Abstract
Infectious bronchitis virus (IBV) produces six subgenomic (sg) mRNAs, each containing a 64 nucleotide (nt) leader sequence, derived from the 5′ end of the genome by a discontinuous process. Several putative functional domains such as a papain-like proteinase (PLpro), main protease (Mpro), RNA-dependent RNA polymerase (RdRp), and RNA helicase encoded by the replicase gene are important for virus replication. We have sequenced four regions of the replicase genes corresponding to the 5′-terminal sequence, PLpro, Mpro, and RdRp domains from 20 heterologous IBV strains, and compared them with previously published coronavirus sequences. All the coronavirus 5′-termini and PLpro domains were divergent, unlike the Mpro and the RdRp domains that were highly conserved with 28% and 48% conserved residues, respectively. Among IBV strains, the 5′ untranslated region including the leader sequence was highly conserved (>94% identical); whereas, the N-terminal coding region and the PLpro domains were highly variable ranging from 84.6% to 100%, and 77.6% to 100% identity, respectively. The IBV Mpro and RdRp domains were highly conserved with 82.7% and 92.7% conserved residues, respectively. The BJ strain was the most different from other IBVs in all four regions of the replicase. Phylogeny-based clustering based on replicase genes was identical to the antigen-based classification of coronaviruses into three groups. However, the IBV strain classification based on replicase gene domains did not correlate with that of the type-specific antigenic groups. The replicase gene sequences of many IBVs recovered from infected chickens were identical to those of vaccine viruses irrespective of serotype, suggesting that either there has been an exchange of genetic material among vaccine and field isolates or that there is a convergent evolution to a specific replicase genotype. There was no correlation between the genotype of any region of the replicase gene and pathotype, suggesting that the replicase is not the sole determinant of IBV pathogenicity.
Keywords: Coronaviruses, Infectious bronchitis virus, Leader sequence, Main protease, Papain-like proteinase, Recombination, Replicase gene, RNA-dependent RNA polymerase, 5′ untranslated region
Introduction
Avian infectious bronchitis virus (IBV), the prototype species of the family Coronaviridae, causes an acute and highly contagious disease of chickens. Numerous IBV strains exist, and clinical signs of the disease they cause vary from mild to severe. More than 20 IBV serotypes have been recognized worldwide, which are defined by cross-reactivity among the structural proteins Cowen and Hitchner, 1975, Hopkins, 1974. Live vaccines containing strains of IBV from multiple serotypes are routinely used in commercial chicken flocks. Disease outbreaks in commercial flocks occur when new viruses emerge from existing viruses by mutations, insertions, deletions, or recombination Cavanagh et al., 1992, Gelb et al., 1991, Kusters et al., 1990. However, because these conclusions are based on available sequence from structural genes, viral evolution has only been described for this small part of the genome. Because only two IBVs have been fully sequenced, it has been impossible to fully understand the viral evolutionary process including the extent to which vaccine viruses have contributed genetic material to pathogenic field strains.
Six subgenomic (sg) mRNA species are transcribed from the IBV genome in virus-infected cells. This includes a genome-length mRNA (mRNA 1) of 27.6 kb and five sg species (mRNA 2–6) with sizes ranging from 2 to 7 kb. These mRNAs form a 3′-co-terminal nested structure (Stern and Sefton, 1984), each containing a short nontranslated leader sequence (64 nucleotides (nt)), derived from the 5′ end of the genome by a discontinuous process. The phenomenon of leader switching has been described for several coronaviruses including IBV Chang et al., 1994, Makino et al., 1988, Stirrups et al., 2000. The mRNA 1 contains two large overlapping open reading frames (ORF 1a and 1b) encoding 441- and 300-kDa polyproteins, respectively (Boursnell et al., 1987). By a unique frameshifting mechanism, the downstream ORF 1b is produced as a fusion protein of 741 kDa with 1a Brierley et al., 1987, Brierley et al., 1989. These polyproteins are proteolytically processed into smaller products required for RNA synthesis and other aspects of viral replication. Four main structural proteins are encoded by sgmRNAs: the spike (S) glycoprotein (mRNA 2), the small envelope (E) protein (mRNA 3), the membrane (M) glycoprotein (mRNA 4), and the nucleocapsid (N) protein (mRNA 6) Lai, 1990, Spaan et al., 1988, Sutou et al., 1988. The S1 fragment of the spike protein carries antigenic epitopes that induce virus-neutralizing (VN) antibody, and also determines virus serotype Cavanagh and Davis, 1986, Cavanagh et al., 1986.
Two overlapping papain-like proteinase (PLpro) domains encoded between nucleotides (nt) 4243 and 5553 (amino acids (aa) 1239–1675), and a picornavirus 3C-like proteinase (3C-LP) domain encoded between nt 8937 and 9357 (aa 2804–2943), are involved in the production of mature viral proteins required for replication Liu and Brown, 1995, Liu et al., 1994, Liu et al., 1995, Liu et al., 1997. Proteolytic processing of the polyproteins by equivalent protease activities has been demonstrated in murine and human coronaviruses (reviewed in Lim et al., 2000, Ng and Liu, 2000). Despite the conservation of function, there are some organizational differences in PLpro domains between IBV and some other coronaviruses Herold et al., 1993, Lee et al., 1991. Because of its key role in replicase gene expression, 3C-LP has been termed the coronavirus main protease (Mpro; Ziebuhr et al., 2000). The IBV PLpro domain-1 (PL1pro), encoded between nt 4243 and 5019 (aa 1239–1497), is responsible for cleavage of the N-terminal regions of 1a and 1a/1b polyproteins in IBV-infected cells, whereas the PLpro domain-2 (PL2pro) encoded between nt 4681–5553 (aa 1385–1675) is reported to be functionally inactive in IBV Lim and Liu, 1998, Lim et al., 2000, Liu et al., 1995. A more important role in the processing of polyproteins is played by the Mpro, which is responsible for the cleavage of other regions of the ORF 1a and 1a/1b polyproteins Liu et al., 1994, Liu et al., 1997, Liu et al., 1998, Ng and Liu, 1998, Ng and Liu, 2002. The catalytic center of both proteinases consists of His and Cys residues in all coronaviruses (reviewed in Lim et al., 2000, Ng and Liu, 2000). The processed products constitute a group of replicative enzymes, including the RNA-dependent RNA polymerase (RdRp) and RNA helicase. Therefore, the significance of the coronavirus leader sequence and several putative functional domains essential for viral replication made them logical choices for sequence analysis and phylogenetic comparisons to correlate with the serological or pathological classification of coronaviruses.
Phylogeny-based clustering based on replicase genes is identical to the original antigen-based classification of coronaviruses Chouljenko et al., 2001, Hegyi and Ziebuhr, 2002, Stephensen et al., 1999. However, within the group 3 coronaviruses, no comparisons of replicase-based and antigen-based phylogenies have been made. One report has described the alignment of the 5′-proximal region of replicase genes from a limited number of IBV strains (Stirrups et al., 2000), and a few reports describe IBV Mpro and RdRp domains from the Beaudette strain only Hegyi and Ziebuhr, 2002, Stephensen et al., 1999. Here, we have sequenced four regions of the replicase gene corresponding to the 5′-terminal end, PLpro, Mpro, and RdRp domains from 20 heterologous IBV strains, and compared then with previously published IBV and other coronavirus sequences. We present evidence that (i) the Mpro and RdRp regions are highly conserved among all coronaviruses unlike the 5′-terminal sequence and PLpro domains, (ii) the clustering of heterologous IBV strains based on the replicase gene sequences does not correlate with the antigen-based S1 phylogeny, (iii) several widely used IBV vaccines and field strains isolated over the past decade have closely related replicase genes suggestive of a possible common ancestry or a converging evolution, and (iv) the replicase gene is unlikely to be the sole determinant of IBV pathogenicity.
