Abstract
Diseases caused by pathogenic microorganisms including bacteria and viruses can cause serious medical issues including death and result in huge economic losses. Despite the myriad of recent advances in the rapid and accurate detection of pathogens, large volume clinical samples with a low concentration of pathogens continue to present challenges for diagnosis and surveillance. We here report a simple and label-free approach via homobifunctional imidoesters (HIs) with a microfluidic platform (SLIM) to efficiently enrich and extract pathogens at low concentrations from clinical samples. The SLIM system consists of an assembled double microfluidic chip for streamlining large volume processing and HIs for capturing pathogens and isolating nucleic acids by both electrostatic and covalent interaction without a chaotropic detergent or bulky instruments. The SLIM system significantly increases the enrichment and extraction rate of pathogens (up to 80% at 10 CFU (colony forming unit) in a 1 mL volume within 50 min). We demonstrated its clinical utility in large sample volumes from 46 clinical specimens including environmental swabs, saliva, and blood plasma. The SLIM system showed higher sensitivity with these samples and could detect pathogens that were below the threshold of detection with other methods. Finally, by combining our SLIM approach with an isothermal optical sensor, pathogens could be detected at a very high sensitivity in blood plasma samples within 80 min via enrichment, extraction and detection steps. Our SLIM system thus provides a simple, reliable, cost-effective and ultrasensitive pathogen diagnosis platform for use with large volume clinical samples and would thus have significant utility for various infectious diseases.
Keywords: Pathogen diagnosis, Enrichment, Microfluidic system, Clinical specimens, Point-of-care testing
Highlights
-
•
SLIM system significantly increases the enrichment and extraction rate of pathogens.
-
•
Demonstrated its clinical utility in large sample volumes from 46 clinical specimens.
-
•
A simple, reliable, cost-effective and ultrasensitive pathogen diagnosis platform.
1. Introduction
Pathogenic bacteria and viruses can cause significant levels of disease even at very low blood concentrations and ultimately lead to unexpected deaths and huge economic losses (Fauci and Morens, 2012, Pennington, 2014). Past outbreaks such as Severe Acute Respiratory Syndrome have resulted in a total of 775 deaths since 2002 and Middle East Respiratory Syndrome infected 186 cases and caused 39 deaths within one month in 2015 in Korea. Emerging infectious diseases such as these caused by new or unknown pathogens will continue to significantly threaten human life because of the absence of appropriate diagnostic tools (Marra et al., 2003, Park et al., 2015, Peiris et al., 2004, Xu et al., 2010). Innovative clinical techniques for the rapid detection and efficient monitoring of both known and unknown pathogens are thus highly desirable to manage and minimize their spread (Hanash et al., 2011, Pennington, 2014).
In general, the pathogen concentrations are extremely low at the early stages of disease and require highly sensitive and specific detection systems for early diagnostic and surveillance purposes. Recent advances with various PCR based techniques and next-generation sequencing methods have assisted with this (Fricke and Rasko, 2014, Gardy and Loman, 2018, Kim et al., 2018). However, these approaches are labor-intensive, require bulky instruments, are expensive to perform, and cannot readily be applied to large volume samples (Mardis, 2008). In real clinical settings, only volumes of 100–200 μL can be effectively analyzed for pathogen infection with a PCR or sequencing method. When the concentration of pathogen is low and the original sample is over 1 mL, the threshold of detection may not be reached for these methods. Techniques for simultaneous pathogen enrichment, concentration, and extraction are therefore needed for clinical applications.
To date, if the required sample volume is large and a pathogen needs to be enriched for detection, the conventional sample preparation assays use immunological capture or physical separation (Rosseel et al., 2015, Yeh et al., 2014). Immunological capture requires antibodies based on previous knowledge of the targets and it therefore limited when identifying new or emerging virus strains. Ultracentrifugation is the most commonly used method for enriching pathogens from clinical samples but is an expensive method requiring a long processing time and bulky equipment. These techniques cannot therefore be used at the point-of-care due to their complexity and equipment requirements (de Mello and Beard, 2003, Ganesh et al., 2016). The gold standard method in hospitals for the diagnosis of pathogens is a culture-based method, but this is also very time-consuming (from several hours to several days) and cannot be used for culture-negative bacteria (Duncan et al., 2007). Although several recent methods for enriching pathogens using nanoparticles and magnetic beads have now been introduced, the preprocessing steps are lengthy and costly, require specific antibodies for pathogen capture and have stability issues under harsh sample conditions (Mannoor et al., 2010, Pérez et al., 1998).
An ideal pathogen diagnostic system that can be used at the point-of-care needs to be rapid, accurate, low-cost, and be able to process the entire clinical sample volume from the patient, including urine, saliva, and blood. However, these fluids usually contain elements that inhibit the amplification of DNA and RNA and rapid and simple techniques that can accurately isolate and enrich pathogens are thus needed to properly diagnose infectious diseases (Lien et al., 2007). A combination of such an enrichment and concentration method with sample preparation would significantly increase the pathogen detection sensitivity in a clinical application.
