Abstract
Between 2003 and 2009, the prevalence of extensively drug-resistant Pseudomonas aeruginosa (XDR-PA) increased significantly in northern Taiwan from 1.0% to 2.1%. Molecular methods were used to investigate the genetic relatedness and carbapenem resistance mechanisms of a collection of 203 non-repetitive XDR-PA isolates available for study. Using pulsed-field gel electrophoresis (PFGE), 52 genotypes were observed; one predominant genotype (pulsotype 1) was found in 57.6% of the isolates. Polymerase chain reaction (PCR), sequencing and quantitative reverse-transcriptase PCR analyses demonstrated that one horizontally acquired mechanism [metallo-β-lactamase (MBL) genes] and two mutational mechanisms (efflux and porins) accounted for the carbapenem resistance. The most predominant horizontally acquired mechanism was carriage of blaVIM-3, which was found in 61.1% of isolates. Decreased expression of oprD was the most prevalent mutational mechanism and was found in 70.0% of the XDR-PA isolates, whereas overexpression of mexA was found in 27.6% of the isolates. The highlight of this study was the discovery of statistically significant relationships between certain horizontally acquired and mutational resistance mechanisms and their contribution to carbapenem susceptibility. MBL-producers expressed significantly lower MexAB and higher OprD than non-MBL-producers. Amongst isolates without an acquired β-lactamase gene, oprD expression was significantly reduced, whilst expression of efflux pumps was increased. Reduced OprD expression alone or the production of VIM-type MBLs showed similar contributions to a low to intermediate MIC50 (minimum inhibitory concentration for 50% of the organisms) for carbapenems. Isolates with reduced OprD expression that simultaneously harboured blaVIM exhibited high levels of resistance to carbapenems, which implied that these two mechanisms had a synergistic effect on the MICs.
Keywords: Extensively drug-resistant Pseudomonas aeruginosa (XDR-PA), Carbapenem resistance, Metallo-β-lactamase, Efflux pump, Porin, MIC
1. Introduction
Pseudomonas aeruginosa is an opportunistic pathogen that is commonly encountered in nosocomial infections, especially in immunocompromised patients or in patients with cystic fibrosis (CF), in the Western world [1]. The emergence of multidrug-resistant P. aeruginosa (MDR-PA) or even extensively drug-resistant P. aeruginosa (XDR-PA) is of great concern because of limited treatment options. The term ‘extensive drug resistance’ should be defined as resistance to all but one or two classes of antimicrobial agents [2]. Previous studies have shown that two major mechanisms are involved in drug resistance, including a change in intrinsic chromosomally encoded resistance-related genes and acquisition of resistance determinants from other bacteria [3]. The mode of horizontally acquired resistance is mainly associated with mobile genetic elements that encode a variety of antibiotic-hydrolysing enzymes. In contrast, chromosomally encoded (mutational) resistance in P. aeruginosa is mostly achieved by increasing the production of drug-inactivating enzymes, generating mutations in target genes, disabling the function of outer membrane porins and overproducing efflux pumps.
Carbapenems, such as imipenem, meropenem and doripenem, are the drugs of choice for severe infections caused by MDR-PA. However, the development of carbapenem resistance is usually multifactorial. Previous reports using MDR-PA have identified several metallo-β-lactamases (MBLs), including IMP, VIM, SPM and GIM [4]. Overexpression of some AmpC enzymes with broadened hydrolytic activity towards imipenem as well as extended-spectrum cephalosporinases have also been recently identified [5]. However, their effects may vary between different carbapenems [6]. It has been assumed that AmpC overproduction alone does not significantly alter carbapenem susceptibility in P. aeruginosa [3]. The P. aeruginosa porin OprD is a channel that allows the diffusion of basic amino acids as well as carbapenems into the cell [7]. Reduced expression of OprD should therefore lead to carbapenem resistance [6], [8]. In addition, constitutive expression of the MexAB–OprM efflux pump contributes to resistance to meropenem but not imipenem [9].
In the Chang Gung Memorial Hospital (CGMH) system in Taiwan, surveillance of XDR-PA strains has been continuously performed. According to their data, a significant increase in XDR-PA infections was noted at two CGMHs in northern Taiwan from 2003 to 2006, after the severe acute respiratory syndrome (SARS) outbreak, which was followed by a significant decrease from 2006 to 2008. A large number of XDR-PA isolates were also identified. The aim of the present study was to investigate the molecular epidemiology of XDR-PA isolates and the mechanisms of carbapenem resistance amongst the XDR-PA isolates.
2. Materials and methods
2.1. Locations and laboratory-based surveillance
Two hospitals located in northern Taiwan (CGMH-Linkou and CGMH-Keelung) were involved in this study. CGMH-Linkou is a university-affiliated medical centre with 4000 beds, whereas CGMH-Keelung is an 800-bed regional teaching hospital.