Results
The 5′-terminal sequences (nt 1–1263)
The nt sequence alignment data obtained from the IBV 5′-termini are presented in Table 1 , Fig. 1, Fig. 2 . The leader sequences (nt 1–64) of H52, CU705, CU510, Ark99, ArkDPI, CU805, CU570, GA98, Cal99, CU994, K1699, Gray, and BJ strains are identical to each other and each have 3 nt substitutions compared to the Beaudette strain (Fig. 1). The DE072 virus has four, the vaccine and challenge M41 strains, and CU-T2 have three, Ma5, K0751, and Florida have two, and Conn has a single nt substitution compared to the Beaudette strain (Fig. 1). The leader junction sequences termed as leader transcription-regulating sequences (TRSs) are absolutely conserved in all of the IBV strains compared. The sequence distance data (Table 1) obtained from the whole 5′ UTR (nt 1–528) indicate that nt sequence identities were 94–100% among all IBV strains in this region. The N-terminal coding region (nt 529–1263) of the 5′ terminal sequence is more variable with the lowest identity represented by the BJ strain at 84.6–86% identity with the other strains. Pairwise comparisons of the N-terminal sequences from other strains revealed 89.7–100% identity irrespective of serotype, geographic source, and virulence (Table 1). The challenge strain NV M41 is identical to the vaccine strain M41 in the noncoding region but in the coding region has a single nt change that results in an amino acid change (data not shown). These relationships are graphically shown in the phylogenetic tree created from the nt sequences of the entire 5′ terminal region (Fig. 2a) . No detectable sequence similarity was found when IBV 5′ terminal sequences were compared to those of coronaviruses from other groups (data not shown).
Table 1.
Nucleotide identities in the 5′ UTR (nt 1–528: right-hand side) and the N-terminal coding region (nt 529–1263) of the replicase gene of 21 IBV strains
| Virus | 5′ UTR |
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beau | M41a | Ma5 | H52 | K0 751 | CU 705 | Conn | CU 510 | Flori | CU T2 | Ark 99 | Ark DPI | DE 072 | CU 805 | CU 570 | GA 98 | Cal 99 | CU 994 | K1 699 | Gra | BJ | |
| Beau | 98.7 | 97.2 | 96.2 | 96.6 | 96.4 | 97.0 | 96.2 | 96.4 | 96.2 | 96.0 | 96.2 | 95.8 | 96.2 | 96.4 | 96.0 | 95.6 | 96.0 | 94.3 | 95.8 | 94.7 | |
| M41a | 92.9 | 96.8 | 97.2 | 97.5 | 97.3 | 97.0 | 97.2 | 96.8 | 97.2 | 96.6 | 97.2 | 96.8 | 97.2 | 97.3 | 97.0 | 96.6 | 97.0 | 95.3 | 96.8 | 94.9 | |
| Ma5 | 94.6 | 93.3 | 99.6 | 99.2 | 98.7 | 97.2 | 99.2 | 96.6 | 98.5 | 97.2 | 99.6 | 99.2 | 98.9 | 99.4 | 99.4 | 99.1 | 99.4 | 96.0 | 99.2 | 95.1 | |
| H52 | 94.6 | 93.3 | 100 | 99.6 | 99.1 | 97.2 | 99.6 | 97.0 | 98.9 | 97.5 | 100 | 99.6 | 99.2 | 99.8 | 99.8 | 99.4 | 99.8 | 96.4 | 99.6 | 95.5 | |
| K0751 | 94.1 | 93.2 | 99.3 | 99.3 | 99.1 | 97.5 | 99.2 | 97.3 | 99.2 | 97.5 | 99.6 | 99.2 | 99.2 | 99.4 | 99.4 | 99.1 | 99.4 | 96.2 | 99.2 | 95.3 | |
| CU705 | 94.0 | 92.8 | 98.6 | 98.6 | 98.0 | 97.3 | 98.7 | 97.2 | 99.1 | 97.7 | 99.1 | 98.7 | 99.4 | 98.9 | 98.9 | 98.5 | 98.9 | 96.8 | 98.7 | 95.8 | |
| Conn | 94.4 | 93.3 | 98.2 | 98.2 | 97.6 | 97.1 | 97.2 | 99.4 | 97.2 | 97.3 | 97.2 | 96.8 | 97.2 | 97.0 | 97.0 | 96.6 | 97.0 | 95.1 | 96.8 | 94.2 | |
| CU510 | 94.4 | 93.2 | 99.7 | 99.7 | 99.0 | 98.4 | 98.0 | 97.0 | 98.5 | 97.2 | 99.6 | 99.2 | 98.9 | 99.4 | 99.4 | 99.2 | 99.4 | 96.0 | 99.2 | 95.1 | |
| Florida | 94.4 | 93.3 | 98.2 | 98.2 | 97.6 | 97.1 | 99.7 | 98.0 | 97.0 | 97.2 | 97.0 | 96.6 | 97.0 | 96.8 | 96.8 | 96.4 | 96.8 | 94.9 | 96.6 | 94.0 | |
| CUT2 | 93.9 | 92.9 | 99.0 | 99.0 | 98.4 | 97.7 | 97.6 | 98.8 | 97.6 | 97.2 | 98.9 | 98.5 | 98.9 | 98.7 | 98.7 | 98.3 | 98.7 | 96.2 | 98.5 | 95.3 | |
| Ark99 | 94.8 | 92.8 | 97.0 | 97.0 | 96.6 | 95.6 | 97.4 | 96.6 | 97.4 | 96.3 | 97.5 | 97.2 | 97.7 | 97.3 | 97.3 | 97.0 | 97.3 | 96.0 | 97.2 | 94.9 | |
| ADPI | 94.7 | 93.2 | 99.9 | 99.9 | 99.2 | 98.8 | 98.1 | 99.6 | 98.1 | 98.9 | 96.9 | 99.6 | 99.2 | 99.8 | 99.8 | 99.4 | 99.8 | 96.4 | 99.6 | 95.5 | |
| DE072 | 94.6 | 93.3 | 100 | 100 | 99.3 | 98.6 | 98.2 | 99.7 | 98.2 | 99.0 | 97.0 | 99.9 | 98.9 | 99.4 | 99.7 | 99.1 | 99.4 | 96.0 | 99.2 | 95.1 | |
| CU805 | 93.5 | 92.5 | 97.8 | 97.8 | 97.1 | 96.7 | 96.3 | 97.7 | 96.3 | 96.9 | 95.6 | 97.7 | 97.8 | 99.1 | 99.1 | 98.7 | 99.1 | 96.6 | 98.9 | 96.0 | |
| CU570 | 94.0 | 92.8 | 99.2 | 99.2 | 98.5 | 98.6 | 97.4 | 98.9 | 97.4 | 98.2 | 96.2 | 99.3 | 99.2 | 97.3 | 99.6 | 99.2 | 99.6 | 96.2 | 99.5 | 95.5 | |
| GA98 | 94.6 | 93.1 | 99.7 | 99.7 | 99.0 | 98.6 | 98.2 | 99.5 | 98.2 | 98.8 | 97.0 | 99.9 | 99.7 | 97.6 | 99.2 | 99.2 | 99.6 | 96.2 | 99.3 | 95.3 | |
| Cal99 | 94.8 | 93.6 | 99.7 | 99.7 | 99.0 | 98.4 | 98.5 | 99.1 | 98.5 | 98.8 | 97.3 | 99.6 | 99.7 | 97.6 | 98.9 | 99.5 | 99.2 | 95.8 | 99.1 | 95.1 | |
| CU994 | 94.4 | 93.2 | 99.6 | 99.6 | 98.9 | 98.2 | 98.1 | 99.3 | 98.1 | 98.6 | 96.9 | 99.5 | 99.6 | 97.4 | 98.8 | 99.3 | 99.6 | 96.2 | 99.4 | 95.3 | |
| K1699 | 90.8 | 89.7 | 92.1 | 92.1 | 91.9 | 92.1 | 91.5 | 92.0 | 91.5 | 91.3 | 91.2 | 92.3 | 92.1 | 91.6 | 92.3 | 92.1 | 92.3 | 92.0 | 96.0 | 95.7 | |
| Gray | 94.6 | 92.8 | 99.5 | 99.5 | 98.8 | 98.4 | 98.0 | 99.2 | 98.0 | 98.5 | 96.7 | 99.6 | 99.5 | 97.3 | 98.9 | 99.7 | 99.2 | 99.0 | 91.9 | 95.1 | |
| BJ | 84.8 | 84.6 | 85.8 | 85.8 | 85.5 | 84.6 | 85.6 | 85.8 | 85.6 | 85.4 | 85.0 | 85.6 | 85.8 | 85.2 | 85.2 | 95.5 | 85.9 | 85.6 | 86.0 | 85.2 | |
| N-terminal coding region |
|||||||||||||||||||||
Sequences with >95% identity are indicated in bold letters.
Because of the overall sequence identity of the vaccine and challenge strains of M41, for brevity, only the vaccine strain is included in this table.
Fig. 1.