We here describe a simple and label-free pathogen enrichment technique via the use of homobifunctional imidoesters (HIs) using a microfluidic (SLIM) system for ultrasensitive pathogen detection in various clinical specimens. HI reagents including dimethyl adipimidate (DMA), dimethyl pimelimidate (DMP) and dimethyl suberimidate (DMS) with assembled double microfluidic disposable chips (ADC) are used to effectively capture pathogens (bacteria and viruses) from the clinical samples within 10 min. All HI reagents consist of positive charge groups that can directly and selectively bind negatively charged pathogens in a 1–2 mL volume without the need for detergents or bulky instruments. In addition, we used the ADC microfluidic platform to streamline the processing of larger sample volumes and thereby save time and expense. Nucleic acids were extracted with the SLIM system using a solution of HI reagents with lysis buffer from the pathogens enriched on this same system. DMA and DMP are known as cross-linking reagents and contain two imidoester groups, which react with amine groups of nucleic acids, proteins and cells to form amidine bonds (Jin et al., 2017, Shin et al., 2015). Accordingly, the SLIM system can be used for enrichment and extraction within 50 min on a single platform with a reduced risk of external contamination, and without the need for complicated procedures or separate equipment. We demonstrate the clinical utility of SLIM system to process 46 clinical specimens including environmental swabs, saliva, and blood plasma which showed at least a 2-fold higher sensitivity for pathogen detection than existing methods. Furthermore, we synergistically combined the SLIM system with an isothermal solid-phase nucleic acid amplification and detection (iNAD) sensor for ultrasensitive detection of pathogen in clinical specimens. The iNAD is a refractive index sensor for pathogen diagnosis and combines an isothermal amplification method with a silicon micro ring resonator sensor to confirm the wavelength shift that changes in a label-free and real-time manner, in accordance with the combination of primer and target nucleic acids on the sensor (Kim et al., 2018). We validated the clinical utility of this combination system for the enrichment and detection of pathogens in 17 plasma samples including 14 herpes zoster (HZ) and 3 non-HZ controls. Our combined approach not only enriched pathogens in a sample by at least 10-fold, but also successfully detected nucleic acids via the iNAD to enable the rapid and ultrasensitive detection of pathogens at the point of care (POC) in the clinic.
2. Materials and methods
2.1. Fabrication and operation of SLIM system
A low-cost SLIM device for use as a microfluidic system was fabricated with a CO2 laser-cutting machine (VLS3.50 (610 × 305 mm); Universal Laser Systems, Scottsdale, AZ) and comprised microfluidic chambers in a single microchannel in combination with HI (DMA, DMP and DMS) reagents for pathogen enrichment. Two 36 slot-type microwell chips were assembled into five layers on the SLIM system to enrich various pathogens from different sources. The design of the microfluidic chip was made using AutoCAD (Autodesk, Inc., San Rafael, CA) and then cut out with a laser-cutting machine (10.6 μ CO2 laser sources ranging in power from 10 to 50 W and speed 20 mm/sec). This enabled the rapid fabrication as a rapid prototyping device simply, rapidly and at a low cost. The 3D disposable chip was assembled with two layers comprising 300-μm-thick double-sided tape (Adhesive 300LSE-9495LE, 3 M, Minnesota, US) and one 100-μm layer of thin-film (Kemafoil hydrophilic film, HNW-100, COVEME, Italy) as inner layers, and two 100-μm thin-films as outer layers (Supplementary Fig. S1). The thin films were attached to the permanent adhesive surfaces at the top and bottom of the inner layers to generate a 3D disposable chip for the streamlining of large sample volume processing. Accordingly, the chamber height was set to approximately 600 µm, and the total volume was 600 μL. Moreover, to control the reactions in the SLIM system, a cast acrylic sheet, tygon tubing, and epoxy were used as previously reported (Shin et al., 2015).
To use HI non-chaotropic reagents with the thin-film platform for pathogen enrichment, the surface modification protocol for the inside of the microfluidic channel was optimized. Briefly, the surface was treated with oxygen plasma (Covance Model, Femtoscience, Korea) for 10 min to change the surface properties of the inner film from hydrophobic to hydrophilic, and then immersed in a solution of 2% 3-aminopropyl triethoxysilane (APTES; Sigma-Aldrich, St. Louis, MO) for 60 min at 65 °C, followed by thorough rinsing with deionized water. After silanization with APTES on the inner surface, the surface hydrophilicity was increased (contact angle approximately 30–40°), as reported previously (Shin et al., 2015). The chips can then be stored at room temperature until. We next optimized the assay conditions and solution used to enrich pathogens using the SLIM system. We tested all HI groups such as DMA, DMS and DMP to compare their ability to directly bind pathogens. All HI reagents were purchased from Sigma-Aldrich. To conduct the reaction, a 1 mL preparation of Escherichia coli (ATCC 25922) diluted in PBS was mixed with 300 μL of each HI solution (100 mg mL−1) and then injected into the SLIM system using a syringe pump (KD Scientific, Holliston, MA) at 100 μL min−1. The system was then placed at room temperature for 10 min to enable pathogen capture followed by elution with 10 mM sodium bicarbonate (pH < 10.6) at a flow rate of 50 μL min−1. To extract nucleic acids from the enriched pathogen solution, a QIAamp® DNA mini kit (Qiagen, Germany) or nucleic acid extraction using our microfluidic platform with HIs was conducted as described previously (Shin et al., 2015). Briefly, the enriched pathogen sample was mixed with proteinase K, in-house lysis buffer (100 mM Tris-HCl (pH 8.0), 10 mM ethylenediaminetetraacetic acid, 1% sodium dodecyl sulfate, and 10% Triton X-100) and HI reagent (100 mg mL−1). After incubating the system at 56 °C (for DNA) or room temperature (for RNA) for 20 min, PBS was used to wash the system to remove debris from the samples. Elution buffer (pH > 10.6) was used to collect the extracted nucleic acids within a few minutes. The quantity and purity of the extracted nucleic acids were measured using both Qiagen kit and the microfluidic platform, based on the ratio of the optical densities of the samples at 260 nm and 280 nm measured with a Nano Drop spectrometer (Thermo Fisher Scientific, Waltham, MA).
