Abstract
PEDV remains one of the most important swine diseases that infects pigs of all ages. It causes devastating viral enteric disease in piglets with a high mortality rate, leading to significant threats and huge economic loss to the pork industry. In this study, a transcriptomic shotgun sequencing (RNA-Seq) procedure was used to study gene responses against PEDV infection. Genome-wide analysis of differentially expressed genes (DEGs) was performed in Vero E6 cells post-PEDV infection. mTOR signaling pathway activator-MHY1485, and inhibitor-PP242 were used to study the antiviral function. Results revealed that the IRF3 was significantly up-regulated post-PEDV infection. Although most of the IFN-regulatory and –related genes evaluated in this study were either down-regulated or remained unchanged, IL11 behaved significantly up-regulated, with the peak at 16 hpi. Nearly 90% of PEDV infections were suppressed in the PP242 pretreated cells whereas the reverse effect was observed in the MYH1485 pretreated cells. Results indicated that the mTOR signaling pathway played a vital role in the PEDV antiviral regulation in the Vero E6 cells. Future studies will contribute to better understand the cellular antiviral mechanism against PEDV.
Keywords: PEDV, RNA-Seq, mTOR signaling pathway, Antiviral
Highlights
-
•
RNA-Seq was used to study gene responses against PEDV infection.
-
•
Genome-wide analysis of DEGs was performed in Vero E6 cells post-PEDV infection.
-
•
The mTOR signaling pathway activator and inhibitor can affect the PEDV infection rate.
1. Introduction
Porcine epidemic diarrhea virus (PEDV) belongs to the genus Alphacoronavirus in the family Coronaviridae [1]. The virus was first reported in the growing and fattening pigs in England in 1971, and this agent was further identified as a coronavirus-like particle in Belgium in 1978 [2]. Since then, outbreaks of PEDV have been documented in several countries. PEDV was first recorded in China in 1980s. However, in 2010, several cases were characterized as infection of virulent strains, causing high fatality rates and enormous economic losses [3]. In May 2013, a virulent strain emerged suddenly in US, and phylogenetic analyses indicated a close relationship with the Chinese PEDV strain AH2012 [4]. Subsequently, severe outbreaks of virulent PEDV strains were reported in American and Asian countries [5].
PEDV genome consists of a single-stranded, positive sense RNA genome of approximately 28kb. It comprises at least seven open reading frames (ORFs) that encode four structural proteins—spike (S, 150–220kDa), envelope (E, 7kDa), membrane (M, 20–30kDa), nucleocapsid (N, 58kDa) and three non-structural proteins—replicase 1a, 1b, ORF3, flanked by 5′ untranslated region (UTR) and 3′ UTR. These are arranged on the genome in the order of 5′-replicase(1a/1b)-S-ORF3-E-M-N-3’ [6]. The S protein is the major envelope glycoprotein responsible for receptor binding, virus attachment, cell membrane fusion and entry [7]. M protein, the most abundant surface protein, is co-expressed with E protein to form pseudo-particles which results in interfering genic activity [8]. N protein is highly conserved and the epitopes may be important for induction of cell-mediated immunity [9]. The replicases 1a and 1b of the nonstructural proteins play a pivotal role in viral genome replication. Studies have shown that the ORF3 gene has unexpected genetic variability and is thought to affect the virulence [10].
Interferons (IFNs) are class II cytokines characterized by the cellular antiviral effects and are also key components of innate immunity in response to viral infection. It is generally believed that the type I (IFN-α, IFN-β) and type III IFNs (IFN-λ2, IFN-λ3) play essential roles in antiviral defense [11]. Many viruses, including coronaviruses, have evolved strategies to counteract the signaling cascades of the innate immune system, especially through inhibiting IFN signaling and reduction of IFN production [12]. Viral proteins from different viruses, including influenza virus [13], Herpes virus [14], Coronaviruses severe acute respiratory syndrome (SARS) [15], Middle east respiratory syndrome (MERS) [16] and PEDV [17] have been identified as IFN antagonists. The mechanism involved in viral infection can be reflected through interactions between host-encoded pattern recognition receptors (PRRs) and pathogen-associated molecular patterns (PAMPs) at the cellular level [18]. Although the Vero E6 cell line lacks the ability to transcribe type I IFNs as these cells possess a genetic deletion, research has indicated that the cells mount a weak initial IRF3-dependent response to various ligands, inducing a weak innate response to viruses [19]. With respect to coronaviruses, upon IFNs binding to the corresponding receptors, a downstream signaling is initiated and acts to block one or more steps in the virus life cycle [20]. Mammalian target of rapamycin (mTOR) plays a vital role in monitoring the availability of nutrients, mitogenic signals and cellular energy, and is a necessary part of the phosphatidylinositol 3-kinase (PI3K) cell survival pathway [21]. It has been shown that the genes related to the mTOR signaling pathway are differentially expressed at different times post-PEDV infection stages [22,23]. However, the function of the mTOR signaling pathway in Vero E6 cells post-PEDV infection remains unknown. In this study, we have used a transcriptomic shotgun sequencing (RNA-Seq) procedure in Vero E6 cells to study gene responses against PEDV infection. This technique has previously been well documented in the study of PRRSV [24]. Genome-wide analysis of differentially expressed genes (DEGs) was performed in PEDV-infected Vero E6 cells. The analysis of genes in the mammalian target of Rapamycin (mTOR) signaling pathway suggested that the mTOR signaling pathway enacted a critical role in antiviral response against PEDV infection in Vero E6 cells. Results indicated that both mTOR activator-MHY1485 and inhibitor-PP242 modulated the PEDV infection rate in Vero E6 cells.
2. Materials and methods
2.1. Cells and viruses
Vero E6 cells were initially obtained from the ATCC (American Type Culture Collection, Manassas, VA) and cultured in Dulbecco's Modified Eagle Medium (DMEM) (GBICO-BRL, Grand Island, NY) containing 10% fetal bovine serum (FBS), at 37 °C and 5% CO2 atmosphere. The PEDV JS201603 strain used in this study was successfully isolated and serially passaged in our laboratory [25]. The PEDV isolate was propagated in Vero E6 cells supplemented with 10 μg/mL trypsin.
