Abstract
The ability of viruses to transfer macromolecules between cells makes them attractive starting points for the design of biological delivery vehicles. Virus-based vectors and sub-viral systems are already finding biotechnological and medical applications for gene, peptide, vaccine and drug delivery. Progress has been made in understanding the cellular and molecular mechanisms underlying virus entry, particularly in identifying virus receptors. However, receptor binding is only a first step and we now have to understand how these molecules facilitate entry, how enveloped viruses fuse with cells or non-enveloped viruses penetrate the cell membrane, and what happens following penetration. Only through these detailed analyses will the full potential of viruses as vectors and delivery vehicles be realised. Here we discuss aspects of the entry mechanisms for several well-characterised viral systems. We do not attempt to provide a fully comprehensive review of virus entry but focus primarily on enveloped viruses.
Keywords: Endocytosis, Enveloped viruses, Membrane fusion, Virus entry, Viruses, Virus fusion, Virus receptors
1. Introduction
Currently, viruses are used as delivery vehicles in two different ways. Firstly, viral genomes have been manipulated so that foreign genes can be inserted and the modified genome packaged to generate virus particles that resemble the original viruses. These virions then follow the pathways used by the parental virus to infect a target cell. Such systems are well established for retroviral vectors, and for a variety of other virus systems including adeno-, adeno-associated virus, alpha-, vaccinia, and herpes virus. As an alternative strategy, components deemed to be required for viral entry have been isolated and used to facilitate delivery. For example, the influenza envelope glycoprotein, haemagglutinin (HA), has been used to make virosomes (liposomes containing reconstituted viral membrane proteins that can be loaded with peptides, drugs, DNA, etc.) that are as effective for fusion as the original virions 27, 199, 217, 225. Peptides representing an HA fusion motif have been coupled to DNA and used to deliver genes to tissue culture cells 169, 221. Similarly, inactivated adenovirus–ligand–DNA conjugates, in which the virus acts as a carrier and not an infectious particle, have been developed [145]. Although these systems work well in tissue culture, and in some cases in vivo, they lack the specificity and efficiency required for clinical applications. In many cases these limitations are associated with the process of entry of the vehicle into cells. Thus a more detailed understanding of virus entry may suggest ways in which existing delivery systems can be improved or novel ones developed.
2. Strategies for virus entry
Virus entry involves binding of virions to the surface of an appropriate cell and delivery of the viral nucleic acid to the cytoplasm or nucleus of that cell. Frequently viruses are taken up by endocytosis. However, virions within membrane-bound endocytic vesicles remain topologically outside the cytoplasm until the events that lead to penetration are triggered. In this review we will use the term `penetration' to describe the process whereby viral nucleic acid is transferred across a cellular membrane into the cytosol of the target cell. The term `internalisation' is reserved for the endocytosis of virions, a process that may or may not lead to productive infection but does not itself constitute viral entry.
2.1. Enveloped viruses
Viruses have evolved many strategies to invade cells. In a few cases the cellular and molecular details are reasonably well understood, but for most viruses little is known. Nevertheless, the well-studied systems have established paradigms against which other viruses can be compared. The entry strategies used by different viruses are determined primarily by the structure of the virus. Viruses are either enveloped or non-enveloped. Enveloped viruses contain the viral genome and core proteins wrapped within one or more membranes. These membranes are acquired from the host cell during virus assembly and budding [166]. Many enveloped viruses, such as the orthomyxo- (e.g., influenza), paramyxo- (e.g., Sendai), rhabdo- (e.g., vesicular stomatitis virus), retro- (e.g., human immunodeficiency, [HIV]) and alphaviruses [e.g., Semliki Forest virus (SFV)]), contain a single membrane. Other viruses, such as herpes virus, may undergo several budding and fusion steps with different intracellular membrane compartments before finally acquiring a single membrane from the exocytic pathway. In contrast, vaccinia virus (a member of the Poxviruses) acquires several membranes by interaction with different membrane compartments in the infected cell 183, 192, 214. The composition of the viral membrane(s) varies for different viruses and is determined by (i) the complement of virally encoded membrane or envelope proteins and (ii) the content of host cell lipid and proteins. The latter reflects (i) the degree to which cellular proteins are incorporated into the budding virion and (ii) the composition of the membrane system where budding occurs [different viruses assemble and bud in different cellular locations, for example the endoplasmic reticulum (ER), intermediate compartment, Golgi apparatus and plasma membrane [166]].
Enveloped viruses use membrane fusion to penetrate a cell [239]. The viral membrane fuses with a cellular membrane so that the genome-containing viral capsid or core is transferred to the cytosol. Membrane fusion is one of the most frequent reactions occurring in eukaryotic cells. Cell-to-cell fusions, such as sperm–oocyte or myoblast fusion, are topologically closely related to virus entry [240]. However, membrane fusion events occur at higher frequency inside cells as vesicles transporting membrane and cargo between intracellular organelles are formed and consumed [177]. In principle, the cycle of virus assembly and fusion is similar to that of transport vesicle formation and consumption. Recently membrane proteins on the transport vesicle and the target membrane (the so-called vesicle (v)- and target (t)-SNARES) have been implicated as receptors that ensure transport vesicles fuse with the correct compartment [177]. Similarly, the membrane of enveloped viruses must also contain the components required for (i) binding to, and (ii) fusing with a target cell.
2.2. Non-enveloped viruses
Per definition, non-enveloped viruses do not contain membranes (though rotaviruses may transiently acquire a membrane by budding into the ER, only to lose the membrane during subsequent maturation [211]). The viral genome is incorporated into a protein shell in the cytoplasm or nucleoplasm of the infected cell and the assembled virions are then released by cell lysis or, for Rotaviruses, secretion.
Like viral envelopes, the outer protein shell of non-enveloped viruses must contain the necessary molecular equipment for getting into cells. However, unlike enveloped viruses, non-enveloped viruses must rely on strategies other than fusion. In general, proteins and protein complexes are not spontaneously transported across membranes but require complex molecular machines for translocation. The mechanisms through which the protein shells of non-enveloped viruses interact with cell membranes are not well understood, and there is no single principle analogous to fusion. Picornaviruses bind to cell-surface components that can dock into `canyon-like' depressions on the surface of the virion 155, 176. Following endocytosis, the virions appear to generate pores in the endosome membrane that allow the viral RNA to exit from the virion and penetrate to the cytoplasm without complete disassembly of the capsid or disruption of the endosomal membrane [171]. Receptor binding may initiate changes in the capsid structure that facilitate pore formation. In contrast, adenoviruses employ fibres that project from the virion surface to bind to cellular receptors. After initial adsorption to the cell surface, adenoviruses bind to vitronectin-binding integrins and undergo endocytosis through coated vesicles. Within endosomes the low pH activates a lysin on the virus that disrupts the endosome membrane allowing the virion and other endosomal contents into the cytoplasm where the viral capsid is subsequently disassembled 78, 79.
2.3. Cellular sites for penetration: Acid-dependent versus acid-independent penetration
Although enveloped and non-enveloped viruses use different mechanisms to enter cells, penetration can occur either at the cell surface or from endocytic organelles (mainly early and late endosomes). Which of these sites is used varies for different virus families. The fusion reactions of enveloped viruses are mediated by specific virally encoded envelope proteins. These fusion proteins are displayed on the virion in metastable forms that must be activated to initiate fusion. For alpha-, orthomyxo-, and rhabdoviruses, at least, fusion is triggered by exposure of the fusion proteins to low pH and thus usually occurs only after virions have undergone endocytosis into acidic endocytic organelles. Fusion is activated in early (pH>6.0) or late endosomes (pH<6.0) depending on the pH requirement of the specific envelope protein [102]. For these viruses endocytosis is essential for infection.
Not all enveloped viruses are pH-dependent for entry. Coronaviruses and paramyxoviruses, as well as most retroviruses, including the primate immunodeficiency viruses [HIV-1, HIV-2 and the simian immunodeficiency viruses (SIV)] do not require low pH to activate fusion. The fusion triggers in these cases are unknown but may involve interaction of the viral envelope proteins with specific receptors (see below). Some pH-independent viruses appear to be capable of fusion across a range of pH values 129, 239and may potentially fuse at the cell surface or within endocytic organelles. The pH optimum for cell-to-cell fusion mediated by the envelope protein (Env) of a T cell line-adapted HIV-1 strain is around pH 7.5, well above the pH of endosomes [70], and such strains of HIV-1 enter HeLa-CD4 cells by fusion at the cell surface 156, 165. However, the pH-dependencies of other strains of this virus are unknown and, given the observations of HIV in endocytic organelles in various cell types 22, 74, 173, it remains possible that some may use the endocytic route for infection.
Epstein–Barr virus appears to use an endocytic route to infect lymphocytes, but fuses at the surface of epithelial cells [137]. Similarly, a murine ecotropic retrovirus is apparently pH-independent for entry on some cell types but pH-dependent on others [130]. The causes of these differences are unclear. Penetration may depend on conditions other than the pH of intracellular organelles. For example, endosomal proteases have been suggested to activate Moloney murine leukemia virus fusion [4]. Thus although pH-dependent viruses only penetrate from endocytic organelles, it should not be assumed that pH-independent viruses only penetrate at the cell surface.
2.4. Weak bases and other agents that raise the pH of acidic organelles
For many pH-dependent viruses entry has been investigated using weak bases, ionophores or specific inhibitors of vacuolar-type H+-ATPases (vATPases, the enzymes responsible for acidification of intracellular organelles) such as Bafilomycin A and Concanamycin A. Though their modes of action differ, all these agents raise the pH of acidic organelles and inhibit pH-dependent viral penetration 85, 100. They do not directly affect pH-independent viruses [129], though they may influence infection indirectly. Modulation of endosomal pH, for example, can affect vesicular transport between endocytic compartments. The weak base primaquine interferes with recycling from endosomes 89, 204, while neutralisation of endosomes appears to inhibit transport from early to late endosomes under some circumstances [39]. Blockade of this latter step, using specific antibodies, can inhibit vesicular stomatitis virus infection [242]. Thus perturbation of intracellular transport by agents that neutralise intracellular organelles may prevent viruses from reaching sites necessary for penetration or infection. The pH-neutralising agents can also perturb the assembly of viruses by neutralising exocytic compartments in cells (see, for example, Ref. [129]). Thus entry experiments that use release of infectious virus as a read-out should be designed so that the agents are only present during the time of virus entry.
3. Receptors and adsorption
Virus entry is initiated when a virion first binds to the surface of a potential host cell. The events that occur during this first encounter are unclear and likely to vary for different viruses. Viral attachment is mediated on the one hand by binding proteins exposed on the surface of the virus particle (we refer to these as viral attachment proteins—VAPs), and on the other by `virus receptors' expressed on the target cell (Fig. 1 ). The idea that specific cell surface components could be used as `virus receptors' was initially proposed to explain viral tropism—the capacity of a virus to replicate in specific types of cells, i.e. to enter them and subsequently produce infectious progeny virus. Although the use of specific receptors accounts for viral tropism in many instances, post entry effects can also be responsible (see, for example, Refs. 23, 178).
Fig. 1.
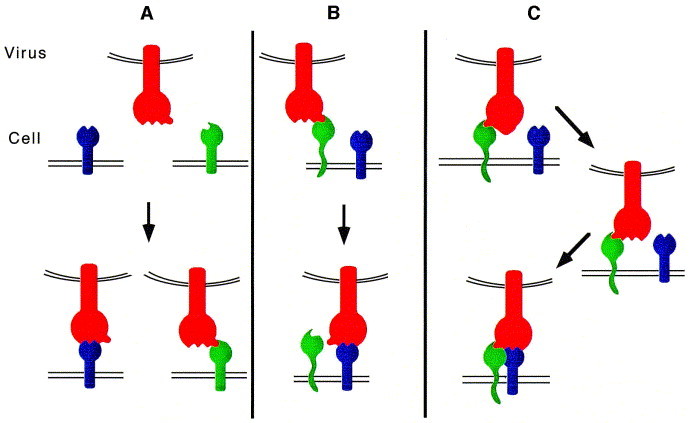
Attachment of virus particles to cell surface molecules. (A) Direct high affinity binding of a viral attachment protein (VAP) (red) to primary receptors (blue and green). The VAP depicted here has two receptor-binding sites, which allows it to attach to alternative receptors, expressed on different cell types. Examples: HIV-1 which can bind to both CD4 and galactosyl ceramide and SFV which can bind to MHC class I and an unknown receptor. (B) Adsorption of virus particles to the target cell surface by binding of the VAP to `pre-receptor' molecules (green), followed by high affinity binding to a primary receptor molecule (blue). Examples: recruitment of HSV by heparan sulphate and subsequent transfer to the tumour necrosis factor/nerve growth factor receptor homologue, and recruitment of adenovirus by an unknown receptor and transfer to the vitronectin receptor. (C) High affinity binding of a VAP to a receptor (green) induces conformational changes leading to the exposure of binding sites for a co-receptor (blue). Examples: primate lentiviruses (HIV-1, HIV-2 and SIV) which bind to CD4 and chemokine receptor co-receptors (NB: some tissue culture variants of HIV may be able to by-pass the CD4 binding step and interact directly with the chemokine receptor molecule (see Fig. 2; [62])
3.1. Viral attachment proteins (VAPs)
Enveloped viruses attach to cell surface receptors through one or more of their envelope glycoproteins. Many viruses contain just a single species of envelope glycoprotein complex that must mediate both the receptor binding and fusion functions, e.g. the spike glycoprotein complex of alphaviruses, and the trimeric glycoprotein (G protein) of rhabdoviruses. The primate lentiviruses carry one species of virally encoded envelope protein (Env—gp120/gp41), but pick up a range of cellular proteins during assembly, including MHC class II antigens [9]. Although not essential, these additional proteins may facilitate binding to a host cell and may be targets for antibodies with apparent antiviral activities [9]. In the paramyxoviruses, binding and fusion are performed by different proteins, and more complex viruses such as herpes, may carry several different VAPs and fusion proteins [184].
To date the best-characterised VAP is influenza virus HA. A pocket in the globular head of the HA1 subunit binds the terminal sialic-acid residue of oligosaccharides attached to cell surface glycolipids and glycoproteins. The 3D structure of HA (from the X-31 strain of influenza virus), complexed with sialyllactose, has been resolved by X-ray diffraction. In this structure sialic acid completely fills the pocket and is co-ordinated by residues that line the cavity [231]. The receptor interactions with canyon-like sites on the surfaces of picornaviruses, in particular rhinoviruses, have also been structurally determined by crystallography and electron microscopy 155, 176. For many other viruses, however, information on receptor binding is less direct. For example, extensive studies of HIV-1 Env using soluble forms of the major receptor for the virus, CD4, and anti-Env monoclonal antibodies have identified continuous and discontinuous epitopes that are likely to be involved in CD4 binding (Ref. [142]and references therein). These studies suggest the CD4 binding site is assembled from different regions of the gp120 molecule, some of which are far apart on the linear sequence.
VAP–receptor binding interactions have been explored using intact virus and soluble forms of the VAPs and of the receptors. Of these the HIV gp120–CD4 interaction is among the most intensely studied. With a number of different gp120 preparations the dissociation constant for monomeric gp120(SU) binding to CD4 is in the nanomolar range [191]. For a soluble form of the receptor, sCD4, binding to oligomeric SU anchored to TM on a cell 53, 104or virion surface [140], it is somewhat higher. However, as yet there is no method to translate these binding constants into the functional affinity of whole virions for receptor-positive cells because of the potential multivalency of that interaction.
3.2. Virus receptors
Table 1 lists the cell surface components for which a role in enveloped virus entry has been established. This table indicates that viruses can use a variety of different surface moieties including glycoproteins, glycolipids and phospholipids, though the majority are glycoproteins. Of these, representatives of most classes of protein can be used; single pass type 1 integral membrane proteins (e.g., CD4), multispanning proteins (e.g., amino acid transporters) and glycophosphatidylinositol (GPI)-linked proteins [e.g., the low density lipoprotein receptor (LDLR)-related protein]. Many virus receptors are members of the immunoglobulin super family, molecules that are often implicated in recognition events; others are transporters, components of the glycocalyx, adhesion molecules, etc. Among the receptors for retroviruses alone, molecules such as CD4, multimembrane-spanning transporters and a glycolipid-anchored homologue of the LDLR are used by viruses with similar SU–TM complexes. This disparity notwithstanding, one unifying trait among these interactions has been suggested: three crucial residues in CD4, the ecotropic MuLV receptor (cationic amino acid transporter) and the ALV receptor (LDLR-related protein) encompass one aromatic residue and at least one charged residue which may reflect similarities in well-adapted modes of viral receptor binding 233, 256, 261.