2.2. The iNAD for ultrasensitive pathogen detection
The iNAD sensor was combined with an isothermal nucleic acid amplification technique to enable the detection of pathogenic nucleic acids in clinical samples within 30 min ( Fig. 1A, right). The principle of iNAD and the detailed structure of this bio-optical sensor have been described previously (Kim et al., 2018). The bio-optical sensor was then placed on a thermal pad to maintain the temperature. Wavelength shifts were acquired every 5 min for up to 30 min to monitor the amplification and detection of pathogenic nucleic acids.
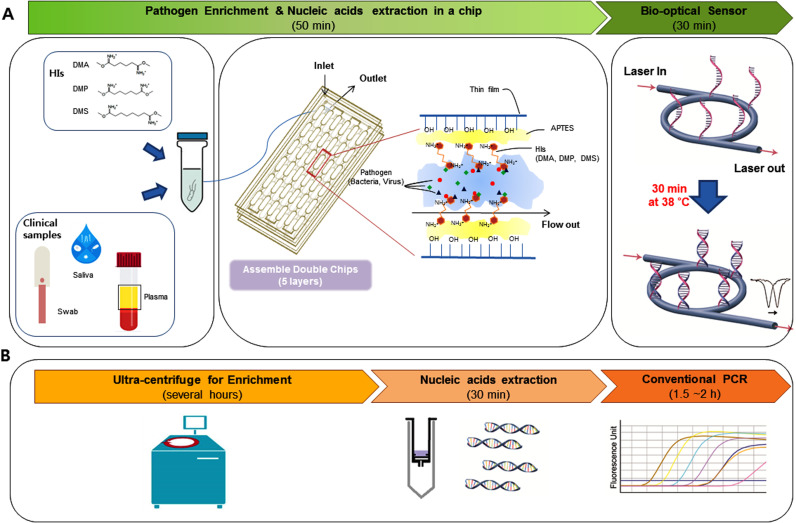
Fig. 1.
Schematic representation of the principle of ultrasensitive pathogen detection in clinical specimens. (A) Ultrasensitive pathogen detection system combining a simple and label-free system via homobifunctional imidoesters (HIs) with a microfluidic (SLIM) platform and an isothermal solid-phase nucleic acid amplification and detection sensor (iNAD). A mixture clinical sample and HI reagent is added to the SLIM system to enrich the pathogen and extract the DNA/RNA within 50 min without the need for detergents or bulky instruments. The extracted nucleic acids using the SLIM system were then used to detect a pathogen via an iNAD sensor. (B) Work flow of conventional methods for detection of pathogens with enrichment via ultracentrifugation, nucleic acid extraction on a column, and pathogen detection by real-time PCR.
3. Results and discussion
3.1. Simple and label-free pathogen enrichment via homobifunctional imidoesters using a microfluidic (SLIM) system for ultrasensitive pathogen detection
The principle of the SLIM system for ultrasensitive pathogen detection in large clinical specimen volumes (1–2 mL) via pathogen enrichment and the use of a single microfluidic chip are illustrated in Fig. 1. The source of the pathogen, such as an environmental swab, saliva, or plasma, needs to be mixed with HI reagents which are used as non-chaotropic reagents to enrich and extract these organisms onto the assembled double chips (ADC). HI consists of several methylene groups and bifunctional imidoester groups (Fig. 1A, left) that combine with the ADC platform consisting of 5 layers, including 3 thin films (top, middle and bottom) and 2 double-side tapes with 36 microwells (Supplementary Fig. S1) for streamlining of the processing. The microwells including channels and chambers were used as binding reservoirs for rapid mixing with high flow rate of sample volume during the sample loading, washing and elution (Yoon et al., 2018). The simple and low-cost ADC platform can be readily fabricated as a rapid prototype device using a CO2 laser cutting machine.
First of all, to enrich and concentrate the pathogens from a large volume samples via the SLIM system, a pre-mixed solution of sample and HI is introduced onto the ADC platform which has been pre-activated with amine reactive groups on its surface to electrostatically capture complexes of pathogens and HI reagents. The positively charged HI reagents by the amino group of HI capture the negatively charged pathogens by electrostatic adsorption (Fig. 1A, middle). As the suspension flows through the platform, pathogen capture occurs on the inner surface while other substances flow through (Fig. 1A, middle). Second, after the enrichment process at room temperature to selectively capture pathogens, a lysis buffer with additional HIs is added to isolate nucleic acids (DNA and RNA) from the captured pathogens by covalent bonding on the same platform at 56 °C for 20 min. Elution buffer is added to collect the nucleic acids by breaking the interaction between the complex and the inner surface of the platform. The SLIM system can thus capture and extract nucleic acids from pathogens from 1 or 2 mL samples without the need for other instruments such as centrifuges and vortexes. Finally, a rapid and sensitive detection of the pathogenic organisms is made using the isothermal solid phase nucleic acid amplification and detection (iNAD) optical sensor (Fig. 1A, right). A single primer is covalently immobilized on the sensor surface to which a mixture of the other primer, the target nucleic acids, and recombinase polymerase amplification (RPA) is added. The target nucleic acid sequences are exponentially amplified on the sensor surface at 38 °C for 30 min, which is monitored in real-time using the wavelength shift on the optical sensor without the need for labeling. Using the SLIM system, pathogens in a large samples can be rapidly and sensitively detected within 80 min via enrichment and extraction compared with conventional assays (several hours) such as ultra-centrifugation for enrichment and extraction, and detection with PCR (Fig. 1B). The clear advantages of the SLIM system are effective and direct pathogen enrichment from large clinical samples with HI reagents followed by a streamlined processing of large volumes with double microfluidic chips with no loss of the pathogens. In general, a large sample volume will increase the time, cost and effort involved in testing it. However, the SLIM system significantly decreases the time, cost, and complex instrument requirements and with a high sensitivity and specificity for pathogen diagnosis in clinical specimens.
