Abstract
Background: Synthetic probes that mimic natural substrates can enable the detection of enzymatic activities in a cellular environment. One area where such activity-based probes have been applied is the ubiquitin–proteasome pathway, which is emerging as an important therapeutic target. A family of reagents has been developed that specifically label deubiquitylating enzymes (DUBs) and facilitate characterization of their inhibitors. Scope of review: Here we focus on the application of probes for intracellular DUBs, a group of specific proteases involved in the ubiquitin proteasome system. In particular, the functional characterization of the active subunits of this family of proteases that specifically recognize ubiquitin and ubiquitin-like proteins will be discussed. In addition we present the potential and design of activity-based probes targeting kinases and phosphatases to study phosphorylation. Major conclusions: Synthetic molecular probes have increased our understanding of the functional role of DUBs in living cells. In addition to the detection of enzymatic activities of known members, activity-based probes have contributed to a number of functional assignments of previously uncharacterized enzymes. This method enables cellular validation of the specificity of small molecule DUB inhibitors. General significance: Molecular probes combined with mass spectrometry-based proteomics and cellular assays represent a powerful approach for discovery and functional validation, a concept that can be expanded to other enzyme classes. This addresses a need for more informative cell-based assays that are required to accelerate the drug development process. This article is part of a Special Issue entitled: Ubiquitin Drug Discovery and Diagnostics.
Abbreviations: ABP, Activity-based probe; ADP, Adenosine diphosphate; ATP, Adenosine triphosphate; DUB, Deubiquitylating enzyme; E1, Ubiquitin activating enzyme; E2, Ubiquitin conjugating enzyme; E3, Ubiquitin-protein ligase; HAUbBr2, HA-tagged ubiquitin ethyl bromide; HAUbVME, HA-tagged ubiquitin vinyl methyl ester; HAUbVS, HA-tagged ubiquitin vinyl sulfone; IP, Immunoprecipitation; MS, Mass Spectrometry; PTM, Post-Translational Modification; PTP, Protein Tyrosine Phosphatase; SDS-PAGE, Sodium dodecylsulfate polyacrylamide gel electrophoresis; SPPS, Solid Phase Peptide Synthesis; Ub, Ubiquitin; Ubl, Ubiquitin-like protein; UCH-L1, Ubiquitin carboxyl terminal hydrolase isozyme L1; UCH-L3, Ubiquitin carboxyl terminal hydrolase isozyme L3; UCH-L5, Ubiquitin carboxyl terminal hydrolase isozyme L5; USP7, Ubiquitin specific processing protease 7
Keywords: Ubiquitin, Small molecular inhibitor, Deubiquitinating enzyme, Ubiquitin specific protease, Proteomics, Active site-directed molecular probe
Highlights
► Activity-based chemical probe screen for specificity of small molecule deubiquitylating enzyme inhibitors in cells. ► Cell-based profiling of inhibitors specific for deubiquitylating enzymes by mass spectrometry. ► Overview of proteasome, deubiquitylating enzyme, kinase, and phosphatase probes.
1. Introduction
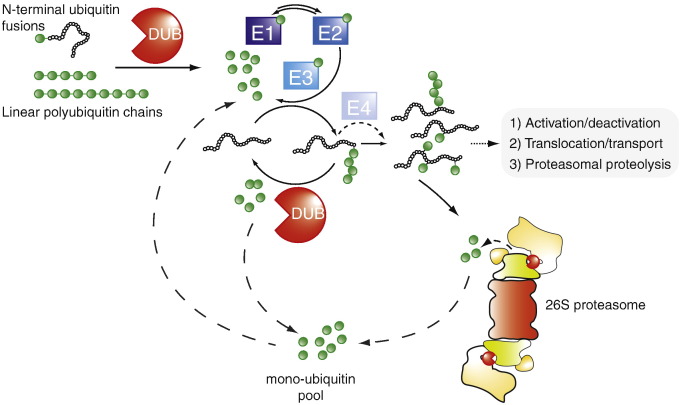
The last two decades have witnessed the development and application of activity based probes for the detection of enzymatic activities related to a wide range of protein post-translational modifications (PTMs). These tools have allowed for the detection of many enzymatic activities in relation to conjugation and deconjugation of PTMs. The greatest success and widest application of this approach have probably been found within the ubiquitin proteasome pathway [1]. The ubiquitin proteasome system (UPS) consists of three different elements with partially opposing, but complementary roles; the ubiquitin conjugating cascade, the deubiquitylating enzymes and the proteasome complex (Fig. 1 ). The ubiquitin conjugating cascade of enzymes (E1, E2, E3 and E4) is responsible for conjugating ubiquitin (Ub)/ubiquitin-like proteins (Ubls) to other proteins as signaling molecules in several biological processes [2], [3]. The deubiquitylating enzymes (DUBs) are responsible for removing, trimming or editing the Ub/Ubl modifications from proteins [2]. The proteasome complex, composed of at least three different proteolytically active subunits β1, β2 and β5, degrades the majority of soluble proteins within the cell following polyubiquitylation [4]. The UPS not only enables the targeted destruction of proteins no longer required by the cell, but also enables the selective deactivation of signaling molecules, removal of pathogenic proteins and production of peptides for presentation to the immune system. The entire pathway comprises close to 1000 genes, is vital for many normal cellular functions and is frequently disrupted in cellular pathologies [5], [6].
Fig. 1.
Schematic illustration key components of ubiquitin proteasome system; from the activation of newly transcribed ubiquitin (Ub), modification of proteins with Ub by the Ub enzyme cascade and deubiquitinating enzymes to the fate of ubiquitinated proteins.
A likely explanation for the remarkable success of development of activity-based probes in the UPS when compared to small molecule probes related to other PTMs, is the ability to target cysteine and threonine proteases successfully by covalent inhibition. These represent the major enzymatic activities of DUBs and the proteasome respectively. Improved recognition of the protein-based ubiquitin and peptide-derived proteasome probes as compared with small molecule based probes may also contribute. The generation of activity based probes for the ubiquitin pathway as important tools to obtain insights into the ubiquitin proteasome system has been reviewed previously [7], [8], [9], [10]. The focus of this review is the application of activity-based probes in the context of inhibitor development and characterization. This process represents a critical step in arriving at a better understanding of fundamental cellular processes and enables the development of novel targeted therapeutic agents.
2. Activity-based proteomics probes
Activity-based probes (ABPs) are defined by their ability to target subsets of a complex proteome based on labeling of specific enzyme classes in a covalent fashion. The incorporation of specific tags for detection and enrichment allows for visualization and further analysis for example by tandem mass spectrometry. The ABP approach is highly complementary to mass spectrometry-based proteomics due to the ability to enrich for specifically captured substrates with the aid of an affinity tag. This also distinguishes the method from the use of radiolabeled covalent inhibitors that support detection but not affinity enrichment. A critical parameter is also the choice of reactive group for covalent labeling of the target enzyme at or close to the enzyme active site. Depending on the mechanism of enzymatic catalysis different reactive groups can be chosen for covalent capture. The alkylation of nucleophilic residues (Cys, Ser, Thr) present in protease active sites by reactive electrophiles has been a very successful realization of this method. The utility of ABPs is particularly evident in the study of enzyme systems involved in the attachment and removal of PTMs. Due to the enzyme active site chemistries involved ABP development has been generally more fruitful for the targeting of deconjugating enzymes that usually contain strong nucleophiles at their active sites when compared to the respective ligases/transferases.
3. Proteasome active site probes
The discovery and development of proteasome inhibitors have been previously covered by comprehensive reviews [10], [11]. Here, we will therefore only give a brief introduction to the proteasome complex and proteasome active site probes.
