Abstract
Feline calicivirus (FCV) is a small non-enveloped virus containing a single-stranded, positive-sense RNA genome of approximately 7.7 kb. FCV is a highly infectious pathogen of cats and typically causes moderate, self-limiting acute oral and upper respiratory tract diseases or chronic oral diseases. In addition, in recent years, virulent, systemic FCV (vs-FCV) strains causing severe systemic diseases with a high mortality rate of up to 67% have been reported in cats. Although FCV vaccines are commercially available, their efficacy is limited due to antigenic diversity of FCV strains and short duration of immunity. In this study, we identified fexaramine as a potent inhibitor of FCV including vs-FCV strains in cell culture and demonstrated that fexaramine is a entry blocker for FCV by using various experiments including time-of-addition studies, generation of resistant viruses in cell culture and the reverse genetics system. A fexaramine resistant FCV mutant has a single amino acid change in the P2 domain of VP1 (the major capsid), and the importance of this mutation for conferring resistance was confirmed using the reverse genetics system. A comparative analysis of viral resistance was also performed using a peptidyl inhibitor (NPI52) targeting FCV 3C-like protease. Finally, the effects of combination treatment of fexaramine and NPI52 against FCV replication and emergence of resistant viruses were investigated in cell culture.
Keywords: Feline calicivirus, Fexaramine, Entry blocker, Virus resistance, Combination treatment
Highlights
-
•
Fexaramine inhibits the replication of feline calicivirus in cells as an entry blocker.
-
•
A single mutation in the capsid protein of FCV confers resistance to fexaramine.
-
•
Combined treatment of fexaramine and a viral protease inhibitor substantially delays the emergence of virus resistance.
1. Introduction
Feline calicivirus (FCV) is a small non-enveloped virus that contains a positive-sense single-stranded RNA genome of approximately 7.7 kb (Clarke et al., 2012). FCV belongs to the Caliciviridae, a family of viruses that includes important pathogens of humans and animals such as noroviruses, sapoviruses, vesicular exanthema swine virus and rabbit hemorrhagic disease virus (Green, 2007, Pesavento et al., 2008). There are at least six genera in Caliciviridae: Norovirus, Sapovirus, Vesivirus, Lagovirus, Nebovirus and Recovirus (Clarke et al., 2012). Genera Norovirus and Sapovirus contain human caliciviruses that are the major causes of viral gastroenteritis (Green, 2007). FCV belongs to the genus Vesivirus and is a highly important infectious pathogen of cats. FCV causes acute, self-limiting oral and upper respiratory tract disease or lameness but chronic infection also occurs often. In recent years, virulent, systemic FCV (vs-FCV) strains that cause severe systemic diseases with a high mortality of up to 67% were reported in the US and the EU (Foley et al., 2006, Pesavento et al., 2008). FCV vaccines are available for cats but the antigenic diversity of FCV strains and short-living immunity limits the efficacy of vaccines. In addition, symptomatic or non-symptomatic carriers pose a continuous challenge to the control of FCV infection. Therefore the development of effective therapeutic agents for various FCV strains is expected to provide significant benefit to the management and control of FCV infection in cats.
FCV genome has three open reading frames (ORFs) 1, 2 and 3 (Pesavento et al., 2008). Viral polyproteins consisting of non-structural proteins, such as viral 3C-like protease (3CLpro) and viral polymerase, are produced from ORF1 and processed by 3CLpro into functional mature proteins. A capsid precursor is translated from ORF2 and subsequently cleaved by viral 3CLpro into the major capsid protein (VP1) and the leader protein of the capsid (Sosnovtsev et al., 1998). The ORF3 encodes the minor structural protein (VP2), which interacts with VP1 to form the capsid and is associated with RNA genome packaging (Sosnovtsev et al., 2005). FCV capsid is composed of 180 copies of VP1 and only a few copies of VP2 (Prasad et al., 1994, Sosnovtsev and Green, 2000). The VP1 is composed of S, P1 and P2 subdomain (Pesavento et al., 2008, Sosnovtsev et al., 1998) and the P2 subdomain is involved in antigenicity and receptor interactions (Bhella and Goodfellow, 2011, Ossiboff et al., 2010). The process of cell entry by FCV and other non-enveloped virus is generally not well understood, but viral binding to the cellular receptor is believed to trigger a conformational change in the capsid leading to capsid disassembly and release of its contents for replication (Bhella et al., 2008, Johnson, 2010). Recently, virus entry has become one of the targets for therapeutic intervention, but only a few small molecule inhibitors targeting the virus capsid have been reported for non-enveloped viruses including picornaviruses (Buontempo et al., 1997, De Palma et al., 2008, Ho et al., 2016, Kim et al., 2017, Ma et al., 2017, Romero, 2001, Thys et al., 2008, Tijsma et al., 2014, Torres-Torres et al., 2015).
In the present study, we identified fexaramine as a highly effective inhibitor against FCV in cell culture. We determined that fexaramine, a synthetic agonist of the farnesoid X receptor (FXR), functions as an entry blocker by using various in vitro assays and identified a mutation conferring viral resistance to fexaramine in cell culture. In addition, the effects of combination treatment of fexaramine and NPI52, an inhibitor of 3CLpro that was previously reported by our group (Kim et al., 2012, Kim et al., 2015, Prior et al., 2013), in the replication of FCV and the emergence of viral resistance were investigated.
2. Materials and Method
2.1. Cells and virus
Crandell-Rees feline kidney cells (CRFK) and feline infectious peritonitis virus (FIPV) 1146 strain, a feline coronavirus, were purchased from American Type Culture Collection (Manassas, VA). Non-vs-FCV strains including Urbana, F9 and 131, and vs-FCV strains including Jengo and Deuce were kind gifts from Dr. Green at National Institutes of Health (Sosnovtsev and Green, 1995) or Dr. Parker at Cornell University (Ossiboff and Parker, 2007). FCV and FIPV were propagated with Eagle's Minimal Essential Medium (MEM) supplemented with 5% fetal bovine serum, 100 U/ml penicillin and 100 μg/ml streptomycin in CRFK cells.