2.2. Bacterial strains
From 1999 to 2009, a total of 45 805 P. aeruginosa clinical isolates were identified at CGMH-Linkou and CGMH-Keelung by standard methods, and their susceptibilities to antimicrobial agents were determined by a routine disk diffusion method according to Clinical and Laboratory Standards Institute (CLSI) guidelines [10]. A total of 203 non-repetitive XDR-PA clinical isolates were collected between May 2005 and December 2008. XDR-PA strains were defined as isolates that were resistant or intermediately resistant to all antimicrobial agents tested (amikacin, gentamicin, ceftazidime, aztreonam, ciprofloxacin, cefepime, piperacillin, piperacillin/tazobactam, meropenem and imipenem). For strains of interests (n = 47), minimum inhibitory concentrations (MICs) of imipenem and meropenem (both by Etest and agar dilution methods) and doripenem (by Etest only) were determined and were interpreted according to CLSI guidelines [10]. MIC50 and MIC90 values were defined as the MICs that inhibited the growth of 50% and 90% of the organisms, respectively. All of these 47 isolates were susceptible to colistin.
2.3. Genomic fingerprinting by pulsed-field gel electrophoresis (PFGE)
Total DNA was prepared and PFGE analysis of SpeI-digested DNA fragments was performed as described previously [11]. Thiourea (50 μM) was added in 0.5 × TBE running buffer [45 mM Tris, 45 mM boric acid, 1.0 mM ethylene diamine tetra-acetic acid (EDTA), pH 8.0] to avoid DNA degradation during PFGE. An auto-algorithm mode was chosen for running molecular weights ranging from 50 kb to 500 kb. The resulting band patterns were compared visually and were classified as indistinguishable (clonal), closely related (clonal variants, three band differences or less), possibly related (four to six band differences) or unrelated according to previously described criteria [12].
2.4. Polymerase chain reaction (PCR) amplification and DNA sequencing
DNA extraction was performed using a QIAamp DNA Mini Kit (QIAGEN, Hilden, Germany) according to the manufacturer's instructions. To detect antimicrobial resistance genes, primer sets previously described for detecting bla IMP-1-, bla IMP-2-, bla VIM-1-, bla VIM-2-, bla OXA-group I-, bla OXA-group II-, bla OXA-group III-, bla OXA-23- and bla OXA-24-related genes were used in the amplification procedure [13], [14], [15]. Amplicons were purified using Microcon PCR Centrifugal Filter Devices (Millipore, Billerica, MA) and were sequenced using an ABI 3100-Avant Genetic Analyzer (Applied Biosystems, Foster City, CA). A search for homologous sequences was conducted in the GenBank database using the Basic Local Alignment Search Tool (BLAST) through the National Center for Biotechnology Information (NCBI) website (http://www.ncbi.nlm.nih.gov/BLAST/).
2.5. Quantitative real-time reverse transcriptase polymerase chain reaction (qRT-PCR)
Expression of the efflux gene mexA was determined by qRT-PCR. Briefly, total RNA was isolated from exponential-phase cultures using a NucleoSpin RNA Kit (Macherey-Nagel, Duren, Germany) according to the manufacturer's instructions. A total of 1 μg of RNA from all strains was reverse transcribed into cDNA using SuperScript™ III reverse transcriptase (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. mexA cDNA was subsequently quantified in a LightCycler® System (Roche, Mannheim, Germany) with a FastStart SYBR Green I Kit (Roche). The primer set (mexA-F1, 5′-CTGGAGGACGGTAGCCAATA-3′; and mexA-R1, 5′-GAGGATGGCCTTCTGCTTGA-3′) was designed according to the published sequence (GenBank accession no. AE004091). All amplifications were performed in triplicate. The level of mRNA was normalised to that of the housekeeping gene rpsL [16]. Analysis of the results was done in Microsoft Excel (Microsoft Corp., Redmond, WA) using the Pfaffl equation [17]. Results are presented as fold-change expression level relative to the wild-type strain P. aeruginosa PAO1. An effect on gene expression was considered significant when the ratio was ≥3-fold as previously described [18]. To determine the transcription level of the oprD gene, the previously described specific primers oprD1-f and oprD2-r were used for qRT-PCR [19]. Normalised expression of the oprD gene was calibrated against the corresponding mRNA expression in the PAO1 strain. An effect on gene expression was considered significant when the ratios were >2.5 or <0.4. Expression levels of ampC genes were determined using previously described primers and were considered hyperproductive if the mRNA levels were >10-fold higher than that of PAO1 [18].
2.6. Statistical analysis
The χ 2 test and non-parametric tests were used to determine the significance of differences. A difference was considered statistically significant at a P-value of <0.05.