The proteolytically active sites of the proteasome complex are located on distinct subunits (for example β1, β2 and β5 in the constitutive proteasome) and this is reflected by the labeling pattern obtained by active site-directed proteasome probes [12]. The three subunits responsible for proteolysis also correspond to different activity profiles with respect to substrate specificity which were named after prototypic proteases: chymotrypsin-like activity for β5 (CT-L), trypsin-like activity for β2 (T-L) and petidylglutamyl peptide hydrolyzing-like (PGPH-L) activity (also referred to as caspase-like) for β1 [13]. Exchange of the active subunits with close homologues can be induced in immune cells [14] and the thymus [15]. This allows for the tuning of the peptide repertoire produced by proteasomal proteolysis, which underscores the importance of presentation of these peptides (after further processing steps) in immune surveillance.
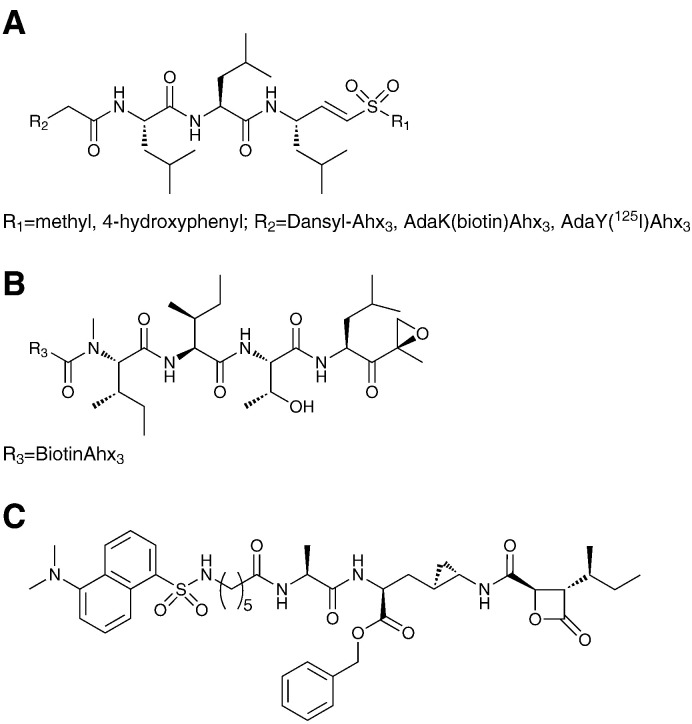
Proteolytically active subunits of the proteasome have been targeted by probes based on peptide scaffolds bearing a C-terminal electrophile (Fig. 2 ). The most popular designs have been based on the trileucine vinylsulfone (L3VS) sub-structure and on functionalized epoxomicin derivatives. Profiling with these probes is enabled by detection with dansyl [16] and biotin [17], [18] tags and with strong fluorophores [19], [20] as well as by radiolabels [12], [21]. Alternatively, a two step protocol can be applied whereby the tag is attached in a bioorthogonal ligation step following labeling of active proteasome subunits in a cell lysate [22]. This selective ligation is possible through application of the Staudinger ligation [23] or 1,4-triazole formation also known as "click" chemistry [24], [25]. A more recent addition to the arsenal of proteasome-directed probes has been the Belacostin A derived dansyl-KF33955 probe [26]. This β-lactone containing probe was used successfully in the labeling of β1, β2 and β5 subunits of the 26S proteasome in HeLa cells as well as the 20S proteasome from erythrocytes.
Fig. 2.
Design of proteasome active site probes; peptide-based probes for labeling of active proteasome subunits have been based on trileucine vinyl sulfones (A), the epoxyketone containing epoxomicin (B) and a belacostin A based derivative Dansyl-KF33955 (C). Abbreviations: Ahx: 6-aminohexanoyl, Ada: Adamantaneacetyl, Dansyl: 5-(dimethylamino)naphthalene-1-sulfonyl.
4. Ubiquitin-derived probes
Ubiquitin derived ABPs [27], [28] have been used for the profiling of the active DUB population in cell lines, primary cells and tissues [29]. Subsequently this concept has been extended to the study of proteases specific for ubiquitin-like proteins (Ubls) [30]. A complex proteome such as a cell or tissue lysate is labeled by incubation with ubiquitin probe, followed by SDS-PAGE separation and immunoblotting against the epitope tag genetically encoded in the probe construct. Thereby a read-out, which is dependent on DUB abundance and activity, is obtained. This approach was also extended to the investigation of cancer cell lines [29], [31] and patient biopsies [31]. Particularly fruitful was the study of in vitro infection models of viral [32], [33], [34], [35], parasitic [36], [37] and bacterial [38], [39] pathogens. In this way previously unknown DUBs encoded by pathogens were identified and other interactions of pathogen encoded factors with the host ubiquitin proteasome system were elucidated. DUB related pathogen–host interactions have been reviewed in detail elsewhere [40], [41], [42], [43]. Ubiquitin probes have further enabled structural studies on DUB-ubiquitin complexes [44]. In these investigations the crystallization and X-ray diffraction of covalent adducts formed between enzyme and the ubiquitin probe allowed insights into some of the molecular determinants of interactions of DUBs with ubiquitylated substrates. These types of studies have also been successfully carried out with complexes involving ubiquitin aldehyde [45], [46], [47] or ubiquitin probes devoid of an epitope tag [48], [49], [50].
In addition to the utility of ubiquitin ABPs targeting Cys protease DUBs, MS-based proteomics experiments of ubiquitin probes have also been shown to retrieve other regulators of ubiquitylation, for example Metalloprotease DUBs were retrieved under native pull-down conditions [51]. This finding might be explained by the formation of non-covalent complexes which can be enriched in immunoprecipitations (IPs) under non-denaturing conditions. The targeting and retrieval of ubiquitin ligases by ubiquitin probes have also been observed [52]. Usually the reduced nucleophilicity of Cys residues in this enzyme class (when compared to Cys as part of the catalytic triad in DUB proteases) results in strongly reduced capture. This can apparently be compensated for to a certain extend by strong C-terminal electrophiles conjugated to the Ub probes [52].
5. Activity-based probes allow enhanced inhibitor screening
In addition to the above-mentioned applications activity-based proteomics probes have also enabled the development of pharmacologically active enzyme inhibitors. In one format, competition assays between an inhibitor candidate compound and the activity-based probe can be performed in cell lysates. In this type of assay format the competition between the small molecule inhibitor and the probe leads to a reduced labeling profile for the activity-based probe. The loss of signal for labeling of specific target enzymes allows assessment of the specificity of inhibition, while titration of concentration ranges provides a measure of IC50 values for the respective inhibitor. Several features make this type of assay format particularly attractive: The approach represents a cell-based assay in which treatment by the inhibitor candidate can be performed on intact cells. In contrast to the majority of in vitro enzyme assays, a range of active cellular enzymes can be assayed simultaneously. This is only limited by the number of enzymes successfully labeled by the activity-based probe and the representation of active enzymes in the cellular proteome. Quantifiable results can be obtained using various readout formats including immunoblotting, in-gel fluorescence detection or IP followed by quantitative MS-based proteomics.
Such an approach has been employed successfully for a number of ABPs including in the identification and characterization of selective inhibitors of the proteasome [53], [54], of cysteine proteases in the DUB enzyme family [51] and serine hydrolases in the fatty acid amide hydrolase class [55]. Promiscuous inhibitors can be tested in this way in order to demonstrate their broad inhibition profile [56] and competition assays can be applied to the characterization of the specificity of known drugs.
6. Characterization of the subunit specificity of proteasome inhibitors
The development of novel cancer therapeutics is a prime example where the success of bortezomib [16], [57] for treatment of multiple myeloma [58] has provided proof of concept for the value of interventions targeting the ubiquitin proteasome system. The effects of Bortezomib (PS-341) on the activity and composition of proteasomes in multiple myeloma cells were investigated by labeling with proteasome probe Ada-[125I]-Y-Ahx3L3VS demonstrating that the inhibitor selectively targets β5 and β1 subunits (as well as immunoproteasome subunits β5i and β1i) [57]. The corresponding experiments were carried out in the presence or absence of cytokines, which induce the formation of immunoproteasomes under conditions that may be encountered in the bone marrow.
