Abstract
Respiratory viral diseases can be spread when a virus-containing particle (droplet) from one individual is aerosolized and subsequently comes into either direct or indirect contact with another individual. Increasing numbers of studies are examining the occupational risk to healthcare workers due to proximity to patients. Selecting the appropriate air sampling method is a critical factor in assuring the analytical performance characteristics of a clinical study. The objective of this study was to compare the physical collection efficiency and virus collection efficiency of a 5 mL compact SKC BioSampler®, a gelatin filter, and a glass fiber filter, in a laboratory setting. The gelatin filter and the glass fiber filter were housed in a home-made filter holder. Submersion (with vortexing and subsequent centrifugation) was used for the gelatin and glass fiber filters. Swabbing method was also tested to retrieve the viruses from the glass fiber filter. Experiments were conducted using the H1N1 influenza A virus A/Puerto Rico/8/1934 (IAV-PR8), and viral recovery was determined using culture and commercial real-time-PCR (BioFire and Xpert). An atomizer was used to aerosolize a solution of influenza virus in PBS for measurement, and two Scanning Mobility Particle Sizers were used to determine particle size distributions. The SKC BioSampler demonstrated a U-shaped physical collection efficiency, lowest for particles around 30–50 nm, and highest at 10 nm and 300–350 nm within the size range examined. The physical collection efficiency of the gelatin filter was strongly influenced by air flow and time: a stable collection across all particle sizes was only observed at 2 L/min for the 9 min sampling time, otherwise, degradation of the filter was observed. The glass fiber filter demonstrated the highest physical collection efficiency (100% for all sizes) of all tested samplers, however, its overall virus recovery efficiency fared the worst (too low to quantify). The highest viral collection efficiencies for the SKC BioSampler and gelatin filter were 5% and 1.5%, respectively. Overall, the SKC BioSampler outperformed the filters. It is important to consider the total concentration of viruses entering the sampler when interpreting the results.
Keywords: SKC BioSampler®, Gelatin filter, Glass fiber filter, Physical collection efficiency, Virus collection efficiency
Highlights
-
•
A 5 mL SKC BioSampler®, gelatin filter, and glass fiber filter were compared in a laboratory.
-
•
The physical collection and influenza A viral recovery efficiencies were examined.
-
•
The glass fiber filter exhibited 100% physical collection efficiency for all sizes.
-
•
The SKC BioSampler® exhibited the highest virus collection efficiency (up to 5%).
-
•
All samplers collected only a small fraction of the total atomized viruses.
1. Introduction
Respiratory viral diseases can be spread when a virus-containing particle is aerosolized, frequently through coughing, sneezing, and talking, and subsequently comes into either direct or indirect contact with another individual via the mouth, eyes or nose, or by being inhaled into the lungs (Gao et al., 2009, Verreault et al., 2008, Belser et al., 2014). The infectivity of an airborne virus depends on factors such as relative humidity (RH), temperature, aerosolization medium, and residence time in the air (Verreault et al., 2008). Common diseases that can be transmitted through the air include chickenpox, measles, tuberculosis (TB), and influenza virus (Gao et al., 2009). In an age of globalization, increasingly mobile populations exacerbate the potential health risks from airborne infectious diseases by both increasing their spatial influence, and decreasing the time it takes to reach them (Charu et al., 2017, Fidler, 2004). This has led to several well-known pandemics including the emergence of Severe Acute Respiratory Syndrome (SARS) in 2003, which was associated with more than 700 fatalities in only a few months (WHO, 2004), and the influenza A (H1N1) virus, which by May 2009, had over 10,000 laboratory-confirmed cases across 41 countries (WHO, 2009).
The influenza virus has been the subject of much research since its discovery more than 70 years ago, and debate continues over the relative importance of its different potential transmission routes: airborne, droplet, or contact (Brankston et al., 2007, Teunis et al., 2010, Weber and Stilianakis, 2008). Currently, the airborne route followed by inhalation at a close proximity is considered to have high infectivity, and may be a pathway for influenza transmission in an indoor environment (Teunis et al., 2010, Weber and Stilianakis, 2008). Many studies attempted to assess the importance of this route within a clinical setting. Bischoff, Swett, Leng, and Peters (2013) used three Anderson air samplers to detect the aerosolized influenza virus (RNA) in 26 of the 61 symptomatic patients admitted to the emergency department of a medical center, with the highest concentrations occurring within 1 foot from the patient's head; however, the virus was also detected in particles (<4.7 µm) up to 6 feet away. Lindsley et al. (2010) detected viable influenza virus with a NIOSH two-stage bioaerosol cyclone sampler and an SKC BioSampler® from the coughs of 38 of the 58 patients presenting with symptoms at a student health clinic; the majority (42%) of viruses were detected in particles <1 µm, while 35% were detected in particles >4 µm. Lednicky and Loeb (2013) sampled influenza H3N2 with a Sioutas Personal Cascade Impactor and an SKC BioSampler and suggested that viable virus may be produced by influenza patients. Marchand, Duchaine, Lavoie, Veillette, and Cloutier (2016) sampled the surrounding air during bronchoscopy procedures with a wet wall cyclonic sampler and an impactor to examine whether the aerosolized particles contained pathogens that were dangerous to healthcare workers. Although the influenza virus was not detected, several bacteria were, leading authors to conclude that aerosolized pathogens could possibly pose an occupational health risk. Another study by Leung et al. (2016) sampled with cyclone samplers in the rooms of hospital patients with confirmed influenza virus. Although no aerosolizing procedures were conducted during the measurement period, they detected the virus in 50% of collected air samples by PCR, and highlighted the need for additional studies that collected air samples during routine patient procedures, in order to gain further understanding regarding the risks posed to healthcare workers.
Although influenza is generally considered to be spread by larger droplets, these findings support the possibility that influenza transmission can also occur via an airborne route, especially within the immediate vicinity of an influenza patient. This poses significant challenges for healthcare workers, who currently adopt face masks to prevent transmission, with special ventilation controls only for certain circumstances (Bischoff et al., 2013, Brankston et al., 2007). Nevertheless, there have been numerous cases of healthcare workers being infected during routine healthcare procedures (Lau, 2004), and calls for further research on the viability of airborne influenza viruses and the risk of transmission have been made (Lindsley et al., 2010).
Airborne influenza virus is highly infectious. As such, it is imperative that efficient samplers are used to collect and quantify these pathogens in order to determine their spread. This will not only benefit aerobiological research, but also enable us to evaluate whether standard precautions currently undertaken in a clinical setting are adequate to protect patients and health-workers from infection. Samplers currently used to collect pathogens include: solid impactors (the Anderson sampler, slit sampler, and cyclone sampler); liquid impactors (All-glass Impinger (AGI) and SKC BioSampler); and filters (gelatin filter, polytetrafluoroethylene (PTFE) filter, and glass fiber filter). The Anderson sampler is most efficient at capturing larger particles (0.65–7.5 µm), and also at providing size distributions (Verreault et al., 2008). Slit samplers are capable of determining the aerosol concentration of bacteria as a function of time (Verreault et al., 2008). The AGI and SKC BioSamplers operate on similar principles and demonstrate comparable performances (Hogan et al., 2005). Filters are widely used because of their high physical collection efficiencies, for example, the gelatin filter was reported to have a high physical collection efficiency (>93%) for MS2 virus (Burton, Grinshpun, & Reponen, 2007). Burton et al. (2007) recommend 0.3-µm PTFE filters for long-term virus sampling. The glass fiber filter was evaluated for capturing endotoxin and influenza virus during air sampling (Blachere et al., 2007, Thorne et al., 1997). In this study, we evaluated the performance of the 5 mL SKC BioSampler (also referred to as an impinger), gelatin filter, and glass fiber filter in a laboratory setting with aerosolized solutions of influenza virus.