3.2. Characterization of the SLIM system for pathogen enrichment
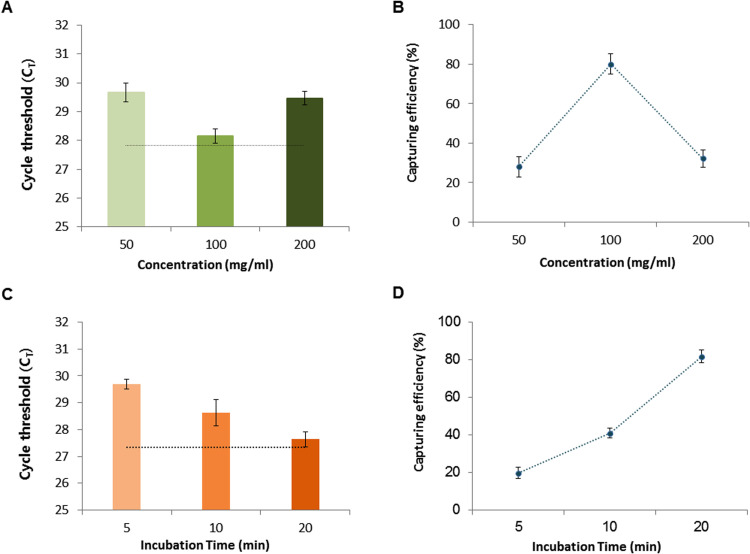
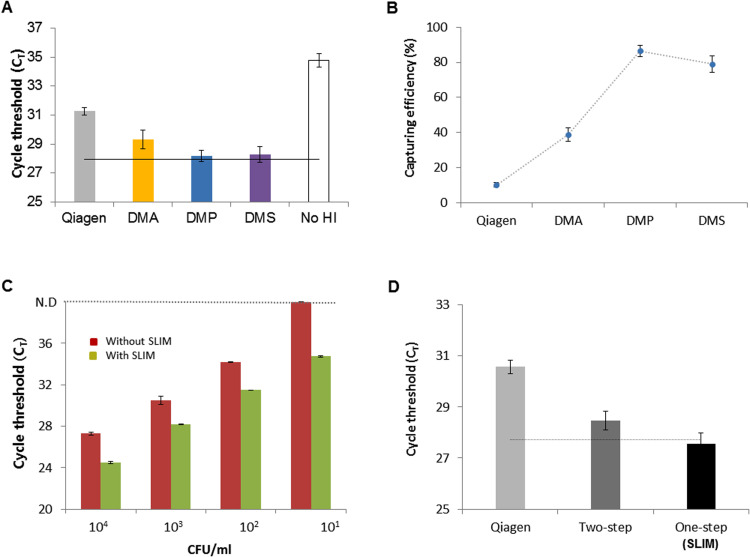
Commonly used pathogen detection assays for clinical samples include conventional PCR and pathogen cultures. A limitation of these established approaches however is their ability to detect true pathogens at very low concentrations in the sample. In actual clinical settings in which the patient sample volumes often range from at least 1 mL to as high as 10 mL depending on sources such as saliva, urine, and blood, researchers have typically used maximum volumes of only 200 μL due to the capacity limitations of available methods. Thus, the detection of low concentration pathogens has remained a challenge. We investigated whether the SLIM system could enrich the pathogens from a large volume simply and rapidly and with high sensitivity and specificity. We tested this by loading 1 mL of E. coli samples (104 CFU mL−1) onto the SLIM platform and enriched and re-suspended these organisms with 200 μL of elution buffer. The enriched sample was then subjected to nucleic acid extraction with a Qiagen kit. We tested different concentrations of DMA (one of the HIs) and various incubation times to optimize this enrichment step and then amplified the rodA gene of the enriched E. coli using real-time PCR ( Fig. 2). The real-time PCR CT value and capture rate, depending on the concentration of DMA (50, 100 or 200 mg mL−1), were measured for the 1 mL E. coli preparation (104 CFU mL−1). A 100 μL volume of sample (105 CFU mL−1) without pathogen enrichment was tested in the real-time PCR as a reference absolute value. Following SLIM enrichment, we observed higher amplification efficiency at 100 mg mL−1 concentration of DMA (Fig. 2A). The capture rate of E. coli was about 80% of the absolute value at this DMA level (Fig. 2B). We also tested the CT value and capture efficiency of the system at different incubation times (5, 10 and 20 min). We observed a better amplification efficiency with the 20 min incubation time that was almost 80% enriched compared to the absolute value (Fig. 2C, D). We next evaluated the optimal HI reagent (DMA, DMP or DMS) to use for pathogen enrichment again using a 1 mL preparation of E. coli (104 CFU mL−1). DMP was found to be the most efficient of these reagents for pathogen enrichment and gave real-time result about 4 cycles earlier than that of the Qiagen kit without any enrichment ( Fig. 3A and B). Our system protocol was thus optimized for the concentration (100 mg mL−1; Fig. 2A, B) of reagent, incubation time (20 min; Fig. 2C, D) and selection of HI reagent (DMP; Fig. 3A, B) for enriching pathogens from a large sample volume.
Fig. 2.
Characterization of the SLIM system for pathogen enrichment. (A) Amplification efficiency for a 1 mL E. coli test sample (104 CFU) following its enrichment on the SLIM system using the DMA HI reagent. The black dotted line represents the CT value for E. coli DNA from the absolute concentration (104 CFU) reference control extracted with a Qiagen kit. (B) The E. coli capture rate using the SLIM system depends on the HI concentration. (C) The amplification efficiency following E. coli (104 CFU/mL) enrichment depends on the incubation time on the platform. The black dotted line denotes the CT value of the absolute concentration (104 CFU) E. coli reference control DNA extracted using a Qiagen kit. (D) The E. coli capture rate using the SLIM system also depends on the incubation time. The error bars indicate the standard deviation from the mean, based on at least three independent experiments.
