Highlights
-
•
We identified novel feline norovirus (FNoV) M49-1 strain.
-
•
Based on the analysis of VP1, FNoV M49-1 strain was classified into genogroup GVI.
-
•
FNoV M49-1 strain seems to be produced by recombination between GIV and GVI NoV.
-
•
Cats inoculated with FNoV gene-positive-fecal samples showed clinical symptoms.
Keywords: Enteric virus, Norovirus, Feline, Genogroup, Gastroenteritis
Abstract
Norovirus (NoV) has been classified into 6 genogroups, GI-GVI. In the present study, we identified novel feline NoV (FNoV) M49-1 strain. The C-terminal of RNA-dependent RNA polymerase of the FNoV M49-1 strain was highly homologous with GIV FNoV and GIV lion norovirus, whereas VP1 was highly homologous with GVI canine NoV (CNoV). Based on the results of the Simplot analysis, the FNoV M49-1 strain may have been produced by recombination between GIV.2 FNoV and GVI.1 CNoV. In addition, specific pathogen-free cats inoculated with FNoV gene-positive-fecal samples developed diarrhea symptoms, and the viral gene was detected in their feces and blood.
1. Introduction
Norovirus (NoV) is an approximately 7.5-Kbp single positive-strand RNA virus that has been classified into the Caliciviridae, genus Norovirus (Clarke et al., 2011). There are 3 open reading frames (ORFs) in the NoV genome. ORF1 encodes NTPase (helicase), Vpg and RNA-dependent RNA polymerase (RdRp), ORF2 encodes the major capsid protein, VP1, and ORF3 encodes the minor capsid protein, VP2. (Clarke et al., 2011, Zheng et al., 2006). NoV has been classified into 6 genogroups, GI-GVI (Clarke et al., 2011, White, 2014), based on the deduced amino acid sequence of the full-length VP1 region. NoV genogroups are further classified into subgenogroups. For example, in GIV NoV, GIV HNoV is classified into subgenogroup 1 (GIV.1) while GIV FNoV and GIV CNoV are classified into subgenogroup 2 (GIV.2). GI, GII, and GIV (GIV.1) have been identified in humans, GII in pigs, GIII in ruminants including cattle, GIV (GIV.2) in cats and lions, GV in mice, and GIV(GVI.2) and GVI(GVI.1 and GVI.2) in dogs (Clarke et al., 2011). The first NoV in carnivores was detected in captive lion cubs with severe enteritis (Martella et al., 2007).
Human NoV (HNoV) is main cause of viral gastroenteritis, and it is also the main cause of epidemic diarrhea especially in infants and the elderly (Glass et al., 2009, Harris et al., 2008, Huhti et al., 2011). HNoV infection occurs in hospitals, schools, and hotels (Hutson et al., 2004). In cats, feline NoV (FNoV) was detected in domestic cats in 2012 (Pinto et al., 2012). FNoV infection occur in animal shelter. FNoV have been detected in 2–3-month-old cats that developed gastroenteritis. However, it remains unclear whether gastroenteritis can be reproduced by inoculating cats with FNoV.
In the present study, we identified novel FNoV M49-1 strain. Based on the genome analysis of the full-length VP1, this virus was classified into genogroup GVI, which has not previously been reported in cats. This novel GVI FNoV was suggested to be produced by recombination between genogroups GIV NoV and GVI NoV.
2. Materials and methods
2.1. FNoV-positive rectal swab samples
Rectal swab samples were collected from cats in an animal shelter in Japan. The FNoV gene was detected in samples collected from Cat No. 49-2012, as confirmed by RT-PCR described below, and these samples were subsequently used in this study. The age of Cat No. 49-2012 was unclear because it was a stray (a young, mixed breed cat), but it had a healthy appearance.