3. Results
3.1. Epidemiology and genotypes of multidrug-resistant P. aeruginosa
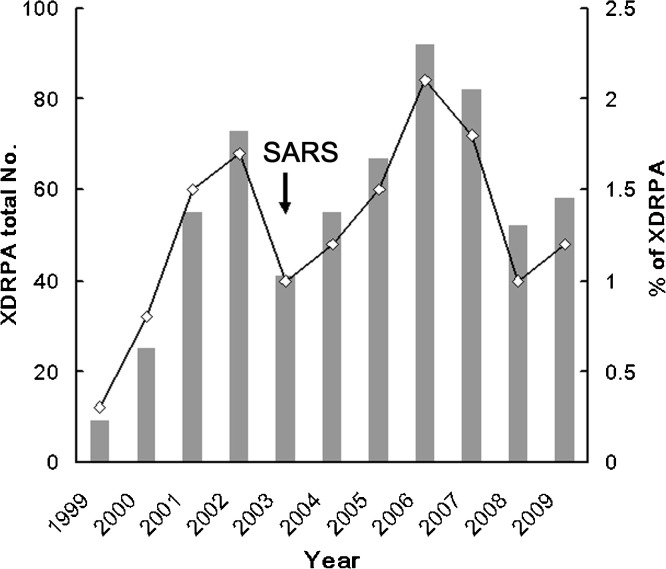
Of a total of 45 805 P. aeruginosa clinical isolates, 603 (1.3%) were XDR-PA strains. The yearly prevalence of XDR-PA is shown in Fig. 1 . A significant increase in XDR-PA was noted from 1.0% in 2003 to 2.1% by 2006 (P < 0.005), which was followed by a significant drop to 1.0% by 2008 (P < 0.01). Of the 603 XDR-PA isolates, 203 non-repetitive clinical isolates collected between 2005 and 2008 were further selected to investigate their epidemiological correlation by PFGE. The distribution of the various genotypes amongst different wards and recovery sites is shown in Table 1 . Amongst the 52 pulsotypes identified by PFGE, multiple isolates were found in pulsotypes 1 to 13, whereas only one isolate was observed in each of the remaining pulsotypes. Pulsotype 1 was the most predominant (57.6%), followed by pulsotype 2 (6.9%). Pulsotype 1 isolates were persistently observed throughout the study period, whereas pulsotype 2 isolates were only identified between October 2005 and April 2007. Most of the XDR-PA isolates were collected from Intensive Care Units and inpatient departments. The majority of isolates were collected from the respiratory tract (51.7%), followed by urine (28.1%) and wounds (6.9%).
Fig. 1.
Secular trend in the annual number (histograms) and prevalence (lines) of extensively drug-resistant Pseudomonas aeruginosa (XDR-PA) isolates identified between 1999 and 2009 at Chang Gung Memorial Hospital, Taiwan. SARS, indicates the severe acute respiratory syndrome outbreak.
Table 1.
Distribution of various genotypes amongst extensively drug-resistant Pseudomonas aeruginosa isolated from different wards and specimens and association with various resistance mechanisms.
| Specification | Genotype [n (%)a of isolates] |
Total number (%)b | |||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6–52 | ||
| No. (%) of tested | 117 (57.6) | 14 | 7 | 4 | 4 | 57 (28.1) | 203 |
| Ward | |||||||
| ICU | 27 (40.9) | 7 (10.6) | 2 | 3 | 4 | 23 (34.8) | 66 (32.5) |
| IPD | 82 (66.7) | 7 | 5 | 1 | 0 | 28 (22.8) | 123 (60.6) |
| OPD | 8 (57.1) | 0 | 0 | 0 | 0 | 6 (42.9) | 14 |
| Specimen | |||||||
| Respiratory tract | 55 (52.4) | 6 | 3 | 3 | 3 | 35 (33.3) | 105 (51.7) |
| Urine | 38 (66.7) | 3 | 4 | 0 | 1 | 11 (19.3) | 57 (28.1) |
| Wounds | 9 (64.3) | 1 | 0 | 0 | 0 | 4 (28.6) | 14 |
| Others | 15 (55.6) | 4 (14.8) | 0 | 1 | 0 | 7 (25.9) | 27 (13.3) |
| Acquired resistance genes | |||||||
| blaVIM-2 | 0 | 0 | 1 (33.3) | 0 | 0 | 2 (66.7) | 3 |
| blaVIM-3 | 117 (94.4) | 2 | 0 | 0 | 0 | 5 | 124 (61.1) |
| Altered expression of porins and efflux pumpsc | |||||||
| oprD | 66 (46.5) | 12 | 7 | 4 | 4 | 49 (34.5) | 142 (70.0) |
| mexA | 20 (35.7) | 1 | 1 | 4 | 2 | 28 (50.0) | 56 (27.6) |
ICU, Intensive Care Unit; IPD, inpatient department; OPD, outpatient department.
Percentage of isolates of genotype amongst isolates with particular characteristic (percentages given only for those >10%).
Percentage of isolates with particular characteristic amongst total isolates (percentages given only for those >10%).
Altered expression means diminished expression of oprD of <40% or increased expression of mexA three times higher relative to the control strain P. aeruginosa PAO1.
3.2. Evaluation of the mechanisms of carbapenem resistance
Amongst the 203 XDR-PA isolates, 127 isolates (62.6%) harboured MBLs. VIM-type β-lactamase was the predominant mechanism and was the only MBL detected in these XDR-PA isolates, which included 124 and 3 isolates carrying bla VIM-3 and bla VIM-2, respectively. All pulsotype 1 isolates were bla VIM-3-carriers (P < 0.0001) (Table 1). For oprD expression, the expression level of the isolates ranged from 2 × 10−7- to 2.73-fold difference (mean 0.33) compared with the control PAO1. A significant reduction of oprD expression of ≥60% compared with the control PAO1 was observed in 142 isolates (70.0%) (Table 1). In the evaluation of the MexAB–OprM efflux pump system, differences in expression levels of mexA compared with the control PAO1 ranged from 0.03- to 24.17-fold (mean 3.09). Significantly increased mexA expression (≥3-fold change) was found in 56 isolates (27.6%) (Table 1). A concomitant reduction of OprD expression and bla VIM production was identified in 72 isolates (35.5%), whereas a concomitant reduction of OprD expression and overexpression of MexA was found in 16 isolates (7.9%). Two isolates overexpressed MexA only and 36 isolates (17.7%) produced VIM-3 only.
