on genetic distances calculated by the Kimura two-parameter model. Numbers on branches are percentage bootstrap values (>50%) from 1000 replicates.
4. Discussion
This work constitutes the first extensive study in which the presence of Giardia and Cryptosporidium was investigated in alpacas in their natural habitat (Peruvian altiplano). Currently, published data about these two protozoan parasites in alpacas is limited, involving mainly descriptions of clinical cases (Bidewell and Cattell, 1998, Shapiro et al., 2005, Starkey et al., 2007, Waitt et al., 2008); isolated cases (Ey et al., 1997, Spano et al., 1997, Spano et al., 1998, Morgan et al., 1998, Ryan et al., 2003, Stewart et al., 2005) or epidemiological studies performed at farms located in countries such as the USA and UK where these animals have been introduced (Cebra et al., 2003, Trout et al., 2008, Twomey et al., 2008).
In SAC, the first case of Giardia infection was described in 1987 in an asymptomatic young llama in Wisconsin (Kiorpes et al., 1987). Subsequently, Giardia was identified as the enteropathogen involved in 18% of diarrhoea cases in unweaned llama and alpaca crias in Oregon (Cebra et al., 2003). In addition, data collected between the years 1999 and 2004 from 58 clinical cases of diarrhoea in alpacas less than 4 months of age at Ohio State University revealed that 32.8% of the crias were positive for Giardia spp. (Whitehead and Anderson, 2006). Recently, Trout et al. (2008) reported the detection of Giardia by PCR analysis in 4.9% of the faecal samples from 61 adult alpacas and crias from two farms in Maryland. The 3 positive samples were obtained from 14 and 27 month old females and from a 13 week old male. In our study, immunofluorescence microscopy and PCR analysis of faecal samples from 274 alpacas originating from 12 herds has revealed a global prevalence of 50.0%, reaching 75.0% in some herds, and highlighting the elevated prevalence of Giardia in these domestic animals. Overall, high percentages (14.3–80.0%) of positive animals were found in all age groups, except 9 weeks in which the 2 samples analysed were negative. Infection rates over 50% were observed in animals of ages 5–10 weeks, and were also elevated in animals as young as 1 week old (42.9%) as well as in older animals, >10 weeks (20.0%). These results are in agreement with data reported in a previous alpaca study for the same age group, 32.8% in animals <8–9 weeks of age (Whitehead and Anderson, 2006), as well as with the 15.6% found in llamas and alpacas between 4 and 17 weeks of age (Cebra et al., 2003), although the percentages found here are superior. However, in llamas detection of Giardia was higher in animals 0–2 months of age (25.0%), followed by groups of 2–4 (12.5%), 4–12 (5.3%) and 13–24 (3.2%) months, not detecting the parasite in animals 25 to >96 months of age (Rulofson et al., 2001).
Cryptosporidium, as Giardia, has been reported in alpacas in the few clinical reports or epidemiological studies performed outside South America. A prevalence of 8.8% was reported by the Veterinary Laboratory Agency Service in England and Wales in samples from alpaca and llama crias submitted for a diagnosis of diarrhoea between 1999 and 2005 (Twomey et al., 2008). In addition, prevalences of 6.7% and 25.9% were observed in llama and alpaca crias with diarrhoea aged between 1–2 and 1–14 weeks, respectively, from USA (Cebra et al., 2003, Whitehead and Anderson, 2006). In the present study, the total prevalence for Cryptosporidium was 4.4%, being present in 58.3% of the herds analysed although at very low intensity of infection. The low number of oocysts observed in the faecal samples (<1 oocyst per microscopic field at 400× magnification) and the small amount of faecal sample available could account for the differences in detection observed in the present study between immunofluorescence microscopy and PCR. Cryptosporidium spp. oocysts were detected by microscopy in 12 faecal samples, whereas molecular characterisation could only be achieved in 3 of these samples. The highest intra-herd prevalence was 13.6%. In contrast to other studies, positive animals were found in age groups 5 (9.1%), 6 (6.6%), 8 (20%) and >10 (5.7%) weeks old. However, Cryptosporidium was not detected in any of the 61 alpaca samples from two Maryland farms (Trout et al., 2008). This may be due to the age of the animals examined, since in this study, the age range was 10 weeks to 10 years of age.