A similar characterization of the subunit specificity of proteasome inhibition was also carried out for the orally active inhibitor NPI-0052 (Salinosporamide A) in multiple myeloma cells [59]. The results of competitive labeling with the proteasome probe Dansyl-Ahx3L3VS indicated that the inhibitor targets the β1, β2 and β5 subunits. This result was consistent with the finding that cleavage of fluorogenic substrates showed inhibition of the CT-L, T-L and PGPH-L activities of the proteasome.
In a comparable study the specificity of the three proteasome inhibitors BzLLLCOCHO, Bortezomib (PS-341) and MG132 was assessed by labeling with the cell permeable Dansyl-Ahx3L3VS probe following treatment with individual inhibitors for 24 h [60]. This demonstrated clearly distinct labeling pattern of the proteolytically active proteasome subunits β1, β2 and β5 for the three inhibitors. The observed labeling patterns were positively correlated with the inhibition of the CT-L, T-L and PGPH-L activities of the proteasome by hydrolysis of suitable fluorogenic substrates in the presence of inhibitors.
The selective inhibition of the immunoproteasome subunit β1i (LMP2) by synthetic Dihydroeponemycin analogs was demonstrated by competitive labeling experiments in EL4 cells using biotin-epoxomicin and biotin-eponemycin as a proteasome active site probes [61].
These examples highlight the value of utilizing active site-directed probes in the development and characterization of proteasome inhibitors. This is enabled by their ability to provide a rapid assay of the corresponding proteolytically active subunits in different cell types and in the presence or absence of added competitive inhibitors the specificities of which can thereby be assessed. This approach has been informative to basic science as well as to the development of lead compounds for novel therapeutics.
7. Competitive ubiquitin probe labeling for DUB inhibitor development
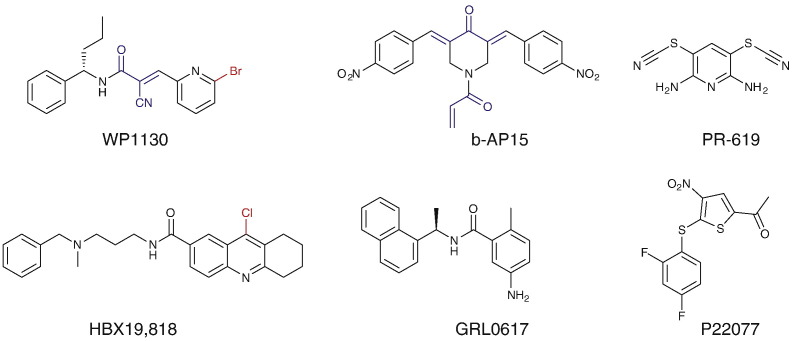
Due to the large number of cellular DUBs there is a need for innovative assay designs that allow assessment of DUB activity in a cellular context. Competitive labeling with Ub probes provides characterization of lead compounds in a cell culture model thereby providing valuable data for assessment of both compound potency and selectivity of different DUBs relevant to a given cell type. A number of DUB inhibitors have been characterized and developed into lead compounds for medicinal chemistry programs. Generally the initial hit has been obtained by high throughput screening (HTS) of compound libraries followed by further optimization using structure–activity relationships. It is noteworthy that many of the identified lead compounds and inhibitors have the ability to covalently label the active site cysteine residue present in the majority of DUBs (Fig. 3 ). This may occur by conjugate addition, nucleophilic aromatic substitution or disulfide bond formation depending on the chemistry of the small molecule inhibitor.
Fig. 3.
Molecular structures of selected DUB and Ubl-protease inhibitors characterized by competitive probe labeling; sub-structures with Michael acceptor functionality (capable of conjugate addition to Cys) are highlighted in blue and aryl halides which are capable of nucleophilic aromatic substitution are highlighted in red.
One example of Ub probe labeling in the presence of a small molecule inhibitor has been the characterization of WP1130 as a partially selective DUB inhibitor which induces aggresome formation and tumor cell apoptosis by Kapuria et al. [62]. The selectivity of inhibition for different members of the DUB family of enzymes was also investigated by in vitro inhibition assays using UbAMC as a substrate in combination with recombinant purified DUBs. The total cellular DUB activity in lysates treated with inhibitors was tested by UbAMC hydrolysis. Labeling of treated versus untreated cell lysates with the activity-based probe HAUbVS was performed in order to assess the DUB inhibitory profile of WP1130. In this assay format reduced labeling by the HAUbVS probe as detected in anti-HA immunoblotting of lysates from cells treated with WP1130 was used as readout of selectivity of DUB inhibition. A further focus of this work was the observation of accumulation of polyubiquitylated protein material upon treatment with WP1130 despite a lack of inhibition of proteasome activity.
Altun et al. described a more extensive application of activity-based probes to the characterization and development of novel DUB inhibitors [51]. In this study two DUB inhibitors, PR-619 and P22077, were characterized by in vitro enzyme inhibition assays and in cell culture models. Labeling of HEK293T cell lysates treated with inhibitors at varying concentrations was performed with the ubiquitin probes HAUbVME and HAUbBr2 to investigate DUB inhibition profiles. As a read-out of these assays immunoblotting against HA-tag or specific DUBs of interest was carried out enabling a rapid assessment of inhibitor specificity. In order to obtain an alternative quantitative read-out format, anti-HA immunoprecipitation (IP) was combined with identification and label-free quantification by mass spectrometry based proteomics (Fig. 4 ). Using this approach, quantitative data illustrating the degree of inhibition of 25 cellular DUBs was obtained. These data were consistent with the in vitro enzyme characterization data, demonstrating that PR-619 exhibits a broad inhibition profile whereas P22077 is a selective inhibitor of USP7 and USP47. Both USP7 and USP47 are considered oncology targets and P22077 and related analogs are the subject of further development as anti-cancer therapeutics [63], [64].
Fig. 4.
Small molecule inhibitors affect DUBs in living cells; (A) Workflow of DUB inhibition profile in living cells revealed by activity-based quantitative mass spectrometry as described in [51]. (B) Workflow of profiling inhibitor capacity by immunoblot as described in [51]. (C) HEK293T cells were incubated with 50 µM PR-619, P22077, 0.1% DMSO or 10 µM MG132, lysed and analyzed by SDS-PAGE and anti-HA immunoblotting.
Another application of Ub probe labeling for the development of USP7 inhibitors was recently demonstrated by Reverdy et al. [65]. The screening hit HBX 19,818, which had been identified by high throughput screening (HTS) of a compound library employing the UbAMC assay, was characterized by HAUbVS labeling of HCT116 cells treated with increasing concentrations of the inhibitor. The readout of the assay was performed by anti-HA immunoblotting and showed reduced probe labeling of USP7 while leaving the competitive labeling of other cellular DUBs unaffected. This result was consistent with the in vitro enzyme inhibition data that also indicated a lack of inhibition of other tested cysteine proteases, DUBs and Ubl specific proteases, but the activity of HBX 19,818 towards the closely related DUB USP47 was not discussed. The mechanism of inhibition was investigated by the authors and covalent labeling of the active site Cys223 in USP7 was demonstrated, indicating that alkylation of this residue leads to irreversible inactivation of the DUB. Furthermore the cellular effects of the inhibitor were characterized as reduction in cell proliferation, induction of apoptosis and G1 arrest in a dose-dependent fashion.
A novel DUB inhibitor, b-AP15 was recently described by D'Arcy et al. [66]. DUB profiling was carried out in the presence of ubiquitin probe HAUbVS in HCT-116 cells after 3 h treatment with 1.0 μM inhibitor; demonstrating that USP14 and to a lesser extend UCHL5 were inhibited at this concentration. Treatment of cells with b-AP15 led to tumor cell apoptosis, which was independent of TP53 status. Additionally an accumulation of high molecular weight Ub conjugates and reduced levels of free ubiquitin were observed and ascribed to the inhibition of the 19S associated DUBs USP14 and UCHL5. The authors concluded that the compound does not act as a broad spectrum DUB inhibitor at the concentration employed, rather it inhibits the proteasome associated DUBs USP14 and UCH-L5. Thus, b-AP15 effectively inhibits proteasomal activity by inhibiting the DUBs that process polyubiquitylated proteins prior to their engagement by the proteasome.