2.2. Virus infection
Confluent monolayers of Vero E6 cells in Five T25 flasks were inoculated with PEDV JS201603 isolate dissolved in DMEM (without EDTA) to reach a multiplicity of infection [MOI] of 0.1 in the presence of 10 μg/mL trypsin (GBICO, USA). At 8 h, 16 h, 24 h, 32 h, 40 h post PEDV-infection (hpi), the supernatant was removed and the cells were washed with 1 × phosphate buffered saline. The cells were then lysed with 1mL Trizol Reagent (Life Technology, USA) for RNA extraction. The mock group (non-infected cells) was set as 0 hpi, and the cells were lysed under the conditions previously described.
2.3. RNA-seq (transcriptomic shotgun sequencing) pipeline
For RNA-Seq, total RNA was extracted with TRIZOL (Invitrogen, US) following the manufacturer's instructions and tested for a RIN number to inspect RNA integrity by an Agilent Bioanalyzer 2100 (Agilent technologies, Santa Clara, CA, US). Qualified total RNA was further purified by RNeasy micro kit (Cat#74004, QIAGEN, GmBH, Germany) and RNase-Free DNase Set (Cat#79254, QIAGEN, GmBH, Germany) following manufacturer's instructions. Messenger RNA purification, fragmentation, construction of sequencing libraries and sequencing were performed using the Illumina Pipeline (BGI Americas, Cambridge, MA) [22]. Approximately 3–10M clean reads per sample were generated for genome-wide transcriptomic analyses. The trimmed reads were further assembled and mapped to the UniGene (https://www.ncbi.nlm.nih.gov/unigene/?term = % 20) and RefSeq (ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF_000409795.2 _Chlorocebus_ sabeus_1.1). Values of reads per kilobase per million mapped reads (RPKM) were generated. The total number of expressed genes and DEGs identified with an edgeR procedure were compared with the mock group. The DEGs between two stages were analyzed based on an algorithm as previously described [26]. Furthermore, quantitative real-time PCR assays were performed to confirm the mTOR signaling pathway related DEGs. Functional classification of genes was analyzed with DAVID web tools (https://david.ncifcrf.gov). To compared with genome background, every term gene numbers were calculated and significantly enriched GO terms in DEGs was summarized. The calculating formula is shown below with
i, N and n representing the number of successes, the number of genes with GO annotation, and the number of DEGs in N, respectively. M is the number of all genes which are annotated to the certain GO terms while m is the number of DEGs in M. GO terms in agreement with the condition P ≤ 0.05 were defined as significantly enriched GO terms in DEGs [27].
2.4. The effect of mTOR signaling pathway activator and inhibitor on the PEDV replication
Vero E6 cells were pretreated with the mTOR inhibitor PP242, or the mTOR activator MHY1485, at 2 μM concentration for 24 h, followed by infection with PEDV JS201603 strain at a MOI of 0.1. 24 h post-infection, the cells were fixed with 80% ice-cold ethanol for 30 min at −20 °C. A monoclonal antibody specially for PEDV was added to the cells at a dilution of 1:100 for 40 min at 37 °C. As a secondary antibody, FITC-conjugated donkey anti-mouse IgG antibody (Life Technologies, Carlsbad, CA, USA) was used at 1:2000 dilution. Cells were observed using a Nikon fluorescent microscopy (Shinjuku, Tokyo, Japan) at a magnification of 20 × , and the viral infection rate was also quantified. All chemicals used in immunofluorescence were dissolved in dimethyl sulfoxide (DMSO, cell culture grade, Sigma).
2.5. Data analysis
The quality of RNA-Seq data was analyzed in terms of the proportion of gene-mapped reads and the saturation/randomness for genome-wide gene coverage using previously established protocols [28]. DEGs were profiled with an edgeR package (genes with a false discovery rate [FDR] of ≤0.001, a fold change of >2) [29].
3. Results
3.1. Comparing significant DEGs of infection stages with the mock group
Quality control (QC) was applied to each stage of data handling to ensure that all data were processed correctly and met the criteria for genome-wide transcriptomic analysis [30]. We compared each stage with the mock group, more than 22000 DEGs were normalized and 1826–4015 significant DEGs were filtered out. For example, the highest number of 4015 significant DEGs including 484 up-regulated and 3531 down-regulated genes, was detected at 24 h post PEDV-infection (24hpi) status compared with the mock group. The results showed that the DEGs failed to follow a linear regulation over the time of infection, this is the case for genes up- or down-regulated(data not shown).