3.3. Interplay between metallo-β-lactamases and the mutational resistance mechanism
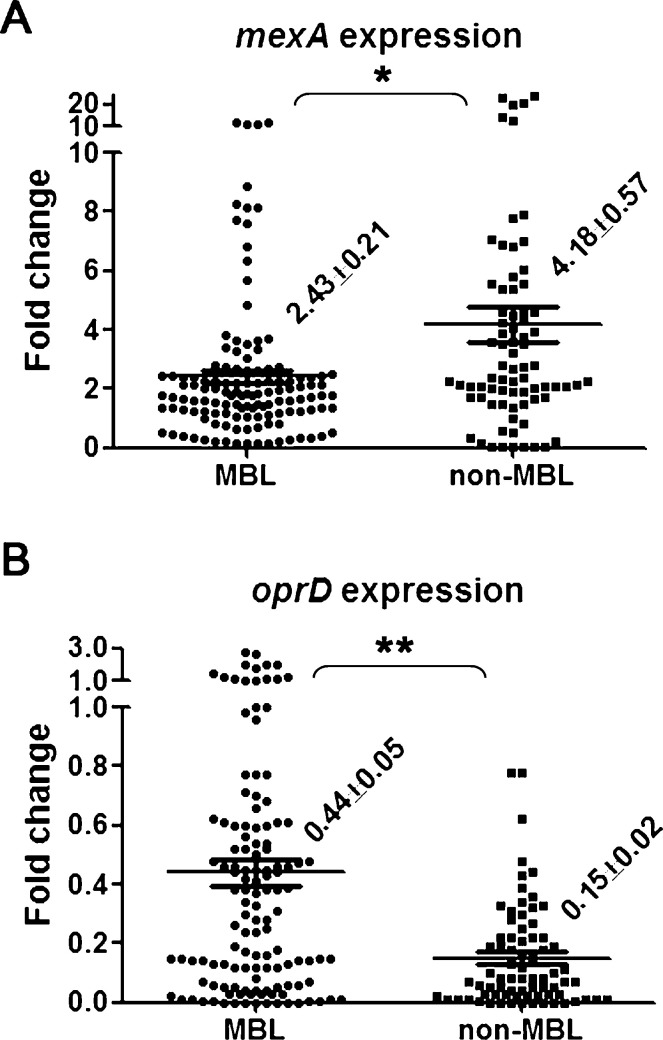
The correlation between acquired MBLs and expression of oprD or mexA was analysed and the results are shown in Fig. 2 . Isolates with MBL production showed significantly lower mexA expression than non-MBL-producers (mean 2.43 vs. 4.18, respectively; P < 0.005) (Fig. 2A). Isolates without MBL production displayed significantly lower oprD expression than their counterparts (mean 0.15 vs. 0.44, respectively; P < 0.0001) (Fig. 2B).
Fig. 2.
Average expression levels of (A) mexA and (B) oprD transcripts relative to the control strain Pseudomonas aeruginosa PAO1 in extensively drug-resistant P. aeruginosa isolates with and without acquired metallo-β-lactamases (MBLs). Mean values ± standard error of the mean are indicated. Significant differences are indicated by * (P < 0.005) or ** (P < 0.0001).
To reveal the contribution of different resistance mechanisms to the resistance levels to various carbapenems, 47 isolates were selected for further MIC determination, including: 7 isolates with significant reduction of OprD expression but without other known resistance mechanisms, including overproduction of efflux pumps and production of MBLs, extended-spectrum β-lactamases or penicillinases; 12 bla VIM-carrying isolates with normal OprD expression; and 28 randomly selected isolates showing concomitant reduction of OprD expression and production of VIM MBLs. The results are shown in Table 2 . The MIC50 values of both imipenem and meropenem were 32 μg/mL amongst isolates with either reduced OprD expression only or the presence of bla VIM. However, different levels (16 μg/mL and >32 μg/mL) were observed for the MIC50 of doripenem when the respective group of isolates were tested. Significantly higher MIC90 values for imipenem (4-fold; 128 μg/mL to 32 μg/mL) and meropenem (2-fold; 128 μg/mL to 64 μg/mL) were observed amongst VIM-producers compared with OprD-reduced isolates. A remarkably increased MIC50 was noted for both imipenem (256 μg/mL) and meropenem (128 μg/mL) amongst isolates with concomitant reduction of OprD expression and production of VIM MBLs (OprD + VIM). The imipenem MIC90 in OprD + VIM isolates was 8-fold and 2-fold higher than isolates expressing reduced OprD only and harbouring bla VIM, respectively. A 2-fold higher meropenem MIC90 was detected in OprD + VIM isolates compared with OprD-reduced isolates, but there was no difference between OprD + VIM isolates and VIM-producers. The contribution of MexA to carbapenems, especially to meropenem resistance, was investigated by assaying the MICs amongst OprD + VIM isolates with and without MexA overexpression. Isolates with overexpressed MexA showed lower MIC50 values of both imipenem and meropenem compared with isolates with unaltered MexA expression. MIC90 values of the two carbapenems were the same between the two populations.
Table 2.