2.2. Reagents and antibodies
Fexaramine was purchased from Sigma-Aldrich (St. Louis, MO) and dissolved in DMSO to 50 mM. Synthesis of the protease inhibitor NPI52 was previously reported by our group (Kim et al., 2012). Hyperimmune guinea pig serum raised against FCV was previously described (Sosnovtsev and Green, 1995). Monoclonal antibody against β-actin was purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
2.3. Antiviral effects of compounds in cell culture
To determine the 50% effective concentrations (EC50) of fexaramine and NPI52 for FCV, three-day-old confluent CRFK cells in 12- or 24-well plates (∼0.2 × 106 or ∼0.4 × 106 cells per well, respectively) were inoculated with FCV at a multiplicity of infection (MOI) of 0.05. The virus infected cells were then incubated in the presence of fexaramine or NPI52 at various concentrations (0.01–10 μM) for up to 2 days. Those compounds were also tested against FIPV because NPI52 was previously reported to have a strong antiviral effects for FIPV and FCV (Kim et al., 2015). The infected cells were freeze-thawed three times and virus titers were determined by the standard TCID50 method with the 10-fold dilution of each sample (Reed and Muench, 1938). The EC50 of each compound was calculated by nonlinear regression analyses of dose-dependent curves of virus titers against log inhibitor concentrations (variable slope) using GraphPad Prism 6 (GraphPad Software, Inc, San Diego, CA). At least 2 independent samples were used for determining EC50 values.
2.4. Nonspecific cytotoxic effect
Confluent CRFK cells grown in 24-well plates were incubated with fexaramine or NPI52 at various concentrations up to 100 μM for up to 2 days. Cell cytotoxicity was measured by CytoTox 96 nonradioactive cytotoxicity assay kit (Promega, Madison, WI) after 2 day-incubation, and the 50% cytotoxic concentration (CC50) of fexaramine or NPI52 was determined using GraphPad Prism 6. The in-vitro therapeutic indices (CC50/EC50) were also calculated for each virus strain.
2.5. Western blot analysis
Confluent CRFK cells were mock-treated or treated with various concentrations of fexaramine or NPI52, and immediately infected with FCV Urbana strain at an MOI of 2. Following incubation at 37 °C for 12 h, the cells were lysed with SDS-PAGE sample buffer containing 1% β-mercaptoethanol. The proteins were resolved on 10% Novex Tris-Bis gels (Invitrogen, Carlsbad, CA) and transferred to nitrocellulose membranes and the viral proteins were probed using FCV antibody (1:1000 dilution) (Sosnovtsev and Green, 1995) and then with peroxidase-conjugated, goat anti-guinea pig IgG. Following incubation with a chemiluminescent substrate (1:5000 dilution) (Pierce Biotechnology, Rockford, IL), the chemiluminescent signals were detected using a Fotodyne Transilluminator/Digital Camera System (Fotodyne/FX, Hartland, WI). β-actin was used as a loading control.
2.6. Time-of-addition study with fexaramine or NPI52 in cell culture
Confluent CRFK cells were washed with PBS and infected with FCV Urbana strain at an MOI of 10 for 1 h at 37 °C. The cells were then washed with PBS three times to remove residual viruses. Fexaramine or NPI52 was added to the cells at 1 h pre-infection, or 0, 1, 2, 4 or 6 h post-infection. The cells were then further incubated at 37 °C in the presence of each compound. At 12 h post-infection, cells were freeze thawed three times and virus titers were determined by the TCID50 method (Reed and Muench, 1938). In a separate experiment, confluent CRFK cells were preincubated with fexaramine (2 μM) or NPI52 (2 μM) for 1 or 4 h and washed with PBS prior to virus infection. Cells were then incubated at 37 °C without any compound for 12 h and viral replication was evaluated as described above.
2.7. Generation of FCV resistant to fexaramine or NPI52 in cell culture
Sequential cell culture passages of FCV Urbana strain were performed in the presence of fexaramine or NPI52 to generate resistant viruses. The synthesis and antiviral effects of NPI52, a 3CLpro inhibitor, were previously reported by our group, but FCV resistance to NPI52 has not been determined. CRFK cells were infected with FCV at an MOI of 0.05 in the presence of fexaramine or NPI52 at concentrations near their EC50 values. At each passage, supernatants containing viruses were passed on to fresh cells in the presence of increasing concentrations of fexaramine or NPI52. Alternatively, cells were infected with FCV at an MOI of 2 in the presence of fexaramine or NPI52 at 0.05 to 5 μM, and supernatants were passaged with the same concentration of each compound. As a control, FCV Urbana (WT FCV) virus was passaged in the absence of compound following the same procedure. Virus titers at select passage numbers were determined by the TCID50 assay and the fold changes in the EC50 values relative to WT FCV were determined. Total viral RNA was isolated using the RNeasy Mini kit (Invitrogen) at passage number 4 (fexaramine) or 12 (NPI52) and the full length FCV genome was sequenced following amplification by RT-PCR for the identification of mutations.
2.8. Generation of FCV infectious clones carrying fexaramine- or NPI52-resistant mutations
To construct an infectious clone carrying a single amino acid mutation identified in fexaramine-resistant FCV, A539T mutation was introduced into the VP1 gene of an Urbana strain-based infectious clone, pQ14 (Sosnovtsev and Green, 1995) by site-directed mutagenesis and the resulting clone was designated as pQ14CapA539T. Another infectious clone carrying a single amino acid mutation identified in NPI52-resistant FCV (pQ14ProT138S) was also generated by introducing the mutation into 3CLpro in a pQ14 clone. The primers for the sequence analysis and the in vitro mutagenesis are listed in Table 1 . The WT pQ14 and the mutant clones were verified by sequence analysis and tested for their replication capability, as previously described (Chang et al., 2008). Briefly, one-day old semi-confluent CRFK cells were infected with vaccinia virus encoding T7 polymerase (MVA-T7) for 1 h, washed with PBS and added with fresh medium. The pQ14, pQ14CapA539T or pQ14ProT138S plasmid was then transfected into the cells using Lipofectamine 2000, according to the manufacturer's protocol. Following incubation of the cells at 37 °C for 48 h, the cell supernatant was transferred to a monolayer of CRFK cells. The cells were then incubated at 37 °C and observed daily for appearance of cytopathic effects (CPE) for 96 h. The recovered WT or each mutant FCV (M Cap A539T or M Pro T138S) was examined for amino acid change in the genome by sequencing analysis. For each WT and mutant FCV, growth kinetics and EC50 values of fexaramine or NPI52 were determined as described above.
Table 1.
