Abstract
A novel lineage C betacoronavirus, originally named human coronavirus EMC/2012 (HCoV-EMC) and recently renamed Middle East respiratory syndrome coronavirus (MERS-CoV), that is phylogenetically closely related to Tylonycteris bat coronavirus HKU4 and Pipistrellus bat coronavirus HKU5, which we discovered in 2007 from bats in Hong Kong, has recently emerged in the Middle East to cause a severe acute respiratory syndrome (SARS)-like infection in humans. The first laboratory-confirmed case, which involved a 60-year-old man from Bisha, the Kingdom of Saudi Arabia (KSA), who died of rapidly progressive community-acquired pneumonia and acute renal failure, was announced by the World Health Organization (WHO) on September 23, 2012. Since then, a total of 70 cases, including 39 fatalities, have been reported in the Middle East and Europe. Recent clusters involving epidemiologically-linked household contacts and hospital contacts in the Middle East, Europe, and Africa strongly suggested possible human-to-human transmission. Clinical and laboratory research data generated in the past few months have provided new insights into the possible animal reservoirs, transmissibility, and virulence of MERS-CoV, and the optimal laboratory diagnostic options and potential antiviral targets for MERS-CoV-associated infection.
Keywords: coronavirus, EMC, human, Middle East, SARS
Introduction: the “new SARS”?
Ten years after the devastating epidemic of severe acute respiratory syndrome (SARS) caused by SARS coronavirus (SARS-CoV), which resulted in a total of 774 deaths among more than 8000 confirmed cases in over 30 countries, the world is facing a new challenge posted by a “SARS-like” infection caused by another novel coronavirus emerging from the Middle East, which was originally named human coronavirus EMC/2012 (HCoV-EMC) and recently renamed by the Coronavirus Study Group of the International Committee for Taxonomy of Viruses as Middle East respiratory syndrome coronavirus (MERS-CoV).1, 2, 3, 4, 5, 6, 7, 8 The complete genome of the virus was sequenced and released in October 2012 after the isolation of the virus from two patients with severe community-acquired pneumonia in Bisha, the Kingdom of Saudi Arabia (KSA), and Doha, Qatar, first announced by the World Health Organization (WHO) on 23 September 2012.9 As of May 12, 2013, the total number of laboratory-confirmed cases of MERS-CoV infection has increased to 34 with 20 deaths, including two cases confirmed retrospectively from a Jordanian cluster of severe respiratory disease reported by the Ministry of Health of Jordan in April 2012 (Table 1 ).6, 7, 10, 11, 12, 13, 14 Although the number of laboratory-confirmed cases remains limited, the severe clinical manifestations with an unusually high mortality rate of over 50%, the spread of the infection beyond the geographical confinement in the Middle East, and the epidemiological evidence of human-to-human transmission arising from the recent clusters of cases in a family in the United Kingdom (Cases 10 to 12), and in hospitals in KSA (Cases 18 to 30, 32 and 33) and France (Cases 31 and 34), have raised significant concerns on the possible emergence of another SARS-like epidemic in the near future. In anticipation of the potential spread of this highly pathogenic virus from the Middle East and Europe to other parts of the world, especially the densely populated Southeast Asia, an updated review of the latest research findings on MERS-CoV and their implications on the clinical management of MERS-CoV infection is essential.
Table 1.
Characteristics of patients with laboratory-confirmed MERS-CoV infection as of 12 May 2013a.6, 7, 10, 11, 12, 13, 14
| Case (WHO report date) | Demographics and epidemiological links |
Clinical manifestations |
Laboratory diagnosis (sample & date) | Treatment | |||||
|---|---|---|---|---|---|---|---|---|---|
| Sex/age | Comorbidities | Place of symptom onset (date) | Exposure Hx | Pneumonia / respiratory failure | ARF | Death (date) | |||
| 1 (23 Sep 12) |
M/60 | None | Bisha/KSA (6 Jun 12) | ? | Yes | Yes | Yes (24 Jun 2012) | 1. Viral culture (sputum 13 Jun 12 in LLC-MK2 & Vero cells) 2. Serum IgG (22 & 23 Jun 12; 1:20) 3. Pan-CoV RT-PCR & sequencing |
ICU: MV, oseltamivir, antibiotics |
| 2 (23 Sep 12) |
M/49 | None | Doha/Qatar (3 Sep 12) |
Kept camel and sheep in a farm | Yes | Yes | No (under Rx) |
Pan-CoV RT-PCR & sequencing (nose & throat swab 13 Sep 12, sputum 17 Sep 12, TA 19 Sep 12) | ICU: ECMO, oseltamivir, antibiotics, corticosteroid |
| 3 (23 Nov 12) |
M/45 | Heavy smoker, Atrophic R kidney DM, IHD |
Riyadh/KSA (9 Oct 12) |
Contacted farm animals (06/10/12) | Yes | Yes | No (recovered) | RT-PCR (URT 13 Oct 12, TA 23 Oct 12) | ICU: MV, RRT, oseltamivir, antibiotics |