Contribution of different resistance mechanisms to the minimum inhibitory concentrations (MICs) of various carbapenems amongst 47 extensively drug-resistance Pseudomonas aeruginosa isolates.
| Resistance mechanisma | Number of isolates | MIC (μg/mL) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Imipenem |
Meropenem |
Doripenemb |
||||||||
| Range | MIC50 | MIC90 | Range | MIC50 | MIC90 | Range | MIC50 | MIC90 | ||
| OprD | 7 | 32–32 | 32 | 32 | 8–64 | 32 | 64 | 4 to >32 | 16 | >32 |
| VIM | 12 | 32 to >256 | 32 | 128 | 8–256 | 32 | 128 | 4 to >32 | >32 | >32 |
| OprD + VIM | ||||||||||
| Overall | 28 | 32–256 | 256 | 256 | 8–256 | 128 | 128 | 12 to >32 | >32 | >32 |
| Altered MexA | 5 | 32–256 | 32 | 256 | 8–256 | 32 | 128 | 32 to >32 | >32 | >32 |
| Normal MexA | 23 | 32–256 | 256 | 256 | 32–256 | 128 | 128 | 12 to >32 | >32 | >32 |
MIC50/90, MICs that inhibited the growth of 50% and 90% of the organisms, respectively.
Isolates with either reduced OprD expression only or the presence of blaVIM only, or both, were designated as ‘OprD’, ‘VIM’ and ‘OprD + VIM’, respectively.
MICs of doripenem were determined using Etest only.
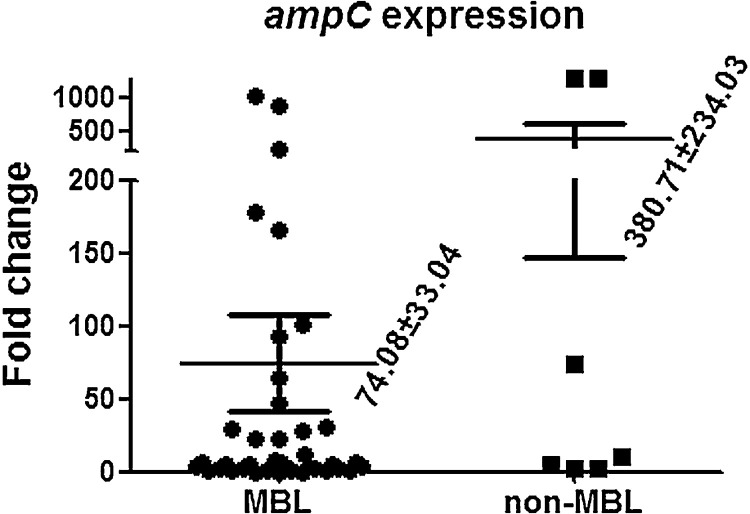
The expression level of AmpC has been shown to play a role in carbapenem resistance of P. aeruginosa [18] and thus was further examined amongst the 47 isolates. Compared with the control strain, the expression level of ampC ranged from 1.4-fold to 1295-fold (average 381-fold) in the seven OprD-reduced isolates (non-MBL-producers). Overexpression of ampC (>10-fold change) was found in three (42.9%) of the isolates. In comparison, the expression level of ampC ranged from 0.12-fold to 1012-fold (average 74-fold) in the remaining 40 MBL-producing isolates, and 15 (37.5%) of them were AmpC hyperproducers. Although the expression level of ampC was higher in the non-MBL-producing group, no significant difference was found (Supplementary Fig. S1).
4. Discussion
This study aimed to investigate the mechanisms underlying carbapenem resistance in a collection of XDR-PA isolates. To our knowledge, this is the first time a statistically significant interplay between acquired and intrinsic resistance mechanisms and their contribution to carbapenem susceptibility has been demonstrated, which was determined by analysing a large number of XDR-PA clinical isolates. From this study, the overall prevalence of XDR-PA reported herein was 1.3%. However, a previous study from one medical centre in Taiwan indicated that the prevalence of MDR-PA infections increased significantly from 1.74% in 2001 to 6.31% in 2004 [20]. The reported prevalence of MDR-PA varies amongst Asian countries, from 1.62% in Japan to 4.49% in India and 5.46% in Iran [21], [22], [23]. The rate is similar (4.7%) in Europe but is much higher (10%) in the USA [24], [25]. Although the reported prevalence rates of such multiresistant isolates were quite different, it should also be noted that different criteria for the definition of multidrug resistance used in these studies may also contribute to the difference in the results. For instance, the prevalence of MDR-PA in the USA was 10% in isolates resistant to three classes of antimicrobial agents; the rate would reduce to 2% if only isolates resistant to four classes of antimicrobial agents (nearly XDR-PA) were compared [25]. None the less, a common observation from all reports, including ours, is that most MDR-PA isolates were recovered from respiratory tract specimens.