Primers used in the sequence analysis and mutagenesis study.
| Primer | Sequence (5′ to 3′) |
|---|---|
| FCV-5E-F | GTAAAAGAAATTTGAGACAATG |
| FCV-1K-F | FTTTAGATCAGAGGATGTGGC |
| FCV-1K-R | ATGTGGTTGTTGGGTTATTCAGC |
| FCV-2K-F | TTCTGAACATTGACTCCTTAGC |
| FCV-2K-R | ATGCCATGTTTTTCAGTGTGAAAT |
| FCV-3K-F | AAAATTGGACCTGTCAGTTGAG |
| FCV-3K-R | GCTCCCAATGCCGCGCGGTGA |
| FCV-4K-F | AAGATGTTAATTGACCATTTATC |
| FCV-4K-R | CATAGAGGTTTCTGAGGAAATGTT |
| FCV-5K-F | AATTCTACTACATCAAGGGTG |
| FCV-5K-R | GGTTGGGTCTATACTTTTTGGG |
| FCV-5K-F | AAGCTTGCTGCTATAGTTGTG |
| FCV-5K-R | GCAGCATCGAGGTACTTTGGAT |
| FCV-7K-F | ATCCAATATTTTACAAGAACTC |
| FCV-3E-R | GAAAACATCAATTGATCTAATCAC |
| FCV CapA538T-F | GGCATTGGAGAAGAAACCATTGGTGCTAATAG |
| FCV CapA538T-R | CTATTAGCACCAATGGTTTCTTCTCCAATGCC |
| FCV Pro T138S-F | GGTAACTGGATTGACTCCGGGTCTGGTGGTCC |
| FCV Pro T138S-R | GGACCACCAGACCCGGAGTGCAATCCAGTTACC |
2.9. Three-dimensional model structures of VP1 and 3CLpro of FCV
A three-dimensional structure of FCV VP1 was generated with Pymol (http://www.pymol.org) using PDB ID 3M8L (DeLano, 2010). A three-dimensional structure of FCV 3CLpro was built by EasyModeller 4.0 software using rhinovirus 3Cpro, poliovirus 3Cpro, human norovirus 3CLpro, and hepatitis A virus 3Cpro (PDB ID: 1CQQ, 1L1N, 2LNC, and 1QA7, respectively) as templates as previously described (Kim et al., 2015).
2.10. Combination treatment of fexaramine and NPI52
First, combination treatment of fexaramine and NPI52 was conducted to determine whether it has synergistic, antagonistic or additive effects in FCV replication. FCV Urbana strain was inoculated into confluent CRFK cells at an MOI of 0.05 for 1 h and then the medium was replaced with mock-medium, or medium containing NPI52 or fexaramine, or a combination of fexaramine and NPI52 at various concentrations. The infected cells were further incubated for 24 h, and virus titers were determined by the TCID50 assay. For drug–drug interactions, the data were analyzed at the 95% confidence level using the MacSynergy II software (Suhnel, 1992). First, theoretical additive interactions were calculated from the dose–response curve for each compound. Then the calculated additive surface was subtracted from the experimentally determined dose–response surface to yield regions of synergistic or antagonistic interactions. The resulting horizontal plane is where the interactions of two compounds are additive (0% synergy). Synergy or antagonism is shown as peaks above or below the horizontal plane, respectively.
In a separate experiment, the effect of combination treatment of fexaramine and NPI52 in the emergence of viral resistance to these compounds was determined. The same procedure as used for generating resistant viruses was followed: sequential cell culture passages of FCV Urbana strain were conducted in the presence of fexaramine and NPI52 at 0.05 to 5 μM. Virus titers at select passage numbers were determined by the TCID50 assay and the fold changes in EC50 values for fexaramine or NPI52 relative to WT FCV were calculated for up to 20 virus passages.
2.11. Statistics
The student t-test was used to compare the significance of the unpaired sample means. P values < 0.01 were considered significant.
3. Results
3.1. Fexaramine inhibits the replication of multiple FCV strains including vs-FCV strains
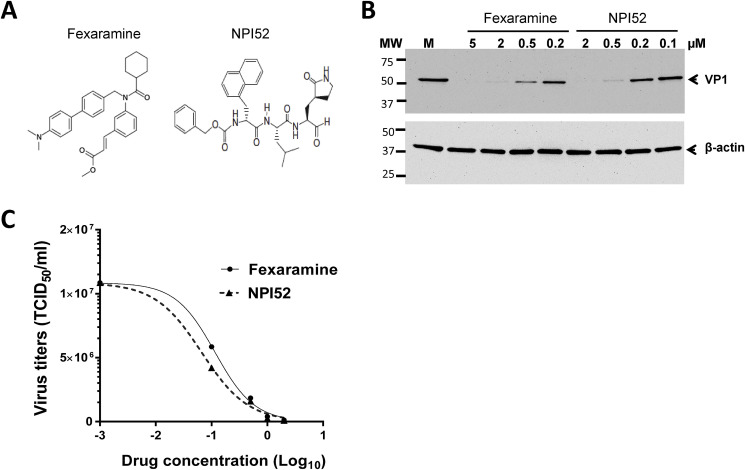
The structures of fexaramine and NPI52 are shown in Fig. 1 A. Fexaramine inhibited the replication of non-vs-FCV and vs-FCV strains with EC50 values ranging 0.18 to 0.49 μM (Table 2 ). The CC50 value of fexaramine in CRFK cells was determined to be 46.3 μM (Table 2). The calculated in-vitro therapeutic indices (CC50/EC50) of fexaramine ranged from 46.3 to 94.5 for FCV, and those of NPI52 ranged from 319.5 to 3514.5 for FCV or FIP. While NPI52 showed significant antiviral effects, fexaramine had little effect on the replication of FIPV 1146 at up to 10 μM (Table 2). We have previously reported that NPI52 has a strong broad-spectrum antiviral effect against FCV and FIPV 1146 (Kim et al., 2012, Kim et al., 2015) (Table 2). The antiviral effects of fexaramine or NPI52 against FCV Urbana strain was confirmed with Western blot analysis, which showed a significant reduction of VP1 by each compound in a dose-dependent manner (Fig. 1B). The dose-dependent inhibition of fexaramine and NPI52 against FCV Urbana determined by viral titration (TCID50) is shown in Fig. 1C.
Fig. 1.
Structures of fexaramine and NPI52 (A) and the inhibitory activities of fexaramine and NPI52 against FCV (B–C). The replication of FCV Urbana strain with various concentrations of each compound was monitored by Western blot analysis (B) and the TCID50 assay (C) as described in the Materials and Method section. M in Fig. 1B indicates the mock-treatment.
Table 2.
