Highlights
-
•
Bats are natural reservoirs of many coronaviruses that can infect humans.
-
•
Mechanisms of cross-species transmission of coronaviruses are important scientific questions.
-
•
The coronaviral spike protein is an important viral determinant of cross-species transmission.
-
•
Receptor-binding characteristics and cleavage priming of the spike protein are summarized.
Keywords: coronavirus, interspecies transmission, viral and host determinants, spike (S), SARS-CoV, MERS-CoV
Abstract
Both severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV) are zoonotic pathogens that crossed the species barriers to infect humans. The mechanism of viral interspecies transmission is an important scientific question to be addressed. These coronaviruses contain a surface-located spike (S) protein that initiates infection by mediating receptor-recognition and membrane fusion and is therefore a key factor in host specificity. In addition, the S protein needs to be cleaved by host proteases before executing fusion, making these proteases a second determinant of coronavirus interspecies infection. Here, we summarize the progress made in the past decade in understanding the cross-species transmission of SARS-CoV and MERS-CoV by focusing on the features of the S protein, its receptor-binding characteristics, and the cleavage process involved in priming.
Coronavirus spike protein: a major viral determinant in interspecies transmission
Coronaviruses (CoVs) are large, enveloped, positive-sense, single-stranded RNA viruses that can infect both animals and humans [1]. The viruses are further subdivided, based on genotypic and serological characters, into four genera: Alpha-, Beta-, Gamma-, and Deltacoronavirus 2, 3. Thus far, all identified CoVs that can infect humans belong to the first two genera. These include the alphacoronaviruses (alphaCoVs) hCoV-NL63 and hCoV-229E and the betacoronaviruses (betaCoVs) HCoV-OC43, HKU1, SARS-CoV, and MERS-CoV 1, 4, 5. Special attention has been paid to betaCoVs, which have caused two unexpected coronaviral epidemics in the past decade [6]. In 2002–2003, SARS-CoV first emerged in China and swiftly spread to other parts of the world, leading to >8000 infection cases and ∼800 deaths [6]. In 2012, a novel CoV, named MERS-CoV, was identified in the Middle East 4, 5. The virus managed to spread to multiple countries despite intense human interventions, causing 1110 infections and 422 related deaths as of 29 April 2015 (http://www.who.int/csr/disease/coronavirus_infections/archive_updates/en/). Both SARS-CoV and MERS-CoV are zoonotic pathogens originating from animals. They are believed to have been transmitted from a natural host, possibly originating from bats, to humans through some intermediate mammalian hosts 7, 8. Thus, determining how these viruses evolved to cross species barriers and to infect humans is an active area of CoV research.
The key determinant of the host specificity of a CoV is the surface-located trimeric spike (S) glycoprotein, which can be further divided into an N-terminal S1 subunit and a membrane-embedded C-terminal S2 region [1]. S1 specializes in recognizing host-cell receptors and is normally more variable in sequence among different CoVs than is the S2 region 1, 9. Two discrete domains that can fold independently are located in the S1 N- and C-terminal portions, both of which can be used for receptor engagement [10]. The N-terminal domain (NTD), functioning as the entity involved in receptor recognition, is exemplified by murine hepatitis virus (MHV), which utilizes carcinoembryonic antigen cell-adhesion molecules (CEACAMs) for cell entry 11, 12. In most CoVs, however, the receptor-binding domain (RBD) is found in the S1 C-terminus 10, 13, 14, 15, 16, 17. In such cases, the NTD might facilitate the initial attachment of the virus to the cell surface by recognizing specific sugar molecules 18, 19, 20, 21. The S1–receptor interaction is therefore a key factor determining the tissue tropism and host range of CoVs.
Following receptor binding via S1, the CoV S2 functions to mediate fusion between the viral and the cellular membranes [1]. With characteristics of type I fusion proteins, CoV S2 normally contains multiple key components, including one or more fusion peptides and two conserved heptad repeats (HRs), driving membrane penetration and virus–cell fusion [1]. The fusion peptides are proposed to insert into, and perturb, the targeted membranes 22, 23. The HRs can trimerize into a coiled-coil structure and drag the virus envelope and the host cell bilayer into close proximity, preparing for fusion to occur 24, 25, 26, 27, 28. It is notable that the CoV S protein is commonly cleaved by host proteases to liberate S2 and the fusion peptides from the otherwise covalently-linked S1 subunit. This so-called priming process is highly dependent on the spatiotemporal patterns of the host enzymes, which is another key factor affecting cell tropism and the entry route of CoVs [29].
In this review, we first summarize the features of the S protein, the receptor-binding characteristics, the priming cleavage process, and the interspecies transmission mechanisms of SARS-CoV. Previous research on these topics has made SARS-CoV one of the best studied natural models of a viral disease emerging from zoonotic sources. Special attention will then be paid to MERS-CoV, focusing on the progress of the research made in the past several years regarding each of these items. We also retrospectively review several recent studies on bat coronaviruses (BatCoVs), which could implicate a zoonotic origin of MERS-CoV.
The SARS-CoV S glycoprotein, its cleavage priming and interaction with ACE2, and viral interspecies transmission
SARS-CoV S is a 1255-residue glycoprotein; it is suggested to be cleaved either between R667 and S668 by trypsin, or between T678 and M679 by endosomal cathepsin L, into S1 and S2 subunits 30, 31, although the functional relevance of T678 in virus–cell fusion remains to be fully investigated. Several important modules in both S1 and S2 have been systematically characterized thus far (Figure 1A,B). The SARS-CoV RBD is found in the C-terminal portion of S1, which spans ∼220 amino acids (Figure 1A). It is composed of two subdomains: a core and an external subdomain [13]. The core has a center β-sheet composed of five antiparallel strands, which are further surrounded by the polypeptide loops connecting the strands and several surface helices, together forming a globular fold. The external region consists mainly of two small β-strands and a large interstrand loop and is located distally to the terminal side of the domain. A portion of the interstrand loop extends extensively over the surface of the core subdomain, and, together with the two β-strands, anchors the external region to the core like a clamp (Figure 1B). It is interesting that one structure of the free SARS-CoV RBD unexpectedly revealed the possible dimerization of the protein through its terminal side [32]. The biological relevance of this structural observation, however, remains to be investigated. The authors suggest that RBD dimerization might cross-link S trimers on the viral surface, thereby affecting virus stability and infectivity. With systematic structural studies on SARS-CoV RBD, the structure of the SARS-CoV S NTD is still not known. It should be noted that this NTD, unlike its counterparts in bovine coronavirus (BCoV) or HCoV-OC43 20, 21, cannot recognize sugar moieties on mucin [12].
Figure 1.
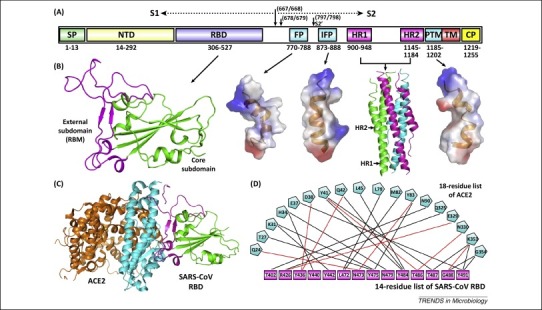
Severe acute respiratory syndrome coronavirus (SARS-CoV) spike features. (A) Schematic representation of the SARS-CoV spike protein (S). The individual components of S that were either experimentally characterized in previous studies – including receptor-binding domain (RBD), fusion peptide (FP), internal fusion peptide (IFP), heptad repeat 1/2 (HR1/2), and pretransmembrane domain (PTM) 13, 27, 35 – or are based on bioinformatics analyses, for example, N-terminal domain (NTD), are marked with the boundary-residue numbers listed below. The S1/S2 cleavage sites and the S2’-recognition site are highlighted. Other abbreviations: SP, signal peptide; TM, transmembrane domain; and CP, cytoplasmic domain. (B) Atomic structures of SARS-CoV spike RBD, FP, IFP, HR1/HR2 complex, and PTM (from left to right). The crystal structures of RBD (core subdomain in green and external subdomain in magenta) and the six-helix bundle fusion core (consisting of three HR1/HR2 helical hairpins in green, cyan, and magenta, respectively) are shown as ribbons, while the solution NMR structures of FP, IFP, and PTM are contoured using the electrostatic surface. (C) The complex structure between SARS-CoV RBD and its receptor ACE2. The core and external subdomains of RBD and the N- and C-terminal lobes of ACE2 are colored green, magenta, cyan, and orange, respectively. (D) The amino acid interactions at the RBD–ACE2 interface. According to a previous study [13], this binding network involves at least 18 residues in the receptor and 14 residues in SARS-CoV RBD, which are listed and connected with solid lines. Black lines indicate van der Waals contacts, and red lines represent H-bond or salt-bridge interactions.