3.2. Potential signature genes response to the PEDV infection
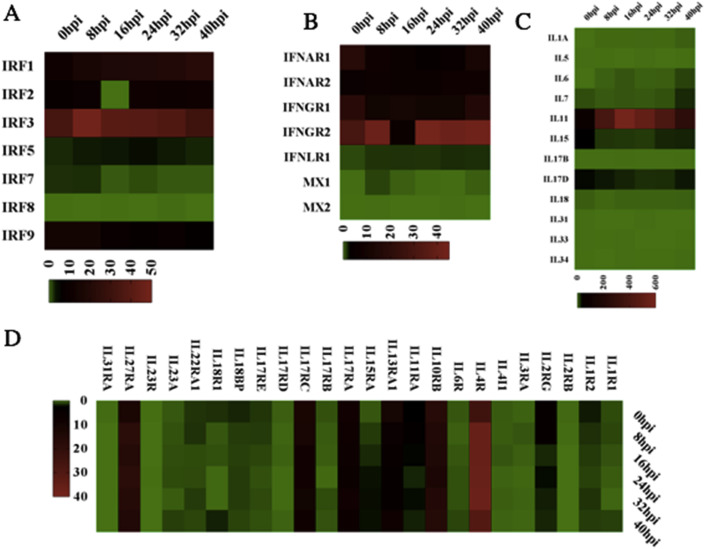
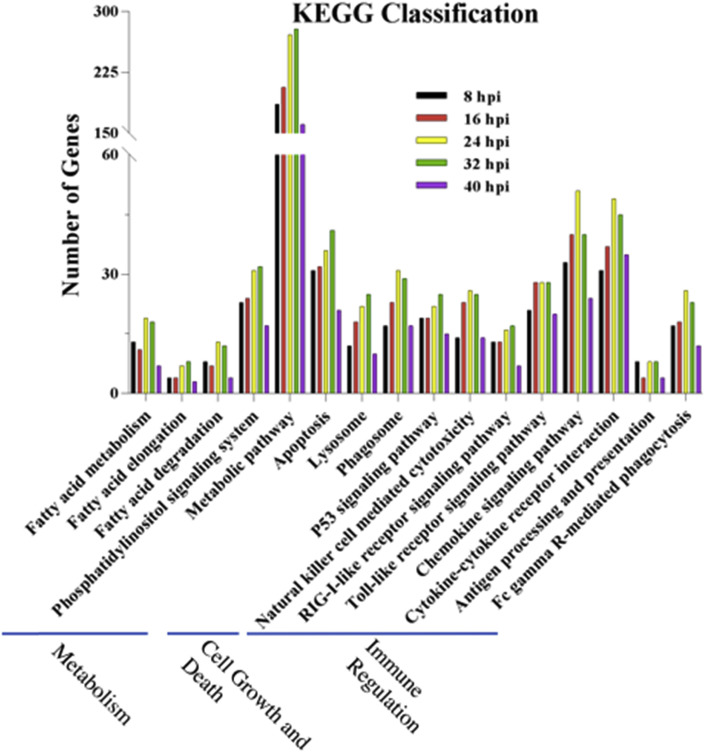
To investigate genes playing a role in the response to the PEDV infection in Vero E6 cells, DEGs of the infection stages were compared with the mock group. The comparison resulted in totally 15226 non-redundant genes that were significantly up- or down-regulated at different times post-PEDV infection stages. The antiviral effect of interferons (IFNs) upon coronavirus infection has been reported [31]. To profile potential signature IFNs-related, interleukin (IL) genes, significant up or down-regulated DEGs were clustered. For example, for each infection stage, IFNAR1 was identified down-regulated before 24 hpi but it was up-regulated at 32 hpi and 40 hpi. The IL4R expression was up-regulated before 32 hpi but down-regulated at 40 hpi (Fig. 1 ). Although other genes also showed up-regulation or down-regulation in other aspects of functional regulation, most genes have not been studied. Based on the KEGG database, pathway analysis was performed to evaluate the biological and ontological significance of DEGs. Compared with the mock group, more than 200 pathways showed to have significant DEGs post PEDV infection. Most of the pathways belonged to the metabolism regulation, signaling transduction and the immune, digestive and endocrine system. As shown in Fig. 2 , 16 different pathways were selected that might be relevant to the protection against PEDV infections. Results showed that there was a significant change in the regulation of the metabolic pathway post-PEDV infection, and a significant change was also observed in the toll-like receptor, chemokine, cytokine-cytokine receptor in the immune regulation pathways.
Fig. 1.
RNA-seq analysis comparing the different times post-PEDV infection stages of Interleukin and IFN-related DEGs.
A and B: Heat maps of significant IFN-related DEGs.
C and D: Heat maps of significant interleukin-related DEGs.
Fig. 2.
Pathway analysis of DEGs was annotated against the KEGG database.
Three selected categories were shown at the bottom. The vertical axis represented the number of significant DEGs and the horizontal axis was the related pathway. Results showed that there was a significant change in the regulation of the metabolic pathway post-PEDV infection, and a significant change was also observed in the toll-like receptor, chemokine, cytokine-cytokine receptor in the immune regulation pathways.
3.3. The expression of interleukin and IFN-related gene upon the PEDV infection at different times post-PEDV infection stages
Compared with the mock group, 36 genes in the interleukin family were identified significantly regulated at different times post-PEDV infection stages. Among them, IL4R was significantly up-regulated whereas IL5 was down-regulated. IL11 is a multi-potent cytokine which can induce three major signaling pathways—Jak/STAT, ERK and PI3K/Akt, playing a vital role in anti-apoptosis, pro-proliferation upon cell injury [32]. Wang et al. found that IL11 activates Jak/STAT signaling pathway, up-regulated Bcl-2 and down-regulated Bax expression, maintaining intestinal epithelial homeostasis and regeneration [33]. In this study, the IL11 kept up-regulation and peaked at 24 hpi, but showed down-regulation at 32 hpi and 40 hpi. IL-4 is a main regulatory cytokine which binds to the ectodomain of IL-4 receptor alpha chain (IL4-BP) with high affinity and specificity [34]. The up-regulation of IL4R would contribute to the transmembrane signaling and the induction of inflammatory response (Fig. 1).
Vero E6 cells are characteristically interferon-deficient. Jan Desemyter ever found that the Vero cells failed to produce interferon when infected with Newcastle disease, Sendai and rubella viruses [35]. The whole gene sequence of Vero cells showed that the homozygous 9-Mb deletion on chromosome 12 resulted the loss of the type I interferon gene cluster and cyclin-dependent kinase inhibitor genes [36]. In this study, the same phenomenon was observed. None of the interferon-production genes were detected in the experiment.
Vero cells possess the Interferon- α/β receptor (IFNAR), which binds type I interferons including interferon-α and -β. It also activates the JAK-STAT signaling pathway, which transmits information from extracellular signals to the nucleus and is involved in immunity, proliferation and other functional regulations [37,38]. Upon PEDV infection, up-regulation of IFNAR2 was detected at different times post-infection stage. Compared with IFNAR1, IFNAR2, and IFNGR1, interestingly, IFNGR2 showed strikingly significant up-regulation upon PEDV-infection. IFNGR1 encodes the ligand-binding chain of the heterodimeric gamma interferon receptor while the IFNGR2 encodes the non-ligand-binding part [39]. Functional studies have shown that the IFN-γ receptors are secreted from the vaccinia and cowpox virus infected cells, binding to the IFN-γ with a wide range of diverse species and also inhibiting IFN-γ from binding to its cellular receptor [40]. Interferon lambda receptor (IFNLR), a single heterodimeric receptor, contains the IFNLR1 signaling chain and the accessory chain [41]. However, according to the data analysis in this study, no change in the IFNLR1 gene was behaved post-PEDV infection and there was also no IFN-λ secretion (Fig. 1).