| 4 (23 Nov 12) |
M/70 | ? | KSA (Oct 12) |
? | Yes | Yes | Yes | RT-PCR | Hospitalized |
| 5 (23 Nov 12) |
M/40+ | Heavy smoker | Qatar (5 Oct 12) |
Contacted an ill animal caretaker (camel and ill goats) | Yes | Yes | No (recovered) | RT-PCR (respiratory sample 17 Oct 12 & BAL 25 Oct 12) | ICU |
| 6 (23 Nov 12) |
? | ? | KSA (Oct 12) |
Household family member of case 4 | Yes | No | No (recovered) | RT-PCR | Hospitalized |
| 7 (30 Nov 12) |
? | ? | KSA (Oct 12) |
Household family member of case 4 | Yes | Yes | Yes | RT-PCR | Hospitalized |
| 8 (30 Nov 12) |
? | ? | Zarga/Jordan (Apr 12) |
? | Yes | No | Yes | ?RT-PCR | ICU |
| 9 (30 Nov 12) |
? | Yes | Zarga/Jordan (Apr 12) | ICU nurse of case 8 | Yes | No | Yes | ?RT-PCR | ? |
| 10 (11 Feb 13) |
M/60 | None | KSA (24 Jan 13) |
Travelled to Pakistan for 5 weeks and then to KSA (20-28 Jan 2012) before returning to UK (28 Jan 2012) No animal or patient contact | Yes | ? | Yes (Mar 13) | RT-PCR (throat swab 7 Feb 13) Coinfection: influenza A(H1N1)pdm09 (1 Feb 13) | ICU: ECMO |
| 11 (13 Feb 13) | M/38 | Malignancy with Rx | UK (6 Feb 13) |
Relative of case 10 | Yes | ? | Yes (17 Feb 2013) | RT-PCR (nose & throat swab 10 Feb 13) Coinfection: type 2 PIF (nose & throat swab 10 Feb 13) | ICU, ECMO |
| 12 (16 Feb 13) |
F/30 | None | UK (5 Feb 13) |
Relative of case 10 (total 2.5 h of contact) | No | No | No (recovered) | RT-PCR (sputum 13 Feb 13) Coinfection: type 2 PIF (nose & throat swab 15 Feb 13) |
Self-recovery (15 Feb 13) |
| 13 (21 Feb 13) |
? | ? | KSA (Jan 13) |
? | ? | ? | Yes (10 Feb 13) | ?RT-PCR (18 Feb 13) | Hospitalized |
| 14 (6 Mar 13) |
M/69 | ? | KSA (Feb 13) |
No patient contact ?animal contact |
? | ? | Yes (19 Feb 13) | ?RT-PCR (18 Feb 13) | Hospitalized |
| 15 (12 Mar 13) |
M/39 | ? | KSA (Feb 13) |
No patient contact ?animal contact |
? | ? | Yes (2 Mar 13) | ?RT-PCR (18 Feb 13) | Hospitalized |
| 16 (23 Mar 13) |
? | ? | KSA | Contact of case 15 | No | ? | No (recovered) | ?RT-PCR | Self-recovery |
| 17 (26 Mar 13) |
M/73 | ? | UAE (10 Mar 13) |
? | Yes | ? | Yes (26 Mar 13) | ?RT-PCR | Hospitalized |
| 18 (2 May 13) |
M/59 | Yes | Al-Hasa/KSA (14 Apr 13) |
Hospital cluster | Yes | ? | Yes (19 Apr 13) | ?RT-PCR | ICU |
| 19 (2 May 13) |
M/24 | Yes | Al-Hasa/KSA (17 Apr 13) |
Hospital cluster | Yes | ? | ?∗ | ?RT-PCR | ICU |
| 20 (2 May 13) |
M/87 | Yes | Al-Hasa/KSA (17 Apr 13) |
Hospital cluster | Yes | ? | Yes (28 Apr 13) | ?RT-PCR | ICU |
| 21 (2 May 13) |
M/58 | Yes | Al-Hasa/KSA (22 Apr 13) |
Hospital cluster | Yes | ? | ?∗ | ?RT-PCR | ICU |
| 22 (2 May 13) |
M/94 | Yes | Al-Hasa/KSA (22 Apr 13) |
Hospital cluster | Yes | ? | Yes (26 Apr 13) | ?RT-PCR | ICU |
| 23 (2 May 13) |
M/56 | Yes | Al-Hasa/KSA (22 Apr 13) |
Hospital cluster | Yes | ? | Yes (30 Apr 13) | ?RT-PCR | ICU |
| 24 (2 May 13) |
M/56 | Yes | Al-Hasa/KSA (22 Apr 13) |
Hospital cluster | Yes | ? | Yes (29 Apr 13) | ?RT-PCR | ICU |
| 25 (3 May 13) |
F/58 | Yes | Al-Hasa/KSA (27 Apr 13) |
Hospital cluster | Yes | ? | ?∗ | ?RT-PCR | ICU |
| 26 (3 May 13) |
M/50 | Yes | Al-Hasa/KSA (30 Apr 13) |
Hospital cluster | Yes | ? | ?∗ | ?RT-PCR | Hospitalized |
| 27 (3 May 13) |
M/33 | Yes | Al-Hasa/KSA (28 Apr 13) |
Hospital cluster, family contact of a deceased case | Yes | ? | ?∗ | ?RT-PCR | Hospitalized |
| 28 (6 May 13) |
F/62 | Yes | Al-Hasa/KSA (19 Apr 13) |
Hospital cluster | Yes | ? | Yes (3 May 13) | ?RT-PCR | ICU |
| 29 (6 May 13) |
M/71 | Yes | Al-Hasa/KSA (15 Apr 13) |
Hospital cluster | Yes | ? | Yes (3 May 13) | ?RT-PCR | ICU |
| 30 (6 May 13) |
F/58 | Yes | Al-Hasa/KSA (1 May 13) |
Hospital cluster | Yes | ? | ?∗ | ?RT-PCR | ICU |
| 31 (8 May 13) |
M/65 | Yes | Valenciennes/France (23 Apr 13) |
Travelled to Dubai, UAE in Apr 2013 | Yes | ? | ?∗ | RT-PCR: (BAL 7 May 13) | ICU |
| 32 (9 May 13) |
M/48 | Yes | Al-Hasa/KSA (29 Apr 13) |
Hospital cluster | Yes | ? | ?∗ | ?RT-PCR (8 May 13) | Hospitalized |
| 33 (9 May 13) |
M/58 | Yes | Al-Hasa/KSA (3 May 13) |
Hospital cluster | Yes | ? | No (recovered) |
?RT-PCR (8 May 13) | Discharged (3 May 13) |
| 34 | ?/? | ? | Valenciennes/France | Stayed in the same hospital room with case 31 | ? | ? | ? | ?RT-PCR (11-12 May 13) | ? |
Abbreviations: ARF, acute renal failure; BAL, bronchoalveolar lavage; CoV, coronavirus; DM, diabetes mellitus; F, female; h, hour; Hx, history; ICU, intensive care unit; IgG, immunoglobulin G; IHD, ischemic heart disease; LRT, lower respiratory tract; M, male; MV, mechanical ventilation; PIF, parainfluenza virus; R, right; RRT, renal replacement therapy; RT-PCR, reverse transcription polymerase chain reaction; Rx, treatment; TA, tracheal aspirate; UAE, United Arab Emirates; URT, upper respiratory tract.