2.2. Isolation of RNA from samples
Viral RNA was extracted from samples using the High Pure Viral RNA Isolation Kit (Roche, Switzerland) following the manufacturer’s instructions. cDNA was synthesized using an RNA preparation from samples (rectal swabs and fecal and serum samples). Template RNA (1 μg), random primers (final concentration of 50 pM), and a dNTP mixture (final concentration of 10 mM each) were mixed, and the volume was adjusted to 10 μL with ddH2O. This mixture was heated at 65 °C for 5 min and rapidly cooled on ice. After mixing 10 μL of the mixture and 4 μL of 5X PrimerScript Buffer (TaKaRa, Japan), an RNase Inhibitor (final concentration of 20U; TaKaRa, Japan) and PrimeScript Reverse Transcriptase (final concentration of 100 U; TaKaRa, Japan) were added, and the volume was adjusted to 20 μL with ddH2O. The resultant solution was heated at 30 °C for 10 min and then reacted at 42 °C for 60 min. After heating at 70 °C for 15 min, the reaction was rapidly cooled on ice. PCR was performed using the synthesized cDNA.
2.3. PCR for screening of the FNoV and other viral gene
Two microliters of sample cDNA was mixed with 25 μl of Quick Taq HS DyeMix (Toyobo, Japan), 1 μl of 20 μM primer mix (P290d: 5′-GATTACTCCASSTGGGAYTCMAC-3′, P289d: 5′-TGACGATTTCATCATCMCCRTA-3′) (Pinto et al., 2012), and 22 μl of distilled water. Using a PCR Thermal Cycler Dice (TaKaRa, Japan), the DNA was amplified at 94 °C for 5 min, followed by 36 cycles of denaturation at 94 °C for 30 s, primer annealing at 45 °C for 30 s, and synthesis at 72 °C for 60 s, with a final extension at 72 °C for 10 min. PCR products (5 μl) were electrophoresed with a DNA marker on a 1.5% agarose gel. The agarose gel was incubated with Midori Green DNA Stain (Nippon Genetics, Japan), and bands of 319 bp (in case of NoV) and 331 bp (in case of Vesuvius and Sapovirus) were visualized using a UV Transilluminator. The methods used to detect viral genes other than FNoV were described previously (Chung et al., 2013, Elschner et al., 2002, Martella et al., 2012, Pinto et al., 2012, Schunck et al., 1995, Takano et al., 2013).
2.4. PCR for FNoV gene sequencing
Two microliters of sample cDNA was mixed with 10-μl of 5-fold PrimeSTAR Buffer (TaKaRa, Japan), 4 μl of dNTP Mixture (TaKaRa, Japan) containing 2.5 mM of each dNTP, 1 μl of 20 μM primer mix (the primer sequences are shown in Table 1, Table 2 ), 0.5 μl of PrimeSTAR HS DNA Polymerase (2.5 U/ml; TaKaRa, Japan), and 32.5 μl of distilled water. Using a thermal cycler, DNA was amplified at 98 °C for 1 min, followed by 30 cycles of denaturation at 98 °C for 10 s, primer annealing at 55 °C for 15 s, and synthesis at 72 °C for 1 min, with a final extension at 72 °C for 5 min. Gel electrophoresis of the PCR products and stain of gels were performed as described above.
Table 1.
Primer sequences used for RT-PCR and sequencing of the ORF1 gene.
| Oligonucleotide | Orientation | Nucleotide sequence | Locationa | Length | Reference |
|---|---|---|---|---|---|
| P290d | Forward | 5′-GATTACTCCASSTGGGAYTCMAC-3′ | 4445 | 319 | Pinto et al. (2012) |
| P289d | Reverse | 5′-TGACGATTTCATCATCMCCRTA-3′ | 4763 | ||
| FNoV5f | Forward | 5′-AACACTGCCTACGTCCAACC-3′ | 89 | 746 | Pinto et al. (2012), JF781268b |
| FNoV5r | Reverse | 5′-CAGGACTGGTAGGGGTCGTA-3′ | 834 | ||
| FNoV3f | Forward | 5′-CTTTTTGCTGCCAAGTCCTC-3′ | 741 | 1736 | Pinto et al. (2012), JF781268b |
| FNoV3r | Reverse | 5′-CACCTGATCTTGGACCCTGT-3′ | 2476 | ||
| FNoV8f | Forward | 5′-GCCTTCATCAAAACCCTTGA-3′ | 1424 | 2085 | Pinto et al. (2012), JF781268b |
| FNoV8r | Reverse | 5′-ACCCTGCACCTTCATAGTGG-3′ | 3508 | ||
| FNoV2f | Forward | 5′-CCTCCATCTGGTCGAGGATA-3′ | 3183 | 1696 | Pinto et al. (2012), JF781268b |
| FNoV2r | Reverse | 5′-GAGAGTTCCCTGCTGACGAC-3′ | 4878 | ||
| FNoV/CNoV10f | Forward | 5′-GTTGCAGCAAGTGATGCGGG-3′ | 3892 | 1948 | Pinto et al. (2012), Martella et al. (2009), JF781268b, FJ875027b |
| FNoV/CNoV10r | Reverse | 5′-CAGGACACCGTGAACACATC-3′ | 5839 |
The oligonucleotide locations were referred to the full-genome sequence of the FNoV CU81210E strain (JF781268; Pinto et al., 2012).