This approach has utility for viral DUBs as well. The ubiquitin probe HAUbVS has been successfully applied to demonstrate the selectivity of inhibitor GRL0617 for the SARS Coronavirus protease and isopeptidase Papain-like protease (Plpro) in Vero E6 cell lysate [67]. No change in labeling profile was observed for cellular DUBs at inhibitor concentrations of up to 40 μM. In contrast, the labeling of added Plpro was almost completely abolished at the same inhibitor concentration.
8. Utility of activity based proteomics in inhibitor characterization outside the UPS
The usefulness of activity based probes (ABPs) in the development and characterization of enzyme inhibitors has also been recognized in fields outside the ubiquitin proteasome system. Below we describe the advances made in the application of this concept to kinases and phosphatases involved in protein phosphorylation. ABPs for the targeting of kinases and phosphatases have received considerable attention due to the central role of protein phosphorylation in cellular signaling, protein translocation and ultimately its aberrations in disease.
9. Kinase targeted activity-based probes
Initial kinase probes have utilized ATP and ADP analogs as molecular scaffolds thereby targeting the essential kinase co-factor in probe design. Later approaches were built on modification of known kinase inhibitors of natural product origin or related to licensed drugs. Kinase probes based on acyl phosphate ATP and ADP analogs which lead to the transfer of a biotin to amino acid residues on the interacting protein resulted in the labeling and identification of a range of kinases, ATPases, nucleotide biosynthesis-related and other proteins [68]. The lack of selectivity for kinases in a cellular environment is understandable given the wide range of proteins that utilize ATP as a cofactor.
A tetramethylrhodamine-labeled Wortmannin analog AX7503 allowed labeling in mammalian cell lysates and readout of the assay by in gel fluorescence [69]. This fluorescent kinase probe labeled DNA dependent protein kinase (DNA-PK), phosphoinositide 3 kinase and mammalian Polo-like kinase 1 (PLK1). Previously it had not been appreciated that PLK1 is inhibited by Wortmannin at relevant concentrations. Further Wortmannin derivatives labeled with borondipyrromethene (BODIPY), tetramethylrhodamine or biotin were prepared successfully [70]. It was shown that the BODIPY derivative is cell-permeable thereby enabled labeling of intact cells while the biotin derivative allows isolation of labeled protein material from cell lysates. In a follow-up study to this work Liu et al. also demonstrated that Polo-like kinase 3 (PLK3) is a further molecular target of Wortmannin as shown by competitive labeling between AX7503 and Wortmannin [71]. The labeling sites of AX7503 for both PLK1 and PLK3 were determined by tandem mass spectrometry and were found to represent conserved lysine residues in the ATP binding site of each of the kinases.
A bisindolylmaleimide motif was chosen as the scaffold for the design of a further kinase probe AX4697 [72]. This recognition motif with a high affinity for protein kinase C (PKC) was combined with a chloroacetamide electrophile and a tetramethylrhodamine fluorophore. Successful labeling of α and β PKC in Jurkat cells was demonstrated as well as loss of signal in staurosporine-pretreated cells.
A recent design of a probe for the Abelson Tyr kinase (Abl) was based on the anti-cancer drug imatinib [73]. The reversible mode of enzyme inhibition necessitated the incorporation of a photoreactive benzophenone moiety or a dialdehyde motif for covalent labeling. Detection was enabled by the incorporation of a terminal alkyne into the structure allowing attachment of a rhodamine azide dye by Cu(I) catalyzed triazole formation (click chemistry) [24], [25]. Targeting and detection of Abl kinase in the presence of added cell lysate occurred, but the labeling efficiency diminished with increasing amounts of lysate and the approach did not extend to labeling and detection at endogenous levels.
The reversible and non-selective kinase inhibitor staurosporine has also been utilized as the recognition motif in probe design by several groups. In these probe designs covalent labeling is enabled by incorporation of photoreactive phenylazide [74] or diazirine [75] groups and irradiation by UV light. Enrichment of the trapped proteins is performed by a further incorporated biotin moiety [74] or a terminal alkyne for attachment of either a biotin group or a fluorophore for detection following the covalent capture by click chemistry [75].
10. Phosphatase targeted activity-based probes
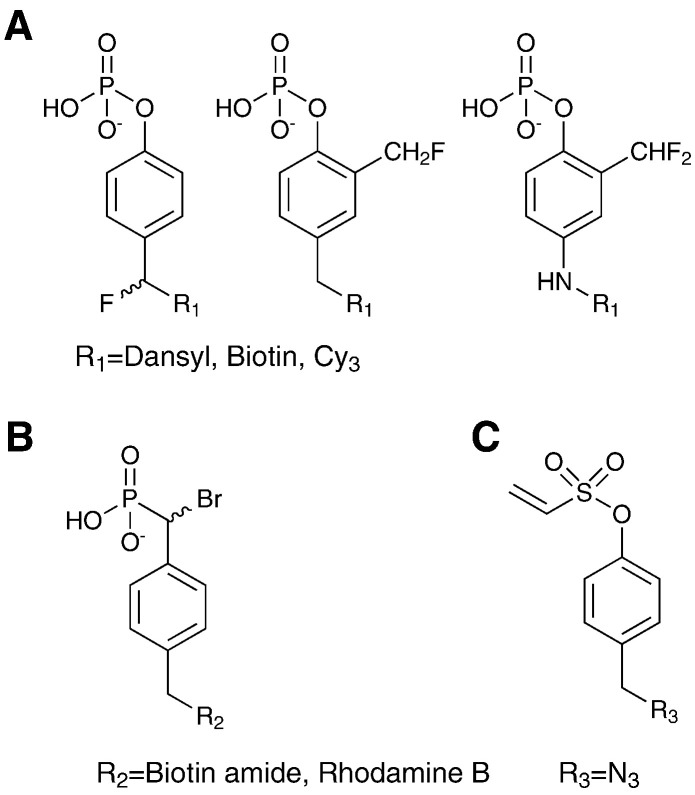
Activity based probes for protein tyrosine phosphatases (PTPs) were developed on several scaffolds that allow mimicry of phosphotyrosine motifs while incorporating reactive electrophiles for the covalent capture of active phosphatases (Fig. 5 ). Several groups exploited the generation of quinone methides for the generation of reactive electrophiles for trapping of PTPs [76], [77], [78], [79]. These are generated from substituted phenyl phosphates with benzylic fluoride substituents in ortho or para position. In a recent publication this concept was extended to the design of the unnatural amino acid 2-fluoromethyl phosphotyrosine which was incorporated into peptides by solid phase peptide synthesis (SPPS) [80]. In this way peptidic activity based probes were produced based on known PTP target sequences.
Fig. 5.
Molecular structures of activity-based protein tyrosine phosphatase probes; depending on the nature of the active electrophile these are divided into (A) quinone methide precursors (B) bromobenzyl phosphonates and (C) vinyl sulfonates.
In earlier work Kumar et al. combined a minimal motif incorporating a bromomethyl phosphonate-substituted phenyl derivative attached to either a fluorophore or biotin for detection [81]. Successful labeling of PTPs was demonstrated using both isolated enzymes in vitro and in cell lysates of different cancer cell lines.
A further alternative approach exploited conjugate addition to substituted aryl vinyl sulfonates and sulfones for the covalent labeling of active PTPs [82]. This methodology was then applied to a structural study of a covalent complex between the bacterial PTP YopH and its mechanism based inhibitor.
Recently a novel probe design was implemented that enables microscopy-based read out through the use of quenched activity based probes [83]. In this strategy the quencher is released upon quinone methide formation and reaction with the target enzyme. The methodology has also been applied to one- and two-photon imaging experiments.