Of the acquired β-lactamase genes investigated, bla VIM-3 was the most prevalent type amongst the XDR-PA isolates. VIM-3 was first reported in Taiwan and has since become the most prevalent MBL amongst MDR-PA isolates in Taiwan, accounting for 53–77% of the population [15], [26]. Similarly, 61.1% of the XDR-PA isolates in the present study were found to harbour the VIM-3 gene (1.5% were VIM-2-producers). However, MBL production is not the major resistance mechanism for MDR-PA or carbapenem-resistant strains in Europe or the USA [27], [28], [29]. Considering that ca. 60% of the XDR-PA isolates in the present report were bla VIM-3-carrying/pulsotype 1 isolates, the increase in the prevalence of XDR-PA observed herein may be associated with the spread of the pulsotype 1 strain in Taiwan. Furthermore, when only isolates of the other pulsotypes are analysed, the prevalence of MBL-producing isolates is greatly reduced to 11.6% (10/86), which is similar to that reported in other countries [27], [28], [29]. Although we were not aware of any evidence of the spread of pulsotype 1 elsewhere in Taiwan, the predominant pulsotype 1 strain in this study is very similar to the major VIM-3-producing clone in the first report from Taiwan (data not shown) [15]. An island-wide study may be needed to determine whether the pulsotype 1 XDR-PA strain has already spread across Taiwan. Previous studies have indicated that carbapenem resistance may involve decreased expression of oprD with or without overexpression of mexAB [3], [27], [30]. In the present study, decreased expression of oprD was found in 70.0% of the XDR-PA isolates regardless of the presence of other factors. In 16 (11.3%) of such OprD-reduced isolates, expression levels of oprD could be reduced by more than 1000 times compared with the control strain. Such extremely low expression of oprD may imply the absence of OprD, which could occur through the presence of some inactivating mutations in the oprD sequence, such as partial or total deletions and insertions [3]. Overexpression of mexA was found in 27.6% of the XDR-PA isolates in this study, but made little to no contribution to the multidrug resistance of P. aeruginosa in France [27].
This study showed that MBL-producers had significantly lower expression of mexA and higher expression of oprD compared with non-MBL-producers, suggesting that MBL production alone was able to confer carbapenem resistance. In contrast, overexpression of mexA and/or depression of oprD may compensate for the lack of MBL in non-MBL-producers (Fig. 2). Reduced OprD expression alone or the production of VIM-type MBLs contributed equally to the MIC50 of imipenem and meropenem. A lower MIC50 of doripenem was found amongst isolates with reduced OprD expression compared with MBL-producers. This result suggests that loss of OprD porins may not play a major role in conferring high levels of doripenem resistance. However, production of MBLs alone could confer intermediate to high levels of resistance to carbapenems. Notably, isolates with reduced OprD expression that simultaneously harboured bla VIM exhibited a high level of resistance to carbapenems, implying that a synergistic effect of these two mechanisms contributed to the MICs (Table 2). However, owing to the detection limit of Etest strips and the unavailability of doripenem standard powder, MICs of doripenem above 32 μg/mL could not be determined in this study. Whether or not the simultaneous presence of bla VIM and reduced expression of OprD may also demonstrate a synergistic effect in doripenem resistance may require further studies. Although the expression level of mexA was correlated with the existence of bla VIM, the contribution of mexA overexpression to carbapenem (especially meropenem) resistance was ambiguous in this study. Strains overexpressing mexA showed a lower MIC50 (both for imipenem and meropenem) than the unaltered mexA populations amongst the OprD + VIM isolates, and an equal MIC90 of these two agents was observed (Table 2). A recent study indicated that amongst imipenem-resistant P. aeruginosa clinical isolates, non-MBL-producing strains exhibited a low level of resistance to carbapenems, with the MIC50 of imipenem and meropenem reported to be 16 μg/mL and 8 μg/mL, respectively [31]. Sohn et al. [32] have demonstrated that MDR-PA isolates harbouring bla VIM-2 (17/40) exhibited high levels of imipenem resistance (MIC50 = 512 μg/mL), but the expression of OprD was not examined. Of the 32 carbapenem-non-susceptible clinical isolates studied in France, all had reduced OprD expression, and the MIC50 for imipenem and meropenem was 16 μg/mL and 8 μg/mL, respectively, although 25 of the isolates were AmpC-overproducers [6]. Similar results were found amongst extensively drug-resistant clinical isolates in neighbouring areas [27], [30]. Thus, carbapenem resistance was largely due to reduced OprD expression, which resulted in a low level of carbapenem resistance in Europe, regardless of the carbapenem- or multidrug-resistant P. aeruginosa background. A relatively high percentage of the P. aeruginosa isolates were recovered from CF patients in Caucasian populations; however, the prevalence of CF is low in Asian countries. Therefore, we propose that the production of MBLs together with reduction of OprD expression may play an important role in producing intermediate to high levels of carbapenem resistance amongst P. aeruginosa isolates from non-CF patients in Asia. The contribution of ampC overexpression to multidrug resistance in the absence of transferable β-lactamases is dubious in this study. Cabot et al. [18] recently showed that ampC overexpression (24.2%) was the most prevalent resistance mechanism amongst 190 P. aeruginosa isolates recovered from bloodstream infections, and only 2 of them produced acquired MBLs. Moreover, overexpression of ampC was found amongst all isolates that were resistant to all β-lactams and aminoglycosides in the same populations. The prevalence of isolates showing ampC overexpression was also high for isolates non-susceptible to meropenem (60%) [18]. In the present study, although the percentage of isolates showing ampC overproduction was higher in the non-MBL-producers (42.9%) than in the MBL-producers (37.5%), the difference was not statistically significant. The discrepancy may be due to the small number of non-MBL-producers evaluated in the present study. Future studies including more such isolates may be able to provide more definite information.
In conclusion, we have statistically demonstrated the interplay amongst the OprD porin, the MexAB efflux pump and acquired MBLs either by studying the relative expression of the associated genes or their contribution to drug susceptibility amongst XDR-PA isolates. Although the acquisition of VIM-type MBLs was the most predominant acquired resistance mechanism for carbapenem resistance amongst XDR-PA isolates, it was not found amongst Klebsiella pneumoniae isolates from the same hospital that had high levels of carbapenem resistance [33]. The mechanism underlying the restriction and transfer of bla VIM genes will be further studied. A great challenge lies ahead for the limitation of therapeutic options owing to the complicated interplay amongst different mechanisms.