GenBank accession number.
Table 2.
Primer sequences used for RT-PCR and sequencing of ORF2 and 3 genes.
| Oligonucleotide | Orientation | Nucleotide sequence | Locationa | Length | Reference |
|---|---|---|---|---|---|
| CNoVGVI22f | Forward | 5′-GCTTGAGCCCATAGTCTTGC-3′ | 1244 | 998 | Martella et al. (2009), FJ875027b |
| CNoVGVI22r | Reverse | 5′-AACCTTGAGTTGGACGTTGG-3′ | 1939 | ||
| CNoVGVI23f | Forward | 5′-CCAACGTCCAACTCAAGGTT-3′ | 1920 | 696 | Martella et al. (2009), FJ875027b |
| CNoVGVI23r | Reverse | 5′-AGTTGAGAGCTGGGTGGAGA-3′ | 2710 |
The oligonucleotide locations were referred to the genome sequence of the CNoV Bari-91 strain (FJ875027; Martella et al., 2009).
GenBank accession number.
2.5. Sequencing and phylogenetic analyses
Thirty microliters of PCR products were electrophoresed as described above. Singlet bands were excised and transferred to microtubes, and DNA was purified using a QIAquick Gel Extraction Kit (QIAGEN GmbH, Germany). The purified DNA was subjected to TA-cloning using the Mighty TA-cloning Reagent Set for PrimeSTAR (TaKaRa, Japan) following the manufacturer’s instructions. The purified plasmid inserting PCR products was sent to Sigma–Aldrich (Japan) for sequencing. Sequences of PCR product were determined by sequencing at least 3 clones. The sequences of the virus genomes were determined and phylogenetic trees were analyzed using the MEGA software (version 6). Phylogenetic relationships were determined using the neighbor-joining algorithm, and branching order reliability was evaluated by 1000 replications of a bootstrap resampling analysis. The constitution of the NoV genome was analyzed using SimPlot (version 3.5.1).
2.6. Animal experiments
The animal experiments described below were performed in accordance with the Guidelines for Animal Experiments of Kitasato University (the number of approval is 13-105). SPF cats were maintained in a temperature-controlled isolated facility.
2.6.1. Administration of FNoV-positive rectal swab samples to SPF cats
The FNoV gene-positive-rectal swab sample from Cat No. 49-2012 (150 μL) and 150 μl of phosphate-buffered saline (PBS) were mixed. This mixture was filtered using a syringe-driven filter unit (SLLGH04NL, pore size 0.2 μm; Merck Millipore, Germany). The filtered sample was administered orally to a two-month-old female SPF cat (Cat No. 1-1). This cat was examined daily for clinical signs, and body temperature and weight were measured for 60 days. When the cat defecated, feces were collected and stored at −80 °C. The collected feces were subjected to RT-PCR using the primer pair of P290d/P289d.