Two additional deaths were reported among these cases. The others are under treatment.
As of 23 June 2013, after the acceptance of the article, the total number of laboratory-confirmed cases have increased to 70 with 39 fatalities.
Viral genomic studies reveal the first lineage C betacoronavirus associated with human infection
MERS-CoV belongs to the genus Betacoronavirus in the family Coronaviridae under the order Nidovirales. Coronaviruses are enveloped viruses with positive-sense single-stranded RNA genomes. Studies in their biodiversity, comparative genomics and phylogeny in the past 10 years have improved our understanding of this family of viruses.15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30 According to the most recent classification by the Coronavirus Study Group of the International Committee for Taxonomy of Viruses, there are four genera in Coronaviridae, namely Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus.28, 31 The genus Alphacoronavirus contains two human coronaviruses, HCoV-229E and HCoV-NL63, which are associated with the common cold. No human coronavirus has been discovered in Gammacoronavirus and Deltacoronavirus, which mainly contain avian coronaviruses with just a few mammalian coronaviruses. The genus Betacoronavirus is comprised of four lineages (A, B, C and D) which contain four coronaviruses associated with human infections: HCoV-OC43 and HCoV-HKU1 (lineage A), SARS-CoV (lineage B), and MERS-CoV (lineage C).15 MERS-CoV is the first known lineage C betacoronavirus associated with human infection and is phylogenetically closely related to the other lineage C betacoronaviruses including Tylonycteris bat CoV HKU4 (Ty-BatCoV-HKU4) and Pipistrellus bat CoV HKU5 (Pi-BatCoV-HKU5), which were discovered in lesser bamboo bats (Tylonycteris pachypus) and Japanese Pipistrelle bats (Pipistrellus abramus), respectively, captured in Hong Kong, China.9, 22, 32 Analysis of the genome of MERS-CoV revealed that it has a genome size of 30,106 bases with the RdRp and S genes having over 90% and around 70% amino acid identities with those of Ty-BatCoV-HKU4 and Pi-BatCoV-HKU5.9, 32 Molecular clock analysis using the RdRp gene showed that MERS-CoV might have diverged from the most recent common ancestor of lineage C betacoronaviruses in year ∼941 AD (529 BC to 1878 AD).32 It is postulated that the emergence of MERS-CoV represents another series of interspecies transmission events in coronaviruses, from bats to possibly other animals and then to humans, a scenario similar to the SARS epidemic.
Epidemiology reports and cell line susceptibility studies suggest possible animal reservoirs and human-to-human transmissions
Epidemiological linkage with animals, including camels, goats, sheep, and farm animals or their caretakers before symptom onset in some of the reported cases supported the hypothesis of MERS-CoV being a zoonotic agent (Table 1). Furthermore, in vitro data from cell line susceptibility studies showed that the virus had a broad species tropism and was able to replicate in bat, primate, porcine, rabbit, and civet cell lines.33, 34 As in the case of SARS-CoV, which likely emerged from its natural animal reservoir the horseshoe bats (Rhinolophus sp.), and jumped to other mammals during their caging in the wild life markets of South China and then to humans, MERS-CoV might have also originated from bats to these susceptible animal species before adapting to humans.16, 35, 36 In addition to Pipistrellus bats, which are the natural hosts of the closely related Pi-BatCoV-HKU5 and are also found in the Middle East, Rousettus, Rhinolophus, Myotis, and Carollia bat cell lines are also susceptible to MERS-CoV infection in vitro.34 Active surveillance of different bat and animal species predominantly found in the Middle East would help to delineate the natural bat reservoir and the evolutionary pathway of MERS-CoV among other susceptible animal species. Indeed, a recent study showed that a number of different bat species in Ghana and Europe are infected with coronaviruses, which also share close homologies with MERS-CoV.37 Determination of the virus' animal hosts might in turn facilitate the control of the outbreak as in SARS, where closure of the wet markets likely contributed to the cessation of the epidemic.
Human-to-human transmission represents a new stage in the evolution of MERS-CoV infection. There are, so far, six clusters of laboratory-confirmed MERS-CoV infection (Table 1). The first cluster occurred in an intensive care unit in Zarga, Jordan in April 2012, where 11 individuals, including seven nurses, one doctor, and a brother of one of the nurses, developed severe pneumonia of unknown etiology. A nurse with “underlying medical conditions” and the index case died (Cases 8 and 9). On November 30, 2012, WHO reported that the stored specimens from these two fatal cases were positive for MERS-CoV, while the details of the other nine patients' test results were not described. The second cluster occurred in October 2012 and involved three family members living within the same household in KSA (Cases 4, 6 and 7) with two fatalities. Another small cluster occurred in KSA in February to March 2013 (Cases 15 and 16). As the clinical and epidemiological details were not available for these three clusters, it was suggested that exposure to a common source in the Middle East was another possible explanation besides human-to-human transmission of the virus. However, recent clusters of MERS-CoV infection occurring among household contacts in the United Kingdom in January 2013, which involved three patients (Cases 10–12), two of whom were without history of traveling to the Middle East, and among hospital contacts in KSA (Cases 18–30, 32 and 33) and France (Cases 31 and 34), provided strong evidence of continuing adaptations of MERS-CoV for human-to-human transmission.12
The seroprevalence and transmissibility of MERS-CoV remain undetermined, although animal-to-human and human-to-human transmissions both appear to be limited at this stage (Fig. 1 ). Among 2400 persons seeking medical attention in a hospital in Jeddah, KSA, none had MERS-CoV-specific IgG detectable by indirect immunofluorescence assay, suggesting that the current situation is likely to be different from those of other human coronaviruses which are endemic in humans.6, 38 The lack of secondary cases among nearly 200 contacts of Cases 2 and 5 who did not practice optimal infection control measures before the diagnosis of MERS-CoV infection was confirmed, implied that this novel virus might be less efficient than SARS-CoV in human-to-human transmission.7, 13, 39 However, the findings of these studies should be interpreted with cautions for two reasons. First, only a relatively small number of subjects (10/64 in Case 2 and 85/123 in Case 5) were tested by reliable laboratory tests. Second, the timing of testing was not clearly described and might be suboptimal for the purpose of excluding the diagnosis. It has been proposed that a second test should be performed in symptomatic patients with initially negative results by reverse transcription-polymerase chain reaction (RT-PCR), within the first 10 days of symptom onset, based on the observation in SARS where viral load peaked at day 10 of symptom onset.5, 40, 41, 42 Therefore, the apparently limited spread of MERS-CoV at present might be an underestimation and ongoing transmission of the virus should be cautiously monitored.