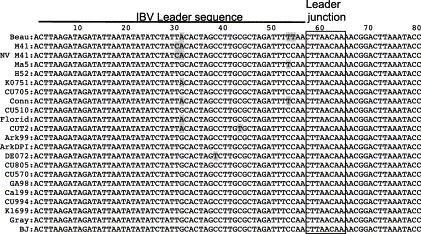
Alignment of the first 80 nucleotides from cDNA sequences derived from the 5′ end of 22 heterologous IBV strains. The gray shading represents differences between sequences. The leader sequence is denoted by a solid black line above the sequence and the leader junction sequence is boxed.
Fig. 2.
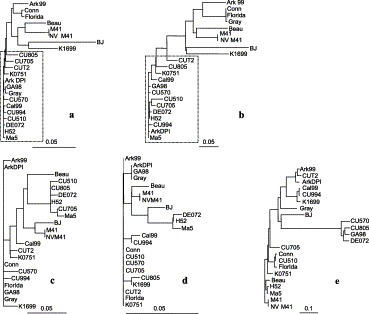
Phylogenetic trees showing the relatedness among the 5′-terminal sequences (a), PLpro (b), Mpro (c), RdRp (d) domains and S1 glycoprotein genes (e) of 22 heterologous IBV strains. A cluster of contemporary field isolates and vaccine strains is indicated with a dashed box (⋯ ⋯) in a and in b. The neighbor-joining tree was constructed from the pairwise nucleotide differences for 5′-termini and deduced amino acid differences in all other respective genes. The length of each pair of branches represents the distance between sequence pairs. The scale at the bottom indicates the number of substitution events.
The PLpro domains (nt 4243–5553: aa 1239–1675)
Sequence distance data derived from multiple alignments of the two overlapping PLpro domains of IBV strains revealed 77.6–100% identity in PL1pro and 79–100% in PL2pro, with the former being more variable than the latter (Table 2) . The BJ strain differed the most from other strains by 18.1–22.4% in PL1pro and 16.5–21% in PL2pro. The catalytic Cys1274 and His1437 residues in the PLpro domains are conserved in all IBV strains (Fig. 3) . In the overlapping area of the PLpro domains, a region of 24 residues with more heterogeneity has been identified, where only two residues (Phe1470 and Lys1485) are conserved among all IBV strains (Fig. 3). In this heterogeneous region, the vaccine and challenge M41 and BJ strains each differed by 12 aa (50%), Conn, CU510, Florida, CU805, and Gray each differed by 11 aa (45%), CU705, Ark99, and Cal99 strains each differed by 10 aa (40%), and all other strains are identical to each other and differed by 9 aa (37.5%) when compared to the Beaudette strain sequence. In the complete PLpro domain, the NV M41 challenge strain has 1 nt and 1 aa difference from the M41 vaccine strain. The relationships between the different sequences are shown in the phylogenetic tree derived from analysis of the aa sequences of the entire PLpro region (aa 1239–1675) (Fig. 2b). No detectable sequence similarity was found when IBV PLpro sequences were compared to the PLpro sequences of coronaviruses from other groups (data not shown).
Table 2.
Amino acid identities in PL1pro (aa 1239–1497) and PL2pro (aa 1385–1675) of the replicase gene of 21 IBV strains
| Virus | PLPD-1 |
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beau | M41a | Ma5 | H52 | K0 751 | CU 705 | Conn | CU 510 | Flori | CUT2 | Ark99 | Ark DPI | DE 072 | CU 805 | CU 570 | GA 98 | Cal99 | CU 994 | K1 699 | Gra | BJ | |
| Beau | 88.8 | 83.4 | 83.4 | 82.6 | 81.5 | 87.3 | 81.9 | 87.3 | 85.3 | 87.3 | 83.0 | 83.0 | 82.2 | 83.0 | 83.0 | 81.9 | 83.0 | 85.7 | 87.3 | 80.7 | |
| M41a | 89.0 | 84.6 | 84.6 | 83.4 | 82.6 | 85.7 | 82.2 | 85.7 | 86.9 | 90.7 | 84.2 | 84.2 | 82.6 | 84.2 | 84.2 | 83.0 | 84.2 | 84.9 | 85.7 | 81.9 | |
| Ma5 | 84.2 | 86.9 | 100 | 96.5 | 98.1 | 84.2 | 96.5 | 84.2 | 91.1 | 84.9 | 99.6 | 99.2 | 95.8 | 99.6 | 99.6 | 97.3 | 99.6 | 86.9 | 84.2 | 80.3 | |
| H52 | 83.8 | 86.6 | 99.7 | 96.5 | 98.1 | 84.2 | 96.5 | 84.2 | 91.1 | 84.9 | 99.6 | 99.2 | 95.8 | 99.6 | 99.6 | 97.3 | 99.6 | 86.9 | 84.2 | 80.3 | |
| K0751 | 83.8 | 86.6 | 99.3 | 99.0 | 95.4 | 86.9 | 93.8 | 86.9 | 93.8 | 86.1 | 96.1 | 95.8 | 97.7 | 96.7 | 96.1 | 96.1 | 96.1 | 89.6 | 86.9 | 79.9 | |
| CU705 | 83.2 | 85.9 | 99.0 | 99.3 | 98.3 | 82.2 | 96.1 | 82.2 | 89.2 | 83.8 | 97.9 | 97.3 | 94.6 | 97.7 | 97.7 | 95.4 | 97.7 | 84.9 | 82.2 | 79.9 | |
| Conn | 87.3 | 87.3 | 84.9 | 84.5 | 84.5 | 83.8 | 81.7 | 100 | 91.1 | 88.0 | 83.8 | 83.8 | 86.1 | 84.6 | 83.3 | 84.9 | 83.8 | 95.3 | 100 | 77.6 | |
| CU510 | 82.8 | 84.9 | 96.6 | 96.9 | 95.9 | 96.9 | 82.8 | 81.9 | 87.6 | 83.8 | 96.1 | 98.5 | 93.1 | 96.1 | 96.1 | 93.8 | 96.1 | 83.8 | 81.9 | 78.4 | |
| Florida | 87.3 | 87.3 | 84.9 | 84.5 | 84.5 | 83.8 | 99.9 | 82.8 | 91.1 | 88.0 | 83.8 | 83.8 | 86.1 | 94.6 | 83.3 | 84.9 | 83.8 | 95.3 | 100 | 77.6 | |
| CUT2 | 84.9 | 87.3 | 97.6 | 97.9 | 96.9 | 97.3 | 85.2 | 94.8 | 85.2 | 88.0 | 90.7 | 90.7 | 93.1 | 91.5 | 90.7 | 90.7 | 90.7 | 93.1 | 91.1 | 80.7 | |
| Ark99 | 87.6 | 89.7 | 84.5 | 84.2 | 84.2 | 83.5 | 92.8 | 82.8 | 92.8 | 84.9 | 84.6 | 84.6 | 84.6 | 85.3 | 84.6 | 84.9 | 84.9 | 87.6 | 88.0 | 81.5 | |
| ADPI | 84.2 | 86.9 | 100 | 99.7 | 99.3 | 99.0 | 84.9 | 96.6 | 84.9 | 97.6 | 84.5 | 98.8 | 95.4 | 99.2 | 99.2 | 96.9 | 99.2 | 86.5 | 83.8 | 80.3 | |
| DE072 | 83.5 | 86.3 | 99.0 | 99.3 | 98.3 | 98.6 | 84.2 | 96.2 | 84.2 | 97.6 | 83.8 | 99.0 | 95.0 | 98.8 | 99.3 | 96.5 | 98.8 | 86.5 | 83.8 | 79.9 | |
| CU805 | 82.5 | 84.9 | 97.3 | 97.3 | 96.6 | 97.6 | 82.8 | 95.5 | 82.8 | 95.5 | 82.5 | 97.3 | 96.9 | 96.1 | 95.4 | 95.4 | 95.4 | 88.8 | 86.1 | 78.8 | |
| CU570 | 83.8 | 86.6 | 99.7 | 99.3 | 99.0 | 98.6 | 84.5 | 96.2 | 84.5 | 97.3 | 84.2 | 99.7 | 98.6 | 96.9 | 99.2 | 96.9 | 99.2 | 87.3 | 84.6 | 79.9 | |
| GA98 | 83.8 | 86.6 | 99.7 | 99.3 | 99.0 | 98.6 | 84.5 | 96.2 | 84.5 | 97.3 | 84.2 | 99.7 | 98.6 | 96.9 | 99.3 | 97.7 | 99.2 | 86.5 | 83.8 | 79.9 | |
| Cal99 | 83.8 | 86.6 | 99.3 | 99.0 | 98.6 | 98.3 | 85.2 | 95.9 | 85.2 | 96.9 | 84.9 | 99.3 | 98.3 | 96.6 | 99.0 | 99.0 | 96.9 | 86.9 | 84.9 | 78.0 | |
| CU994 | 83.2 | 85.9 | 99.0 | 98.6 | 98.3 | 97.9 | 84.5 | 95.5 | 84.5 | 96.6 | 83.5 | 99.0 | 97.9 | 96.2 | 98.0 | 98.6 | 98.3 | 86.9 | 83.8 | 80.3 | |
| K1699 | 81.4 | 82.8 | 84.2 | 83.8 | 83.6 | 83.2 | 82.8 | 82.1 | 82.8 | 84.9 | 83.5 | 84.2 | 83.5 | 82.5 | 83.8 | 83.8 | 83.8 | 84.2 | 90.3 | 80.7 | |
| Gray | 86.9 | 86.9 | 84.5 | 84.2 | 84.2 | 83.5 | 99.7 | 82.5 | 99.7 | 84.9 | 92.4 | 84.5 | 83.8 | 82.8 | 84.2 | 84.2 | 84.9 | 84.2 | 82.5 | 77.6 | |
| BJ | 81.1 | 83.5 | 81.4 | 81.1 | 80.8 | 80.4 | 80.8 | 79.0 | 80.8 | 82.1 | 82.5 | 81.4 | 80.8 | 79.7 | 81.1 | 81.1 | 81.1 | 81.4 | 82.1 | 80.4 | |
| PLPD-2 |
|||||||||||||||||||||
Sequences with >95% identity are indicated in bold letters.