2.6.2. Experimental FNoV infection in SPF cats
Cat No. 1-1-derived FNoV gene-positive-fecal samples (sample No. 1-1) were suspended with PBS to prepare a 20% (w/v) emulsion, and 2 mL of this emulsion was orally administered to four 2-3-month-old SPF cats (Cat Nos. 2-1, 2-2, 2-3, and 2-4). These cats were examined daily for clinical signs, and their body temperatures and weights were measured for 90 days. Peripheral venous blood was collected every 7 days, and sera were isolated and stored at −30 °C. When the cats defecated, feces were collected and stored at −80 °C. The collected sera and feces were subjected to RT-PCR using the primer pair of P290d/P289d.
3. Results
3.1. Detection of the FNoV M49-1 strain in rectal swab samples
Rectal swab samples from Cat No. 49-2012 (sample No. 49) were subjected to RT-PCR using the primer pair of P290d/P289d, and PCR products were subjected to sequencing. A ClustalW analysis revealed that the PCR products showed high homologies with the 3′-terminal of the ORF1 gene fragments of GIV.2 FNoV and GIV.2 lion NoV (Fig. 1 ), confirming that the PCR products were derived from NoV. Full-length VP1 gene sequencing was attempted in order to genotype FNoV (FNoV M49-1 strain) detected in sample No. 49; however, we could not amplify that of the FNoV M49-1 strain.
Fig. 1.

Nucleotide sequence alignment of the 3′-terminal of the ORF1 gene of NoV. The nucleotide sequence of the 3′-terminal of the FNoV ORF1 gene detected in sample No. 49 was compared with those of FNoV and lion NoV. Asterisks indicate identical nucleotides with those of the FNoV M49-1 strain.
3.2. Administration of the FNoV M49-1 strain to a SPF cat
We attempted to grow the FNoV M49-1 strain in vivo. We administered sample No. 49 to a 2-month-old SPF cat (Cat No. 1-1), collected fecal samples from the cat daily. Cat No. 1-1 increased body temperature, and showed low-spirited on days 3–5 after the inoculation. Fecal samples from Cat No. 1-1 were tested by RT-PCR using using the primer pair of P290d/P289d. FNoV ORF1 gene remained detectable until day 21. When the FNoV gene-positive PCR products of fecal samples collected on days 4, 9, 18, and 21 were subjected to a sequencing analysis, all were identical with the ORF1 gene of the FNoV M49-1 strain.
3.3. Analysis of the C-terminal of RdRp and full-length of VP1 from the FNoV M49-1 strain
We successfully amplified the ORF1 gene, excluding a part of the N-terminal, in a fecal sample of Cat No. 1-1 (sample No. 1-1) collected 9 days after inoculation with sample No. 49. By referring to the sequence of the amplified ORF1 gene, the amino acid sequence of the RdRp C-terminal was deduced. A phylogenetic tree analysis based on the amino acid sequence of the C-terminal of RdRp (268-aa) revealed high homologies with GIV.2/Lion/Pistoia-387/2006/ITA and GIV.2/Feline/CU081210E/2010/USA (Fig. 2 ). We also successfully amplified the full-length ORF2 gene in sample No. 1-1. A phylogenetic tree analysis based on the amino acid sequence of the full-length VP1 revealed high homologies with GVI.1/CanineBari-91/2007/ITA and GVI.1/Canine/FD210/2007/ITA were detected. That is, the FNoV M49-1 strain was classified into the genogroup GVI cluster, which was different from the genogroup GIV FNoV (Fig. 3 ).
Fig. 2.
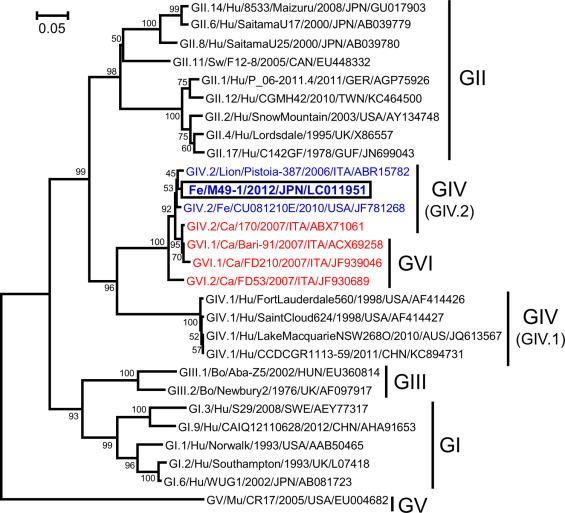
A phylogenetic tree prepared based on the amino acid sequence of the C-terminal of RdRp. GI, GII, GIII, GIV, GV, and GVI represent NoV genogroups. Blue letters represent FNoV and lion NoV, and red letters represent CNoV. Hu: human; Sw: swine; Fe: feline; Ca: canine; Bo: bovine; Mu: murine. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 3.