To enter host cells, SARS-CoV needs to first bind to the cell-surface receptor ACE2 [33] via the viral RBD [13]. ACE2 is a type I membrane glycoprotein and contains a large N-terminal ectodomain built of two α-helical lobes 13, 34. The complex structure of SARS-CoV RBD bound to ACE2 revealed that the viral RBD utilizes its external subdomain to exclusively engage the N-terminal lobe of the receptor (Figure 1C). Residues 424–494 (which are also referred to as the receptor-binding motif or RBM because they make all of the contacts with the receptor) in the RBD external region present an elongated and gently concave outer surface, cradling the most N-terminal helix in ACE2. In addition, the two ridges of this RBM further interact with the receptor by contacting the α2/α3 interhelical loops on one side and a β-hairpin and a small helix on the other [13]. The buried surface area upon complex formation is 927.8 Å2 in the SARS-CoV RBD and 884.7 Å2 in ACE2, respectively. The interface involves at least 18 residues in the receptor and 14 residues in RBD, forming a network of hydrophilic contacts that are suggested to predominate in the RBD/ACE2 interactions (Figure 1D) [13].
After binding to ACE2, fusion between the SARS-CoV envelope and the host cell membrane is executed by the S2 subunit. Multiple fusion-related components in SARS-CoV S2 have been extensively studied thus far (Figure 1A,B). These include the fusion core composed of HR1 and HR2 27, 28 and at least three membranotropic regions that are denoted as the fusion peptide (FP), internal fusion peptide (IFP), and pretransmembrane domain (PTM), respectively [35]. The two HR modules are separately dispatched in S2 and are separated from each other by ∼200 residues. They form a coiled-coil structure built of three HR1–HR2 helical hairpins (Figure 1B) 27, 28, presenting as a canonical six-helix bundle, as observed in other typical type I fusion proteins such as HIV gp41 [36] and Ebola GP [37]. The HR regions are further flanked by the three membranotropic components. Both FP and IFP are located upstream of HR1, spanning residues 770–788 and 873–888, respectively, while PTM is distally downstream of HR2 and directly precedes the transmembrane domain of SARS-CoV S. All of these three components are able to partition into the phospholipid bilayer to disturb membrane integrity [38], and their structural features have recently been elucidated [35]. FP assumes an α-helical conformation but shows significant distortion at its center. In contrast, IFP exhibits a straight α-helical structure. PTM assumes a helix–loop–helix fold. It should be noted that all three components can create a hydrophobic side-surface (Figure 1B), explaining their bilayer-binding capacities [35]. The exact role of these putative fusion peptides in virus–cell fusion, however, remains to be fully examined; for example, it is currently unknown whether FP, IFP, and PTM function individually or in a synergistic manner. The evolutionary reservation of these hydrophobic amino acid sequences in SARS-CoV S highlights their potential participation in the viral entry process.
The priming process of SARS-CoV S by host proteases is likely one of the best characterized so far for viral envelope proteins. Indeed, the proteolytic activation mechanisms are summarized in several excellent reviews 29, 39, 40. What has been astonishing is that this viral protein can be primed via a diverse array of proteases. Due to the lack of a furin-recognizable site, SARS-CoV S is largely uncleaved after biosynthesis [30]. It can be later processed by endosomal cathepsin L during entry, enabling SARS-CoV infection via the endocytosis pathway [41]. In addition, the viral S can also be activated by extracellular enzymes such as trypsin, thermolysin, and elastase, which are shown to induce syncytia formation and virus entry, possibly at the plasma surface [42]. Other proteases that are of potential biological relevance in potentiating SARS-CoV S include TMPRSS2, TMPRSS11a, and HAT 43, 44, 45, which are localized on the cell surface and are highly expressed in the human airway [46]. It is also noteworthy that TMPRSS2 can associate with ACE2 to form a receptor–protease complex, enabling efficient virus entry directly at the cell surface [47]. Echoing the important role of TMPRSS2 in SARS-CoV infection, a recent study further indicated that serine proteases (e.g., TMPRSS2) but not cysteine proteases (e.g., cathepsin L) are required for SARS-CoV spread in vivo [48]. Furthermore, TMPRSS2 as well as other host enzymes, such as HAT and ADAM17, are also indicated in the shedding of human ACE2 receptor, which, in turn, was shown to promote the uptake of virus particles 49, 50. Remarkably, SARS-CoV S also contains an S2′ cleavage site downstream of the S1/S2 boundary 51, 52, 53. This second cleavage event is believed to be crucial for the final activation of S, and the sequence directly C-terminal to S2′ displays characteristics of a viral-fusion peptide and plays an important role in mediating fusion [54]. It is still unknown how the cleavage of S at S1/S2 or S2′, the insertion of the fusion peptides into target membranes, and the assembly of HR regions are combined together as concerted events to complete membrane fusion (e.g., whether these events occur following specific spatiotemporal patterns). It should be noted that SARS-CoV FP, which spans residues 770–788, would be separated from the HR regions after proteolytic cleavage at S2′. This indicates a scenario of membrane fusion with chronological steps such that FP initially targets the host cell membranes to facilitate the following bilayer insertion of IFP, which remains conjugated with the HR regions after S2′ proteolysis. Such a scenario also highlights the importance of including multiple fusion peptides in SARS-CoV S for virus entry.
The interspecies transmission route of SARS-CoV is well established. Mounting evidence shows that the natural hosts of the virus are bats 55, 56, 57. This notion was initially supported by the successful identification of SARS-like coronaviruses (SL-CoVs) in bats. Nevertheless, these viruses contain amino acid deletions in the S-RBM region and are unable to interact with human ACE2 55, 56. Recently, Ge et al. successfully isolated an infectious SL-CoV in Chinese horseshoe bats that shows far more sequence conservation in S to SARS-CoV than previously identified SL-CoVs do [56] and can recognize both bat and human ACE2 as the receptor [57], providing solid evidence for the bat origin of SARS-CoV. Palm civets and raccoon dogs were identified as the replication hosts for SARS-CoV [58], although it is still a matter of debate whether the virus is transmitted from bats to humans directly or via these intermediate animals. The ACE2 receptors of civets and raccoon dogs, however, can faithfully be recognized by SARS-CoV S 59, 60, 61. Mouse ACE2 can also be utilized by SARS-CoV but with much less efficiency than the human receptor [62]. This is because the mouse receptor contains a Lys-to-His mutation at position 353 and is therefore devoid of a key hydrophilic interaction rendered by the lysine residue [13]. Rat ACE2 also harbors this K353H mutation. In addition, it has an extra glycosylation site at position 82. The linked carbohydrate moieties are proposed to sterically occlude binding of SARS-CoV RBD to the rat receptor [13]. In support of this, deletion of the glycan, together with the H353K substitution, restores RBD-binding to the rat receptor 63, 64. In light of the inefficiency of SARS-CoV RBD in recognizing the mouse and rat receptors, it is unlikely that these two species are involved in the SARS-CoV zoonosis.
It is noteworthy that, of the 18 ACE2 residues interfacing with SARS-CoV RBD, multiple (≥7) amino acid substitutions are observed in the civet and raccoon receptors, in contrast to the receptors in other infection-permissive species [such as monkey (African green monkey), macaque, marmoset, hamster, and cat] (reviewed in [65]) that contain ≤4 mutations in the region (Table 1 ). Furthermore, ferret ACE2 (with nine substitutions relative to the human homologue) was mutated for half of the interface residues (Table 1) but can still be recognized by SARS-CoV S [66]. These observations indicate plastic RBD/ACE2 interactions which can ‘tolerate’ relatively large variations in the receptor. The inability of ACE2 of a certain species functioning as the SARS-CoV receptor, therefore, likely arises from combinations of certain mutations. For example, the mutation incorporating a potential N-glycosylation site at N82 in conjugation with the K353H substitution in rat ACE2, but not a single M82N mutation as observed in hamster ACE2, abrogate the receptor's binding capacity for SARS-CoV S. It is also notable that ACE2s of different bat species behave differently regarding serving as the receptor for SARS-CoV [59]. ACE2 of Chinese rufous horseshoe bat Rhinolophus sinicus, but not that of Pearson's horseshoe bat Rhinolophus pearsonii, supports S-mediated SARS-CoV infection [59], although the receptor proteins of the two species both contain seven mutations in the RBD-interfacing region (Table 1). The structural basis underlying this observed difference remains to be illustrated.
Table 1.