The SKC BioSampler is being increasingly used in clinical settings to capture viruses and bacteria due to its relatively higher collection efficiency for viable virus capture compared to the gelatin filter. It has also been used to capture the bacteriophage MS2, which was generated from a vomiting simulation machine to study the transmission of the human noroviruses (Tung-Thompson, Libera, Koch, Francis III, & Jaykus, 2015). The SKC BioSampler was chosen by Cao et al. as the reference with which to evaluate the NIOSH two-stage cyclone bioaerosol sampler and the SKC AirCheck TOUCH personal air sampler (Cao et al., 2011, Nguyen et al., 2017). Fabian, McDevitt, Houseman, and Milton (2009) reported that the gelatin filter, cascade impactor, and Teflon filter recovered around 7–22% of the amount of infectious virus recovered from the SKC BioSampler. This may be attributed to the liquid media in the collection vessel, which provides a favorable condition for viral preservation (Lindsley et al., 2010). However, while it has been reported to efficiently collect submicrometer particles, it was unable to adequately collect bioaerosols in the 30–100 nm size range (Hogan et al., 2005).
The gelatin filter has a high physical collection efficiency in the 100–900 nm size range (Burton et al., 2007), and it even outperformed the SKC BioSampler for the influenza A virus when sampling over a short period of time (under two minutes) (Wu, Shen, & Yao, 2010).
The glass fiber filter has been used for a variety of applications, for example, ambient air sampling of trace elements in particulate matter (Jena and Singh, 2017, Tian et al., 2016), and aromatic amines in cigarette smoke (Zhang, Bai, Zhou, Liu, & Zhou, 2017). Although high collection efficiencies have been reported for fine particles (VanOsdell, Liu, Rubow, & Pui, 1990), there are only limited studies that have applied glass fiber filter technology to aerobiology research. For viral aerosols, the available data on glass fiber filter performance is almost exclusively limited to bacteriophages (Harstad, 1965, Harstad et al., 1967). It is therefore important to extend this research to other viruses, especially given the increasing interest among researchers and industry in low-cost filtration methods to recover or remove virus aerosols. In addition, the glass fiber filter was a good counterpart to the gelatin filter for the experiments described in this paper.
The collection efficiency of each sampler is critical for the reliability of laboratory and field measurements. This is determined by examining two critical parameters: the physical collection efficiency of the sampler, and the recovery rate of infectious particles. The physical collection efficiency describes how many particles can be sustained on the filter or in the collection vessel, regardless of whether these particles are still infectious. This can be determined with microorganisms or small pellets, such as polystyrene latex (PSL) particles or sodium chloride particles. For the SKC BioSampler, the evaporation of the liquid may increase particle bounce or lead to reaerosolization of the collected microorganisms, which decreases the collection efficiency of the device (Grinshpun et al., 1997, Lin et al., 1997). The sampling devices and collection process itself can also have negative effects on the recovery rate of collected microorganisms, for example, desiccation of the microorganism may occur in a filter medium (Dabisch et al., 2012). Dramatic changes to a microorganism's surroundings may also shock and kill them (Lindsley et al., 2010). Therefore, the ability to recover infectious particles, which is measured by the ratio of viable viruses to the total viruses sent to the sampler, is also used to estimate the portion of microorganism that remains infectious after sampling.
Existing research has evaluated and characterized different samplers according to different standards. Fabian et al. (2009) compared the T/I value (total virus concentration / infectious concentration) of the SKC BioSampler, cascade impactor, Teflon filters and gelatin filters with nebulized virus particles whose size is above 1 µm. Turgeon, Toulouse, Martel, Moineau, and Duchaine (2014) compared the collection efficiency of the NIOSH sampler and the SKC BioSampler for five different bacteriophages. Despite theses studies that incorporate different samplers in their sampling portfolio, uncertainty remains regarding the percentage of the total aerosolized viruses, which are subsequently recovered. This percentage has been reported for the 20 mL SKC BioSampler with MS2 virus (Hogan et al., 2005; Lednicky et al., 2016); however, this percentage is still unknown for 5 mL SKC BioSampler with the real influenza virus.
The objective of this study was to estimate the amount of influenza virus that each sampler was able to recover compared to the total aerosolized particles. The size of interest was 10–400 nm, which is closer to the size of the airborne influenza virus when comparing with the size range (above 1 µm) reported by Fabian et al. (2009). The performance of the glass fiber filter and the gelatin filter as for viral recovery was further evaluated. Room sampling was also simulated by comparing direct sampling with indirect chamber sampling.
2. Materials and methods
2.1. Test virus
The H1N1 influenza A virus A/Puerto Rico/8/1934 (IAV-PR8) was used as the test virus in this study and obtained from St. Jude Children's Research Hospital. It was propagated in the allantoic cavities of 10-day-old embryonic chicken eggs (Boon et al., 2010). The allantoic fluid containing IAV-PR8 was diluted in sterile phosphate buffered saline (PBS) at the indicated doses of 105–10 tissue culture infectious doses per mL (TCID50/mL).
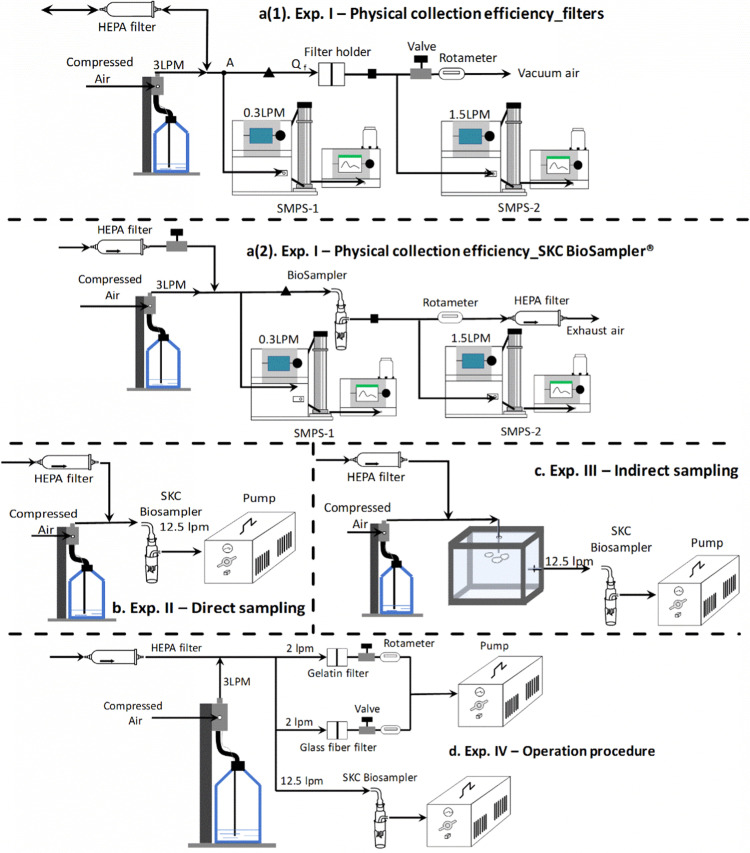
2.2. Test samplers
Three samplers were compared: the SKC BioSampler (5 mL, SKC Inc., Eighty-Four, PA, USA), the gelatin filter (3.0-μm pore size, Sartorius AG, Göttingen, Germany), and the glass fiber filter (Grade EPM 2000, 47 mm, Whatman®, USA) (Fig. S1). Each of these collection samplers possess different structures, working principles, and recovery methodologies. The SKC BioSampler uses inertial impaction to collect and subsequently entrain particles in its sampling media. It consists of three parts: an inlet, a critical orifice section, and a 5 mL collection vessel. These parts were autoclaved separately under a 30-min sterilization cycle and a 30-min dry cycle before sampling to minimize potential cross-contamination. The temperature for the autoclave was approximately 122 °C, and it was maintained at a pressure of 1.24 pbar. The critical orifice section contains three 0.63 mm tangential critical orifices (Hogan et al., 2005). Connecting the outlet to a vacuum pump creates a negative pressure over 0.5 atm (15 in Hg) downstream of the critical orifices. This high-pressure drop across the critical orifices maintains a stable 12.5 L/min flowrate and creates a vortex in the media in the collection vessel into which the particles are subsequently entrained.