Table 1.
Receptors for enveloped viruses
| Protein family/class | Receptor | Virus | Virus family | Ref. |
| Immuno-globulin super-family | CD4 | HIV-1, -2, SIV | Retroviridae | 46, 105 |
| HHV7 | Herpesviridae | [117] | ||
| CEA-like, murine biliary glycoprotein | Murine hepatitis virus | Coronaviridae | [250] | |
| Class I MHC | SFV | Togaviridae | [86] | |
| MCV | Herpesviridae | [254] | ||
| Coronavirus OC43 | Coronavirdae | [41] | ||
| Class II MHC | LDV | Arteriviridae | [92] | |
| Visnavirus | Retroviridae | [47] | ||
| Integrins | High-affinity laminin receptor | Sindbis virus | Togaviridae | [228] |
| Regulators of complement activation | CR2 | EBV | Herpesviridae | 66, 68, 148 |
| CD46 | Measles virus | Paramyxoviridae | 55, 146 | |
| Multimembrane-spanning transporters | ||||
| Phosphate transporters | Glvr-1 | GALV, FeLV-B, SSAV | Retroviridae | 93, 233 |
| Ram-1 | MLV-A | Retroviridae | [136] | |
| Cationic amino-acid transporter | ecoR | MLV-E | Retroviridae | [2] |
| 4-span transmembrane protein family | CD9 | FIV | Retroviridae | [248] |
| 7-span transmembrane G-protein | Chemokine receptors | HIV-1, HIV-2, SIV, FIV | Retroviridae | 16, 33, 35, 54 |
| coupled receptors | (CCR5, CCR3, CCR2b, CXCR4) | 57, 62, 65, 120, 249 | ||
| LDL-receptor protein family | ALVA-R, ALSV-A | Retroviridae | [13] | |
| Receptor tyrosine kinase family | EGF receptor | Vaccinia virus | Poxviridae | [63] |
| TNF/NGF receptor homologue | HSV | Herpesviridae | [139] | |
| (also contains Ig-like | ||||
| extra-cellular domain) | ||||
| Zinc-dependent protease | CD13 (aminopeptidase) | Human corona virus 229E (TGEV) | Coronaviridae | 51, 255 |
| Transmitter-gated channel | Acetylcholine receptor | Rabiesvirus | Rhabdoviridae | 108, 109 |
| Others | BLV Receptor 1 | BLV | Retroviridae | [12] |
| Carbohydrate moieties of glycoproteins | Sialic acid | Sendai virus | Paramyxoviridae | [208] |
| and glycolipids | Influenzavirus | Orthomyxoviridae | 87, 88 | |
| Heparan sulphate | HSV | Herpesviridae | [253] | |
| HCMV | [42] | |||
| Galactosyl-ceramide (alternative receptor) | HIV-1 | Retroviridae | 17, 82 | |
Abbreviations: AL(S)V, avian leukaemia (sarcoma) virus; BLV, bovine leukaemia virus; CCR/CXCR, CC and CXC chemokine receptors; CD4,9, etc., cluster of differentiation antigen; CEA, carcinoembryonic antigen; CR2, complement receptor 2; EBV, Epstein–Barr virus; EGF, epidermal growth factor; FeLV, feline leukaemia virus; FIV, feline immunodeficiency virus; HCMV, human cytomegalovirus; HHV7, human herpes virus 7; HIV, human immunodeficiency virus; HSV, herpes simplex virus; GALV, gibbon ape leukaemia virus; LDV, lactate dehydrogenase virus; MCV, murine cytomegalovirus; MHC, major histocompatibility complex; MLV, murine leukaemia virus; NGF, nerve growth factor; SFV, Semliki Forest virus; SIV, simian immunodeficiency virus; SSAV, simian sarcoma-associated virus; TGEV, transmissible gastroenteritis virus; TNF, tumour necrosis factor.
Overall there are no clear patterns in the usage of particular classes of cell surface molecules by specific viruses or virus families, and it is perhaps surprising that pH-dependent viruses are not restricted to using receptors containing well characterised endocytosis motifs. Indeed, poliovirus infection, which is pH-dependent, can be mediated by a poliovirus receptor that lacks the cytoplasmic tail [107]. This suggests that cell surface molecules, regardless of whether they contain an endocytosis signal, will be internalised when cross-linked by multiple VAPs.
In some cases the viral binding site on a receptor may coincide with the binding site for the physiological ligand, e.g. Epstein–Barr virus and complement factor C3dg bind to overlapping sites on complement receptor 2 144, 147. Chemokines that bind the co-receptors for HIV (see below) can inhibit virus entry 19, 40, 52, 57, 152, though ligand-induced receptor endocytosis may be involved in this process [187]. The partial inhibition of the physiological function of virus receptors by VAP binding has been described for the amino-acid and phosphate transporter molecules that serve as receptors for several retroviruses 95, 153, 227.
Virus receptors may just tether virions to the cell surface. However, in a number of cases specific VAP–receptor interactions are required to facilitate viral entry. Specific receptor interactions may be required to initiate conformational changes, or other events, leading to fusion and/or penetration. For example, the interaction of poliovirus with a soluble form of its receptor leads to partial dissociation of the viral capsid structure [107], suggesting that docking to the receptor may have some role in unlocking the capsid structure to allow the viral RNA out.
In addition to the use of cell surface components in entry, there are examples of VAPs that bind specifically to cellular receptors that do not function in viral entry. For example, the Friend erythroleukemia virus VAP binding to the erythropoietin receptor can activate the receptor, but cannot mediate infection [111].
3.3. Alternative receptors
In many systems, viruses appear to bind directly to the cell surface molecules that mediate internalisation or penetration. These can be considered `primary receptors'. In the absence of these primary receptors viruses may use other cell surface components for entry (Fig. 1). These components may be less efficient than the primary receptor. Nevertheless, they are competent for entry. Such molecules can be regarded as `alternative receptors'. For SFV, MHC class I antigens have been proposed as receptors [86], yet this virus clearly infects MHC class I negative cells [154], suggesting that at least one alternative receptor exists. For the related Sindbis virus the laminin receptor has been implicated in virus attachment and entry [228]but it is unclear whether this molecule also binds SFV.
For most primate lentiviruses, CD4 and a chemokine receptor are required for entry. CD4 is sufficient to allow virus binding to cells, but a chemokine receptor is required for fusion and infection and can be regarded as a co-receptor for entry (see below). Some strains of HIV, however, are able to infect CD4 negative cells. These viruses can use CXCR4, CCR3 or an orphan chemokine receptor V28 for entry 62, 174. In these situations the chemokine receptors could be regarded as alternative receptors (provided they alone mediate binding and fusion). HIV-1 can also use galactosylceramide to infect some neural and intestinal CD4 negative cell lines 17, 82. Whether this glycolipid mediates entry itself or requires additional factors is unclear. However, it is regarded as an alternative, albeit inefficient, HIV receptor.
3.4. Co-receptors
For some viruses it is apparent that an initial receptor interaction has a tethering role and that additional cell surface molecules must then be recruited for subsequent steps in entry (Fig. 1, Fig. 2 ). The exact nature of these additional interactions again varies for different viruses. For HIV, interactions with chemokine receptors post-CD4 binding appears to be essential for fusion (see below). For Herpes simplex virus (HSV), heparan-sulphate glycosaminoglycans can function to recruit the virus [253]but a novel tumour necrosis factor/nerve growth factor receptor homologue appears to be required for entry [139]. Similarly, initial high affinity binding of adenovirus to unknown receptors is mediated by the viral fibre protein. The virus then uses Arg–Gly–Asp repeats in the penton base protein to bind to vitronectin receptors (αvβ3 and αvβ5). Conformational changes in the vitronectin receptor are required for ligand-induced receptor internalisation 157, 158. It is unclear whether the penton base protein induces similar changes or triggers endocytosis by an alternative mechanism [243]. Thus although the virus is able to adhere to the cell surface, a second receptor is essential for endocytosis and infection. In cases such as these, we suggest that the term `co-receptor' is used when there is evidence for direct interaction with the virions, and `co-factor' when the role of the second component is indirect or unknown.
Fig. 2.
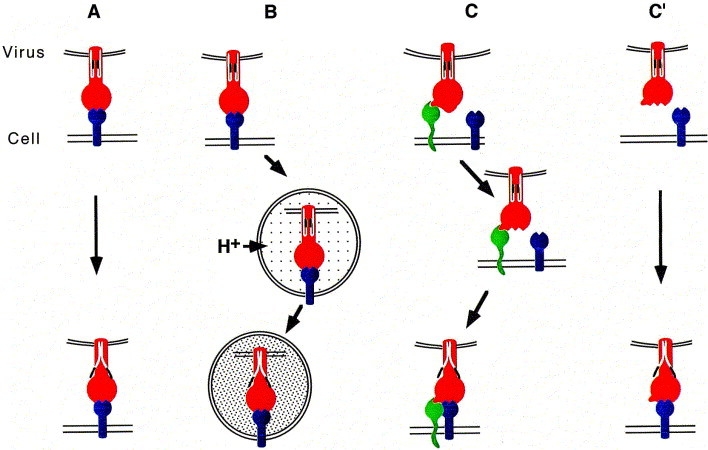
Transition of VAP/fusion proteins to the fusogenic state after VAP binding. (A) Conformational changes in the VAP/fusion protein are triggered through interaction with the receptor. These changes induce fusion competent states of the fusion protein, as illustrated by the exposure of N-terminal fusion peptides (black). Examples: induction of avian leukaemia virus (ALV) fusion through binding to the LDLR-related protein. (B) Low pH-induced fusion: pH-dependent viruses (e.g., influenza virus, VSV and SFV) undergo receptor-mediated endocytosis after binding to cell surface receptors and fuse within intracellular organelles. These organelles are acidified by vacuolar H+ATPases. (C) Two step activation of a VAP/fusion protein. Interaction with a primary receptor exposes binding sites for a co-receptor. Upon binding of the co-receptor the VAP/fusion protein changes to the fusogenic state. Example: primate lentiviruses (HIV-1, -2 and SIV). (C′) In contrast to (C), some HIV Env proteins [62]have a capacity to interact with the co-receptor (blue) alone without the requirement of the primary receptor. Binding to the primary receptor can still occur but is not obligatory.
3.4.1. Co-receptors for the primate lentiviruses
Soon after the identification of CD4 as a receptor for HIV-1 it became apparent that additional cell surface components were required for fusion and infection. Expression of human CD4 in murine NIH 3T3 cells, for example, failed to elicit infection, whereas expression in HeLa cells rendered these cells permissive. Together the data suggested that HIV-1 required at least one other factor for infection 10, 34, 56, 119. It was also recognised that there might be multiple accessory factors as the primate lentiviruses were shown to differ in their capacity to infect different CD4+ve cells. HIV-1 isolated early in the course of infection can usually infect macrophages and primary T cells, but seldom T-cell lines. In contrast, many T-cell line-adapted strains of virus, or viruses isolated from patients with advanced disease, do not infect macrophages [143].
The accessory factors were identified during 1996 and found to be members of a family of seven-transmembrane, G-protein-coupled, chemokine receptors (see Ref. [234]). The initial suggestion that chemokine receptors might be involved in virus entry came from the identification of CC (β) chemokines (RANTES, MIP-1α, and MIP-1β) as HIV-suppressive factors secreted by CD8+ve T cells [40]. Furthermore, lymphocytes from individuals who had been multiply exposed to HIV-1 but remained uninfected showed a resistance to infection by macrophage tropic forms of HIV-1 that was associated with increased secretion of RANTES, MIP-1α, and MIP-1β [164]. Subsequently, the chemokine receptor now called CXCR4 (previously termed LESTR, HUMSTR and Fusin) was identified as an entry co-factor for T cell line-adapted HIV-1 16, 65. CXCR4 has since been shown to bind the CXC (α) chemokine stromal cell derived factor 1[SDF-1] 19, 153. Other chemokine receptors were later identified as co-receptors for various strains of HIV-1, HIV-2 and SIV. In particular, CCR5, which binds RANTES, MIP-1α, and MIP-1β, is used by primary macrophage tropic strains of HIV-1 and by SIV viruses 33, 35, 54, 57, 120, while CCR3 and CCR2b can be used by dualtropic virus (virus which can infect both lymphocytes and macrophages). For a discussion of chemokines and chemokine receptors see Refs. 170, 172.
The viruses that can be isolated early in infection, and that appear to be responsible for transmission, are macrophage tropic and utilise CCR5 as a co-receptor 35, 52, 57, 179, 180. A mutant allele of CCR5 has been identified. This mutation causes a frameshift that truncates CCR5 after the fourth transmembrane domain and the mutant protein is not expressed on the cell surface. Thus CD4-positive cells from homozygotes for the CCR5 mutation are generally not susceptible to infection by macrophage tropic viruses 113, 182(though they can be infected by T cell line-adapted viruses). Together, these findings indicate that CCR5 is important for HIV-1 transmission, and that infection of a population of CCR5 expressing cells is usually required for the virus to establish infection.
The precise mode through which these molecules function together with CD4 to facilitate virus entry is unclear. As discussed above, the primate lentiviruses are all pH-independent for entry 129, 130, 201. The organisation of the fusogenic envelope glycoprotein (Env) of these viruses resembles that of influenza HA (see below). During fusion Env (which contains a membrane bound subunit—gp41 or TM, and a non-covalently associated soluble subunit—gp120 or SU) must undergo a conformational change which at least functionally resembles the acid-induced conformational changes that trigger HA mediated fusion. However, the conformational changes in Env occur at neutral pH. CD4 binding can induce conformational changes in HIV-1 Env that lead to exposure of epitopes associated with gp41 and the V3 loop of gp120 61, 183, and may modulate the interaction between gp120 and gp41 [141]. How these changes relate to the fusion is still under investigation. However, a view is emerging that CD4-induced conformational changes in Env, including reorientation of the V3 loop, allow the Env/CD4 complex to bind to a given chemokine receptor 215, 252and that the chemokine receptor can then complete the conformational changes in Env required for fusion (Fig. 2). In many respects it is likely that the chemokine receptors are essential for virus entry, as some tissue culture strains of virus can dispense with the need for CD4 but still require an appropriate chemokine receptor [62]. Thus the role of CD4 is to recruit the virus to the cell surface and induce the formation of the chemokine receptor binding site on gp120/SU. While it is now well established that Env, CD4 and a chemokine receptor are generally essential in HIV entry, a specific role for additional cell-surface molecules cannot yet be ruled out.
4. Post-binding events
The adsorption of virus to the cell surface and interaction with specific receptors initiates the events that lead to fusion and penetration. For viruses that undergo fusion at the cell surface, receptor engagement may initiate the molecular rearrangements leading to penetration. For viruses that fuse intracellularly, binding should result in uptake of virions into endocytic vesicles.
4.1. Penetration at the cell surface
The plasma membrane is the principal barrier a virus must cross to access the cell. However, this membrane may not be the only obstacle that the virus has to tackle. Many cells contain a highly developed cortical cytoskeleton below the plasma membrane (see Ref. [123]). This complex of actin filaments and actin binding proteins can be up to 100 nm in depth and has the capacity to exclude ribosomes and other organelles from the area immediately adjacent to the plasma membrane. Eukaryotic ribosomes have sedimentation coefficients of 80S and are approximately 4.2×106 kD in size; by comparison the pre-integration complex of HIV has a sedimentation coefficient of about 160–300S. Thus the cortical cytoskeleton has the potential to restrict the movement of incoming viral capsids to regions deeper in the cell. SFV that has fused at the surface of baby hamster kidney cells (after a transient drop in the pH of the medium to activate fusion) will infect these cells 85, 123. However, when the same manipulation is performed on Chinese hamster ovary cells the virus does not infect, though it can do so through the endocytic pathway [123]. The cortical structures are developed to different extents in different cell types. For example, in BHK cells the cortex is not well developed but in CHO cells it can be seen in electron micrographs.
If the cortex is a barrier to penetration, viruses will have developed strategies to by-pass this structure. The endocytic route may provide one such mechanism. Though the molecular basis is not established, endocytic vesicles can pass through the cortex. In general, the endocytic pathway is a hostile environment for a virus, lysosomes being a primary site of cellular degradation. The need to penetrate the cortex may be one of the driving forces that has led viruses to use low pH as a means to delay triggering fusion until after endocytosis. Thus even for pH-independent viruses, endocytosed virions may be at an advantage, as far as traversing the cortex is concerned, over virions that fuse at the cell surface.