Fig. 3.
Application of the SLIM system to bacterial enrichment and extraction. (A) The amplification efficiency for a sample of E. coli (104 CFU/mL) following bacterial enrichment using the SLIM system is dependent on the type of HI reagents (DMA; yellow, DMP; blue, DMS; purple). The control experiment without HI reagents is examined as negative control (No HI). The black dotted line denotes the CT value for the reference control E. coli DNA extracted from the absolute concentration (104 CFU) sample control of using a Qiagen kit. (B) The E. coli capture rate using the SLIM system depends on the HI concentration. The error bars indicate the standard deviation from the mean, based on at least three independent experiments. (C) Capacity of the SLIM system to process test E. coli samples in concentrations ranging from 1 × 101 to 1 × 104 CFU/mL using both the Qiagen kit (red) and the SLIM system (light green) for DNA extraction. The error bars indicate the standard deviation from the mean, based on at least three independent experiments. (D) Use of the SLIM system for simultaneous enrichment and extraction with a test E. coli sample of 1 × 104 CFU/mL. Qiagen (light gray); DNA extraction only without enrichment, Two-step (dark gray); DNA extraction with Qiagen kit after bacterial enrichment with the SLIM platform, and One-step (black); simultaneous enrichment and extraction on the same system. The black dotted line denotes the CT value of the absolute (104 CFU) E. coli reference control DNA extracted using a Qiagen kit. The error bars indicate the standard deviation from the mean, based on at least three independent experiments. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
3.3. Bacterial enrichment and extraction with the SLIM system
After optimizing the SLIM protocol for pathogen enrichment, we evaluated its capacity to analyze 1 mL E. coli samples ranging in concentration from 1 × 101 to 1 × 104 CFU mL−1. Once the bacteria had been process using the SLIM system, we used a Qiagen kit for nucleic acid extraction followed by real-time PCR detect the bacteria. The PCR results indicated that DNAs from the enriched E. coli were amplified gradually and that this depended on the serial dilution of the pathogens. At all sample concentrations, much earlier CT values were obtained for the enriched DNA than from DNA extracted using the Qiagen kit (Fig. 3C). The limit of detection limit was 10-fold higher using the SLIM system. We next investigated whether the SLIM system could be used to both enrich the bacteria and perform the extraction on the same platform. To compare the detection efficiency of Qiagen without enrichment, a two-step process (enrichment and extraction in two separate chips) or the proposed one-step method (enrichment and extraction in a same chip), we again used E. coli (104 CFU mL−1) and real-time PCR. The CT values when using Qiagen without enrichment were 3–4 cycles delayed compared with the absolute value of the pathogen. With the SLIM system with a 1 mL volume, the amplification efficiency over a two-step process was 1–2 cycles longer than that of the absolute value. However, when both enrichment and extraction were done on the same chip, the amplification efficiency was similar to the absolute value of the pathogen (Fig. 3D). Therefore, the SLIM system can significantly improve both the bacterial enrichment rate and detection limit within 50 min.
3.4. Validation of the SLIM system for use with RNA virus in environmental swab samples
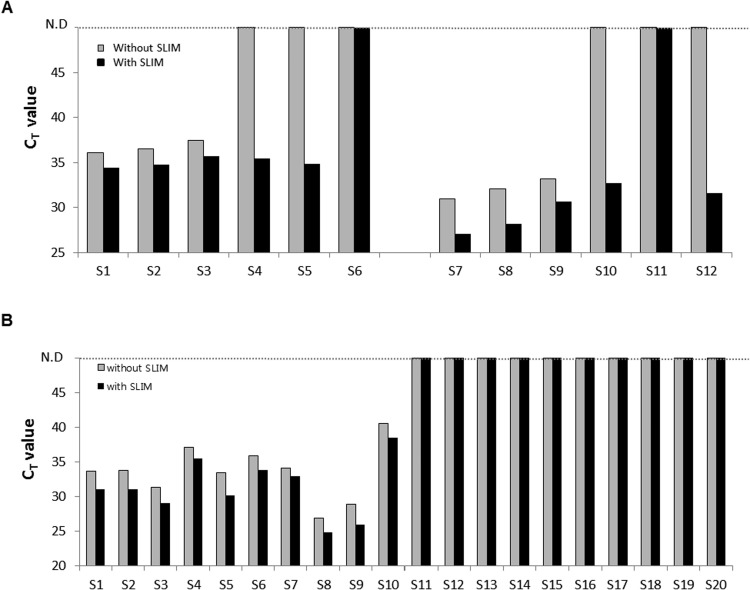
To validate our new SLIM system for real samples, we applied swab samples taken from environmental surfaces in contact with patients during an hPIV-3 outbreak between May and June 2016 in South Korea ( Fig. 4 and Supplementary Fig. S2). Detecting viral nucleic acids from environmental samples is important as environmental contamination by hPIV-3 is known to precede the nosocomial transmission of hPIV-3 in hospital wards (Kim et al., 2017). For the detection of hPIV-3, 140 μL aliquots from 2 mL environmental swab samples were used to extract viral RNA and amplify it by qRT-PCR. Due to limitations in existing nucleic acid extraction methods, the 2 mL entire sample volume could not be directly used for the detection of viral RNA. Using conventional methods, it has been reported that successful hPIV-3 detection the environmental swabs is only about 49% by qRT-PCR (Kim et al., 2017). Using our SLIM system however, the entire 2 mL volume of the environmental swab sample was used for hPIV-3 virus enrichment and viral RNA extraction (Supplementary Fig. S2A). First, we one positive and three negative samples determined by real-time PCR from 49 swabs to test the virus enrichment ability of the SLIM system. As shown in Supplementary Fig. S2B, one if the three negative samples as determined by real-time PCR was in fact found to be hPIV-3 positive using the SLIM system. The positive sample determined by real-time PCR showed an enhanced the amplification efficiency for the virus compared with the Qiagen kit (Supplementary Fig. S2B). We further tested 6 positive and 6 negative swabs from a patient with hPIV-3 virus and found using the SLIM system that the amplification efficiency of the 6 positives was 1–2 cycles earlier than conventional approaches. Moreover, 4 out of the 6 negative swabs determined by real-time PCR showed CT values with SLIM system (Fig. 4A and Supplementary Fig. S2C-D). The detection sensitivity with SLIM system was calculate at 83.3% (10/12) compared to 50% without the SLIM system (6/12) (Fig. 4A and Supplementary Table S2-S3). The positive samples were further confirmed by Sanger sequencing for the hemagglutinin-neuraminidase (HN) gene of hPIV-3.