Effects of fexaramine and NPI52 in the replication of various strains of FCV or FIPV in CRFK cells.
| EC50 (μM) |
CC50 (μM) | ||||||
|---|---|---|---|---|---|---|---|
| FCV |
FIPV |
||||||
| Urbana | F9 | 131 | Deucea | Jengoa | 1146 | ||
| Fexaramine | 0.18 ± 0.1 | 0.36 ± 0.2 | 0.10 ± 0.03 | 0.33 ± 0.25 | 0.49 ± 0.16 | >10 | 46.3 ± 3.5 |
| NPI52 | 0.02 ± 0.01 | 0.05 ± 0.02 | 0.05 ± 0.02 | 0.22 ± 0.2 | 0.03 ± 0.03 | 0.02 ± 0.03 | 70.29 ± 5.1 |
EC50 and CC50 indicates the 50% effective concentration and 50% cytotoxic concentration, respectively. Values are average ± standard deviation from at least 2 independent experiments.
Virulent, systemic strains of FCV. FIPV 1146 strain was included as a control. The EC50 and CC50 values of NPI52 were previously reported in our publication (Kim et al., 2015) and included in the table for comparative purposes.
3.2. Fexaramine inhibits the early stage of viral infection
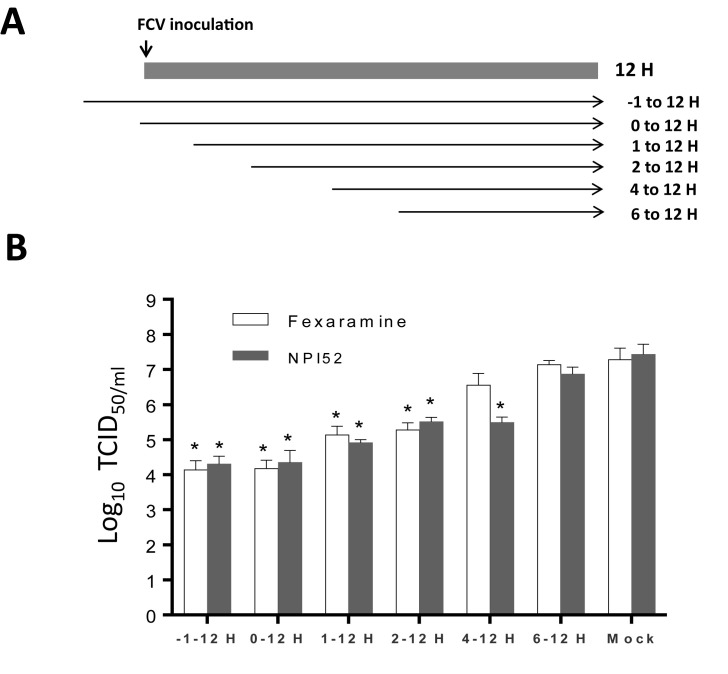
Our time-of-addition study showed that significant viral inhibition occurred when fexaramine was added to the cell culture prior to virus infection (−1 h) or within 2 h post-infection (PI), but not after 4 h PI (Fig. 2 B). NPI52 that blocks cleavage of newly produced viral polyproteins (Kim et al., 2012) did not inhibit virus replication when added after 6 h PI (Fig. 2B). The preincubation of CRFK cells with fexaramine (2 μM) for 1 or 4 h prior to virus infection had little effects on the virus replication (data not shown). These results show that fexaramine works at a virus entry step before virus replication occurs.
Fig. 2.
Time-of-addition studies of fexaramine or NPI52 using FCV Urbana strain. Confluent CRFK cells were infected with FCV, and fexaramine or NPI52 was added at 1 h prior to virus infection or 0, 1, 2, 4 or 6 h following virus infection (A). At 12 h post-infection, virus titers were measured by the TCID50 assay (B). The asterisk indicates statistical significance (p < 0.05) between the indicated group and the mock-treatment (no compound).
3.3. Generation of FCV strains resistant to fexaramine or NPI52 in cell culture for identification and analysis of mutations
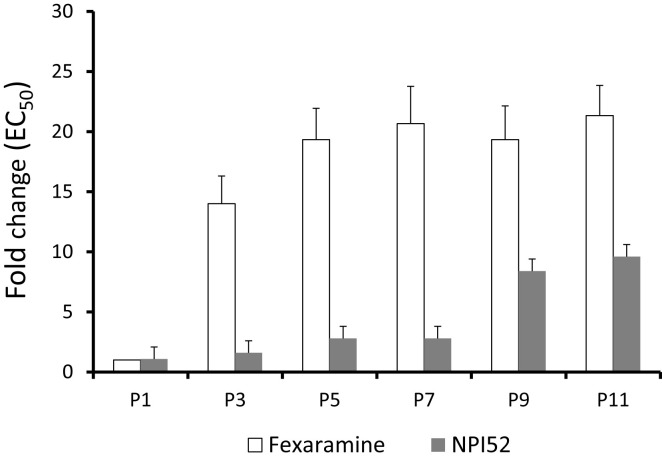
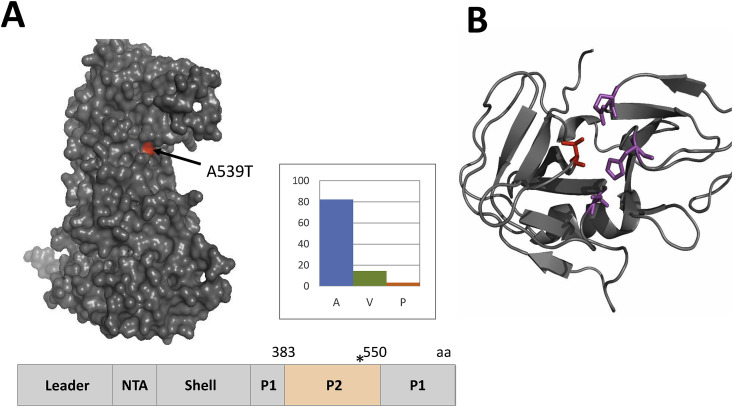
FCVs resistant to fexaramine or NPI52 were generated by passaging FCV Urbana strain in the presence of each compound in CRFK cells. The fold increases in the EC50 values of serially passaged viruses compared to WT FCV are shown in Fig. 3 . Rapid emergence of fexaramine-resistant virus was observed with 18.6- to 28.6-fold increase in the EC50 values for P3 and P11. However, in contrast to fexaramine, NPI52-resistant viruses appeared at later passages with a 9.4-fold increase in the EC50 value at P11 (Fig. 3). Analysis of the full-genome sequences of viruses resistant to fexaramine or NPI52 at P4 and P12 led to the identification of a single amino acid substitution in VP1 (A539T) and 3CLpro gene (T138S), respectively. The A539T substitution in P4 FCV is located in the P2 subdomain of VP1 (Fig. 4 A). The alanine at amino acid position 539 in the VP1 is highly conserved among FCV VP1 sequences reported in GenBank (270 strains) and comprises 82.2% of FCV VP1 sequences, followed by valine (14.4%) and proline (3.3%) (Fig. 4A). However, none of FCV VP1 sequences was reported to have threonine at that position. The T138S substitution in FCV P12 3CLpro is located in the C-terminal domain near the active site (Fig. 4B). The threonine residue at the position of 138 in 3CLpro is conserved in all reported FCV 3CLpro sequences (48 strains).