Because of the overall sequence identity of the vaccine and challenge strains of M41, for brevity, only the vaccine strain is included in this table.
Fig. 3.

Amino acid sequence alignment of the papain-like proteinase domains of 22 IBV strains indicating the catalytic Cys1274 and His1437 residues marked as asterisks (*) and the more variable region marked by thick bar (—) below the alignment. Residue numbers are given above the sequence; four-digit numbers are ended at the labeled residue. The internal sequences not presented in this alignment are indicated by dashes (–––).
The Mpro (aa ORF1a 2804–2943) and the RdRp (aa ORF1b 548–780) domains
The alignments of the deduced aa sequences of IBV Mpro and RdRp domains are summarized in Table 3 . The overall sequences are highly conserved in which there are 82.7% (116 aa) absolutely conserved residues in Mpro and 92.7% (216 aa) absolutely conserved residues in RdRp (data not shown). The remaining 24 aa differences found in Mpro and 17 aa differences in RdRp are distributed throughout the domains (Table 3). The Mpro and RdRp domains of ArkDPI, GA98, and Gray strains are identical to each other and those of the challenge NV M41 and vaccine M41 strains are also identical to each other. These relationships are graphically shown in the phylogenetic tree created from the aa sequences of the Mpro and RdRp domains (Figs. 2c and d).
Table 3.
Amino acid exchanges in the main protease (aa ORF1a 2804–2943) and the RNA-dependent RNA polymerase (aa ORF1b 548–780) domains among 21 IBV strains
| Strains | Exchange of aa in the Mpro at positiona |
Exchange of aa in the RdRp at positiona |
|||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2814 | 2815 | 2829 | 2831 | 2835 | 2840 | 2845 | 2847 | 2848 | 2849 | 2851 | 2855 | 2860 | 2874 | 2881 | 2894 | 2898 | 2900 | 2906 | 2925 | 2928 | 2929 | 2935 | 2937 | 569 | 572 | 592 | 596 | 606 | 608 | 611 | 674 | 678 | 681 | 698 | 708 | 744 | 752 | 754 | 756 | 765 | |
| Beau | T | I | Q | N | N | H | T | Q | H | G | T | V | K | E | I | A | T | V | T | V | N | I | N | F | E | I | A | V | S | S | I | I | N | S | P | L | V | F | A | S | E |
| M41b | S | . | . | G | . | . | V | . | N | . | . | . | . | . | V | S | . | I | . | . | . | . | . | . | . | . | . | . | T | . | V | D | . | . | . | . | . | . | . | . | |
| Ma5 | . | . | . | . | . | Y | V | . | N | N | . | . | . | . | V | S | . | . | I | P | . | . | . | Y | . | . | . | . | T | P | . | . | D | D | S | . | I | Y | . | P | . |
| H52 | . | . | . | . | . | . | V | . | N | N | . | . | . | . | V | S | . | . | . | P | . | . | . | Y | . | . | . | . | T | P | . | . | D | D | S | . | I | Y | S | . | . |
| K0751 | . | . | . | S | . | Y | V | . | N | . | . | . | R | D | V | S | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | D | . | . | . | . | . | S | . | . |
| CU705 | . | . | . | . | . | . | V | . | N | N | . | . | . | . | V | S | . | . | I | P | . | . | . | Y | . | . | . | . | T | . | . | . | D | . | . | . | . | . | S | . | . |
| Conn | . | . | . | S | . | . | A | . | N | . | . | . | R | D | V | S | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | D | . | . | . | . | . | S | . | . | |
| CU510 | . | . | . | . | . | . | V | . | N | N | . | . | . | . | V | S | . | . | . | P | . | V | . | Y | . | . | . | . | T | . | . | . | D | . | . | . | . | . | S | . | . |
| Florida | . | . | . | S | . | . | A | . | N | . | . | . | R | D | V | S | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | D | . | . | . | . | . | S | . | . |
| CUT2 | . | . | H | S | . | . | V | . | N | . | . | . | R | D | V | S | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | D | . | . | . | . | . | S | . | . |
| Ark99 | . | . | . | S | . | . | A | . | N | . | I | . | R | D | V | S | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | D | . | . | . | . | . | S | . | . |
| ArkDPI | . | . | . | S | . | . | A | . | N | . | . | . | R | D | V | S | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | D | . | . | . | . | . | S | A | . |
| DE072 | . | . | . | . | Y | . | V | . | N | N | . | . | . | . | V | S | . | . | . | P | . | . | . | Y | . | . | . | . | T | P | . | . | D | D | S | . | I | Y | . | . | G |
| CU805 | . | . | . | . | . | . | V | . | N | N | . | . | . | . | V | S | . | . | . | P | . | . | . | Y | . | . | . | I | T | . | . | V | D | . | . | . | . | . | S | . | . |
| CU570 | . | . | . | S | . | . | A | . | N | . | . | . | R | D | V | S | S | . | . | . | . | . | S | . | . | . | . | . | T | . | . | . | D | . | . | . | . | . | S | . | . |
| GA98 | . | . | . | S | . | . | A | . | N | . | . | . | R | D | V | S | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | D | . | . | . | . | . | S | A | . |
| Cal99 | . | . | . | . | . | . | V | . | N | N | . | . | R | D | V | S | . | . | . | . | . | . | . | . | A | . | V | . | T | . | . | . | D | . | . | . | . | . | S | . | . |
| CU994 | . | . | . | S | . | . | A | . | N | . | . | R | D | V | S | . | . | . | . | T | . | . | . | . | . | V | . | T | . | . | . | D | . | . | . | . | . | S | . | . | |
| K1699 | . | V | . | S | . | . | A | . | N | . | . | I | R | D | V | S | . | . | . | . | . | . | . | . | . | . | . | I | T | . | . | . | D | . | . | . | . | . | S | . | . |
| Gray | . | . | . | S | . | . | A | . | N | . | . | . | R | D | V | S | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | D | . | . | . | . | . | S | A | . |
| BJ | S | . | . | D | . | . | V | G | N | . | . | . | R | . | M | S | . | I | . | . | . | . | . | . | . | M | . | . | T | . | . | . | D | . | . | M | I | Y | . | . | . |
Amino acid residues identical to that of Beaudette virus are indicated as dots (.).
Because of the overall sequence identity of the vaccine and challenge strains of M41, for brevity, only the vaccine strain is included in this table.
By aligning with the corresponding regions of prototype members of other coronavirus groups, all Mpro had 28% (40 aa) absolutely conserved residues including catalytic Cys-His dyad and three substrate-binding sites (Tyr-Met-His), and the RdRp had 47.6% (111 aa) absolutely conserved residues (data not shown). The four RdRp motifs (DXXXD, SGXXXTXXXN, SDD, and K) appear to be fully conserved in all coronaviruses. The IBV Mpro domains have 38.7–40.1% similarity with 229E, 45.1–45.8% with TGEV, 38.5–41.5% with MHV, 39.2–41.5% with BoCV, and 43.4–44.8% with the SARS coronavirus in pairwise comparisons. Whereas the IBV RdRp has 60.9–62.7% similarity with 229E, 60.4–61.8% with TGEV, 66.5–68.7% with MHV, 66.1–68.2% with BoCV, and 66.1–68.2% with SARS coronavirus. The phylogenetic trees created from both Mpro and RdRp of various coronaviruses cluster into the three major antigenic groups (data not shown).