Comparison among different species of the ACE2 residues interfacing with severe acute respiratory syndrome coronavirus (SARS-CoV) receptor-binding domain (RBD)a
| Position Species |
24 | 27 | 31 | 34 | 37 | 38 | 41 | 42 | 45 | 79 | 82 | 83 | 90 | 325 | 329 | 330 | 353 | 354 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | Q | T | K | H | E | D | Y | Q | L | L | M | Y | N | Q | E | N | K | G |
| African green monkey | Q | T | K | H | E | D | Y | Q | L | L | M | Y | N | Q | E | N | K | G |
| Macaque | Q | T | K | H | E | D | Y | Q | L | L | M | Y | N | Q | E | N | K | G |
| Marmoset | Q | T | K | H | E | D | H | E | L | L | T | Y | N | Q | E | N | K | Q |
| Hamster | Q | T | K | Q | E | D | Y | Q | L | L | N | Y | N | Q | E | N | K | G |
| Cat | L | T | K | H | E | E | Y | Q | L | L | T | Y | N | Q | E | N | K | G |
| Civet | L | T | T | Y | Q | E | Y | Q | V | L | T | Y | D | Q | E | N | K | G |
| Raccoon | L | T | N | N | E | E | Y | Q | L | Q | T | Y | D | Q | E | N | K | G |
| Ferret | L | T | K | Y | E | E | Y | Q | L | H | T | Y | D | E | Q | N | K | R |
| Mouse | N | T | N | Q | E | D | Y | Q | L | T | S | F | T | Q | A | N | H | G |
| Bat (R. sinicus) | R | T | E | S | E | N | Y | Q | L | L | N | Y | N | E | N | N | K | G |
| Rat | K | S | K | Q | E | D | Y | Q | L | I | N | F | N | P | T | N | H | G |
| Bat (R. pearsonii) | R | T | K | H | E | D | H | E | L | L | D | Y | N | E | N | N | K | D |
The 18 residues in human ACE2 that are identified to interface with SARS-CoV RBD were listed and compared for the conservatism in different species. The letters in red highlight the amino acid mutations at the corresponding positions, which are based on human ACE2 numbering. The ACE2 receptors that can be recognized by the SARS-CoV S protein include those from human, monkey (African green monkey), macaque, marmoset, hamster, cat, civet, raccoon dog, ferret, mouse, and bat (Rhinolophus sinicus, R. sinicus), although the mouse and bat (R. sinicus) ACE2s are utilized inefficiently. The rat and bat (Rhinolophus pearsonii, R. pearsonii) receptors, however, are unable to be used by SARS-CoV. Accession numbers: human (AY623811), monkey (AY996037), macaque (NM_001135696), marmoset (XM_008988993), hamster (XM_005074209), cat (NM_001039456), civet (AY881174), raccoon (AB211998), ferret (AB208708), mouse (EF408740), bat (R. sinicus) (GQ999936), rat (AY881244), bat (R. pearsonii) (EF569964).
The S adaptation for binding to the human receptor is also well recorded for SARS-CoV. Comparison of the RBD sequences of SARS-CoV isolated from humans and civets revealed six residue-substitutions [67], among which three (at positions 472, 479, and 487, respectively) belong to the 14-interfacing-residue list (Figure 1D). K479N and S487T mutations have been reported in several studies 64, 68, 69 as the key changes in adapting SARS-CoV RBD for the human receptor. S protein with the civet-specific K479 and S487 residues can efficiently recognize civet ACE2 but interacts with human ACE2 much less efficiently [64]. Substitution of these two amino acids with the human-specific N479 and T487, either individually or in combination, dramatically increases the affinity of S for the human receptor 64, 68. This increased binding affinity is believed to be related to the elimination of unfavorable free charges at the interface upon mutation [70] and the extra contacts established by the methyl group of T487 [71]. Residue changes at other positions in the RBM might also be related to the SARS-CoV adaption. For instance, a virus bearing the civet S with the K479N mutation was passaged on human airway epithelial cells. Adaptive substitution occurred at residues 442 and 472, rather than at the 487 site identified in the epidemic strains [69]. The changes in SARS-CoV S required for interspecies transmission are also exemplified in two independent studies on mouse-adapted viruses. Two groups identified the same S-substitution at position 436, which is believed to be directly linked to the enhanced infectivity and pathogenesis in the murine host 72, 73.
MERS-CoV S, its cleavage priming and interaction with CD26, and viral interspecies transmission
MERS-CoV S is composed of 1353 residues and displays a remarkably similar domain arrangement to its SARS-CoV homologue (Figure 2A), although the overall sequence identity between the two viral proteins is rather limited. However, unlike SARS-CoV S, the MERS-CoV S protein can be readily processed into S1 and S2 subunits upon expression 74, 75, 76. In S1, the receptor-recognizing RBD is localized to the C-terminal portion, spanning ∼240 residues 16, 17, 77. These amino acids fold into a structure consisting of two subdomains, as reported in the SARS-CoV equivalent. The core subdomain presents remarkable similarities to that of the SARS-CoV RBD, but the external subdomain is structurally distinct from the SARS-CoV RBD external region and comprises mainly four antiparallel β-strands (Figure 2B). In S2, the HR regions are also well studied 26, 78. As expected, the HR1 and HR2 of MERS-CoV also form an intra-hairpin helical structure that can trimerically assemble into a six-helix bundle (Figure 2B), demonstrating a canonical membrane-fusion mechanism as reported for other type I fusion proteins [24]. These studies provide insight into the characteristics of MERS-CoV S. Nevertheless, other S-components of this novel CoV remain largely uninvestigated. For example, it is still unknown whether the RBD-preceding NTD of MERS-CoV S1 might similarly fold into a galectin-like structure (as in MHV [12]) and function to facilitate the initial viral attachment to the cell surface by recognizing certain sugar molecules (as in BCoV and HCoV-OC43 20, 21). In addition, the S2 fusion peptides of MERS-CoV must also be experimentally investigated, although similar concentration of hydrophobic residues to the SARS-CoV FP, IFP, and PTM can be individually identified in the equivalent regions of MERS-CoV S (Figure 2B).
Figure 2.
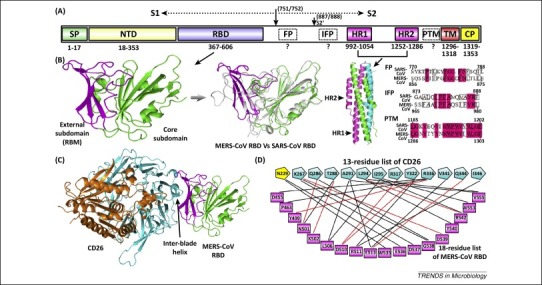
Middle East respiratory syndrome coronavirus (MERS-CoV) spike features. (A) Schematic representation of the MERS-CoV spike protein. The boundaries for the individual components, as well as the S1/S2 and S2’ cleavage sites, are marked. Abbreviations: SP, signal peptide; NTD, N-terminal domain; RBD, receptor-binding domain; FP, fusion peptide; IFP, internal fusion peptide; HR1/2, heptad repeat 1/2; PTM, pre-transmembrane domain; TM, transmembrane domain; and CP, cytoplasmic domain. Question marks highlight the fusion peptides (FP, IFP, and PTM) of MERS-CoV that still await structural and functional characterization. (B) Crystal structures of the MERS-CoV spike RBD and HR1/HR2 fusion core. Left panel: the RBD structure with its core subdomain highlighted in green and external subdomain in magenta. Middle-left panel: a structural superimposition between MERS-CoV RBD (core and external subdomains in green and magenta, respectively) and severe acute respiratory syndrome coronavirus (SARS-CoV) RBD (in gray). Middle-right panel: the fusion core structure with the three HR1/HR2 chains in green, cyan, and magenta, respectively. Right panel: sequence comparison between SARS-CoV and MERS-CoV highlighting the spike regions of SARS-CoV FP, IFP, and PTM, respectively. Important hydrophobic residues are marked in boxes. (C) The complex structure between MERS-CoV RBD and the receptor CD26/DPP4. MERS-CoV RBD is colored as in panel (B), and the receptor is highlighted in cyan for the β-propeller domain and in orange for the α/β-hydrolase domain, respectively. The inter-blade helix referred to in the text is marked. (D) Atomic binding-network between MERS-CoV RBD and CD26 [16]. The RBD–CD26 interface includes 13 amino acids from the receptor and 18 residues from the virus RBD, which are individually connected with either black lines, for van der Waals contacts, or red lines, for H-bond or salt-bridge interactions. The CD26 residue N229 contributes to the RBD-binding via its linked sugar moieties rather than directly engaging RBD, and is therefore highlighted in yellow.