The interferon regulatory factors (IRFs) are transcriptional mediators in signaling pathways induced by pathogenic infection, and are implicating in the regulation of antiviral defense, immune response, cell growth regulation and apoptosis [42]. The IRF genes include IRF-1, IRF-2, IRF-3, IRF-4/Pip/ICSAT, IRF-5, IRF-6, IRF-7, ICSBP/IRF-8 and ISGF3 γ/p48/IRF-9 as well as virus-encoded analogues have been identified [43]. By using RNA-Seq data, Sang et al. found that porcine IRF1 was highly stimulated in M1-IFN γ and M1-LPS statuses while IRF8 was only observed in M1-IFN-γ status [22]. Research analysis of interferon-stimulated genes response in endothelial cells during hantaan virus infection found two patterns of gene expression: the first pattern was identified by early induction and short action, such as type I IFNs; the other was characterized as IRF-7, MxA and TAP-1/2 by delayed induction and long duration [44]. Our data analysis showed that IRF-3 was highly stimulated at 8 hpi and then decreased gradually. Because the Vero E6-encoded IRF-3 was relatively inefficient, this could lead to a muted initial response to virus infection [18,45]. IRF5 behaved nearly unchanged throughout the experiment. On the contrary, a pronounced suppression was found at 16 hpi in IRF2. Furthermore, the highest suppression of expression for IRF1, IRF2, IRF3, IRF5, IRF7 and IRF8 was found at 40 hpi (Fig. 1).
3.4. Genes related to the mTOR signaling pathway were differentially expressed in PEDV infected Vero E6 cells at different times post-PEDV infection stages and modulated the antiviral responses
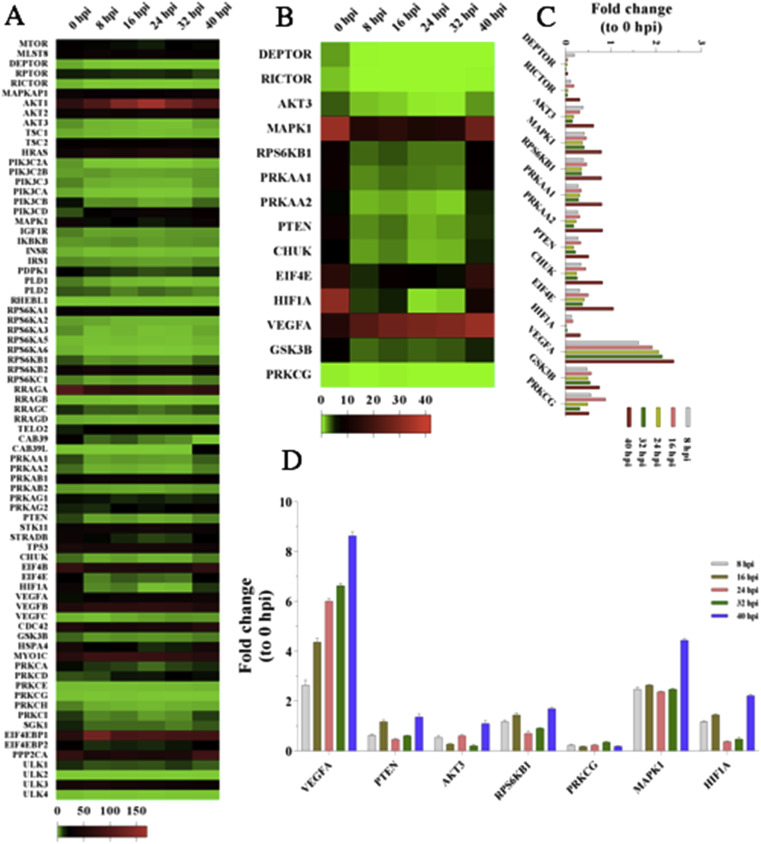
mTOR mediates its activity through two functionally distinct multi-protein complexes, mTOR complex1 (mTORC1) and mTOR complex 2 (mTORC2). The mTORC1 regulates mRNA translation. On the other part, mTORC2 contributes to the maintenance of cellular size and viability, among other functions [46]. These two signaling pathways integrate intracellular and extracellular signals to regulate various cellular processes [47]. In the study, 78 DEGs in the mTOR signaling pathway were normalized, and 14 significant DEGs were filtered out (Fig. 3 A). Compared with the mock group, most of the mTOR upstream regulators (AKT3, MAPK1, RPS6KB1, PRKAA1, PRKAA2, and PTEN) and downstream effectors (CHUK, EIF4E, HIF1A, GSK3B, and PRKCG) were significantly up-regulated or down-regulated at 40 hpi, respectively (Fig. 3B and C). However, one of the downstream effector-VEGFA exerted significantly up-regulated compared with the other statuses including the mock group. To confirm the RNA-Seq expressional analysis, totally seven upstream regulators or downstream effectors were selected for the RT-PCR assay. It was observed that the expression levels at different times post-PEDV infection stages matched with the RNA-Seq data (Fig. 3D).
Fig. 3.
Compared the DEGs of mTOR signaling pathway at different times post-PEDV infection stages with the mock group.
(A) 78 DEGs concerned with the mTOR signaling pathway were normalized.
(B), (C) 14 significant DEGs were filtered out.
(D) 7 significant DEGs were selected to re-analyze using the RT-PCR and the results matched with the RNA-Seq data.