In comparison, the targeting of Ser/Thr phosphatases by activity-based probes has been less frequently realized. A successful example is that of a microcystin derived probe which was prepared and applied to the labeling and isolation of endogenous levels of Ser/Thr protein phosphatases from Jurkat cell lysate [84].
11. Characterization of kinase inhibitors by competition assays and targeted proteomics
Acylphosphate kinase probes based on the ATP cofactor [68] were used recently in an extensive study evaluating the specificity profiles of a range of kinase inhibitors [85]. In this ‘in situ native kinase profiling’ (KiNativ) approach, labeling of mammalian cell lysates with the kinase probes was conducted in the presence or absence of the respective inhibitor. The complex samples were then digested with trypsin and biotinylated peptides enriched with streptavidin resin. Following elution the captured peptides were analyzed by quantitative proteomics experiments using a targeted LC–MS2 approach. This detection method significantly improved the signal to noise ratios and allows detection of lower amounts of analyte. Quantitative inhibition data for over 200 kinases with six kinase inhibitors was obtained by this method. This approach provides access to quantitative inhibition profiles that are not easily obtained by other experimental approaches and exploits the strengths of both activity-based probe profiling and targeted mass spectrometry-based proteomics.
12. Future perspectives
The introduction of activity-based probes into the development of enzyme inhibitors enables the assessment of compound selectivity in a cellular context. This ideally complements more conventional in vitro enzyme assays and allows implementation of this assay format at an early stage in drug development. Furthermore competitive labeling experiments have greatly aided the definition of specificities of inhibitors used in cellular research. New therapies in the UPS will employ the manipulation of DUB activity in a highly selective manner to achieve the desired outcomes. Therefore inhibition of selected intracellular DUB activities will receive increasing attention as a strategy for pharmacological intervention in disease. DUBs with a validated role in oncology are expected to be the first in line for future DUB inhibitors. Provided that challenges of developing specific inhibitors with desirable compound profiles can be overcome, it is expected that this will enable successful intervention in other pathologies such as inflammatory diseases.
13. Summary
Activity-based probe designs have provided insights into multiple cellular pathways and have proven extremely valuable as tools for the study of PTM conjugating and deconjugating enzymes. The investigation of the ubiquitin proteasome system has benefited from the development of probes for proteasomal proteolysis as well as deubiquitylating enzymes. Specificities and overlapping reactivities of ubiquitin-like proteases were investigated by analogous Ubl probes. Inhibitor development has been facilitated through competitive labeling assays enabling elucidation of inhibition profiles in a cellular environment. In addition, natural product derived kinase probes have demonstrated the protein targets of these covalent inhibitors, indicating previously unknown molecular targets and providing information in situations where specificities of the respective compounds are not always fully elucidated. The importance of access to selective inhibitors for basic cell biology and drug development makes this area highly suited for successful industry-academia collaborations.
Acknowledgements
M.A. is supported by the Swedish Research Council and the Foundation for Geriatric Diseases at Karolinska Institutet. B.M.K. is supported by the Biomedical Research Centre (NIHR), Oxford, UK, and H.B.K. is supported by the Wellcome Trust OXION Initiative in Ion Channels and Disease.
Footnotes
This article is part of a Special Issue entitled: Ubiquitin Drug Discovery and Diagnostics.
References
- 1.Ciechanover A. The ubiquitin–proteasome pathway: on protein death and cell life. EMBO J. 1998;17:7151–7160. doi: 10.1093/emboj/17.24.7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amerik A.Y., Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim. Biophys. Acta, Mol. Cell Res. 2004;1695:189–207. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 3.Glickman M.H., Ciechanover A. The ubiquitin–proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 4.Rock K.L., Gramm C., Rothstein L., Clark K., Stein R., Dick L., Hwang D., Goldberg A.L. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 5.Schwartz A.L., Ciechanover A. Targeting proteins for destruction by the ubiquitin system: implications for human pathobiology. Annu. Rev. Pharmacol. Toxicol. 2009;49:73–96. doi: 10.1146/annurev.pharmtox.051208.165340. [DOI] [PubMed] [Google Scholar]
- 6.Lecker S.H., Goldberg A.L., Mitch W.E. Protein degradation by the ubiquitin–proteasome pathway in normal and disease states. J. Am. Soc. Nephrol. 2006;17:1807–1819. doi: 10.1681/ASN.2006010083. [DOI] [PubMed] [Google Scholar]
- 7.Hemelaar J., Galardy P.J., Borodovsky A., Kessler B.M., Ploegh H.L., Ovaa H. Chemistry-based functional proteomics: mechanism-based activity-profiling tools for ubiquitin and ubiquitin-like specific proteases. J. Proteome Res. 2004;3:268–276. doi: 10.1021/pr0341080. [DOI] [PubMed] [Google Scholar]
- 8.Kessler B.M. Putting proteomics on target: activity-based profiling of ubiquitin and ubiquitin-like processing enzymes. Expert Rev. Proteomics. 2006;3:213–221. doi: 10.1586/14789450.3.2.213. [DOI] [PubMed] [Google Scholar]
- 9.Ovaa H. Active-site directed probes to report enzymatic action in the ubiquitin proteasome system. Nat. Rev. Cancer. 2007;7:613–620. doi: 10.1038/nrc2128. [DOI] [PubMed] [Google Scholar]
- 10.Verdoes M., Florea B.I., Van Der Marel G.A., Overkleeft H.S. Chemical tools to study the proteasome. Eur. J. Org. Chem. 2009:3301–3313. [Google Scholar]
- 11.de Bettignies G., Coux O. Proteasome inhibitors: dozens of molecules and still counting. Biochimie. 2010;92:1530–1545. doi: 10.1016/j.biochi.2010.06.023. [DOI] [PubMed] [Google Scholar]
- 12.Bogyo M., Shin S., McMaster J.S., Ploegh H.L. Substrate binding and sequence preference of the proteasome revealed by active-site-directed affinity probes. Chem. Biol. 1998;5:307–320. doi: 10.1016/s1074-5521(98)90169-7. [DOI] [PubMed] [Google Scholar]
- 13.Heinemeyer W., Fischer M., Krimmer T., Stachon U., Wolf D.H. The active sites of the eukaryotic 20S proteasome and their involvement in subunit precursor processing. J. Biol. Chem. 1997;272:25200–25209. doi: 10.1074/jbc.272.40.25200. [DOI] [PubMed] [Google Scholar]