Fig. 3.
Fold changes in the EC50 values of fexaramine against FCV serially passaged with each compound compared to that of WT FCV in CRFK cells. FCV Urbana strain was serially passaged in CRFK cells in the presence of fexaramine or NPI52 and the EC50 values of each compound for viruses at selected passages was determined and compared to that for WT FCV.
Fig. 4.
Resistant mutations against fexaramine or NPI52. A. A single residue substitution A539T (red) is located in the P2 subdomain of the VP1 gene of FCV passaged in the presence of fexaramine. The A539T substitution (asterisk) is also shown on the schematic representation of FCV VP1 gene. The P2 domain is located between amino acids 383 and 550 of the VP1. B. A ribbon representation of FCV 3CLpro built from the 3Cpro of rhinovirus, poliovirus, hepatitis A virus and human norovirus by the EasyModeller program. The residues in the catalytic triad (H39, E60 and C122) are shown as purple sticks. The red stick indicates the amino acid substitution (T138) on FCV 3CLpro. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.4. Mutant FCVs carrying the identified mutation generated by the reverse genetics are resistant to fexaramine or NPI52
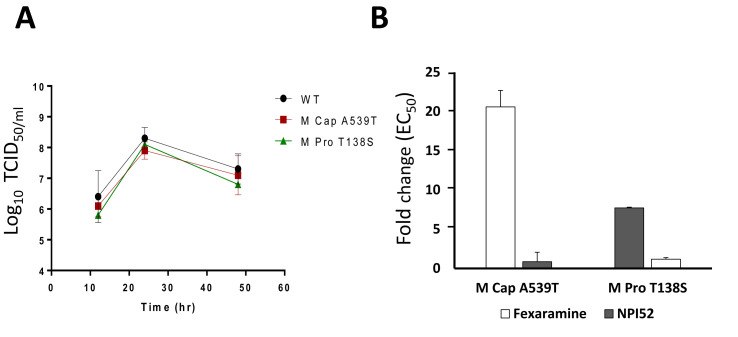
Using the reverse genetics system of FCV, M Cap A539T (recombinant FCV with A539T mutation in VP1) and M Pro T138S (recombinant FCV with P138S mutation in 3CLpro) were generated. The growth kinetics of M Cap A539T and M Pro T138S were comparable to that of WT FCV (Fig. 5 A), indicating that the replication efficiencies of those mutant FCVs are not compromised by the mutation. The EC50 values of M Cap A539T against fexaramine and M Pro T138S against NPI52 were increased by 20.6- or 7.7-fold, compared to that of the WT FCV (Fig. 5B), respectively. These fold increases are comparable to those observed with resistant viruses generated by serial passages (Fig. 3). As expected, M Cap A539T and M Pro T138S retained its susceptibility to NPI52 or fexaramine, respectively, in cell culture (Fig. 5B).
Fig. 5.
The growth kinetics (A) and susceptibility (B) of FCV mutants to fexaramine or NPI52. An infectious recombinant FCV full-genome clone carrying A539T in the VP1 gene or T138S in the 3CLpro gene was generated using the reverse genetics system of FCV. A. The growth kinetics of WT- and the mutant FCVs were compared. WT, M Cap A539T and M Pro T138S indicate WT, mutant FCV carrying A539T in the VP1 and recombinant FCV carrying T138S in the 3CLpro, respectively. B. The fold changes in the EC50 values of fexaramine or NPI52 against each mutant FCV compared to WT FCV were determined in cell culture.
3.5. Combination treatment of fexaramine and NPI52 showed synergistic antiviral effects and delayed the emergence of viral resistance
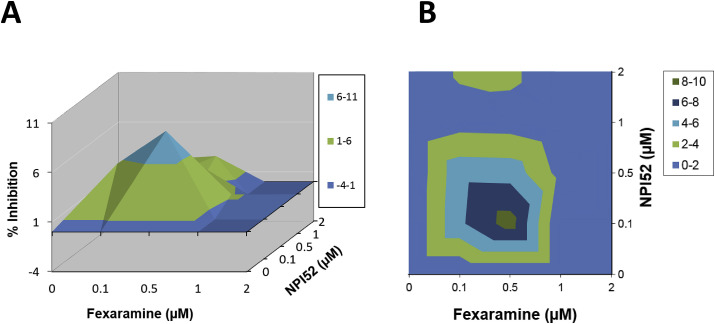
When virus-infected cells were treated with combinations of fexaramine (0–2 μM) and NPI52 (0–2 μM), synergistic effects were observed with a maximal synergy at concentrations of 0.1–0.5 μM (Fig. 6 A and B). Synergy volumes determined at the 95% confidence interval were 33.4 μM2% with a log volume of 8.35. Cytotoxicity was not observed at any concentrations of combination treatment. Emergence of resistant viruses against fexaramine or NPI52 was not observed up to 20 sequential cell culture passages of FCV with combination treatment of fexaramine and NPI52.
Fig. 6.