Fig. 4.
Validation of the SLIM system in real samples. (A) SLIM system in environmental swab samples. 12 environmental swab samples taken from areas containing hPIV-3 infected patients were collected and enriched using the SLIM system. Real-time PCR fluorescence signals for amplified hPIV-3 RNAs from the same 6 specimens extracted using a Qiagen kit (without SLIM enriched) or enriched using the SLIM system from patient A (S1-S6) and patient B (S7-S12). (B) SLIM system in saliva samples from HZ patients. 20 saliva samples from HZ infected patients were collected and enriched with and without the SLIM system. Fluorescence signals obtained by real-time PCR amplification of VZV DNAs extracted from 20 Hz patient samples using a Qiagen kit (without SLIM system) (gray) and enriched from the same 20 specimens with the SLIM system (black). The sample numbers represent positive (S1-S10) and negative (S11-S20) samples that were confirmed by end-point PCR.
3.5. Validation of the SLIM system in human specimens
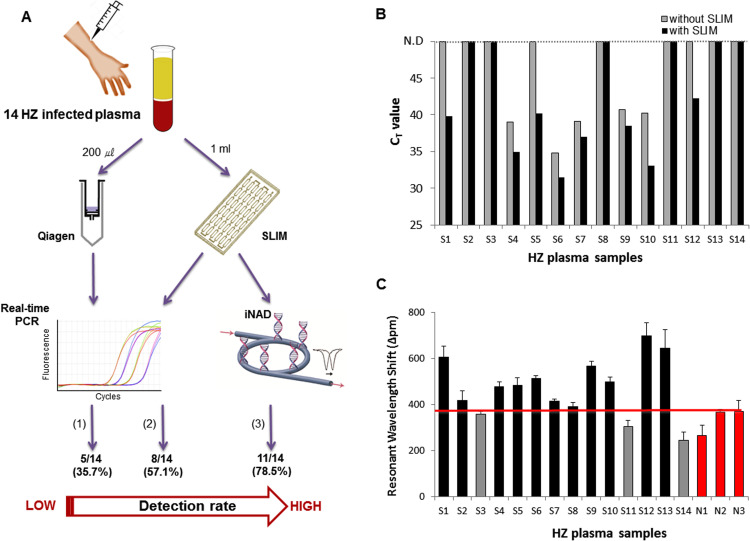
We applied our SLIM system to saliva samples from patients with HZ ( Figs. 4B and 5). Chickenpox and HZ are caused by varicella-zoster virus (VZV). Although the HZ rash is generally considered sufficient for clinical diagnosis, better diagnostic assays for saliva and plasma specimens are needed to distinguish HZ and HZ-mimicking diseases such as herpes simplex virus (Park et al., 2017). Recently, one study described the diagnostic usefulness of PCR for detecting VZV infection in saliva and plasma obtained from HZ patients. Saliva sample volumes of 200 μL of a total 1 mL sample have previously been used for PCR analysis without any enrichment process. The sensitivity of PCR analysis detecting VZV is far higher in saliva DNA (88%) than in plasma DNA (28%) (Park et al., 2017). However, although, plasma samples from HZ patients are less sensitive than salivary DNA for PCR detection, false-positive results can be obtained from saliva and saliva sampling is relatively painful for the patients suffering from the respiratory disease (Mannoor et al., 2010, Granger et al., 2007). For these reasons, we tested the use of both saliva and plasma samples to diagnose HZ and determine the efficiency of enrichment in both cases with the SLIM system. We first used 1 mL saliva sample volumes for VZV enrichment and viral DNA extraction (Supplementary Fig. S3A). We selected 10 saliva-positive samples as previously determined by real-time PCR. All PCR-positive samples that had been confirmed using the ORF62 region of VZV had earlier CT values with the SLIM system (Fig. 4B and Supplementary Fig. S3B). On the other hand, none of the previously determined PCR-negative samples showed a positive result for either group with the SLIM system (Fig. 4B and Supplementary Fig. S3C). These results are consistent with the previous findings that saliva samples have greater utility for the detection of VZV (Park et al., 2017, Supplementary Table S4). We further validated this SLIM system with 14 plasma samples obtained from patients with HZ as previously determined by PCR analysis of saliva samples (Fig. 5). When a 200 μL plasma sample volume was used for viral DNA extraction without the SLIM system, the sensitivity for subsequent VZV detection by real-time PCR was only 35.7% (5/14) (Fig. 5A, B and Supplementary Table S5). Using the SLIM system however, this improved to 57.1% (8/14) by real-time PCR (Fig. 5A, B and Supplementary Table S5-S6). In addition, all 8 samples enriched with the SLIM system showed better amplification efficiencies and higher virus detection sensitivities than samples that were not processed with the SLIM system (Fig. 5B). Finally, as an alternative to real-time PCR, we applied our rapid and accurate SLIM detection system to proof-of concept testing at the point-of-care. The iNAD is an optical sensing system for simultaneous nucleic acid amplification and detection. We tested plasma DNA samples from 14 patients with HZ as positives and 3 patients with human adenovirus (HAdV) as negatives to validate our combined SLIM and iNAD sensor system. We again used 1 mL of the plasma samples for VZV enrichment and viral DNA extraction (Fig. 5A). As shown in Fig. 5C, DNAs obtained via the SLIM system were simultaneously amplified and detected within 30 min in a label-free and real-time manner. Based on the HAdV negative samples, the cut-off line for detecting a VZV positive/negative sample was determined to be a shift of more than 380 p.m. at 30 min. Based on this criterion, we could identify VZV in 11 positive samples from 14 Hz samples within 80 min including the enrichment, extraction and detection steps. Although 3 samples deemed to be positive by previous saliva testing did not show a positive result, the detection sensitivity for HZ was significantly enhanced up to 78.6% on the SLIM system, indicating that plasma samples have utility for the diagnosis of HZ if using our combination system.