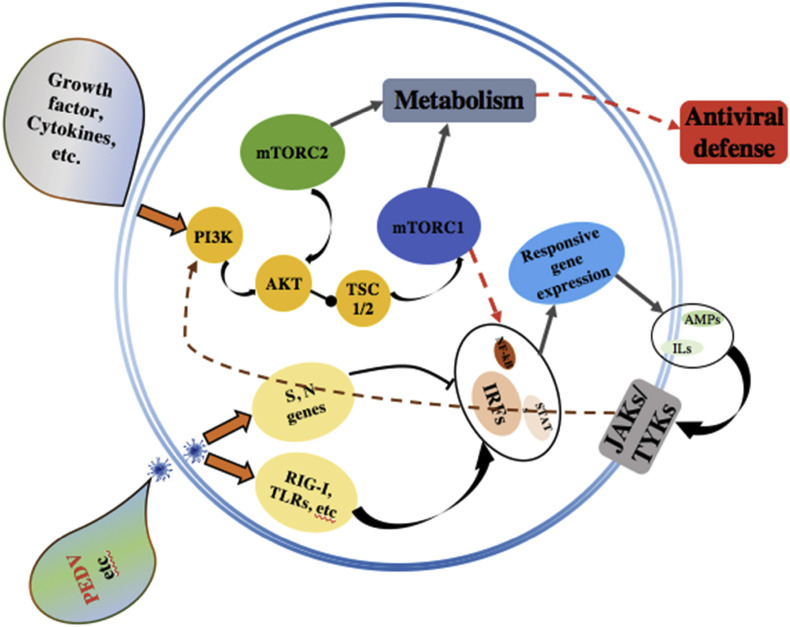
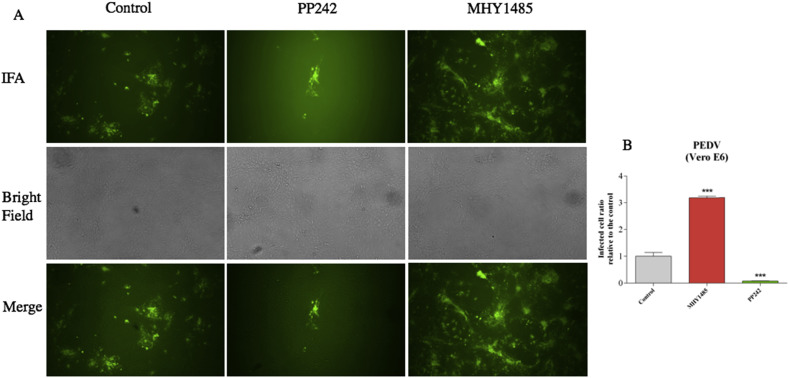
The significant DEGs at different times post-infection stages revealed that the mTOR signaling pathway could be involved in the PEDV infection. As is shown in the Fig. 4 , the viral proteins and cellular cytokines could regulate the mTOR signaling pathway through the PI3/Akt interactive signaling pathway. However, whether the mTOR signaling pathway plays a vital role in anti-PEDV infection is still unknown. In this study, the mTOR inhibitor-PP242 and mTOR activator-MHY1485 were selected to modulate the mTOR signaling pathway. The PP242 suppresses both the mTORC1 and mTORC2 activities. Compared with the control (Fig. 5 ), results revealed that nearly 90% of PEDV infection was suppressed in the PP242 pretreated cells. This could be due to the inhibition of mTOR signaling pathway, resulting in an increase in cell proliferation and survival rate, because the virus propagation depends on specialized host cells to supply the genetic materials for replication and protein biosynthesis. However, the reverse effect was found in the MYH1485 pretreated cells. The virus infection rate was two times higher than the mock group. The MHY1485 might suppress autophagy signaling by activating the mTOR signaling pathway and also contributed to the maintenance of persistent infection of PEDV.
Fig. 4.
mTOR signaling pathway plays a vital role in modulating the pathogenic infection.
Figure depicting how the IRFs worked as the transcriptional mediators post-PEDV infection, and the corresponding responsive gene expression. Some ILs, for example IL11, a multi-potent cytokine which can induce the signaling pathways Jak/STAT and PI3K/Akt and regulate the mTOR signaling pathway.
Fig. 5.
Regulation of PEDV infection rate by pretreated the Vero E6 cells with mTOR signaling pathway activator and inhibitor.
(A) Vero E6 cells pretreated with the 2 μM PP242 and MHY1485 for 24 h and later infected with the PEDV. 24 h post-PEDV infection, cells were visualized by fluorescence microscopy.
(B) The viral infection rate was quantified. The MHY1485 pretreated group was twice higher than the mock group while three times higher than the PP242 pretreated group.
4. Discussion
During viral infection, sensing of foreign nucleic acids induces both innate and adaptive immune responses. Usually, the innate immune system acts as the first line of defense, producing type I IFNs and proinflammatory cytokines through germ-line encoded pattern-recognition receptors (PRRs) that sense and respond to the pathogen-associated molecular patterns (PAMPs) of virus. In this sense, triggering the activation of retinoic acid-inducible gene I (RIG-I) or melanoma differentiation gene 5 (MDA5) results in the expression of antiviral genes [48]. Although an early innate response is required to shape an adequate downstream adaptive immune response, an overactive innate immune response can also result in deleterious consequences for the organism, such as immune pathology and subsequent tissue damage [49]. Most viruses have evolved to acquire the mechanisms to circumvent the host innate immune responses during infection, at least to a certain extent [50]. Therefore, detailed knowledge of the interaction between virus and the immune systems will greatly contribute to a comprehensive understanding of such antiviral mechanisms. In this study, Vero E6 cells were selected to study the genome-wide differentially expressed genes at different times post-PEDV infection stages. Although IFNs were not detected throughout the experiment, it was ever observed that the Hantaan virus (HTNV) could induce the type III IFNs(IFN-λ) in the Vero E6 cells. This suggests a specific PRR and signaling pathway responsible for the HTNV-driven induction of IFN-λ1, which needs further research [51].
Compared with the mock group, nearly all of the detected IRFs were down-regulated. Especially IRF2, a protein coding gene usually binds to the upstream regulatory region of IFN-inducible MHC class I genes, showing significantly down-regulation at 16 hpi. However, IRF3 was significantly up-regulated after PEDV infection. IRF3 is recognized as the key transcriptional regulator of type I IFN-dependent immune responses, playing a vital role in the innate immune response against virus infection when it becomes phosphorylated by different kinases such as IKBKE and TAK1 kinases [52]. Upon phosphorylation, IRF3 undergoes a conformational change that leads to its self-dimerization and nuclear localization. Once in the nucleus, IRF3 associates with the CREB protein to form the dsRNA-activated factor 1, regulating the transcription of type I IFN. However, due to the loss of type I IFN gene cluster in Vero E6 cells, IRF3 failed to up-regulate the production of type I IFN.