How do viruses that fuse at the cell surface penetrate the cortex? At present there is little information. In HIV, the virion-associated protease is known to cleave cellular proteins in addition to its own Gag gene products. Among these cellular proteins are actin and several actin-binding proteins 1, 185, 186, 187, 213. Perhaps lentiviruses use their protease to penetrate the cortex.
4.2. Endocytosis
Most cells have the capacity for endocytosis through several distinct mechanisms. Although endocytic routes for virus entry have long been proposed, it only became clear that endocytosis could be essential for virus infection when the pathway of SFV entry was determined [84].
4.2.1. Clathrin-coated vesicles
Clathrin-coated vesicles are about 100 nm in diameter and form by invagination of coated pits—plasma membrane domains coated with a complex of clathrin and AP2 adaptors. Depending on the cell type, many hundreds of these vesicles may form each minute through most of the cell cycle and are the main conduit for receptor-mediated endocytosis, fluid-phase endocytosis and constitutive membrane turn over [229]. Following budding, the clathrin coat is removed through the action of an uncoating ATPase and the vesicles fuse with early endosomes. From endosomes, membrane and content can be recycled to the cell surface or delivered to other cellular destinations, including late endosomes and lysosomes, the Golgi apparatus and, in specialised cells, synaptic vesicles, MHC class II-containing compartments and alternative plasma membrane domains (see Ref. [80]for review). Early endosomes are acidified to approximately pH 6, through the membrane-associated vATPase, and late endosomes and lysosomes can be more acidic. Internalised ligands en route to lysosomes will transit increasingly acidic compartments. Viruses may exploit this pH gradient to facilitate their delivery to specific sites in the cell [102].
The size of particles that can be internalised through coated vesicles is likely to be limited. SFV virions which are 65 nm in diameter are easily accommodated in coated vesicles. The larger virions of influenza virus and HIV (approx. 100 nm diameter) also enter these vesicles, as do those of rhabdoviruses such as VSV. However, these latter elliptical virions, for which the long axis is approximately 150 nm, tend to be internalised slowly compared to SFV, possibly reflecting some difficulty in entering coated vesicles [127].
Non-clathrin mediated endocytosis has also been implicated in constitutive endocytosis [219]. The molecular mechanisms involved in the formation of these vesicles are not well characterised. In some situations this pathway appears to be upregulated in response to inhibition of clathrin-mediated endocytosis 45, 48. It is unclear what the basal level and function of the pathway is in non-perturbed cells. In BHK at least, clathrin-coated vesicles mediate the bulk of constitutive endocytosis [124].
4.2.2. Phagocytosis
Many cells can take up particulate ligands by phagocytosis [15]. Although most prominent in specialised cells, such as monocytes and neutrophils, phagocytosis is not limited to these cells. The process is receptor-mediated and is initiated when appropriate cell surface receptors are complexed by polyvalent ligands. Receptor ligation leads to the activation of a signalling cascade involving tyrosine kinases and other enzymes that drive the polymerisation of actin on the cytosolic face of the membrane adjacent to the adsorbed particle. This actin polymerization pushes membrane around the particle, allowing the engagement of more receptors and culminating in the engulfment of the particle. Phagocytosis is strictly ATP dependent and can be blocked by inhibitors of tyrosine kinase activity and actin polymerisation.
In general, phagocytosis is involved in the uptake of large particles, such as cells and opsonised bacteria, that cannot be included within clathrin-coated vesicles. Whether it has a role in the entry of viruses is unclear. Influenza virions have been seen in uncoated vesicles with closely opposed vesicle membranes reminiscent of phagocytic vesicles [163], and it is possible that larger virions such as the poxviruses are internalised through phagocytic vesicles. Following phagosome formation these vesicles fuse with endosomes and lysosomes to acquire vATPase and enzymes capable of hydrolysing the phagocytic content. The acquisition of the vATPase again causes the phagocytic vesicles to become acidic, thereby enabling the fusion potential of acid-dependent viruses to be activated.
4.2.3. Macropinocytosis
Macropinocytosis involves the formation of, usually, large vesicles in regions of plasma membrane ruffling [209]. The processes can be induced by treating cells with growth factors or phorbol esters, but appears to be constitutively active in macrophages and dendritic cells, where it has been implicated in a pathway for presentation of exogenous antigens on MHC class I molecules 150, 151. As with phagocytosis the formation of macropinocytic vesicles involves actin polymerization, and can lead to delivery of internalised ligands to late endosomes and lysosomes. However, the process is not ligand-driven nor does it appear to require activation of specific receptors. The role of macropinocytosis in viral infection is unknown. However, constitutive macropinocytic activity may predispose monocytes and dendritic cells to infection by viruses such as HIV 30, 106.
4.2.4. Caveolae
Caveolae are small 50 nm flask-shaped plasma membrane invaginations that have long been recognised in capillary endothelia and other cells. Recent evidence suggests these structures are more widespread and that they have a role in the clustering and signalling pathways associated with GPI-linked and other membrane proteins [112]. The integrity of caveolae appears to depend on the presence of cholesterol and a cholesterol-binding protein Vip21/caveolin [161]. Initially, it was proposed that these structures might transiently invaginate, in a process termed photocytosis [6]. More recently it has become apparent that caveolae can undergo actin-dependent internalisation in cells treated with the phosphatase inhibitor okadaic acid [162]. The role of this internalisation is uncertain and there is no evidence that caveolae are delivered to the endocytic pathway. However, internalised caveolae appear to be relocated to the perinuclear region adjacent to the microtubule-organising centre and may interact with the ER [190].
Morphological studies indicated that SV40 is taken up in small vesicles and delivered directly to the ER/nuclear membrane [94]. Recent studies have shown that these vesicles are caveolae 5, 196. SV40 is believed to use MHC class I antigens as receptors 11, 25. Early morphological studies showed that class I antigens could be associated with small non-clathrin coated invaginations of the plasma membrane [91]. In retrospect, these invaginations might have been caveolae. However, class I antigens have also been implicated as receptors for SFV [86], a virus that clearly uses the clathrin-mediated route for endocytosis [124]. Thus MHC class I antigens might internalise through different endocytic routes [118], or different viruses might direct these receptors to specific endocytic carriers, possibly through association with other co-receptor molecules. Although implicated in SV40 entry, a role for caveolae in the uptake of other viruses remains to be established.
4.3. Fusion mechanisms
Regardless of the location, the critical event in penetration for enveloped viruses is fusion. Currently, the fusion reactions triggered by influenza HA in particular but also the spike glycoprotein of SFV have been dissected in considerable detail. The influenza virus system provides the best model for cellular membrane fusion (for a more extensive discussion of viral fusion mechanisms see Ref. 14, 21).
4.3.1. Influenza virus fusion
4.3.1.1. Structure of the influenza virus particle.
The influenza virus envelope carries three different transmembrane proteins (i) HA (approx. 500 copies per virion), (ii) the neuraminidase (NA—approx. 100 copies per virion), and (iii) the M2 protein (a small tetrameric protein present in low copy number per virion) 90, 206. Of these, HA alone is required for fusion.
4.3.1.2. Haemagglutinin (HA) synthesis and structure.
HA is assembled in the ER of infected cells from a single precursor polypeptide (HAO) of approximately 560 amino acids. The protein undergoes N-linked glycosylation, trimerisation and folding within the ER, and only when the protein has folded correctly is it released by the ER quality control system for transport to the Golgi and plasma membrane. The folding and assembly of HA have been studied extensively and involve the combined activities of a set of ER chaperones 24, 122, 210. En route to the cell surface the oligosaccharides are modified to complex forms, and each HAO molecule is proteolytically cleaved by furin or furin-type enzymes in the trans Golgi cisternae or trans Golgi network to generate the HA1 and HA2 subunits. The X-ray structure of HA shows that the mature protein contains two domains (i) the globular heads assembled from HA1 containing the sialic acid-binding sites and (ii) a stalk domain composed primarily of HA2 which supports the head domains. After cleavage, the newly generated N terminus of HA2 contains a stretch of conserved hydrophobic residues that is termed the `fusion peptide'. This segment is crucial for fusion; non-cleaved HA molecules are not fusion-competent. In the neutral pH form of HA, the fusion peptide is buried away from the solvent within the stalk structure about 3.5 nm from the viral membrane. The cleavage in part provides a mechanism to allow the HA trimer to transit the acidic compartments of the exocytic pathway, but it is also likely that correct folding and assembly of trimers can only be achieved with the full length precursor protein. The HA2 sequences form two long anti parallel α helices linked by a loop. This region and its flanking α helices contain several leucine-zipper heptad repeats. In other proteins similar sequences show a propensity to form coiled coils. As discussed below it appears that at least part of the HA conformational change during fusion involves the refolding of this loop region to form a coiled coil. Thus the HA protein can be regarded as being `spring-loaded', i.e. the full length HAO is assembled in a metastable conformation that can refold, after cleavage, when a critical stimulus (exposure to low pH) is applied.
4.3.1.3. HA-mediated membrane fusion.
HA thus forms 13.5 nm rod-like structures that are expressed on the cell surface and incorporated into virions during assembly. Many features have made this protein an ideal tool to study protein-induced membrane fusion. The protein can be purified to homogeneity from virus suspensions and is fusion-competent when reconstituted into artificial membranes or when expressed alone in cells in the absence of other viral proteins. Furthermore, a water soluble bromelain-cleaved fragment of HA (BHA; the fragment initially used to derive the crystal structure of the neutral pH conformation), though not fusion-competent, will undergo a number of the conformational changes associated with fusion.
Studies directly correlating the kinetics of the pH-induced changes in the conformation of HA with membrane fusion suggest the following sequence of events. Exposure to low pH induces a rapid change in the tertiary structure of HA involving at least two steps. The first is the exposure of the HA2 fusion peptide which may insert into the target membrane [198](and possibly in the viral membrane as well 230, 237). After a short lag phase, lipid mixing can be detected. Finally, the HA1 subunits dissociate, as detected by (i) the loss of epitopes formed by adjacent HA1 subunits, (ii) the exposure of epitopes initially facing the inside of the stem, and (iii) the unmasking of proteolytic sites in HA1 76, 96, 199, 241.
How exactly the changes in HA occur and how they allow the molecule to interact with membranes to induce fusion is still unclear. The position of the fusion peptide at neutral pH close to the viral membrane initially posed a topological problem: How could the fusion peptide interact with the target membrane, when it is located 100 Å away from it? Modelling studies suggested that during fusion, when the loop structures of adjacent HA2 molecules are released by the protonation of residues involved in maintaining the trimeric structure, they would refold to form an energetically more favourable triple stranded coiled coil. This change would create a long extended helix and thereby relocate the fusion peptide 100 Å from within the stalk to the end of the rod where it could potentially interact with a target membrane [31]. Support for this model came with the resolution of the 3D structure of the acid form of a fragment of HA2 [29]. This structure showed reorganisation of the loop to an α-helix and relocation of the fusion peptide to the tip of the molecule. However, additional changes occur at the juxta-membrane end of the molecule. Part of the long helix in the neutral-pH structure is relocated such that it becomes anti-parallel to the rest of the helix. Thus the longitudinal helix of HA2 is prolonged and straightened out at the N terminus and truncated by bending at the C terminus [29]. Evidence supporting these changes in native HA has been gained from electron microscopy [237]. However, information on the structure of the C terminal region of HA2 is still missing and the orientation of the acid form of the protein with respect to the viral membrane is uncertain. The EM studies, like earlier photo-affinity labelling studies [230], suggested that the fusion peptide might interact with the HA-containing membrane, i.e. that at some stage at least the acid form of the protein might be inverted.
Together the data suggest that exposure to low pH might allow the fusion peptides to swing out from their locations within the HA2 stalk, perhaps driven by the initial events involved in refolding the loop region. These events may culminate in the form of the protein seen in the low-pH crystal structure, but the fusion-competent form could be an intermediate in this series of changes. If, after exposure of the fusion peptide, the HA protein is tipped, the fusion peptides could interact with both the target membrane and the virion membrane, in keeping with labelling studies 197, 231, i.e. the protein may adopt a tilted conformation parallel to both the viral and the target membrane 81, 197, 200, 251. Electron paramagnetic resonance studies have suggested that some or all of the coiled coil could insert into membranes [257]. However, photo affinity labelling indicates that only the fusion peptide is inserted [59].
How changes in HA induce the reorganisation of the opposing lipid bilayers is also still unclear. It is unlikely that fusion pores are formed from a single HA trimer 38, 60. Current data suggest that a minimum of three HA trimers is required to form a functional fusion complex [49]and that the transmembrane domain of HA is crucial for fusion. Using cell-to cell fusion assays, an HA molecule anchored to the membrane by a GPI moiety, rather than its usual transmembrane domain, will induce hemi-fusion—a state in which the outer but not the inner leaflets of the bilayers are fused [97]. Experiments with amphipathes that selectively insert into either the inner or outer leaflet of membranes and change the curvature of that membrane, suggest that the normal transmembrane domain is required to exert pressure on the bilayer to drive a hemi-fusion intermediate to full fusion [131]. Full fusion proceeds through the formation of a narrow (1-2 nm diameter) pore that can be detected by the patch clamp technique 194, 195, 216. The initial pore formation follows after a short lag, but precedes lipid mixing [216]. The initial fusion pore flickers open and closed 194, 195before widening further to allow first the movement of lipids and subsequently the aqueous contents of the fusing cells to mix [20].
4.3.2. Semliki forest virus
4.3.2.1. Structure and synthesis.
Limited X-ray crystallographic data exist for the alphaviruses. Nevertheless, structural information has been gained from image analysis of cryo-preserved viruses by EM 72, 220. Alphaviruses, such as SFV and the closely related Sindbis virus, are uniform in size (approximately 65 nm diameter) and regularly shaped. The SFV nucleocapsid comprises one RNA molecule and 240 copies of the capsid (C) protein. The capsid proteins interact with the viral RNA and each other to form an icosahedral nucleocapsid with a T=4 surface lattice 36, 159, 160. The SFV envelope contains 80 spike glycoproteins, each spike consisting of three E2E1 heterodimers [a small peptide, E3, resulting from proteolytic cleavage of P62 (see below) is lost from Sindbis virus spikes but remains non-covalently attached to the SFV spike complex]. The spikes are triangular and form, like the nucleocapsid, an icosahedral shell with a T=4 surface lattice 71, 220, suggesting that each spike glycoprotein heterodimer directly interacts with a capsid protein 132, 207. This interaction appears to be mediated through a conserved tyrosine in the cytoplasmic domain of E2 and a hydrophobic pocket on the capsid protein 188, 189. Chemical cross-linking studies on Sindbis virus suggest that each spike contains an inner core of the three E1 subunits and an outer shell formed by the three E2 subunits, an arrangement confirmed by cryo-electron microscopy 8, 72. A receptor-binding site in the spike complex has not yet been identified. However, a hydrophobic domain in E1 between amino acids 79 and 97 (for SFV) is highly conserved in alphaviruses [98], and is believed to be involved in fusion 58, 110.
As with HA, alphavirus spike proteins are assembled in the ER, and transported to the cell surface when folding is completed. These spike proteins are also synthesised in precursor form. The E1 protein assembles together with the precursor of E2, the P62 protein. Three such dimers subsequently assemble to form a spike protein complex that is transported to the cell surface. Just prior to its arrival at the plasma membrane, the P62 protein is cleaved by a cellular protease to generate the E2 protein found in the mature spike glycoprotein [50]. The properties of the mature E2E1 heterodimers are distinctly different from the precursor P62E1 dimers. Exposure of E2E1 complexes to mildly acidic conditions results in dissociation of the subunits, whereas P62E1 complexes dissociate only upon exposure to pH values of 4.5–5.0 114, 115, 181, 222. Thus P62 appears to stabilise the immature spike complex and prevent premature activation during transport through the mildly acidic compartments of the exocytic pathway.
The cleavage of P62 may also induce a conformational change in the heterodimers, resulting in the formation or exposure of receptor-binding sites: SFV particles, carrying spikes with an uncleaved P62, have a reduced capacity to bind to cells [180]. Thus, proteolytic cleavage of the SFV spike glycoprotein controls both the activation of the viral fusion potential and the viral receptor-binding capacity. The interaction of Sindbis virus with its cell surface receptor may initiate additional conformational changes in the spike glycoprotein 67, 133, 134. Although these changes might have some role in enhancing the binding, or in preparing the spike protein for fusion, they are unlikely to be essential for fusion: SFV, at least, will fuse with pure lipid membranes when the pH is lowered appropriately [238].