Discussion
The findings of the present study can be summarized as: (i) the Mpro and RdRp regions are highly conserved among all coronaviruses unlike the 5′-terminal sequence and PLpro domains, (ii) the clustering of heterologous IBV strains based on the replicase gene sequences does not correlate with the antigen-based S1 phylogeny, (iii) several widely used IBV vaccines and field strains isolated over the past decade have closely related replicase genes suggestive of a possible common ancestry or a converging evolution, and (iv) the replicase gene is unlikely to be the sole determinant of IBV pathogenicity.
Coronaviruses fall into three antigenic groups (Dea et al., 1990), which are defined by cross-reactivity among the structural proteins. Unlike the structural proteins, the coronavirus replicase gene is not subjected to immune selective pressure. Therefore, one would expect that the replicase gene would be highly conserved among coronavirus groups. In fact, this is the case when the Mpro and RdRp sequences are compared (Table 3) and as others have reported for other coronavirus groups Chouljenko et al., 2001, Gonzalez et al., 2003, Hegyi and Ziebuhr, 2002, Stephensen et al., 1999. We found a high percentage of sequence similarities in coronavirus Mpro (with 28% absolutely conserved residues), including the substrate recognition site, and an even higher similarity in the RdRp (with 48% absolutely conserved residues), including the polymerase motifs. However, two-way sequence comparisons of the coronavirus 5′-termini and the PLpro domains of ORF1a had very low sequence identity between coronaviruses of different groups as has been reported by others (Herold et al., 1999). Despite the fact that these regions have varying degrees of genetic diversity, the relationships between the coronaviruses of different groups are the same, no matter what region of the genome is compared. When recombination occurs (Cavanagh and Davis, 1988), it would seem to occur within the major coronaviruses antigenic groups, but not between them. This is expected because in order for recombination to occur, two viruses must be present in the same cell, which is unlikely between coronaviruses adapted to different host species. The highly conserved Mpro-mediated processing pathways as well as the substrate binding sites of this enzyme in all coronaviruses Anand et al., 2003, Hegyi and Ziebuhr, 2002 make this proteinase an attractive target for the development of drugs directed against coronaviruses. The active domains of RdRp also maintain a high degree of homology among all coronaviruses, as has been found in other RNA viruses Koonin and Dolja, 1993, Otsuka et al., 1999, making this proteinase another potential target for drugs controlling coronaviruses.
With IBV, the phylogenies of the four regions of the replicase gene did not correlate with those of the S1 gene, which correspond to the type-specific antigenic groups (Fig. 2). For example, unlike the S1 genes, the replicase genes of the Ma5 and H52 Mass serotype viruses consistently clustered with those of non-Mass strains (Figs. 2a–d). The common clustering of replicase genes of these heterologous IBV strains could be due to an introduction of a replicase gene from a common source through introduction. Others have speculated that IBV strains substitute large genomic fragments in multiple genes (Lee and Jackwood, 2000) because the viability of the progeny viruses depends on the specific interactions of more than one gene working in concert to maintain structural and replicative integrity. Alternatively, it is probably more likely that the smaller S1 genes are exchanged between viruses through recombination and the stable replicase backbone is maintained. It is entirely possible that recombination events have occurred to introduce new replicase or new S1 genes into these viruses.
Leader acquisition in mRNA synthesis appears to be TRS dependent, and the leader-to-body joining is guided by a base-pairing interaction involving leader and body TRSs Hiscox et al., 1995, Pasternak et al., 2001. The phenomenon of leader switching involves a recombination event over a region comprising the leader sequence and the adjacent 5′ UTR (Chang et al., 1996). Previous reports have suggested that the leader sequences of IBV strains may contain higher (17.2%) substitution rates when compared to the complete 5′ UTR (4.3%; Stirrups et al., 2000). In our study, the 5′ UTR of all 22 IBV strains are highly conserved (94–100% identity), indicating the exchange of leader or adjacent 5′ UTR fragments has occurred in mixed infections. This observation is supported by experimental evidence of the exchange of leader sequences among heterologous IBV strains (Stirrups et al., 2000) and in mixed MHV infections (Makino et al., 1986). The sequence comparison scores are more variable in the N-terminal coding region (Table 1), and the most in PLpro domains (Table 2) including a more variable region in the latter (Fig. 3) than the IBV Mpro and RdRp domains (Table 3). The reason for sequence variation in the N-terminal half of ORF1a among IBV strains is not clear.
The genetic variations in the 5′ terminal region and PLpro gene suggest that there is some selective pressure on these regions. We speculate that there are some replicase genotypes that may confer a selective advantage to the virus. We examined both the 5′ terminal region and the PLpro gene by year and location of isolation, but excluded the Mpro and RdRp genes because they are so highly conserved that no clear differences between isolates can be discerned. In the 5′ terminal region and the PLpro gene, the IBV strains isolated from commercial chickens over the past decade cluster with three specific vaccine virus strains, ArkDPI, H52, and Ma5, no matter where in the United States they were isolated. There are three exceptions to this clustering, the 5′ terminal region of the Gray virus isolated in 1960 (Winterfield and Hitchner, 1962) clusters with modern field strains (Fig. 2a), although its PLpro gene is more like the Conn and Florida reference viruses isolated in 1956 and 1971, respectively Jungherr et al., 1956, Winterfield et al., 1971 (Fig. 2b). In addition, there are two modern field isolates, one from China (BJ) and one from California (K1699) isolated in 2003 and 2001, respectively, that are outliers and do not appear to have 5′ termini or PLpro genes that are related to other viruses (Figs. 2a and b). This common clustering of three widely used vaccines and IBV field isolates from the past decade suggests to us that there may be a common evolutionary progression in the 5′ terminal region and in the PLpro gene. Although the 5′ terminal region genotype found in modern isolates was present in 1960, when the Gray virus was isolated, it appears to be more prevalent today. The question arises as to why this type of progression to a specific genotype would have occurred and how it could have happened. As to the question of why, we speculate that the more modern genotype must offer some fitness advantage to the virus. As to how it could have occurred, we consider the widespread use of multiple types of modified live vaccines in both the broiler and egg layer commercial poultry industries as the possible mechanism by which this modern replicase gene genotype was introduced. The Ark, Conn, and Mass (Ma5, H52) strains have been used simultaneously as live vaccines among commercial poultry (Cavanagh and Naqi, 2003). Although we cannot prove or disprove that recombination has occurred, we can point to the evolutionary trends in the S1 gene as compared to the evolutionary trends in the replicase gene. The trends in S1 evolution have been toward ever increasing diversity (Jia et al., 2002), while it appears that the replicase gene is evolving to less variety between isolates. These juxtaposed evolutionary trends suggest that (1) these two regions are under very different selective pressures and thus undergo much different rates and modes of change, and (2) the exchange of genetic material may have occurred between vaccine strains and field isolates.
The genetic basis of IBV pathogenicity is not known. The role of the replicase gene in the virulence of IBV has not been established, and it is likely that some changes in the Mpro or viral polymerase would influence viral replication rate, and thus the pathogenic potential of a virus. In some RNA viruses, replicase genes are known to contain the determinants of species tropism and virulence factors Brandt et al., 2001, Hatta et al., 2001, Yao et al., 2001. In our study, there were no differences in any essential residues in Mpro and RdRp domains. In addition, we found that there was no clear relationship between viral pathotype and replicase genotype. As an example, the H52 vaccine strain is very closely related to the virulent DE072 isolate in all regions of the replicase (99.8% identity in the 5′ terminus, 99.5% in PLpro, 99.3% in Mpro, 99.6% in RdRp) despite their differences in pathotype. In addition, a direct comparison of the replicase genes of the M41 challenge and vaccine strains where there were very few aa differences between the two strains with clearly different pathotypes. None of the aa changes between the two M41 strains were consistently observed in either all virulent or all avirulent strains. These findings would support our contention that the genotype of the replicase is not the sole determinant of pathogenicity among strains of IBV.