The gelatin and glass fiber filters must be placed in a filter holder (Fig. S1). When air is drawn through the holder, particles collect on the dry film of the filter through the processes of diffusion, interception, and impaction. Because they trap viruses on a dry film rather than in a liquid, additional extraction processes are required. For the gelatin filter, this poses less of an issue, as the filter can be fully dissolved in a liquid (universal viral transport media, UTM (Becton, Dickson and Company, Sparks, MD), was used in the described experiments). In contrast, the glass fiber filter is insoluble and requires extra processing to extract the captured viruses. Two processing methods, submersion and swab, were tested separately on the glass fiber filters. For the submersion method (Blachere et al., 2007), the glass fiber filter was torn into four pieces before being crumpled into a 3 mL vial of UTM solution. To reduce the mechanical agitation that damaged the viral particles, we shortened the vortex time to thirty seconds and prolonged the submersion time to 15 min. Then, the filter was removed and the liquid was centrifuged (1000×g, 10 min) at 4 °C. For the swab method, the glass fiber filter was brushed with a Copan FLOQ swab (Becton, Dickinson and Company) using both vertical and horizontal strokes. Next, the flocked swab was placed into a 3 mL vial of UTM for further analysis.
2.3. Biological analysis with viral culture and real-time PCR
Culture and real-time PCR techniques were used to evaluate the amount of virus collected from each sampler. For the culture method, virus titers were determined in Madin-Darby canine kidney (MDCK) cells as described previously (Boon et al., 2010). Briefly, confluent monolayers of MDCK cells were grown overnight in 96 well-plates. The following day, the cells were washed with phosphate buffered saline (PBS) and inoculated with ten-fold serial dilutions (10−1 to 10−8) of allantoic fluid or sample in Minimal Essential Medium containing penicillin, streptomycin, L-glutamine, and vitamins plus 0.1% bovine serum albumin (M0.1B) for one hour at 37 °C and 5% CO2. After one hour, the cells were washed once with PBS and 200 µl of M0.1B with 1 µg/mL TPCK-trypsin was added to each well. After 72 h at 37 °C and 5% CO2, the presence of influenza A virus was determined by hemagglutination assay using 0.5% turkey red blood cells (Boon et al., 2010, Williams et al., 2016). The 50% Tissue Culture Infectious Dose (TCID50) was determined by the Reed-Muench method and presented as TCID50/mL (Reed & Muench, 1938).
For real-time PCR based detection, the BioFire FilmArray Respiratory Panel (bioMerieux, Durham, NC) and the Xpert Flu/RSV Assay (Cepheid, Sunnyvale, CA) were used to detect the virus. It is important to note that neither of these instruments can provide a quantitative assay. While the Biofire presents results as either positive or negative, the Xpert demonstrates decreasing cycle threshold values with increasing viral concentration ( Table 1). Both tests were performed according to manufacturer recommendations, with 300uL used for Xpert testing and 300uL used for Biofire analysis.
Table 1.
Correlation between the virus suspension concentration and the cycle value from the Xpert for qualitative assessment.
| Virus suspension concentration (TCID50/mL) | Cycle value |
|---|---|
| 100 | 21 |
| 100 | 20.4 |
| 1000 | 17.6 |
| 1000 | 17.2 |
| 10,000 | 13.9 |
| 10,000 | 14 |
2.4. Experimental set-up
Table 2 provides details on the experiments that were performed in this study, characterizing both the physical collection efficiency (Experiment I) and the virus sampling efficiencies (Experiments II, III, and IV). Virus concentrations, test sampling times, and flowrate specifications are also presented in the table. The experimental set-up of each test is shown in Fig. 1. For all experiments, a constant output atomizer (TSI 3076), operating at a flowrate of 3 L/min and a pressure of 35 psi, was used to aerosolize particles. The particle number size distributions both upstream (triangle in Fig. 1a) and downstream (square in Fig. 1a) of the samplers were measured using two scanning mobility particle sizers (SMPS). A HEPA filtered air inlet was included in the setup, to create an open system and allow for the removal of any extra air flow. All tests were conducted in a fume hood and the exhaust air was ventilated directly from the fume hood to a sterilizing exhaust.
Table 2.
Summary of the experimental plan.
| Experiment | Test | Sampler | Atomized solution (TCID50/mL) | Run length (minutes) | Specifications | |
|---|---|---|---|---|---|---|
| I. Efficiency | 1 | Gelatin | PBS | 3×3 min | Flowrate | 2, 3, 4 L/min |
| 2 | Glass | 3×3 min | 2, 3, 4 L/min | |||
| 3 | SKC | 3×3 min | 12.5 L/min | |||
| II. Direct sampling | 1 | SKC | 10 | 20 | SKC solution | 5 mL UTM |
| 2 | 10 | 5 | 5 mL UTM | |||
| 3 | 10 | 4 | 4 mL UTM | |||
| 4 | 10 | 5 | 4 mL PBS | |||
| 5 | 100 | 10 | 4 mL PBS | |||
| 6 | 1000 | 10 | 4 mL PBS | |||
| 7 | 10,000 | 10 | 4 mL PBS | |||
| 8 | 100,000 | 10 | 4 mL PBS | |||
| III. Indirect sampling | 1 | SKC | PBS | 10 | SKC solution | 4 mL PBS |
| 2 | 10 | 10 | 4 mL PBS | |||
| 3 | 10 | 10 | 4 mL PBS | |||
| 4 | 100 | 10 | 4 mL PBS | |||
| 5 | 1000 | 10 | 4 mL PBS | |||
| 6 | 10,000 | 10 | 4 mL PBS | |||
| 7 | 100,000 | 10 | 4 mL PBS | |||
| IV. Operation procedure | 1 | SKC, Gelatin Glass | 1000 | 10 | Glass retrieve method* | submersion |
| 2 | 1000 | 10 | surface swab | |||
| 3 | 100,000 | 10 | submersion | |||
| 4 | 100,000 | 10 | surface swab | |||
SKC = SKC BioSampler; Gelatin = gelatin filter; Glass = glass filter; Samples from the gelatin filter and SKC were retrieved using the same method. The gelatin filter was dissolved in 3 mm of UTM solution. The SKC solution was manually removed from the collection vessel. *Only the Glass fiber filter had different retrieval methods presented in this table.
Fig. 1.
Experimental set-up. *Exp. = experiment; ▲ = the upstream of the sampler; ■ = downstream of the sampler.
2.4.1. Physical collection efficiency
To measure the physical collection efficiency (Experiment I), the atomizer was filled with 200 mL PBS solution and was used to generate nanometer-size particles of PBS, with an average Geometric Mean Diameter (GMD) of 41.71±0.29 nm, ranging from 9.82 to 414.20 nm. The physical collection efficiency (), defined in Eq. (1), was determined from Experiment I, where is the particle diameter, and and are the size distributions at the exit of the nebulizer and downstream of the test samplers.
| (1) |
Fig. 1a(1) depicts the setup for the gelatin and glass fiber filters. The filters were operated at three different flowrates (Q f = 2, 3, and 4 L/min), and controlled using a valve and rotameter. In the SMPS-1 settings, in order to achieve a wide measurement size range, while keeping the particle concentration below the CPC saturation concentration, a sheath flowrate of 6 L/min and a CPC flowrate of 0.3 L/min were used. The SMPS-2 was operated under a high-flow mode (1.5 L/min) to ensure the stability of the flow entering the instrument. When the filter was operated at a flowrate of 2 L/min, the total flowrate before node A was 2.3 L/min (2 L/min from the filter and 0.3 L/min from the SPMS-1). The addition of a HEPA filter permitted the removal of 0.7 L/min of filtered air, which balanced the flowrate in the system. In contrast, when the filter was operated at flowrates of 3 and 4 L/min, the total flowrate before node A was 3.3 and 4.3 L/min, respectively. This time, the HEPA filter was used to supply additional filtered air to the system (0.3 and 1.3 L/min, respectively). During this experiment, the physical collection efficiency of the gelatin and glass fiber filters were averaged over 9 min. The SKC BioSampler was operated in much the same way, except for the higher flowrate (12.5 L/min), depicted in Fig. 1a(2).