MERS-CoV initiates human infection by first specifically interacting with its receptor CD26 (also known as dipeptidyl peptidase 4 or DPP4) [79]. CD26 is a membrane-bound peptidase with a type II topology and can form homodimers on the cell surface 80, 81, 82. Its ectodomain structurally comprises two domains, an α/β-hydrolase domain and an eight-bladed β-propeller 81, 82. The MERS-CoV RBD specifically recognizes, via its external subdomain, the β-propeller of the receptor for engagement (Figure 2C) 16, 17. The four external β-strands of the RBD create a relatively flat surface to interact with the propeller blades IV and V. Large surface areas of 1203.4 Å2 in CD26 and 1113.4 Å2 in MERS-CoV RBD are buried to form an extended binding interface [16], in which 13 residues of the receptor and 18 amino acids of the RBD play important roles in the binding by providing either H-bond/salt-bridge interactions or multiple van-der-Waals contacts (Figure 2D). Among these, a strong network of hydrophilic contacts is created mainly with the interface-residue side-chains. In addition, a small hydrophobic depression in RBD further cradles the bulged inter-blade helix in the receptor, which presents several apolar side-chains (Figure 2C). Finally, the RBD and CD26 binding also involves a receptor-linked carbohydrate entity interacting with several solvent-exposed residues in the RBD (Figure 2D), drawing parallels between MERS-CoV and the alphaCoV porcine respiratory coronavirus. The latter also recognizes a sugar component in the receptor [15]. What has been unexpected regarding the MERS-CoV binding to CD26 is its competitive interference with the interaction between CD26 and adenosine deaminase (ADA), which has been suggested to deliver an important costimulatory signal in immune activation [80]. A majority of the CD26 residues interfacing with MERS-CoV RBD are also shown to engage ADA 16, 17, 83.
The host proteases involved in the priming of MERS-CoV S have also been broadly studied thus far. A pioneering study demonstrated that MERS-CoV S, unlike its SARS-CoV counterpart, can be efficiently cleaved after biosynthesis in HEK-293T cells [74]. It was recently demonstrated that the cleavage occurs at R751/S752, separating S into S1 and S2 subunits by furin [76]. In addition, a second furin cleavage site (S2′) was identified in S2, upstream of the putative fusion peptide that likely corresponds to SARS-CoV IFP, between R887 and S888 (Figure 2A) [76]. With mounting evidence showing that processing at S2′ is an essential determinant of the intracellular site of fusion [84], a two-step activation mechanism for MERS-CoV entry [76] has been proposed such that the former cleavage occurs between S1 and S2 during the secretion of S protein in the endoplasmic reticulum (ER)-Golgi compartments, where furin is localized, and the latter at S2′ during virus entry into target cells. The other reported proteases involved in MERS-CoV S-activation include TMPRSS2 74, 85, TMPRSS4 [86], and endosomal cathepsin B and/or L 74, 85. It is noteworthy that MERS-CoV, similar to SARS-CoV, might use different activation pathways for cell entry depending on the spatiotemporal patterns of the host priming enzymes [87]. For example, the presence of TMPRSS2 or trypsin treatment can bypass the endosomal entry pathway to initiate membrane fusion at the cell surface 85, 87.
The cross-species transmission route of MERS-CoV remains not well known. Nevertheless, mounting evidence indicates that the virus is a zoonotic pathogen which likely originated first in bats and was then transmitted to other animals (dromedary). Despite several studies documenting the interhuman transmission of MERS-CoV 88, 89, a large portion of the cases of infection cannot be directly linked to contacts with index patients. The genome diversity of human MERS-CoV isolates is highly suggestive of human infections from several independent zoonotic events from animal reservoirs 90, 91. The dromedary camel has thus far been well documented as an intermediate host. Both MERS-CoV-specific antibodies and RNAs can be detected in dromedary sera and milk 92, 93, 94, and live viruses were recently isolated from infected camels [95]. Additional direct evidence of dromedary-to-human transmission comes from the isolation of MERS-CoVs with almost identical genomic sequences from patients and from their breeding dromedaries 96, 97. Viral gene fragments identical or quite similar to those of MERS-CoV have also been recovered in bats 98, 99, 100, raising again the possibility that the bat acts as the natural reservoir of MERS-CoV. An evolutionary analysis of bat CD26 genes indicates a long-term arms race between bats and MERS-related CoVs, suggesting that MERS-CoV ancestors circulated in bats for a substantial period of time [101]. It is also interesting to note that a recent study indicates that MERS-CoV may have jumped from bats to camels up to 20 years ago in Africa, with the camels then being imported into the Arabian peninsula [102].
Multiple cells (primary or cell lines) derived from different species have been investigated for susceptibility to MERS-CoV infection. The results show that cells of rhesus macaque, marmoset, goat, horse, rabbit, pig, civet, camel, and bat – but not of mouse, hamster, and ferret – are permissive to MERS-CoV replication 87, 103, 104, 105, 106, 107, 108, 109, 110. By focusing on the list of the 13 residues that were identified as key interface amino acids in the receptor, it is noteworthy that the receptor in species of the permissive group is either identical to the human receptor or varies from it by only one or two residues, whereas the receptor of species in the resistant group is more variant, showing multiple (≥5) substitutions (Table 2 ). The inability of MERS-CoV to infect mouse, hamster, and ferret should therefore be attributed to the inability of the virus to recognize the CD26s of these species, which contain too many mutations in the RBD-binding region. In support of this, expression of hamster CD26 whose variant residues are substituted with the equivalent human amino acids in otherwise nonpermissive baby hamster kidney (BHK) cells restores the viral infection by MERS-CoV [109]. These results demonstrate that the binding capacity by MERS-CoV RBD is a key factor determining the host susceptibility to MERS-CoV infection. It has yet to be determined whether dog and cat, which clearly belong to the second group, are resistant to the virus. It would be of more interest to investigate the 13-residue list in the future for the amino acid combinations that are least required for interaction with MERS-CoV RBD.
Table 2.
Comparison among different species of the CD26 residues interfacing with Middle East respiratory syndrome coronavirus (MERS-CoV) receptor-binding domain (RBD)a
| Position Species |
229 | 267 | 286 | 288 | 291 | 294 | 295 | 317 | 322 | 336 | 341 | 344 | 346 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | N | K | Q | T | A | L | I | R | Y | R | V | Q | I |
| Macaque | N | K | Q | T | A | L | I | R | Y | R | V | Q | I |
| Marmoset | N | K | Q | T | A | L | I | R | Y | R | V | Q | I |
| Cattle | N | K | Q | V | G | L | I | R | Y | R | V | Q | I |
| Horse | N | K | Q | T | A | L | I | R | Y | R | V | Q | I |
| Goat | N | K | Q | V | G | L | I | R | Y | R | V | Q | I |
| Pig | N | K | Q | V | A | L | I | R | Y | R | V | Q | I |
| Camel | N | K | Q | V | A | L | I | R | Y | R | V | Q | I |
| Sheep | N | K | Q | V | G | L | I | R | Y | R | V | Q | I |
| Rabbit | N | R | Q | T | A | L | I | R | Y | R | V | Q | I |
| Bat (Pipistrellus) | N | K | Q | T | A | L | T | R | Y | K | V | Q | I |
| Cat | N | K | E | T | A | L | T | R | Y | K | A | E | I |
| Dog | N | K | E | S | L | L | T | R | Y | – | S | K | I |
| Ferret | N | K | E | T | D | S | T | R | Y | S | E | E | T |
| Hamster | N | K | Q | T | E | L | T | R | Y | T | L | Q | V |
| Rat | N | K | Q | T | A | T | T | R | Y | V | T | E | I |
| Mouse | N | K | Q | P | A | A | R | R | Y | T | S | Q | V |
The 13 residues in human CD26 that are identified to be key interfacing amino acids for MERS-CoV RBD binding were listed and compared for the conservatism in different species. The letters in red highlight the amino acid mutations at the corresponding positions, which are based on human CD26 numbering. Two groups can be identified: the former (permissive), including human, macaque, marmoset, cattle, horse, goat, pig, camel, sheep, rabbit and bat, has accumulated small numbers (0–2) of mutations in the 13-residue list; whereas the latter (resistant), with cat, dog, ferret, hamster, rat and mouse, contains multiple (≥ 5) substitutions in the region. Accession numbers: human (NP_001926), macaque (NP_001034279), marmoset (XM_002749392), cattle (NM_174039), horse (XP_001494049), goat (KF574265), pig (NM_214257), camel (AHK13386), sheep (XP_004004709), rabbit (XP_002712206), Bat (Pipistrellus) (AGF80256), cat (NP_001009838), dog (XP_535933), ferret (KF574264), hamster (XP_007608372), rat (NP_036921), and mouse (NP_034204).
It should also be noted that sheep and bovine CD26s contain the same two residue-variances as goat and are shown to mediate MERS-CoV infection of BHK cells upon expression [109]. Nevertheless, another study demonstrated that cells derived from sheep and cattle are resistant to MERS-CoV [106], and accordingly, no MERS-CoV-specific antibodies were detected in the sera of 80 tested cattle and 40 sheep in an epidemiologic survey [93]. The discrepancy in these results might reflect the difference in the priming-protease system between sheep/cattle cells and BHK cells. Although MERS-CoV can recognize sheep/cattle CD26, the lack of appropriate proteases for S-activation would incapacitate the membrane fusion and the subsequent virus entry. The hamster-derived BHK cells, on the other hand, are able to prime MERS-CoV S and therefore become infection-permissive after gaining the capacity to interact with MERS-CoV RBD. A similar scenario is also observed in mice, which can be effectively infected by MERS-CoV after ectopic expression of human CD26 in the animal [111]. Characterization in different species of the spatiotemporal patterns of the enzymes that prime MERS-CoV S represents an interesting and as-yet-unresolved issue.