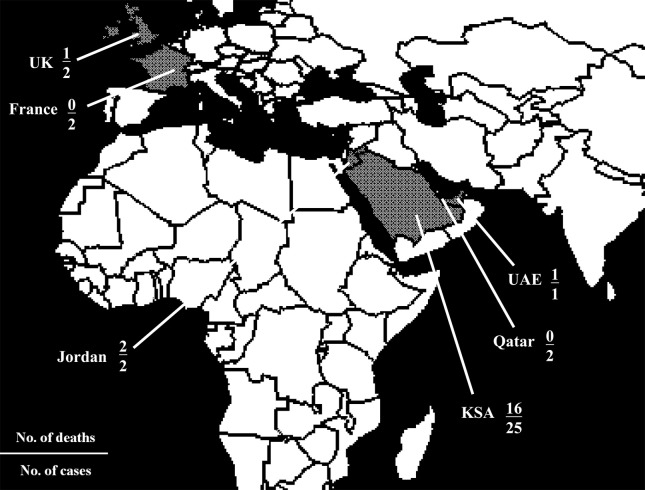
Figure 1.
The world map showing countries with laboratory-confirmed cases of MERS-CoV infection according to the patients' date of symptom onset as of 12 May 2013 (in grey): the Kingdom of Saudi Arabia (KSA), Qatar, Jordan, the United Arab Emirates (UAE), the United Kingdom (UK), and France. After the article was accepted, additional local or imported cases were reported in KSA, UK, Italy, Germany, Tunisia, and Morocco. As of 23 June 2013, the total number of laboratory-confirmed cases was 70 with 39 fatalities.
In vitro viral characteristics and their correlations with the unusual clinical features of MERS-CoV infection
Unlike its close relatives in bats, MERS-CoV is highly pathogenic in humans. The most common clinical presentation among the 34 laboratory-confirmed cases of MERS-CoV infection is acute severe community-acquired pneumonia with acute renal failure following an estimated incubation period of 1–9 days (Table 1).12, 43 This is unusual among human infections caused by coronaviruses, in which severe pneumonia with respiratory failure is seldom seen, except in SARS. The other human coronaviruses, namely HCoV-229E, HCoV-NL63, HCoV-OC43, and HCoV-HKU1, predominantly cause acute self-limited upper respiratory tract infections, and only occasionally cause lower respiratory tract infections in the elderly and immunocompromized populations. Only two of 34 patients (Cases 12 and 16) had a self-limiting, mild, influenza-like illness not requiring hospitalization. Asymptomatic or mild infections have otherwise not been detected among the other contacts of confirmed cases by RT-PCR of upper respiratory tract specimens and/or serological tests, 2400 Saudi residents in Jeddah by indirect immunofluorescent antibody testing of archived sera, and 169 French Hajj pilgrims with upper respiratory symptoms and RT-PCR of prospectively collected nasal swabs.6, 7, 13, 44 The other 32 (94.1%) patients, many of whom had no underlying medical condition, developed rapid clinical deterioration with lower respiratory tract involvement and respiratory failure requiring ventilator support within a few days to 1 week after initial systemic symptoms of fever, myalgia, and malaise, and upper respiratory tract symptoms such as rhinorrhea and sore throat. This correlated with the finding in our recent cell line susceptibility study, which showed that MERS-CoV replicated much better in the lower respiratory tract cell lines including Calu-3 (polarized airway epithelium), A549 (lung adenocarcinoma), and HFL (embryonic lung fibroblasts), than the upper respiratory tract cell line Hep-2 (laryngeal epidermoid carcinoma).33 Another in vitro study also showed that pseudostratified human bronchial epithelium cultures are highly permissive to MERS-CoV infection.45 In ex vivo organ cultures, MERS-CoV productively replicated in both human bronchial and lung tissues, whereas SARS-CoV only productively replicated in lung tissue.46 Recently, a macaque model showed that MERS-CoV caused acute, localized to widespread pneumonia, resulting in mild to moderate clinical disease resembling the illness observed in humans.47 Radiologically, pneumonia is evident by focal consolidations involving single or multiple lobes, with progressive involvement of bilateral lung fields, especially the lower zones. In Case 1, thoracic computed tomography scans revealed mediastinal hilar lymphadenopathies, airspace opacities with air bronchograms, scattered ground-glass opacities, interstitial septal thickenings, and nodularities in the upper lobes without significant pleural and pericardial effusions.6
Acute renal failure was the other dominant clinical feature in MERS-CoV infection and is seen in at least six of the 34 reported cases. Many of the remaining 28 severe cases probably also developed renal impairment, although their clinical details were not available. This was unusual, even when compared to SARS in which 28.8% of the patients had abnormal urinalysis and detectable viral load by quantitative RT-PCR in urine, but only 6.7% developed acute renal failure with histological evidence of acute tubular necrosis and most did not require renal replacement therapy.48 This clinical presentation correlates with the in vitro finding of efficient replication of MERS-CoV in kidney cell lines, including HEK 293, Vero, LLC-MK2, and 769P.33, 34, 49 More importantly, the presence of renal involvement appeared to be a poor prognostic factor, as those with renal failure either died or required renal replacement therapy, while two cases without renal impairment survived (Table 1). The lack of extrapulmonary lesions observed in the macaque model of MERS-CoV infection suggested that acute renal failure was more likely due to hypoxic damage than a direct viral cytopathic effect.47