A phylogenetic tree prepared based on the amino acid sequence of full-length VP1. This figure was prepared based on the deduced amino acid sequences of GIV.2/Lion/Pistoia-387/2006/ITA using a computer program. GI, GII, GIII, GIV, GV, and GVI represent NoV genogroups. Blue letters represent FNoV and lion NoV, and red letters represent CNoV. Hu: human; Sw: swine; Fe: feline; Ca: canine; Bo: bovine; Mu: murine. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.4. Genome constitution of the FNoV M49-1 strain
A Simplot analysis was performed to compare gene sequences including the recombination breaking point among the FNoV M49-1 strain, GIV FNoV, GIV lion norovirus, GIV canine NoV (CNoV), and GVI CNoV. ORF1 of the FNoV M49-1 strain was classified into the GIV.2/CU081210E/USA cluster, whereas ORF2 and ORF3 were classified into the GVI.1/Bari-91/ITA and GVI.1/FD53/ITA cluster (Fig. 4 ), suggesting that the FNoV M49-1 strain was produced by recombination between cat-derived GIV NoV and dog-derived GVI NoV.
Fig. 4.

Similarity plot analysis of the FNoV M49-1 strain. The base sequence homology of the C-terminal of ORF1 through to the N-terminal of ORF3 was compared among CNoV, FNoV and lion NoV. The bold horizontal bars represent the positions of the open reading frames of the NoV genome. Nucleotide sequences were analyzed in a window of 200 mer and steps of 20 mer. The horizontal axis represents nucleotide positons, and the vertical axis represents the similarity (%) of the NoV genome.
3.5. Infectivity and pathogenicity of the FNoV M49-1 strain for SPF cats
FNoV M49-1 strain-positive fecal samples were administered to the SPF cats. The presence of viruses other than FNoV (feline coronavirus, feline calicivirus, feline astrovirus, feline rotavirus, feline kobuvirus, and feline panleukopenia virus) was investigated in the FNoV-positive fecal samples collected from Cat No. 1-1 (sample No. 1-1). All these viruses were not detect in sample No. 1-1. Shedding of FNoV into feces started 2 days after the inoculation with the FNoV-positive fecal samples in all cats, and the virus was continuously shed until 7–8 days after the inoculation (Fig. 5 ). FNoV was also shed 22 and 25 days after the inoculation in Cat No. 2-3. The induction of viremia by the inoculation with the FNoV M49-1 strain was investigated. The FNoV gene was detected in the serum of all cats 7 days after the inoculation. The FNoV gene was detected in serum of all cats, excluding Cat No. 2-2, 14 and 28 days after the inoculation. The FNoV gene was detected in the serum of Cat No. 2-4 on days 7, 14, 21, 28, 49, and 56 (Table 3 ).
Fig. 5.

Changes in body temperatures in FNoV-infected cats and detection of the FNoV gene in feces. Arrows indicate days with FNoV gene detection in feces.
Table 3.
Detection of the FNoV ORF1 gene in serum of FNoV-infected cats by RT-PCR.
| Cat No. | Days after the inoculation |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 7 | 14 | 21 | 28 | 35 | 42 | 49 | 56 | 63 | 70 | |
| 2-1 | − | + | + | − | + | − | − | − | − | − | − |
| 2-2 | − | + | − | − | − | − | − | − | − | − | − |
| 2-3 | − | + | + | − | + | − | − | − | − | − | − |
| 2-4 | − | + | + | + | + | − | − | + | + | − | − |
+: FNoV ORF1 gene positive; −: FNoV ORF1 gene negative.