Effects of combination treatment of fexaramine and NPI52 on the replication of FCV in cell culture. Confluent CRFK cells were infected with FCV Urbana strain and the medium was replaced with fresh medium containing mock or the combinations of fexaramine and NPI52 (0–2 μM). The cells were incubated for 24 h and virus replication was measured by the TCID50 assay. A and B. Drug-drug interactions were analyzed by the MacSynergy II software at 95% confidence limits. The colors indicate the level of synergy or anatogonism and the peaks above the plane of 0% synergy in the plot indicate synergy. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
Fexaramine is a synthetic agonist of the farnesoid X receptor (FXR), a member of a nuclear receptor family, which plays an important role in metabolic pathways such as lipid and glucose metabolism (Downes et al., 2003). The effects of FXR in modulating metabolism and increasing energy expenditure on food intake has suggested the potential use of fexaramine in treatment of obesity and metabolic syndrome such as insulin resistance (Fang et al., 2015). The effects of FXR agonists in lipid metabolism have also shown to play a role in the replication of viruses that requires close interaction with lipid contents (Honke et al., 2017, Kim and Chang, 2012, Radreau et al., 2016). We previously reported that FXR agonists including bile acids and fexaramine inhibit the replication of rotavirus and their inhibitory effects are mediated by reduction of cellular lipid contents by the activation of FXR (Kim and Chang, 2012). In this study, we demonstrated that the replication of FCV is potently inhibited by fexaramine in cell culture. Interestingly, investigation on the mode of FCV inhibition by fexaramine by various experiments including the time-of-addition studies and analysis of viral resistance showed that, in contrast with inhibition of rotavirus (Kim and Chang, 2012), fexaramine acts as an entry inhibitor of FCV. Virus entry inhibitors have been developed or reported for enveloped viruses including human immunodeficiency virus (HIV) (Haqqani and Tilton, 2013, Woollard and Kanmogne, 2015), influenza virus (Palese and Shaw, 2007), Measles virus (Plemper et al., 2005) and Dengue virus (Scaturro et al., 2014). Virus entry inhibitors have also been reported for non-enveloped viruses including picornaviruses including poliovirus (Thys et al., 2008), enterovirus (Buontempo et al., 1997, Ho et al., 2016, Ma et al., 2017, Torres-Torres et al., 2015) and rhinoviruses (De Palma et al., 2008, Kim et al., 2017, Tijsma et al., 2014). However, entry inhibitors of any calicivirus have not been reported and this is the first report on a small molecule entry inhibitor of feline calicivirus. We also tested fexaramine against murine norovirus and porcine enteric calicivirus (sapovirus) in a preliminary study but it had minimal inhibitory activity at up to 10 μM in cell culture against those viruses (data not shown).
In our study, different field strains of FCV, including vs-FCV strains that are causative agents of fatal, systemic infection among cats, were susceptible to antiviral effects of fexaramine in cell culture. Although potent, we found that the antiviral effect of fexaramine against FCV is slightly less than that of NPI52, an inhibitor of FCV 3CLpro in cell culture (Kim et al., 2012, Kim et al., 2015). It is of note that fexaramine has a low genetic barrier to resistance as fexaramine-resistant viruses were readily obtained within 3–5 serial passages (Fig. 3). This was in contrast to the case with NPI52 against FCV where NPI52-resistant viruses were obtained at later viral passages and the decrease in drug susceptibility was of less magnitude compared to fexaramine (Fig. 3).
Our analysis of fexaramine-resistant FCV showed that a single mutation in VP1 (A539T) confers viral resistance to fexaramine. The alanine at the 539 site of the P2 subdomain of VP1 is located in the surface-exposed loop of the P2 subdomain of VP1 and is highly conserved among FCV field strains (Fig. 4A). The A539T mutation in fexaramine-resistant virus sits adjacent to a mutation (I535V) that was previously identified in some FCV mutants conferring resistance to the soluble ectodomain of the FCV receptor (Ossiboff et al., 2010), feline functional adhesion molecule A (fJAM-A) (Makino et al., 2006, Ossiboff and Parker, 2007). Analysis of resistant mutants generated by soluble fJAM-A suggested that binding of FCV to fJAM leads to conformational change of FCV VP1 (Ossiboff et al., 2010), and the binding and subsequent conformation changes seem to be mediated by residues located in the surface-exposed or hidden regions in VP1, including the P2 subdomain, respectively (Ossiboff et al., 2010). Other non-enveloped viruses, such as human rhinoviruses (HRVs), are reported to undergo conformational changes of virus capsids during virus entry, and inhibition of capsid conformational changes by a small molecule inhibitor was reported for HRV and other enteroviruses. Pleconaril, an entry inhibitor, binds to a hydrophobic pocket in the VP1 of HRV, inducing conformational changes that lead to altered receptor binding and viral uncoating (Romero, 2001). Although we did not further investigate the specific mechanism underlying the inhibition by fexaramine in this study, it is plausible that fexaramine interacts with the residue in the P2 subdomain of FCV VP1 to interfere with viral receptor binding and/or uncoating during virus entry. We plan to conduct co-crystallization with FCV (or FCV VP1)-fexaramine complex to understand the inhibitory mechanism, which may generate valuable information for potential optimization of fexaramine to improve its antiviral effects. We also found that the mutation in FCV 3CLpro that confers resistance to NPI52 (T138S) is located at the active site of the enzyme (Fig. 4B). Threonine at 138 position is conserved among FCV strains and corresponds to threonine at 134 position in human norovirus 3CLpro, a virus protease structurally and functionally closely related to FCV 3CLpro. Previous crystallographic study on the complex of human norovirus 3CLpro-dipeptidyl protease inhibitor showed that threonine at 134 position forms a hydrogen bond with the inhibitor that shares the core structure with NPI52 (Kim et al., 2012, Takahashi et al., 2013). Interestingly, the growth kinetics of the mutant FCVs carrying A539T or T138S were comparable to that of WT FCV, suggesting that these mutations do not affect the efficiency of virus replication in cell culture.
Combination of antiviral agents with differing viral targets has been used to reduce the risk of viral resistance and improve efficacy for human immunodeficiency virus and hepatitis C virus infections (Asselah et al., 2016, Pirrone et al., 2011). Therefore we studied the effects of combination treatment of fexaramine and NPI52 in cell culture and found that combination treatment shows synergistic antiviral effects at the tested drug concentrations (Fig. 6). More importantly, combination treatment substantially delayed the emergence of resistant viruses to fexaramine or NPI52 up to 20 passages. These results provide a strong rationale for further investigation of combinational treatment of antiviral agents targeting different virus proteins to overcome resistance to single antiviral agent and enhance antiviral activity for FCV infection. In conclusion, our study reports the first entry inhibitor of FCV and potential combination use of an entry inhibitor and viral protease inhibitor against FCV infection.
Acknowledgements
This work was generously supported by the National Institutes of Health (R01AI109039) and Morris Animal Foundation, Denver, CO (D14FE-012 and D16FE-512). We thank David George for technical assistance.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.antiviral.2018.02.009.
Contributor Information
Yunjeong Kim, Email: ykim@vet.ksu.edu.