4.3.2.2. SFV fusion.
Following exposure to low pH the rearrangements in the SFV spike protein are initiated by the dissociation of the E2 subunits from E1 115, 180, 224. The released E1 monomers then reorganise into homotrimers 28, 223, 224. The masking of trypsin cleavage sites on E1, and changes in the antigenicity of epitopes on E1 28, 100, 101, 180, 224parallel the reorganisation of the spike protein observed morphologically. Time-resolved images of SFV after pH activation indicate that a cavity in the neutral pH spike complex is narrowed by the centripedal movement of the E1 subunit which is extended towards a target membrane. In contrast, the E2 subunits separate [72]. A role for these changes is supported by the ability of a monoclonal antibody that recognises an epitope exposed on the low pH form of E1 to inhibit fusion and infection 72, 223, 224.
The structural changes precede the onset of fusion, as monitored by lipid mixing [28]. Following the conformational changes described above, the virus acquires the capacity to bind to cholesterol-containing lipid vesicles 99, 149, 167, 238. Though SFV will bind to cholesterol-containing liposomes at low pH, it will not fuse with these membranes unless they contain low amounts (about 1 mol%) of sphingolipids 43, 138, 149. Ceramide is the minimal structural component of the sphingolipids that support fusion [149]. SFV E1 interacts with both cholesterol [99]and sphingolipids [138]in a stereo-specific manner. How these lipids promote SFV fusion is not known, but they may promote additional conformational changes that are required for fusion or prevent the inactivation of the fusion-active form of E1.
4.3.3. Other viruses
A pattern that emerges from studies on SFV and influenza virus is that conformational changes induced by low pH lead to the formation of stable trimeric complexes of subunits of the envelope proteins (E1 and HA2, respectively). Studies of the Flavi virus tick-borne encephalitis virus (TBE), for which a 3D structure of the VAP/fusion protein has been resolved [175], suggest that this molecule also rearranges to form stable trimers during low pH-induced fusion 3, 175. In contrast to SFV and influenza virus, in which the spike protein and HA respectively form trimeric structures that project out from the membrane, the TBE envelope protein is a dimer that is oriented parallel to the viral membrane. Thus for this virus, trimer formation during fusion involves extensive reorganisation of the fusion protein subunits.
4.3.3.1. Retrovirus fusion.
In most cases the retroviruses are pH-independent for fusion. As discussed above for HIV, it is likely that some aspect of the envelope protein–receptor interaction activates fusion. Binding of the ALV-A receptor to the viral envelope protein complex has been shown to induce conformational changes in the SU–TM complex [75]. Several of the primary receptors for retroviruses, as well as the co-receptors for the primate lentiviruses, are multi-membrane spanning proteins, suggesting that some property of this class of protein might facilitate retroviral fusion. However, at least two retroviruses (bovine leukaemia virus and avian leukosis virus) do not use multi-membrane spanning protein receptors (Table 1).
Similarities in the organisation of retroviral envelope proteins and influenza HA suggest that aspects of retrovirus and orthomyxovirus fusion may be similar. However, the 3D structure of a native retroviral envelope protein has not yet been solved and current comparisons to HA should be regarded with some caution. Nevertheless, several studies have suggested that the retroviral envelope proteins form trimers 18, 64, 232. The retroviral transmembrane proteins contain N terminal hydrophobic sequences that resemble the HA fusion peptide (Fig. 3 ). Furthermore, domains with heptad repeats have been implicated in fusion 244, 247. Synthetic peptides representing sequences C-terminal to the fusion peptide have a strong propensity to form α helices in solution 103, 246. Hence, it has been speculated that receptor binding can unleash conformational changes that involve the formation of coiled coils and exposure of the TM fusion peptide 18, 64, 116, 245. Synthetic peptides representing the HIV heptad repeats can block virus entry, perhaps by interfering with the ability of these regions to oligomerise during the fusion reaction [128]. The structure of an alpha helical domain from the extracellular portion of HIV-1 TM has been solved crystallographically [32]. This domain forms trimers of two interacting peptides. The N-terminal peptide, which contains heptad repeats, forms a trimeric coiled coil. Around this coiled coil the C-terminal peptides are wound such that they fit into the hydrophobic grooves on the coiled coil. This structure might position the HIV fusion peptide at the top of a protruding six helix bundle such that the fusion peptides could insert into a membrane. The structure shares some features with that of the low pH fragment of influenza virus HA and is suggested to represent the core of the fusion active gp41. Alternatively, the complex may represent the terminal structure of the TM/gp41 ectodomain achieved after a metastable intermediate has initiated membrane fusion.
Fig. 3.
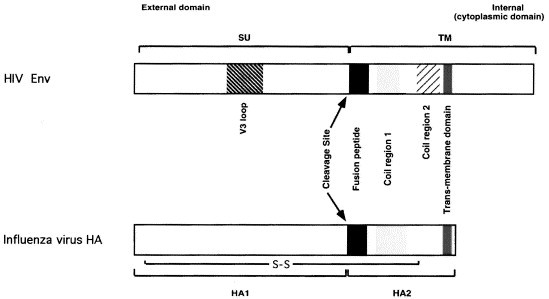
Comparison of the VAP/fusion proteins of HIV and influenza virus. The precursors of the HIV Env protein (top panel) and the influenza virus haemagglutonin (HA) (bottom panel) are aligned schematically to illustrate some hypothetical structure–functional similarities. The two precursor polypeptide chains are represented from their N-termini to the left to their C-termini to the right. The HIV Env encompasses approximately 860 residues and influenza HA 560. The C-termini of both molecules are cytoplasmic, or intravirional, but the cytoplasmic domain of HIV Env is considerably longer than that of HA (as represented by the segments to the right of the trans-membrane domains). During transport to the cell surface both precursors are cleaved as a prerequisite for subsequent fusion activity. The two resulting polypeptide chains of HIV Env, SU/gp120 (surface) and TM/gp41 (transmembrane), are non-covalently associated. Those of influenza virus, HA1 and HA2, are linked by a disulphide bond (intrachain disulphide bonds are not marked). SU and HA1 are responsible for the binding of the respective viruses to cell surface receptors, CD4 and sialic acid. Both receptor-binding sites are created by the juxtaposition in space of conserved residues that are non-contiguous in the amino-acid sequence. In SU of HIV these residues are situated in conserved regions on both sides of the V3 loop. The V3 loop influences the interactions with the chemokine-receptor co-receptors. The cleavage of the precursors creates novel N-termini in the transmembrane proteins. These N-terminal peptides share similar hydrophobic sequences and have been implicated in membrane fusion. C-terminal to the fusion peptides are potential `coil regions', two in the HIV TM extracellular domain (approx. residues 550–580 and 630–660 in the precursor), and one region in HA2 (residues 40–105). These regions have a propensity to form alpha helices and may form coiled-coils as a step in a series of events that activate the membrane-fusing capacities of the two proteins.
5. Early post-penetration events
After fusion the fate of the nucleocapsid varies according to the destination of the virus within the cell. Multiple mechanisms, illustrated by the systems discussed below, demonstrate the ability of different viruses to exploit the properties of host cells for their own special requirements. For example, the DNA genome of HSV must be delivered to the nucleus. Although many of the molecular details remain to be established, it seems that following fusion some of the tegument proteins are removed but the capsid remains intact, binds cytoplasmic dynein and uses the cellular microtubule system to translocate itself to the perinuclear region. In this location the capsids bind directly to the nuclear pore and the DNA is delivered to the nucleus without complete disassembly of the capsid [193].
5.1. Influenza virus
In the case of influenza virus acidification of the viral core is required for cellular infection. The envelopes of influenza virions contain small numbers of M2 protein molecules that function as proton channels 26, 37, 126, 205, 257, 258. On exposure of influenza virions to low pH, proton influx through M2 acidifies the interior of the virion. Acidification of the viral core is presumed to disrupt the pH-sensitive interaction of matrix protein (M1) and the RNA binding nucleoprotein (NP) 126, 259, 260, thereby allowing the viral RNPs to be transported to the nucleus 83, 125, 126. Amantadine, a weak base that accumulates in endosomes, interrupts this sequence of events by blocking the channel function of M2 77, 83, 126, 168, 202, 205, 206, 226.
5.2. SFV
The intracellular part of the alphavirus replication cycle takes place entirely in the cytoplasm of infected cells. Following fusion the nucleocapsids are uncoated through interaction of the viral capsid protein with 60S ribosomal subunits 187, 235, 236. Morphological analysis of infected cells indicates that replication occurs in association with membranes of the late endosomes, lysosomes and ER [69]. Thus the viral RNA appears to remain associated with the same compartments from which the virus fuses.
5.3. HIV
In contrast to most retrovirus, HIV will infect non-dividing cells [203]. This indicates that the HIV proviral DNA, which has been reverse-transcribed from the RNA genome, is able to penetrate the intact nuclear membrane. The HIV pre-integration complex contains the viral matrix (MA) protein [203]. Recent studies have identified nuclear targeting signals in MA similar to those previously characterised in other cellular and viral proteins. MA is linked to the plasma membrane via an N-terminal myristic acid group. However, MA can be phosphorylated on multiple serine and tyrosine residues. Phosphorylation displaces MA from membranes, in a manner similar to that demonstrated for the MARCKS protein [212]. Thus MA-containing viral pre-integration complexes are likely to be released from the plasma membrane by phosphorylation of MA. The nature of the kinase(s) involved in these phosphorylation events is unclear, but serine/threonine and tyrosine kinase activity has been detected in highly purified HIV virions [203]. Thus the virus may package cellular kinases during assembly to facilitate its subsequent targeting to the nucleus.
How the released complex is transported to the nuclear membrane where the nucleophilic signals presumably allow interaction with the nuclear pore is unclear. As discussed above the cortical cytoskeleton may present a barrier that the viral capsid has to traverse (Section 4.1). Other virally encoded proteins including integrase and the product of the vpr gene are also present in the pre-integration complex, though the capsid protein is not. Vpr also appears to be required for infection of non-dividing cells. Nuclear targeting signals have not been identified in this protein, although it is karyophilic [73]. It is not impossible that Vpr and/or other proteins in the pre-integration complex function in bringing the incoming viral pre-integration complex from the plasma membrane to the nucleus through interaction with cellular transport pathways [203].
6. Conclusions
Delivery systems based on retro-, adeno-, adeno-associated, herpes and other viruses have been developed for gene-replacement therapy, intracellular `immunisation', vaccination and other therapeutic purposes. A number of these systems have now reached the stage of clinical trials (for reviews see Refs. 7, 135). Vectors based on retroviruses, adeno- and herpesviruses, which will allow stable integration and expression of a given gene, have proved useful in vitro, but their therapeutic applicability has yet to be demonstrated. The molecular and cellular basis of viral entry and infection is now becoming increasingly well understood, and the knowledge gained from these studies is being used to develop improved vectors. For example, altered tropisms of C-type retroviruses have been attempted by creating chimeras between the outer envelope protein, SU, and ligands with the desired receptor specificity. While some of these viruses are not infectious because of blocks at post-receptor-binding steps of entry [44], others have given expanded viral tropism to, for example, integrin- and human MHC class I-expressing cells 121, 218. Virosomes containing reconstituted influenza, Sendai, VSV or EBV fusion proteins have proven to be effective delivery vehicles for DNA, drugs, peptides and toxins in tissue culture. Initial studies suggest some of these also work in animal models for the delivery of peptides to the cytoplasm of cells and presentation on MHC class I antigens. Future studies will undoubtedly allow more effective and more useful vectors to be developed.
Acknowledgements
The authors are supported by grants from the United Kingdom Medical Research council.
References
- 1.Adams L.D, Tomasselli A.G, Robbins P, Moss B, Heinrikson R.L. HIV-1 protease cleaves actin during acute infection of human T-lymphocytes. AIDS Res. Hum. Retroviruses. 1992;8:291–295. doi: 10.1089/aid.1992.8.291. [DOI] [PubMed] [Google Scholar]
- 2.Albritton L.M, Tseng L, Scadden D, Cunningham J.M. A putative murine retrovirus receptor gene encodes a multiple membrane spanning protein and confers susceptibility to virus infection. Cell. 1989;57:659–666. doi: 10.1016/0092-8674(89)90134-7. [DOI] [PubMed] [Google Scholar]
- 3.Allison S.L, Schalich J, Stiasny K, Mandl C.W, Kunz C, Heinz F.X. Oligomeric rearrangement of tick-borne encephalitis virus envelope proteins induced by an acidic pH. J. Virol. 1995;69:695–700. doi: 10.1128/jvi.69.2.695-700.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andersen K.B, Skov H. Retrovirus-induced cell fusion is enhanced by protease treatment. J. Gen. Virol. 1989;70:1921–1927. doi: 10.1099/0022-1317-70-7-1921. [DOI] [PubMed] [Google Scholar]
- 5.Anderson H.A, Chen Y, Norkin L.C. Bound simian virus 40 translocates to caveolin-enriched membrane domains, and its entry is inhibited by drugs that selectively disrupt caveolae. Mol. Biol. Cell. 1996;7:1825–1834. doi: 10.1091/mbc.7.11.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderson R.G.W. Potocytosis of small molecules and ions by caveolae. Trends Cell Biol. 1993;3:69–72. doi: 10.1016/0962-8924(93)90065-9. [DOI] [PubMed] [Google Scholar]
- 7.Anderson W.F. Human gene therapy. Science. 1992;256:808–813. doi: 10.1126/science.1589762. [DOI] [PubMed] [Google Scholar]
- 8.Anthony R.P, Brown D.T. Protein–protein interactions in an alphavirus membrane. J. Virol. 1991;65:1187–1194. doi: 10.1128/jvi.65.3.1187-1194.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arthur L.O, Bess J.W, Jr., Sowder R.C.d, Benveniste R.E, Mann D.L, Chermann J.C, Henderson L.E. Cellular proteins bound to immunodeficiency viruses: Implications for pathogenesis and vaccines. Science. 1992;258:1935–1938. doi: 10.1126/science.1470916. [DOI] [PubMed] [Google Scholar]
- 10.Ashorn P.A, Berger E.A, Moss B. Human immunodeficiency virus envelope glycoprotein/CD4-mediated fusion of nonprimate cells with human cell. J. Virol. 1990;64:2149–2156. doi: 10.1128/jvi.64.5.2149-2156.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Atwood W.J, Norkin L.C. Class I major histocompatibility proteins as cell surface receptors for simian virus 40. J. Virol. 1989;63:4474–4477. doi: 10.1128/jvi.63.10.4474-4477.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ban J, Portetelle D, Altaner C, Horion B, Milan D, Krchnak V, Burny A, Kettmann R. Isolation and characterization of a 2.3-kilobase-pair cDNA fragment encoding the binding domain of the bovine leukemia virus cell receptor. J. Virol. 1993;67:1050–1057. doi: 10.1128/jvi.67.2.1050-1057.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bates P, Young J.A, Varmus H.E. A receptor for subgroup A Rous sarcoma virus is related to the low density lipoprotein receptor. Cell. 1993;74:1043–1051. doi: 10.1016/0092-8674(93)90726-7. [DOI] [PubMed] [Google Scholar]
- 14.J. Bentz, Viral Fusion Mechanisms, CRC Press, Boca Raton, FL, 1993.