Giardia and Cryptosporidium have been included amongst the most common pathogens causing diarrhoea in neonatal camelids (Cebra et al., 2003, Whitehead and Anderson, 2006, Waitt et al., 2008). In the present work, it was not possible to establish this association although the prevalence for both Giardia and Cryptosporidium detected in diarrhoeic samples was slightly higher. The average intensity of infection observed in diarrhoeic samples, however, was equal to or lower than that observed in non-diarrhoeic samples. This may be due to the age of the animals (>5 weeks in the case of Cryptosporidium) or to the dilution of the diarrhoeic faecal samples. Subclinical infection with Giardia and Cryptosporidium has been previously reported in alpacas (Trout et al., 2008, Twomey et al., 2008). In addition, G. duodenalis was reported in 3.4% of 354 asymptomatic llamas (Rulofson et al., 2001). Trout et al. (2008) suggested that the absence of clinical signs could indicate that Giardia might not necessarily be pathogenic in alpacas. However, shedding of the resistant forms of these parasites can extend beyond the period of diarrhoea and excretion is intermittent (Buret et al., 1990, Thompson et al., 2008), therefore the time at which samples are collected and the number of samples analysed per animal are critical to establish any association with diarrhoea. Nevertheless, subclinical shedding of cysts or oocysts has important implications at herd management level, since apparently healthy animals can be a source of infection to other animals in their vicinity.
Early molecular analysis of Giardia from alpacas in Australia (4 isolates) suggested that this host could be infected by assemblages A and E of G. duodenalis (Ey et al., 1997). Later, Trout et al. (2008) identified G. duodenalis assemblage A in 3 of 61 animals examined and no assemblage E. In the present study, DNA sequence analysis revealed that amongst the 92 samples positive by PCR, 67 samples contained G. duodenalis assemblage A; 24 samples G. duodenalis assemblage E; and in the remaining sample there was an inconsistency between the two molecular markers used. These results show a clear predominance of the widely distributed zoonotic assemblage A in all age groups over assemblage E, which shows specificity to artiodactyl hosts.
The first studies in which Cryptosporidium isolates were characterised by molecular methods were those performed with the A1 alpaca isolate originating from Peru (Spano et al., 1997, Spano et al., 1998, Morgan et al., 1998) and with an alpaca isolate from the Czech Republic (Ryan et al., 2003), which showed consistent PCR, PCR-RFLP, and sequence analysis results for multiple loci with C. parvum. Later, this result was confirmed using several isolates from alpaca crias with and without diarrhoea in USA and UK (Starkey et al., 2007, Twomey et al., 2008). Similarly, in the present study, C. parvum was identified in two animals. In addition, we describe here the presence, for the first time in this host, of C. ubiquitum in a female alpaca older than 10 weeks of age. This recently named Cryptosporidium species was previously identified as Cryptosporidium cervine genotype and occasionally as the cervid, W4 or genotype 3 genotype, and has been shown by molecular testing to infect the greatest number of host species of any species of Cryptosporidium, being found in domestic and wild ruminants, rodents and primates. C. ubiquitum has also been involved in human infections worldwide, including Peru (Fayer et al., 2010).
In summary, this is the first study carrying out the molecular characterisation of a large number of Giardia isolates from alpacas, identifying for the first time G. duodenalis assemblages A and E in alpacas from the Peruvian altiplano. We also report for the first time the presence of C. ubiquitum in these populations. Because humans can be infected by G. duodenalis assemblage A, the high prevalence of Giardia detected, together with the identification of C. parvum and C. ubiquitum in these samples, suggests that zoonotic transmission of giardiosis and cryptosporidiosis between alpaca and humans can occur, and therefore these animals should be considered as potential sources of infection for humans. In fact, an outbreak of human cryptosporidiosis caused by C. parvum attributable to an alpaca source has been reported (Starkey et al., 2007). Moreover, and due to the high prevalence of these two protozoan infections in humans in Peru (Checkley et al., 1997, Pérez Cordón et al., 2008), the inverse situation may also occur. Further studies are therefore needed to investigate the epidemiology of these diseases in Peruvian alpacas.
Acknowledgements
HG-C is funded by USC through the Programme Angeles Alvariño of Xunta de Galicia (Government of the Autonomous Region of Galicia). For this specific study, HG-C was awarded a mobility grant by Xunta de Galicia and the European Social Fund. We would like to thank the Centro de Investigaciones de la Raya, University of the Altiplano-Puno for enabling the sample collection and Karol Guzmán and Jorge Maximiliano for their help and assistance. We also thank Dr. Jane Wheeler for the careful reading of this manuscript and helpful comments.