Kyeong-Ok Chang, Email: kchang@vet.ksu.edu.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- Asselah T., Boyer N., Saadoun D., Martinot-Peignoux M., Marcellin P. Direct-acting antivirals for the treatment of hepatitis C virus infection: optimizing current IFN-free treatment and future perspectives. Liver Int. 2016;36(Suppl. 1):47–57. doi: 10.1111/liv.13027. [DOI] [PubMed] [Google Scholar]
- Bhella D., Gatherer D., Chaudhry Y., Pink R., Goodfellow I.G. Structural insights into calicivirus attachment and uncoating. J. Virol. 2008;82:8051–8058. doi: 10.1128/JVI.00550-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhella D., Goodfellow I.G. The cryo-electron microscopy structure of feline calicivirus bound to junctional adhesion molecule A at 9-angstrom resolution reveals receptor-induced flexibility and two distinct conformational changes in the capsid protein VP1. J. Virol. 2011;85:11381–11390. doi: 10.1128/JVI.05621-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buontempo P.J., Cox S., Wright-Minogue J., DeMartino J.L., Skelton A.M., Ferrari E., Albin R., Rozhon E.J., Girijavallabhan V., Modlin J.F., O'Connell J.F. SCH 48973: a potent, broad-spectrum, antienterovirus compound. Antimicrob. Agents Chemother. 1997;41:1220–1225. doi: 10.1128/aac.41.6.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang K.O., George D.W., Patton J.B., Green K.Y., Sosnovtsev S.V. Leader of the capsid protein in feline calicivirus promotes replication of Norwalk virus in cell culture. J. Virol. 2008;82:9306–9317. doi: 10.1128/JVI.00301-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke I.N., Estes M.K., Green K.Y., Hansman G.S., Knowles N.J. Family Caliciviridae. In: King A.M.Q., Adams M.J., Carstens E.B., editors. Virus Taxonomy Classification and Nomenclature of Viruses Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press; Lefkowitz, EJ: 2012. [Google Scholar]
- De Palma A.M., Vliegen I., De Clercq E., Neyts J. Selective inhibitors of picornavirus replication. Med. Res. Rev. 2008;28:823–884. doi: 10.1002/med.20125. [DOI] [PubMed] [Google Scholar]
- DeLano W.L. DeLano Scientific LLC; San Carlos, CA: 2010. The PyMOL Molecular Graphics System.http://www.pymol.org [Google Scholar]
- Downes M., Verdecia M.A., Roecker A.J., Hughes R., Hogenesch J.B., Kast-Woelbern H.R., Bowman M.E., Ferrer J.L., Anisfeld A.M., Edwards P.A., Rosenfeld J.M., Alvarez J.G., Noel J.P., Nicolaou K.C., Evans R.M. A chemical, genetic, and structural analysis of the nuclear bile acid receptor FXR. Mol. Cell. 2003;11:1079–1092. doi: 10.1016/s1097-2765(03)00104-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S., Suh J.M., Reilly S.M., Yu E., Osborn O., Lackey D., Yoshihara E., Perino A., Jacinto S., Lukasheva Y., Atkins A.R., Khvat A., Schnabl B., Yu R.T., Brenner D.A., Coulter S., Liddle C., Schoonjans K., Olefsky J.M., Saltiel A.R., Downes M., Evans R.M. Intestinal FXR agonism promotes adipose tissue browning and reduces obesity and insulin resistance. Nat. Med. 2015;21:159–165. doi: 10.1038/nm.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley J., Hurley K., Pesavento P.A., Poland A., Pedersen N.C. Virulent systemic feline calicivirus infection: local cytokine modulation and contribution of viral mutants. J. Feline Med. Surg. 2006;8:55–61. doi: 10.1016/j.jfms.2005.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green K.Y. fifth ed. Lippincott Williams & Wilkins; Philadelphia: 2007. Caliciviruses: the Noroviruses. [Google Scholar]
- Haqqani A.A., Tilton J.C. Entry inhibitors and their use in the treatment of HIV-1 infection. Antivir. Res. 2013;98:158–170. doi: 10.1016/j.antiviral.2013.03.017. [DOI] [PubMed] [Google Scholar]
- Ho J.Y., Chern J.H., Hsieh C.F., Liu S.T., Liu C.J., Wang Y.S., Kuo T.W., Hsu S.J., Yeh T.K., Shih S.R., Hsieh P.W., Chiu C.H., Horng J.T. In vitro and in vivo studies of a potent capsid-binding inhibitor of enterovirus 71. J. Antimicrob. Chemother. 2016;71:1922–1932. doi: 10.1093/jac/dkw101. [DOI] [PubMed] [Google Scholar]
- Honke N., Shaabani N., Hardt C., Krings C., Haussinger D., Lang P.A., Lang K.S., Keitel V. Farnesoid X receptor in mice prevents severe liver immunopathology during lymphocytic choriomeningitis virus infection. Cell. Physiol. Biochem. 2017;41:323–338. doi: 10.1159/000456168. [DOI] [PubMed] [Google Scholar]
- Johnson J.E. Springer; Heidelberg ; New York: 2010. Cell Entry by Non-enveloped Viruses. [Google Scholar]
- Kim J., Jung Y.K., Kim C., Shin J.S., Scheers E., Lee J.Y., Han S.B., Lee C.K., Neyts J., Ha J.D., Jung Y.S. A novel series of highly potent small molecule inhibitors of rhinovirus replication. J. Med. Chem. 2017;60:5472–5492. doi: 10.1021/acs.jmedchem.7b00175. [DOI] [PubMed] [Google Scholar]
- Kim Y., Chang K.O. Inhibitory effects of bile acids and synthetic farnesoid X receptor agonists on rotavirus replication. J. Virol. 2012;85:12570–12577. doi: 10.1128/JVI.05839-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Lovell S., Tiew K.C., Mandadapu S.R., Alliston K.R., Battaile K.P., Groutas W.C., Chang K.O. Broad-spectrum antivirals against 3C or 3C-like proteases of picornaviruses, noroviruses, and coronaviruses. J. Virol. 2012;86:11754–11762. doi: 10.1128/JVI.01348-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Shivanna V., Narayanan S., Prior A.M., Weerasekara S., Hua D.H., Kankanamalage A.C., Groutas W.C., Chang K.O. Broad-spectrum inhibitors against 3C-Like proteases of feline coronaviruses and feline caliciviruses. J. Virol. 2015;89:4942–4950. doi: 10.1128/JVI.03688-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Abdelnabi R., Delang L., Froeyen M., Luyten W., Neyts J., Mirabelli C. New class of early-stage enterovirus inhibitors with a novel mechanism of action. Antivir. Res. 2017;147:67–74. doi: 10.1016/j.antiviral.2017.10.004. [DOI] [PubMed] [Google Scholar]