Other clinical, laboratory and microbiological findings reported in MERS-CoV infection included pericarditis, disseminated intravascular coagulation, leukocytosis with neutrophilia and lymphopenia, thrombocytopenia, anemia, hyponatremia, hypoalbuminemia, elevated liver enzymes, lactate dehydrogenase, C-reactive protein, and procalcitonin levels, possible secondary bacterial pneumonia caused by Klebsiella pneumoniae, Staphylococcus aureus, and Acinetobacter sp., and coinfection with other respiratory viruses, including influenza A(H1N1)pdm09 and type 2 parainfluenza virus.6, 7, 10, 11, 12, 13, 50 It is interesting to note the absence of watery diarrhea in the acute phase of MERS-CoV infection, despite the in vitro finding of viral replication in the colonic cell line Caco-2 (colorectal adenocarcinoma), in contrast to SARS in which around 20% of patients developed enterocolitis.1, 40 It remains to be seen in future case cohorts whether this is due to under-reporting, or a genuine difference in clinical presentations between the two diseases. Together with severe acute respiratory and renal failure, these clinical features underscore the success of the innate immune evasion mechanisms of MERS-CoV in humans, leading to overwhelming infection and possible cytokine dysregulation, as reflected by the lack of interferon through inhibition of interferon regulatory factor family 3 (IRF-3).45, 49 The discovery of dipeptidyl peptidase 4 (DPP4), a multifunctional 766-amino-acid-long type-II transmembrane glycoprotein exopeptidase expressed on human non-ciliated bronchial, renal, enteric, hepatic and prostatic epithelial cells, which is important in the regulation of hormone and chemokine bioactivity, glucose metabolism, T-cell activation, chemotaxis modulation, cell adhesion, apoptosis and regulation of tumorigenicity, as a functional receptor for MERS-CoV, provides further insights into the unique pattern of organ involvement and unusually severe clinical presentation of this emerging infection.51
Clinical utility and practical concerns of published laboratory diagnostic options
Several laboratory methods are available for establishing a virological diagnosis of MERS-CoV infection. The most definitive tests are viral culture from respiratory, fecal, urine, or tissue specimens, and/or a fourfold rise in the serum neutralizing antibody titers taken at 14 to 21 days apart. The first clinical isolate of MERS-CoV was cultured on monkey kidney cells, like Vero and LLC-MK2, which showed cytopathic effects of syncytium formation, rounding and detachment of cells.6 Subsequent studies have identified various human cell types, including bronchial epithelial, colonic, hepatic, renal, and neuronal cells, monocytes and histiocytes that support the replication of MERS-CoV. However, the use of viral culture is limited by the requirement of a biosafety level three setting, which is not available in most clinical microbiology laboratories. A serum neutralizing antibody test has the disadvantage of requiring convalescent sera and is therefore mainly used for diagnosis in the convalescent instead of acute phase.13, 52 Furthermore, the sera of SARS patients may contain low-titer cross-reactive neutralizing antibodies against MERS-CoV, which may lead to the wrong serodiagnosis, especially in countries where the general population had previous exposure.53 It remains to be seen whether neutralizing antibodies against MERS-CoV might also cross-react with other closely related lineage C betacoronaviruses, like Ty-BatCoV-HKU4 and Pi-BatCoV-HKU5.54 In most of the laboratory-confirmed cases, diagnosis was established by the detection of nucleic acid by RT-PCR of respiratory tract samples, including combined nose and throat swab, sputum, tracheal aspirate, bronchoalveolar lavage, and/or urine taken between Day 5 (Cases 3 and 11) and Day 20 (Case 5).51 Two highly sensitive real-time RT-PCR assays targeting regions upstream of the E gene (upE), with sensitivity of up to 3.4 RNA copies per reaction and within an open reading frame (ORF) 1b, with sensitivity of up to 64 RNA copies per reaction, have been proposed for screening and confirmation of MERS-CoV infection, respectively, and are available in about half of the countries in the WHO European Region.55, 56 Additional testing of other gene targets with partial or whole genome sequence analysis may also be used for confirmation.55, 56 For example, pan-coronavirus RT-PCR targeting the RdRp gene was used in Case 2 and a real-time quantitative RT-PCR assay targeting the N gene has been used in in vitro studies, and may also be useful for clinical diagnosis, although further evaluations are needed.33 Other potential diagnostic options which might be used in areas without RT-PCR are N or S protein-based enzyme-linked immunosorbent assays, which were shown to be highly sensitive and/or specific for the diagnosis of SARS. Of note, the detection of other respiratory viruses in the respiratory tract specimen does not preclude the need for specific MERS-CoV testing in patients with epidemiological risk factors, or unusually severe disease, despite antiviral treatment as exemplified by the presence of coinfections in Cases 10–12.