Fig. 5.
Clinical utility of the ultrasensitive pathogen diagnostic system integrated with the SLIM system and iNAD sensor to test blood plasma samples from HZ patients. (A) Fourteen plasma samples from HZ infected patients were collected and enriched with and without the SLIM platform and detected by either PCR or with the bio-optical iNAD sensor as follows: (1): real-time PCR detection without enrichment, (2): real-time PCR detection after processing on the SLIM system, (3): iNAD detection after processing on the SLIM system. (B) Fluorescence signals obtained by real-time PCR amplification of viral DNA from 14 specimens extracted using either a Qiagen kit (gray) or the SLIM system (black). (C) Clinical utility of the ultrasensitive pathogen diagnostic SLIM system in analyzing samples from 14 human HZ patients (S1–S14, black and gray) and 3 non-HZ patients (human adenovirus) (N1–N3, red) over a 30 min duration. The red line represents the cut-off (criterion) for reporting viral positivity. The error bars indicate the standard deviation from the mean, based on at least three independent experiments. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
4. Conclusions
Pathogenic diseases caused by both known and unknown bacteria and by viruses can have devastating public health consequences. Indeed, newly emerging diseases caused by unknown pathogens have caused lethal outbreaks in recent decades. Various approaches have thus been introduced for rapid and sensitive pathogen detection and to identify unknown pathogens. However, along with the advances in detection approaches, improved pathogen sample preparation techniques including enrichment and concentration are desired to process large volume clinical samples. In real clinical settings at present, only small amounts of sample (100–200 μL) are typically used from a 1–2 mL sample, due to the technical limitations of existing assays. Although several conventional assays for virus separation have been established such as antibody-based capture and centrifuge-based separation, these are not appropriate for clinical identification of unknown pathogens as they require known antibodies, bulky equipment, and long processing times.
To overcome the aforementioned limitations in using the entire volume of a valuable clinical sample from a patient, we have devised a simple and label-free pathogen enrichment via homobifunctional imidoesters with microfluidic (SLIM) system for ultrasensitive pathogen detection in clinical specimens. The SLIM system provides an innovative platform that combines HI reagents with a microfluidic device for pathogen enrichment and nucleic acid extraction. Using the HI reagents which bind with negatively charged molecules through an electrostatic interaction, pathogens that are negatively charged can be readily (>80%) captured from large volume samples. Our system does not require detergents for cell lysis, bulky instruments to perform amplification reactions, or specific antibodies or probes and thus offers the distinct advantage of being able to diagnose an emerging pathogen in a clinical setting. In addition, we used a laser cutting machine to make the microfluidic device. The assembled double chips that we used to streamline the sample processing are potentially scalable at low-cost and increase the surface area for pathogen capture without increasing the size of the device, thereby performing pathogen enrichment in a highly efficient way. We demonstrated that our SLIM system significantly improved the detection sensitivity compared with real-time PCR by enabling pathogen enrichment and nucleic acid extraction on a single platform. We next validated the clinical utility of the SLIM system to process different types of clinical specimens within 50 min, including environmental swabs, saliva, and blood plasma. Environmental swabs containing low concentrations of hPIV-3 were collected from environmental surfaces in a hospital ward that had come in contact with infected patients. Following enrichment and extraction with the SLIM system, the detection sensitivity was found to be 83.3% compared to 50% without using SLIM. In addition, the VZV detection sensitivity without SLIM in plasma samples was 35.7%, but increased to 57.1% using SLIM. Finally, to the best of our knowledge, we are the first group to combine sample preparation including pathogen enrichment and nucleic acid extraction, and bio-optical sensor for rapid and accurate analysis within 80 min. When we tested 17 blood plasma samples including 14 positives and 3 negatives using this combination system, the VZV detection sensitivity increased to 78.6% from 35.7% in the conventional assays. Notwithstanding the great potential of our SLIM system however, we still do not have a fully integrated and automated system on a single device that can detect various pathogens in real clinical settings. This could potentially improve the clinical utility of our prototype system. Moreover, use of more complex body fluids, such as whole blood, would be a notable limitation of the SLIM system at present. HI reagents recognize negatively charged molecules and would non-specifically bind to a number of molecules in complex samples. Therefore, further study for the validation of the real chemical reaction between HI and pathogens as well as further development of this process for use with various types of clinical samples will be needed. Nevertheless, we believe that an integrated system such as this has considerable potential a POC based pathogen diagnostic system that could have diverse clinical applications in humans and also in animal healthcare.