It is of note that IFNGR2, a downstream receptor for IFN-γ, was up-regulated after PEDV infection. All other IFN-regulatory and –related genes evaluated in this study were either down-regulated or unchanged. The results are in accordance with the fact that most of coronaviruses have evolved the mechanisms to inhibit one or more steps in the IFN response [53]. IL11 was significantly up-regulated with the peak at 16 hpi. IL11 playes a critical role in the signal transduction upon binding with IL-11Ralpha and gp130. This allows the homodimerization of the gp130 molecule and the subsequent activation of Janus kinases (JAK) and regulation of the mTOR activity (Fig. 4). Various as well as multiple extra-and intracellular molecular signals are integrated into the mTOR signaling pathway. Viral infection is a significant stress which can activate, reduce or even suppress the mTOR signaling pathway. Furthermore, viruses have evolved a plethora of different mechanisms to attack and co-opt the mTOR pathway in order to make a hospitable environment for replication in host cells [54]. In our study, upon PEDV infection, most of the genes in the mTOR signaling pathway underwent down-regulation compared with the mock group. However, the VEGFA was up-regulated at different times post-PEDV infection stages and reached its highest level at 40 hpi. Possibly the cells infected with PEDV could indirectly stimulate the production of VEGFA. Todd Wuest et al. observed that the herpes simplex virus-1 drived the expression of VEGFA by mimicking the function of the VEGFA promoter [55]. Tomomi Takano also found that the production of VEGF was associated with the proliferation of Feline infectious peritonitis virus [56]. MHY1485 is an activator of the mTOR signaling pathway while the PP242 acts as an inhibitor. However, it has not been previously reported whether the activator or inhibitor can affect the PEDV replication. This study showed that the MHY1485 promoted the PEDV infection. MHY1485 could inhibit the autophagy process by suppressing the fusion between autophagosomes and lysosomes, eventually contributing to the viral replication. Diverse studies have found that MHY1485 has an inhibitory effect on the autophagic process that leads to the accumulation of LC3 II protein and enlarged autophagosomes [57]. Interestingly, the cells pre-treated with the PP242 showed an opposite effect. PP242 is a selective inhibitor of mTOR signaling pathway which targets the ATP domain. The PP242 treatment in BT242 cells exerted an inhibition of the phosphorylation of Akt, p70S6 and the downstream target S6 [58]. Bao Hoang et al. also found that PP242 is an effective agent against primary multiple myeloma cells in vitro and growth of 8226 cells in mice [59]. The phosphorylation of viral and cellular proteins can exert major impacts on viral infection and replication. By inducing the phosphorylation of Akt, EBV LMP1 can activate the PI3K/Akt pathway, which is involved in the actin cytoskeleton reorganization of EBV-infected cells [60]. Inhibition of the phosphorylation of the cellular proteins in Vero E6 cells results in a reduction of PEDV replication.
5. Conclusion
In this study, RNA-Seq was used to profile signature genes in PEDV infected Vero E6 cells. Numerous(n = 15226) non-redundant genes were significantly up- or down-regulated at different times post-PEDV infection stages. More than 200 pathways were classified to have significant DEGs post PEDV infection. However, none of the interferons were detected throughout the experiment. The mTOR signaling pathway played a vital role in modulating the PEDV infection. Nearly 90% of PEDV infection was suppressed in the PP242 pretreated cells while the reverse effect was found in the MYH1485 pretreated cells. The virus infection rate was two times higher than the control group. These efforts will contribute to the further study of PEDV mechanism.
Conflicts of interest
The authors declare that they have no conflict of interests.
Acknowledgements
Thanks to Dr. Francisco Dominguez (Postdoc Reseacher, KSVDL, Kansas State University) for his help to improve the English Writing. We also thank Xiaotao Cheng (Shanghai Biotechnology Corporation) for helping in data analysis. Thanks to the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD) to ZC. This study was supported by Jilin Provincial Major Science and Technology Development Project Fund (No.20150201006NY).
References
- 1.Pensaert M., De Bouck P. A new coronavirus-like particle associated with diarrhea in swine. Arch. Virol. 1978;58:243–247. doi: 10.1007/BF01317606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wood E. An apparently new syndrome of porcine epidemic diarrhoea. Vet. Rec. 1977;100(243) doi: 10.1136/vr.100.12.243. [DOI] [PubMed] [Google Scholar]
- 3.Pan Y., Tian X., Li W., Zhou Q., Wang D., Bi Y. Isolation and characterization of a variant porcine epidemic diarrhea virus in China. Virol. J. 2012;9:195. doi: 10.1186/1743-422X-9-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin C.-M., Gao X., Oka T., Vlasova A.N., Esseili M.A., Wang Q. Antigenic relationships among porcine epidemic diarrhea virus and transmissible gastroenteritis virus strains. J. Virol. 2015;89:3332–3342. doi: 10.1128/JVI.03196-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vlasova A.N., Marthaler D., Wang Q., Culhane M.R., Rossow K.D., Rovira A. Distinct characteristics and complex evolution of PEDV strains, North America, May 2013–February 2014. Emerg. Infect. Dis. 2014;20:1620. doi: 10.3201/eid2010.140491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bridgen A., Kocherhans R., Tobler K., Carvajal A., Ackermann M. Coronaviruses and Arteriviruses. Springer; 1998. Further analysis of the genome of porcine epidemic diarrhoea virus; pp. 781–786. [DOI] [PubMed] [Google Scholar]