Insights into the potential targets for antiviral and vaccine development from laboratory experiments
The exceptionally high crude mortality rate of 60% in MERS-CoV infection is partly due to the lack of specific anti-coronavirus treatment and effective vaccine. In most of the cases, intensive supportive treatment with extracorporeal membrane oxygenation, renal replacement therapy, and empirical broad-spectrum antibacterial and antiviral agents were used. Case 2, who is still in a critical condition nearly 6 months after symptom onset, also received a corticosteroid in the initial phase of treatment, but its efficacy is unknown. Its use might be limited by serious side effects and the availability of ECMO for organ support during the critical phase. Alternatively, type I interferons appear to be promising therapeutic options, as MERS-CoV has been shown to be much more sensitive than SARS-CoV to the antiviral action of interferon in vitro.49 The replication of MERS-CoV was shown to be reduced in human lung ex vivo organ cultures treated with type I interferons.46 A recent study showed that while both interferon-α2b and ribavirin reduced the replication of MERS-CoV in Vero and LLC-MK2 cells, the combination of the two drugs achieved the same endpoints at a much lower concentration, which might facilitate their clinical applications.57 Other potential specific antiviral targets include type II transmembrane serine proteases (TMPRSS2) and endosomal cathepsins, which are responsible for MERS-CoV S protein activation required for virus-cell fusion and entry of the virus into host cells.54 The identification of DPP4, but not angiotensin-converting enzyme 2 (ACE2), aminopeptidase N, and carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1), as a functional receptor for MERS-CoV implies that in vivo manipulation of DPP4 levels and development of inhibitors against the S1 domain-DPP4 interface might have potential therapeutic roles in MERS-CoV infection, as with ACE2 analogues in the case of SARS.51 Finally, the recognition of the predicted receptor-binding domain /critical neutralizing domain at residues 377 to 662 in the MERS-CoV S protein might facilitate vaccine development for this emerging infection.58
Conclusions: more questions than answers
In the past few months, following the WHO's announcement of the first two cases of MERS-CoV infection on September 23, 2012, clinicians and scientists worldwide have collaborated closely in an attempt to prevent a SARS-like epidemic from happening again. The discovery of certain important characteristics of MERS-CoV including its genome arrangement, phylogenetic relatedness with other coronaviruses, in vitro tissue and species tropism, and functional receptor, enhanced our understanding of the clinical presentation, pathogenesis, epidemiology, and design of diagnostic and therapeutic options of this emerging novel human coronavirus. However, key questions concerning the definitive animal reservoirs of the virus, evolutionary process, transmissibility, and the prognostic factors and optimal treatment modalities of the infection, remain elusive. More importantly, the increasing number of laboratory-confirmed cases in the Middle East, and the recent evidence of human-to-human transmission in Europe are suggestive of continuing viral adaptations in humans, which might precede a large-scale epidemic. Indeed, as of 23 June 2013, after the acceptance of the article, the total number of laboratory-confirmed cases have increased to 70 with 39 fatalities.59 Additional clusters of cases involving household and/or hospital contacts were reported in KSA, Italy, and Tunisia. Collaborations between global and local health authorities and their sustained support for further research on MERS-CoV are crucial to control this “new SARS”.
Acknowledgments
This work is partly supported by the Research Grant Council Grant, Theme-based Research Scheme, University Grant Council; HKSAR Health and Medical Research Fund; Strategic Research Theme Fund and University Development Fund, the University of Hong Kong; and Consultancy Service for Enhancing Laboratory Surveillance of Emerging Infectious Disease for the HKSAR Department of Health.
References
- 1.Cheng V.C., Lau S.K., Woo P.C., Yuen K.Y. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin Microbiol Rev. 2007;20:660–694. doi: 10.1128/CMR.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peiris J.S., Lai S.T., Poon L.L., Guan Y., Yam L.Y., Lim W. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peiris J.S., Yuen K.Y., Osterhaus A.D., Stöhr K. The severe acute respiratory syndrome. N Engl J Med. 2003;349:2431–2441. doi: 10.1056/NEJMra032498. [DOI] [PubMed] [Google Scholar]
- 4.Woo P.C., Lau S.K., Tsoi H.W., Chan K.H., Wong B.H., Che X.Y. Relative rates of non-pneumonic SARS coronavirus infection and SARS coronavirus pneumonia. Lancet. 2004;363:841–845. doi: 10.1016/S0140-6736(04)15729-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan J.F., Li K.S., To K.K., Cheng V.C., Chen H., Yuen K.Y. Is the discovery of the novel human betacoronavirus 2c EMC/2012 (HCoV-EMC) the beginning of another SARS-like pandemic? J Infect. 2012;65:477–489. doi: 10.1016/j.jinf.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 7.Bermingham A., Chand M.A., Brown C.S., Aarons E., Tong C., Langrish C. Severe respiratory illness caused by a novel coronavirus, in a patient transferred to the United Kingdom from the Middle East, September 2012. Euro Surveill. 2012;17:20290. [PubMed] [Google Scholar]