Funding: This work was supported by the National Science Council, Taiwan (grant no. NSC95-2320-B-182A-003-MY3).
Competing interests: None declared.
Ethical approval: Not required.
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.ijantimicag.2011.09.023.
Appendix A. Supplementary data
Supplementary Fig. S1.
Average expression levels of ampC transcripts relative to the control strain Pseudomonas aeruginosa PAO1 in extensively drug-resistant P. aeruginosa isolates with and without acquired metallo-β-lactamases (MBLs). Mean values ± standard error of the mean are indicated.
References
- 1.O'Sullivan B.P., Freedman S.D. Cystic fibrosis. Lancet. 2009;373:1891–1904. doi: 10.1016/S0140-6736(09)60327-5. [DOI] [PubMed] [Google Scholar]
- 2.Falagas M.E., Karageorgopoulos D.E. Pandrug resistance (PDR), extensive drug resistance (XDR), and multidrug resistance (MDR) among Gram-negative bacilli: need for international harmonization in terminology. Clin Infect Dis. 2008;46:1121–1122. doi: 10.1086/528867. [DOI] [PubMed] [Google Scholar]
- 3.Lister P.D., Wolter D.J., Hanson N.D. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev. 2009;22:582–610. doi: 10.1128/CMR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fritsche T.R., Sader H.S., Toleman M.A., Walsh T.R., Jones R.N. Emerging metallo-β-lactamase-mediated resistances: a summary report from the worldwide SENTRY Antimicrobial Surveillance Program. Clin Infect Dis. 2005;41(Suppl. 4):S276–S278. doi: 10.1086/430790. [DOI] [PubMed] [Google Scholar]
- 5.Rodríguez-Martínez J.M., Poirel L., Nordmann P. Extended-spectrum cephalosporinases in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2009;53:1766–1771. doi: 10.1128/AAC.01410-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodríguez-Martínez J.M., Poirel L., Nordmann P. Molecular epidemiology and mechanisms of carbapenem resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2009;53:4783–4788. doi: 10.1128/AAC.00574-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trias J., Nikaido H. Outer membrane protein D2 catalyzes facilitated diffusion of carbapenems and penems through the outer membrane of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1990;34:52–57. doi: 10.1128/aac.34.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Livermore D.M. Interplay of impermeability and chromosomal β-lactamase activity in imipenem-resistant Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1992;36:2046–2048. doi: 10.1128/aac.36.9.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quale J., Bratu S., Gupta J., Landman D. Interplay of efflux system, ampC, and oprD expression in carbapenem resistance of Pseudomonas aeruginosa clinical isolates. Antimicrob Agents Chemother. 2006;50:1633–1641. doi: 10.1128/AAC.50.5.1633-1641.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clinical and Laboratory Standards Institute . CLSI; Wayne, PA: 2010. Performance standards for antimicrobial susceptibility testing; twentieth informational supplement. Document M100-S20. [Google Scholar]
- 11.Su L.H., Leu H.S., Chiu Y.P., Chia J.H., Kuo A.J., Sun C.F. Molecular investigation of two clusters of hospital-acquired bacteraemia caused by multi-resistant Klebsiella pneumoniae using pulsed-field gel electrophoresis and in frequent restriction site PCR. Infection Control Group. J Hosp Infect. 2000;46:110–117. doi: 10.1053/jhin.2000.0815. [DOI] [PubMed] [Google Scholar]
- 12.Tenover F.C., Arbeit R.D., Goering R.V., Mickelsen P.A., Murray B.E., Persing D.H. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee S., Park Y.J., Kim M., Lee H.K., Han K., Kang C.S. Prevalence of Ambler class A and D β-lactamases among clinical isolates of Pseudomonas aeruginosa in Korea. J Antimicrob Chemother. 2005;56:122–127. doi: 10.1093/jac/dki160. [DOI] [PubMed] [Google Scholar]
- 14.Vahaboglu H., Ozturk R., Akbal H., Saribas S., Tansel O., Coskunkan F. Practical approach for detection and identification of OXA-10-derived ceftazidime-hydrolyzing extended-spectrum β-lactamases. J Clin Microbiol. 1998;36:827–829. doi: 10.1128/jcm.36.3.827-829.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan J.J., Hsueh P.R., Ko W.C., Luh K.T., Tsai S.H., Wu H.M. Metallo-β-lactamases in clinical Pseudomonas isolates in Taiwan and identification of VIM-3, a novel variant of the VIM-2 enzyme. Antimicrob Agents Chemother. 2001;45:2224–2228. doi: 10.1128/AAC.45.8.2224-2228.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Llanes C., Hocquet D., Vogne C., Benali-Baitich D., Neuwirth C., Plesiat P. Clinical strains of Pseudomonas aeruginosa overproducing MexAB–OprM and MexXY efflux pumps simultaneously. Antimicrob Agents Chemother. 2004;48:1797–1802. doi: 10.1128/AAC.48.5.1797-1802.