- 15.Beron W, AlvarezDominguez C, Mayorga L, Stahl P.D. Membrane trafficking along the phagocytic pathway. Trends Cell Biol. 1995;5:101–104. doi: 10.1016/s0962-8924(00)88958-8. [DOI] [PubMed] [Google Scholar]
- 16.Berson J.F, Long D, Doranz B.J, Rucker J, Jirik F.R, Doms R.W. A seven-transmembrane domain receptor involved in fusion and entry of T-cell-tropic human immunodeficiency virus type 1 strains. J. Virol. 1996;70:6288–6295. doi: 10.1128/jvi.70.9.6288-6295.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhat S, Spitalnik S.L, Gonzalez-Scarano F, Silberberg D.H. Galactosylceramide or a molecule derived from it is an essential component of the neural receptor for HIV-1 envelope glycoprotein gp120. Proc. Natl. Acad. Sci. USA. 1991;88:7131–7139. doi: 10.1073/pnas.88.16.7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blacklow S.C, Lu M, Kim P.S. A trimeric subdomain of the simian immunodeficiency virus envelope glycoprotein. Biochemistry. 1995;34:14955–14962. doi: 10.1021/bi00046a001. [DOI] [PubMed] [Google Scholar]
- 19.Bleul C.C, Farzan M, Choe H, Parolin C, Clark-Lewis I, Sodroski J, Springer T.A. The lymphocyte chemoattractant SDF-1 is a ligand for Lestr/fusin and blocks HIV-1 entry. Nature (London) 1996;382:829–832. doi: 10.1038/382829a0. [DOI] [PubMed] [Google Scholar]
- 20.Blumenthal R, Sarkar D.P, Durell S, Howard D.E, Morris S.J. Dilation of the influenza hemagglutinin fusion pore revealed by the kinetis of individual cell–cell fusion events. J. Cell Biol. 1996;135:63–71. doi: 10.1083/jcb.135.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blumenthal R, Schoch C, Puri A, Clague M.J. A dissection of steps leading to viral envelope protein-mediated membrane fusion. Ann. N.Y. Acad. Sci. 1991;635:285–296. doi: 10.1111/j.1749-6632.1991.tb36499.x. [DOI] [PubMed] [Google Scholar]
- 22.Bourinbaiar A.S, Phillips D.M. Transmission of human immunodeficiency virus from monocytes to epithelia. J. AIDS. 1991;4:56–63. [PubMed] [Google Scholar]
- 23.Boycott R, Klenk H.D, Ohuchi M. Cell tropism of influenza virus mediated by hemagglutinin activation at the stage of virus entry. Virology. 1994;203:313–319. doi: 10.1006/viro.1994.1489. [DOI] [PubMed] [Google Scholar]
- 24.Braakman I, Hoover-Litty H, Wagner K.R, Helenius A. Folding of Influenza hemagglutinin in the endoplasmic reiculum. J. Cell Biol. 1991;114:401–411. doi: 10.1083/jcb.114.3.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Breau W.C, Atwood W.J, Norkin L.C. Class I major histocompatibility proteins are an essential component of the simian virus 40 receptor. J. Virol. 1992;66:2037–2045. doi: 10.1128/jvi.66.4.2037-2045.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bron R, Kendal A.P, Klenk H.D, Wilschut J. Role of the M2 protein in influenza virus membrane fusion: Effects of amantadine and monensin on fusion kinetics. Virology. 1993;195:808–811. doi: 10.1006/viro.1993.1435. [DOI] [PubMed] [Google Scholar]
- 27.Bron R, Ortiz A, Wilschut J. Cellular cytoplasmic delivery of a polypeptide toxin by reconstituted influenza virus envelopes (virosomes) Biochemistry. 1994;33:9110–9117. doi: 10.1021/bi00197a013. [DOI] [PubMed] [Google Scholar]
- 28.Bron R, Wahlberg J.M, Garoff H, Wilschut J. Membrane fusion of Semliki Forest virus in a model system: Correlation between fusion kinetics and structural changes in the envelope glycoprotein. EMBO J. 1993;12:693–701. doi: 10.1002/j.1460-2075.1993.tb05703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bullough P.A, Hughson F.M, Skehel J.J, Wiley D.C. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 30.Cameron P.U, Lowe M.G, Crowe S.M, O'Doherty U, Pope M, Gezelter S, Steinman R.M. Susceptibility of dendritic cells to HIV-1 infection in vitro. J. Leukocyte Biol. 1994;56:257–265. doi: 10.1002/jlb.56.3.257. [DOI] [PubMed] [Google Scholar]
- 31.Carr C, Kim P. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell. 1993;73:823–832. doi: 10.1016/0092-8674(93)90260-w. [DOI] [PubMed] [Google Scholar]
- 32.Chan D.C, Fass D, Berger J.M, Kim P.S. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 33.Chen Z, Zhou P, Ho D.D, Landau N.R, Marx P.A. Genetically divergent strains of simian immunodeficiency virus use CCR5 as a coreceptor for entry. J. Virol. 1997;71:2705–2714. doi: 10.1128/jvi.71.4.2705-2714.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chesebro B, Buller R, Portis J, Wehrly K. Failure of human immunodeficiency virus entry and infection in CD4-positive human brain and skin cells. J. Virol. 1990;64:215–221. doi: 10.1128/jvi.64.1.215-221.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P.D, Wu L, LaRosa M.C.R.G, Newman W, Gerard N, Gerard C, Sodroski J. The β-Chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 36.Choi H.-K, Tong L, Minor W, Dumas P, Boege U, Rossmann M.G, Wengler G. Structure of Sindbis virus core protein reveals a chymotrypsin-like serine proteinase and the organisation of the virion. Nature. 1991;354:37–43. doi: 10.1038/354037a0. [DOI] [PubMed] [Google Scholar]
- 37.Ciampor F, Bayley P.M, Nermut M.V, Hirst E, Sugrue R.J, Hay A.J. Evidence that the amantadine-induced, M2-mediated conversion of influenza-A virus hemagglutinin to the low pH conformation occurs in an acidic trans-golgi compartment. Virology. 1992;188:14–24. doi: 10.1016/0042-6822(92)90730-d. [DOI] [PubMed] [Google Scholar]
- 38.Clague M.J, Schoch C, Blumenthal R. Delay time for influenza virus hemagglutinin-induced membrane fusion depends on hemagglutinin surface density. J. Virol. 1991;65:2402–2407. doi: 10.1128/jvi.65.5.2402-2407.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.M.J. Clague, S. Urbe, F. Aniento, J. Gruenberg, Vacuolar ATPase activity is required for endosomal carrier vesicle formation, J. Biol. Chem. (1994) 21–24. [PubMed]
- 40.Cocchi F, DeVico A.L, Garzino-Demo A, Arya S.K, Gallo R.C, Lusso P. Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells. Science. 1995;270:1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 41.Collins A.R. HLA class I antigen serves as a receptor for human coronavirus OC43. Immunol. Invest. 1993;22:95–103. doi: 10.3109/08820139309063393. [DOI] [PubMed] [Google Scholar]
- 42.Compton T, Nowlin D.M, Cooper N.R. Initiation of human cytomegalovirus infection requires initial interaction with cell surface heparan sulfate. Virology. 1993;193:834–841. doi: 10.1006/viro.1993.1192. [DOI] [PubMed] [Google Scholar]
- 43.Corver J, Moesby L, Erukulla R.K, Reddy K.C, Bittman R, Wilschut J. Sphingolipid-dependent fusion of Semliki Forest virus with cholesterol-containing liposomes requires both the 3-hydroxyl group and the double bond of the sphingolipid backbone. J. Virol. 1995;69:3220–3223. doi: 10.1128/jvi.69.5.3220-3223.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cosset F.L, Morling F.J, Takeuchi Y, Weiss R.A, Collins M.K, Russell S.J. Retroviral retargeting by envelopes expressing an N-terminal binding domain. J. Virol. 1995;69:6314–6322. doi: 10.1128/jvi.69.10.6314-6322.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cupers P, Veithen A, Baudhuin P, Courtoy P.J. Clathrin polymerization is not required for bulk-phase endocytosis in rat foetal fibroblasts. J. Cell Biol. 1994;127:725–736. doi: 10.1083/jcb.127.3.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dalgleish A.G, Beverley P.C.L, Clapham P.R, Crawford D.H, Greaves M.F, Weiss R.A. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature. 1984;312:763–767. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- 47.Dalziel R.G, Hopkins J, Watt N.J, Dutia B.M, Clarke H.A, McConnell I. Identification of a putative cellular receptor for the lentivirus visna virus. J. Gen. Virol. 1991;72:1905–1911. doi: 10.1099/0022-1317-72-8-1905. [DOI] [PubMed] [Google Scholar]
- 48.Damke H, Baba T, Warnock D.E, Schmid S.L. Induction of mutant dynamin specifically blocks endocytic coated vesicle formation. J. Cell Biol. 1994;127:915–934. doi: 10.1083/jcb.127.4.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Danieli T, Pelletier S.L, Henis Y.I, White J.M. Membrane fusion mediated by the influenza virus hemagglutinin requires the concerted action of at least three hemagglutinin trimers. J. Cell Biol. 1996;133:559–569. doi: 10.1083/jcb.133.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de Curtis J, Simons K. Dissection of Semliki Forest virus glycoprotein delivery from the trans-Golgi network to the cell surface in permeabilized BHK cells. Proc. Natl. Acad. Sci. USA. 1988;85:8052–8056. doi: 10.1073/pnas.85.21.8052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Delmas B, Gelfi J, L'Haridon R, Vogel L.K, Sjöström H, Norén O, Laude H. Aminopeptidase N is a major receptor for the enteropathogenic coronavirus TGEV. Nature. 1992;357:417–420. doi: 10.1038/357417a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R.E, Hill C.M, Davis C.B, Peiper S.C, Schall T.J, Littman D.R, Landau N.R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 53.Dimitrov D.S, Hillman K, Manischewitz J, Blumenthal R, Golding H. Correlation between kinetics of soluble CD4 interactions with HIV-1-Env-expressing cells and inhibition of syncytia formation: Implications for mechanisms of cell fusion and therapy for AIDS. AIDS. 1992;6:249–256. doi: 10.1097/00002030-199203000-00001. [DOI] [PubMed] [Google Scholar]
- 54.Doranz B.J, Rucker J, Yi Y, Smyth R.J, Samson M, Peiper S, Parmentier M, Collman R.J, Doms R.W. A dual-tropic, primary HIV-1 isolate that uses Fusin and the β-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 55.Dorig R.E, Marcil A, Chopra A, Richardson C.D. The human CD46 molecule is a receptor for measles virus (Edmonston strain) Cell. 1993;75:295–305. doi: 10.1016/0092-8674(93)80071-l. [DOI] [PubMed] [Google Scholar]
- 56.Dragic T, Charneau P, Clavel F, Alizon M. Complementation of murine cells for human immunodeficiency virus envelope/CD4-mediated fusion in human/murine heterokaryons. J. Virol. 1992;66:4794–4802. doi: 10.1128/jvi.66.8.4794-4802.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dragic T, Litwin V, Allaway G.P, Martin S.R, Huang Y, Nagashima K.A, Cayanan C, Maddon P.J, Koup R.A, Moore J.P, Paxton W.A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 58.Duffus W.A, Levymintz P, Klimjack M.R, Kielian M. Mutations in the putative fusion peptide of Semliki forest virus affect spike protein oligomerization and virus assembly. J. Virol. 1995;69:2471–2479. doi: 10.1128/jvi.69.4.2471-2479.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Durrer P, Galli C, Hoenke S, Corti C, Gluck R, Vorherr T, Brunner J. H+-induced membrane insertion of influenza virus hemagglutinin involves the HA2 amino-terminal fusion peptide but not the coiled coil region. J. Biol. Chem. 1996;271:13417–13421. doi: 10.1074/jbc.271.23.13417. [DOI] [PubMed] [Google Scholar]
- 60.Ellens H, Bentz J, Mason D, Zhang F, White J.M. Fusion of influenza hemagglutinin-expressing fibroblasts with glycophorin-bearing liposomes—role of hemagglutinin surface density. Biochemistry. 1990;29:9697–9707. doi: 10.1021/bi00493a027. [DOI] [PubMed] [Google Scholar]
- 61.H. Ellens, C. Larsen, CD4-induced changes in gp120/gp41 confomation and its potential relationship to fusion, in: J. Bentz (Ed.), Viral Fusion Mechanisms, CRC Press, Boca Raton, 1993, pp. 291–312.
- 62.Endres M.J, Clapham P.R, Marsh M, Ahuja M, Davis-Turner J, McKnight A, Thomas J, Stoebenau-Haggarty B, Choe S, Vance P.J, Wells T.N.C, Power C.A, Sutterwala S.S, Doms R.W, Landau N.R, Hoxie J.A. CD4-independent infection by HIV-2 is mediated by fusin/CXCR4. Cell. 1996;87:745–756. doi: 10.1016/s0092-8674(00)81393-8. [DOI] [PubMed] [Google Scholar]
- 63.Eppstein D.A, Marsh Y.V, Schreiber A.B, Newman S.R, Todaro G.J, Nestor J.J., Jr. Epidermal growth factor receptor occupancy inhibits vaccinia virus infection. Nature. 1985;318:663–665. doi: 10.1038/318663a0. [DOI] [PubMed] [Google Scholar]
- 64.Fass D, Harrison S.C, Kim P.S. Retrovirus envelope domain at 1.7 angstrom resolution. Nat. Struct. Biol. 1996;3:465–469. doi: 10.1038/nsb0596-465. [DOI] [PubMed] [Google Scholar]
- 65.Feng Y, Broder C.C, Kennedy P.E, Berger E.A. HIV-1 entry cofactor: Functional cDNA cloning of a seven-transmembrane. G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 66.Fingeroth J.D, Weis J.J, Tedder T.F, Strominger J.L, Biro P.A, Fearon D.T. Epstein–Barr virus receptor of human B lymphocytes is the C3d receptor CR2. Proc. Natl. Acad. Sci. USA. 1984;81:4510–4514. doi: 10.1073/pnas.81.14.4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Flynn D.C, Meyer W.J, MacKenzie J.M, Johnston R.E. A conformational change in Sindbis virus glycoproteins E1 and E2 is detected at the plasmamembrane as a consequence of early virus–cell interaction. J. Virol. 1990;64:3643–3653. doi: 10.1128/jvi.64.8.3643-3653.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Frade R, Barel M, Ehlin Henriksson B, Klein G. gp140, the C3d receptor of human B lymphocytes, is also the Epstein–Barr virus receptor. Proc. Natl. Acad. Sci. USA. 1985;82:1490–1493. doi: 10.1073/pnas.82.5.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Froshauer S, Kartenbeck J, Helenius A. Alphavirus RNA replicase is located on the cytoplasmic surface of endosomes and lysosomes. J. Cell Biol. 1988;107:2075–2086. doi: 10.1083/jcb.107.6.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fu Y.K, Hart T.K, Jonak Z.L, Bugelski P.J. Physicochemical dissociation of CD4-mediated syncytium formation and shedding of human immunodeficiency virus type 1 gp120. J. Virol. 1993;67:3818–3825. doi: 10.1128/jvi.67.7.3818-3825.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fuller S.D. The T=4 envelope of Sindbis virus is organized by interactions with a complementary T=3 capsid. Cell. 1987;48:923–934. doi: 10.1016/0092-8674(87)90701-x. [DOI] [PubMed] [Google Scholar]
- 72.Fuller S.D, Berriman J.A, Butcher S.J, Gowen B.E. Low pH induces swiveling of the glycoprotein heterodimers in the Semliki–forest virus spike complex. Cell. 1995;81:715–725. doi: 10.1016/0092-8674(95)90533-2. [DOI] [PubMed] [Google Scholar]
- 73.Gallay P, Stitt V, Mundy C, Oettinger M, Trono D. Role of the karyopherin pathway in human immunodeficiency virus type 1 nuclear import. J. Virol. 1996;70:1027–1032. doi: 10.1128/jvi.70.2.1027-1032.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gelderblom H.R. Fine structure, antigenic composition and functional aspects of the AIDS virus (HIV) Zeiss-Inform. 1987;5:46–50. [Google Scholar]
- 75.Gilbert J.M, Hernandez L.D, Balliet J.W, Bates P, White J.M. Receptor-induced conformational changes in the subgroup A avian leukosis and sarcoma virus envelope glycoprotein. J. Virol. 1995;69:7410–7415. doi: 10.1128/jvi.69.12.7410-7415.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Godley L, Pfeifer J, Steinhauer D, Ely B, Shaw G, Kaufmann R, Suchanek E, Pabo C, Skehel J.J, Wiley D.C, Wharton S. Introduction of intersubunit disulfide bonds in the membrane-distal region of the influenza hemagglutinin abolishes membrane fusion activity. Cell. 1992;68:635–645. doi: 10.1016/0092-8674(92)90140-8. [DOI] [PubMed] [Google Scholar]