- 8.de Groot R.J., Baker S.C., Baric R.S., Brown C.S., Drosten C., Enjuanes L. Middle East Respiratory Syndrome Coronavirus (MERS-CoV); Announcement of the Coronavirus Study Group. J Virol. 2013 doi: 10.1128/JVI.01244-13. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Boheemen S., de Graaf M., Lauber C., Bestebroer T.M., Raj V.S., Zaki A.M. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. MBio. 2012:3. doi: 10.1128/mBio.00473-12. pii:e00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pollack M.P., Pringle C., Madoff L.C., Memish Z.A. Latest outbreak news from ProMED-mail: novel coronavirus – Middle East. Int J Infect Dis. 2013;17:e143–e144. doi: 10.1016/j.ijid.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Albarrak A.M., Stephens G.M., Hewson R., Memish Z.A. Recovery from severe novel coronavirus infection. Saudi Med J. 2012;33:1265–1269. [PubMed] [Google Scholar]
- 12.Health Protection Agency (HPA) UK Novel Coronavirus Investigation Team Evidence of person-to-person transmission within a family cluster of novel coronavirus infections, United Kingdom, February 2013. Eruo Surveill. 2013:18. doi: 10.2807/ese.18.11.20427-en. pii:20427. [DOI] [PubMed] [Google Scholar]
- 13.Buchholz U., Müller M.A., Nitsche A., Sanewski A., Wevering N., Bauer-Balci T. Contact investigation of a case of human novel coronavirus infection treated in a German hospital, October-November 2012. Euro Surveill. 2013;18 pii:20406. [PubMed] [Google Scholar]
- 14.World Health Organization . WHO; Geneva: 2012. Global alert and response: novel coronavirus infection–update.http://www.who.int/csr/don/2013_05_09_ncov/en/index.html [accessed 11.05.13] [Google Scholar]
- 15.Woo P.C., Lau S.K., Huang Y., Yuen K.Y. Coronavirus diversity, phylogeny and interspecies jumping. Exp Biol Med (Maywood) 2009;234:1117–1127. doi: 10.3181/0903-MR-94. [DOI] [PubMed] [Google Scholar]
- 16.Lau S.K., Woo P.C., Li K.S., Huang Y., Tsoi H.W., Wong B.H. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci U S A. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woo P.C., Lau S.K., Chu C.M., Chan K.H., Tsoi H.W., Huang Y. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woo P.C., Lau S.K., Li K.S., Poon R.W., Wong B.H., Tsoi H.W. Molecular diversity of coronaviruses in bats. Virology. 2006;351:180–187. doi: 10.1016/j.virol.2006.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lau S.K., Woo P.C., Yip C.C., Tse H., Tsoi H.W., Cheng V.C. Coronavirus HKU1 and other coronavirus infections in Hong Kong. J Clin Microbiol. 2006;44:2063–2071. doi: 10.1128/JCM.02614-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woo P.C., Lau S.K., Yip C.C., Huang Y., Tsoi H.W., Chan K.H. Comparative analysis of 22 coronavirus HKU1 genomes reveals a novel genotype and evidence of natural recombination in coronavirus HKU1. J Virol. 2006;80:7136–7145. doi: 10.1128/JVI.00509-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woo P.C., Lau S.K., Yuen K.Y. Infectious diseases emerging from Chinese wet-markets: zoonotic origins of severe respiratory viral infections. Curr Opin Infect Dis. 2006;19:401–407. doi: 10.1097/01.qco.0000244043.08264.fc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woo P.C., Wang M., Lau S.K., Xu H., Poon R.W., Guo R. Comparative analysis of twelve genomes of three novel group 2c and group 2d coronaviruses reveals unique group and subgroup features. J Virol. 2007;81:1574–1585. doi: 10.1128/JVI.02182-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lau S.K., Woo P.C., Li K.S., Huang Y., Wang M., Lam C.S. Complete genome sequence of bat coronavirus HKU2 from Chinese horseshoe bats revealed a much smaller spike gene with a different evolutionary lineage from the rest of the genome. Virology. 2007;367:428–439. doi: 10.1016/j.virol.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang Y., Lau S.K., Woo P.C., Yuen K.Y. CoVDB: a comprehensive database for comparative analysis of coronavirus genes and genomes. Nucleic Acids Res. 2008;36:D504–D511. doi: 10.1093/nar/gkm754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woo P.C., Lau S.K., Lam C.S., Lai K.K., Huang Y., Lee P. Comparative analysis of complete genome sequences of three avian coronaviruses reveals a novel group 3c coronavirus. J Virol. 2009;83:908–917. doi: 10.1128/JVI.01977-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lau S.K., Li K.S., Huang Y., Shek C.T., Tse H., Wang M. Ecoepidemiology and complete genome comparison of different strains of severe acute respiratory syndrome-related Rhinolophus bat coronavirus in China reveal bats as a reservoir for acute, self-limiting infection that allows recombination events. J Virol. 2010;84:2808–2819. doi: 10.1128/JVI.02219-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lau S.K., Poon R.W., Wong B.H., Wang M., Huang Y., Xu H. Coexistence of different genotypes in the same bat and serological characterization of Rousettus bat coronavirus HKu9 belonging to a novel Betacoronavirus subgroup. J Virol. 2010;84:11385–11394. doi: 10.1128/JVI.01121-10. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Woo P.C., Lau S.K., Lam C.S., Lau C.C., Tsang A.K., Lau J.H. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lau S.K., Woo P.C., Yip C.C., Fan R.Y., Huang Y., Wang M. Isolation and characterization of a novel betacoronavirus subgroup A coronavirus, rabbit coronavirus HKU14, from domestic rabbits. J Virol. 2012;86:5481–5496. doi: 10.1128/JVI.06927-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lau S.K., Li K.S., Tsang A.K., Shek C.T., Wang M., Choi G.K. Recent transmission of a novel alphacoronavirus, bat coronavirus HKU10, from Leschenault's rousettes to pomona leaf-nosed bats: first evidence of interspecies transmission of coronavirus between bats of different suborders. J Virol. 2012;86:11906–11918. doi: 10.1128/JVI.01305-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.International Committee on Taxonomy of Viruses. Virus Taxonomy: 2012 Release (current). http://ictvonline.org/virusTaxonomy.asp?version=2012 [accessed 14.04.13].