Fever developed in Cat No. 2-2 on day 6 after the inoculation; however, no marked change was noted in the body temperatures of the other cats (Fig. 5). In all cats, excluding Cat No. 2-3, stools were loose from days 2 to 6 or 10 after the inoculation. In Cat No. 2-4, vomiting was noted on day 2 (Fig. 6 ), and the FNoV gene was detected in this vomit. Body weights increased in 3 of the cats inoculated with the FNoV M49-1 strain, while low-spirited and transient weight loss were noted on day 7 in Cat No. 2-4, which vomited and persistently defecated loose stools (Fig. 6).
Fig. 6.

Changes in body weights of FNoV-infected cats and clinical symptoms. Asterisks indicate days with soft stools and triangles indicate days with vomiting.
4. Discussion
NoV may recombine genomes between ORF1 and ORF2 (Bull et al., 2007, Jiang et al., 1999). The involvement of recombination in changes in adaptation to the host and pathogenicity of NoV has been reported previously (Phan et al., 2007), and HNoV was confirmed to recombine between genotypes (Jiang et al., 1999); however, recombination between genogroups is probably rare. Based on the results of the Simplot analysis, the FNoV M49-1 strain (GVI.1) may have been produced by recombination between GIV.2 FNoV and GVI.1 CNoV. It currently remains unclear which of these 3 viruses has the same origin. The number of GIV NoV and GVI NoV samples needs to be increased and a gene analysis performed in order to investigate their origins. The high homology of a part of VP1 between GVI.1 CNoV and oyster-derived NoV was previously reported (Martella et al., 2009). Cat No. 49, which carried the FNoV M49-1 strain, was captured in a region along the sea in which oyster farming is thriving. Thus, we cannot rule out that the FNoV M49-1 strain was derived from oysters, and needs to be investigated in future studies.
No FNoV infection experiment has been conducted in cats to date. We administered an FNoV gene-positive fecal emulsion to SPF cats and monitored them for clinical symptoms. Three out of 4 cats inoculated with the fecal emulsion developed diarrhea while one started vomiting. The development of these symptoms was almost consistent with the FNoV gene-detecting period, suggesting that FNoV is the cause of infectious gastroenteritis in cats. However, it is difficult to demonstrate the pathogenicity of FNoV based on these results because only 4 cats were used in this infection experiment. Moreover, the presence of unknown pathogens in the fecal emulsion cannot be ruled out. In order to concretely demonstrate that FNoV is the cause of infectious gastroenteritis, an infection experiment needs to be conducted that meets the following conditions: (1) FNoV isolated from a fecal emulsion is used to infect animals, (2) the number of cats is increased, and (3) a negative control is included in the experiment. Furthermore, difficulties are associated with accurately assessing pathogenicity using the FNoV M49-1 strain alone. Therefore, new strains of FNoV need to be isolated from cats in Japan and several FNoV strains used in infection experiments.
The influence of allelic variations in the histo-blood group antigens (HBGAs) gene on human NoV sensitivity has been reported previously (Lindesmith et al., 2003, Tan and Jiang, 2005). Allelic variations in the FUT2 gene encoding flucosyltransferase 2 (FUT2) have also been shown to influence NoV sensitivity (Lindesmith et al., 2003, Tan and Jiang, 2005). Therefore, further studies are needed in order to determine whether FNoV infection is restricted by allelic variations in the HBGAs and FUT2 genes.
Viremia has been noted with shedding of the virus in the feces of HNoV-infected children (Frange et al., 2012, Takanashi et al., 2009). Viremia was also observed early after infection in gnotobiotic animals inoculated with NoV (Souza et al., 2008). However, viremia was not detected in NoV-inoculated chimpanzees even though the virus was shed in their feces (Bok et al., 2011). Transient viremia was noted in all cats with primary infection with the FNoV M49-1 strain, and cats clearly showing clinical symptoms were more likely to intermittently develop viremia; however, we could not identify the significance of viremia in FNoV M49-1 strain infection. The relationship between viremia and clinical symptoms has not yet been elucidated.