- Makino A., Shimojima M., Miyazawa T., Kato K., Tohya Y., Akashi H. Junctional adhesion molecule 1 is a functional receptor for feline calicivirus. J. Virol. 2006;80:4482–4490. doi: 10.1128/JVI.80.9.4482-4490.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossiboff R.J., Parker J.S. Identification of regions and residues in feline junctional adhesion molecule required for feline calicivirus binding and infection. J. Virol. 2007;81:13608–13621. doi: 10.1128/JVI.01509-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossiboff R.J., Zhou Y., Lightfoot P.J., Prasad B.V., Parker J.S. Conformational changes in the capsid of a calicivirus upon interaction with its functional receptor. J. Virol. 2010;84:5550–5564. doi: 10.1128/JVI.02371-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palese P., Shaw M.L. Orthomyxoviridae: the viruses and their replication. In: Knipe D.M.E.A., editor. Fields Virology. Wolters Kluwer/Lippincott Williams & Wilkins; Philadelphia: 2007. pp. 1647–1690. [Google Scholar]
- Pesavento P.A., Chang K.O., Parker J.S. Molecular virology of feline calicivirus. Vet. Clin. North Am. Small Anim. Pract. 2008;38:775–786. doi: 10.1016/j.cvsm.2008.03.002. vii. [DOI] [PubMed] [Google Scholar]
- Pirrone V., Thakkar N., Jacobson J.M., Wigdahl B., Krebs F.C. Combinatorial approaches to the prevention and treatment of HIV-1 infection. Antimicrob. Agents Chemother. 2011;55:1831–1842. doi: 10.1128/AAC.00976-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plemper R.K., Doyle J., Sun A., Prussia A., Cheng L.T., Rota P.A., Liotta D.C., Snyder J.P., Compans R.W. Design of a small-molecule entry inhibitor with activity against primary measles virus strains. Antimicrob. Agents Chemother. 2005;49:3755–3761. doi: 10.1128/AAC.49.9.3755-3761.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad B.V., Matson D.O., Smith A.W. Three-dimensional structure of calicivirus. J. Mol. Biol. 1994;240:256–264. doi: 10.1006/jmbi.1994.1439. [DOI] [PubMed] [Google Scholar]
- Prior A.M., Kim Y., Weerasekara S., Moroze M., Alliston K.R., Uy R.A., Groutas W.C., Chang K.O., Hua D.H. Design, synthesis, and bioevaluation of viral 3C and 3C-like protease inhibitors. Bioorg. Med. Chem. Lett. 2013;23:6317–6320. doi: 10.1016/j.bmcl.2013.09.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radreau P., Porcherot M., Ramiere C., Mouzannar K., Lotteau V., Andre P. Reciprocal regulation of farnesoid X receptor alpha activity and hepatitis B virus replication in differentiated HepaRG cells and primary human hepatocytes. FASEB J. 2016;30:3146–3154. doi: 10.1096/fj.201500134. [DOI] [PubMed] [Google Scholar]
- Reed L.J., Muench H. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 1938;27:493–497. [Google Scholar]
- Romero J.R. Pleconaril: a novel antipicornaviral drug. Expert Opin. Investig. Drugs. 2001;10:369–379. doi: 10.1517/13543784.10.2.369. [DOI] [PubMed] [Google Scholar]
- Scaturro P., Trist I.M., Paul D., Kumar A., Acosta E.G., Byrd C.M., Jordan R., Brancale A., Bartenschlager R. Characterization of the mode of action of a potent dengue virus capsid inhibitor. J. Virol. 2014;88:11540–11555. doi: 10.1128/JVI.01745-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosnovtsev S., Green K.Y. RNA transcripts derived from a cloned full-length copy of the feline calicivirus genome do not require VpG for infectivity. Virology. 1995;210:383–390. doi: 10.1006/viro.1995.1354. [DOI] [PubMed] [Google Scholar]
- Sosnovtsev S.V., Belliot G., Chang K.O., Onwudiwe O., Green K.Y. Feline calicivirus VP2 is essential for the production of infectious virions. J. Virol. 2005;79:4012–4024. doi: 10.1128/JVI.79.7.4012-4024.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosnovtsev S.V., Green K.Y. Identification and genomic mapping of the ORF3 and VPg proteins in feline calicivirus virions. Virology. 2000;277:193–203. doi: 10.1006/viro.2000.0579. [DOI] [PubMed] [Google Scholar]
- Sosnovtsev S.V., Sosnovtseva S.A., Green K.Y. Cleavage of the feline calicivirus capsid precursor is mediated by a virus-encoded proteinase. J. Virol. 1998;72:3051–3059. doi: 10.1128/jvi.72.4.3051-3059.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhnel J. Comment on the paper: a three-dimensional model to analyze drug-drug interactions. Prichard, M.N. and Shipman, C., Jr. (1990) Antiviral Res. 1992;14:181–206. doi: 10.1016/0166-3542(90)90001-n. Antiviral Res 17, 91-98. [DOI] [PubMed] [Google Scholar]
- Takahashi D., Kim Y., Lovell S., Prakash O., Groutas W.C., Chang K.O. Structural and inhibitor studies of norovirus 3C-like proteases. Virus Res. 2013;178:437–444. doi: 10.1016/j.virusres.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thys B., De Palma A.M., Neyts J., Andries K., Vrijsen R., Rombaut B. R75761, a lead compound for the development of antiviral drugs in late stage poliomyelitis eradication strategies and beyond. Antivir. Res. 2008;78:278–281. doi: 10.1016/j.antiviral.2008.01.002. [DOI] [PubMed] [Google Scholar]
- Tijsma A., Franco D., Tucker S., Hilgenfeld R., Froeyen M., Leyssen P., Neyts J. The capsid binder Vapendavir and the novel protease inhibitor SG85 inhibit enterovirus 71 replication. Antimicrob. Agents Chemother. 2014;58:6990–6992. doi: 10.1128/AAC.03328-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Torres S., Myers A.L., Klatte J.M., Rhoden E.E., Oberste M.S., Collett M.S., McCulloh R.J. First use of investigational antiviral drug pocapavir (v-073) for treating neonatal enteroviral sepsis. Pediatr. Infect. Dis. J. 2015;34:52–54. doi: 10.1097/INF.0000000000000497. [DOI] [PubMed] [Google Scholar]
- Woollard S.M., Kanmogne G.D. Maraviroc: a review of its use in HIV infection and beyond. Drug Des. Dev. Ther. 2015;9:5447–5468. doi: 10.2147/DDDT.S90580. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.