- 77.Grambas S, Bennett M.S, Hay A.J. Influence of amantadine resistance mutations on the pH regulatory function of the M2-protein of influenza-A viruses. Virology. 1992;191:541–549. doi: 10.1016/0042-6822(92)90229-i. [DOI] [PubMed] [Google Scholar]
- 78.Greber U.F, Singh I, Helenius A. Mechanisms of virus uncoating. Trends Microbiol. 1994;2:52–56. doi: 10.1016/0966-842x(94)90126-0. [DOI] [PubMed] [Google Scholar]
- 79.Greber U.F, Willetts M, Webster P, Helenius A. Stepwise dismantling of adenovirus 2 during entry into cells. Cell. 1993;75:477–486. doi: 10.1016/0092-8674(93)90382-z. [DOI] [PubMed] [Google Scholar]
- 80.Gruenberg J, Maxfield F.R. Membrane transport in the endocytic pathway. Curr. Opin. Cell Biol. 1995;7:552–563. doi: 10.1016/0955-0674(95)80013-1. [DOI] [PubMed] [Google Scholar]
- 81.Guy H.R, Durell S.R, Schoch C, Blumenthal R. Analyzing the fusion process of influenza hemagglutinin by mutagenesis and molecular modeling. Biophys. J. 1992;62:95–97. doi: 10.1016/S0006-3495(92)81790-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Harouse J.M, Bhat S, Spitalnik S.L, Laughlin M, Stefano K, Silberberg D.H, Gonzalez Scarano F. Inhibition of entry of HIV-1 in neural cell lines by antibodies against galactosyl ceramide. Science. 1991;253:320–323. doi: 10.1126/science.1857969. [DOI] [PubMed] [Google Scholar]
- 83.Helenius A. Unpacking the incoming influenza virus. Cell. 1992;69:577–578. doi: 10.1016/0092-8674(92)90219-3. [DOI] [PubMed] [Google Scholar]
- 84.Helenius A, Kartenbeck J, Simons K, Fries E. On the entry of Semliki Forest virus into BHK-21 cells. J. Cell Biol. 1980;84:404–420. doi: 10.1083/jcb.84.2.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Helenius A, Marsh M, White J. Effect of lysosomotropic weak bases on Semliki Forest virus penetration into host cells. J. Gen. Virol. 1982;58:47–61. doi: 10.1099/0022-1317-58-1-47. [DOI] [PubMed] [Google Scholar]
- 86.Helenius A, Morein B, Fries E, Simons K, Robinson P, Schirrmacher V, Terhorst C, Strominger J.L. Human (HLA-A and HLA-B) and murine (H-2K and H-2D) histocompatibility antigens are cell surface receptors for Semliki Forest virus. Proc. Natl. Acad. Sci. USA. 1978;75:3846–3850. doi: 10.1073/pnas.75.8.3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Herrler G, Rott R, Klenk H.D. Neuraminic acid is involved in the binding of influenza C virus to erythrocytes. Virology. 1985;141:144–147. doi: 10.1016/0042-6822(85)90190-4. [DOI] [PubMed] [Google Scholar]
- 88.Herrler G, Rott R, Klenk H.D, Muller H.P, Shukla A.K, Schauer R. The receptor-destroying enzyme of influenza C virus is neuraminate-O-acetylesterase. EMBO J. 1985;4:1503–1516. doi: 10.1002/j.1460-2075.1985.tb03809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hiebsch R.R, Raub T.J, Wattenberg B.W. Primaquine blocks transport by inhibiting the formation of functional transport vesicles. Studies in a cell-free assay of protein transport through the Golgi apparatus. J. Biol. Chem. 1991;266:20323–20328. [PubMed] [Google Scholar]
- 90.Holsinger L.J, Lamb R.A. Influenza virus M2 integral membrane protein is a homotetramer stabilized by formation of disulphide bonds. Virology. 1991;183:32–43. doi: 10.1016/0042-6822(91)90115-r. [DOI] [PubMed] [Google Scholar]
- 91.Huet C, Ash J.F, Singer S.J. The antibody-induced clustering and endocytosis of HLA antigens on cultured human fibroblsts. Cell. 1980;21:429–438. doi: 10.1016/0092-8674(80)90479-1. [DOI] [PubMed] [Google Scholar]
- 92.Inada T, Mims C.A. Mouse Ia antigens are receptors for lactate dehydrogenase virus. Nature. 1984;309:59–61. doi: 10.1038/309059a0. [DOI] [PubMed] [Google Scholar]
- 93.Johann S.V.G.J.J, O'Hara B. GLVR1, a receptor for gibbon ape leukemia virus, is homologous to a phosphate permease of Neurospora crassa and is expressed at high levels in the brain and thymus. J. Virol. 1992;66:1635–1640. doi: 10.1128/jvi.66.3.1635-1640.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kartenbeck J, Stukenbrok H, Helenius A. Endocytosis of simian virus 40 into the endoplasmic reticulum. J. Cell Biol. 1989;109:2721–2729. doi: 10.1083/jcb.109.6.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kavanaugh M.P, Miller D.G, Zhang W, Law W, Kozak S.L, Kabat D, Miller A.D. Cell-surface receptors for gibbon ape leukemia virus and amphotropic murine retrovirus are inducible sodium-dependent phosphate symporters. Proc. Natl. Acad. Sci. USA. 1994;91:7071–7075. doi: 10.1073/pnas.91.15.7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kemble G.W, Bodian D.L, Rosé J, Wilson I.A, White J.M. Intermonomer disulfide bonds impair the fusion activity of influenza virus hemagglutinin. J. Virol. 1992;66:4940–4950. doi: 10.1128/jvi.66.8.4940-4950.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kemble G.W, Danieli T, White J.M. Lipid-anchored influenza hemagglutinin promotes hemifusion, not complete fusion. Cell. 1994;76:383–391. doi: 10.1016/0092-8674(94)90344-1. [DOI] [PubMed] [Google Scholar]
- 98.M. Kielian, Membrane fusion activity of alphaviruses, in: J. Bentz (Ed.), Viral Fusion Mechanisms, CRC Press, Boca Raton, FL, 1993, pp. 385–412.
- 99.Kielian M, Helenius A. Role of cholesterol in fusion of Semliki forest virus with membranes. J. Virol. 1984;52:281–283. doi: 10.1128/jvi.52.1.281-283.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kielian M, Helenius A. pH-induced alterations in the fusogenic spike protein of Semliki Forest virus. J. Cell Biol. 1985;101:2284–2291. doi: 10.1083/jcb.101.6.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kielian M, Jungerwirth S, Ulrich Sayad K, De Candido S. Biosynthesis, maturation, and acid activation of the Semliki Forest virus fusion protein. J. Virol. 1990;64:4614–4624. doi: 10.1128/jvi.64.10.4614-4624.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kielian M.C, Marsh M, Helenius A. Kinetics of endosome acidification detected by mutant and wild-type Semliki Forest virus. EMBO J. 1986;5:3103–3109. doi: 10.1002/j.1460-2075.1986.tb04616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Klasse P.J, Pipkorn R, Blomberg J, Han K.H, Hilton B, Ferretti J.A. Three-dimensional structure and antigenicity of transmembrane-protein peptides of the human immunodeficiency virus type 1. Effects of a neutralization-escape substitution. FEBS Lett. 1993;323:68–72. doi: 10.1016/0014-5793(93)81450-e. [DOI] [PubMed] [Google Scholar]
- 104.Klasse P.J, Moore J.P. Kinetics of the HIV–CD4 interactions and virus–cell fusion. AIDS. 1992;6:325–327. doi: 10.1097/00002030-199203000-00011. [DOI] [PubMed] [Google Scholar]
- 105.Klatzman D, Champagne E, Chamaret S, Gruest J, Guetard D, Hercend T, Gluckman J.-C, Montagnier L. T-lymphocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature. 1984;312:767–768. doi: 10.1038/312767a0. [DOI] [PubMed] [Google Scholar]
- 106.Knight S.C, Macatonia S.E, Patterson S. HIV I infection of dendritic cells. Int. Rev. Immunol. 1990;6:163–175. doi: 10.3109/08830189009056627. [DOI] [PubMed] [Google Scholar]
- 107.Koike S, Ise I, Sato Y, Mitsui K, Umeyama H, Nomoto A. Early events of poliovirus infection. Semin. Virol. 1992;3:109–115. [Google Scholar]
- 108.Lentz T.L. Rabies virus binding to an acetylcholine receptor alpha-subunit peptide. J. Mol. Recog. 1990;3:82–88. doi: 10.1002/jmr.300030205. [DOI] [PubMed] [Google Scholar]
- 109.Lentz T.L. Structure–function relationships of curaremimetic neurotoxin loop 2 and of a structurally similar segment of rabies virus glycoprotein in their interaction with the nicotinic acetylcholine receptor. Biochemistry. 1991;30:10949–10957. doi: 10.1021/bi00109a020. [DOI] [PubMed] [Google Scholar]
- 110.Levy-Mintz P, Kielian M. Mutagenesis of the putative fusion domain of the Semliki Forest virus spike protein. J. Virol. 1991;65:4292–4300. doi: 10.1128/jvi.65.8.4292-4300.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Li J.P, Hu H.O, Niu Q.T, Fang C. Cell surface activation of the erythropoietin receptor by Friend spleen focus-forming virus gp55. J. Virol. 1995;69:1714–1719. doi: 10.1128/jvi.69.3.1714-1719.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lisanti M.P, Tang Z.L, Scherer P.E, Kubler E, Koleske A.J, Sargiacomo M. Caveolae, transmembrane signalling and cellular transformation. Mol. Membr. Biol. 1995;12:121–124. doi: 10.3109/09687689509038506. [DOI] [PubMed] [Google Scholar]
- 113.Liu R, Paxton W.A, Choe S, Ceradini D, Martin S.R, Horuk R, MacDonald S.R, Stuhlmann H, Koup R.A, Landau N.R. Homozygous defect in HIV-1 co-receptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 114.Lobigs M, Garoff H. Fusion function of the Semliki Forest virus spike is activated by proteolytic cleavage of the envelope glycoprotein precursor p62. J. Virol. 1990;64:1233–1240. doi: 10.1128/jvi.64.3.1233-1240.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lobigs M, Wahlberg J.M, Garoff H. Spike protein oligomerization control of Semliki Forest virus fusion. J. Virol. 1990;64:5214–5218. doi: 10.1128/jvi.64.10.5214-5218.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lu M, Blacklow S.C, Kim P.S. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat. Struct. Biol. 1995;2:1075–1082. doi: 10.1038/nsb1295-1075. [DOI] [PubMed] [Google Scholar]
- 117.Lusso P, Secchiero P, Crowley R.W, Garzino Demo A, Berneman Z.N, Gallo R.C. CD4 is a critical component of the receptor for human herpesvirus 7: Interference with human immunodeficiency virus. Proc. Natl. Acad. Sci. USA. 1994;91:3872–3876. doi: 10.1073/pnas.91.9.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Machy P, Truneh A, Gennaro D, Hoffstein S. Major histocompatibility complex class I molecules internalized via coated pits in T lymphocytes. Nature. 1987;328:724–726. doi: 10.1038/328724a0. [DOI] [PubMed] [Google Scholar]
- 119.Maddon P.J, Dalgleish A.G, McDougal J.S, Clapham P.R, Weiss R.A, Axel R. The T4 gene encodes the AIDS virus receptor and is expressed in the immune system and the brain. Cell. 1986;47:333–348. doi: 10.1016/0092-8674(86)90590-8. [DOI] [PubMed] [Google Scholar]
- 120.Marcon L, Choe H, Martin K.A, Farzan M, Ponath P.D, Wu L, Newman W, Gerard N, Gerard C, Sodroski J. Utilization of C-C chemokine receptor 5 by the envelope glycoproteins of a pathogenic simian immunodeficiency virus, SIVmac239. J. Virol. 1997;71:2522–2527. doi: 10.1128/jvi.71.3.2522-2527.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Marin M, Noel D, Valsesia Wittman S, Brockly F, Etienne Julan M, Russell S, Cosset F.L, Piechaczyk M. Targeted infection of human cells via major histocompatibility complex class I molecules by Moloney murine leukemia virus-derived viruses displaying single-chain antibody fragment-envelope fusion proteins. J. Virol. 1996;70:2957–2962. doi: 10.1128/jvi.70.5.2957-2962.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Marquardt T, Hebert D.N, Helenius A. Post-translational folding of influenza hemagglutinin in isolated endoplasmic reticulum-derived microsomes. J. Biol. Chem. 1993;268:19618–19625. [PubMed] [Google Scholar]
- 123.Marsh M, Bron R. SFV infection in CHO cells: Cell-type specific restrictions to productive virus entry at the cell surface. J. Cell Sci. 1997;110:95–103. doi: 10.1242/jcs.110.1.95. [DOI] [PubMed] [Google Scholar]
- 124.Marsh M, Helenius A. Adsorptive endocytosis of Semliki Forest virus. J. Mol. Biol. 1980;142:439–454. doi: 10.1016/0022-2836(80)90281-8. [DOI] [PubMed] [Google Scholar]
- 125.Martin K, Helenius A. Nuclear transport of influenza virus ribonucleoproteins: The viral matrix protein (M1) promotes export and inhibits import. Cell. 1991;67:117–130. doi: 10.1016/0092-8674(91)90576-k. [DOI] [PubMed] [Google Scholar]
- 126.Martin K, Helenius A. Transport of incoming influenza virus nucleocapsids into the nucleus. J. Virol. 1991;65:232–244. doi: 10.1128/jvi.65.1.232-244.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Matlin K.S, Reggio H, Helenius A, Simons K. Pathway of vesicular stomatitis virus entry leading to infection. J. Mol. Biol. 1982;156:609–631. doi: 10.1016/0022-2836(82)90269-8. [DOI] [PubMed] [Google Scholar]
- 128.Matthews T.J, Wild C, Chen C.H, Bolognesi D.P, Greenberg M.L. Structural rearrangements in the transmembrane glycoprotein after receptor binding. Immunol. Rev. 1994;140:93–104. doi: 10.1111/j.1600-065x.1994.tb00866.x. [DOI] [PubMed] [Google Scholar]
- 129.McClure M.O, Marsh M, Weiss R.A. Human immunodeficiency virus infection of CD4-bearing cells occurs by a pH-independent mechanism. EMBO J. 1988;7:513–518. doi: 10.1002/j.1460-2075.1988.tb02839.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.McClure M.O, Sommerfelt M.A, Marsh M, Weiss R.A. The pH independence of mammalian retrovirus infection. J. Gen. Virol. 1990;71:767–773. doi: 10.1099/0022-1317-71-4-767. [DOI] [PubMed] [Google Scholar]
- 131.Melikyan G.B, Brener S.A, Ok D.C, Cohen F.S. Inner but not outer membrane leaflets control the transition from glycosylphosphatidylinositol-anchored influenza hemagglutinin-induced hemifusion to full fusion. J. Cell Biol. 1997;136:995–1005. doi: 10.1083/jcb.136.5.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Metsikkö K, Garoff H. Oligomers of the cytoplasmic domain of the p62/E2 membrane protein of Semliki Forest virus bind to the nucleocapsid in vitro. J. Virol. 1990;64:4678–4683. doi: 10.1128/jvi.64.10.4678-4683.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Meyer W.J, Gidwitz S, Ayers V.K, Schoepp R.J, Johnston R.E. Conformational alterations of Sindbis virion glycoproteins induced by heat, reducing agents, or low pH. J. Virol. 1992;66:3504–3513. doi: 10.1128/jvi.66.6.3504-3513.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Meyer W.J, Johnston R.E. Structural rearrangement of infecting Sindbis virions at the cell surface—Mapping of newly accessible epitopes. J. Virol. 1993;67:5117–5125. doi: 10.1128/jvi.67.9.5117-5125.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Miller A.D. Human gene therapy comes of age. Nature. 1992;357:455–460. doi: 10.1038/357455a0. [DOI] [PubMed] [Google Scholar]
- 136.Miller D.G, Miller A.D. A family of retroviruses that utilize related phosphate transporters for cell entry. J. Virol. 1994;68:8270–8276. doi: 10.1128/jvi.68.12.8270-8276.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Miller N, Hutt Fletcher L.M. Epstein–Barr virus enters B cells and epithelial cells by different routes. J. Virol. 1992;66:3409–3414. doi: 10.1128/jvi.66.6.3409-3414.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Moesby L, Corver J, Erukulla R.K, Bittman R, Wilschut J. Sphingolipids activate membrane-fusion of semliki-forest-virus in a stereospecific manner. Biochemistry. 1995;34:10319–10324. doi: 10.1021/bi00033a001. [DOI] [PubMed] [Google Scholar]