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cabot G., Ocampo-Sosa A.A., Tubau F., Macia M.D., Rodríguez C., Moya B. Overexpression of AmpC and efflux pumps in Pseudomonas aeruginosa isolates from bloodstream infections: prevalence and impact on resistance in a Spanish multicenter study. Antimicrob Agents Chemother. 2011;55:1906–1911. doi: 10.1128/AAC.01645-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farra A., Islam S., Stralfors A., Sorberg M., Wretlind B. Role of outer membrane protein OprD and penicillin-binding proteins in resistance of Pseudomonas aeruginosa to imipenem and meropenem. Int J Antimicrob Agents. 2008;31:427–433. doi: 10.1016/j.ijantimicag.2007.12.016. [DOI] [PubMed] [Google Scholar]
- 20.Leung C.H., Wang N.Y., Liu C.P., Weng L.C., Hsieh F.C., Lee C.M. Antimicrobial therapy and control of multidrug-resistant Pseudomonas aeruginosa bacteremia in a teaching hospital in Taiwan. J Microbiol Immunol Infect. 2008;41:491–498. [PubMed] [Google Scholar]
- 21.Tsuchimochi N., Takuma T., Shimono N., Nagasaki Y., Uchida Y., Harada M. Antimicrobial susceptibility and molecular epidemiological analysis of clinical strains of Pseudomonas aeruginosa. J Infect Chemother. 2008;14:99–104. doi: 10.1007/s10156-007-0578-8. [DOI] [PubMed] [Google Scholar]
- 22.Jayakumar S., Appalaraju B. Prevalence of multi and pan drug resistant Pseudomonas aeruginosa with respect to ESBL and MBL in a tertiary care hospital. Indian J Pathol Microbiol. 2007;50:922–925. [PubMed] [Google Scholar]
- 23.Shahcheraghi F., Badmasti F., Feizabadi M.M. Molecular characterization of class 1 integrons in MDR Pseudomonas aeruginosa isolated from clinical settings in Iran, Tehran. FEMS Immunol Med Microbiol. 2010;58:421–425. doi: 10.1111/j.1574-695X.2009.00636.x. [DOI] [PubMed] [Google Scholar]
- 24.Souli M., Galani I., Giamarellou H. Emergence of extensively drug-resistant and pandrug-resistant Gram-negative bacilli in Europe. Euro Surveill. 2008;13 pii:19045. [PubMed] [Google Scholar]
- 25.Kallen A.J., Hidron A.I., Patel J., Srinivasan A. Multidrug resistance among Gram-negative pathogens that caused healthcare-associated infections reported to the National Healthcare Safety Network, 2006–2008. Infect Control Hosp Epidemiol. 2010;31:528–531. doi: 10.1086/652152. [DOI] [PubMed] [Google Scholar]
- 26.Tseng S.P., Tsai J.C., Teng L.J., Hsueh P.R. Dissemination of transposon Tn6001 in carbapenem-non-susceptible and extensively drug-resistant Pseudomonas aeruginosa in Taiwan. J Antimicrob Chemother. 2009;64:1170–1174. doi: 10.1093/jac/dkp341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vettoretti L., Floret N., Hocquet D., Dehecq B., Plesiat P., Talon D. Emergence of extensive-drug-resistant Pseudomonas aeruginosa in a French university hospital. Eur J Clin Microbiol Infect Dis. 2009;28:1217–1222. doi: 10.1007/s10096-009-0767-8. [DOI] [PubMed] [Google Scholar]
- 28.Tam V.H., Chang K.T., Abdelraouf K., Brioso C.G., Ameka M., McCaskey L.A. Prevalence, resistance mechanisms, and susceptibility of multidrug-resistant bloodstream isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2010;54:1160–1164. doi: 10.1128/AAC.01446-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Riera E., Cabot G., Mulet X., García-Castillo M., del Campo R., Juan C. Pseudomonas aeruginosa carbapenem resistance mechanisms in Spain: impact on the activity of imipenem, meropenem and doripenem. J Antimicrob Chemother. 2011;66:2022–2027. doi: 10.1093/jac/dkr232. [DOI] [PubMed] [Google Scholar]
- 30.Tomás M., Doumith M., Warner M., Turton J.F., Beceiro A., Bou G. Efflux pumps, OprD porin, AmpC β-lactamase, and multiresistance in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Antimicrob Agents Chemother. 2010;54:2219–2224. doi: 10.1128/AAC.00816-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao W.H., Chen G., Ito R., Hu Z.Q. Relevance of resistance levels to carbapenems and integron-borne blaIMP-1, blaIMP-7, blaIMP-10 and blaVIM-2 in clinical isolates of Pseudomonas aeruginosa. J Med Microbiol. 2009;58:1080–1085. doi: 10.1099/jmm.0.010017-0. [DOI] [PubMed] [Google Scholar]
- 32.Sohn E.S., Yoo J.S., Lee J.K., Lee K.M., Chung G.T., Shin E.S. Investigation of β-lactamase-producing multidrug-resistant Pseudomonas aeruginosa isolated from non-tertiary care hospitals in Korea. J Microbiol Biotechnol. 2007;17:1733–1737. [PubMed] [Google Scholar]
- 33.Chia J.H., Su L.H., Lee M.H., Kuo A.J., Shih N.Y., Siu L.K. Development of high-level carbapenem resistance in Klebsiella pneumoniae among patients with prolonged hospitalization and carbapenem exposure. Microb Drug Resist. 2010;16:317–325. doi: 10.1089/mdr.2009.0048. [DOI] [PubMed] [Google Scholar]