- 14.Griffin T.A., Nandi D., Cruz M., Fehling H.J., Van Kaer L., Monaco J.J., Colbert R.A. Immunoproteasome assembly: cooperative incorporation of interferon γ (IFN-γ)-inducible subunits. J. Exp. Med. 1998;187:97–104. doi: 10.1084/jem.187.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murata S., Sasaki K., Kishimoto T., Niwa S.I., Hayashi H., Takahama Y., Tanaka K. Regulation of CD8+ T cell development by thymus-specific proteasomes. Science. 2007;316:1349–1353. doi: 10.1126/science.1141915. [DOI] [PubMed] [Google Scholar]
- 16.Berkers C.R., Verdoes M., Lichtman E., Fiebiger E., Kessler B.M., Anderson K.C., Ploegh H.L., Ovaa H., Galardy P.J. Activity probe for in vivo profiling of the specificity of proteasome inhibitor bortezomib. Nat. Methods. 2005;2:357–362. doi: 10.1038/nmeth759. [DOI] [PubMed] [Google Scholar]
- 17.Sin N., Kyung B.K., Elofsson M., Meng L., Auth H., Kwok B.H.B., Crews C.M. Total synthesis of the potent proteasome inhibitor epoxomicin: a useful tool for understanding proteasome biology. Bioorg. Med. Chem. Lett. 1999;9:2283–2288. doi: 10.1016/s0960-894x(99)00376-5. [DOI] [PubMed] [Google Scholar]
- 18.Kessler B.M., Tortorella D., Altun M., Kisselev A.F., Fiebiger E., Hekking B.G., Ploegh H.L., Overkleeft H.S. Extended peptide-based inhibitors efficiently target the proteasome and reveal overlapping specificities of the catalytic β-subunits. Chem. Biol. 2001;8:913–929. doi: 10.1016/s1074-5521(01)00069-2. [DOI] [PubMed] [Google Scholar]
- 19.Berkers C.R., Van Leeuwen F.W.B., Groothuis T.A., Peperzak V., Van Tilburg E.W., Borst J., Neefjes J.J., Ovaa H. Profiling proteasome activity in tissue with fluorescent probes. Mol. Pharm. 2007;4:739–748. doi: 10.1021/mp0700256. [DOI] [PubMed] [Google Scholar]
- 20.Verdoes M., Florea B.I., Menendez-Benito V., Maynard C.J., Witte M.D., van der Linden W.A., van den Nieuwendijk A.M.C.H., Hofmann T., Berkers C.R., van Leeuwen F.W.B., Groothuis T.A., Leeuwenburgh M.A., Ovaa H., Neefjes J.J., Filippov D.V., van der Marel G.A., Dantuma N.P., Overkleeft H.S. A fluorescent broad-spectrum proteasome inhibitor for labeling proteasomes in vitro and in vivo. Chem. Biol. 2006;13:1217–1226. doi: 10.1016/j.chembiol.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 21.Greenbaum D., Medzihradszky K.F., Burlingame A., Bogyo M. Epoxide electrophiles as activity-dependent cysteine protease profiling and discovery tools. Chem. Biol. 2000;7:569–581. doi: 10.1016/s1074-5521(00)00014-4. [DOI] [PubMed] [Google Scholar]
- 22.Ovaa H., Van Swieten P.F., Kessler B.M., Leeuwenburgh M.A., Fiebiger E., Van den Nieuwendijk A.M.C.H., Galardy P.J., Van der Marel G.A., Ploegh H.L., Overkleeft H.S. Chemistry in living cells: detection of active proteasomes by a two-step labeling strategy. Angew. Chem. Int. Ed. 2003;42:3626–3629. doi: 10.1002/anie.200351314. [DOI] [PubMed] [Google Scholar]
- 23.Saxon E., Bertozzi C.R. Cell surface engineering by a modified Staudinger reaction. Science. 2000;287:2007–2010. doi: 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- 24.Tornoe C.W., Christensen C., Meldal M. Peptidotriazoles on solid phase: [1,2,3]-triazoles by regiospecific copper(I)-catalyzed 1,3-dipolar cycloadditions of terminal alkynes to azides. J. Org. Chem. 2002;67:3057–3064. doi: 10.1021/jo011148j. [DOI] [PubMed] [Google Scholar]
- 25.Rostovtsev V.V., Green L.G., Fokin V.V., Sharpless K.B. A stepwise huisgen cycloaddition process: copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew. Chem. Int. Ed. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 26.Hasegawa M., Kinoshita K., Nishimura C., Matsumura U., Shionyu M., Ikeda S.i., Mizukami T. Affinity labeling of the proteasome by a belactosin A derived inhibitor. Bioorg. Med. Chem. Lett. 2008;18:5668–5671. doi: 10.1016/j.bmcl.2008.08.073. [DOI] [PubMed] [Google Scholar]
- 27.Borodovsky A., Ovaa H., Kolli N., Gan-Erdene T., Wilkinson K.D., Ploegh H.L., Kessler B.M. Chemistry-based functional proteomics reveals novel members of the deubiquitinating enzyme family. Chem. Biol. 2002;9:1149–1159. doi: 10.1016/s1074-5521(02)00248-x. [DOI] [PubMed] [Google Scholar]
- 28.Borodovsky A., Kessler B.M., Casagrande R., Overkleeft H.S., Wilkinson K.D., Ploegh H.L. A novel active site-directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. EMBO J. 2001;20:5187–5196. doi: 10.1093/emboj/20.18.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ovaa H., Kessler B.M., Rolan U., Ploegh H.L., Masucci M.G. Activity-based ubiquitin-specific protease (USP), profiling of virus-infected and malignant human cells. Proc. Natl. Acad. Sci. U. S. A. 2004;101:2253–2258. doi: 10.1073/pnas.0308411100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hemelaar J., Borodovsky A., Kessler B.M., Reverter D., Cook J., Kolli N., Gan-Erdene T., Wilkinson K.D., Gill G., Lima C.D., Ploegh H.L., Ovaa H. Specific and covalent targeting of conjugating and deconjugating enzymes of ubiquitin-like proteins. Mol. Cell. Biol. 2004;24:84–95. doi: 10.1128/MCB.24.1.84-95.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rolan U., Kobzeva V., Gasparjan N., Ovaa H., Winberg G., Kisseljov F., Masucci M.G. Activity profiling of deubiquitinating enzymes in cervical carcinoma biopsies and cell lines. Mol. Carcinog. 2006;45:260–269. doi: 10.1002/mc.20177. [DOI] [PubMed] [Google Scholar]
- 32.Gredmark S., Schlieker C., Quesada V., Spooner E., Ploegh H.L. A functional ubiquitin-specific protease embedded in the large tegument protein (ORF64) of murine gammaherpesvirus 68 is active during the course of infection. J. Virol. 2007;81:10300–10309. doi: 10.1128/JVI.01149-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gredmark-Russ S., Isaacson M.K., Kattenhorn L., Cheung E.J., Watson N., Ploegh H.L. A gammaherpesvirus ubiquitin-specific protease is involved in the establishment of murine gammaherpesvirus 68 infection. J. Virol. 2009;83:10644–10652. doi: 10.1128/JVI.01017-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kattenhorn L.M., Korbel G.A., Kessler B.M., Spooner E., Ploegh H.L. A deubiquitinating enzyme encoded by HSV-1 belongs to a family of cysteine proteases that is conserved across the family Herpesviridae. Mol. Cell. 2005;19:547–557. doi: 10.1016/j.molcel.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 35.Wang J., Loveland A.N., Kattenhorn L.M., Ploegh H.L., Gibson W. High-molecular-weight protein (pUL48) of human cytomegalovirus is a competent deubiquitinating protease: mutant viruses altered in its active-site cysteine or histidine are viable. J. Virol. 2006;80:6003–6012. doi: 10.1128/JVI.00401-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.White R.R., Miyata S., Papa E., Spooner E., Gounaris K., Selkirk M.E., Artavanis-Tsakonas K. Characterisation of the trichinella spiralis deubiquitinating enzyme, TsUCH37, an evolutionarily conserved proteasome interaction partner. PLoS Negl. Trop. Dis. 2011;5 doi: 10.1371/journal.pntd.0001340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Artavanis-Tsakonas K., Misaghi S., Comeaux C.A., Catic A., Spooner E., Duraisingh M.T., Ploegh H.L. Identification by functional proteomics of a deubiquitinating/deNeddylating enzyme in Plasmodium falciparum. Mol. Microbiol. 2006;61:1187–1195. doi: 10.1111/j.1365-2958.2006.05307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Catic A., Misaghi S., Korbel G.A., Ploegh H.L. ElaD, a deubiquitinating protease expressed by E. coli. PLoS One. 2007;2 doi: 10.1371/journal.pone.0000381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Misaghi S., Balsara Z.R., Catic A., Spooner E., Ploegh H.L., Starnbach M.N. Chlamydia trachomatis-derived deubiquitinating enzymes in mammalian cells during infection. Mol. Microbiol. 2006;61:142–150. doi: 10.1111/j.1365-2958.2006.05199.x. [DOI] [PubMed] [Google Scholar]