Materials and methods
Experimental design
Twenty IBV strains from nine different serotypes, Mass (M41, NV M41, Ma5, H52, K0751, CU705), Conn (Conn, CU510), Florida, CU-T2, Arkansas (Ark-99, ArkDPI), Delaware 072 (DE072, CU805, CU570), Georgia 98 (GA98), California 99 (Cal99, CU994, K1699), and Gray, were used in this study (Table 4) . These virus strains were selected because they are very common vaccine and field strains that are frequently isolated from commercial chickens and are associated with clinical disease. All the reference IBV strains were obtained from Dr. Syed Naqi's laboratory (Cornell University, Ithaca), where they had been propagated in specific pathogen-free (SPF) embryonated chicken eggs to maintain the stock. In our laboratory, all of the viruses were also propagated in 9-day-old embryonated SPF eggs (Charles River SPAFAS, Wilmington, MA). Four regions of the replicase gene from 20 heterologous IBV strains, which correspond to the 5′-termini, and the PLpro, the Mpro, and the RdRp domains (Fig. 4) , were sequenced and compared with those of previously published IBV (Beaudette, BJ) and other coronavirus sequences.
Table 4.
IBV strains examined in this study
| Straina | Serotype | Geographic origin | Original description or source (reference) | GenBank accession no.b |
|---|---|---|---|---|
| Beaudette | Massachusetts | New Jersey, USA | Laboratory strainc | M95169 |
| M41 | Massachusetts | Massachusetts, USA | Jungherr et al. (1956) | AY561711 |
| NV M41 | Massachusetts | USA | Challenge strain (NVSL, USDA) | AY561712 |
| Ma5 | Massachusetts | Europe | Vaccine straind (Intervet Inc.) | AY561713 |
| H52 | Massachusetts | Europe | Vaccine straind (Intervet Inc.) | AF352315 |
| K0751 | Massachusetts | California, USA | Field isolatee (UCDavis, 2001) | AY561714 |
| CU705 | Massachusetts | New York, USA | Field isolatee (Cornell Univ., 2002) | AY561715 |
| Conn | Connecticut | Connecticut, USA | Jungherr et al. (1956) | L18990 |
| CU510 | Connecticut | New York, USA | Field isolatee (Cornell Univ., 2002) | AY561716 |
| Florida | Florida | Florida, USA | Winterfield et al. (1971) | AF027512 |
| CU-T2 | CU-T2 | New York, USA | Jia et al. (1995) | U04739 |
| Ark99 | Arkansas | Arkansas, USA | Johnson et al. (1973) | L10384 |
| ArkDPI | Arkansas | Delmarva, USA | Gelb et al. (1981) | AF006624 |
| DE072 | DE072 | Delmarva, USA | Gelb et al. (1997) | U77298 |
| CU805 | DE072 | New York, USA | Mondal et al. (2001) | AF317215 |
| CU570 | DE072 | New York, USA | Field isolatee (Cornell Univ., 2003) | AY561717 |
| GA98 | GA 98 | Georgia, USA | Lee and Jackwood (2001) | AF274437 |
| Cal99 | Cal99 | California, USA | Martin et al. (2001) | AY514485 |
| CU994 | Cal99 | New Mexico, USA | Field isolatee (Cornell Univ., 2000) | AF317499 |
| K1699 | Cal99 | California, USA | Field isolatee (UCDavis, 2001) | AY561718 |
| Gray | Gray | Delmarva, USA | Winterfield and Hitchner (1962) | L14069 |
| BJ | Unknown | China | GenBank (China, 2003) | AY319651 |
Except for the Gray strain that is nephrotropic and the BJ strain whose tissue tropism is unknown, all other strains are pneumotropic.
Based on S1 gene sequence.
Extensively propagated in vitro since the first isolation (Beaudette and Hudson, 1937) and known to be attenuated.
Introduced to the US market at about 1990 (Ma5) and 1980 (H52).
IBV recovered from infected chicken flocks and associated with clinical disease and their serotype determined by S1 sequence.
Fig. 4.
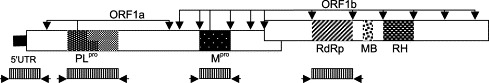
Diagram of the coronavirus replicase gene (encoding ORF 1a and 1b) illustrating the 5′ untranslated region (UTR), papain-like proteinase (PLpro), main protease (Mpro), RNA-dependent RNA polymerase (RdRp), metal-binding (MB), and RNA helicase (RH) domains. The arrows originating from PLpro and Mpro indicate the cleavage sites. The four regions amplified in this study are indicated as shading areas with arrow at each end (bottom).
Viral RNA extraction, RT-PCR amplification and sequencing
Genomic RNA was extracted from virus-inoculated allantoic fluid with TRIzol reagent (Invitrogen Co., Carlsbad, CA) following the manufacturer's instructions. The first strand cDNA synthesis and subsequent PCR were performed using GeneAmp RNA PCR kit (Perkin-Elmer Cetus, Norwalk, CT) following the manufacturer's instructions. The selected regions of the replicase genes were amplified with primers synthesized from conserved regions designed by aligning IBV Beaudette sequence and sequences from other coronaviruses. The PCR profiles involved an initial denaturation for 4 min at 94 °C followed by 35 cycles of annealing at 55 °C for 30 s, extension at 72 °C for 1 min, and melting at 94 °C for 1 min. The amplified products were directly sequenced at a commercial sequencing facility (Davis Sequencing, Davis, CA). The genome sense Pol32 (5′-CACTAGCCTTGCGCTAGA-3′) and anti-sense Pol1501 (5′-GACCAACCTTCTGGTTCAAC-3′) primers were used to amplify the 5′-termini that correspond to nt 32–1501 of the Beaudette sequence (Boursnell et al., 1987). The 5′ ends of all IBV strains were amplified with FirstChoice RLM-RACE cDNA amplification kit (Ambion Inc., Austin, TX) using genomic antisense Pol1263 (5′-GCTTGCAAGACAAGTTCCTGC-3′) primer according to the manufacturer's protocol. The Pol1263 paired with another inner site primer Pol133 (5′-GGCACCTGGCCACCTGTCAGG-3′) selected from conserved areas (by aligning 11 IBV sequences including the Beaudette virus) could amplify nt 133–1263 (according to Beaudette sequence) of all IBV strains tested in our laboratory (data not shown). Three more consensus primer pairs were used to amplify other regions as follows: primers for the overlapping PLpro amplification are PL55 (5′-AGGATAAAGAAATYCTCTTC-3′) and PL58 (5′-GGACCACAYAAAGAACCCTC-3′); primers for Mpro amplification are Mp51F (5′-CGCCACGTTACTCTATTGGT-3′) and Mp33 (5′-GCCGCATAGAGCCATGCTAC-3′); and primers for RdRp amplification are Rp52 (5′-CTACTATGACTAATAGGCAG-3′) and Rp31 (5′-CTGAGAAAGCTCTTGATAGAG-3′).
Sequence analysis
Assembly of contiguous sequences, translation of nt sequence into protein sequence, and initial multiple sequence alignments were performed with Vector NTI Suite 9 software (InforMax, North Bethesda, MD). Selected sequences from GenBank were included in the alignment. After the alignment, the AlignX program constructs a phylogenetic tree employing the neighbor-joining method. Comparisons with published sequences were made by performing BlastN search (GenBank). Pairwise blast searches were also performed when there were no significant hits with BlastN search.
GenBank accession numbers
The accession numbers for IBV replicase gene sequences are as follows: (a) the 5′-termini: M41, AY392047; NV M41, AY561719; Ma5, AY561720; H52, AY392048; K0751, AY561721; CU705, AY561722; Conn, AY392049; CU510, AY561723; Florida, AY392050; CU-T2, AY561724; Ark99, AY392051; ArkDPI, AY392052; DE072, AY392054; CU805, AY561725; CU570, AY561726; GA98, AY392053; Cal99, AY392055; CU994, AY561727; K1699, AY561728; Gray, AY392056; (b) the PLpro: M41, AY392057; NV M41, AY561729; Ma5, AY561730; H52, AY392058; K0751, AY561731; CU705, AY561732; Conn, AY392059; CU510, AY561733; Florida, AY392060; CU-T2, AY561734; Ark99, AY392061; ArkDPI, AY392062; DE072, AY392064; CU805, AY561735; CU570, AY561736; GA98, AY392063; Cal99, AY392065; CU994, AY561737; K1699, AY561738; Gray, AY392066; (c) the Mpro: M41, AY392067; NV M41, AY561739; Ma5, AY561740; H52, AY392068; K0751, AY561741; CU705, AY561742; Conn, AY392069; CU510, AY561743; Florida, AY392070; CU-T2, AY561744; Ark99, AY392071; ArkDPI, AY392072; DE072, AY392074; CU805, AY561745; CU570, AY561746; GA98, AY392073; Cal99, AY392075; CU994, AY561747; K1699, AY561748; Gray, AY392076; (d) the RdRp: M41, AY392077; NV M41, AY561749; Ma5, AY561750; H52, AY392078; K0751, AY561751; CU705, AY561752; Conn, AY392079; CU510, AY561733; Florida, AY392080; CU-T2, AY561754; Ark99, AY392081; ArkDPI, AY392082; DE072, AY392084; CU805, AY561755; CU570, AY561756; GA98, AY392083; Cal99, AY392085; CU994, AY561757; K1699, AY561758; Gray, AY392086.