Acknowledgements
This study was supported by a grant from the Korea Health Technology R & D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (HI16C-0272-010016).
Acknowledgments
Conflict of interest
All authors declare no conflict of interest.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bios.2018.04.001.
Appendix A. Supplementary material
Supplementary material
References
- Duncan A.W., Maggi R.G., Breitschwerdt E.B. J. Microbiol. Methods. 2007;69:273–281. doi: 10.1016/j.mimet.2007.01.010. [DOI] [PubMed] [Google Scholar]
- Fauci A.S., Morens D.M. N. Engl. J. Med. 2012;366:454–461. doi: 10.1056/NEJMra1108296. [DOI] [PubMed] [Google Scholar]
- Fricke W.F., Rasko D.A. Nat. Rev. Genet. 2014;15:49–55. doi: 10.1038/nrg3624. [DOI] [PubMed] [Google Scholar]
- Ganesh I., Tran B.M., Kim Y., Kim J., Cheng H., Lee N.Y., Park S. Biomed. Microdevices. 2016;18:116. doi: 10.1007/s10544-016-0139-y. [DOI] [PubMed] [Google Scholar]
- Gardy J.L., Loman N.J. Nat. Rev. Genet. 2018;19:9–20. doi: 10.1038/nrg.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granger D.A., Kivlighan K.T., Fortunato C., Harmon A.G., Hibel L.C., Schwartz E.B., Whembolua G.L. Physiol. Behav. 2007;92(4):583–590. doi: 10.1016/j.physbeh.2007.05.004. [DOI] [PubMed] [Google Scholar]
- Hanash S.M., Baik C.S., Kallioniemi O. Nat. Rev. Clin. Oncol. 2011;8:142–150. doi: 10.1038/nrclinonc.2010.220. [DOI] [PubMed] [Google Scholar]
- Jin C.E., Lee T.Y., Koo B., Choi K.-C., Chang S., Park S.Y., Kim J.Y., Kim S.-H., Shin Y. Anal. Chem. 2017;89:7502–7510. doi: 10.1021/acs.analchem.7b01193. [DOI] [PubMed] [Google Scholar]
- Kim J.Y., Koo B., Jin C.E., Kim M.C., Chong Y.P., Lee S.-O., Choi S.-H., Kim Y.S., Woo J.H., Shin Y., Kim S.-H. Clin. Chem. 2018;64:556–565. doi: 10.1373/clinchem.2017.280230. [DOI] [PubMed] [Google Scholar]
- Kim T., Jin C.E., Koo B., Park J., Kim S.M., Kim J.Y., Chong Y.P., Lee S.O., Choi S.H., Kim Y.S., Woo J.H., Lee J.H., Lee J.H., Lee K.H., Shin Y., Kim S.-H. J. Hosp. Infect. 2017;97:403–413. doi: 10.1016/j.jhin.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien K.Y., Lin J.L., Liu C.Y., Lei H.Y., Lee G.B. Lab Chip. 2007;7:868–875. doi: 10.1039/b700516d. [DOI] [PubMed] [Google Scholar]
- de Mello A.J., Beard N. Lab Chip. 2003;3:11N–20N. doi: 10.1039/b301019h. [DOI] [PubMed] [Google Scholar]
- Mannoor M.S., Zhang S., Link A.J., McAlpine M.C. Proc. Natl. Acad. Sci. USA. 2010;107:19207–19212. doi: 10.1073/pnas.1008768107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis E.R. Annu. Rev. Genom. Hum. Genet. 2008;9:387–402. doi: 10.1146/annurev.genom.9.081307.164359. [DOI] [PubMed] [Google Scholar]
- Marra M.A., Jones S.J.M., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S.N., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- Park S.Y., Kim J.Y., Kim J.A., Kwon J., Kim S.M., Jeon N.Y., Kim M.C., Chong Y.P., Lee S.O., Choi S.H., Kim Y.S., Woo J.H., Kim S.-H. J. Infect. Dis. 2017;217:51–57. doi: 10.1093/infdis/jix508. [DOI] [PubMed] [Google Scholar]
- Park W.B., Perera R.A.P.M., Choe P.G., Lau E.H.Y., Choi S.J., Chun J.Y., Oh H.S., Song K.-H., Bang J.H., Kim H.B., Park S.W., Kim N.J., Poon L.L.M., Peiris M., Oh M. Emerg. Infect. Dis. 2015;21:2186. doi: 10.3201/eid2112.151421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S.M., Guan Y., Yuen K.Y. Nat. Med. 2004;10:S88. doi: 10.1038/nm1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennington H. Nat. Rev. Microbiol. 2014;2:259–262. doi: 10.1038/nrmicro846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez F.G., Mascini M., Tothill I.E., Turner A.P. Anal. Chem. 1998;70:2380–2386. doi: 10.1021/ac970715t. [DOI] [PubMed] [Google Scholar]
- Rosseel T., Ozhelvaci O., Freimanis G., Borm S.V. J. Virol. Methods. 2015;222:72–80. doi: 10.1016/j.jviromet.2015.05.010. [DOI] [PubMed] [Google Scholar]
- Shin Y., Lim S.Y., Lee T.Y., Park M.K. Sci. Rep. 2015;5:14127. doi: 10.1038/srep14127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R., Ekiert D.C., Krause J.C., Hai R., Crowe J.E., Wilson I.A. Science. 2010;328:357–360. doi: 10.1126/science.1186430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh Y.Y., Nisic M., Yu X., Xia Y., Zheng S.-Y. Ann. Biomed. Eng. 2014;42:2333–2343. doi: 10.1007/s10439-014-1044-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J., Yoon Y.J., Lee T.Y., Park M.K., Chung J., Shin Y. Sens. Actuators B Chem. 2018;255:1491–1499. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material