Abstract
A sensitive reverse-transcription loop-mediated isothermal amplification (RT-LAMP) assay was developed for the rapid visual detection of turkey coronavirus (TCoV) infection. The reaction is performed in one step in a single tube at 65 °C for 45 min, with hydroxynaphthol blue (HNB) dye added prior to amplification. The detection limit of the RT-LAMP assay was approximately 102 EID50/50 μl TCoV genome, and no cross-reaction with other avian viruses was observed. The assay was evaluated further in tissue suspensions prepared from the ileum and ileum–caecal junctions of infected turkey embryos; 100% of these samples were positive in the RT-LAMP assay. All individual feces samples collected in the field were considered positive by both conventional RT-PCR and RT-LAMP. In conclusion, RT-LAMP with HNB dye was shown to be a sensitive, simple assay for the rapid diagnosis of TCoV infection, either directly from feces or in association with virus isolation methods.
Keywords: Turkey coronavirus, Hydroxynaphthol blue, LAMP assay, Spike gene
The Brazilian turkey industry is the second most productive in the world, and last year, more than 198 million carcasses were produced. In 2006, an outbreak of poult enteritis mortality syndrome (PEMS) was detected for the first time and found to be caused by a group 3 Coronavirus [1]. Cases of TCoV enteritis are still reported in the U.S, Canada and several European countries [2], [3], [4].
Prior to more extensive sequence analysis, TCoV had been identified as a member of group 2 of the coronavirus genus, but it has since been definitively classified as group 3 with other avian coronaviruses. It is a pleomorphic enveloped virus with a size of approximately 80–160 nm in diameter and 20 nm-long club-shaped projections around virions. Its genome is linear, positive sense, single stranded RNA. The major structural proteins of TCoV include the glycoprotein spike (S1 and S2), a membrane (M) protein and a nucleocapsid (N) protein. The S protein is highly variable and the spike gene structure remains unclear, despite several genetic studies of TCoV having been conducted [5]. The amino terminal S1 region of several coronaviruses contains receptor binding domains, whereas the S2 region consists of a transmembrane domain that induces cell fusion and pathogenesis [6]. In addition, the S2 gene is more conserved and shares only 40% of similarity with other coronaviruses such as infectious bronchitis virus (IBV) [5], [6].
Conventional virological methods for detecting TCoV include virus isolation (VI), immunofluorescent antibody assay (IFA), reverse transcriptase polymerase chain reaction (RT-PCR) and real-time RT-PCR (RRT-PCR) [4], [7], [8], [12]. Although RT-PCR and RRT-PCR methods are widely applied [4], [12], RT-loop-mediated isothermal amplification (LAMP) is simpler, using only a water bath or heat block, and is highly efficient because the reaction is isothermal and requires no time for thermal changes [11]. Moreover, TCoV isolates can be propagated in turkey embryos, but the virus cannot yet be propagated in cell culture [7].
The objective of this study was to develop and validate a reverse-transcription loop-mediated isothermal amplification (LAMP) method for the direct detection of viral RNA from tissues and feces collected from experimentally and naturally infected birds. The results were directly compared with those obtained from conventional IFA and RT-PCR.
Turkey coronavirus (TCoV/BR/2007) was obtained from a Brazilian turkey farm suffering from severe cases of acute enteritis, described in 2007 [1]. This TCoV strain was successfully propagated in embryonated turkey eggs supplied for a commercial hatchery [8]. Viral purification, RNA isolation and subsequent RT-PCR amplification were carried out using a pooled intestinal tissue suspension of infected intestines harvested from inoculated embryos, as in our previous study [8]. The virus titration was performed with 10-fold dilutions into five groups of five 28-day-old embryonated turkey eggs. The 50% embryos infectious dose (EID50) was calculated according to Reed and Muench (1938) by analyzing macroscopic lesions in the intestinal tract [7], [8]. Intestinal tissues infected at a 2.0 × 104 EID50 titer were homogenized in a 2-fold volume of minimal essential medium (GIBCO-BRL, Invitrogen, Carlsbad, CA), clarified by centrifugation at 2500 × g for 20 min and filtered consecutively through 0.45-μm and 0.22-μm membrane filters (Millipore, Bedford, MA). The presence of TCoV in the filtrate was confirmed using RT-PCR of a portion of the 3′UTR (untranslated) region [2]. Extensive testing of the inoculums excluded the presence of pathogens other than TCoV.