References
- Appelbee A.J., Frederick L.M., Heitman T.L., Olson M.E. Prevalence and genotyping of Giardia duodenalis from beef calves in Alberta, Canada. Vet. Parasitol. 2003;112:289–294. doi: 10.1016/s0304-4017(02)00422-3. [DOI] [PubMed] [Google Scholar]
- Bidewell C.A., Cattell J.H. Cryptosporidiosis in young alpacas. Vet. Rec. 1998;142:287. [PubMed] [Google Scholar]
- Buret A., denHollander N., Wallis P.M., Befus D., Olson M.E. Zoonotic potential of giardiasis in domestic ruminants. J. Infect. Dis. 1990;162:231–237. doi: 10.1093/infdis/162.1.231. [DOI] [PubMed] [Google Scholar]
- Cama V.A., Bern C., Roberts J., Cabrera L., Sterling C.R., Ortega Y., Gilman R.H., Xiao L. Cryptosporidium species and subtypes and clinical manifestations in children, Peru. Emerg. Infect. Dis. 2008;14:1567–1574. doi: 10.3201/eid1410.071273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebra C.K., Mattson D.E., Baker R.J., Sonn R.J., Dearing P.L. Potential pathogens in feces from unweaned llamas and alpacas with diarrhea. J. Am. Vet. Med. Assoc. 2003;223:1806–1808. doi: 10.2460/javma.2003.223.1806. [DOI] [PubMed] [Google Scholar]
- Checkley W., Gilman R.H., Epstein L.D., Suarez M., Diaz J.F., Cabrera L., Black R.E., Sterling C.R. Asymptomatic and symptomatic cryptosporidiosis: their acute effect on weight gain in Peruvian children. Am. J. Epidemiol. 1997;145:156–163. doi: 10.1093/oxfordjournals.aje.a009086. [DOI] [PubMed] [Google Scholar]
- Ey P.L., Mansouri M., Kulda J., Nohynkova E., Monis P.T., Andrews R.H., Mayrhofer G. Genetic analysis of Giardia from hoofed farm animals reveals artiodactyl-specific and potentially zoonotic genotypes. J. Eukaryot. Microbiol. 1997;44:626–635. doi: 10.1111/j.1550-7408.1997.tb05970.x. [DOI] [PubMed] [Google Scholar]
- Fayer R. Taxonomy and species delimitation in Cryptosporidium. Exp. Parasitol. 2010;124:90–97. doi: 10.1016/j.exppara.2009.03.005. [DOI] [PubMed] [Google Scholar]
- Fayer R., Santín M., Macarisin D. Cryptosporidium ubiquitum n. sp. in animals and humans. Vet. Parasitol. 2010;172:23–32. doi: 10.1016/j.vetpar.2010.04.028. [DOI] [PubMed] [Google Scholar]
- Feng Y., Xiao L. Zoonotic potential and molecular epidemiology of Giardia species and giardiasis. Clin. Microbiol. Rev. 2011;24:110–140. doi: 10.1128/CMR.00033-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Kiorpes A.L., Kirkpatrick C.E., Bowman D.D. Isolation of Giardia from a llama and from sheep. Can. J. Vet. Res. 1987;51:277–280. [PMC free article] [PubMed] [Google Scholar]
- Lalle M., Jiménez-Cardosa E., Cacciò S.M., Pozio E. Genotyping of Giardia duodenalis from humans and dogs from Mexico using a beta-giardin nested polymerase chain reaction assay. J. Parasitol. 2005;91:203–205. doi: 10.1645/GE-293R. [DOI] [PubMed] [Google Scholar]
- McLauchlin J., Pedraza-Díaz S., Amar-Hoetzeneder C., Nichols G.L. Genetic characterization of Cryptosporidium strains from 218 patients with diarrhea diagnosed as having sporadic cryptosporidiosis. J. Clin. Microbiol. 1999;37:3153–3158. doi: 10.1128/jcm.37.10.3153-3158.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan U.M., Sargent K.D., Deplazes P., Forbes D.A., Spano F., Hertzberg H., Elliot A., Thompson R.C. Molecular characterization of Cryptosporidium from various hosts. Parasitology. 1998;117:31–37. doi: 10.1017/s0031182098002765. [DOI] [PubMed] [Google Scholar]
- Pérez Cordón G., Córdova Paz Soldan O., Vargas Vásquez F., Velasco Soto J.R., Sempere Bordes L., Sánchez Moreno M., Rosales M.J. Prevalence of enteroparasites and genotyping of Giardia lamblia in Peruvian children. Parasitol. Res. 2008;103:459–465. doi: 10.1007/s00436-008-1007-3. [DOI] [PubMed] [Google Scholar]
- Rulofson F.C., Atwill E.R., Holmberg C.A. Fecal shedding of Giardia duodenalis, Cryptosporidium parvum, Salmonella organisms, and Escherichia coli O157:H7 from llamas in California. Am. J. Vet. Res. 2001;62:637–642. doi: 10.2460/ajvr.2001.62.637. [DOI] [PubMed] [Google Scholar]