- 139.Montgomery R.I, Warner M.S, Lum B.J, Spear P.G. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell. 1996;87:427–436. doi: 10.1016/s0092-8674(00)81363-x. [DOI] [PubMed] [Google Scholar]
- 140.Moore J.P, Klasse P.J. Thermodynamic and kinetic analysis of sCD4 binding to HIV-1 virions and of gp120 dissociation. AIDS Res. Hum. Retroviruses. 1992;8:443–450. doi: 10.1089/aid.1992.8.443. [DOI] [PubMed] [Google Scholar]
- 141.Moore J.P, McKeating J.A, Weiss R.A, Sattentau Q.J. Dissociation of gp120 from HIV-1 virions induced by soluble CD4. Science. 1990;250:1139–1142. doi: 10.1126/science.2251501. [DOI] [PubMed] [Google Scholar]
- 142.Moore J.P, Sodroski J. Antibody cross-competition analysis of the human immunodeficiency virus type 1 gp120 exterior envelope glycoprotein. J. Virol. 1996;70:1863–1872. doi: 10.1128/jvi.70.3.1863-1872.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Moore J.P, Trkola A, Dragic T. Co-receptors for HIV-1 entry. Curr. Biol. Opin. Immunol. 1997;9:551–562. doi: 10.1016/s0952-7915(97)80110-0. [DOI] [PubMed] [Google Scholar]
- 144.Moore M.D, Cannon M.J, Sewall A, Finlayson M, Okimoto M, Nemerow G.R. Inhibition of Epstein–Barr virus infection in vitro and in vivo by soluble CR2 (CD21) containing two short consensus repeats. J. Virol. 1991;65:3559–3565. doi: 10.1128/jvi.65.7.3559-3565.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mulligan R.C. The basic science of gene therapy. Science. 1993;260:926–932. doi: 10.1126/science.8493530. [DOI] [PubMed] [Google Scholar]
- 146.Naniche D, Varior Krishnan G, Cervoni F, Wild T.F, Rossi B, Rabourdin Combe C, Gerlier D. Human membrane cofactor protein (CD46) acts as a cellular receptor for measles virus. J. Virol. 1993;67:6025–6032. doi: 10.1128/jvi.67.10.6025-6032.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Nemerow G.R, Moore M.D, Cooper N.R. Structure and function of the B-lymphocyte Epstein–Barr virus/C3d receptor. Adv. Cancer Res. 1990;54:273–300. doi: 10.1016/s0065-230x(08)60814-3. [DOI] [PubMed] [Google Scholar]
- 148.Nemerow G.R, Wolfert R, McNaughton M.E, Cooper N.R. Identification and characterization of the Epstein–Barr virus receptor on human B lymphocytes and its relationship to the C3d complement receptor (CR2) J. Virol. 1985;55:347–351. doi: 10.1128/jvi.55.2.347-351.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Nieva J.L, Bron R, Corver J, Wilschut J. Membrane fusion of Semliki Forest virus requires sphingolipids in the target membrane. EMBO J. 1994;13:2797–2804. doi: 10.1002/j.1460-2075.1994.tb06573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Norbury C.C, Chambers B.J, Prescott A.R, Ljunggren H.G, Watts C. Constitutive macropinocytosis allows TAP-dependent major histocompatibility complex class I presentation of exogenous soluble antigen by bone marrow-derived dendritic cells. Eur. J. Immunol. 1997;27:280–288. doi: 10.1002/eji.1830270141. [DOI] [PubMed] [Google Scholar]
- 151.Norbury C.C, Hewlett L.J, Prescott A.R, Shastri N, Watts C. Class I MHC presentation of exogenous soluble antigen via macropinocytosis in bone marrow macrophages. Immunity. 1995;3:783–791. doi: 10.1016/1074-7613(95)90067-5. [DOI] [PubMed] [Google Scholar]
- 152.Oberlin E, Amara A, Bacherlerie F, Bessia C, Virelizier J.-L, Arenzana-Seisdedos F, Schwartz O, Heard J.-M, Clark-Lewis I, Legler D.F, Loetscher M, Baggiolini M, Moser B. The CXC chemokine SDF-1 is the ligand for Lestr/Fusin and prevents infection by T-cell-line-adapted HIV-1. Nature (London) 1996;382:833–835. doi: 10.1038/382833a0. [DOI] [PubMed] [Google Scholar]
- 153.Olah Z, Lehel C, Anderson W.B, Eiden M.V, Wilson C.A. The cellular receptor for gibbon ape leukemia virus is a novel high affinity sodium-dependent phosphate transporter. J. Biol. Chem. 1994;269:25426–25431. [PubMed] [Google Scholar]
- 154.Oldstone M.B.A, Tishon A, Dutko F, Kennedy S.I.T, Holland J.J, Lampert P.W. Does the major histocompatibility complex serve as a specific receptor for Semliki Forest virus. J. Virol. 1980;34:256–265. doi: 10.1128/jvi.34.1.256-265.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Olson N.H, Kolatkar P.R, Oliveira M.A, Cheng R.H, Greve J.M, McClelland A, Baker T.S, Rossmann M.G. Structure of a human rhinovirus complexed with its receptor molecule. Proc. Natl. Acad. Sci. USA. 1993;90:507–511. doi: 10.1073/pnas.90.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Orloff G.M, Orloff S.L, Kennedy M.S, Maddon P.J, McDougal J.S. Penetration of CD4 T cells by HIV-1. The CD4 receptor does not internalise with HIV, and CD4-related signal transduction events are not required for entry. J. Immunol. 1991;146:2578–2587. [PubMed] [Google Scholar]
- 157.Panetti T.S, McKeown-Longo P.J. The alpha v beta 5 integrin receptor regulates receptor-mediated endocytosis of vitronectin. J. Biol. Chem. 1993;268:11492–11495. [PubMed] [Google Scholar]
- 158.Panetti T.S, McKeown-Longo P.J. Receptor-mediated endocytosis of vitronectin is regulated by its conformational state. J. Biol. Chem. 1993;268:11988–11993. [PubMed] [Google Scholar]
- 159.Paredes A.M, Brown D.T, Rothnagel R, Chiu W, Schoepp R.J, Johnston R.E, Prasad B.V.V. 3-Dimensional structure of a membrane-containing virus. Proc. Natl. Acad. Sci. USA. 1993;90:9095–9099. doi: 10.1073/pnas.90.19.9095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Paredes A.M, Simon M.N, Brown D.T. The mass of the Sindbis virus nucleocapsid suggests it has T=4 icosahedral symmetry. Virology. 1992;187:329–332. doi: 10.1016/0042-6822(92)90322-g. [DOI] [PubMed] [Google Scholar]
- 161.Parton R.G. Caveolae and caveolins. Curr. Opin. Cell Biol. 1996;8:542–548. doi: 10.1016/s0955-0674(96)80033-0. [DOI] [PubMed] [Google Scholar]
- 162.Parton R.G, Joggerst B, Simons K. Regulated internalization of caveolae. J. Cell Biol. 1994;127:1199–1215. doi: 10.1083/jcb.127.5.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Patterson S, Oxford J.S, Dourmashkin R.R. Studies on the mechanism of influenza virus entry into cells. J. Gen. Virol. 1979;43:223–229. doi: 10.1099/0022-1317-43-1-223. [DOI] [PubMed] [Google Scholar]
- 164.Paxton W.A, Martin S.R, Tse D, O'Brien T.R, Skurnick J, VanDevanter N.L, Padian N, Braun J.F, Kotler D.P, Wolinsky S.M, Koup R.A. Relative resistance to HIV-1 infection of CD4 lymphocytes from persons who remain uninfected despite multiple high-risk sexual exposure. Nat. Med. 1996;2:412–417. doi: 10.1038/nm0496-412. [DOI] [PubMed] [Google Scholar]
- 165.Pelchen-Matthews A, Clapham P, Marsh M. The role of CD4 endocytosis in HIV infection. J. Virol. 1995;69:8164–8168. doi: 10.1128/jvi.69.12.8164-8168.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Pettersson R.F. Protein localization and virus assembly at intracellular membranes. Curr. Top. Microbiol. Immunol. 1991;170:67–106. doi: 10.1007/978-3-642-76389-2_3. [DOI] [PubMed] [Google Scholar]
- 167.Phalen T, Kielian M. Cholesterol is required for infection by Semliki Forest virus. J. Cell Biol. 1991;112:615–623. doi: 10.1083/jcb.112.4.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Pinto L.H, Holsinger L.J, Lamb R.A. Influenza M2 protein has ion channel activity. Cell. 1992;69:517–528. doi: 10.1016/0092-8674(92)90452-i. [DOI] [PubMed] [Google Scholar]
- 169.Plank C, Oberhauser B, Mechtler K, Koch C, Wagner E. The influence of endosome-disruptive peptides on gene transfer using synthetic virus-like gene transfer systems. J. Biol. Chem. 1994;269:12918–12924. [PubMed] [Google Scholar]
- 170.Power C.A, Wells T.N.C. Cloning and characterisation of human chemokine receptors. Trends Pharmacol. Sci. 1996;17:209–213. doi: 10.1016/0165-6147(96)10019-5. [DOI] [PubMed] [Google Scholar]
- 171.Prchla E, Plank C, Wagner E, Blaas D, Fuchs R. Virus-mediated release of endosomal content in vitro: Different behavior of adenovirus and rhinovirus serotype 2. J. Cell Biol. 1995;131:111–123. doi: 10.1083/jcb.131.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Premack B.A, Schall T.J. Chemokine receptors: Gateways to inflammation and infection. Nat. Med. 1996;2:1174–1178. doi: 10.1038/nm1196-1174. [DOI] [PubMed] [Google Scholar]
- 173.Pudney J, Song M.J. Electron microscopic analysis of HIV-host cell interactions. Tissue Cell. 1994;26:539–550. doi: 10.1016/0040-8166(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 174.Reeves J.D, McKnight A, Potempa S, Simmons G, Gray P.W, Power C.A, Wells T, Weiss R.A, Talbot S. CD4-independent infection by HIV-2 (ROD/B): Use of the 7-transmembrane receptors CXCR4, CCR3 and V28 for entry. Virology. 1997;231:130–134. doi: 10.1006/viro.1997.8508. [DOI] [PubMed] [Google Scholar]
- 175.Rey F.A, Heinz F.X, Mandl C, Kunz C, Harrison S. The envelope glycoprotein from the tick-borne encephalitis virus at 2 Å resolution. Nature. 1995;375:291–298. doi: 10.1038/375291a0. [DOI] [PubMed] [Google Scholar]
- 176.Rossmann M.G. Viral cell recognition and entry. Protein Sci. 1994;3:1712–1725. doi: 10.1002/pro.5560031010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Rothman J.E. Mechanisms of intracellular protein transport. Nature. 1994;372:55–63. doi: 10.1038/372055a0. [DOI] [PubMed] [Google Scholar]
- 178.Rott R, Klenk H.D, Nagai Y, Tashiro M. Influenza viruses, cell enzymes, and pathogenicity. Am. J. Respir. Crit. Care Med. 1995;152:S16–S19. doi: 10.1164/ajrccm/152.4_Pt_2.S16. [DOI] [PubMed] [Google Scholar]
- 179.Rucker J, Samson M, Doranz B.J, Libert F, Berson J.F, Yi Y, Collman R.G, Broder C.C, Vassart G, Doms R.W, Parmentier M. Regions in chemokine receptors CCR5 and CCR2b that determine HIV-1 cofactor specificity. Cell. 1996;87:437–446. doi: 10.1016/s0092-8674(00)81364-1. [DOI] [PubMed] [Google Scholar]
- 180.Salminen A, Wahlberg J.M, Lobigs M, Liljestrom P, Garoff H. Membrane fusion process of Semliki Forest virus II: Cleavage-dependent reorganization of the spike protein complex controls virus entry. J. Cell Biol. 1992;116:349–357. doi: 10.1083/jcb.116.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Samson M, Libert F, Doranz B.J, Rucker J, Liesnard C, Farber C.-M, Saragosti S, Lapouméroulie C, Cognaux J, Forceille C, Muyldermans G, Verhofstede C, Burtonboy G, Georges M, Imai T, Rana S, Yi Y, Smyth R.J, Collman R.G, Doms R.W, Vassart G, Parmentier M. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 182.Sattentau Q.J, Moore J.P. Conformational changes induced in the human immunodeficiency virus envelope glycoprotein by soluble CD4 binding. J. Exp. Med. 1991;174:407–415. doi: 10.1084/jem.174.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Schmelz M, Sodeik B, Ericsson M, Wolffe E.J, Shida H, Hiller G, Griffiths G. Assembly of vaccinia virus: The second wrapping cisterna is derived from the trans Golgi network. J. Virol. 1994;68:130–147. doi: 10.1128/jvi.68.1.130-147.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Sears A.E, McGwire B.S, Roizman B. Infection of polarized MDCK cells with herpes simplex virus 1: Two asymmetrically distributed cell receptors interact with different viral proteins. Proc. Natl. Acad. Sci. USA. 1991;88:5087–5091. doi: 10.1073/pnas.88.12.5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Shoeman R.L, Kesselmier C, Mothes E, Honer B, Traub P. Non-viral cellular substrates for human immunodeficiency virus type 1 protease. FEBS Lett. 1991;278:199–203. doi: 10.1016/0014-5793(91)80116-k. [DOI] [PubMed] [Google Scholar]
- 186.Shoeman R.L, Sachse C, Honer B, Mothes E, Kaufmann M, Traub P. Cleavage of human and mouse cytoskeletal and sarcomeric proteins by human immunodeficiency virus type 1 protease. Actin, desmin, myosin, and tropomyosin. Am. J. Pathol. 1993;142:221–230. [PMC free article] [PubMed] [Google Scholar]
- 187.Signoret N, Oldridge J, Pelchen-Matthews A, Klasse P.J, Tran T, Brass L.F, Rosenkilde M.M, Schwartz T.W, Holmes W, Dallas W, Luther M.A, Wells T.N.C, Hoxie J.A, Marsh M. Phorbel esters and SDF-1 induce rapid endocytosis and down modulation of the chemokine receptor CXCR4. J. Cell Biol. 1997;139:651–664. doi: 10.1083/jcb.139.3.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Singh I, Helenius A. Role of ribosomes in Semliki Forest virus nucleocapsid uncoating. J. Virol. 1992;66:7049–7058. doi: 10.1128/jvi.66.12.7049-7058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Skoging U, Vihinen M, Nilsson L, Liljestrom P. Aromatic interactions define the binding of the alphavirus spike to its nucleocapsid. Structure. 1996;4:519–529. doi: 10.1016/s0969-2126(96)00058-5. [DOI] [PubMed] [Google Scholar]
- 190.Smart E.J, Ying Y.S, Conrad P.A, Anderson R.G. Caveolin moves from caveolae to the Golgi apparatus in response to cholesterol oxidation. J. Cell Biol. 1994;127:1185–1197. doi: 10.1083/jcb.127.5.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Smith D.H, Byrn R.A, Marsters S.A, Gregory T, Groopman J.E, Capon D.J. Blocking of HIV-1 infectivity by a soluble, secreted form of the CD4 antigen. Science. 1987;238:1704–1707. doi: 10.1126/science.3500514. [DOI] [PubMed] [Google Scholar]
- 192.Sodeik B, Doms R.W, Ericsson M, Hiller G, Machamer C.E, van't Hof W, van Meer G, Moss B, Griffiths G. Assembly of vaccinia virus: Role of intermediate compartment between the endoplasmic reticulum and the Golgi stacks. J. Cell Biol. 1993;121:521–542. doi: 10.1083/jcb.121.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Sodeik B, Ebersold M.W, Helenius A. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J. Cell Biol. 1997;136:1007–1021. doi: 10.1083/jcb.136.5.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Spruce A.E, Iwata A, Almers W. The first milliseconds of the pore formed by a fusogenic viral envelope protein during membrane fusion. Proc. Natl. Acad. Sci. USA. 1991;88:3623–3627. doi: 10.1073/pnas.88.9.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Spruce A.E, Iwata A, White J.M, Almers W. Patch clamp studies of single cell-fusion events mediated by a viral fusion protein. Nature. 1989;342:555–558. doi: 10.1038/342555a0. [DOI] [PubMed] [Google Scholar]
- 196.Stang E, Kartenbeck J, Parton R.G. Major histocompatibility complex class I molecules mediate association of SV40 with caveolae. Mol. Biol. Cell. 1997;8:47–57. doi: 10.1091/mbc.8.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Stegmann T. Membrane fusion: Anchors aweigh. Curr. Biol. 1994;4:551–554. doi: 10.1016/s0960-9822(00)00123-8. [DOI] [PubMed] [Google Scholar]
- 198.Stegmann T, Delfino J.M, Richards F.M, Helenius A. The HA2 subunit of influenza hemagglutinin inserts into the target membrane prior to fusion. J. Biol. Chem. 1991;266:18404–18410. [PubMed] [Google Scholar]
- 199.Stegmann T, Morselt H.W.M, Booy F.P, van Breemen J.F.L, Scherphof G, Wilschut J. Functional reconstitution of influenza virus envelopes. EMBO J. 1987;6:2651–2659. doi: 10.1002/j.1460-2075.1987.tb02556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Stegmann T, White J.M, Helenius A. Intermediates in influenza induced membrane fusion. EMBO J. 1990;9:4231–4241. doi: 10.1002/j.1460-2075.1990.tb07871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Stein B.S, Gowda S.D, Lifson J.D, Penhallow R.C, Bensch K.G, Engleman E.G. pH-Independent HIV entry into CD4-positive T cells via virus envelope fusion to the plasma membrane. Cell. 1987;49:659–668. doi: 10.1016/0092-8674(87)90542-3. [DOI] [PubMed] [Google Scholar]