Embryo tissues (ileum and ileum–caecal junction portions) were prepared by infecting 25-day-old embryonated turkey eggs with 0.3 ml 102.4 EID50 TCoV via the amniotic sac, as described previously [9], [10]. A control group of 20 turkey embryos at the same age were inoculated with sterile phosphate buffered solution (PBS). Three days after infection, embryos were submitted to an external exam, and samples of ileum and ileum–caecal junction tissues were collected. The tissues were stored at −86 °C, fixed in 10% neutral buffered formalin and embedded in paraffin blocks. IFA was performed in tissue sections, as described previously [8] in order to detect viral antigens along the cell surface. Intestinal suspensions (IS) were prepared by cutting slices from the ileum and ileum–caecal junctions from a total of 25 samples collected from infected and 20 from uninfected embryos. The specimens were homogenized in 2 volumes of minimal essential medium (MEM) pH 7.4 and clarified by centrifugation at 3000 × g for 20 min. The supernatant was first filtered through a 0.75-μm paper filter (Millipore) and then twice through a 0.25-μm syringe filter (Corning®). These suspensions were heated at 100 °C for 30 min in a water bath before total RNA extraction was performed. Fecal samples (n = 100) were obtained from a commercial flock that had experienced an outbreak of PEMS this year. The samples were collected directly from the cloaca and individually stored at −86 °C until use.
Total RNA was extracted from IS and feces with a standard Trizol protocol, based on guanidinium isothiocyanate and acid–phenol, with some modifications [1], [3]. Each 200-μl aliquot of clinical suspension was mixed with 500-μl Trizol reagent and incubated for 10 min at room temperature. After the addition of 200-μl chloroform, the solution was mixed vigorously for 10 s and centrifuged at 13,000 × g for 10 min. The upper aqueous phase was mixed with an equal volume of cold isopropanol and incubated on ice for 10 min. The total RNA precipitate was then pelleted by centrifugation at 13,000 × g for 10 min and washed with ethanol. The RNA was dissolved in 30 μl diethylpyrocarbonate(DEPC)-treated sterile double-distilled water and stored at −20 °C.
The primers were designed based on S2 sequence (1178–2073 position) information obtained from GenBank and assigned the following accession numbers: TCoV-1/BR/2008 (FJ957898); TCoV-2/BR/2008 (FJ9557899), TCoV-3/BR/2008 (FJ957900); TCoV-540 (EU022525); TCoV-ATCC (EU022526); TCoV-Gh (AY342356); TCoV-GI (AY342357); TCoV (Canadian isolate, NC_004718). All primers were designed with PrimerExplorer V.4, a software program for LAMP primer design (http://primerexplorer.jp/e/). The names and sequences of each primer are shown in Table 1 . To test the specificity of the established RT-LAMP, other viruses, including Newcastle disease virus (NDV, La Sota vaccine strain), turkey astrovirus (TAstV-1, GenBank accession number FJ178641) and infectious bronchitis virus (IBV-M41 strain) were tested. The median embryo infectious dose (EID50) of the TCoV used in the sensitivity test was calculated according to the Reed Muench formula. A series of 10-fold dilutions ranging from 1 to 104 EID50/50 μl of titrated virus was used as templates.
Table 1.
Sequences of primers used in this study.
| Primer name | Sequence (5′–3′) |
|---|---|
| F3 (forward outer primer) | CACTGCTAATGTTTTCC |
| B3 (backward outer primer) | GCACTGCCAGTAAATAAAGC |
| FIP (forward inner primer) | AACTAGTATGATTCCAGGGAGTCAGGACACAATTCTTCAAAAAAGG |
| BIP (backward inner primer) | AACACGATGACAGTTACACAAGTGTAAGCACATGGGTTCAGG |
| F2 (loop forward primer) | GGACACAATTCTTCAAAAAAAGG |
| B2 (loop backward primer) | GTAAGCACATGGGTTCAGG |
RT-LAMP was performed in a one-step reaction with 6 primers in a 25 μl mixture containing 2 μl genomic RNA, 40 pmol (each) primers FIB and BIP, 5 pmol (each) primers F3 and B3 and 3 pmol of each primers F2 and B2, 1 U Thermo-X reverse transcriptase (Invitrogen™) and 8 U Bst DNA polymerase (New England BioLabs), with corresponding buffers. Amplification was carried out at 64 °C for 45 min or 75 min and terminated by incubation at 80 °C for 2 min. The products of the reaction were inspected by eye following the addition of 1 μl hydroxynaphthol blue (Sigma–Aldrich®). All samples were also tested by conventional RT-PCR, as described previously [9]. Each assay was conducted in triplicate. A positive amplification was indicated by a color change from violet to sky blue, as shown in Fig. 1A, and verified by agarose gel electrophoresis (Fig. 1C). The detection limit of RT-LAMP and conventional RT-PCR is illustrated in Fig. 1B and C. Ten-fold serial dilution of TCoV demonstrated that RT-LAMP is able to detect 102 EID50/50 μl, whereas the RT-PCR is able to detect 104 EID50/50 μl titrated virus.
Fig. 1.