2.4.2. Virus collection efficiency
To evaluate the virus collection efficiency (Experiments II-IV), different concentrations of viruses, suspended in PBS solution, were atomized, generating particle sizes in similar ranges as mentioned previously. This time; however, the aerosolized particles were comprised of a mixture of viruses and PBS. Similar to the findings from Hogan et al. (2005), the size distributions did not change with increasing virus concentrations. This is due to the low virus mass to PBS solute mass ratio, and the fact that the PBS concentration was identical for each test. In 1 mL of 105 TCID50/mL virus suspension, the mass of the total virus is 10−9 smaller than the mass of the total solutes from the PBS. However, the concentration of each virus suspension will influence the number of viruses carried in each droplet or particle.
Experiments II, III, and IV evaluated the capability of each sampler to collect viruses within the different experimental parameters. In Experiment II (direct sampling) (Fig. 1b), virus suspensions ranging from 10 to 105/mL were aerosolized and sent directly to the SKC BioSampler, which contained either a 4 mL solution of UTM or a 4 mL solution of PBS in its collection vessel. Tests 1–4 compared the performance of the PBS and UTM solution. However, the UTM solution proved an unsuitable collection liquid due to excessive foaming which led to unacceptable evaporation and liquid losses; therefore, the remaining tests only used PBS solution as the collection fluid (Prior testing had demonstrated comparable PCR testing results with viral solutions in PBS and UTM, shown in Table S1). Tests 5–8 examined the sampling capability of the SKC BioSampler at different concentrations of virus suspensions. The test parameters in Experiment III (indirect sampling) (Fig. 1c) echoed those of Experiment II, except for the addition of a chamber measuring 15” ×15” ×15” that was placed between the atomizer and the SKC BioSampler. This forced the virus droplets generated by the atomizer to undergo additional evaporation, diffusion, and convection inside the chamber (Wang et al., 2016), before being sampled by the SKC BioSampler, thus simulating more closely the physical changes of aerosols generated in a clinical setting, such as a hospital room. During the sampling process, the relative humidity levels within the chamber were sustained around 58.3–68.1%. Theoretically, the sampling efficiency for the indirect method should be lower than for the direct method due to particle diffusion losses in the chamber. It is also worth noting that the size of the chamber will influence particle loss and the sampling results. For these experiments, the chamber size was dictated by the space inside the fume hood where the tests were being conducted.
Experiment IV (Fig. 1d) compared the side-by-side sampling performances of the SKC BioSampler, the gelatin filter, and the glass fiber filter. The aerosolized particles were sent to each sampler simultaneously. The flowrates remained the same for each test: 12.5 L/min for the SKC BioSampler, and 2 L/min for the gelatin and glass fiber filters. The collection vessel of the SKC BioSampler was always filled with 4 mL PBS solution. The liquid from the SKC BioSampler and the UTM solution in which the gelatin filters were dissolved, were stored directly as samples. To retrieve the viruses from the glass fiber filters, Tests 1 and 3 used the submersion method, while Tests 2 and 4 used the swab methods (described previously).
3. Results and discussion
3.1. Experiment I: physical collection efficiency
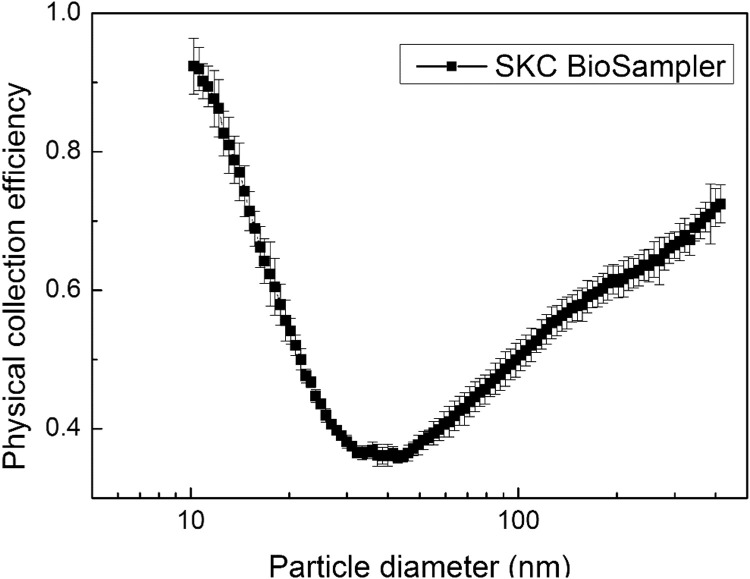
The physical collection efficiencies were calculated for each sampler using Eq. (1) and the particle number size distributions obtained from the 2 SMPSs during Experiment I. Fig. 2, Fig. 3 present the results for the SKC BioSampler and gelatin filter respectively. The SKC BioSampler demonstrated a U-shaped collection efficiency curve (Fig. 2), lowest for particles around 30–50 nm, which is close to the results from previous studies (Hogan et al., 2005, Wei et al., 2010), but higher than the efficiency reported by Hogan et al. (2005). However, while Hogan et al. (2005) tested the 20 mL SKC BioSampler, the 5 mL SKC BioSampler was used in this study. According to Zheng and Yao (2017), SKC BioSamplers with different vessel volumes demonstrated different collection efficiencies in the bacterial size range. Presumably, this would pertain to virus size range also, therefore, differences in physical collection efficiencies may have resulted from the sizes of the collection vessels. In addition, although we suspect that viral particles in the size range of 10–400 nm were not collected, we cannot rule out the mechanism of reaerosolization (Riemenschneider et al., 2010, Grinshpun et al., 1997), given the fierce vortex generated by the high flowrate, and the hydrophobic nature of the test virus. Reaerosolization is determined by multiple factors: sampling time, aerosol flowrate, and the suspension concentrations of the liquid in the collection vessel. Although reaerosolization has not been characterized as a significant limitation of the SKC BioSampler, it may still influence the overall performance (Riemenschneider et al., 2010, Grinshpun et al., 1997). Within the examined size range, the highest efficiencies were also observed at 10 nm (0.9), possibly due to enhanced diffusion inside the collection vessel, and 300–350 nm (~0.7), which has been attributed to enhanced impaction and interception (Hogan et al., 2005). The strong dependence between collection efficiency and particle size emphasizes the importance of knowing the size range of the pathogen being collected, which in the case of spherical IAV-PR8 virus, is 80–120 nm (Rossman, Leser, & Lamb, 2012).
Fig. 2.
The physical collection efficiency of the SKC BioSampler. *The flowrate was kept constant at 12.5 L/min. Results are based on the average of 9 test runs.
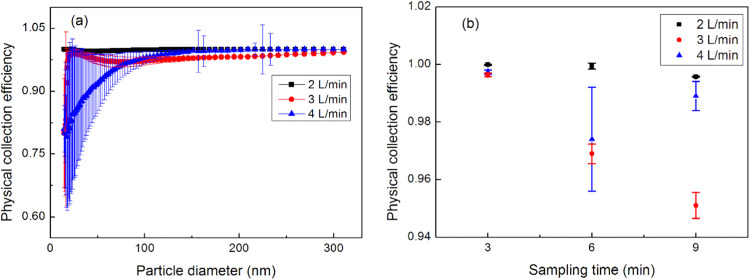
Fig. 3.