The changes in S related to MERS-CoV interspecies adaptation are thus far unknown. Several genetic analyses were recently conducted to characterize the evolutionary status of the virus since its identification in 2012. The results show that the MERS-CoV RBD has largely remained unchanged in sequence in the circulating viruses. In a study focusing on the human MERS-CoV strains, the authors demonstrate that only one codon of spike residue 1020 (located in S2) is under strong positive selection, despite the fact that the overall evolutionary rate of the virus is estimated to be 1.12 × 10−3 substitutions per site per year [112]. Several substitutions have also been detected in the S-RBM region of some MERS-CoV strains, including those at positions 482, 506, 509, and 534. Among these, only L506 plays an important role in CD26 binding (Figure 2D). The identified L506F mutation, however, reduces the receptor-binding capacity and thereby impairs viral fitness [113]. It should be noted that artificial selection of escape mutants with MERS-CoV RBD-specific antibodies can lead to the same L506F substitution [113], raising the possibility that the naturally occurring residue change at this position is the consequence of host immune pressure rather than a result of evolution for a better affinity to CD26. Accordingly, none of the identified S-changes are observed in multiple genomes [112]. A second study analyzed the MERS-CoV sequences of the dromedary isolates and identified only the A520S substitution in the RBD [114]. Although this residue is located in the external subdomain, it does not directly contact the receptor. Therefore, it remains to be investigated whether any residue substitutions in the RBD occur naturally and can facilitate cross-species transmission of MERS-CoV by increasing the S affinity for human CD26. The current data indicate that the combination of the 18 RBD amino acids listed in Figure 2D remains dominant in the circulating strains, both in humans and dromedaries. This seems to favor the notion that the present MERS-CoV RBM sequence represents one of the best CD26-interacting candidates. Residues that are determinant for MERS-CoV S preference for binding to CD26 of a certain species still await identification.
BatCoV HKU4 S protein interaction with CD26 and its implication for the bat origin of MERS-CoV
A large number of coronaviruses have been recorded as having origins in bats (at least for their genomes) [115]. However, their public health relevance and/or evolutionary relatedness to the known human-infecting coronaviruses remain to be examined. BatCoVs HKU4 and HKU5 have recently drawn increasing attention due to their close phylogenetic relationship to MERS-CoV [116]. These CoVs were first identified as genomic sequences in 2005 in lesser bamboo bats and Japanese pipistrelles, respectively [117]. Though isolation of the infectious viruses has thus far been unsuccessful, mounting evidence indicates that these two viruses are still circulating in bats [118]. Recently, Yang et al. [119] and our group [75] concomitantly showed that BatCoV HKU4, but not HKU5, can recognize human CD26 as a functional receptor for cell entry. HKU4 S is composed of 1352 residues (Figure 3A) and can readily interact with human CD26 [75]. But it does not contain a clear furin-recognition site [29] and is expressed as an intact protein in 293T cells, remaining uncleaved upon incorporation into the pseudoviral envelope. Accordingly, the BatCoV HKU4 pseudovirus was unable to infect cells expressing human CD26 [75]. But potential trypsin-cleavage sequences can be identified in two regions homologous to the S1/S2 and S2′ sites of other CoVs [29], and trypsin treatment indeed efficiently primes HKU4 S and leads to sufficient pseudoviral transductions [75]. These observations revealed the fact that the inability of HKU4 S to drive entry into human cells (and thus, potentially, to be transmitted to humans) is due to lack of priming and not to lack of receptor engagement, highlighting once again the indispensability of S cleavage in coronavirus infection. Despite lacking recognizable sites for furin, it remains to be investigated whether HKU4 S might be activated by any other commonly observed priming proteases, such as TMPRSSs and cathepsins. Special attention should be paid to virus variants that are more susceptible to protease cleavage by host enzymes other than trypsin.
Figure 3.
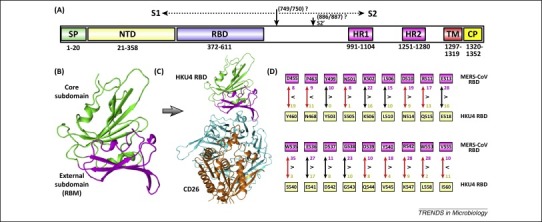
Bat coronavirus (BatCoV) HKU4 spike features. (A) Schematic representation of the HKU4 spike protein. The listed component boundaries are mostly defined according to the bioinformatics analyses, except for the RBD which has been experimentally characterized [75]. The cleavage sites for S1/S2 and S2’ were predicted based on the homology sequence comparison with other coronaviruses and are therefore labeled with question marks. Abbreviations: SP, signal peptide; NTD, N-terminal domain; RBD, receptor-binding domain; HR1/2, heptad repeat 1/2; TM, transmembrane domain; and CP, cytoplasmic domain. (B) Crystal structure of HKU4 RBD. The external and core subdomains are colored magenta and green, respectively. (C) Complex structure between HKU4 RBD and human CD26. The coloring scheme is: RBD core, green; RBD external, magenta; receptor β-propeller domain, cyan; and receptor α/β-hydrolase domain, orange. (D) The HKU4 RBD is suboptimal for CD26 interaction compared to Middle East respiratory syndrome coronavirus (MERS-CoV) RBD [75]. The 18 CD26-interfacing residues in MERS-CoV RBD, as listed in Figure 2D, were individually compared with the equivalent amino acids in HKU4 RBD. The numbers highlight the van der Waals contacts each residue can provide for interacting with CD26. ‘>’ indicates that the MERS-CoV residues are better adapted for CD26-binding, and conversely, ‘<’ implies that the HKU4 amino acids are better adapted. The residue differences are highlighted with red arrows.
The RBD of BatCoV HKU4, which spans residues 372–611 (Figure 3A), has also been structurally characterized [75]. It displays a fold that resembles the MERS-CoV RBD (Figure 3B) and utilizes a conserved receptor binding mode for interaction with CD26 (Figure 3C). Interestingly, of the 18 identified CD26-interfacing residues in MERS-CoV RBD, 11 amino acids are mutated and 15 are suboptimal for receptor interaction in HKU4 RBD (Figure 3D) [75]. Nonetheless, a pseudoviral infection assay demonstrates that HKU4 S is able to mediate virus entry, although less efficiently than MERS-CoV S. These results indicate that dramatic changes at this 18-residue interface do not necessarily abrogate the interaction between viral S and CD26, which in return provides the space for MERS-CoV and the related viruses (e.g., BatCoV HKU4) to evolve to escape from the neutralizing antibodies targeting the RBM and to facilitate interspecies transmission. It is also notable that BatCoV HKU4 exhibits better binding capacity for bat CD26 than for human CD26 [119], but a converse CD26-interaction has been reported for MERS-CoV [119]. This implies a common ancestor in bats for MERS-CoV and BatCoV HKU4, which divergently evolved for better interaction with the human and bat receptors, respectively. These studies also indicate the need for surveillance of HKU4-related viruses for their cross-species potential in the future.
It is notable that SARS-CoV seems to ‘tolerate’ large variations in the receptor (as illustrated in ferret ACE2 with half of the interfacing residues being substituted). Small variations in the viral RBD (with N479K and T487S), however, can lead to altered receptor-binding specificity, dramatically decreasing its affinity for human ACE2. In contrast, MERS-CoV likely only recognizes conserved CD26 sequences with a maximum of two mutations in the RBD-binding region. Nevertheless, the capacity of receptor engagement can still be reserved despite dramatic changes in the viral ligand (as demonstrated in HKU4 RBD). These differences could indicate different evolutionary and interspecies transmission routes between SARS-CoV and MERS-CoV, which would be an interesting issue awaiting answers.
Concluding remarks
The emergence of two betaCoV-related epidemics in the past decade revitalized CoV research, focusing on the interspecies transmission mechanisms of these viruses. The CoV S protein is a key factor in determining viral tissue tropism and host range. Much progress has been made thus far regarding the features of S, the interaction of S with receptors, and the priming of S by host proteases. Although SARS-CoV represents one of the best studied models for which the cross-species transmission route has been well established, many questions related to MERS-CoV interspecies transmission remain unanswered (Box 1 ). These include, but are not limited to, the structure and function of the S NTD, the composition of the fusion peptides, the key determinants in S for CD26 interaction, and the virus/host interplay determining the entry route of the virus. Such questions should be systematically addressed in the future. It is also noteworthy that all current views on CoV S are built on the discrete functional domains. An intact S structure is not available for any CoV, although the low-resolution electron-microscopy structure of SARS-CoV S has been reported 120, 121. Having an intact S structure with high resolution would be an interesting issue deserving even higher priority (Box 1). In summary, this review focused on our understanding of the coronaviral S proteins to illustrate the interspecies transmission basis of SARS-CoV, MERS-CoV, and beyond, the knowledge of which should be able to help prevent or predict further transmission events.