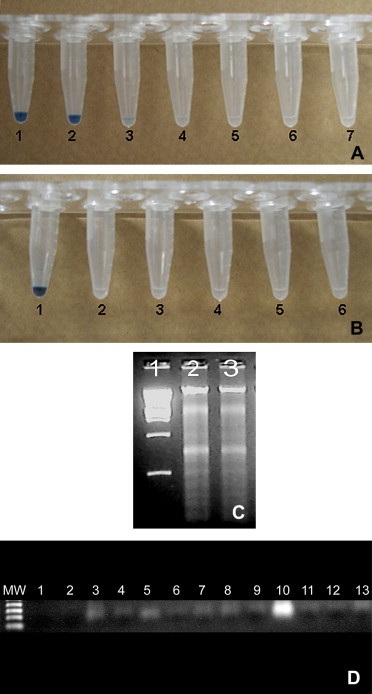
Sensitivity (A) and specificity (B) of RT-LAMP assay evaluated by eye after the addition of hydroxynaphthol blue dye. A) Lanes 1–6: cDNA from the TCoV (TCoV/BR/2007) at different titers: 104 EID50/50μl (1), 103 EID50/50μl (2), 102 EID50/50μl (3), 101 EID50/50μl (4), 100 EID50/50μl (5), 10−1 EID50/50μl (6); lane 7 corresponded to water instead of viral cDNA; B) Lanes 1–6: RNA from TCoV (TCoV/BR/2007) (1), NDV (2), TAstV-1 (3), IBV-M41 (4), IBV-H120 (5), AMPV (6); C) Agarose gel electrophoresis of RT-LAMP products from 102 EID50/50μl (line 2), 103 EID50/50μl (line 3) dilution of TCoV; line 1, DNA marker 2 kb plus; D) Sensitivity and specificity of conventional RT-PCR; Lanes (1) corresponded to water instead of viral RNA, (2) TAstV-1, (3) IBV-41, (4) IBV-H120; Lanes 5–10: RT-PCR products from TCoV at different titers: 10−1 EID50/50μl (5), 100 EID50/50μl (6), 101 EID50/50μl (7), 102 EID50/50μl (8), 103 EID50/50μl (9), 104 EID50/50μl (10); Lanes 11–13 tissue corresponded to RT-PCR amplification from RNA extracted from uninfected poult embryos.
IFA analysis indicated that 20 out of 25 tested samples were positive, whereas 100% of samples tested positive by RT-LAMP. A number of 25 ileum–caecal junction samples from the 25 infected embryos 13 were scored as negative for TCoV infection with IFA (Table 2 ). These false negative results were likely observed because viral antigens are not normally expressed on the cell surface until after viral RNA transcription [6]. When conventional RT-PCR and RT-LAMP performed in the same samples were directly compared, 14 ileum–caecal junction samples negative by RT-PCR were negative by IFA and 5 samples were considered negative with conventional RT-PCR (Table 3 ). This phenomenon could be explained by properties of DNA polymerization offered by the use of four primers in LAMP assay which can amplify few amounts of viral RNA into cDNA [11]. Probably those negative samples generated in conventional RT-PCR were collected from low viral infection samples with titers lower than 104 EID50/50 μl and not visualized on agarose gel electrophoresis system [12]. All negative samples collected from non-infected embryos were negative in all tests (data not shown). A perfect correlation was found between conventional RT-PCR and RT-LAMP results for feces collected in the field (Table 3). No false positives (i.e., cross-reaction) of TCoV RT-LAMP were detected in samples of other avian viruses (Fig. 1B).
Table 2.
Comparison between IFA and RT-LAMP assays in poult tissues.
| IFA assay |
|||||
|---|---|---|---|---|---|
| Ileum (n = 25) |
Ileum–caecal junction (n = 25) |
||||
| Positive | Negative | Positive | Negative | ||
| RT-LAMP assay | Positive | 20 | 5 | 18 | 13 |
| Negative | 0 | 0 | 0 | 0 | |
Table 3.
Comparison between conventional RT-PCR and RT-LAMP assays in detecting viral RNA from feces and tissue suspensions.
| Conventional RT-PCR |
|||||||
|---|---|---|---|---|---|---|---|
| Feces (n = 100) |
Ileum suspension (n = 25) |
Ileum–caecal junction suspension (n = 25) |
|||||
| Positive | Negative | Positive | Negative | Positive | Negative | ||
| RT-LAMP assay | Positive | 100 | 0 | 20 | 5 | 11 | 14 |
| Negative | 0 | 0 | 0 | 0 | 0 | 0 | |
RT-LAMP employs an initial reverse transcriptase step followed by DNA polymerization with a set of four primers specially designed for a specific target, representing a novel nucleic acid amplification method [11]. One of the most attractive features of this RT-LAMP assay is that the results can be observed and determined by hydroxynaphthol blue (HNB) dye-mediated visualization using the naked eye; furthermore, tubes do not have to be opened after amplification. The results reported here indicate that this diagnostic technique could reliably be used to detect TCoV in field samples and may be used to effectively monitor infections, as demonstrated previously for real-time RT-PCR for the same target gene [4]. Although no quantitative data was generated in this study, RT-LAMP associated with spectrophotometric evaluation can yield absolute values, as demonstrated previously [12], [13], [14].