The physical collection efficiency of the gelatin filter with varying flowrates and sampling times.
Fig. 3a depicts the influence of particle size and flowrate on the physical collection efficiency of the gelatin filter, which was generally able to capture larger particles more efficiently. However, the sampling flowrate influenced the efficiency curves: a collection efficiency of almost 100% across all particle sizes was observed at the lowest flowrate, while the collection efficiency increased with particle size at the highest flowrate. In addition, the performance of the gelatin filter was unpredictable under higher flowrate. This trend persisted during repeated tests. To show this phenomenon, the total physical collection efficiency (), defined in Eq. (2), where and represent the upstream and downstream particle number concentrations, was calculated and plotted against time (Fig. 3b).
| (2) |
In general, it is difficult to determine the influence of flowrate on a gelatin filter. In these experiments, the gelatin filter collecting at a flowrate below 2 L/min demonstrated stable performances during three repetitions each lasting 9 min. However, the physical collection efficiencies when collecting between 3 and 4 L/min were unpredictable. The texture of the gelatin filter deteriorated during the sampling period, changing from brittle to ductile as the aerosolized particles were continually introduced (Wu et al., 2010). Fig. S3 is a photo of the gelatin filter dissolving within the filter holder after sampling. The gelatin filter is sensitive to relative humidity, which may impede its performance over extended periods of time (Haig, Mackay, Walker, & Williams, 2016), potentially making it an unpredictable collection medium. Results indicate that a dry flow, and a low flowrate, as well as a short sampling time are the best operating conditions for the gelatin filter, which seems to perform sub-optimally when these conditions are reversed. In contrast, the glass fiber filter demonstrated a very stable physical collection efficiency (100%) across all measured particle sizes, and during the different flowrates. The efficiency remained stable for the entire test duration (~ 2 h).
3.2. Experiments II and III: comparison of direct and indirect sampling
Experiments II and III focused on the collection efficiency of viral particles from the SKC BioSampler only. Three methods were used to detect collected viral particles: 1) BioFire Multiplex Respiratory Panel (Biofire), 2) Xpert Flu/RSV assay, and 3) culture. While culture is able to determine viable virus concentration, Biofire and Xpert, which are both real-time PCR methods, are able to detect the presence of viral RNA but are not able to distinguish whether there are viable viruses or not. Tests 5–8 used PBS solution as the virus collection liquid. For the remainder of this section, the term “suspension” will refer to the solution inside the atomizer, and the term “liquid sample” will refer to the solution inside the collection vessel of the SKC BioSampler. The results from the BioFire and Xpert PCRs are presented in Table 3 and Fig. 4, respectively. In both direct and indirect sampling, the BioFire PCR reported positive influenza A results only from starting suspensions of 10,000 and 100,000 TCID50/mL. The Xpert PCR was able to detect virus from starting suspensions of 1000, 10,000 and 100,000 TCID50/mL in direct sampling, and suspensions of 10,000 and 100,000 TCID50/mL in indirect sampling. However, the Xpert cycle threshold for 1000 TCID50/mL was 35.4 for the liquid sample from both tests, compared to 34 and 32.3 for 10,000 and 28.2 and 27.3 for 100,000 TCID50/mL, respectively, reflecting the relatively lower abundance of virus in the 1000 TCID50/mL sample. Although lower numbers of PCR cycles were needed to detect the virus in the more concentrated suspensions, results still indicated that the relative abundance of virus in these samples was low (refer to Table 1). The viral culture results were similar ( Table 4); a positive well was detected only in the sample liquid for the highest virus suspension (100,000 TCID50/mL). The positive well was detected with a 1:10 dilution ratio, which translates to a virus concentration of the liquid sample at approximately 100 TCID50/mL.
Table 3.
Summary of BioFire results from the direct and indirect sampling.
| Experiment | Atomized virus suspension (TCID50/mL) | Interpretation |
|---|---|---|
| Direct sampling-Experiment II | 100 | Negative |
| 1000 | Negative | |
| 10,000 | Influenza A | |
| 100,000 | Influenza A | |
| Indirect sampling-Experiment III | PBS | Negative |
| 10 | Negative | |
| 100 | Negative | |
| 1000 | Negative | |
| 10,000 | Influenza A | |
| 100,000 | Influenza A |
*Identical results were obtained with all repeated sampling.
Fig. 4.
Summary of Xpert results from a) direct sampling, and b) indirect sampling using the SKC BioSampler. *PBS = only PBS was aerosolized for a control; all other experiments used PBS plus different concentrations of virus; Assay 1 = first batch of results; Assay 2 = second batch of results; Neg = negative (no influenza A virus was detected in the sample).
Table 4.
Summary of culture results from the Experiment IV.
| Experiment (Test number) | Atomized virus suspension (TCID50/mL) | Sampler | Culture results (TCID50/mL) |
|---|---|---|---|
| Experiment IV (1) | 1000 | SKC | Negative |
| Gelatin | Negative | ||
| Glass (submersion) | Negative | ||
| Experiment IV (2) | 1000 | SKC | Negative |
| Gelatin | Negative | ||
| Glass (swab) | Negative | ||
| Experiment IV (3) | 100,000 | SKC | 1000 |
| Gelatin | <31 | ||
| Glass (submersion) | <31 | ||
| Experiment IV (4) | 100,000 | SKC | 100 |
| Gelatin | 47 | ||
| Glass (swab) | <31 |
The Biofire PCR results from indirect sampling (with the chamber) were similar to those of direct sampling, with positive influenza A virus detected in the liquid samples of 10,000 and 100,000 TCID50/mL suspensions (Table 3). However, the Xpert PCR was able to detect virus when the atomized virus suspension concentration was 1000 TCID50/mL or higher during direct sampling, but was only able to detect virus in the 10,000 and 100,000 TCID50/mL suspensions during indirect sampling. Another thing to note is the increased number of Xpert PCR cycles that were required to detect any viral particles for the atomized 100,000 TCID50/mL suspension during indirect versus direct sampling, which indicates lower amounts of virus present in the samples collected during indirect sampling. The particle size distributions for direct sampling and indirect sampling are included in the supplementary material (Fig. S2), showing that smaller particles (<50 nm) may be scavenged due to diffusion loss. This may indicate that fewer viral particles were collected during indirect compared to direct sampling and consideration should be paid regarding the type of sampling (direct versus indirect) that is being conducted in order to interpret the results obtained. These findings were reinforced by the culture results.
3.3. Experiment IV: comparison of different samplers and different operation procedures
First, the extraction rate for the gelatin and glass fiber filters were compared with direct inoculation experiments. Two glass fiber filters and one gelatin filter were placed in individual petri dishes. Virus suspension solution, containing approximately 31,000 live viruses, was then injected onto each filter inside of the petri dish. Next, the gelatin filter was dissolved in 3 mL of UTM solution. One of the glass fiber filters was treated with the submersion method using 3 mL of UTM solution, and the other one was treated with the swab method using another 3 mL of UTM solution. Subsequent culture analysis of the liquid samples detected concentrations of approximately 4700, 470, and <31 TCID50/mL for the dissolved gelatin filter, submerged glass fiber filter, and the swabbed glass fiber filter, respectively. After multiplying the detected concentrations by the volume of the UTM solution (3 mL), the total number of viruses detected from the gelatin filter, the submersion method, and the swab method were calculated to be 14,100, 1410, and <93, respectively. These results indicate that the gelatin filter outperformed the glass fiber filter ten-fold and was able to retrieve around 45% of the total virus loading that had been injected into the petri dish. It is likely that viruses were killed during the processing, which would explain the moderate virus recovery percentage of the gelatin filter. The glass fiber filter fared even worse, with an extraction rate of only 5% and 0.3% for the submersion and swab methods, respectively. A similar concern, regarding the impact of low extraction rates on qPCR results, was previously expressed by Hospodsky, Yamamoto, and Peccia (2010). Either the glass fiber filters may bind viruses tightly to their surfaces, or the recovery process involving mechanical agitation and vibration may deactivate virus.