In this study, we genotyped the FNoV M49-1 strain. When the amino acid sequence of the region from the C-terminal of RdRp to the N-terminal of VP2 was analyzed, the FNoV M49-1 strain was genotyped GVI, which was different from the conventional stain of FNoV. FNoV was detected with the development of clinical symptoms in SPF cats orally inoculated with fecal samples containing the FNoV M49-1 strain. These results may be useful for studies on FNoV and other GIV and GVI NoV.
Acknowledgment
This work was supported by JSPS KAKENHI Grant Number 26850185.
References
- Bok K., Parra G.I., Mitra T., Abente E., Shaver C.K., Boon D., Engle R., Yu C., Kapikian A.Z., Sosnovtsev S.V., Purcell R.H., Green K.Y. Chimpanzees as an animal model for human norovirus infection and vaccine development. Proc. Natl. Acad. Sci. U. S. A. 2011;108:325–330. doi: 10.1073/pnas.1014577107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull R.A., Tanaka M.M., White P.A. Norovirus recombination. J. Gen. Virol. 2007;88:3347–3359. doi: 10.1099/vir.0.83321-0. [DOI] [PubMed] [Google Scholar]
- Chung J.Y., Kim S.H., Kim Y.H., Lee M.H., Lee K.K., Oem J.K. Detection and genetic characterization of feline kobuviruses. Virus Genes. 2013;47:559–562. doi: 10.1007/s11262-013-0953-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke I.N., Estes M.K., Green K.Y., Hansman G.S., Knowles N.J., Koopmans M.K., Matson D.O., Meyers G., Neill J.D., Radford A., Smith A.W., Studdert M.J., Thiel H.J., Vinjé J. Caliciviridae. In: King A.M.Q., Adams M.J., Carstens E.B., Lefkowitz E.J., editors. Virus Taxonomy: Classification and Nomenclature of Viruses: Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier; San Diego: 2011. pp. 977–986e4. [Google Scholar]
- Elschner M., Prudlo J., Hotzel H., Otto P., Sachse K. Nested reverse transcriptase–polymerase chain reaction for the detection of group A rotaviruses. J. Vet. Med. B Infect. Dis. Vet. Public Health. 2002;49:77–81. doi: 10.1046/j.1439-0450.2002.00510.x. [DOI] [PubMed] [Google Scholar]
- Frange P., Touzot F., Debré M., Héritier S., Leruez-Ville M., Cros G., Rouzioux C., Blanche S., Fischer A., Avettand-Fenoël V. Prevalence and clinical impact of norovirus fecal shedding in children with inherited immune deficiencies. J. Infect. Dis. 2012;206:1269–1274. doi: 10.1093/infdis/jis498. [DOI] [PubMed] [Google Scholar]
- Glass R.I., Parashar U.D., Estes M.K. Norovirus gastroenteritis. N. Engl. J. Med. 2009;361:1776–1785. doi: 10.1056/NEJMra0804575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris J.P., Edmunds W.J., Pebody R., Brown D.W., Lopman B.A. Deaths from norovirus among the elderly, England and Wales. Emerg. Infect. Dis. 2008;14:1546–1552. doi: 10.3201/eid1410.080188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huhti L., Szakal E.D., Puustinen L., Salminen M., Huhtala H., Valve O., Blazevic V., Vesikari T. Norovirus GII-4 causes a more severe gastroenteritis than other noroviruses in young children. J. Infect. Dis. 2011;203:1442–1444. doi: 10.1093/infdis/jir039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutson A.M., Atmar R.L., Estes M.K. Norovirus disease: changing epidemiology and host susceptibility factor. Trends Microbiol. 2004;12:279–287. doi: 10.1016/j.tim.2004.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Espul C., Zhong W.M., Cuello H., Matson D.O. Characterization of a novel human calicivirus that may be a naturally occurring recombinant. Arch. Virol. 1999;144:2377–2387. doi: 10.1007/s007050050651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindesmith L., Moe C., Marionneau S., Ruvoen N., Jiang X., Lindblad L., Stewart P., LePendu J., Baric R. Human susceptibility and resistance to Norwalk virus infection. Nat. Med. 2003;9:548–553. doi: 10.1038/nm860. [DOI] [PubMed] [Google Scholar]