- 40.Edelmann M.J., Kessler B.M. Ubiquitin and ubiquitin-like specific proteases targeted by infectious pathogens: emerging patterns and molecular principles. Biochim. Biophys. Acta, Mol. Basis Dis. 2008;1782:809–816. doi: 10.1016/j.bbadis.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Isaacson M.K., Ploegh H.L. Ubiquitination, ubiquitin-like modifiers, and deubiquitination in viral infection. Cell Host Microbe. 2009;5:559–570. doi: 10.1016/j.chom.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rytkanen A., Holden D.W. Bacterial interference of ubiquitination and deubiquitination. Cell Host Microbe. 2007;1:13–22. doi: 10.1016/j.chom.2007.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Randow F., Lehner P.J. Viral avoidance and exploitation of the ubiquitin system. Nat. Cell Biol. 2009;11:527–534. doi: 10.1038/ncb0509-527. [DOI] [PubMed] [Google Scholar]
- 44.Misaghi S., Galardy P.J., Meester W.J.N., Ovaa H., Ploegh H.L., Gaudet R. Structure of the ubiquitin hydrolase UCH-L3 complexed with a suicide substrate. J. Biol. Chem. 2005;280:1512–1520. doi: 10.1074/jbc.M410770200. [DOI] [PubMed] [Google Scholar]
- 45.Hu M., Li P., Li M., Li W., Yao T., Wu J.W., Gu W., Cohen R.E., Shi Y. Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell. 2002;111:1041–1054. doi: 10.1016/s0092-8674(02)01199-6. [DOI] [PubMed] [Google Scholar]
- 46.Hu M., Li P., Song L., Jeffrey P.D., Chenova T.A., Wilkinson K.D., Cohen R.E., Shi Y. Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14. EMBO J. 2005;24:3747–3756. doi: 10.1038/sj.emboj.7600832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnston S.C., Riddle S.M., Cohen R.E., Hill C.P. Structural basis for the specificity of ubiquitin C-terminal hydrolases. EMBO J. 1999;18:3877–3887. doi: 10.1093/emboj/18.14.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Akutsu M., Ye Y., Virdee S., Chin J.W., Komander D. Molecular basis for ubiquitin and ISG15 cross-reactivity in viral ovarian tumor domains. Proc. Natl. Acad. Sci. U. S. A. 2011;108:2228–2233. doi: 10.1073/pnas.1015287108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Capodagli G.C., McKercher M.A., Baker E.A., Masters E.M., Brunzelle J.S., Pegan S.D. Structural analysis of a viral ovarian tumor domain protease from the Crimean-Congo hemorrhagic fever virus in complex with covalently bonded ubiquitin. J. Virol. 2011;85:3621–3630. doi: 10.1128/JVI.02496-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.James T.W., Frias-Staheli N., Bacik J.P., Levingston Macleod J.M., Khajehpour M., GarcÃa-Sastre A., Mark B.L. Structural basis for the removal of ubiquitin and interferon-stimulated gene 15 by a viral ovarian tumor domain-containing protease. Proc. Natl. Acad. Sci. U. S. A. 2011;108:2222–2227. doi: 10.1073/pnas.1013388108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Altun M., Kramer H.B., Willems L.I., McDermott J.L., Leach C.A., Goldenberg S.J., Kumar K.G.S., Konietzny R., Fischer R., Kogan E., MacKeen M.M., McGouran J., Khoronenkova S.V., Parsons J.L., Dianov G.L., Nicholson B., Kessler B.M. Activity-based chemical proteomics accelerates inhibitor development for deubiquitylating enzymes. Chem. Biol. 2011;18:1401–1412. doi: 10.1016/j.chembiol.2011.08.018. [DOI] [PubMed] [Google Scholar]
- 52.Love K.R., Pandya R.K., Spooner E., Ploegh H.L. Ubiquitin C-terminal electrophiles are activity-based probes for identification and mechanistic study of ubiquitin conjugating machinery. ACS Chem. Biol. 2009;4:275–287. doi: 10.1021/cb9000348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bogyo M. Screening for selective small molecule inhibitors of the proteasome using activity-based probes. Methods Enzymol. 2005;399:609–622. doi: 10.1016/S0076-6879(05)99040-X. [DOI] [PubMed] [Google Scholar]
- 54.Greenbaum D.C., Arnold W.D., Lu F., Hayrapetian L., Baruch A., Krumrine J., Toba S., Chehade K., Brömme D., Kuntz I.D., Bogyo M. Small molecule affinity fingerprinting: a tool for enzyme family subclassification, target identification, and inhibitor design. Chem. Biol. 2002;9:1085–1094. doi: 10.1016/s1074-5521(02)00238-7. [DOI] [PubMed] [Google Scholar]
- 55.Leung D., Hardouin C., Boger D.L., Cravatt B.F. Discovering potent and selective reversible inhibitors of enzymes in complex proteomes. Nat. Biotechnol. 2003;21:687–691. doi: 10.1038/nbt826. [DOI] [PubMed] [Google Scholar]
- 56.Kapuria V., Peterson L.F., Showalter H.D.H., Kirchhoff P.D., Talpaz M., Donato N.J. Protein cross-linking as a novel mechanism of action of a ubiquitin-activating enzyme inhibitor with anti-tumor activity. Biochem. Pharmacol. 2011;82:341–349. doi: 10.1016/j.bcp.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 57.Altun M., Galardy P.J., Shringarpure R., Hideshima T., LeBlanc R., Anderson K.C., Ploegh H.L., Kessler B.M. Effects of PS-341 on the activity and composition of proteasomes in multiple myeloma cells. Cancer Res. 2005;65:7896–7901. doi: 10.1158/0008-5472.CAN-05-0506. [DOI] [PubMed] [Google Scholar]
- 58.Richardson P.G., Barlogie B., Berenson J., Singhal S., Jagannath S., Irwin D., Rajkumar S.V., Srkalovic G., Alsina M., Alexanian R., Siegel D., Orlowski R.Z., Kuter D., Limentani S.A., Lee S., Hideshima T., Esseltine D.L., Kauffman M., Adams J., Schenkein D.P., Anderson K.C. A phase 2 study of Bortezomib in relapsed, refractory myeloma. N. Engl. J. Med. 2003;348:2609–2617. doi: 10.1056/NEJMoa030288. [DOI] [PubMed] [Google Scholar]
- 59.Chauhan D., Catley L., Li G., Podar K., Hideshima T., Velankar M., Mitsiades C., Mitsiades N., Yasui H., Letai A., Ovaa H., Berkers C., Nicholson B., Chao T.H., Neuteboom S.T.C., Richardson P., Palladino M.A., Anderson K.C. A novel orally active proteasome inhibitor induces apoptosis in multiple myeloma cells with mechanisms distinct from Bortezomib. Cancer Cell. 2005;8:407–419. doi: 10.1016/j.ccr.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 60.Crawford L.J.A., Walker B., Ovaa H., Chauhan D., Anderson K.C., Morris T.C.M., Irvine A.E. Comparative selectivity and specificity of the proteasome inhibitors BzLLLCOCHO, PS-341, and MG-132. Cancer Res. 2006;66:6379–6386. doi: 10.1158/0008-5472.CAN-06-0605. [DOI] [PubMed] [Google Scholar]