After direct inoculation experiments, the liquid samples from the SKC BioSampler, the gelatin filter, and the glass fiber filter collected during atomization of the viral suspension solutions were compared using culture and Xpert PCR; results are reported in Table 4 and Fig. 5. As previously described, no viable viruses were detected during viral culture of the samples collected for the 1000 TCID50/mL atomized virus suspensions by any of the samplers. For suspensions of 100,000 TCID50/mL, only the SKC BioSampler and the gelatin filter produced positive culture results. The SKC BioSampler retrieved approximately 100–1000 TCID50/mL of 4 mL liquid in the collection vessel using viral culture, which was the highest retrieval rate of all the samplers. This was consistent with the Xpert PCR results, which showed that sample liquid from the SKC BioSampler required the lowest number of cycles to detect (Fig. 5). The gelatin filter retrieved around 30–50 TCID50/mL of 3 mL of UTM solution. The number of viral particles captured by the gelatin filter was around 3 – 35% of what was captured by the SKC BioSampler.
Fig. 5.
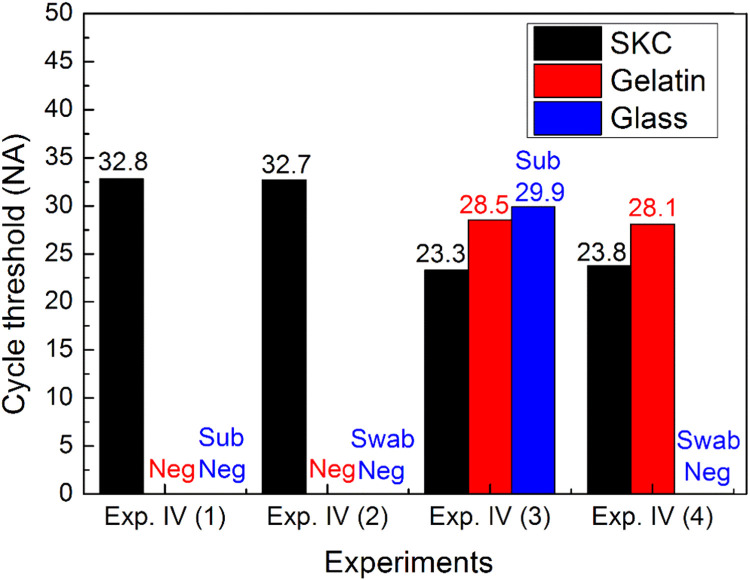
Summary of Xpert results from Experiment IV. *SKC = SKC BioSampler; Gelatin = gelatin filter; Glass = glass fiber filter; Exp = experiment; Sub = submersion method; Swab = swab method; Neg = negative (no influenza A virus was detected in the sample).
For the glass fiber filter, the Xpert reported positive results for the submersion method, and negative results for the swab method for the same virus suspension (100,000 TCID50/mL). Therefore, the submersion extraction method was more effective than the swab method.
It is also important to compare the number of viral particles retrieved to the total number of viral particles that entered the sampler. To estimate this number, the liquid consumption rate of the atomizer was estimated and compared to the viral culture results. In general, the atomizer consumed 7 mL hourly. This means that an atomized virus suspension of 100,000 TCID50/mL would generate a total of 90,000 viral particles in the air flow entering the SKC BioSampler, over a period of 10 min (the length of sampling for the current testing). This was calculated by multiplying the initial virus concentration in the atomizer (100,000 TCID50/mL) by the hourly liquid consumption of the atomizer (7 mL/h), multiplied by the duration of the experiment (1/6 h), multiplied by the SKC BioSampler flowrate (12.5 L/min) divided by the total flowrate for all samplers (16.5 L/min). Since there was 4 mL of liquid sample inside the SKC BioSampler, there would be around 400 to 4000 viable viruses (100–1000 TCID50/mL * 4 mL) (Table 4) captured by the SKC BioSampler. Therefore, the SKC BioSampler was able to collect only 0.5–5% of atomized viruses. This percentage is comparable with the percentage reported by Hogan et al. (2005) and Lednicky et al. (2016) for the 20 mL SKC BioSampler. A low collection efficiency will be a concern in the clinical setting, especially for low virus concentrations. This work indicates that if a positive result is obtained using SKC BioSamplers, the virus concentration in the surroundings is likely to be high. However, negative results can not assure a healthy environment. Further work and improvements are required to extrapolate the results in practical use for quantitatively measuring airborne viruses. The recently developed laminar-flow, water-based viable virus aerosol sampler (VIVAS) may be as a promising technology to improve the viable sampling efficiency (Lednicky et al., 2016; Pan et al., 2016).
Although the virus collection efficiency for the SKC BioSampler is relatively low, it far outperforms both the gelatin (maximum of 1.5%) and glass fiber filter (too low to quantify). There are several reasons for the overall low collection efficiency reported here. The large pressure drop in the atomizer may have killed and deactivated some of the virus particles. The similar T/I values of the prepared viral suspension and the BioSampler® collected liquid implies that the nebulization process did not overly affect the viability of virus (Fabian et al., 2009). However, the T/I value characterizes the viability of the captured influenza virus, rather than quantatively estimates the proportion of the viruses captured by the samplers. So, it is possible that particles are destroyed or deactivated during the atomization process. Furthermore, the droplets from the atomizer evaporated gradually during the sampling process, which may lower the portion of viable viruses (Haig et al., 2016). Equally, increasing sampling time will also desiccate or deteriorate the filters, either of which could compromise the viability of the pathogen (Haig et al., 2016, Wu et al., 2010). This evaporative process may also decrease the risk of viable virus survival in clinical settings. Specific to the SKC BioSampler, the pressure variation at the critical orifice and the shear force due to the violent vortex in the collection vessel could also influence the viability of the viruses (Haig et al., 2016). There may also have been reaerosolization of the particles due to the high sampling flowrate and the low virus concentration of the liquid in the collection vessel (Riemenschneider et al., 2010). Virus particles adhere to the walls of the collection vessel may further decrease the collection efficiency (Haig et al., 2016). Studies have also shown that the volume of the collection liquid can impact viability (Zheng & Yao, 2017). The dry surfaces intrinsic to the filters may not be a suitable environment to sustain virus viability (Dabisch et al., 2012, Lindsley et al., 2010). What is more, the additional extraction processes required for retrieval from the filters could result in even further losses.
4. Conclusions
Results indicated that the gelatin and glass fiber filters demonstrated high physical collection efficiencies. However, concerns over the stability of the gelatin filter were noted. The glass fiber filter maintained high physical collection efficiency across all measured particle sizes, sampling flowrates, and sampling times. The 5 mL compact SKC BioSampler demonstrated a slightly lower physical collection efficiency, especially for particles around 30–50 nm, but had the highest virus collection efficiency compared to either of the filters. This was most likely due to the liquid media inside the collection vessel, which provided a more suitable environment for the preservation of viruses. Although the SKC BioSampler demonstrated the highest retrieval rates, it still only managed to recover at most 5% of the total influenza A virus particles. However, the retrieval rate for the gelatin filter (at most 1.5%) and glass filter (too low to quantify) were still lower. In order to obtain positive results for any of the samplers, the total concentration of viruses entering the sampler must be considered. This poses a challenge when working in the field where there is expected to be substantial variability in viral shedding by a patient depending on a patient's immune status and vaccination status, how long they have been symptomatic and whether they are taking antiviral medications.
Acknowledgements
This project was supported in part by the U.S. Centers for Disease Control and Prevention (CDC) Safety and Healthcare Epidemiology Prevention Research Development (SHEPheRD) program, contract number 200–2011-42070.
Biographies
Jiayu Li finished her BS in Environmental Engineering from Tsinghua University in 2014. She is now a PhD student in the Department of Energy, Environmental and Chemical Engineeirng at Washington University in St. Louis. Her research interests include sensor development and aerosol field measurement.