- Martella V., Campolo M., Lorusso E., Cavicchio P., Camero M., Bellacicco A.L., Decaro N., Elia G., Greco G., Corrente M., Desario C., Arista S., Banyai K., Koopmans M., Buonavoglia C. Norovirus in captive lion cub (Panthera leo) Emerg. Infect. Dis. 2007;13:1071–1073. doi: 10.3201/eid1307.070268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martella V., Decaro N., Lorusso E., Radogna A., Moschidou P., Amorisco F., Lucente M.S., Desario C., Mari V., Elia G., Banyai K., Carmichael L.E., Buonavoglia C. Genetic heterogeneity and recombination in canine norovirus. J. Virol. 2009;83:11391–11396. doi: 10.1128/JVI.01385-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martella V., Moschidou P., Catella C., Larocca V., Pinto P., Losurdo M., Corrente M., Lorusso E., Bànyai K., Decaro N., Lavazza A., Buonavoglia C. Enteric disease in dogs naturally infected by a novel canine astrovirus. J. Clin. Microbiol. 2012;50:1066–1069. doi: 10.1128/JCM.05018-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan T.G., Kaneshi K., Ueda Y., Nakaya S., Nishimura S., Yamamoto A., Sugita K., Takahashi S., Okitsu S., Ushijima H. Genetic heterogeneity, evolution, and recombination in noroviruses. J. Med. Virol. 2007;79:1388–1400. doi: 10.1002/jmv.20924. [DOI] [PubMed] [Google Scholar]
- Pinto P., Wang Q., Chen N., Dubovi E.J., Daniels J.B., Millward L.M., Buonavoglia C., Martella V., Saif L.J. Discovery and genomic characterization of noroviruses from a gastroenteritis outbreak in domestic cats in the US. PLoS One. 2012;7:e32739. doi: 10.1371/journal.pone.0032739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schunck B., Kraft W., Truyen U. A simple touch-down polymerase chain reaction for the detection of canine parvovirus and feline panleukemia virus in feces. J. Virol. Methods. 1995;55:427–433. doi: 10.1016/0166-0934(95)00069-3. [DOI] [PubMed] [Google Scholar]
- Souza M., Azevedo M.S., Jung K., Cheetham S., Saif L.J. Pathogenesis and immune responses in gnotobiotic calves after infection with the genogroup II. 4-HS66 strain of human norovirus. J. Virol. 2008;82:1777–1786. doi: 10.1128/JVI.01347-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takanashi S., Hashira S., Matsunaga T., Yoshida A., Shiota T., Tung P.G., Khamrin P., Okitsu S., Mizuguchi M., Igarashi T., Ushijima H. Detection, genetic characterization, and quantification of norovirus RNA from sera of children with gastroenteritis. J. Clin. Virol. 2009;44:161–163. doi: 10.1016/j.jcv.2008.11.011. [DOI] [PubMed] [Google Scholar]
- Takano T., Katoh Y., Doki T., Hohdatsu T. Effect of chloroquine on feline infectious peritonitis virus infection in vitro and in vivo. Antivir. Res. 2013;99:100–107. doi: 10.1016/j.antiviral.2013.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M., Jiang X. Norovirus and its histo-blood group antigen receptors: an answer to a historical puzzle. Trends Microbiol. 2005;13:285–293. doi: 10.1016/j.tim.2005.04.004. [DOI] [PubMed] [Google Scholar]
- White P.A. Evolution of norovirus. Clin. Microbiol. Infect. 2014;20:741–745. doi: 10.1111/1469-0691.12746. [DOI] [PubMed] [Google Scholar]
- Zheng D.P., Ando T., Fankhauser R.L., Beard R.S., Glass R.I., Monroe S.S. Norovirus classification and proposed strain nomenclature. Virology. 2006;346:312–323. doi: 10.1016/j.virol.2005.11.015. [DOI] [PubMed] [Google Scholar]