- 32.Woo P.C., Lau S.K., Li K.S., Tsang A.K., Yuen K.Y. Genetic relatedness of the novel human lineage C betacoronavirus to Tylonycteris bat coronavirus HKU4 and Pipistrellus bat coronavirus HKU5. Emerging Microbes Infect. 2012;1:e35. doi: 10.1038/emi.2012.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chan J.F., Chan K.H., Choi G.K., To K.K., Tse H., Cai J.P. Differential cell line susceptibility to the emerging novel human betacoronavirus 2C EMC/2012: implications on disease pathogenesis and clinical manifestation. J Infect Dis. 2013;207:1743–1752. doi: 10.1093/infdis/jit123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Müller M.A., Raj V.S., Muth D., Meyer B., Kallies S., Smits S.L. Human coronavirus EMC does not require the SARS-coronavirus receptor and maintains broad replicative capability in mammalian cell lines. MBio. 2012:3. doi: 10.1128/mBio.00515-12. pii:e00515–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 36.Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- 37.Annan A., Baldwin H.J., Corman V.M., Sm Klose, Owusu M., Nkrumah E.E. Human betacoronavirus 2c EMC/2012-related viruses in bats, Ghana and Europe. Emerg Infect Dis. 2013;19:456–459. doi: 10.3201/eid1903.121503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Woo P.C., Lau S.K., Tsoi H.W., Huang Y., Poon R.W., Chu C.M. Clinical and molecular epidemiological features of coronavirus HKU1-associated community-acquired pneumonia. J Infect Dis. 2005;192:1898–1907. doi: 10.1086/497151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pebody R.G., Chand M.A., Thomas H.L., Green H.K., Boddington N.L., Carvalho C. The United Kingdom public health response to an imported laboratory confirmed case of a novel coronavirus in September 2012. Euro Surveill. 2012;18:20292. [PubMed] [Google Scholar]
- 40.Cheng V.C., Hung I.F., Tang B.S., Chu C.M., Wong M.M., Chan K.H. Viral replication in the nasopharynx is associated with diarrhea in patients with severe acute respiratory syndrome. Clin Infect Dis. 2004;38:467–475. doi: 10.1086/382681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hung I.F., Cheng V.C., Wu A.K., Tang B.S., Chan K.H., Chu C.M. Viral loads in clinical specimens and SARS manifestations. Emerg Infect Dis. 2004;10:1550–1557. doi: 10.3201/eid1009.040058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peiris J.S., Chu C.M., Cheng V.C., Chan K.S., Hung I.F., Poon L.L. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361:1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nishiura H., Mizumoto K., Ejima K., Zhong Y., Cowling B., Omori R. Incubation period as part of the case definition of severe respiratory illness caused by a novel coronavirus. Euro Surveill. 2012:17. pii:20296. [PMC free article] [PubMed] [Google Scholar]
- 44.Gautret P., Charrel R., Belhouchat K., Drali T., Benkouiten S., Nougairede A. Lack of nasal carriage of novel corona virus (HCoV-EMC) in French Hajj pilgrims returning from the Hajj 2012, despite a high rate of respiratory symptoms. Clin Microbiol Infect. 2013 doi: 10.1111/1469–0691.12174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kindler E., Jónsdóttir H.R., Muth D., Hamming O.J., Hartmann R., Rodriguez R. Efficient replication of the novel human betacoronavirus EMC on primary human epithelium highlights its zoonotic potential. MBio. 2013;4:e00611–e00612. doi: 10.1128/mBio.00611-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chan R.W., Chan M.C., Agnihothram S., Chan L.L., Kuok D.I., Fong J.H. Tropism and innate immune responses of the novel human betacoronavirus lineage C virus in human ex vivo respiratory organ cultures. J Virol. 2013 doi: 10.1128/JVI.00009-13. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Munster V.J., de Wit E., Feldmann H. Pneumonia from human coronavirus in a macaque model. N Engl J Med. 2013 doi: 10.1056/NEJMc1215691. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chu K.H., Tsang W.K., Tang C.S., Lam M.F., Lai F.M., To K.F. Acute renal impairment in coronavirus-associated severe acute respiratory syndrome. Kidney Int. 2005;67:698–705. doi: 10.1111/j.1523-1755.2005.67130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zielecki F., Weber M., Eickmann M., Spiegelberg L., Zaki A.M., Matrosovich M. Human cell tropism and innate immune system interactions of human respiratory coronavirus EMC compared to those of severe acute respiratory syndrome coronavirus. J Virol. 2013;87:5300–5304. doi: 10.1128/JVI.03496-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.World Health Organization . WHO; Geneva: 2012. Global alert and response: Novel coronavirus infection– update.http://www.who.int/csr/disease/coronavirus_infections/update_20121221/en/index.html [accessed 22.12.12] [Google Scholar]
- 51.Raj V.S., Mou H., Smits S.L., Dekkers D.H., Müller M.A., Dijkman R. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Corman V.M., Müller M.A., Costabel U., Timm J., Binger T., Meyer B. Assays for laboratory confirmation of novel human coronavirus (hCoV-EMC) infections. Euro Surveill. 2012:17. doi: 10.2807/ese.17.49.20334-en. pii:20334. [DOI] [PubMed] [Google Scholar]
- 53.Chan K.H., Chan J.F., Tse H., Chen H., Lau C.C., Cai J. Cross-reactive antibodies in convalescent SARS patients' sera against the emerging novel human coronavirus EMC (2012) by both immunofluorescent and neutralizing antibody tests. J Infect. 2013 doi: 10.1016/j.jinf.2013.03.015. [Epub ahead of print] S0163-4453(13)00071-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gierer S., Bertram S., Kaup F., Wrensch F., Heurich A., Krämer-Kühl A. The spike-protein of the emerging betacoronavirus EMC uses a novel coronavirus receptor for entry, can be activated by TMPRSS2 and is targeted by neutralizing antibodies. J Virol. 2013 doi: 10.1128/JVI.00128-13. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Corman V.M., Eckerle I., Bleicker T., Zaki A., Landt O., Eschbach-Bludau M. Detection of a novel human coronavirus by real-time reverse-transcription polymerase chain reaction. Euro Surveill. 2012;17 doi: 10.2807/ese.17.39.20285-en. pii:20285. [DOI] [PubMed] [Google Scholar]
- 56.Palm D., Pereyaslov D., Vaz J., Broberg E., Zeller H., Gross D. Laboratory capability for molecular detection and confirmation of novel coronavirus in Europe, November 2012. Euro Surveill. 2012:17. doi: 10.2807/ese.17.49.20335-en. pii:20335. [DOI] [PubMed] [Google Scholar]
- 57.Falzarano D., de Wit E., Martellaro C., Callison J., Munster V.J., Feldmann H. Inhibition of novel β coronavirus replication by a combination of interferon-α2b and ribavirin. Sci Rep. 2013;3:1686. doi: 10.1038/srep01686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jiang S., Lu L., Du L., Debnath A.K. A predicted receptor-binding and critical neutralizing domain in S protein of the novel human coronavirus HCoV-EMC. J Infect. 2013;66:464–466. doi: 10.1016/j.jinf.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.World Health Organization . WHO; Geneva: 2013. Global alert and response (GAR): Middle East respiratory syndrome coronavirus (MERS-CoV) – update.http://www.who.int/csr/don/2013_06_23/en/index.html [accessed 24.06.13] [Google Scholar]