Anna Levey finished her PhD in Atmospheric Sciences from The University of Manchester in 2010. She is now a Research Scientist at the Department of Energy, Environmental and Chemical Engineering at Washington University in St. Louis. Her research interests include urban aerosol sampling, environmental and occupational health, and epidemiology.
Yang Wang finished his BS in Thermal Engineering from Tsinghua University in 2012. He is now a PhD student in the Department of Energy, Environmental and Chemical Engineeirng at Washington University in St. Louis. His research interests include nanoparticle flame synthesis, particle formation mechanisms, and aerosol instrumentation.
Caroline O’Neil earned a MA from the University of Missouri in 2006 and a MPH from St. Louis University in 2008. She is employed as a research coordinator for the Infectious Diseases division at Washington University School of Medicine. She has worked on projects focusing on aerosol generation during healthcare procedures, respiratory viruses in long term care settings, central-line associated bloodstream infection, ventilator-associated pneumonia, accidental falls among hospital patients, hospital readmissions, surgical site infections, and infections in outpatient hemodialysis patients.
Meghan A. Wallace received her Bachelors of Science in Clinical Laboratory Science at St. Louis University. She continued on to work at St. Louis Children's Hospital in the microbiology laboratory. She currently conducts research in the laboratory of Dr. Carey-Ann Burnham in the Pathology department at Washington University in St. Louis.
Carey-Ann D. Burnham PhD, D(ABMM) is an Associate Professor of Pathology & Immunology, Molecular Microbiology, and Pediatrics at Washington University School of Medicine in St. Louis, MO. She is also the Medical Director of Clinical Microbiology for Barnes Jewish Hospital. Burnham is the Program Director for the Medical and Public Health Microbiology Fellowship at Washington University, the Co-Editor of Medical Microbiology Question of the Day, and the Section Editor for "The Brief Case" for the Journal of Clinical Microbiology.
Adrianus CM Boon earned his Ph.D. in 2003 from The Erasmus University in The Netherlands. He is now an Assistant Professor in the Department of Medicine at Washington University School of Medicine in St. Louis. His research interests are influenza virology and viral pathogenesis.
Hilary Babcock is an Associate Professor of Medicine in the Infectious Diseases division at Washington University School of Medicine in St Louis. She received her medical degree from University of Texas Southwestern Medical Center in Dallas in 1994 and completed a Master’s degree in Public Health from St Louis University in 2006. She is the Medical Director for the Infection Prevention and Epidemiology Consortium of BJC HealthCare, a multi-hospital system in St Louis, and also the Medical Director for Occupational Health for Infectious Diseases for Barnes-Jewish Hospital and St Louis Children’s Hospitals. Her research interests include healthcare associated infections, transmission of pathogens in healthcare settings and protection of healthcare personnel.
Pratim Biswas received his Ph. D. in 1985 from California Institute of Technology. He is now the Lucy and Stanley Lopata Professor, Chair of the Department of Energy, Environmental and Chemical Engineering, and also the Director of the McDonnell Academy Global Energy and Environment Partnership (MAGEEP) at Washington University in St. Louis. His research interests include aerosol science and engineering, air quality engineering, environmentally benign energy production, and thermal sciences.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.jaerosci.2017.08.007.
Appendix A. Supplementary material
Supplementary material
References
- Bischoff W.E., Swett K., Leng I., Peters T.R. Exposure to influenza virus aerosols during routine patient care. Journal of Infectious Diseases. 2013;207(7):1037–1046. doi: 10.1093/infdis/jis773. [DOI] [PubMed] [Google Scholar]
- Belser J.A., Gustin K.M., Katz J.M., Maines T.R., Tumpey T.M. Influenza virus infectivity and virulence following ocular-only aerosol inoculation of ferrets. Journal of Virology. 2014;88(17):9647–9654. doi: 10.1128/JVI.01067-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blachere F.M., Lindsley W.G., Slaven J.E., Green B.J., Anderson S.E., Chen B.T., Beezhold D.H. Bioaerosol sampling for the detection of aerosolized influenza virus. Influenza Other Respir Viruses. 2007;1(3):113–120. doi: 10.1111/j.1750-2659.2007.00020.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon A.C., Krauss S., Rubrum A., Webb A.D., Webster R.G., McElhaney J., Webby R.J. Cross-reactive neutralizing antibodies directed against pandemic H1N1 2009 virus are protective in a highly sensitive DBA/2 mouse influenza model. Journal of Virology. 2010;84(15):7662–7667. doi: 10.1128/JVI.02444-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brankston G., Gitterman L., Hirji Z., Lemieux C., Gardam M. Transmission of influenza A in human beings. Lancet Infectious Disease. 2007;7:257–265. doi: 10.1016/S1473-3099(07)70029-4. [DOI] [PubMed] [Google Scholar]
- Burton N.C., Grinshpun S.A., Reponen T. Physical collection efficiency of filter materials for bacteria and viruses. Annals of Occupational Hygiene. 2007;51(2):143–151. doi: 10.1093/annhyg/mel073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao G., Noti J., Blachere F., Lindsley W., Beezhold D. Development of an improved methodology to detect infectious airborne influenza virus using the NIOSH bioaerosol sampler. Journal of Environmental Monitoring. 2011;13(12):3321–3328. doi: 10.1039/c1em10607d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charu V., Zeger S., Gog J., Bjørnstad O.N., Kissler S., Simonsen L., Viboud C. Human mobility and the spatial transmission of influenza in the United States. PLOS Computational Biology. 2017;13(2):e1005382. doi: 10.1371/journal.pcbi.1005382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabisch P., Yeager J., Kline J., Klinedinst K., Welsch A., Pitt M.L. Comparison of the efficiency of sampling devices for aerosolized Burkholderia pseudomallei. Inhalation Toxicology. 2012;24(5):247–254. doi: 10.3109/08958378.2012.666682. [DOI] [PubMed] [Google Scholar]
- Fabian P., McDevitt J., Houseman E., Milton D. Airborne influenza virus detection with four aerosol samplers using molecular and infectivity assays: Considerations for a new infectious virus aerosol sampler. Indoor Air. 2009;19(5):433–441. doi: 10.1111/j.1600-0668.2009.00609.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidler D. Springer; 2004. SARS, governance and the globalization of disease. [Google Scholar]
- Gao X., Li Y., Leung G.M. Ventilation control of indoor transmission of airborne diseases in an urban community. Indoor and Built Environment. 2009;18(3):205–218. [Google Scholar]
- Grinshpun S.A., Willeke K., Ulevicius V., Juozaitis A., Terzieva S., Donnelly J., Brenner K.P. Effect of impaction, bounce and reaerosolization on the collection efficiency of impingers. Aerosol Science and Technology. 1997;26(4):326–342. [Google Scholar]