Box 1. Outstanding questions.
-
•
The fusion peptides of MERS-CoV S still await structural and functional characterization. Could any of these fusion peptides be targeted by small molecules to inhibit virus infection?
-
•
What will be revealed by systematic and comparative studies on the spatiotemporal characteristics of the enzymes potentially involved in MERS-CoV S-priming among different species?
-
•
In the list of the 13 CD26 residues that interface with the MERS-CoV RBD, what residue combination(s) constitute the key component that is indispensable in RBD-binding? The answers to this and the second point would enable us to predict the infection and transmission capacity of MERS-CoV in a specific species.
-
•
Is the dromedary camel the only intermediate host of MERS-CoV, or are other animals also involved in the interspecies transmission of the virus from its natural host, possibly bat, to humans? Special attention should be paid to the livestock animals in the first group (Table 2) whose CD26 receptors are able to be recognized by MERS-CoV, although no evidence of these animals being infected by MERS-CoV has come to light thus far. In addition, pets such as cats and dogs in the second group (Table 2) are in close contact with humans and should be investigated to ensure that they do not carry MERS-CoV.
-
•
What S-substitutions are involved in the interspecies adaptation of MERS-CoV? A large-scale genomic characterization of the MERS-CoV isolates from human and dromedaries, and of the MERS-CoV-related viruses from bats, should be conducted, focusing on the residue changes in the receptor-binding region, to determine whether there are any naturally occurring mutations that enhance or decrease its binding capacity for human or camel CD26. It is of equal importance to identify, via artificial substitutions, the key residues determining the preference of MERS-CoV S for the CD26 of a certain species.
-
•
What is the role of the SARS-CoV and MERS-CoV NTD in virus infection? Do they share structural features with galectin, as reported in betaCoVs such as HCoV-OC43 and BCoV?
-
•
What do we expect to observe at the atomic level in an intact S trimer? An intact S structure has not been solved for any CoV.
Acknowledgments
Work on coronavirus in the laboratory of G.F.G. is supported by the National Natural Science Foundation of China (NSFC, grant numbers 81461168030 and 31400154) and the China National Grand S&T Special Project (number 2014ZX10004-001-006). G.F.G. is a leading principal investigator of the NSFC Innovative Research Group (grant number 81321063). G.L. is supported by the Excellent Young Scientist Grant from the Chinese Academy of Sciences.
Contributor Information
Guangwen Lu, Email: luguangwen2001@126.com.
George F. Gao, Email: gaof@im.ac.cn.
References
- 1.Lai M.M. Coronaviridae. In: Knipe D.M., editor. Fields Virology. 5th edn. Lippincott Williams & Wilkins; 2007. pp. 1305–1336. [Google Scholar]
- 2.International Committee on Taxonomy of Viruses, King A.M.Q. Academic Press; 2012. Virus Taxonomy: Classification and Nomenclature of Viruses: Ninth Report of the International Committee on Taxonomy of Viruses. [Google Scholar]
- 3.Adams M.J., Carstens E.B. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2012) Arch. Virol. 2012;157:1411–1422. doi: 10.1007/s00705-012-1299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bermingham A. Severe respiratory illness caused by a novel coronavirus, in a patient transferred to the United Kingdom from the Middle East, September 2012. Euro. Surveill. 2012;17:20290. [PubMed] [Google Scholar]
- 5.Zaki A.M. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 6.Weinstein R.A. Planning for epidemics – the lessons of SARS. N. Engl. J. Med. 2004;350:2332–2334. doi: 10.1056/NEJMp048082. [DOI] [PubMed] [Google Scholar]
- 7.Bolles M. SARS-CoV and emergent coronaviruses: viral determinants of interspecies transmission. Curr. Opin. Virol. 2011;1:624–634. doi: 10.1016/j.coviro.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Al-Tawfiq J.A., Memish Z.A. Middle East respiratory syndrome coronavirus: transmission and phylogenetic evolution. Trends Microbiol. 2014;22:573–579. doi: 10.1016/j.tim.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Masters P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006;66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li F. Evidence for a common evolutionary origin of coronavirus spike protein receptor-binding subunits. J. Virol. 2012;86:2856–2858. doi: 10.1128/JVI.06882-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams R.K. Receptor for mouse hepatitis virus is a member of the carcinoembryonic antigen family of glycoproteins. Proc. Natl. Acad. Sci. U.S.A. 1991;88:5533–5536. doi: 10.1073/pnas.88.13.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peng G. Crystal structure of mouse coronavirus receptor-binding domain complexed with its murine receptor. Proc. Natl. Acad. Sci. U.S.A. 2011;108:10696–10701. doi: 10.1073/pnas.1104306108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li F. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 14.Wu K. Crystal structure of NL63 respiratory coronavirus receptor-binding domain complexed with its human receptor. Proc. Natl. Acad. Sci. U.S.A. 2009;106:19970–19974. doi: 10.1073/pnas.0908837106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reguera J. Structural bases of coronavirus attachment to host aminopeptidase N and its inhibition by neutralizing antibodies. PLoS Pathog. 2012;8:e1002859. doi: 10.1371/journal.ppat.1002859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu G. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature. 2013;500:227–231. doi: 10.1038/nature12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang N. Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res. 2013;23:986–993. doi: 10.1038/cr.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwegmann-Wessels C., Herrler G. Sialic acids as receptor determinants for coronaviruses. Glycoconjugate J. 2006;23:51–58. doi: 10.1007/s10719-006-5437-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krempl C. Point mutations in the S protein connect the sialic acid binding activity with the enteropathogenicity of transmissible gastroenteritis coronavirus. J. Virol. 1997;71:3285–3287. doi: 10.1128/jvi.71.4.3285-3287.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schultze B. The S-protein of bovine coronavirus is a hemagglutinin recognizing 9-O-acetylated sialic-acid as a receptor determinant. J. Virol. 1991;65:6232–6237. doi: 10.1128/jvi.65.11.6232-6237.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kunkel F., Herrler G. Structural and functional analysis of the surface protein of human coronavirus OC43. Virology. 1993;195:195–202. doi: 10.1006/viro.1993.1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Epand R.M. Fusion peptides and the mechanism of viral fusion. Biochim. Biophys. Acta. 2003;1614:116–121. doi: 10.1016/s0005-2736(03)00169-x. [DOI] [PubMed] [Google Scholar]
- 23.Peisajovich S.G., Shai Y. Viral fusion proteins: multiple regions contribute to membrane fusion. Biochim. Biophys. Acta. 2003;1614:122–129. doi: 10.1016/s0005-2736(03)00170-6. [DOI] [PubMed] [Google Scholar]
- 24.Gao G.F. Peptide inhibitors targeting virus-cell fusion in class I enveloped viruses. In: Torrence P.F., editor. Combating the Threat of Pandemic Influenza: Drug Discovery Approaches. John Wiley; 2007. pp. 226–246. [Google Scholar]
- 25.Eckert D.M., Kim P.S. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 2001;70:777–810. doi: 10.1146/annurev.biochem.70.1.777. [DOI] [PubMed] [Google Scholar]
- 26.Gao J. Structure of the fusion core and inhibition of fusion by a heptad repeat peptide derived from the S protein of Middle East respiratory syndrome coronavirus. J. Virol. 2013;87:13134–13140. doi: 10.1128/JVI.02433-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu Y. Crystal structure of severe acute respiratory syndrome coronavirus spike protein fusion core. J. Biol. Chem. 2004;279:49414–49419. doi: 10.1074/jbc.M408782200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu J. Following the rule: formation of the 6-helix bundle of the fusion core from severe acute respiratory syndrome coronavirus spike protein and identification of potent peptide inhibitors. Biochem. Biophys. Res. Commun. 2004;319:283–288. doi: 10.1016/j.bbrc.2004.04.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Millet J.K., Whittaker G.R. Host cell proteases: Critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2014;202:120–213. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simmons G. Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc. Natl. Acad. Sci. U.S.A. 2004;101:4240–4245. doi: 10.1073/pnas.0306446101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bosch B.J. Cathepsin L functionally cleaves the severe acute respiratory syndrome coronavirus class I fusion protein upstream of rather than adjacent to the fusion peptide. J. Virol. 2008;82:8887–8890. doi: 10.1128/JVI.00415-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hwang W.C. Structural basis of neutralization by a human anti-severe acute respiratory syndrome spike protein antibody, 80R. J. Biol. Chem. 2006;281:34610–34616. doi: 10.1074/jbc.M603275200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li W. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Towler P. ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis. J. Biol. Chem. 2004;279:17996–18007. doi: 10.1074/jbc.M311191200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mahajan M., Bhattacharjya S. NMR structures and localization of the potential fusion peptides and the pre-transmembrane region of SARS-CoV: Implications in membrane fusion. Biochim. Biophys. Acta. 2015;1848:721–730. doi: 10.1016/j.bbamem.2014.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tan K. Atomic structure of a thermostable subdomain of HIV-1 gp41. Proc. Natl. Acad. Sci. U.S.A. 1997;94:12303–12308. doi: 10.1073/pnas.94.23.12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malashkevich V.N. Core structure of the envelope glycoprotein GP2 from Ebola virus at 1.9-A resolution. Proc. Natl. Acad. Sci. U.S.A. 1999;96:2662–2667. doi: 10.1073/pnas.96.6.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guillen J. Membrane insertion of the three main membranotropic sequences from SARS-CoV S2 glycoprotein. Biochim. Biophys. Acta. 2008;1778:2765–2774. doi: 10.1016/j.bbamem.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simmons G. Different host cell proteases activate the SARS-coronavirus spike-protein for cell-cell and virus-cell fusion. Virology. 2011;413:265–274. doi: 10.1016/j.virol.2011.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simmons G. Proteolytic activation of the SARS-coronavirus spike protein: cutting enzymes at the cutting edge of antiviral research. Antiviral Res. 2013;100:605–614. doi: 10.1016/j.antiviral.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simmons G. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. U.S.A. 2005;102:11876–11881. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matsuyama S. Protease-mediated enhancement of severe acute respiratory syndrome coronavirus infection. Proc. Natl. Acad. Sci. U.S.A. 2005;102:12543–12547. doi: 10.1073/pnas.0503203102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bertram S. Cleavage and activation of the severe acute respiratory syndrome coronavirus spike protein by human airway trypsin-like protease. J. Virol. 2011;85:13363–13372. doi: 10.1128/JVI.05300-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Glowacka I. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 2011;85:4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kam Y.W. Cleavage of the SARS coronavirus spike glycoprotein by airway proteases enhances virus entry into human bronchial epithelial cells in vitro. PLoS ONE. 2009;4:e7870. doi: 10.1371/journal.pone.0007870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choi S.Y. Type II transmembrane serine proteases in cancer and viral infections. Trends Mol. Med. 2009;15:303–312. doi: 10.1016/j.molmed.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 47.Shulla A. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J. Virol. 2011;85:873–882. doi: 10.1128/JVI.02062-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou Y. Protease inhibitors targeting coronavirus and filovirus entry. Antiviral Res. 2015;116:76–84. doi: 10.1016/j.antiviral.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haga S. Modulation of TNF-alpha-converting enzyme by the spike protein of SARS-CoV and ACE2 induces TNF-alpha production and facilitates viral entry. Proc. Natl. Acad. Sci. U.S.A. 2008;105:7809–7814. doi: 10.1073/pnas.0711241105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heurich A. TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein. J. Virol. 2014;88:1293–1307. doi: 10.1128/JVI.02202-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Watanabe R. Entry from the cell surface of severe acute respiratory syndrome coronavirus with cleaved S protein as revealed by pseudotype virus bearing cleaved S protein. J. Virol. 2008;82:11985–11991. doi: 10.1128/JVI.01412-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun X. Molecular architecture of the bipartite fusion loops of vesicular stomatitis virus glycoprotein G, a class III viral fusion protein. J. Biol. Chem. 2008;283:6418–6427. doi: 10.1074/jbc.M708955200. [DOI] [PubMed] [Google Scholar]
- 53.Belouzard S. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. U.S.A. 2009;106:5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Madu I.G. Characterization of a highly conserved domain within the severe acute respiratory syndrome coronavirus spike protein S2 domain with characteristics of a viral fusion peptide. J. Virol. 2009;83:7411–7421. doi: 10.1128/JVI.00079-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li W. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 56.Lau S.K. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. U.S.A. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ge X.Y. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guan Y. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- 59.Hou Y. Angiotensin-converting enzyme 2 (ACE2) proteins of different bat species confer variable susceptibility to SARS-CoV entry. Arch. Virol. 2010;155:1563–1569. doi: 10.1007/s00705-010-0729-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu L. Angiotensin-converting enzyme 2 (ACE2) from raccoon dog can serve as an efficient receptor for the spike protein of severe acute respiratory syndrome coronavirus. J. Gen. Virol. 2009;90:2695–2703. doi: 10.1099/vir.0.013490-0. [DOI] [PubMed] [Google Scholar]
- 61.Sheahan T. Pathways of cross-species transmission of synthetically reconstructed zoonotic severe acute respiratory syndrome coronavirus. J. Virol. 2008;82:8721–8732. doi: 10.1128/JVI.00818-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li W. Efficient replication of severe acute respiratory syndrome coronavirus in mouse cells is limited by murine angiotensin-converting enzyme 2. J. Virol. 2004;78:11429–11433. doi: 10.1128/JVI.78.20.11429-11433.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Holmes K.V. Structural biology. Adaptation of SARS coronavirus to humans. Science. 2005;309:1822–1823. doi: 10.1126/science.1118817. [DOI] [PubMed] [Google Scholar]
- 64.Li W. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J. 2005;24:1634–1643. doi: 10.1038/sj.emboj.7600640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shi Z., Hu Z. A review of studies on animal reservoirs of the SARS coronavirus. Virus Res. 2008;133:74–87. doi: 10.1016/j.virusres.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Martina B.E. Virology: SARS virus infection of cats and ferrets. Nature. 2003;425:915. doi: 10.1038/425915a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Graham R.L., Baric R.S. Recombination, reservoirs, and the modular spike: mechanisms of coronavirus cross-species transmission. J. Virol. 2010;84:3134–3146. doi: 10.1128/JVI.01394-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Qu X.X. Identification of two critical amino acid residues of the severe acute respiratory syndrome coronavirus spike protein for its variation in zoonotic tropism transition via a double substitution strategy. J. Biol. Chem. 2005;280:29588–29595. doi: 10.1074/jbc.M500662200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sheahan T. Mechanisms of zoonotic severe acute respiratory syndrome coronavirus host range expansion in human airway epithelium. J. Virol. 2008;82:2274–2285. doi: 10.1128/JVI.02041-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li F. Structural analysis of major species barriers between humans and palm civets for severe acute respiratory syndrome coronavirus infections. J. Virol. 2008;82:6984–6991. doi: 10.1128/JVI.00442-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Reguera J. A structural view of coronavirus-receptor interactions. Virus Res. 2014;194:3–15. doi: 10.1016/j.virusres.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Roberts A. A mouse-adapted SARS-coronavirus causes disease and mortality in BALB/c mice. PLoS Pathog. 2007;3:e5. doi: 10.1371/journal.ppat.0030005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Day C.W. A new mouse-adapted strain of SARS-CoV as a lethal model for evaluating antiviral agents in vitro and in vivo. Virology. 2009;395:210–222. doi: 10.1016/j.virol.2009.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gierer S. The spike protein of the emerging betacoronavirus EMC uses a novel coronavirus receptor for entry, can be activated by TMPRSS2, and is targeted by neutralizing antibodies. J. Virol. 2013;87:5502–5511. doi: 10.1128/JVI.00128-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang Q. Bat origins of MERS-CoV supported by bat coronavirus HKU4 usage of human receptor CD26. Cell Host Microbe. 2014;16:328–337. doi: 10.1016/j.chom.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Millet J.K., Whittaker G.R. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc. Natl. Acad. Sci. U.S.A. 2014;111:15214–15219. doi: 10.1073/pnas.1407087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen Y. Crystal structure of the receptor-binding domain from newly emerged Middle East respiratory syndrome coronavirus. J. Virol. 2013;87:10777–10783. doi: 10.1128/JVI.01756-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lu L. Structure-based discovery of Middle East respiratory syndrome coronavirus fusion inhibitor. Nat. Commun. 2014;5:3067. doi: 10.1038/ncomms4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Raj V.S. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gorrell M.D. CD26: a multifunctional integral membrane and secreted protein of activated lymphocytes. Scand. J. Immunol. 2001;54:249–264. doi: 10.1046/j.1365-3083.2001.00984.x. [DOI] [PubMed] [Google Scholar]