The complete genome sequences of Beaudette (M95169) and BJ (AY319651) strains of IBV, HcoV 229E (NC_002645), TGEV (NC_002306), MHV (NC_001846), BoCV (NC_003045), and SARS (NC_004718) were obtained from GenBank. The S1 sequences of IBV strains were also obtained from GenBank (Table 4).
Acknowledgements
We thank Drs. Syed A. Naqi and Udeni Balasuriya for their important suggestions in preparing the manuscript. We also thank Ms. Kelly Weaver for her excellent technical assistance.
References
- Anand K, Ziebuhr J, Wadhwani P, Mesters J.R, Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300:1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- Beaudette F.R, Hudson C.B. Cultivation of the virus of infectious bronchitis. J. Am. Vet. Med. Asso. 1937;90:51–60. [Google Scholar]
- Boursnell M.E, Brown T.D, Foulds I.J, Green P.F, Tomley F.M, Binns M.M. Completion of the sequence of the genome of the coronavirus avian infectious bronchitis virus. J. Gen. Virol. 1987;68:57–77. doi: 10.1099/0022-1317-68-1-57. [DOI] [PubMed] [Google Scholar]
- Brandt M, Yao K, Liu M, Heckert R.A, Vakharia V.N. Molecular determinants of virulence, cell tropism, and pathogenic phenotype of infectious bursal disease virus. J. Virol. 2001;75:11974–11982. doi: 10.1128/JVI.75.24.11974-11982.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I, Boursnell M.E, Binns M.M, Bilimoria B, Blok V.C, Brown T.D, Inglis S.C. An efficient ribosomal frame-shifting signal in the polymerase-encoding region of the coronavirus IBV. EMBO J. 1987;6:3779–3785. doi: 10.1002/j.1460-2075.1987.tb02713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I, Digard P, Inglis S.C. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell. 1989;57:537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D, Davis P.J. Coronavirus IBV: removal of spike glycopolypeptide S1 by urea abolishes infectivity and haemagglutination but not attachment to cells. J. Gen. Virol. 1986;67:1443–1448. doi: 10.1099/0022-1317-67-7-1443. [DOI] [PubMed] [Google Scholar]
- Cavanagh D, Davis P.J. Evolution of avian coronavirus IBV: sequence of the matrix glycoprotein gene and intergenic region of several serotypes. J. Gen. Virol. 1988;69:621–629. doi: 10.1099/0022-1317-69-3-621. [DOI] [PubMed] [Google Scholar]
- Cavanagh D, Naqi S. Infectious bronchitis. In: Saif Y.M, McDougald L.R, editors. Diseases of Poultry. 11th ed. Iowa State Univ. Press; Ames, IA: 2003. pp. 101–119. [Google Scholar]
- Cavanagh D, Davis P.J, Pappin D.J. Coronavirus IBV glycopolypeptides: locational studies using proteases and saponin, a membrane permeabilizer. Virus Res. 1986;4:145–156. doi: 10.1016/0168-1702(86)90038-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D, Davis P.J, Cook J.K.A, Li D, Kant A, Koch G. Location of the amino acid differences in the S1 spike glycoprotein subunit of closely related serotypes of infectious bronchitis virus. Avian Pathol. 1992;21:33–43. doi: 10.1080/03079459208418816. [DOI] [PubMed] [Google Scholar]
- Chang R.Y, Hofmann M.A, Sethna P.B, Brian D.A. A cis-acting function for the coronavirus leader in defective interfering RNA replication. J. Virol. 1994;68:8223–8231. doi: 10.1128/jvi.68.12.8223-8231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang R.Y, Krishnan R, Brian D.A. The UCUAAAC promoter motif is not required for high-frequency leader recombination in bovine coronavirus defective interfering RNA. J. Virol. 1996;70:2720–2729. doi: 10.1128/jvi.70.5.2720-2729.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouljenko V.N, Lin X.Q, Storz J, Kousoulas K.G, Gorbalenya A.E. Comparison of genomic and predicted amino acid sequences of respiratory and enteric bovine coronaviruses isolated from the same animal with fatal shipping pneumonia. J. Gen. Virol. 2001;82:2927–2933. doi: 10.1099/0022-1317-82-12-2927. [DOI] [PubMed] [Google Scholar]
- Cowen B.S, Hitchner S.B. Serotyping of avian infectious bronchitis viruses by the virus-neutralization test. Avian Dis. 1975;19:583–595. [PubMed] [Google Scholar]
- Dea S, Verbeek A.J, Tijssen P. Antigenic and genomic relationships among turkey and bovine enteric coronaviruses. J. Virol. 1990;64:3112–3118. doi: 10.1128/jvi.64.6.3112-3118.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelb J, Jr., Perkins B.E, Rosenberger J.K, Allen P.H. Serologic and cross-protection studies with several infectious bronchitis virus isolates from Delmarava-reared broiler chickens. Avian Dis. 1981;25:655–666. [PubMed] [Google Scholar]
- Gelb J, Jr., Wolff J.B, Moran C.A. Variant serotypes of infectious bronchitis virus isolated from commercial layer and broiler chickens. Avian Dis. 1991;35:82–87. [PubMed] [Google Scholar]
- Gelb J, Jr., Keeler C.L, Jr., Nix W.A, Rosenberger J.K, Cloud S.S. Antigenic and S-1 genomic characterization of the Delaware variant serotype of infectious bronchitis virus. Avian Dis. 1997;41:661–669. [PubMed] [Google Scholar]
- Gonzalez J.M, Gomez-Puertas P, Cavanagh D, Gorbalenya A.E, Enjuanes L. A comparative sequence analysis to revise the current taxonomy of the family Coronaviridae. Arch. Virol. 2003;148:2207–2235. doi: 10.1007/s00705-003-0162-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatta M, Gao P, Halfmann P, Kawaoka Y. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science. 2001;293:1840–1842. doi: 10.1126/science.1062882. [DOI] [PubMed] [Google Scholar]
- Hegyi A, Ziebuhr J. Conservation of substrate specificities among coronavirus main proteases. J. Gen. Virol. 2002;83:595–599. doi: 10.1099/0022-1317-83-3-595. [DOI] [PubMed] [Google Scholar]
- Herold J, Raabe T, Schelle-Prinz B, Siddell S.G. Nucleotide sequence of the human coronavirus 229E RNA polymerase locus. Virology. 1993;195:680–691. doi: 10.1006/viro.1993.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold J, Siddell S.G, Gorbalenya A.E. A human RNA viral cysteine proteinase that depends upon a unique Zn2+-binding finger connecting the two domains of a papain-like fold. J. Biol. Chem. 1999;274:14918–14925. doi: 10.1074/jbc.274.21.14918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscox J.A, Mawditt K.L, Cavanagh D, Britton P. Investigation of the control of coronavirus subgenomic mRNA transcription by using T7-generated negative-sense RNA transcripts. J. Virol. 1995;69:6219–6227. doi: 10.1128/jvi.69.10.6219-6227.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins S.R. Serological comparisons of strains of infectious bronchitis virus using plaque purified isolants. Avian Dis. 1974;18:231–239. [PubMed] [Google Scholar]
- Jia W, Karaca K, Parrish C.R, Naqi S.A. A novel variant of avian infectious bronchitis virus resulting from recombination among three different strains. Arch. Virol. 1995;140:259–271. doi: 10.1007/BF01309861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia W, Mondal S.P, Naqi S.A. Genetic and antigenic diversity in avian infectious bronchitis virus isolates of the 1940s. Avian Dis. 2002;46:437–441. doi: 10.1637/0005-2086(2002)046[0437:GAADIA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Johnson R.B, Marquardt W.W, Newman J.A. A new serotype of infectious bronchitis virus responsible for respiratory disease in Arkansas broiler flocks. Avian Dis. 1973;17:518–523. [PubMed] [Google Scholar]
- Jungherr E.L, Chomiak T.W, Luginbuhl R.E. Proceedings of the 60th Annual Meeting of the U.S. Livestock Sanitary Association. 1956. Immunological differences in strains of infectious bronchitis virus; pp. 203–209. Chicago, Illinois. [Google Scholar]
- Koonin E.V, Dolja V.V. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit. Rev. Biochem. Mol. Biol. 1993;28:375–430. doi: 10.3109/10409239309078440. [DOI] [PubMed] [Google Scholar]