- Haig C., Mackay W., Walker J., Williams C. Bioaerosol sampling: Sampling mechanisms, bioefficiency and field studies. Journal of Hospital Infection. 2016;93(3):242–255. doi: 10.1016/j.jhin.2016.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harstad J.B. Sampling submicron T1 bacteriophage aerosols. Applied Microbiology. 1965;13(6):899–908. doi: 10.1128/am.13.6.899-908.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harstad J.B., Decker H.M., Buchanan L., Filler M.E. Air filtration of submicron virus aerosols. American Journal of Public Health and the Nation's Health. 1967;57(12):2186–2193. doi: 10.2105/ajph.57.12.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hospodsky D., Yamamoto N., Peccia J. Accuracy, precision, and method detection limits of quantitative PCR for airborne bacteria and fungi. Appl Environ Microbiol. 2010;76(21):7004–7012. doi: 10.1128/AEM.01240-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan C., Kettleson E., Lee M.H., Ramaswami B., Angenent L., Biswas P. Sampling methodologies and dosage assessment techniques for submicrometre and ultrafine virus aerosol particles. Journal of Applied Microbiology. 2005;99(6):1422–1434. doi: 10.1111/j.1365-2672.2005.02720.x. [DOI] [PubMed] [Google Scholar]
- Jena S., Singh G. Human health risk assessment of airborne trace elements in Dhanbad, India. [Article] Atmospheric Pollution Research. 2017;8(3):490–502. [Google Scholar]
- Lau, J. (2004). SARS Transmission among Hospital Workers in Hong Kong-Volume 10, Number 2—February 2004-Emerging Infectious Disease journal-CDC. [DOI] [PMC free article] [PubMed]
- Lednicky J., Pan M., Loeb J., Hsieh H., Eiguren-Fernandez A., Hering S., Wu C.-Y. Highly efficient collection of infectious pandemic influenza H1N1virus (2009) through laminar-flow water based condensation. Aerosol Science and Technology. 2016;50(7):i–iv. [Google Scholar]
- Lednicky J.A., Loeb J.C. Detection and isolation of airborne influenza A H3N2 virus using a Sioutas Personal Cascade Impactor Sampler. Influenza Research and Treatment. 2013:2013. doi: 10.1155/2013/656825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung N.H.L., Zhou J., Chu D.K.W., Yu H., Lindsley W.G., Beezhold D.H., Cowling B.J. Quantification of influenza virus RNA in aerosols in patient rooms. PLoS ONE. 2016;11(2) doi: 10.1371/journal.pone.0148669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X., Willeke K., Ulevicius V., Grinshpun S.A. Effect of sampling time on the collection efficiency of all-glass impingers. American Industrial Hygiene Association Journal. 1997;58(7):480–488. [Google Scholar]
- Lindsley W.G., Blachere F.M., Thewlis R.E., Vishnu A., Davis K.A., Cao G., Khakoo R. Measurements of airborne influenza virus in aerosol particles from human coughs. PloS One. 2010;5(11):e15100. doi: 10.1371/journal.pone.0015100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand G., Duchaine C., Lavoie J., Veillette M., Cloutier Y. Bacteria emitted in ambient air during bronchoscopy—a risk to health care workers? American Journal of Infection Control. 2016;44(12):1634–1638. doi: 10.1016/j.ajic.2016.04.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T.T., Poh M.K., Low J., Kalimuddin S., Thoon K.C., Ng W.C., Gray G.C. Bioaerosol sampling in clinical settings: A promising, noninvasive approach for detecting respiratory viruses. Open Forum Infectious Diseases. 2017;4(1) doi: 10.1093/ofid/ofw259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan M., Eiguren-Fernandez A., Hsieh H., Afshar-Mohajer N., Hering S., Lednicky J.…Wu C.Y. Efficient collection of viable virus aerosol through laminar‐flow, water‐based condensational particle growth. Journal of Applied Microbiology. 2016;120(3):805–815. doi: 10.1111/jam.13051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed L.J., Muench H. A simple method of estimating fifty per cent endpoints. American Journal of Epidemiology. 1938;27(3):493–497. [Google Scholar]
- Riemenschneider L., Woo M.H., Wu C.Y., Lundgren D., Wander J., Lee J.H., Heimbuch B. Characterization of reaerosolization from impingers in an effort to improve airborne virus sampling. Journal of Applied Microbiology. 2010;108(1):315–324. doi: 10.1111/j.1365-2672.2009.04425.x. [DOI] [PubMed] [Google Scholar]
- Rossman J.S., Leser G.P., Lamb R.A. Filamentous influenza virus enters cells via macropinocytosis. Journal of Virology. 2012;86(20):10950–10960. doi: 10.1128/JVI.05992-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teunis P.F.M., Brienen N., Kretzschmar M.E.E. High infectivity and pathogenicity of influenza A virus via aerosol and droplet transmission. Epidemics. 2010;2:215–222. doi: 10.1016/j.epidem.2010.10.001. [DOI] [PubMed] [Google Scholar]
- Thorne P.S., Reynolds S.J., Milton D.K., Bloebaum P.D., Zhang X., Whitten P., Burmeister L.F. Field evaluation of endotoxin air sampling assay methods. American Industrial. 1997;58(11):792–799. doi: 10.1080/15428119791012298. [DOI] [PubMed] [Google Scholar]
- Tian S., Pan Y., Wang J., Wang Y. Concurrent measurements of size-segregated particulate sulfate, nitrate and ammonium using quartz fiber filters, glass fiber filters and cellulose membranes. Atmospheric Environment. 2016;145:293–298. [Google Scholar]
- Tung-Thompson G., Libera D.A., Koch K.L., Francis L., III, Jaykus L.-A. Aerosolization of a human norovirus surrogate, bacteriophage MS2, during simulated vomiting. PLoS One. 2015;10(8):e0134277. doi: 10.1371/journal.pone.0134277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turgeon N., Toulouse M.-J., Martel B., Moineau S., Duchaine C. Comparison of five bacteriophages as models for viral aerosol studies. Applied and Environmental Microbiology. 2014;80(14):4242–4250. doi: 10.1128/AEM.00767-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanOsdell D.W., Liu B.Y., Rubow K.L., Pui D.Y. Experimental study of submicrometer and ultrafine particle penetration and pressure drop for high efficiency filters. Aerosol Science and Technology. 1990;12(4):911–925. [Google Scholar]
- Verreault D., Moineau S., Duchaine C. Methods for sampling of airborne viruses. Microbiology and Molecular Biology Reviews. 2008;72(3):413–444. doi: 10.1128/MMBR.00002-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Li J., Leavey A., O'Neil C., Babcock H.M., Biswas P. Comparative Study on the Size Distributions, Respiratory Deposition, and Transport of Particles Generated from Commonly Used Medical Nebulizers. Journal of Aerosol Medicine and Pulmonary Drug Delivery. 2016 doi: 10.1089/jamp.2016.1340. [DOI] [PubMed] [Google Scholar]
- Weber T.P., Stilianakis N.I. Inactivation of influenza A viruses in the environment and modes of transmission: A critical review. Journal of Infection. 2008;57:361–373. doi: 10.1016/j.jinf.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z., Rosario R.C., Montoya L.D. Collection efficiency of a midget impinger for nanoparticles in the range of 3–100 nm. Atmospheric Environment. 2010;44(6):872–876. [Google Scholar]
- WHO (2004). W. H. O. WHO guidelines for the global surveillance of severe acute respiratory syndrome (SARS). Updated recommendations, October 2004 Retrieved 14 March 2017, from 〈http://www.who.int/csr/resources/publications/WHO_CDS_CSR_ARO_2004_1/en/〉.
- WHO (2009). W. H. O. Weekly epidemiological record Retrieved 14 March 2017, from 〈http://www.who.int/wer/2009/wer8421.pdf?Ua=1〉.
- Williams G.D., Pinto A.K., Doll B., Boon A.C. A North American H7N3 influenza virus supports reassortment with 2009 pandemic H1N1 and induces disease in mice without prior adaptation. Journal of virology. 2016;90(9):4796–4806. doi: 10.1128/JVI.02761-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Shen F., Yao M. Use of gelatin filter and BioSampler in detecting airborne H5N1 nucleotides, bacteria and allergens. Journal of Aerosol Science. 2010;41(9):869–879. [Google Scholar]
- Zhang J., Bai R., Zhou Z., Liu X., Zhou J. Simultaneous analysis of nine aromatic amines in mainstream cigarette smoke using online solid-phase extraction combined with liquid chromatography-tandem mass spectrometry. [Article in Press] Analytical and Bioanalytical Chemistry. 2017:1–13. doi: 10.1007/s00216-017-0245-6. [DOI] [PubMed] [Google Scholar]
- Zheng Y., Yao M. Liquid impinger BioSampler's performance for size-resolved viable bioaerosol particles. Journal of Aerosol Science. 2017;106:34–42. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material