- 81.Engel M. The crystal structure of dipeptidyl peptidase IV (CD26) reveals its functional regulation and enzymatic mechanism. Proc. Natl. Acad. Sci. U.S.A. 2003;100:5063–5068. doi: 10.1073/pnas.0230620100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rasmussen H.B. Crystal structure of human dipeptidyl peptidase IV/CD26 in complex with a substrate analog. Nat. Struct. Biol. 2003;10:19–25. doi: 10.1038/nsb882. [DOI] [PubMed] [Google Scholar]
- 83.Weihofen W.A. Crystal structure of CD26/dipeptidyl-peptidase IV in complex with adenosine deaminase reveals a highly amphiphilic interface. J. Biol. Chem. 2004;279:43330–43335. doi: 10.1074/jbc.M405001200. [DOI] [PubMed] [Google Scholar]
- 84.Burkard C. Coronavirus cell entry occurs through the endo-/lysosomal pathway in a proteolysis-dependent manner. PLoS Pathog. 2014;10:e1004502. doi: 10.1371/journal.ppat.1004502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shirato K. Middle East respiratory syndrome coronavirus infection mediated by the transmembrane serine protease TMPRSS2. J. Virol. 2013;87:12552–12561. doi: 10.1128/JVI.01890-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Qian Z. Role of the spike glycoprotein of human Middle East respiratory syndrome coronavirus (MERS-CoV) in virus entry and syncytia formation. PLoS ONE. 2013;8:e76469. doi: 10.1371/journal.pone.0076469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Barlan A. Receptor variation and susceptibility to Middle East respiratory syndrome coronavirus infection. J. Virol. 2014;88:4953–4961. doi: 10.1128/JVI.00161-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Assiri A. Hospital outbreak of Middle East respiratory syndrome coronavirus. N. Engl. J. Med. 2013;369:407–416. doi: 10.1056/NEJMoa1306742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Memish Z.A. Family cluster of Middle East respiratory syndrome coronavirus infections. N. Engl. J. Med. 2013;368:2487–2494. doi: 10.1056/NEJMoa1303729. [DOI] [PubMed] [Google Scholar]
- 90.Cotten M. Transmission and evolution of the Middle East respiratory syndrome coronavirus in Saudi Arabia: a descriptive genomic study. Lancet. 2013;382:1993–2002. doi: 10.1016/S0140-6736(13)61887-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Drosten C. Clinical features and virological analysis of a case of Middle East respiratory syndrome coronavirus infection. Lancet Infect. Dis. 2013;13:745–751. doi: 10.1016/S1473-3099(13)70154-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hemida M.G. Middle East Respiratory Syndrome (MERS) coronavirus seroprevalence in domestic livestock in Saudi Arabia, 2010 to 2013. Euro. Surveill. 2013;18:20659. doi: 10.2807/1560-7917.es2013.18.50.20659. [DOI] [PubMed] [Google Scholar]
- 93.Reusken C.B. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. Lancet Infect. Dis. 2013;13:859–866. doi: 10.1016/S1473-3099(13)70164-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Reusken C.B. Middle East respiratory syndrome coronavirus (MERS-CoV) RNA and neutralising antibodies in milk collected according to local customs from dromedary camels, Qatar, April 2014. Euro. Surveill. 2014;19:20829. doi: 10.2807/1560-7917.es2014.19.23.20829. [DOI] [PubMed] [Google Scholar]
- 95.Chan R.W. Tropism and replication of Middle East respiratory syndrome coronavirus from dromedary camels in the human respiratory tract: an in-vitro and ex-vivo study. Lancet Respir. Med. 2014;2:813–822. doi: 10.1016/S2213-2600(14)70158-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Memish Z.A. Human infection with MERS coronavirus after exposure to infected camels, Saudi Arabia, 2013. Emerg. Infect. Dis. 2014;20:1012–1015. doi: 10.3201/eid2006.140402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Azhar E.I. Evidence for camel-to-human transmission of MERS coronavirus. N. Engl. J. Med. 2014;370:2499–2505. doi: 10.1056/NEJMoa1401505. [DOI] [PubMed] [Google Scholar]
- 98.Annan A. Human betacoronavirus 2c EMC/2012-related viruses in bats, Ghana and Europe. Emerg. Infect. Dis. 2013;19:456–459. doi: 10.3201/eid1903.121503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ithete N.L. Close relative of human Middle East respiratory syndrome coronavirus in bat, South Africa. Emerg. Infect. Dis. 2013;19:1697–1699. doi: 10.3201/eid1910.130946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Memish Z.A. Middle East respiratory syndrome coronavirus in bats, Saudi Arabia. Emerg. Infect. Dis. 2013;19:1819–1823. doi: 10.3201/eid1911.131172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cui J. Adaptive evolution of bat dipeptidyl peptidase 4 (dpp4): implications for the origin and emergence of Middle East respiratory syndrome coronavirus. Virol. J. 2013;10:304. doi: 10.1186/1743-422X-10-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Corman V.M. Rooting the phylogenetic tree of middle East respiratory syndrome coronavirus by characterization of a conspecific virus from an African bat. J. Virol. 2014;88:11297–11303. doi: 10.1128/JVI.01498-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.de Wit E. The Middle East respiratory syndrome coronavirus (MERS-CoV) does not replicate in Syrian hamsters. PLoS ONE. 2013;8:e69127. doi: 10.1371/journal.pone.0069127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Raj V.S. Adenosine deaminase acts as a natural antagonist for dipeptidyl peptidase 4-mediated entry of the Middle East respiratory syndrome coronavirus. J. Virol. 2014;88:1834–1838. doi: 10.1128/JVI.02935-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Coleman C.M. Wild-type and innate immune-deficient mice are not susceptible to the Middle East respiratory syndrome coronavirus. J. Gen. Virol. 2014;95:408–412. doi: 10.1099/vir.0.060640-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Eckerle I. Replicative capacity of MERS coronavirus in livestock cell lines. Emerg. Infect. Dis. 2014;20:276–279. doi: 10.3201/eid2002.131182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chan J.F. Differential cell line susceptibility to the emerging novel human betacoronavirus 2c EMC/2012: implications for disease pathogenesis and clinical manifestation. J. Infect. Dis. 2013;207:1743–1752. doi: 10.1093/infdis/jit123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Müller M.A. Human coronavirus EMC does not require the SARS-coronavirus receptor and maintains broad replicative capability in mammalian cell lines. Mbio. 2012;3:e00515–e612. doi: 10.1128/mBio.00515-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.van Doremalen N. Host species restriction of Middle East respiratory syndrome coronavirus through its receptor, dipeptidyl peptidase 4. J. Virol. 2014;88:9220–9232. doi: 10.1128/JVI.00676-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Falzarano D. Infection with MERS-CoV causes lethal pneumonia in the common marmoset. PLoS Pathog. 2014;10:e1004250. doi: 10.1371/journal.ppat.1004250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhao J. Rapid generation of a mouse model for Middle East respiratory syndrome. Proc. Natl. Acad. Sci. U.S.A. 2014;111:4970–4975. doi: 10.1073/pnas.1323279111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cotten M. Spread, circulation, and evolution of the Middle East respiratory syndrome coronavirus. Mbio. 2014;5:e01062–e1113. doi: 10.1128/mBio.01062-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tang X.C. Identification of human neutralizing antibodies against MERS-CoV and their role in virus adaptive evolution. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E2018–E2026. doi: 10.1073/pnas.1402074111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Briese T. Middle East respiratory syndrome coronavirus quasispecies that include homologues of human isolates revealed through whole-genome analysis and virus cultured from dromedary camels in Saudi Arabia. MBio. 2014;5 doi: 10.1128/mBio.01146-14. e01146–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chan J.F. Interspecies transmission and emergence of novel viruses: lessons from bats and birds. Trends Microbiol. 2013;21:544–555. doi: 10.1016/j.tim.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lu G., Liu D. SARS-like virus in the Middle East: a truly bat-related coronavirus causing human diseases. Protein Cell. 2012;3:803–805. doi: 10.1007/s13238-012-2811-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Woo P.C. Molecular diversity of coronaviruses in bats. Virology. 2006;351:180–187. doi: 10.1016/j.virol.2006.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lau S.K. Genetic characterization of Betacoronavirus lineage C viruses in bats reveals marked sequence divergence in the spike protein of pipistrellus bat coronavirus HKU5 in Japanese pipistrelle: implications for the origin of the novel Middle East respiratory syndrome coronavirus. J. Virol. 2013;87:8638–8650. doi: 10.1128/JVI.01055-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yang Y. Receptor usage and cell entry of bat coronavirus HKU4 provide insight into bat-to-human transmission of MERS coronavirus. Proc. Natl. Acad. Sci. U.S.A. 2014;111:12516–12521. doi: 10.1073/pnas.1405889111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Beniac D.R. Architecture of the SARS coronavirus prefusion spike. Nat. Struct. Mol. Biol. 2006;13:751–752. doi: 10.1038/nsmb1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Beniac D.R. Conformational reorganization of the SARS coronavirus spike following receptor binding: implications for membrane fusion. PLoS ONE. 2007;2:e1082. doi: 10.1371/journal.pone.0001082. [DOI] [PMC free article] [PubMed] [Google Scholar]