- 202.Steinhauer D.A, Wharton S.A, Skehel J.J, Wiley D.C, Hay A.J. Amantadine selection of a mutant influenza virus containing an acid-stable hemagglutinin glycoprotein: Evidence for virus-specific regulation of the pH of glycoprotein transport vesicles. Proc. Natl. Acad. Sci. USA. 1991;88:11525–11529. doi: 10.1073/pnas.88.24.11525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Stevenson M. Portals of entry: Iuncovering HIV nuclear transport pathways. Trends Cell Biol. 1996;6:9–15. doi: 10.1016/0962-8924(96)81032-4. [DOI] [PubMed] [Google Scholar]
- 204.Stoorvogel W, Geuze H.J, Strous G.J. Sorting of endocytosed transferrin and asialoglycoprotein occurs immediately after internalization in HepG2 cells. J. Cell Biol. 1987;104:1261–1268. doi: 10.1083/jcb.104.5.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Sugrue R.J, Bahadur G, Zambon M.C, Hall S.M, Douglas A.R, Hay A.J. Specific structural alteration of the influenza haemagglutinin by amantadine. EMBO J. 1990;9:3469–3476. doi: 10.1002/j.1460-2075.1990.tb07555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Sugrue R.J, Hay A.J. Structural characteristics of the M2 protein of influenza viruses: Evidence that it forms a tetrameric channel. Virology. 1991;180:617–624. doi: 10.1016/0042-6822(91)90075-M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Suomalainen M, Liljeström P, Garoff H. Spike protein–nucleocapsid interactions drive the budding of alphaviruses. J. Virol. 1992;66:4737–4747. doi: 10.1128/jvi.66.8.4737-4747.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Suzuki Y, Suzuki T, Matsunaga M, Matsumoto M. Gangliosides as paramyxovirus receptor. Structural requirement of sialo-oligosaccharides in receptors for hemagglutinating virus of Japan (Sendai virus) and Newcastle disease virus. J. Biochem. Tokyo. 1985;97:1189–1199. doi: 10.1093/oxfordjournals.jbchem.a135164. [DOI] [PubMed] [Google Scholar]
- 209.Swanson J.A, Watts C. Macropinocytosis. Trends Cell Biol. 1995;5:424–428. doi: 10.1016/s0962-8924(00)89101-1. [DOI] [PubMed] [Google Scholar]
- 210.Tatu U, Helenius A. Interactions between newly synthesized glycoproteins, calnexin and a network of resident chaperones in the endoplasmic reticulum. J. Cell Biol. 1997;136:555–565. doi: 10.1083/jcb.136.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Taylor J.A, O'Brien J.A, Lord V.J, Meyer J.C, Bellamy A.R. The RER-localized rotavirus intracellular receptor: A truncated purified soluble form is multivalent and binds virus particles. Virology. 1993;194:807–814. doi: 10.1006/viro.1993.1322. [DOI] [PubMed] [Google Scholar]
- 212.Thelen M, Rosen A, Nairn A.C, Aderem A. Regulation by phosphorylation of reversible association of a myristoylated protein kinase C substrate with the plasma membrane. Nature. 1991;351:320–322. doi: 10.1038/351320a0. [DOI] [PubMed] [Google Scholar]
- 213.Tomasselli A.G, Hui J.O, Adams L, Chosay J, Lowery D, Greenberg B, Yem A, Deibel M.R, Zurcher N.H, Heinrikson R.L. Actin, troponin C, Alzheimer amyloid precursor protein and pro-interleukin 1 beta as substrates of the protease from human immunodeficiency virus. J. Biol. Chem. 1991;266:14548–14553. [PubMed] [Google Scholar]
- 214.Tooze J, Hollinshead M, Reis B, Radsak K, Kern H. Progeny vaccinia and human cytomegalovirus particles utilize early endosomal cisternae for their envelopes. Eur. J. Cell Biol. 1993;60:163–178. [PubMed] [Google Scholar]
- 215.Trkola A, Dragic T, Arthos J, Binley J.M, Olson W.C, Allaway G.P, Cheng-Mayer C, Robinson J, Maddon P.J, Moore J.P. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature. 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 216.Tse F.W, Iwata A, Almers W. Membrane flux through the pore formed by a fusogenic viral envelope protein during cell fusion. J. Cell Biol. 1993;121:543–552. doi: 10.1083/jcb.121.3.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Vainstein A, Hershkovitz M, Israel S, Rabin S, Loyter A. A new method for reconstitution of highly fusogenic Sendai envelopes. Biochim. Biophys. Acta. 1984;773:181–188. doi: 10.1016/0005-2736(84)90081-6. [DOI] [PubMed] [Google Scholar]
- 218.Valsesia Wittmann S, Drynda A, Deleage G, Aumailley M, Heard J.M, Danos O, Verdier G, Cosset F.L. Modifications in the binding domain of avian retrovirus envelope protein to redirect the host range of retroviral vectors. J. Virol. 1994;68:4609–4619. doi: 10.1128/jvi.68.7.4609-4619.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.van Deurs B, Petersen O.W, Olsnes S, Sandvig K. The ways of endocytosis. Int. Rev. Cytol. 1989;117:131–177. doi: 10.1016/s0074-7696(08)61336-4. [DOI] [PubMed] [Google Scholar]
- 220.R.H. Vogel, S.W. Provencher, C.H. von Bonsdorff, M. Adrian, J. Dubochet, Envelope structure of Semliki Forest virus reconstructed from cryo-electron micrographs, Nature (1986) 533–535. [DOI] [PubMed]
- 221.Wagner E, Plank C, Zatloukal K, Cotten M, Birnstiel M.L. Influenza virus hemagglutinin-HA-2 N-terminal fusogenic peptides augment gene transfer by transferrin-polylysine- DNA complexes—Toward a synthetic virus-like gene-transfer vehicle. Proc. Natl. Acad. Sci. USA. 1992;89:7934–7938. doi: 10.1073/pnas.89.17.7934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Wahlberg J.M, Boere W.A, Garoff H. The heterodimeric association between the membrane proteins of Semliki Forest virus changes its sensitivity to low pH during virus maturation. J. Virol. 1989;63:4991–4997. doi: 10.1128/jvi.63.12.4991-4997.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Wahlberg J.M, Bron R, Wilschut J, Garoff H. Membrane fusion of Semliki Forest virus involves homotrimers of the fusion protein. J. Virol. 1992;66:7309–7318. doi: 10.1128/jvi.66.12.7309-7318.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Wahlberg J.M, Garoff H. Membrane fusion process of Semliki Forest virus I: Low pH-induced rearrangement in spike protein quaternary structure precedes virus penetration into cells. J. Cell Biol. 1992;116:339–348. doi: 10.1083/jcb.116.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.A. Walter, O. Eidelman, M. Ollivon, R. Blumenthal, Functional Reconstitution of Viral Envelopes, Marcel Dekker, New York, 1990, pp. 395–408.
- 226.Wang C, Takeuchi K, Pinto L.H, Lamb R.A. Ion channel activity of influenza-A virus M2 protein—Characterization of the amantadine block. J. Virol. 1993;67:5585–5594. doi: 10.1128/jvi.67.9.5585-5594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Wang H, Dechant E, Kavanaugh M, North R.A, Kabat D. Effects of ecotropic murine retroviruses on the dual-function cell surface receptor/basic amino acid transporter. J. Biol. Chem. 1992;267:23617–23624. [PubMed] [Google Scholar]
- 228.Wang K.S, Kuhn R.J, Strauss E.G, Ou S, Strauss J.H. High-affinity laminin receptor is a receptor for Sindbis virus in mammalian cells. J. Virol. 1992;66:4992–5001. doi: 10.1128/jvi.66.8.4992-5001.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Watts C, Marsh M. Endocytosis: What goes in and how? J. Cell Sci. 1992;103:1–8. doi: 10.1242/jcs.103.1.1a. [DOI] [PubMed] [Google Scholar]
- 230.Weber T, Paesold G, Galli C, Mischler R, Semenza G, Brunner J. Evidence for H+-induced insertion of influenza hemagglutinin HA2 N-terminal segment into viral membrane. J. Biol. Chem. 1994;269:18353–18358. [PubMed] [Google Scholar]
- 231.Weis W, Brown J.H, Cusack S, Paulson J.C, Skehel J.J, Wiley D.C. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature. 1988;333:426–431. doi: 10.1038/333426a0. [DOI] [PubMed] [Google Scholar]
- 232.Weiss C.D, Levy J.A, White J.M. Oligomeric organization of gp120 on infectious human immunodeficiency virus type 1 particles. J. Virol. 1990;64:5674–5677. doi: 10.1128/jvi.64.11.5674-5677.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Weiss R.A, Tailor C.S. Retrovirus receptors. Cell. 1995;82:531–533. doi: 10.1016/0092-8674(95)90024-1. [DOI] [PubMed] [Google Scholar]
- 234.Wells T.N.C, Proudfoot A.E.I, Power C.A, Marsh M. Chemokine receptors—The new frontier for AIDS research. Chem. Biol. 1996;3:603–609. doi: 10.1016/s1074-5521(96)90126-x. [DOI] [PubMed] [Google Scholar]
- 235.Wengler G, Gros C, Wengler G. Analyses of the role of structural changes in the regulation of uncoating and assembly of alphavirus cores. Virology. 1996;222:123–132. doi: 10.1006/viro.1996.0403. [DOI] [PubMed] [Google Scholar]
- 236.Wengler G, Wurkner D, Wengler G. Identification of a sequence element in the alphavirus core protein which mediates interaction of cores with ribosomes and the disassembly of cores. Virology. 1992;191:880–888. doi: 10.1016/0042-6822(92)90263-o. [DOI] [PubMed] [Google Scholar]
- 237.Wharton S.A, Calder L.J, Ruigrok R, Skehel J.J, Steinhauer D.A, Wiley D.C. Electron microscopy of antibody complexes of influenza virus haemagglutinin in the fusion pH conformation. EMBO J. 1995;14:240–246. doi: 10.1002/j.1460-2075.1995.tb06997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.White J, Helenius A. pH-dependent fusion between the Semliki forest virus membrane and liposomes. Proc. Natl. Acad. Sci. USA. 1980;77:3273–3277. doi: 10.1073/pnas.77.6.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.White J, Kielian M, Helenius A. Membrane fusion proteins of enveloped viruses. Q. Rev. Biophys. 1983;16:151–195. doi: 10.1017/s0033583500005072. [DOI] [PubMed] [Google Scholar]
- 240.White J.M. Membrane fusion. Science. 1992;258:917–924. doi: 10.1126/science.1439803. [DOI] [PubMed] [Google Scholar]
- 241.White J.M, Wilson I.A. Anti-peptide antibodies detect steps in a protein conformational change: Low-pH activation of the influenza virus hemagglutinin. J. Cell Biol. 1987;105:2887–2896. doi: 10.1083/jcb.105.6.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Whitney J.A, Gomez M, Sheff D, Kreis T.E, Mellman I. Cytoplasmic coat proteins involved in endosome function. Cell. 1995;83:703–713. doi: 10.1016/0092-8674(95)90183-3. [DOI] [PubMed] [Google Scholar]
- 243.Wickham T.J, Mathias P, Cheresh D.A, Nemerow G.R. Integrins alpha v beta 3 and alpha v beta 5 promote adenovirus internalization but not virus attachment. Cell. 1993;73:309–319. doi: 10.1016/0092-8674(93)90231-e. [DOI] [PubMed] [Google Scholar]
- 244.Wild C, Dubay J.W, Greenwell T, Baird T, Jr., Oas T.G, McDanal C, Hunter E, Matthews T. Propensity for a leucine zipper-like domain of human immunodeficiency virus type 1 gp41 to form oligomers correlates with a role in virus-induced fusion rather than assembly of the glycoprotein complex. Proc. Natl. Acad. Sci. USA. 1994;91:12676–12680. doi: 10.1073/pnas.91.26.12676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Wild C, Greenwell T, Matthews T. A synthetic peptide from HIV-1 gp41 is a potent inhibitor of virus-mediated cell–cell fusion. AIDS Res. Hum. Retroviruses. 1993;9:1051–1053. doi: 10.1089/aid.1993.9.1051. [DOI] [PubMed] [Google Scholar]
- 246.Wild C, Oas T, Mcdanal C, Bolognesi D, Matthews T. A synthetic peptide inhibitor of human-immunodeficiency-virus replication—Correlation between solution structure and viral inhibition. Proc. Natl. Acad. Sci. USA. 1992;89:10537–10541. doi: 10.1073/pnas.89.21.10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Wild C.T, Shugars D.C, Greenwell T.K, McDanal C.B, Matthews T.J. Peptides corresponding to a predictive alpha-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc. Natl. Acad. Sci. USA. 1994;91:9770–9774. doi: 10.1073/pnas.91.21.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Willett B.J, Hosie M.J, Jarrett O, Neil J.C. Identification of a putative cellular receptor for feline immunodeficiency virus as the feline homologue of CD9. Immunology. 1994;81:228–233. [PMC free article] [PubMed] [Google Scholar]
- 249.Willett B.J, Hosie M.J, Neil J.C, Turner J.D, Hoxie J.A. Common mechanism of infection by lentiviruses. Nature. 1997;385:587. doi: 10.1038/385587a0. [DOI] [PubMed] [Google Scholar]
- 250.Williams R.K, Jiang G.S, Holmes K.V. Receptor for mouse hepatitis virus is a member of the carcinoembryonic antigen family of glycoproteins. Proc. Natl. Acad. Sci. USA. 1991;88:5533–5536. doi: 10.1073/pnas.88.13.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.J. Wilschut, R. Bron, The influenza virus hemagglutinin: Membrane fusion activity in whole virions and reconstituted virosomes, in: J. Bentz (Ed.), Viral Fusion Mechanisms, CRC Press, Boca Raton, FL, 1993.
- 252.Wu L, Gerard N.P, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso A.A, Desjardin E, Newman W, Gerard C, Sodroski J. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 253.WuDunn D, Spear P.G. Initial interaction of herpes simplex virus with cells is binding to heparan sulfate. J. Virol. 1989;63:52–58. doi: 10.1128/jvi.63.1.52-58.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Wykes M.N, Shellam G.R, McCluskey J, Kast W.M, Dallas P.B, Price P. Murine cytomegalovirus interacts with major histocompatibility complex class I molecules to establish cellular infection. J. Virol. 1993;67:4182–4189. doi: 10.1128/jvi.67.7.4182-4189.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Yeager C.L, Ashmun R.A, Williams R.K, Cardellichio C.B, Shapiro L.H, Look A.T, Holmes K.V. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature. 1992;357:420–422. doi: 10.1038/357420a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Yoshimoto T, Yoshimoto E, Meruelo D. Identification of amino acid residues critical for infection with ecotropic murine leukemia retrovirus. J. Virol. 1993;67:1310–1314. doi: 10.1128/jvi.67.3.1310-1314.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Yu Y.G, King D.S, Shin Y.K. Insertion of a coiled-coil peptide from influenza virus hemagglutinin into membranes. Science. 1994;266:274–276. doi: 10.1126/science.7939662. [DOI] [PubMed] [Google Scholar]
- 258.Zebedee S.L, Lamb R.A. Influenza A virus M2 protein: Monoclonal antibody restriction of virus growth and detection of M2 in virions. J. Virol. 1988;62:2762–2772. doi: 10.1128/jvi.62.8.2762-2772.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Zhirnov O.P. Solubilization of matrix protein M1/M from virions occurs at different pH for orthomyxo- and paramyxoviruses. Virology. 1990;1776:274–279. doi: 10.1016/0042-6822(90)90253-n. [DOI] [PubMed] [Google Scholar]
- 260.Zhirnov O.P. Isolation of matrix M1 from influenza viruses by acid-dependent extraction with nonionic detergent. Virology. 1992;186:324–330. doi: 10.1016/0042-6822(92)90090-c. [DOI] [PubMed] [Google Scholar]
- 261.Zingler K, Belanger C, Peters R, Agard E, Young J.A. Identification and characterization of the viral interaction determinant of the subgroup A avian leukosis virus receptor. J. Virol. 1995;69:4261–4266. doi: 10.1128/jvi.69.7.4261-4266.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]