- 7.Cruz D.J.M., Kim C.-J., Shin H.-J. The GPRLQPY motif located at the carboxy-terminal of the spike protein induces antibodies that neutralize Porcine epidemic diarrhea virus. Virus Res. 2008;132:192–196. doi: 10.1016/j.virusres.2007.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Su Y., Liu Y., Chen Y., Zhao B., Ji P., Xing G. Detection and phylogenetic analysis of porcine epidemic diarrhea virus in central China based on the ORF3 gene and the S1 gene. Virol. J. 2016;13:192. doi: 10.1186/s12985-016-0646-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Curtis K.M., Yount B., Baric R.S. Heterologous gene expression from transmissible gastroenteritis virus replicon particles. J. Virol. 2002;76:1422–1434. doi: 10.1128/JVI.76.3.1422-1434.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang K., Lu W., Chen J., Xie S., Shi H., Hsu H. PEDV ORF3 encodes an ion channel protein and regulates virus production. FEBS Lett. 2012;586:384–391. doi: 10.1016/j.febslet.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hernández P.P., Mahlakõiv T., Yang I., Schwierzeck V., Nguyen N., Guendel F. Interferon-[lambda] and interleukin 22 act synergistically for the induction of interferon-stimulated genes and control of rotavirus infection. Nat. Immunol. 2015;16:698–707. doi: 10.1038/ni.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo L., Luo X., Li R., Xu Y., Zhang J., Ge J. Porcine epidemic diarrhea virus infection inhibits interferon signaling by targeted degradation of STAT1. J. Virol. 2016;90:8281–8292. doi: 10.1128/JVI.01091-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krug R.M. Functions of the influenza A virus NS1 protein in antiviral defense. Corr. Opin. Virol. 2015;12:1–6. doi: 10.1016/j.coviro.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amsler L., Verweij M.C., DeFilippis V.R. The tiers and dimensions of evasion of the type I interferon response by human cytomegalovirus. J. Mol. Biol. 2013;425:4857–4871. doi: 10.1016/j.jmb.2013.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Totura A.L., Baric R.S. SARS coronavirus pathogenesis: host innate immune responses and viral antagonism of interferon. Corr. Opin. Virol. 2012;2:264–275. doi: 10.1016/j.coviro.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Y., Ye F., Zhu N., Wang W., Deng Y., Zhao Z. Middle East respiratory syndrome coronavirus ORF4b protein inhibits type I interferon production through both cytoplasmic and nuclear targets. Sci. Rep. 2015;5 doi: 10.1038/srep17554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xing Y., Chen J., Tu J., Zhang B., Chen X., Shi H. The papain-like protease of porcine epidemic diarrhea virus negatively regulates type I interferon pathway by acting as a viral deubiquitinase. J. Gen. Virol. 2013;94:1554–1567. doi: 10.1099/vir.0.051169-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prescott J., Hall P., Acuna-Retamar M., Ye C., Wathelet M.G., Ebihara H. New World hantaviruses activate IFNλ production in type I IFN-deficient Vero E6 cells. PLoS One. 2010;5 doi: 10.1371/journal.pone.0011159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chew T., Noyce R., Collins S.E., Hancock M.H., Mossman K.L. Characterization of the interferon regulatory factor 3-mediated antiviral response in a cell line deficient for IFN production. Mol. Immunol. 2009;46:393–399. doi: 10.1016/j.molimm.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 20.de Lang A., Osterhaus A.D., Haagmans B.L. Interferon-γ and interleukin-4 downregulate expression of the SARS coronavirus receptor ACE2 in Vero E6 cells. Virology. 2006;353:474–481. doi: 10.1016/j.virol.2006.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zarogoulidis P., Lampaki S., Turner J.F., Huang H., Kakolyris S., Syrigos K. mTOR pathway: a current, up-to-date mini-review. Oncol. Lett. 2014;8:2367–2370. doi: 10.3892/ol.2014.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sang Y., Brichalli W., Rowland R.R., Blecha F. Genome-wide analysis of antiviral signature genes in porcine macrophages at different activation statuses. PLoS One. 2014;9 doi: 10.1371/journal.pone.0087613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Q., Miller L.C., Blecha F., Sang Y. Reduction of infection by inhibiting mTOR pathway is associated with reversed repression of type I interferon by porcine reproductive and respiratory syndrome virus. J. Gen. Virol. 2017;98:1316–1328. doi: 10.1099/jgv.0.000802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller L.C., Fleming D., Arbogast A., Bayles D.O., Guo B., Lager K.M. Analysis of the swine tracheobronchial lymph node transcriptomic response to infection with a Chinese highly pathogenic strain of porcine reproductive and respiratory syndrome virus. BMC Vet. Res. 2012;8:208. doi: 10.1186/1746-6148-8-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang H., Xia M., Ju D., Wu B., Ning C., Song N. Isolation, molecular characterization and an artificial infection model for a variant porcine epidemic diarrhea virus strain from Jiangsu Province, China. Arch. Virol. 2017:1–8. doi: 10.1007/s00705-017-3518-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou X., Wang K., Lv D., Wu C., Li J., Zhao P. Global analysis of differentially expressed genes and proteins in the wheat callus infected by Agrobacterium tumefaciens. PLoS One. 2013;8 doi: 10.1371/journal.pone.0079390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiao S., Jia J., Mo D., Wang Q., Qin L., He Z. Understanding PRRSV infection in porcine lung based on genome-wide transcriptome response identified by deep sequencing. PLoS One. 2010;5 doi: 10.1371/journal.pone.0011377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sang Y., Rowland R.R., Blecha F. Antiviral regulation in porcine monocytic cells at different activation states. J. Virol. 2014;88:11395–11410. doi: 10.1128/JVI.01714-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hardie D.G., Ross F.A., Hawley S.A. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat. Rev. Mol. Cell Biol. 2012;13(251) doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morgenstern B., Michaelis M., Baer P.C., Doerr H.W., Cinatl J. Ribavirin and interferon-β synergistically inhibit SARS-associated coronavirus replication in animal and human cell lines. Biochem. Biophys. Res. Commun. 2005;326:905–908. doi: 10.1016/j.bbrc.2004.11.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kusuda S., Fujimura M., Uchiyama A., Totsu S., Matsunami K. Trends in morbidity and mortality among very-low-birth-weight infants from 2003 to 2008 in Japan. Pediatr. Res. 2012;72:531–538. doi: 10.1038/pr.2012.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang R., Peng R., Gao Y., Chang G., Xu X., Fu K. Jak/STAT signaling pathway of IL-11 in the protection of intestinal epithelial cells from neutron radiation. Chinese J. Cell. Mol. Immunol. 2009;25:27–30. [PubMed] [Google Scholar]