The major advantage of RT-LAMP over conventional molecular methods is time; only 45 min are required for the reaction and more than 2 h for RT-PCR. In addition, the reaction is conducted in a water bath rather than a thermocycler, and its results can be interpreted by direct visualization rather than gel electrophoresis, which also save time. In summary, the RT-LAMP assay is novel, simple, visual and provides a sensitive and specific tool for the rapid detection and diagnosis of TCoV in simply equipped laboratories and under field conditions in developing countries. Its ease of use will support epidemiological programs in the turkey industry.
Acknowledgments
This work was supported by FAPESP (Fundação Amparo à Pesquisa do Estado de São Paulo) and CNPq. We are indebted to the technical team from SADIA-Unidade Uberlândia, MG Brazil. Tereza Cristina Cardoso is recipient of CNPq council grants.
References
- 1.Teixeira M.C., Luvizotto M.C., Ferrari H.F., Mendes A.R., da Silva S.E., Cardoso T.C. Detection of turkey coronavirus in commercial turkey poults in Brazil. Avian Pathol. 2007;36(1):29–33. doi: 10.1080/03079450601102939. [DOI] [PubMed] [Google Scholar]
- 2.Culver F., Dziva F., Cavanagh D., Stevens M.P. Poult enteritis and mortality syndrome in turkeys in Great Britain. Vet Rec. 2006;159(7):209–210. doi: 10.1136/vr.159.7.209. [DOI] [PubMed] [Google Scholar]
- 3.Pantin-Jackwood M.J., Day J.M., Jackwood M.W., Spackman E. Enteric viruses detected by molecular methods in commercial chicken and turkey flocks in the United States between 2005 and 2006. Avian Dis. 2008;52(2):235–244. doi: 10.1637/8174-111507-Reg.1. [DOI] [PubMed] [Google Scholar]
- 4.Chen Y.-N., Wu C.C., Bryan T., Hooper T., Schrader D., Lin T.L. Specific real-time reverse transcription polymerase chain reaction for detection and quantification of turkey coronavirus RNA in tissues and feces from turkeys infected with turkey coronavirus. J Virol Methods. 2010;163:452–458. doi: 10.1016/j.jviromet.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cavanagh D. Coronaviruses in poultry and other birds. Avian Pathol. 2005;34(6):439–448. doi: 10.1080/03079450500367682. [DOI] [PubMed] [Google Scholar]
- 6.Masters P. The molecular biology of coronavirus. Adv Virus Res. 2006;66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guy J.S. Isolation and propagation of coronaviruses in embryonated eggs. Methods Mol Biol. 2008;454:109–117. doi: 10.1007/978-1-59745-181-9_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cardoso T.C., Castanheira T.L., Teixeira M.C.B., Rosa A.C.G., Hirata K.Y., Astolphi R.D. Validation of an immunohistochemistry assay to detect turkey coronavirus: a rapid and simple screening tool for limited resource settings. Poult Sci. 2008;87(7):1347–1352. doi: 10.3382/ps.2008-00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gomes D.E., Hirata K.S., Saheki K., Rosa A.C.G., Luvizotto M.C.R., Cardoso T.C. Pathology and virus tissue distribution of turkey coronavirus (TCoV) in experimentally infected chicks and turkey poults. J Comp Pathol. 2010;147:8–13. doi: 10.1016/j.jcpa.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jackwood M.W., Boynton T.O., Hilt D.A., McKinley E.T., Kissinger J.C., Paterson A.H. Emergence of a group 3 coronavirus through recombination. Virology. 2010;398(1):98–108. doi: 10.1016/j.virol.2009.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:e63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen H.T., Zang J., Ma Y.P., Ma L.N., Ding Y.Z., Liu X.T. Reverse transcription loop-mediated isothermal amplification for rapid detection of infectious bronchitis virus in infected chicken tissues. Mol Cell Probes. 2010;24:104–106. doi: 10.1016/j.mcp.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xie Q.-M., Ji J., Pickens T.T., Du L.-Q., Cao Y.-C., Li H.-M. Rapid detection of infectious laryngotracheitis virus isolates by loop-mediated isothermal amplification. J Virol Methods. 2010;165:71–75. doi: 10.1016/j.jviromet.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 14.Pham H.M., Nakajima C., Ohashi K., Onuma M. Loop-mediated isothermal amplification for rapid detection of Newcastle disease virus. J Clin Microbiol. 2005;43:1646–1650. doi: 10.1128/JCM.43.4.1646-1650.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]