- 61.Ho Y.K., Bargagna-Mohan P., Wehenkel M., Mohan R., Kim K.B. LMP2-specific inhibitors: chemical genetic tools for proteasome biology. Chem. Biol. 2007;14:419–430. doi: 10.1016/j.chembiol.2007.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kapuria V., Peterson L.F., Fang D., Bornmann W.G., Talpaz M., Donato N.J. Deubiquitinase inhibition by small-molecule WP1130 triggers aggresome formation and tumor cell apoptosis. Cancer Res. 2010;70:9265–9276. doi: 10.1158/0008-5472.CAN-10-1530. [DOI] [PubMed] [Google Scholar]
- 63.Nicholson B., Suresh Kumar K.G. The multifaceted roles of USP7: new therapeutic opportunities. Cell Biochem. Biophys. 2011;60:61–68. doi: 10.1007/s12013-011-9185-5. [DOI] [PubMed] [Google Scholar]
- 64.Parsons J.L., Dianova I.I., Khoronenkova S.V., Edelmann M.J., Kessler B.M., Dianov G.L. USP47 is a deubiquitylating enzyme that regulates base excision repair by controlling steady-state levels of DNA polymerase beta. Mol. Cell. 2011;41:609–615. doi: 10.1016/j.molcel.2011.02.016. [DOI] [PubMed] [Google Scholar]
- 65.Reverdy C., Conrath S., Lopez R., Planquette C., Atmanene C., Collura V., Harpon J., Battaglia V., Vivat V., Sippl W., Colland F. Discovery of specific inhibitors of human USP7/HAUSP deubiquitinating enzyme. Chem. Biol. 2012;19:467–477. doi: 10.1016/j.chembiol.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 66.D'Arcy P., Brnjic S., Olofsson M.H., Fryknas M., Lindsten K., De Cesare M., Perego P., Sadeghi B., Hassan M., Larsson R., Linder S. Inhibition of proteasome deubiquitinating activity as a new cancer therapy. Nat. Med. 2011 doi: 10.1038/nm.2536. [DOI] [PubMed] [Google Scholar]
- 67.Ratia K., Pegan S., Takayama J., Sleeman K., Coughlin M., Baliji S., Chaudhuri R., Fu W., Prabhakar B.S., Johnson M.E., Baker S.C., Ghosh A.K., Mesecar A.D. A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication. Proc. Natl. Acad. Sci. U. S. A. 2008;105:16119–16124. doi: 10.1073/pnas.0805240105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Patricelli M.P., Szardenings A.K., Liyanage M., Nomanbhoy T.K., Wu M., Weissig H., Aban A., Chun D., Tanner S., Kozarich J.W. Functional interrogation of the kinome using nucleotide acyl phosphates. Biochemistry. 2007;46:350–358. doi: 10.1021/bi062142x. [DOI] [PubMed] [Google Scholar]
- 69.Liu Y., Shreder K.R., Gai W., Corral S., Ferris D.K., Rosenblum J.S. Wortmannin, a widely used phosphoinositide 3-kinase inhibitor, also potently inhibits mammalian polo-like kinase. Chem. Biol. 2005;12:99–107. doi: 10.1016/j.chembiol.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 70.Yee M.C., Fas S.C., Stohlmeyer M.M., Wandless T.J., Cimprich K.A. A cell-permeable, activity-based probe for protein and lipid kinases. J. Biol. Chem. 2005;280:29053–29059. doi: 10.1074/jbc.M504730200. [DOI] [PubMed] [Google Scholar]
- 71.Liu Y., Jiang N., Wu J., Dai W., Rosenblum J.S. Polo-like kinases inhibited by Wortmannin: labeling site and downstream effects. J. Biol. Chem. 2007;282:2505–2511. doi: 10.1074/jbc.M609603200. [DOI] [PubMed] [Google Scholar]
- 72.Liu Y., Wu J., Weissig H., Betancort J.M., Gai W.Z., Leventhal P.S., Patricelli M.P., Samii B., Szardenings A.K., Shreder K.R., Kozarich J.W. Design and synthesis of AX4697, a bisindolylmaleimide exo-affinity probe that labels protein kinase C alpha and beta. Bioorg. Med. Chem. Lett. 2008;18:5955–5958. doi: 10.1016/j.bmcl.2008.08.045. [DOI] [PubMed] [Google Scholar]
- 73.Kalesh K.A., Sim D.S.B., Wang J., Liu K., Lin Q., Yao S.Q. Small molecule probes that target Abl kinase. Chem. Commun. 2010;46:1118–1120. doi: 10.1039/b919888a. [DOI] [PubMed] [Google Scholar]
- 74.Fischer J.J., Graebner O.Y., Dalhoff C., Michaelis S., Schrey A.K., Ungewiss J., Andrich K., Jeske D., Kroll F., Glinski M., Sefkow M., Dreger M., Koester H. Comprehensive identification of staurosporine-binding kinases in the hepatocyte cell line HepG2 using capture compound mass spectrometry (CCMS) J. Proteome Res. 2010;9:806–817. doi: 10.1021/pr9007333. [DOI] [PubMed] [Google Scholar]
- 75.Shi H., Cheng X., Sze S.K., Yao S.Q. Proteome profiling reveals potential cellular targets of staurosporine using a clickable cell-permeable probe. Chem. Commun. 2011;47:11306–11308. doi: 10.1039/c1cc14824a. [DOI] [PubMed] [Google Scholar]
- 76.Lo L.C., Chiang Y.L., Kuo C.H., Liao H.K., Chen Y.J., Lin J.J. Study of the preferred modification sites of the quinone methide intermediate resulting from the latent trapping device of the activity probes for hydrolases. Biochem. Biophys. Res. Commun. 2004;326:30–35. doi: 10.1016/j.bbrc.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 77.Lo L.C., Pang T.L., Kuo C.H., Chiang Y.L., Wang H.Y., Lin J.J. Design and synthesis of class-selective activity probes for protein tyrosine phosphatases. J. Proteome Res. 2002;1:35–40. doi: 10.1021/pr015506a. [DOI] [PubMed] [Google Scholar]
- 78.Zhu Q., Huang X., Chen G.Y.J., Yao S.Q. Activity-based fluorescent probes that target phosphatases. Tetrahedron Lett. 2003;44:2669–2672. [Google Scholar]
- 79.Kumar S., Zhou B., Liang F., Wang W.Q., Huang Z., Zhang Z.Y. Activity-based probes for protein tyrosine phosphatases. Proc. Natl. Acad. Sci. U. S. A. 2004;101:7943–7948. doi: 10.1073/pnas.0402323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kalesh K.A., Tan L.P., Lu K., Gao L., Wang J., Yao S.Q. Peptide-based activity-based probes (ABPs) for target-specific profiling of protein tyrosine phosphatases (PTPs) Chem. Commun. 2010;46:589–591. doi: 10.1039/b919744c. [DOI] [PubMed] [Google Scholar]
- 81.Kumar S., Zhou B., Liang F., Yang H., Wang W.Q., Zhang Z.Y. Global analysis of protein tyrosine phosphatase activity with ultra-sensitive fluorescent probes. J. Proteome Res. 2006;5:1898–1905. doi: 10.1021/pr050449x. [DOI] [PubMed] [Google Scholar]
- 82.Liu S., Zhou B., Yang H., He Y., Jiang Z.X., Kumar S., Wu L., Zhang Z.Y. Aryl vinyl sulfonates and sulfones as active site-directed and mechanism-based probes for protein tyrosine phosphatases. J. Am. Chem. Soc. 2008;130:8251–8260. doi: 10.1021/ja711125p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hu M., Li L., Wu H., Su Y., Yang P.Y., Uttamchandani M., Xu Q.H., Yao S.Q. Multicolor, one- and two-photon imaging of enzymatic activities in live cells with fluorescently quenched activity-based probes (qABPs) J. Am. Chem. Soc. 2011;133:12009–12020. doi: 10.1021/ja200808y. [DOI] [PubMed] [Google Scholar]
- 84.Shreder K.R., Liu Y., Nomanhboy T., Fuller S.R., Wong M.S., Gai W.Z., Wu J., Leventhal P.S., Lill J.R., Corral S. Design and synthesis of AX7574: a microcystin-derived, fluorescent probe for serine/threonine phosphatases. Bioconjug. Chem. 2004;15:790–798. doi: 10.1021/bc0499580. [DOI] [PubMed] [Google Scholar]
- 85.Patricelli M.P., Nomanbhoy T.K., Wu J., Brown H., Zhou D., Zhang J., Jagannathan S., Aban A., Okerberg E., Herring C., Nordin B., Weissig H., Yang Q., Lee J.D., Gray N.S., Kozarich J.W. In situ kinase profiling reveals functionally relevant properties of native kinases. Chem. Biol. 2011;18:699–710. doi: 10.1016/j.chembiol.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]