- 34.Hage T., Sebald W., Reinemer P. Crystal structure of the interleukin-4/receptor α chain complex reveals a mosaic binding interface. Cell. 1999;97:271–281. doi: 10.1016/s0092-8674(00)80736-9. [DOI] [PubMed] [Google Scholar]
- 35.Desmyter J., Melnick J.L., Rawls W.E. Defectiveness of interferon production and of rubella virus interference in a line of African green monkey kidney cells (Vero) J. Virol. 1968;2:955–961. doi: 10.1128/jvi.2.10.955-961.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Osada N., Kohara A., Yamaji T., Hirayama N., Kasai F., Sekizuka T. The genome landscape of the African green monkey kidney-derived Vero cell line. DNA Res. 2014;21:673–683. doi: 10.1093/dnares/dsu029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uze G., Schreiber G., Piehler J., Pellegrini S. The receptor of the type I interferon family. Interferon: The 50th Anniversary. 2007:71–95. doi: 10.1007/978-3-540-71329-6_5. [DOI] [PubMed] [Google Scholar]
- 38.Aaronson D.S., Horvath C.M. A road map for those who don't know JAK-STAT. Science. 2002;296:1653–1655. doi: 10.1126/science.1071545. [DOI] [PubMed] [Google Scholar]
- 39.Platanias L.C. Mechanisms of type-I-and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 40.Alcami A., Smith G.L. Vaccinia, cowpox, and camelpox viruses encode soluble gamma interferon receptors with novel broad species specificity. J. Virol. 1995;69:4633–4639. doi: 10.1128/jvi.69.8.4633-4639.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sommereyns C., Paul S., Staeheli P., Michiels T. IFN-lambda (IFN-) is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paun A., Pitha P. The IRF family, revisited. Biochimie. 2007;89:744–753. doi: 10.1016/j.biochi.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barnes B., Lubyova B., Pitha P.M. On the role of IRF in host defense. J. Interferon Cytokine Res. 2002;22:59–71. doi: 10.1089/107999002753452665. [DOI] [PubMed] [Google Scholar]
- 44.Kim I.-W., Hwang J.-Y., Kim S.-K., Kim J.-K., Park H.-S. Interferon-stimulated genes response in endothelial cells following Hantaan virus infection. J. Kor. Med. Sci. 2007;22:987–992. doi: 10.3346/jkms.2007.22.6.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Au W.-C., Moore P.A., LaFleur D.W., Tombal B., Pitha P.M. Characterization of the interferon regulatory factor-7 and its potential role in the transcription activation of interferon A genes. J. Biol. Chem. 1998;273:29210–29217. doi: 10.1074/jbc.273.44.29210. [DOI] [PubMed] [Google Scholar]
- 46.Ding X., Bloch W., Iden S., Rüegg M.A., Hall M.N., Leptin M. mTORC1 and mTORC2 regulate skin morphogenesis and epidermal barrier formation. Nat. Commun. 2016;7:13226. doi: 10.1038/ncomms13226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Laplante M., Sabatini D.M. mTOR signaling at a glance. J. Cell Sci. 2009;122:3589–3594. doi: 10.1242/jcs.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Q., Shi K., Yoo D. Suppression of type I interferon production by porcine epidemic diarrhea virus and degradation of CREB-binding protein by nsp1. Virology. 2016;489:252–268. doi: 10.1016/j.virol.2015.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frieman M., Heise M., Baric R. SARS coronavirus and innate immunity. Virus Res. 2008;133:101–112. doi: 10.1016/j.virusres.2007.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang D., Fang L., Shi Y., Zhang H., Gao L., Peng G. Porcine epidemic diarrhea virus 3C-like protease regulates its interferon antagonism by cleaving NEMO. J. Virol. 2016;90:2090–2101. doi: 10.1128/JVI.02514-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stoltz M., Klingström J. Alpha/beta interferon (IFN-α/β)-independent induction of IFN-λ1 (interleukin-29) in response to Hantaan virus infection. J. Virol. 2010;84:9140–9148. doi: 10.1128/JVI.00717-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Challa S., Guo J.-P., Ding X., Xu C.-X., Li Y., Kim D. IKBKE is a substrate of EGFR and a therapeutic target in non–small cell lung cancer with activating mutations of EGFR. Canc. Res. 2016;76:4418–4429. doi: 10.1158/0008-5472.CAN-16-0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Siu K.-L., Chan C.-P., Kok K.-H., Woo P.C.-Y., Jin D.-Y. Suppression of innate antiviral response by severe acute respiratory syndrome coronavirus M protein is mediated through the first transmembrane domain. Cell. Mol. Immunol. 2014;11:141–149. doi: 10.1038/cmi.2013.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Le Sage V., Cinti A., Amorim R., Mouland A.J. Adapting the stress response: viral subversion of the mTOR signaling pathway. Viruses. 2016;8(152) doi: 10.3390/v8060152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wuest T., Zheng M., Efstathiou S., Halford W.P., Carr D.J. The herpes simplex virus-1 transactivator infected cell protein-4 drives VEGF-A dependent neovascularization. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takano T., Ohyama T., Kokumoto A., Satoh R., Hohdatsu T. Vascular endothelial growth factor (VEGF), produced by feline infectious peritonitis (FIP) virus-infected monocytes and macrophages, induces vascular permeability and effusion in cats with FIP. Virus Res. 2011;158:161–168. doi: 10.1016/j.virusres.2011.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Choi Y.J., Park Y.J., Park J.Y., Jeong H.O., Kim D.H., Ha Y.M. Inhibitory effect of mTOR activator MHY1485 on autophagy: suppression of lysosomal fusion. PLoS One. 2012;7 doi: 10.1371/journal.pone.0043418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Apsel B., Blair J.A., Gonzalez B., Nazif T.M., Feldman M.E., Aizenstein B. Targeted polypharmacology: discovery of dual inhibitors of tyrosine and phosphoinositide kinases. Nat. Chem. Biol. 2008;4:691–699. doi: 10.1038/nchembio.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hoang B., Frost P., Shi Y., Belanger E., Benavides A., Pezeshkpour G. Targeting TORC2 in multiple myeloma with a new mTOR kinase inhibitor. Blood. 2010;116:4560–4568. doi: 10.1182/blood-2010-05-285726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dawson C.W., Tramountanis G., Eliopoulos A.G., Young L.S. Epstein-Barr virus latent membrane protein 1 (LMP1) activates the phosphatidylinositol 3-kinase/Akt pathway to promote cell survival and induce actin filament remodeling. J. Biol. Chem. 2003;278:3694–3704. doi: 10.1074/jbc.M209840200. [DOI] [PubMed] [Google Scholar]