- Ryan U., Xiao L., Read C., Zhou L., Lal A.A., Pavlasek I. Identification of novel Cryptosporidium genotypes from the Czech Republic. Appl. Environ. Microbiol. 2003;69:4302–4307. doi: 10.1128/AEM.69.7.4302-4307.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro J.L., Watson P., McEwen B., Carman S. Highlights of camelid diagnoses from necropsy submissions to the Animal Health Laboratory, University of Guelph, from 1998 to 2004. Can. Vet. J. 2005;46:317–318. [PMC free article] [PubMed] [Google Scholar]
- Spano F., Putignani L., McLauchlin J., Casemore D.P., Crisanti A. PCR-RFLP analysis of the Cryptosporidium oocyst wall protein (COWP) gene discriminates between C. wrairi and C. parvum, and between C. parvum isolates of human and animal origin. FEMS Microbiol. Lett. 1997;150:209–217. doi: 10.1016/s0378-1097(97)00115-8. [DOI] [PubMed] [Google Scholar]
- Spano F., Putignani L., Crisanti A., Sallicandro P., Morgan U.M., Le Blancq S.M., Tchack L., Tzipori S., Widmer G. Multilocus genotypic analysis of Cryptosporidium parvum isolates from different hosts and geographical origins. J. Clin. Microbiol. 1998;36:3255–3259. doi: 10.1128/jcm.36.11.3255-3259.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starkey S.R., Johnson A.L., Ziegler P.E., Mohammed H.O. An outbreak of cryptosporidiosis among alpaca crias and their human caregivers. J. Am. Vet. Med. Assoc. 2007;231:1562–1567. doi: 10.2460/javma.231.10.1562. [DOI] [PubMed] [Google Scholar]
- Stewart W.C., Pollock K.G., Browning L.M., Young D., Smith-Palmer A., Reilly W.J. Survey of zoonoses recorded in Scotland between 1993 and 2002. Vet. Rec. 2005;157:697–702. doi: 10.1136/vr.157.22.697. [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011 doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson R.C., Palmer C.S., O’Handley R. The public health and clinical significance of Giardia and Cryptosporidium in domestic animals. Vet. J. 2008;177:18–25. doi: 10.1016/j.tvjl.2007.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trout J.M., Santín M., Fayer R. Detection of assemblage A, Giardia duodenalis and Eimeria spp. in alpacas on two Maryland farms. Vet. Parasitol. 2008;153:203–208. doi: 10.1016/j.vetpar.2008.02.006. [DOI] [PubMed] [Google Scholar]
- Twomey D.F., Barlow A.M., Bell S., Chalmers R.M., Elwin K., Giles M., Higgins R.J., Robinson G., Stringer R.M. Cryptosporidiosis in two alpaca (Lama pacos) holdings in the South-West of England. Vet. J. 2008;175:419–422. doi: 10.1016/j.tvjl.2007.01.017. [DOI] [PubMed] [Google Scholar]
- Waitt L.H., Cebra C.K., Firshman A.M., McKenzie E.C., Schlipf J.W., Jr. Cryptosporidiosis in 20 alpaca crias. J. Am. Vet. Med. Assoc. 2008;233:294–298. doi: 10.2460/javma.233.2.294. [DOI] [PubMed] [Google Scholar]
- Whitehead C.E., Anderson D.E. Neonatal diarrhea in llamas and alpacas. Small Rumin. Res. 2006;61:207–215. doi: 10.1016/j.smallrumres.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L. Molecular epidemiology of cryptosporidiosis: an update. Exp. Parasitol. 2010;124:80–89. doi: 10.1016/j.exppara.2009.03.018. [DOI] [PubMed] [Google Scholar]
- Xiao L., Fayer R. Molecular characterisation of species and genotypes of Cryptosporidium and Giardia and assessment of zoonotic transmission. Int. J. Parasitol. 2008;38:1239–1255. doi: 10.1016/j.ijpara.2008.03.006. [DOI] [PubMed] [Google Scholar]
- Xiao L., Alderisio K., Limor J., Royer M., Lal A.A. Identification of species and sources of Cryptosporidium oocysts in storm waters with a small-subunit rRNA-based diagnostic and genotyping tool. Appl. Environ. Microbiol. 2000;66:5492–5498. doi: 10.1128/aem.66.12.5492-5498.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]