- Kusters J.G, Jager E.J, Niesters H.G, van der Zeijst B.A. Sequence evidence for RNA recombination in field isolates of avian coronavirus infectious bronchitis virus. Vaccine. 1990;8:605–608. doi: 10.1016/0264-410X(90)90018-H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.M. Background paper. Transcription and replication of coronavirus RNA: a 1989 update. Adv. Exp. Med. Biol. 1990;276:327–333. doi: 10.1007/978-1-4684-5823-7_44. [DOI] [PubMed] [Google Scholar]
- Lee C.W, Jackwood M.W. Evidence of genetic diversity generated by recombination among avian coronavirus IBV. Arch. Virol. 2000;145:2135–2148. doi: 10.1007/s007050070044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.W, Jackwood M.W. Origin and evolution of Georgia 98 (GA98), a new serotype of infectious bronchitis virus. Virus Res. 2001;80:33–39. doi: 10.1016/s0168-1702(01)00345-8. [DOI] [PubMed] [Google Scholar]
- Lee H.J, Shieh C.K, Gorbalenya A.E, Koonin E.V, La Monica N, Tuler J, Bagdzhadzhyan A, Lai M.M. The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology. 1991;180:567–582. doi: 10.1016/0042-6822(91)90071-I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim K.P, Liu D.X. Characterization of the two overlapping papain-like proteinase domains encoded in gene 1 of the coronavirus infectious bronchitis virus and determination of the C-terminal cleavage site of an 87-kDa protein. Virology. 1998;245:303–312. doi: 10.1006/viro.1998.9164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim K.P, Ng L.F, Liu D.X. Identification of a novel cleavage activity of the first papain-like proteinase domain encoded by open reading frame 1a of the coronavirus Avian infectious bronchitis virus and characterization of the cleavage products. J. Virol. 2000;74:1674–1685. doi: 10.1128/jvi.74.4.1674-1685.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.X, Brown T.D. Characterisation and mutational analysis of an ORF 1a-encoding proteinase domain responsible for proteolytic processing of the infectious bronchitis virus 1a/1b polyprotein. Virology. 1995;209:420–427. doi: 10.1006/viro.1995.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.X, Brierley I, Tibbles K.W, Brown T.D. A 100-kilodalton polypeptide encoded by open reading frame (ORF) 1b of the coronavirus infectious bronchitis virus is processed by ORF 1a products. J. Virol. 1994;68:5772–5780. doi: 10.1128/jvi.68.9.5772-5780.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.X, Tibbles K.W, Cavanagh D, Brown T.D, Brierley I. Identification, expression, and processing of an 87-kDa polypeptide encoded by ORF 1a of the coronavirus infectious bronchitis virus. Virology. 1995;208:48–57. doi: 10.1006/viro.1995.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.X, Xu H.Y, Brown T.D. Proteolytic processing of the coronavirus infectious bronchitis virus 1a polyprotein: identification of a 10-kilodalton polypeptide and determination of its cleavage sites. J. Virol. 1997;71:1814–1820. doi: 10.1128/jvi.71.3.1814-1820.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.X, Xu H.Y, Lim K.P. Regulation of mRNA 1 expression by the 5′-untranslated region (5′-UTR) of the coronavirus infectious bronchitis virus (IBV) Adv. Exp. Med. Biol. 1998;440:303–311. doi: 10.1007/978-1-4615-5331-1_40. [DOI] [PubMed] [Google Scholar]
- Makino S, Stohlman S.A, Lai M.M. Leader sequences of murine coronavirus mRNAs can be freely reassorted: evidence for the role of free leader RNA in transcription. Proc. Natl. Acad. Sci. U.S.A. 1986;83:4204–4208. doi: 10.1073/pnas.83.12.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S, Shieh C, Keck J.G, Lai M.M.C. Defective-interfering particles of murine coronavirus: mechanism of synthesis of defective viral RNAs. Virology. 1988;163:104–111. doi: 10.1016/0042-6822(88)90237-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M.P, Waknell P.S, Woolcock P. Evaluation of commercially produced infectious bronchitis virus vaccines against an IBV field isolate obtained from broilers in California. Proceedings of the fiftieth Western Poultry Disease Conference, Sacramento, CA, March 2001; 2001. [Google Scholar]
- Mondal S.P, Lucio-Martinez B, Naqi S.A. Isolation and characterization of a novel antigenic subtype of infectious bronchitis virus serotype DE072. Avian Dis. 2001;45:1054–1059. [PubMed] [Google Scholar]
- Ng L.F, Liu D.X. Identification of a 24-kDa polypeptide processed from the coronavirus infectious bronchitis virus 1a polyprotein by the 3C-like proteinase and determination of its cleavage sites. Virology. 1998;243:388–395. doi: 10.1006/viro.1998.9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng L.F, Liu D.X. Further characterization of the coronavirus infectious bronchitis virus 3C-like proteinase and determination of a new cleavage site. Virology. 2000;272:27–39. doi: 10.1006/viro.2000.0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng L.F, Liu D.X. Membrane association and dimerization of a cysteine-rich, 16-kilodalton polypeptide released from the C-terminal region of the coronavirus infectious bronchitis virus 1a polyprotein. J. Virol. 2002;76:6257–6267. doi: 10.1128/JVI.76.12.6257-6267.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka J, Kikuchi N, Kojima S. Similarity relations of DNA and RNA polymerases investigated by the principal component analysis of amino acid sequences. Biochim. Biophys. Acta. 1999;1434:221–247. doi: 10.1016/S0167-4838(99)00187-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak A.O, van den Born E, Spaan W.J.M, Snijder E.J. Sequence requirements for RNA strand transfer during nidovirus discontinuous subgenomic RNA synthesis. EMBO J. 2001;20:7220–7228. doi: 10.1093/emboj/20.24.7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaan W, Cavanagh D, Horzinek M.C. Coronaviruses: structure and genome expression. J. Gen. Virol. 1988;69:2939–2952. doi: 10.1099/0022-1317-69-12-2939. [DOI] [PubMed] [Google Scholar]
- Stephensen C.B, Casebolt D.B, Gangopadhyay N.N. Phylogenetic analysis of a highly conserved region of the polymerase gene from 11 coronaviruses and development of a consensus polymerase chain reaction assay. Virus Res. 1999;60:181–189. doi: 10.1016/S0168-1702(99)00017-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D.F, Sefton B.M. Coronavirus multiplication: locations of genes for virion proteins on the avian infectious bronchitis virus genome. J. Virol. 1984;50:22–29. doi: 10.1128/jvi.50.1.22-29.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirrups K, Shaw K, Evans S, Dalton K, Cavanagh D, Britton P. Leader switching occurs during the rescue of defective RNAs by heterologous strains of the coronavirus infectious bronchitis virus. J. Gen. Virol. 2000;81:791–801. doi: 10.1099/0022-1317-81-3-791. [DOI] [PubMed] [Google Scholar]
- Sutou S, Sato S, Okabe T, Nakai M, Sasaki N. Cloning and sequencing of genes encoding structural proteins of avian infectious bronchitis virus. Virology. 1988;165:589–595. doi: 10.1016/0042-6822(88)90603-4. [DOI] [PubMed] [Google Scholar]
- Winterfield R.W, Hitchner S.B. Etiology of an infectious nephritis–nephrosis syndrome of chickens. Am. J. Vet. Res. 1962;23:1273–1279. [PubMed] [Google Scholar]
- Winterfield R.W, Fadly A.M, Hanley J.E. Characteristics of an isolate of infectious bronchitis virus from chickens in Florida. Avian Dis. 1971;15:305–311. [PubMed] [Google Scholar]
- Yao Y, Mingay L.J, McCauley J.W, Barclay W.S. Sequences in influenza A virus PB2 protein that determine productive infection for an avian influenza virus in mouse and human cell lines. J. Virol. 2001;75:5410–5415. doi: 10.1128/JVI.75.11.5410-5415.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J, Snijder E.J, Gorbalenya A.E. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 2000;81:853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]


